Abstract
Genetic germline variants of PPP2R5D (encoding: phosphoprotein phosphatase 2 regulatory protein 5D) result in PPP2R5D-related disorder (Jordan’s Syndrome), which is characterized by intellectual disability, hypotonia, seizures, macrocephaly, autism spectrum disorder, and delayed motor skill development. The disorder originates from de novo single nucleotide mutations, generating missense variants that act in a dominant manner. Pathogenic mutations altering 13 different amino acids have been identified, with the E198K variant accounting for ∼40% of reported cases. However, the generation of a heterozygous E198K variant cell line to study the molecular effects of the pathogenic mutation has been challenging. Here, we use CRISPR-PRIME genomic editing to introduce a transition (c.592G>A) in a single PPP2R5D allele in HEK293 cells, generating E198K-heterozygous lines to complement existing E420K variant lines. We generate global protein and phosphorylation profiles of WT, E198K, and E420K cell lines and find unique and shared changes between variants and WT cells in kinase- and phosphatase-controlled signaling cascades. We observed ribosomal protein S6 (RPS6) hyperphosphorylation as a shared signaling alteration, indicative of increased ribosomal protein S6-kinase activity. Treatment with rapamycin or an RPS6-kinase inhibitor (LY2584702) suppressed RPS6 phosphorylation in both, suggesting upstream activation of mTORC1/p70S6K. Intriguingly, our data suggests ERK-dependent activation of mTORC1 in both E198K and E420K variant cells, with additional AKT-mediated mTORC1 activation in the E420K variant. Thus, although upstream activation of mTORC1 differs between PPP2R5D-related disorder genotypes, inhibition of mTORC1 or RPS6 kinases warrants further investigation as potential therapeutic strategies for patients.
Keywords: PPP2R5D, PP2A, phosphatase, PPP2R5D-related neurodevelopmental disorder, proteomics, phosphoproteomics, mTOR, rapamycin, LY2584702, Jordan’s syndrome
Phosphoprotein phosphatase 2 regulatory protein 5D (PPP2R5D)-related intellectual disability (ID) and developmental delay disorder (OMIM#616355) is a syndrome characterized by mild to severe neurodevelopmental delay. The disorder, also known as Jordan’s Syndrome, is characterized by ID, epilepsy, macrocephaly, autism-spectrum-disorder (ASD), hypotonia, and delayed motor skill development (1, 2, 3, 4). PPP2R5D-related developmental disorder is autosomal dominant, arising from de novo germline missense mutations in the PPP2R5D gene (1, 2, 5).
PPP2R5D encodes a regulatory subunit of type 2A serine/threonine phosphoprotein phosphatase (PP2A). Most PP2A phosphatases function as heterotrimeric holoenzymes, which are ubiquitously expressed. To generate each PP2A holoenzyme, a unique regulatory-targeting (B) subunit is assembled with a common core dimer consisting of a scaffolding (A) subunit and a catalytic (C) subunit (6, 7) (Fig. 1A). The A-C core dimer is expressed in most, if not all, human cells, and humans express two highly similar isoforms of both the C and A subunits (PPP2CA/PPP2CB and PPP2R1A/PPP2R1B, respectively). There are four families of B-subunits (B, B’, B’’, and B’’’/Striatin), and each family has several members (Fig. 1A). Some B subunits are widely expressed, while the expression of others is restricted to a subset of cell types (8). PPP2R5D encodes the delta isoform of the B’-family subunit and is called B’δ, B56δ, PR61, and PPP2R5D in the literature. PPP2R5D/B56δ is ubiquitously expressed, with slightly higher levels reported in the brain, breast, testis, and gastrointestinal tissue (9, 10, 11). In addition to each B-family having several isoforms, additional isoforms are generated by alternate splicing (8, 12). Therefore, although commonly referred to as PP2A in the literature, combinatorially, the PP2A family may include over a 100 unique holoenzymes (Fig. 1A).
Figure 1.
PP2A holoenzyme structure, PPP2R5D variants, and E198K versus E420K variant activity profiling.A, diagram of PP2A holoenzyme structure and components. B, number of cases corresponding to pathogenic PPP2R5D variants based on SFARI database. C, homology model of the PP2A holoenzyme containing PPP2R5D showing the locations of the common PPP2R5D variants. This PPP2R5D model is superimposed onto the known structure of the PPP2R5C-containing holoenzyme (PDB ID: 2IAE). PPP2R5D is colored in yellow, the scaffold subunit in green, and the catalytic subunit in cyan. Metal ions of the catalytic site are pink spheres. Red surfaces correspond to the variant sites such as E198 and E420. D, a zoomed in view of E198 shows an interaction with R89 of the catalytic subunit. E, a zoomed in view of E420 shows potential interactions with aromatic ring hydrogens of F416 and the sidechain of T461. F, Western analysis of total cell lysates (input) and endogenous PPP2R5D IPs probed for PPP2R5D or for the PP2A catalytic subunit (PP2AC). Relative amounts of PP2AC from WT, E198K-het, or E420K-het cells normalized to total PPP2R5D in IPs and represented as the mean percent of WT. The control (CTR) is bead only. Coomassie stain was used as a loading control. (N = 3 independent biological replicates, mean ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) G, relative phosphatase activity of endogenous PPP2R5D IPs. Endogenous PPP2R5D was immunoprecipitated from WT, E198K-het, and E420K-het cells, and hydrolysis of DiFMUP was measured at Ex 360 nm and at Em 460 nm. Activity was normalized to total levels of PPP2R5D detected in IPs through immunoblotting shown in (F). WT activity (black) was normalized at 30 min (endpoint) to 1, and variant activity (E198K-het = red and E420K-het = blue) is shown relative to WT (N = 3 independent biological replicates of cells, each performed as n = 2 technical replicates). H, proliferation assay of WT (black), E198K (red), and E420K (blue) variant cells. (N = 3 independent biological replicates, each performed as n = 2 technical replicates). Error bars represent the mean ± SD, two-way ANOVA Tukey’s, ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, NS: non-significant). PP2A, phosphoprotein phosphatase 2A; PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D.
To date, over 20 germline mutations in the PPP2R5D coding region have been reported, generating pathogenic missense mutations of 13 amino acids (13) (Fig. 1B and Table S1). Many pathogenic variants are charge reversal changes, in which a negatively charged acidic amino acid (e.g., glutamic acid, E) is mutated to a positively charged basic amino acid (e.g., lysine or arginine, K or R) due to a single base genomic alteration (e.g., glutamate (E), encoded by GAA or GAG, is converted to lysine (K), encoded by AAA or AAG). The most frequently observed pathogenic changes are p.Glu198Lys (E198K), p.Glu200Lys (E200K), p.Glu420Lys (E420K), and p.Asp251X (D251X, with X representing Ala, His, Val, or Tyr). Patients with E198K or E420K variants, both predicted to be “probably damaging” mutations using PolyPhen-2 (14), often display severe clinical symptoms (1, 15).
The E198 residue, its surrounding region, and much of the protein core are conserved between PPP2R5C/B56γ and PPP2R5D/B56δ (Fig. S1), and the crystal structure of a trimeric PP2A holoenzyme containing PPP2R5C (B56γ) has been solved (1, 16, 17, 18). In the PPP2R5C structure, the amino acids corresponding to E197, E198, and E200 in PPP2R5D reside in an acidic loop between α-helices 3 and 4. This loop is in close contact with the catalytic metals contained in the C-subunit of the PP2A-core dimer (1, 16, 17, 18), suggesting a role in the regulation of catalytic activity (Fig. 1, C–E). The prevalence of pathogenic variants in the corresponding loop in PPP2R5D indicates that the acidic loop is important for holoenzyme function (Fig. 1, B–E). Homology models also suggest that other pathogenic variants (e.g., E197K, E200K, P201R, W207R, and E420K) are positioned on the surface, facing the active site of the catalytic subunit (Fig. 1, C–E) (1, 4, 15). The greater severity of symptoms observed in patients with the E198K variant compared to patients with the E200K variant may indicate a more significant role for E198 in PP2A function (1, 4, 15). Previous biochemical studies have reported that when expressed in vitro as a tagged fusion protein in HEK298T (1), overexpression of PPP2R5D E198K increases the phosphorylation of PP2A-B56δ substrates, such as serine 9 of GSK-3β (1). These observations are consistent with dominant-negative suppression of PPP2R5D-dependent PP2A activities (1).
To decipher the molecular events associated with clinically relevant PPP2R5D variants, we adapted genomic editing methods to establish human cell lines recapitulating known pathogenic variants in PPP2R5D, allowing the study of how the expression and actions of endogenous variant proteins alter normal biological processes. Employing a fourth-generation single-base editing system (BE4-Gam), we previously generated heterozygous E420K variant cell lines (19). We reported that the endogenous E420K variant in HEK293 cells is associated with increased phosphorylation levels of several PPP2R5D substrates (19). However, the most common pathogenic variant (E198K) could not be generated using BE4-Gam without also causing nonsynonymous bystander mutations, likely because BE4-Gam has a multibase editing window and the desired c.592G>A mutation is located in a region surrounded by multiple cytosines encoding E197, D199, and E200. To overcome this technical challenge, we adapted a CRISPR-PRIME system, which utilizes a catalytically impaired Cas9 nickase (H840A mutant) fused to reverse transcriptase (RT). CRISPR-PRIME editing uses a synthetic prime editing guide RNA (pegRNA), which both specifies the target site and encodes a template encoding the desired edit (Figs. S2 and S3). The methods described herein proved useful for introducing the desired nucleotide change (c.592G>A: p.E198K) in the genome of HEK293 cells and can likely be adapted to generate cell lines with almost any single base change.
To investigate changes in PP2A-PPP2R5D function in E198K heterozygous cells, we performed quantitative proteomic and phosphoproteomic analyses to determine protein and phosphorylation abundance changes in WT, E198K, and E420K HEK293 cells. Our studies reveal both unique and common deregulated phosphorylation events in PPP2R5D E198K and E420K heterozygous cell lines. Intriguingly, we identify increased phosphorylation of an activating threonine (T202) and tyrosine (Y204) on extracellular signal-regulated kinase (ERK) (p44/p42 MAPK), an activating threonine (T389/T412) in p70 ribosomal protein S6 kinase (p70S6K/RPS6KB1/2), activating serines (S221 and S380) of p90 ribosomal protein S6 kinase 1 (RSK1/RPS6KA1), and (S232 and S389) of p90 ribosomal protein S6 kinase 6 (RSK4/RPS6KA6) with a corresponding increase in the phosphorylation of a common substrate (RPS6) as shared in both variant cell lines (20). RPS6 plays a key role in global protein synthesis, ribosomal biogenesis, maturation of pre-rRNA, and translation (21, 22). RPS6 has also been implicated in several other cellular processes including apoptosis, the response to DNA damage, cellular proliferation and migration, and more (20). Yet, while it is well-established that RPS6 is indispensable, the precise role of RPS6 phosphorylation has yet to be elucidated (22).
