Abstract
Is Cannabis a boon or bane? Cannabis sativa has long been a versatile crop for fiber extraction (industrial hemp), traditional Chinese medicine (hemp seeds), and recreational drugs (marijuana). Cannabis faced global prohibition in the twentieth century because of the psychoactive properties of ∆9-tetrahydrocannabinol; however, recently, the perspective has changed with the recognition of additional therapeutic values, particularly the pharmacological potential of cannabidiol. A comprehensive understanding of the underlying mechanism of cannabinoid biosynthesis is necessary to cultivate and promote globally the medicinal application of Cannabis resources. Here, we comprehensively review the historical usage of Cannabis, biosynthesis of trichome-specific cannabinoids, regulatory network of trichome development, and synthetic biology of cannabinoids. This review provides valuable insights into the efficient biosynthesis and green production of cannabinoids, and the development and utilization of novel Cannabis varieties.
Introduction
Cannabis sativa (Cannabaceae) is a diploid (2n = 20) dicotyledonous annual herbaceous plant and is distributed worldwide [1]. Historically, Cannabis has been grown as an important food, fiber, and medicinal crop. The whole Cannabis plant is used for various purposes. For example, its stems have been used for fiber production, which has been practiced for millennia. Its fruit, known as ‘huomaren’ in traditional Chinese medicine, functions as both medicine and food as it contains over 90 types of polyunsaturated fatty acids [2, 3]. Additionally, Cannabis roots are used to treat inflammation and pain [4]. Cannabinoids, abundant in secretory glandular trichomes (GTs) densely inlaid in the female inflorescence of Cannabis [5], exhibit medicinal or recreational properties.
Cannabinoids, a group of C21 terpenophenolic compounds, are the most abundant in Cannabis. Among them, the well-known psychoactive cannabinoid ∆9-tetrahydrocannabinol (Δ9-THC) has addictive properties but is efficacious as an analgesic, antiemetic, and antispastic agent. Further, cannabidiol (CBD), a nonpsychoactive cannabinoid, has been clinically validated to treat specific medical conditions, such as epilepsy, glaucoma, and depressive disorder. Moreover, CBD shows potential as an anti-inflammatory treatment for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [6]. Cannabis has traditionally been classified into two primary types according to the concentration of THC: industrial hemp (THC <0.3%) and marijuana (THC ≥0.3%) [7]. Owing to the high economic value of CBD, an updated classification has emerged for medicinal Cannabis, with THC content <0.3% and high CBD levels [8]. However, the legal status and regulations surrounding the use of Cannabis and its cannabinoids vary by different countries. Therefore, it is essential to seek guidance from medical professionals and adhere to local laws and regulations when considering the medical applications of Cannabis. The global legal Cannabis market is projected to reach a value of US$102.2 billion by 2030, driven by the increasing medical and economic demands (https://www.giiresearch.com/publisher/GRVI/). This growth also presents significant challenges for Cannabis cultivation and cannabinoid production.
Remarkable advancements in the cultivation of Cannabis plants with high-yielding cannabinoids and the reconstruction of cannabinoid production in microorganisms via metabolic engineering have been achieved [9]. The integration of multi-omics methodologies, including genomics, transcriptomics, and metabolomics, has provided comprehensive insights into the genetic composition, gene expression patterns, and regulation of cannabinoid biosynthesis. Further, the relaxation of regulations and the authorization of Cannabis research have expanded the opportunities for studying this plant. Here, we summarize Cannabis historical usage, cannabinoid biosynthesis, trichome development regulatory network, and cannabinoid metabolic engineering. This review aims to enhance the understanding of Cannabis development and utilization, paving the way for further exploration and innovation in Cannabis research.
History and domestication of cannabis
Cannabis: ancient significance, enduring influence
Cannabis has a long history of human use, with documented evidence from around the world [10, 11] (Fig. S1). The medicinal use of Cannabis was first documented in ‘Shen Nong