Although the dysregulation of the upstream regulatory mechanisms differs between the E198K and E420K variant cell lines, inhibition of mTORC1 with rapamycin or of p70S6K with LY2584702 reduces RPS6 phosphorylation in both variant cell lines. These results suggest inappropriate mTORC1 activation may be a common deregulation in PPP2R5D-related neurodevelopmental disorders. Thus, further investigation with rapamycin analogs, such as Everolimus, which has proven safe and effective in treating neuropsychiatric disorders (e.g., tuberous sclerosis complex syndromes (TSC)), as well as LY2584702 treatment, which has been shown helpful to patients with ASD and fragile X, may be warranted to evaluate their potential as therapeutic options for patients with the most common PPP2R5D variants (23, 24, 25).
Results
Generation of PPP2R5D E198K variant cell lines
Our previous studies with PPP2R5D E420K variant cell lines revealed alterations in phosphorylation-regulated signaling cascades. However, it is unclear if the different pathogenic variants alter similar or distinct signaling pathways (19). To address this key question, we employed PE3b, a third-generation prime editor (Figs. S2 and S3), to introduce a single transition (c.592G>A) into the genome of HEK293 cells, generating E198K cell lines containing the most common pathogenic PPP2R5D variant, for comparison with a previously generated E420K line (26).
For PRIME editing, we designed a pegRNA that positions a nicking Cas9 mutant (nCas9) to target the to-be-edited strand at a specific genomic locus and also encodes the desired edit via a RT template and primer binding site contained within its sequence. We synthesized a plasmid expressing both the pegRNA and a secondary nicking sgRNA containing a guide sequence designed to target nicking of the unedited strand only after the pegRNA/RT–mediated single strand edit was made. Upon expression of the PRIME editor system, the sense strand 5′ of the E198 coding sequence is nicked in the PPP2R5D exon 5 (pegRNA nick in Fig. S2), and the K198 mutation is introduced via reverse transcription. Following flap resolution, the sgRNA selectively guides nicking of the unedited antisense strand in edit-containing duplexes (sgRNA nick in Fig. S2) to stimulate mismatch repair using the edited strand as the template. We used electroporation to introduce the plasmid into the cells. The cells were grown for 48 h to ensure cell replication had occurred, which made the mutation permanent in the genome. The cells were then single-cell sorted, and the emerging colonies were screened for the desired c.592G>A base change in a single allele (E198K-heterozygous) by Sanger sequencing (Fig. S3). To ensure the clonal cell lines with the desired mutation were homogenous, each confirmed E198K line was single-cell sorted two additional times before further analysis. Complete genomic exon sequencing of parental, E198K-het-, and E420K-het-variant lines was performed to detect potential off-target editing and spontaneous somatic mutations that could have occurred during repeated single-cell sorting, as previously described (19). Cell lines with off-target or enriched somatic mutations in protein-coding regions were discarded.
Expression, holoenzyme assembly, activity, and proliferation of the PPP2R5D E198K variant
Both mass spectrometry (MS) and Western blot analysis of total lysates from the parental (WT), the E198K, and E420K cell lysates revealed similar PPP2R5D expression (Figs. 1F, S4 and S5). Furthermore, the amount of PPP2R2A, PPP2R5A, and the PP2A scaffolding subunit were not significantly changed (Fig. S4). However, by Western blotting, we detected a slight increase in the levels of PP2AC (Fig. S4). To determine if the expression of PPP2R5D variants affects PP2A holoenzyme stability or assembly, we immunoprecipitated endogenous PPP2R5D and probed the precipitates for PPP2R5D and PP2AC (Figs. 1F and S5). We found that in cells expressing PPP2R5D variants, the amount of catalytic subunit that co-IPs with PPP2R5D was reduced by 25.97 ± 7.5% and 56.1 ± 1.1% for the E198K and E420K variants, respectively (Figs. 1F and S5).
We also compared the phosphatase activity of PP2A holoenzymes containing WT or variant PPP2R5D (Fig. 1G) using an established fluorescent assay that employs 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) as a substrate (27, 28, 29, 30). For these studies, we immunoprecipitated endogenous PPP2R5D using a surplus of a previously characterized PPP2R5D-specific antibody that recognizes a near C-terminal epitope unique to PPP2R5D (19). We observed dephosphorylation of DiFMUP in all samples. When normalized to the amount of PPP2R5D immunoprecipitated, less activity was observed for both PPP2R5D variants. These results are consistent with the reduced levels of catalytic subunit in PPP2R5D IPs generated from the variant cell lines. When normalized to the amount of PP2A catalytic subunit detected in the IPs, the phosphatase activity was similar between WT, E198K, and E420K samples. Together, our data is consistent with the reduction in phosphatase activity observed in the IPs of variant cell extracts arising from reduced retention or reduced incorporation of PP2AC into PPP2R5D variant holoenzymes. Analysis of growth rates using a cell proliferation assay, in which cells were plated at equal density and the number of cells were counted over consecutive days, revealed that PPP2R5D variants alter cell growth. When compared to WT HEK293 cells, we observed slower rates of growth in cells heterozygous for PPP2R5D E198K or E420K, with a significant reduction in growth in E198K and E420K variant cells observed by 96 h and 48 h, respectively (Fig. 1H).
Quantitative changes in the proteome and phosphoproteome of PPP2R5D E198K variant cells
To obtain an unbiased assessment of how the E198K and E420K PPP2R5D variants affect cellular signaling, we employed quantitative proteomics and phosphoproteomics using the tandem mass tag (TMT)-MS3 approach, as reported previously (19). HEK293 WT, E198K, and E420K variant cells were grown as asynchronous populations, collected, lysed, and digested into peptides with trypsin to analyze protein abundance (Fig. 2A). To distinguish peptides from different cell lines and replicates, peptides were bar-coded by labeling with TMT reagent. After labeling, peptides were combined, fractionated, and analyzed by LC-MS3 (Fig. 2A). In this analysis, we identified and quantified 8830 proteins, of which only 34 proteins (0.4%) were significantly increased or decreased at least two-fold in E198K variant cells compared to WT (Fig. 2B and Table S2). Consistent with our previous observations, only 312 (3.5%) proteins were significantly increased or decreased at least two-fold in E420K variant cells compared to WT (Fig. S6, A and B). Importantly, we did not observe significant changes in the abundance of PP2A catalytic, scaffolding, and other regulatory subunits in either PPP2R5D variant cell line (Table S3).
Figure 2.
Quantitative changes in the proteome and phosphoproteome in PPP2R5D E198K variant cells. Workflow for the quantitative proteomics (A) and phosphoproteomics (C) protocol used. Volcano plots for proteomic (B) and protein-corrected phosphoproteomic changes (D) in E198K variant cells (N = 3 and N = 4 independent biological replicates for WT and E198K-het, respectively.) Volcano plots show log2 fold change versus the negative log10 of the p value of the fold change. Statistical significance corresponding to a p value of <0.05 is shown by peptides with a −log10 value of 1.3 or greater. Peptides shown in blue or red are two-fold or more decreased or increased in abundance, respectively. PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D.
To identify changes in phosphorylation abundances, we performed phosphopeptide enrichment, TMT labeled the phosphopeptides, identified the proteins from which the peptides were derived, and quantified the phosphopeptide intensities (Figs. 2C and S6C). In this analysis, we identified and quantified 31,714 phosphorylation sites (Table S2). Although we did not observe large changes in protein abundance, we corrected phosphopeptides for changes in the abundance of the respective protein to reveal changes due to phosphorylation at a specific residue. We identified and quantified the corresponding protein for 28,892 of the 31,714 phosphorylation sites, allowing us to perform protein correction on most phosphopeptides (Table S2). After protein correction, we observed 612 (2.1%) phosphorylation sites that were significantly increased or decreased by at least a two-fold log2 change for the E198K variant (Fig. 2D and Table S2). The majority of these sites, 70%, were increased, and 30% were decreased. For the E420K variant, we identified 1751 (6.1%) phosphorylation sites to be significantly different, of which 78% were increased in phosphorylation (Fig. S6D and Table S2). Comparison with our previously published dataset (19) revealed high correlation for these independent analyses done years apart (Fig. S6, E and F). The higher number of phosphorylation sites with an increase in occupancy in E420K versus E198K variant cells might be due to the greater decrease in catalytic subunit association and enzymatic activity that was observed for the E420K variant PP2A holoenzyme, measured using immunoprecipitation and in vitro phosphatase activity analysis, respectively.
Alterations in phosphorylation signaling in PPP2R5D variant cells
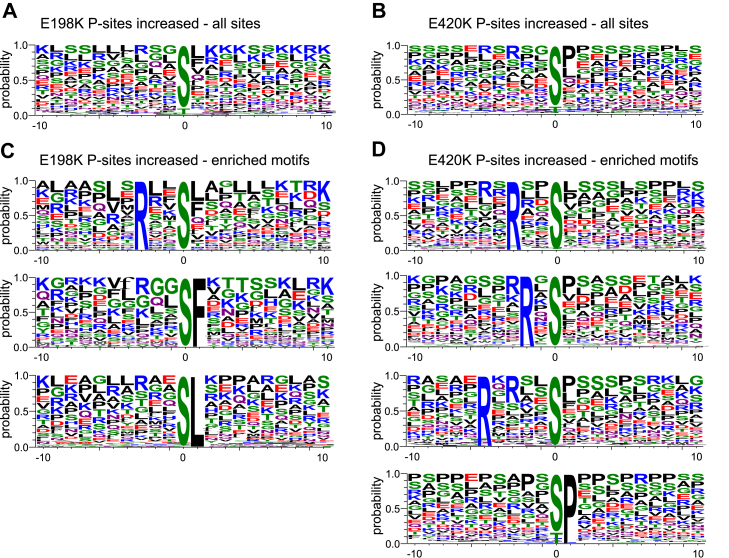
To determine which signaling pathways are altered in the variant cells, we investigated the nature of fully-localized, singly phosphorylated sites in an attempt to reveal the kinases that might be responsible for their phosphorylation. First, we performed phosphorylation site motif enrichment analysis of sites that were significantly increased (Fig. 3, A and B) or decreased (Fig. S7, A and B) by two-fold or more in each variant. For sites that increased in the E198K variant cells, we observed an enrichment of arginine in the −3 position and phenylalanine or leucine in the +1 (Fig. 3C). We found the same arginine enrichment in the −3 position for E420K variant cells and a preference for arginine in the −5 or proline in the +1 position (Fig. 3D). Phosphorylation sites that significantly decreased were characterized by acidic amino acids downstream of the phosphorylation site in both variants (Fig. S7, A and B).
Figure 3.
Phosphorylation site motif enrichment analysis of sites that were significantly increased in the E198K and E420K variant cell line.A, weblogos showing enrichment of amino acids surrounding the phosphorylated residue for sites significantly increased by at least two-fold in PPP2R5D E198K (A) and E420K (B) variant cells. C, enriched motifs among significantly increased phosphorylation sites in the E198K variant. D, enriched motifs among significantly increased phosphorylation sites in the E420K variant. PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D.
To learn more about the kinases that phosphorylate these sites, we used KinomeXplorer to predict upstream kinases of the regulated phosphorylation sites (31). Analysis of phosphorylation sites that increased in variant cells revealed a prevalence of PKC-dependent phosphorylation sites in both the E198K and E420K variant cells (Table S5), which is consistent with an enrichment of arginines in the −3 and larger hydrophobics (leucine or phenylalanine) in the +1 position (Fig. 3, C and D). In E420K variant cells, we also observed protein kinase B- (also known as AKT) and ERK 1/2–dependent motifs and enrichment of consensus sequences of proline-directed kinases, such as cyclin-dependent kinases, glycogen synthase kinase-3, and mitogen-activated protein kinases (MAPKs) (Fig. 3D) (32). Of note, the enrichment of phosphorylation sites with arginine in the −3 and −5 positions (RxRxxS/T) is consistent with the phosphorylation site preference of AKT. This enrichment was previously observed in our studies of E420K variant HEK293 cells (19) but was not apparent in the E198K data. For phosphorylation sites that decreased, we detected a preference for serine-directed sites with proline in the +1 position and the enrichment of acidic amino acids (e.g., E, D) downstream of the phosphorylation site in both datasets. This may indicate dysregulation of casein kinase 2 in the variants (Table S5) (32), which may have relevance because casein kinase 2 has been implicated in a number of neuropsychiatric disorders and the regulation of AKT activity (33).
Pronounced AKT activation occurs in the E420K but not the E198K variant
Studies using inhibitors (e.g., okadaic acid) of the common catalytic subunit (C subunit) or siRNA targeting the common core scaffold (A subunit) have revealed that, as a family, PP2A holoenzymes influence nearly all signaling cascades controlled by reversible phosphorylation (8). Previously, we found that E420K variant cells are characterized by altered insulin-, mTOR-, AMP-activated protein kinase, EGF receptor family (ErbB) signaling, and additional downstream effectors. Notably, in the E420K variant, we observed constitutively active AKT signaling associated with hyperphosphorylation of the downstream protein RPS6 (19). Our phosphoproteomic data confirmed hyperphosphorylation of RPS6 on S235/236 and S240/244 in E420K and revealed a similar increase in E198K variant cells (Fig. 4A). However, although we found an enrichment of an AKT consensus motif (RxRxxS/T) in phosphorylation sites with significantly increased abundance in E420K cells, we did not observe this motif in phosphorylation sites increased in the E198K variant (Fig. 3C). This prompted us to interrogate the AKT signaling pathway further.
Figure 4.
Pronounced AKT activation occurs in the E420K but not the E198K variant.A, LC-MS3 data for sites S240/244 and S235/236 of RPS6 in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines. Value is the log2 of the TMT intensities detected via LC-MS3. B, activating phosphorylations of AKT serine-threonine protein Kinase 1 (AKT1) by 3-phosphoinositide-dependent protein kinase 1 (PDPK1) and mechanistic target of rapamycin kinase complex 2 (mTORC2). C, Log2 LC-MS3 TMT intensities of S473 of AKT1 and S474 of AKT2 in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines. D, representative immunoblots and their quantification showing the levels of P-AKT1/2 at S473/4 and T308/9, of P-RPS6 at S235/236 and S240/244, total AKT protein (Pan AKT) and total RPS6 protein in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lysates. For all plots, mean is represented by the column height, and SD is shown in plots as black bars. Statistical significance (p < 0.05) determined using unpaired, two-tailed t test where an asterisk (∗) indicates a significant result. MS, mass spectrometry; PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D; RPS6, ribosomal protein S6; TMT, tandem mass tag.
Human cells express three isoforms of AKT (AKT1/2/3), and their kinase activity is activated by phosphorylation at T308/9/5 and S473/4/2 by 3-phosphoinositide-dependent protein kinase 1 (PDPK1, also called PDK1) and mTORC2, respectively (Fig. 4B) (34, 35). In our phosphoproteomic data, the amino acid sequence of phosphopeptides allows us to distinguish the AKT isoforms. Compared to WT, we observed an increase in AKT1/2 phosphorylation at S473/4 in E420K but not E198K variant cells, which is consistent with the results of the motif analysis (Figs. 4C and S8). For orthogonal validation of this observation, we performed Western blot analysis of WT, E420K, and E198K variant cells using site-specific phosphoantibodies (Fig. 4D) and observed an increase in AKT1 S473 phosphorylation in E420K but not in E198K variant cells compared to WT (Fig. 4D). Western analysis also revealed increased phosphorylation of the activating T308 site in E420K variant cells (Fig. 4D). Both AKT1 T308 and S473 phosphorylation was increased by greater than 20-fold in the E420K variant cells than WT or E198K. These data revealed that the aberrant AKT activation observed in the E420K variant does not occur in E198K variant, suggesting the observed hyperphosphorylation of RSP6 in E198K variant line may have a different origin.
For orthogonal validation of the AKT phosphorylation/activation unique to E420K variants, we further explored the AKT–mTORC1 pathway by analyzing activating phosphorylations of downstream AKT targets (Fig. 5A). Upon activation, AKT has been reported to phosphorylate and activate mTOR on S2448, when mTOR is part of the mTORC1 complex (36, 37, 38). However, the phosphoproteomic analysis did not reveal a significant increase in this phosphorylation site in either variant (Figs. 5B and S9). mTORC1 activity is also regulated by the small GTPase Rheb, and the TSC subunits 1 and 2 act as a dimer to negatively regulate Rheb-mediated mTORC1 activation (39, 40). AKT phosphorylation of TSC2 at S1130 and S1132 is known to inactivate the TSC1/TSC2 complex preventing the negative regulation of Rheb (40, 41). We found that TSC2 phosphorylation at both S1130 and S1132 is increased in E420K but not E198K variant cells (Fig. 5C). These data are consistent with the activation of mTORC1 in E420K variant cells via an indirect mechanism in which altered AKT-dependent phosphorylation of TSC2 leads to the inactivation of the inhibitory TSC1/2 complex.
Figure 5.
E198K and E420K PPP2R5D variants display aberrant phosphorylation of ribosomal protein S6 and upstream kinases.A, diagram of the AKT-mTOR-RPS6 and Ras-MAPK pathways and their points of convergence. Yellow phosphorylation sites are increased in both variants, whereas red phosphorylation sites are increased in E420K but not E198K. LC-MS3 expression data in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines is shown for phosphorylation of S2448 of mTOR (B) and S1130/1132 of TSC2 (C). Values are log2 TMT intensities detected via LC-MS3. D, representative immunoblot showing the level of P-p70S6K T389 in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines and associated graph of P-T389 of p70S6K normalized to total p70S6K. LC-MS3 TMT intensities in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines are shown for phosphorylation of S241 of PDPK1 (E), S221 of RSK1 and S232 of RSK4 (F), S380 of RSK1 and S389 of RSK4 (G), and S359 and T507 of PKCδ (H). Values are log2 TMT intensities detected via LC-MS3. For all plots, mean is represented by the column height, and SD is shown in plots as black bars. Statistical significance (p < 0.05) was determined using unpaired, two-tailed t test where an asterisk (∗) indicates a significant result. MAPK, mitogen-activated protein kinase; MS, mass spectrometry; PDPK1, 3-phosphoinositide-dependent protein kinase 1; PKCδ, PKC delta; PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D; RPS6, ribosomal protein S6; TSC, tuberous sclerosis complex; TMT, tandem mass tag.
E198K and E420K PPP2R5D variants display aberrant phosphorylation of RPS6 and upstream kinases
RPS6 is phosphorylated by a family of ribosomal protein S6 kinases (S6Ks), PKC delta (PKCδ), PKA, and other kinases affecting growth control (42, 43). For the S6 kinases (RSKs), there are two subfamilies: the p70 ribosomal protein S6 kinases (p70S6Ks: p70S6K-alpha = RPS6KB1, and p70S6K-beta = RPS6KB2) and the p90 ribosomal protein S6 kinases (p90S6Ks: RPS6KA1 = RSK1, RPS6KA2 = RSK3, RPS6KA3 = RSK2, and RPS6KA6 = RSK4). Most members are regulated by reversible phosphorylation of two conserved domains, in which activation requires a “priming” phosphorylation(s) that acts in concert with activating T-loop phosphorylation(s) (44, 45). Together, these phosphorylations lead to the release of autoinhibition and the realigning of active-site residues, allowing both the capture of substrates and the catalysis of phosphorylation.
p70S6Ks are primarily activated via the AKT–mTORC1 pathway (46, 47). Consistent with upstream AKT-mTORC1 activation in the E420K variant, we observed a significant increase in activating p70S6K phosphorylation (T389) in E420K cell lysates at a site known to be phosphorylated by mTORC1 (46, 47). However, we also observed T389 hyperphosphorylation in the E198K variant cells without apparent activation of AKT (Fig. 5D).
The p90S6Ks/RSKs are activated via the PDPK1 and Ras–MAPK pathways (44, 48) (Fig. 5A). Our data revealed that both priming and T-loop activating phosphorylations on RSK1 (S221 T-loop, S380 linker) and RSK4 (S232 T-loop, S389 linker) increased in both the E198K and E420K variants (Fig. 5, F and G). RSK1 S221 and RSK4 S232 are known targets of PDPK1, and phosphorylation of these residues is required for kinase activation (49). However, we did not observe increased phosphorylation of S241 on PDPK1, which is believed to be necessary for its activation (50, 51) (Fig. 5E). In addition, we did not observe altered phosphorylation of other known regulatory residues on several other established substrates for PDPK1 (e.g., RPS6KA3/RSK2, PRKACA, PRKCZ, PAK1, PKN1, or PKN2), suggesting aberrant activation of PDPK1 is not participating in the hyperphosphorylation of p90S6Ks. We also observed increased phosphorylation of two known activating sites on PKCδ: S359 and T507 (Fig. 5H). S359 of PKCδ is an activating phosphorylation site occurring in response to oxidative stress; this site was significantly increased in both the E198K and E420K variants (52). T507 is phosphorylated by PDPK1 (53) and was only significantly increased in the E198K variant. Taken together, although we only observed activating phosphorylation of AKT in E420K variant cells, downstream kinases responsible for RPS6 phosphorylation, including p70S6K, p90S6Ks/RSKs, and PKCδ, are all hyperphosphorylated on sites known to activate kinase activity in both PPP2R5D variants.
E198K variant shows increases in MAPK pathway signaling
To further interrogate the signaling alterations in the variants, specifically the increase in phosphorylation of RSK, S6K, and RPS6, we focused on the Ras–MAPK pathway. RSKs are activated by MAPK and PDPK1 phosphorylation. ERK1 and ERK2, also known as MAPK3 and MAPK1, are activated by phosphorylation at both T202/Y204 and T185/Y187, respectively (54). We observed a marked increase in the phosphorylation of T202/Y204 of ERK1 for both E198K and E420K via Western blot (Figs. 6A and S9). Western blot also confirmed that RSK1 S380 was hyperphosphorylated in both variants, which provides orthogonal validation of the MS data (Figs. 5, F and G and S9). Protein scaffolds have been shown to be essential for efficient ERK signal transduction. One key scaffold is Kinase Suppressor of Ras (KSR1). KSR1 colocalizes with MEK (also known as MAPKK1) and ERK and is phosphorylated by Cdc25C-associated kinase 1 (C-TAK1) at S406 in humans, causing it to bind 14-3-3 proteins and remain sequestered in the cytoplasm (55). It has been shown that in response to growth factor stimulation, PP2A dephosphorylates KSR1 at S406 (S392 in mice), causing KSR1 and associated MAPK complex components to release 14-3-3 proteins, and translocate to the plasma membrane to engage Ras and promote activation of the MAPK pathway (Fig. 6B) (56, 57, 58). Interestingly, we observe a decrease in KSR1 S406 (S392 in mice) phosphorylation in E198K variant cells, possibly contributing to the observed increase in MAPK pathway activity in this variant (Fig. 6C).
Figure 6.
Phosphorylation changes in Ras-MAPK signaling cascade.A, representative immunoblots showing levels of P-p44/p42 MAPK (ERK1/2) T202/Y204 and P-RSK S380 in WT, PPP2R5D E420K-het, and E198K-het HEK293 cell lines. Quantification of P-ERK1/2 (T202/Y204) normalized to total ERK1/2 and of P-RSK (S380) normalized to total level of RSK is shown. B, model depicting KSR1 scaffold leading to coordination of Ras-MAPK signaling following its dephosphorylation of S406 by PP2A. C, LC-MS3 data for S406 of KSR1 in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines. Value is the log2 of the TMT intensities detected via LC-MS3. For all plots, mean is represented by the column height, and SD is shown in plots as black bars. Statistical significance (p < 0.05) was determined using unpaired, two-tailed t test where an asterisk (∗) indicates a significant result. KSR1, Kinase Suppressor of Ras; MAPK, mitogen-activated protein kinase; MS, mass spectrometry; PP2A, phosphoprotein phosphatase 2A; PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D; TMT, tandem mass tag.
Inhibition of RPS6 phosphorylation in variant cells using mTOR and p70S6K inhibitors
In both variant cell lines, we observed increased p70S6K T389 phosphorylation, a direct substrate of mTORC1, and its downstream target RPS6 (S240/244). However, only E420K variant cells displayed hyperactivation of AKT, which is known as a principle mTORC1-activating kinase and can activate mTORC1 via direct and indirect mechanisms (59). Thus, to determine if RPS6 phosphorylation responds to upstream kinase inhibition in both variant cell lines, we treated WT and variant cells with rapamycin (10 nM for 1 h), an mTORC1 inhibitor, or LY2584702 (20 nM for 3 h), a p70S6K inhibitor. We observed that the hyperphosphorylation of p70S6K T389 and RPS6 S240/244 was reduced in WT as well as the E420K and E198K variants by rapamycin treatment (Figs. 7, A–C and S10). Furthermore, inhibition of p70S6K with LY2584702 reduced RPS6 phosphorylation at S235/236 and S240/244 in both the E198K and E420K variant cell lines (Fig. 7, D–F). Of note, after treatment with LY2584702, we observed an increase in p70S6K T389 phosphorylation in WT and variant cell lines and in AKT S473 phosphorylation in WT and E198K variant lines (Fig. S11). Previous reports also observed increased phosphorylation of p70S6K T389 upon LY2584702 treatment, which was explained by an increased association of mTOR with p70S6K (60). Thus, our studies indicate that, although AKT is only hyperphosphorylated at activating residues in E420K PPP2R5D variant cells, activation of mTORC1 and p70S6K signaling occurs in both E198K and E420K variant cells. Furthermore, the downstream aberrant RSP6 hyperphosphorylation is suppressed by either rapamycin or LY2584702 treatment.
Figure 7.
Reversal of aberrant mTOR signaling by rapamycin and p70S6K inhibitor (LY2584702) in E198K and E420K variant cells.A and D, representative immunoblot showing levels of P-p70S6K T389 and P-RPS6 in WT, PPP2R5D E420K-het, and PPP2R5D E198K-het HEK293 cell lines treated with rapamycin (10 nm for 1 h) or LY2584702 (20 nM for 3 h), a p70S6K inhibitor. Quantification of P-T389 of p70S6K normalized to total p70S6K for rapamycin treatment (B) and LY2584702 treatment (E). Quantification of P-S240/244 of RPS6 normalized to total RPS6 for rapamycin (C) and LY2584702 treatment (F). All treatments were performed as N = 3 independent cell experiments, mean ± SD, unpaired t test ∗p < 0.05. PPP2R5D, phosphoprotein phosphatase 2 regulatory protein 5D; RPS6, ribosomal protein S6.
Discussion
Germline mutations in PPP2R5D have been linked to a developmental disorder (OMIM#616355) called Jordan’s Syndrome/PPP2R5D-related neurodevelopment disorder (15). In addition, pathogenic variants in PP2A-scaffolding (PPP2R1A), PP2A-catalytic (PPP2CA), and other regulatory subunits of PP2A holoenzymes have been recently reported to cause neurodevelopmental disorders (61). Now the challenge is to determine the etiology associated with these genomic mutations and to develop interventions for the medical management of the associated symptoms.
The PP2A family, consisting of >100 holoenzymes, is responsible for a large percentage of phosphoserine and phosphothreonine dephosphorylation within a cell (8, 62, 63). Nonetheless, the roles of individual holoenzymes are poorly characterized. To investigate the etiology of PPP2R5D variant disorders, we developed a CRISPR-genomic single-base editing method that allowed the generation of HEK293 cell lines containing a heterozygous transition (c.1258G>A) in PPP2R5D on chromosome 6, precisely recapitulating a clinically relevant E420K variant. Characterization of the HEK293-E420K cell lines revealed that the E420K variant altered an AKT-mTOR–mediated signaling pathway that controls cell size and growth (19). Here, we expanded this analysis to include the most common PPP2R5D variant, E198K, to determine if the different variants alter the same, similar, or different biological processes. To develop cell lines needed for our studies, we adapted a CRISPR Prime Editor 3b (PE3b) strategy described in the methods (26) (Fig. S2). This method allowed us to introduce a single transition c.592G>A in one allele, generating the desired E198K-heterozygous mutation in the genome of HEK293 cells. Parental E198K-het and E420K-het variant lines underwent complete genomic exon sequencing to detect off-target editing and spontaneous somatic mutations (19).
For the study of variant PP2A-holoenzyme complexes, an advantage of employing genomic editing versus exogenous expression is that with genomic editing, the expression, translation, and biogenesis of the variant holoenzymes are controlled by the endogenous cellular machinery. For PP2A holoenzymes, this is particularly important because their biogenesis involves the coordinated actions of many proteins (e.g., alpha4/IGBP1, PPME1, PTPA/PPP2R4, TIPRL, and LCMT1) (8, 64). During biogenesis, the common catalytic subunit is initially inactive. In a poorly understood process, it then undergoes several conformational changes and modifications before it is incorporated into the common A/C core dimer (65). PP2A holoenzymes become functional after a unique B-subunit is integrated with the A/C core dimer. The proteins involved in the biogenesis of PPP2R5D holoenzymes are common for all PP2A holoenzymes found in most cells (12, 66). Thus, in studies where B-regulatory subunits are expressed exogenously, the interpretation of the data can be biased by the possibility that the biogenesis of other PP2A holoenzymes may be altered (e.g., overexpression of a B subunit may overload the biogenesis complex or sequester a limited number of A/C-subunits from other PP2A holoenzymes).
The studies presented here revealed that the PPP2R5D WT and both E198K and E420K variants are expressed at levels that only varied slightly. However, when PPP2R5D is immunoprecipitated, the IPs of both E198K and E420K cell lysates contain less catalytic subunit (PP2AC). This might be due to reduced stability of the variant holoenzymes, altered efficiency of the biogenesis process, altered “recycling” of variant-containing holoenzymes, or a pool of unbound variant PPP2R5D in the cells. Using a homology model of the PP2A holoenzyme, the locations of the common PPP2R5D variants associated with Jordan’s Syndrome are predicted to concentrate at the interface of PPP2R5D and catalytic subunits near the catalytic metals (Fig. 1, C–E). Therefore, charge switching glutamic acid to lysine substitutions may directly or indirectly affect holoenzyme assembly or function. For instance, we recently showed using quantum-based modeling that in the WT holoenzyme, arginine 89 (R89) of the catalytic subunit engages the substrate’s phosphate group for optimal catalytic efficiency while it forms a weak salt bridge with E198 of PPP2R5D when a substrate is absent (67). R89 and R268 in PP2AC act as gatekeepers of the active center by controlling the access of the substrates to the active sites. A lysine substitution of E198 would likely disrupt the interaction with R89 and repel it, possibly also altering local loop conformation. In contrast, E420 is not located within a subunit:subunit interface and only has weak interactions with threonine 461 (T461) and the aromatic ring hydrogens on phenylalanine 416 (F416). It is possible that in the E420K variant, the local structure is altered in some way (e.g., altering orientations of HEAT repeats), weakening the subunit interactions. A cryo-EM study in progress will likely reveal additional insights into the structural effects of these mutations (68). Nonetheless, this aspect of PPP2R5D biology warrants further investigation in future studies of the PPP2R5D-related neurodevelopmental disorder.
However, we also observed a significant reduction in the phosphorylation of PPP2R5D itself at serines 88, 89, and 90 (Table S4). Phosphorylation of these sites was recently predicted to alter the conformation of the PP2A-PPP2R5D holoenzyme leading to its activation (68), which may provide an alternate explanation for our observation of reduced activity. Nonetheless, it should be noted here that the function of these sites is only a prediction, and future studies are needed to experimentally test how PPP2R5D phosphorylation affects holoenzyme activity.
As previously reported for the E420K variant, the global proteome of the PPP2R5D E198K variant cells was stable, with only 0.4% of protein being either increased or decreased compared to WT. Analysis of phosphorylation site levels confirmed our findings related to the E420K variant, revealing ∼6% of the phosphopeptides were significantly increased or decreased by two-fold or more in the E420K variant cell line compared to WT. The E198K variant also had an increase in phosphopeptide occupancy with fewer (only 2.1%) phosphopeptides that significantly increased or decreased by two-fold. In both cell lines, the abundance of the catalytic, scaffolding, and other regulatory subunits of PP2A were unchanged, suggesting there is not an increase in the expression of another subunit to compensate for the aberrant actions of the PPP2R5D variants.
To identify potential therapeutic strategies for the management of PPP2R5D-related disorders, we interrogated signaling cascades altered by PPP2R5D variants. Previously, we found that the E420K PPP2R5D variant was associated with increased AKT–mTORC1 axis signaling leading to RPS6 hyperphosphorylation at S235/236 and S240/244 downstream of constitutive activation of AKT (19). Our results in this study were consistent with our prior observations related to altered AKT activation in E420K based on increased T308 and S473 phosphorylation, which are known to be activating (34). In addition, our dataset clarifies a pathway by which mTORC1 is activated in the E420K variant cells. Notably, our data is not consistent with direct AKT-dependent phosphorylation of mTOR. Instead, we observed indirect regulation, in which TSC2 phosphorylation at S1130 and S1132 was increased. AKT is known to phosphorylate these sites, leading to the inability of TSC1 and TSC2 to act as a dimer in the negative regulation of Rheb-mediated mTORC1 activation (40, 41). Thus, our observations are consistent with mTORC1 activation in the E420K variant occurring by an indirect mechanism in which aberrant AKT activation activates mTORC1 via phosphorylation of TSC2, leading to the inactivation of the inhibitory TSC1/2 complex.
A major observation in this study was that the activating phosphorylations on AKT observed in the E420K variants do not occur in the E198K variant cells. Nonetheless, increased phosphorylation of RPS6 on both S235/236 and S240/244 was observed in both variant cells. RPS6 was the first indispensable ribosomal protein shown to become phosphorylated nearly 50 years ago (42, 69). Studies have since revealed that five evolutionarily conserved and clustered sites are sequentially phosphorylated, starting with S236 and followed by S235, S240, S244, and S247 (42). The initial phosphorylation at S235/236 is mediated by p90S6Ks (RSK1-4), PKC, PKA, protein kinase G, and death-associated protein kinase. P70S6K can also phosphorylate S235/236 in addition to S240/244. Importantly, p70S6K is primarily activated by mTORC1 activation, and p90S6K is activated by ERK/MAPK activation, rendering RPS6 phosphorylation highly responsive to nutrient availability and signaling by growth factors (70). Genetic mouse models displaying altered RPS6 phosphorylation indicate that dysregulation is associated with neurological and neurodevelopmental disorders, including TSC1/TSC2, Fragile X syndrome, Rett syndrome, and others, with common pathologies of autism, ID, and seizures. Altered RPS6 phosphorylation has also been genetically linked to neurodegenerative diseases (e.g., Huntington disease) and mouse models of schizophrenia (42) Mutant mice with nonphosphorylatable RPS6 have impaired glucose tolerance, hypotonia, and growth impairment, whereas children with PPP2R5D variants and RPS6 hyperphosphorylation display overgrowth phenotypes (2, 21, 71). Still despite decades of studies that have clarified the phenotypic consequences of RPS6 phosphorylation at the cellular level, at a molecular level, the functional consequences of RPS6 phosphorylation are not clear.
Because the phenotypic consequences associated with altered RPS6 phosphorylation in animal models share similarities with children afflicted with Jordan’s Syndrome, we further interrogated pathways that lead to RPS6 phosphorylation in our PPP2R5D variant cell lines. Notably, we observed protein phosphorylation alterations in E198K that align with an increase in ERK/MAPK signaling. Of note, we observed an increase in ERK/MAPK phosphorylation at T202/Y204 (activating) and p90RSK phosphorylations: RSK1 (S221 T-loop, S380 linker; activating) and RSK4 (S232-T-loop, S389 linker; activating) in both the E198K and E420K variant lines (Fig. 6). We also observed a significant decrease in KSR1 phosphorylation at S406. KSR1 is part of a multiprotein signaling complex that promotes the activation of MAP kinases downstream of Ras, which has been linked to increased Ras-Raf-MEK-ERK signaling (55, 72). This is particularly interesting given that KSR1 has been reported to be dephosphorylated by PP2A, but our understanding of how growth factor stimulation recruits specific regulatory B subunits to KSR1 is incomplete (57, 73). Inspection of the KSR1 primary amino acid sequence revealed that it contains two predicted short linear docking motifs that have been implicated in the recognition of B56 substrates (74). Together, these observations reveal that increased activity in the Ras–MAPK–ERK pathway activity contributes to RPS6 hyperphosphorylation in PPP2R5D variants.
It is important to note that there is extensive crosstalk between the Ras/ERK and AKT-mTORC1 pathways (59, 75, 76). This crosstalk is context-dependent and highly dynamic. For example, pathway cross-activation is seen when hyperactivity of Ras/ERK pathway leads to direct activation of mTORC1 by ERK/MAPK and p90RSK, as well as indirect activation via inhibition of the TSC2 complex (Fig. 5A) (59). The ability of rapamycin to suppress p70S6K phosphorylation at the activating T389 and RPS6 phosphorylation at S240/244 in the E198K variant cells, without apparent activation of AKT, is consistent with ERK/MAPK activating mTORC1 directly and indirectly via inactivation of TSC2. Here, it is important to note that AKT, ERK, and RSK all phosphorylate different inhibitory sites on TSC2, leading to the disruption and inactivation of the TSC1/TSC2 complex (43, 77). mTORC1 activity has also been shown to be increased through ERK and RSK phosphorylation of Raptor in the mTORC1 complex (78, 79). Deciphering the precise pathway will be difficult because p90RSKs, AKT, p70S6Ks, and PKC are all AGC kinases that share a catalytic kinase domain and have some substrate promiscuity (45) and are capable of phosphorylating the same substrates with the same intensity and duration (59).
Our observations linking both E198K and E420K variants to altered mTOR signaling suggest Jordan’s syndrome should be added to a growing list of disorders associated with the dysregulation of mTOR pathways. Notably, dysregulation of mTOR has been implicated in ASD and other neurological and neurodegenerative disorders (38, 80, 81). Genetic syndromes, such as fragile X syndrome, TSC, PTEN-mediated ASD, neurofibromatosis (NF1), and others (80) are often heterogeneous at a clinical and genetic level, yet numerous studies have shown convergence of key biochemical pathways involving PI3K, AKT, mTOR, ERK (80, 81, 82), and now PPP2R5D. The PI3K/AKT/mTORC1 pathway is also known to regulate neuronal processes, including cell growth and survival, protein synthesis, excitatory synapse formation, and dendritic outgrowth (80). Dysregulation of the pathway also results in seizures, megacephaly, and other symptoms, which are associated with Jordan’s Syndrome and other ASDs (15, 80).
The development of PI3K/mTORC1/p70S6K inhibitors raises the possibility that specific targeting of this pathway may lead to therapeutic advances, and the use of rapamycin and rapalogs has already been widely studied in clinical settings. The ability of both rapamycin and LY2584702 to suppress aberrant RPS6 hyperphosphorylation in PPP2R5D variants suggests that further studies should be conducted to determine if other PPP2R5D variants display similar sensitivities and if these agents will prove helpful in the medical management of Jordan’s Syndrome. Rapamycin has been used for the management of multiple types of cancer, neurodegenerative diseases, including Alzheimer’s and Parkinson’s, and allograft rejection (25, 83, 84). In a mouse model of TSC, rapamycin reversed behavioral abnormalities and reduced epileptic activity (80, 81). However, the use of more specific drugs, such as those that target p70S6K or RSKs, may be better treatment options due to the broad side effects of mTOR inhibition (85, 86). In fragile X mice, seizures that are characterized by elevated RSK signaling can be prevented with RSK inhibitors (86). Several p70S6K inhibitors are in development or clinical trials, and the use of rapamycin or p70S6K inhibitors during early development of PTEN-ASD in mice was able to prevent the emergence ASD-like behavior (77). Similarly, rapamycin and p70S6K inhibitors have been shown to normalize dendritic spine density and reverse neurocognitive deficits in a mouse model of Angelman syndrome (87, 88).
In summary, our studies clarify an AKT-dependent (E420K) and reveal an AKT-independent, ERK/MAPK-mediated (E198K and E420K) activation of mTORC1 and p70S6K as key signaling alterations in PPP2R5D-variant cells. Although the affected pathways are complex and the variant PPP2R5D actions are not yet clearly defined, the pathways identified can be targeted with known drugs that inhibit mTORC1, such as rapamycin or rapamycin analogs and drugs that inhibit p70S6K (e.g., LY2584702). Based on our observations, these agents should be further tested to determine if they can restore normal signaling in a beneficial manner. Exploring mTOR activation in the context of other PPP2R5D variants will help us to gain insight into their complex signaling and determine if targeting this pathway would be of clinical benefit to patients with PPP2R5D-related disorders.
Experimental procedures
Cell culture
HEK293 (Clontech #C3003-1; lot #7030396) cell lines were cultured in Dulbecco’s Modified Eagle’s Medium, containing 25 mM glucose, 4 mM L-glutamine, 1 mM sodium pyruvate, 10 mM MEM nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin (Gibco, Life Technologies), and 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Lot# K19151) at 37 °C with 5% CO2 in a humidified incubator. Cells were routinely passed at 70 to 90% confluency.
pCMV-PE2 (Addgene plasmid #132775; https://www.addgene.org/132775/) was provided as a generous gift from Dr David Liu. This plasmid was amplified in Escherichia coli DH5α and isolated using a Qiagen plasmid DNA purification kit (12123) according to the manufacturer’s protocol and was sequence verified. A dual RNA expression cassette containing sequences for the pegRNA (driven by an hU6 promoter) and the secondary nicking sgRNA (driven by a 7SK promoter) was synthesized, subcloned into pUC57-kan, and sequence verified by GenScript (Piscataway), which also performed plasmid preparation services to provide transfection grade DNA. In addition to the common sgRNA scaffold sequence, the pegRNA incorporated a target (protospacer) sequence (AGGGGCTGAGTTTGACCCAG), a primer-binding site (GGTCAAACTCAGC), and an RT template (TCATCTTTCTCGG). The secondary nicking sgRNA protospacer was GGGTGGGCTCATCTTTCTCG. Two lakh WT HEK293 cells were electroporated using the Neon transfection system. After electroporation, cells were clonally isolated by single-cell sorting into 96-well plates using BD FACSAria II. After clonal expansion, genomic DNA was isolated and regions flanking exon 5 were PCR-amplified using Q5 high-fidelity DNA polymerase (NEB) (oligonucleotide primers F: 5′-TTC GTG TCA GAC CCA CTC AGT G-3′; R: 5′-TTG GCT ATG TTT GGC TGG AAA TC-3′: IDT). Sanger sequencing was employed to detect single-base mutations. Prior to further use, cell lines with the desired mutations were single-cell sorted three times to ensure each cell line represented a homogenous population.
Immunoprecipitations and phosphatase activity assay
Parental, E420K, and E198K variant HEK293 cells were plated and allowed to grow for 48 h. Cells were lysed by the addition of ice-cold lysis buffer [50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA], containing protease inhibitor cocktail (Thermo Fisher Scientific) and passaged several times through a 28G syringe needle. Lysates were clarified by centrifugation at 14,000g for 15 min. The supernatant was transferred to a new tube, and protein concentrations were determined using RC DC Protein Assay (Bio-Rad 5000121). Dynabeads Protein A (100001D) were used to immunoprecipitate (IP) PPP2R5D (ab188323). For the phosphatase assay, beads were incubated with protein lysates for 2 h at room temperature with rotation, and immunoprecipitated samples were eluted into 50 mM Tris–HCl pH 7.4, 150 mM NaCl. The phosphatase activity assay was performed as described previously (19). Briefly, 20 μl of IPs were dispensed in 1.33× assay buffer [0.15 M NaCl, 30 mM Hepes pH 7.0, 1 mM DTT, 0.1 mg/ml BSA, 1 mM ascorbate, 1 mM MnCl2 with a final assay concentration of 75 μM DiFMUP (Invitrogen D6567). Generation of 6,8-difluro-4-methylumbelliferone was measured with a BioTek Synergy (Ex 360 nm, Em 460 nm). Aliquots of the IPs were eluted with near-boiling 2× SDS-PAGE sample buffer and were used for Western analysis to detect total levels of PPP2R5D, which was used to normalize measured activity. To analyze the interaction of the PP2A catalytic subunit, membranes were treated with 0.2 M NaOH for 30 min at 30 °C before probing for PP2AC subunit (Millipore Sigma 05-421, clone 1D6).
Proliferation assays
For analysis of growth curves, cells were counted twice before initial plating. HEK293 WT, E198K-het, and E420K-het cells were plated into 60 mm dishes in three replicates of 1 × 105 cells. After 24, 48, 72, and 96 h, detached or dying cells in the media were collected. Cells were then washed once with PBS, and cells in PBS were added to the tube with cells collected from the media. Adherent cells were detached from the dish with trypsin and combined with the collected cells from the media and PBS. Cells were resuspended and stained with trypan blue for live cell counting. Each plate was counted two times on a TC20 Automated Cell Counter (Bio-Rad 1450102), and the average was recorded for the cell number of that plate.
Western analysis
Cells were grown to confluent monolayers and lysed by scraping cells in near boiling 2× SDS-PAGE sample buffer (62.5 mM Tris–HCl, pH 6.8, 20% glycerol, 4% SDS, 0.0025% bromophenol blue, and 0.02% β-mercaptoethanol) followed by mechanical shearing with a 28G syringe needle. Samples were centrifuged at 14,000g for 15 min, the supernatant was transferred, and protein concentrations were determined using RC DC Protein Assay (Bio-Rad 5000121). Twenty micrograms of each protein sample was separated by electrophoresis and proteins were then transferred onto a Trans-Blot Turbo PVDF Membrane (Biorad 10026933) at 2.5 Å for 3 min using Trans-Blot Turbo Transfer System (Biorad 1704150). The membranes were blocked at room temperature for 1 h in Odyssey blocking buffer (LiCor) and then incubated overnight with primary antibodies at 4 °C. The next day, membranes were incubated with secondary horseradish peroxidase–conjugated antibodies for 1 h at room temperature. Protein bands were visualized with Clarity Western ECL Substrate (Bio-Rad 1705060) using Bio-Rad ChemiDoc MP Imaging System.
Antibody to PPP2R5D was purchased from Abcam (ab188323). Antibody to PP2AC subunit clone 1D6 (05-421) was purchased from Millipore Sigma. For detection of the PP2A catalytic subunit, membranes were treated with 0.2 M NaOH for 30 min at 30 °C before overnight incubation with a primary PP2AC antibody. Primary antibodies to Pan AKT (4691), P-AKT (T308) (13038), P-AKT (S473) (4060), p70 S6 Kinase (34475), P-p70 S6 Kinase (T389) (9205), RPS6 (2217), P-RPS6 (S235/236) (2211), P-RPS6 (S240/244) (5364), PPP2R2A (PP2A B Subunit) (2290), PPP2R1A (PP2A A Subunit) (2041), mTOR (2983), P-mTOR (S2448) (5536), p44/p42 MAPK (ERK1/2) (9102), P-p44/p42 MAPK (ERK1/2) (T202/Y204) (9101), RSK1/RSK2/RSK3 (9355), and P-p90RSK (S380) (11989) were purchased from Cell Signaling Technology. Primary antibody for PPP2R5A (PA5-90634) was purchased from Invitrogen.
Secondary ECL donkey Anti-Rabbit IgG, horseradish peroxidase–linked whole antibodies were purchased from GE Healthcare UK Limited (NA934V), and secondary goat anti-mouse IgG peroxidase-conjugate was purchased from Millipore Sigma (A-5278).
Quantitative proteomic and phosphoproteomic analyses
Cell pellets were resuspended in ice-cold lysis buffer (8 M urea, 25 mM Tris–HCl pH 8.6, 150 mM NaCl, containing phosphatase inhibitors and protease inhibitors (Roche Life Sciences)) and were lysed by sonication. Lysates were subjected to centrifugation (15,000g for 30 min at 4 °C), and supernatants were transferred to a new tube, and the protein concentration was determined using a bicinchoninic acid assay (Pierce/Thermo Fisher Scientific). DTT and iodoacetamide were added to reduce and alkylate, respectively. Samples were incubated overnight at 37 °C with 1:100 (w/w) trypsin. The next day, the trypsin digest was stopped by the addition of 0.25% TFA (final v/v). Precipitated lipids were removed by centrifugation (3500g for 15 min), and the peptides in the supernatant were desalted over an Oasis HLB 60 mg plate (Waters). An aliquot containing ∼100 μg of peptides was removed and labeled with TMT reagent (Thermo Fisher Scientific). Once labeling efficiency was confirmed to be at least 95%, each reaction was quenched by the addition of hydroxylamine to a final concentration of 0.25% for 10 min, mixed, acidified with TFA to a pH of about 2, and desalted over an Oasis HLB 10 mg plate (Waters).
The desalted multiplex was dried by vacuum centrifugation and separated by offline pentafluorophenyl-based reversed-phase HPLC fractionation as previously described (89). TMT-labeled peptides were analyzed on an Orbitrap Lumos mass spectrometer (Thermo Fisher Scientific) equipped with an Easy-nLC 1200 (Thermo Fisher Scientific), and raw data was searched and processed as previously described (19). In brief, samples were analyzed on the Orbitrap Lumos operating in data-dependent, SPS-MS3 quantification mode wherein an Orbitrap MS1 scan was taken (scan range = 350–1400 m/z, R = 120K, AGC target = 2.5e5, max ion injection time = 50 ms), followed by data-dependent ion trap MS2 scans on the most abundant precursors for 1.8 s. Ion selection; charge state = 2: minimum intensity 2e5, precursor selection range 600 to 1400 m/z; charge state 3 to 5: minimum intensity 4e5. Quadrupole isolation = 0.8 m/z, CID collision energy = 35%, CID activation time = 10 ms, activation Q = 0.25, scan range mode = m/z normal, ion trap scan rate = rapid, AGC target = 4e3, max ion injection time = 40 ms. Orbitrap MS3 scans for quantification (R = 50K, AGC target = 5e4, max ion injection time = 100 ms, HCD collision energy = 65%, scan range = 100–500 m/z, synchronous precursors selected = 10). The raw data files were searched using COMET with a static mass of 229.162932 Da on peptide N-termini and lysines and 57.02146 Da on cysteines and a variable mass of 15.99491 Da on methionines against the target-decoy version of the human proteome sequence database (UniProt; downloaded 2/2013, 40,482 entries of forward and reverse protein sequences), maximum of three missed cleavages allowed, precursor ion mass tolerance 1 Da, fragment ion mass tolerance ±8 ppm, and filtered to a <1% FDR at the peptide level. Quantification of LC-MS/MS spectra was performed using in-house developed software. Peptide intensities were adjusted based on total TMT reporter ion intensity in each channel and log2 transformed. p-values were calculated using a two-tailed Student’s t test, assuming unequal variance.
Phosphopeptide enrichment was achieved using an Fe-NTA phosphopeptide enrichment kit (Thermo Fisher Scinetific) according to instructions provided by the manufacturer and desalted over an Oasis HLB 10 mg plate (Waters). Phosphopeptides were then labeled with TMT reagents, offline separated as described above, and analyzed on the Orbitrap Lumos. The probability of phosphorylation site localization was determined by PhosphoRS (90). Quantification and data analysis were carried out as described above.
Bioinformatics analysis
Motif analysis was performed using singly phosphorylated sites with a phosphorylation localization score ≥0.95 that significantly increased or decreased by at least 2-fold after protein correction (91). Upstream kinase prediction was carried out using KinomeXplorer (31).
Statistical analysis
GraphPad Prism was used for all statistical analyses and is reported with SD. Unpaired t-tests were performed for analysis between two sample groups or Kruskal–Wallis with Dunn’s multiple comparisons for multiple sample groups and with Dunnett’s for comparison of multiple sample groups compared to WT, as indicated in figure legends. The proliferation assay was analyzed as two-way ANOVA Tukey’s. N is defined as independent biological replicate cell samples, and n is defined as independent technical replicates. Proteomics and phosphoproteomics data were analyzed using Perseus (92, 93).
Data availability
Raw MS data for this study are available at MassIVE (MSV000090346) and PRIDE accession (PXD036848). Reviewer password: p1163.
Supporting information
This article contains supporting information (19, 26).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
pCMV-PE2 was a gift from David Liu (Addgene plasmid # 132775; http://n2t.net/addgene:132775; RRID:Addgene_132775). We appreciate obtaining access to the genetic data on SFARI Base. We would also like to thank the labs of Dr Prachee Avasthi and Dr Bryan Luikart for their use of antibodies for Western blotting of ERK and mTOR, respectively.
Author contributions
K. A. S., M. R. S., R. E. H., and A. N. K. conceptualization; K. A. S., C. M. P., M. R. S., A. M., C. L., E. A. S., and A. D. C. validation; K. A. S., C. M. P., M. R. S., A. M., C. L., and A. D. C. investigation; K. A. S. and M. R. S. writing–original draft; K. A. S. and C. M. P. visualization; K. A. S. formal analysis; C. M. P., R. E. H., and A. N. K. writing–review and editing; R. E. H. and A. N. K. supervision; R. E. H. and A. N. K. funding acquisition.
Funding and additional information
The research was supported by Jordan’s Guardians Angels; The State of California Regents of the University of California (UC Davis); Project Numbers: 2021 SB 129 #44 and 2018 SB 840 (Nolta, Jan A. (PI); subaward A19-3376-S003 and A22-2855_S003 to R. E. H., Frederic P. Whiddon College of Medicine fellowship to C. M. P., NIH/NIGMS R35GM119455 to A. N. K., and F30GM145149 to K. A. S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Phillip A. Cole
Contributor Information
Richard E. Honkanen, Email: rhonkanen@southalabama.edu.
Arminja N. Kettenbach, Email: Arminja.N.Kettenbach@dartmouth.edu.
Supporting information
References
- 1.Houge G., Haesen D., Vissers L.E.L.M., Mehta S., Parker M.J., Wright M., et al. B56δ-related protein phosphatase 2A dysfunction identified in patients with intellectual disability. J. Clin. Invest. 2015;125:3051–3062. doi: 10.1172/JCI79860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loveday C., Tatton-Brown K., Clarke M., Westwood I., Renwick A., Ramsay E., et al. Mutations in the PP2A regulatory subunit B family genes PPP2R5B, PPP2R5C and PPP2R5D cause human overgrowth. Hum. Mol. Genet. 2015;24:4775–4779. doi: 10.1093/hmg/ddv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirzaa G., Foss K., Nattakom M., Chung W.K. In: GeneReviews® [Internet] Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., et al., editors. University of Washington, Seattle; 1993–2020; Seattle, WA: 2019. PPP2R5D-related neurodevelopmental disorder. [Google Scholar]
- 4.Shang L., Henderson L.B.B., Cho M.T.T., Petrey D.S.S., Fong C.T.C.-T., Haude K.M.M., et al. De novo missense variants in PPP2R5D are associated with intellectual disability, macrocephaly, hypotonia, and autism. Neurogenetics. 2016;17:43–49. doi: 10.1007/s10048-015-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald T.W., Gerety S.S., Jones W.D., van Kogelenberg M., King D.A., McRae J., et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brautigan D.L. Protein Ser/Thr phosphatases--the ugly ducklings of cell signalling. FEBS J. 2013;280:324–345. doi: 10.1111/j.1742-4658.2012.08609.x. [DOI] [PubMed] [Google Scholar]
- 7.Brautigan D.L., Shenolikar S. Protein serine/threonine phosphatases: keys to unlocking regulators and substrates. Annu. Rev. Biochem. 2018;87:921–964. doi: 10.1146/annurev-biochem-062917-012332. [DOI] [PubMed] [Google Scholar]
- 8.Janssens V., Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCright B., Rivers A.M., Audlin S., Virshup D.M. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 1996;271:22081–22089. doi: 10.1074/jbc.271.36.22081. [DOI] [PubMed] [Google Scholar]
- 10.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt T., Samaras P., Frejno M., Gessulat S., Barnert M., Kienegger H., et al. ProteomicsDB. Nucleic Acids Res. 2018;46:D1271–D1281. doi: 10.1093/nar/gkx1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virshup D.M. Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 2000;12:180–185. doi: 10.1016/s0955-0674(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 13.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas D., Cary W., Nolta J.A. PPP2R5D-related intellectual disability and neurodevelopmental delay: a review of the current understanding of the genetics and biochemical basis of the disorder. Int. J. Mol. Sci. 2020;21:1286. doi: 10.3390/ijms21041286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., Fan E., et al. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Cho U.S., Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., Chen Y., Zhang P., Jeffrey P.D., Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol. Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papke C.M., Smolen K.A., Swingle M.R., Cressey L., Heng R.A., Toporsian M., et al. A disorder-related variant (E420K) of a PP2A-regulatory subunit (PPP2R5D) causes constitutively active AKT-mTOR signaling and uncoordinated cell growth. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruvinsky I., Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Bohlen J., Roiuk M., Teleman A.A. Phosphorylation of ribosomal protein S6 differentially affects mRNA translation based on ORF length. Nucleic Acids Res. 2021;49:13062–13074. doi: 10.1093/nar/gkab1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi Y.W., You K.S., Park J.S., Lee S.G., Seong Y.S. Ribosomal protein S6: a potential therapeutic target against cancer? Int. J. Mol. Sci. 2021;23:48. doi: 10.3390/ijms23010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya A., Mamcarz M., Mullins C., Choudhury A., Boyle R.G., Smith D.G., et al. Targeting translation control with p70 S6 kinase 1 inhibitors to reverse phenotypes in fragile X syndrome mice. Neuropsychopharmacology. 2016;41:1991–2000. doi: 10.1038/npp.2015.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolcher A., Goldman J., Patnaik A., Papadopoulos K.P., Westwood P., Kelly C.S., et al. A phase I trial of LY2584702 tosylate, a p70 S6 kinase inhibitor, in patients with advanced solid tumours. Eur. J. Cancer. 2014;50:867–875. doi: 10.1016/j.ejca.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 25.Salussolia C.L., Klonowska K., Kwiatkowski D.J., Sahin M. Genetic etiologies, diagnosis, and treatment of tuberous sclerosis complex. Annu. Rev. Genomics Hum. Genet. 2019;20:217–240. doi: 10.1146/annurev-genom-083118-015354. [DOI] [PubMed] [Google Scholar]
- 26.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choy M.S., Swingle M., D'Arcy B., Abney K., Rusin S.F., Kettenbach A.N., et al. PP1:Tautomycetin complex reveals a path toward the development of PP1-specific inhibitors. J. Am. Chem. Soc. 2017;139:17703–17706. doi: 10.1021/jacs.7b09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chattopadhyay D., Swingle M.R., Salter E.A., Wood E., D'Arcy B., Zivanov C., et al. Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C. Biochem. Pharmacol. 2016;109:14–26. doi: 10.1016/j.bcp.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni L., Swingle M.S., Bourgeois A.C., Honkanen R.E. High yield expression of serine/threonine protein phosphatase type 5, and a fluorescent assay suitable for use in the detection of catalytic inhibitors. Assay Drug Dev. Technol. 2007;5:645–653. doi: 10.1089/adt.2007.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegner A.M., McConnell J.L., Blakely R.D., Wadzinski B.E. An automated fluorescence-based method for continuous assay of PP2A activity. Methods Mol. Biol. 2007;365:61–69. doi: 10.1385/1-59745-267-X:61. [DOI] [PubMed] [Google Scholar]
- 31.Horn H., Schoof E.M., Kim J., Robin X., Miller M.L., Diella F., et al. KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods. 2014;11:603–604. doi: 10.1038/nmeth.2968. [DOI] [PubMed] [Google Scholar]
- 32.Miller M.L., Jensen L.J., Diella F., Jørgensen C., Tinti M., Li L., et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008;1 doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borgo C., D'Amore C., Sarno S., Salvi M., Ruzzene M. Protein kinase CK2: a potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021;6:183. doi: 10.1038/s41392-021-00567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu W., Hall M.N. Regulation of mTORC2 signaling. Genes (Basel) 2020;11:1045. doi: 10.3390/genes11091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosner M., Siegel N., Valli A., Fuchs C., Hengstschläger M. MTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids. 2010;38:223–228. doi: 10.1007/s00726-008-0230-7. [DOI] [PubMed] [Google Scholar]
- 37.Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 38.Hoeffer C.A., Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H., Jiang X., Li B., Yang H.J., Miller M., Yang A., et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J., Manning B.D. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castel P., Ellis H., Bago R., Toska E., Razavi P., Carmona F.J., et al. PDK1-SGK1 signaling sustains AKT-independent mTORC1 activation and confers resistance to PI3Kα inhibition. Cancer Cell. 2016;30:229–242. doi: 10.1016/j.ccell.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biever A., Valjent E., Puighermanal E. Ribosomal protein S6 phosphorylation in the nervous system: from regulation to function. Front. Mol. Neurosci. 2015;8:75. doi: 10.3389/fnmol.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roux P.P., Shahbazian D., Vu H., Holz M.K., Cohen M.S., Taunton J., et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacinto E., Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 2008;410:19–37. doi: 10.1042/BJ20071518. [DOI] [PubMed] [Google Scholar]
- 45.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 46.Pearson R.B., Dennis P.B., Han J.W., Williamson N.A., Kozma S.C., Wettenhall R.E., et al. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnuson B., Ekim B., Fingar D.C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 48.Keshwani M.M., von Daake S., Newton A.C., Harris T.K., Taylor S.S. Hydrophobic motif phosphorylation is not required for activation loop phosphorylation of p70 ribosomal protein S6 kinase 1 (S6K1) J. Biol. Chem. 2011;286:23552–23558. doi: 10.1074/jbc.M111.258004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anjum R., Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 50.Casamayor A., Morrice N.A., Alessi D.R. Phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo. Biochem. J. 1999;342:287–292. [PMC free article] [PubMed] [Google Scholar]
- 51.Levina A., Fleming K.D., Burke J.E., Leonard T.A. Activation of the essential kinase PDK1 by phosphoinositide-driven trans-autophosphorylation. Nat. Commun. 2022;13:1874. doi: 10.1038/s41467-022-29368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong J., Yao Y., Zhang P., Udayasuryan B., Komissarova E.V., Chen J., et al. The C2 domain and altered ATP-binding loop phosphorylation at Ser 359 mediate the redox-dependent increase in protein kinase C-δ activity. Mol. Cell. Biol. 2015;35:1727–1740. doi: 10.1128/MCB.01436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong J., Park M., Steinberg S.F. Cleavage alters the molecular determinants of protein kinase C-δ catalytic activity. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00324-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klomp J.E., Klomp J.A., Der C.J. The ERK mitogen-activated protein kinase signaling network: the final frontier in RAS signal transduction. Biochem. Soc. Trans. 2021;49:253–267. doi: 10.1042/BST20200507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller J., Ory S., Copeland T., Piwnica-Worms H., Morrison D.K. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell. 2001;8:983–993. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 56.Lewis R., Frodyma D., Neilsen B., Costanzo-Garvey D., Fisher K. Coordinating ERK signaling via the molecular Scaffold kinase suppressor of Ras. F1000Res. 2017;6:1621. doi: 10.12688/f1000research.11895.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ory S., Zhou M., Conrads T.P., Veenstra T.D., Morrison D.K. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr. Biol. 2003;13:1356–1364. doi: 10.1016/s0960-9822(03)00535-9. [DOI] [PubMed] [Google Scholar]
- 58.Razidlo G.L., Kortum R.L., Haferbier J.L., Lewis R.E. Phosphorylation regulates KSR1 stability, ERK activation, and cell proliferation. J. Biol. Chem. 2004;279:47808–47814. doi: 10.1074/jbc.M406395200. [DOI] [PubMed] [Google Scholar]
- 59.Mendoza M.C., Er E.E., Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 2011;36:320. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu H., Feng X., Ennis K.N., Behrmann C.A., Sarma P., Jiang T.T., et al. Pharmacologic targeting of S6K1 in PTEN-deficient neoplasia. Cell Rep. 2017;18:2088–2095. doi: 10.1016/j.celrep.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandal P., Jong C.J., Merrill R.A., Song J., Strack S. Protein phosphatase 2A – structure, function and role in neurodevelopmental disorders. J. Cell Sci. 2021;134 doi: 10.1242/jcs.248187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kruse T., Gnosa S.P., Nasa I., Garvanska D.H., Hein J.B., Nguyen H., et al. Mechanisms of site-specific dephosphorylation and kinase opposition imposed by PP2A regulatory subunits. EMBO J. 2020;39 doi: 10.15252/embj.2019103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruvolo P.P. The broken “Off” switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin. 2016;6:87–99. doi: 10.1016/j.bbacli.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kong M., Ditsworth D., Lindsten T., Thompson C.B. Alpha4 is an essential regulator of PP2A phosphatase activity. Mol. Cell. 2009;36:51–60. doi: 10.1016/j.molcel.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sents W., Ivanova E., Lambrecht C., Haesen D., Janssens V. The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J. 2013;280:644–661. doi: 10.1111/j.1742-4658.2012.08579.x. [DOI] [PubMed] [Google Scholar]
- 66.Virshup D.M., Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 67.Salter E.A., Wierzbicki A., Honkanen R.E., Swingle M.R. Quantum-based modeling implies that bidentate Arg89-substrate binding enhances serine/threonine protein phosphatase-2A(PPP2R5D/PPP2R1A/PPP2CA)-mediated dephosphorylation. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1141804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu C.-G., Balakrishnan V.K., Parihar P.S., Konovolov K., Chen Y.-C., Merrill R.A., et al. Extended regulation interface coupled to the allosteric network and disease mutations in the PP2A-B56δ holoenzyme. bioRxiv. 2023 doi: 10.1101/2023.03.09.530109. [preprint] [DOI] [Google Scholar]
- 69.Gressner A.M., Wool I.G. The phosphorylation of liver ribosomal proteins in vivo. Evidence that only a single small subunit protein (S6) is phosphorylated. J. Biol. Chem. 1974;249:6917–6925. [PubMed] [Google Scholar]
- 70.Meyuhas O. Ribosomal protein S6 phosphorylation: four decades of research. Int. Rev. Cell Mol. Biol. 2015;320:41–73. doi: 10.1016/bs.ircmb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Ruvinsky I., Katz M., Dreazen A., Gielchinsky Y., Saada A., Freedman N., et al. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen A., Burack W.R., Stock J.L., Kortum R., Chaika O.V., Afkarian M., et al. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell. Biol. 2002;22:3035–3045. doi: 10.1128/MCB.22.9.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raabe T., Rapp U.R. Ras signaling: PP2A puts Ksr and Raf in the right place. Curr. Biol. 2003;13:R635–R637. doi: 10.1016/s0960-9822(03)00568-2. [DOI] [PubMed] [Google Scholar]
- 74.Davey N.E., Haslam N.J., Shields D.C., Edwards R.J. SLiMFinder: a web server to find novel, significantly over-represented, short protein motifs. Nucleic Acids Res. 2010;38:W534–W539. doi: 10.1093/nar/gkq440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arkun Y. Dynamic modeling and analysis of the cross-talk between insulin/AKT and MAPK/ERK signaling pathways. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aksamitiene E., Kiyatkin A., Kholodenko B.N. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem. Soc. Trans. 2012;40:139–146. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- 77.Huang W.C., Chen Y., Page D.T. Hyperconnectivity of prefrontal cortex to amygdala projections in a mouse model of macrocephaly/autism syndrome. Nat. Commun. 2016;7:1–15. doi: 10.1038/ncomms13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carrière A., Cargnello M., Julien L.A., Gao H., Bonneil É., Thibault P., et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 79.Carriere A., Romeo Y., Acosta-Jaquez H.A., Moreau J., Bonneil E., Thibault P., et al. ERK1/2 phosphorylate raptor to promote ras-dependent activation of mTOR complex 1 (mTORC1) J. Biol. Chem. 2011;286:567–577. doi: 10.1074/jbc.M110.159046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boksha I.S., Prokhorova T.A., Tereshkina E.B., Savushkina O.K., Burbaeva G.S. Protein phosphorylation signaling cascades in autism: the role of mTOR pathway. Biochemistry (Moscow) 2021;86:577–596. doi: 10.1134/S0006297921050072. [DOI] [PubMed] [Google Scholar]
- 81.Hooshmandi M., Wong C., Khoutorsky A. Dysregulation of translational control signaling in autism spectrum disorders. Cell Signal. 2020;75 doi: 10.1016/j.cellsig.2020.109746. [DOI] [PubMed] [Google Scholar]
- 82.Rai S.N., Dilnashin H., Birla H., Singh S.S., Zahra W., Rathore A.S., et al. The role of PI3K/Akt and ERK in neurodegenerative disorders. Neurotox Res. 2019;35:775–795. doi: 10.1007/s12640-019-0003-y. [DOI] [PubMed] [Google Scholar]
- 83.Mohamed M.A., Elkhateeb W.A., Daba G.M. Rapamycin golden jubilee and still the miraculous drug: a potent immunosuppressant, antitumor, rejuvenative agent, and potential contributor in COVID-19 treatment. Bioresour. Bioprocess. 2022;9:65. doi: 10.1186/s40643-022-00554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bové J., Martínez-Vicente M., Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat. Rev. Neurosci. 2011;12:437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 85.Fenton T.R., Gout I.T. Functions and regulation of the 70 kDa ribosomal S6 kinases. Int. J. Biochem. Cell Biol. 2011;43:47–59. doi: 10.1016/j.biocel.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 86.Sawicka K., Pyronneau A., Chao M., Bennett M.V.L., Zukin R.S. Elevated ERK/p90 ribosomal S6 kinase activity underlies audiogenic seizure susceptibility in fragile X mice. Proc. Natl. Acad. Sci. U. S. A. 2016;113 doi: 10.1073/pnas.1610812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun J., Liu Y., Moreno S., Baudry M., Bi X. Imbalanced mechanistic target of rapamycin C1 and C2 activity in the cerebellum of Angelman syndrome mice impairs motor function. J. Neurosci. 2015;35:4706–4718. doi: 10.1523/JNEUROSCI.4276-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun J., Liu Y., Tran J., O’Neal P., Baudry M., Bi X. mTORC1–S6K1 inhibition or mTORC2 activation improves hippocampal synaptic plasticity and learning in Angelman syndrome mice. Cell. Mol. Life Sci. 2016;73:4303–4314. doi: 10.1007/s00018-016-2269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grassetti A.V., Hards R., Gerber S.A. Offline pentafluorophenyl (PFP)-RP prefractionation as an alternative to high-pH RP for comprehensive LC-MS/MS proteomics and phosphoproteomics. Anal. Bioanal. Chem. 2017;409:4615–4625. doi: 10.1007/s00216-017-0407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 91.Wang T., Kettenbach A.N., Gerber S.A., Bailey-Kellogg C. MMFPh: a maximal motif finder for phosphoproteomics datasets. Bioinformatics. 2012;28:1562–1570. doi: 10.1093/bioinformatics/bts195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 93.Yu S.H., Ferretti D., Schessner J.P., Rudolph J.D., Borner G.H.H., Cox J. Expanding the Perseus software for omics data analysis with custom plugins. Curr. Protoc. Bioinformatics. 2020;71:1–29. doi: 10.1002/cpbi.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw MS data for this study are available at MassIVE (MSV000090346) and PRIDE accession (PXD036848). Reviewer password: p1163.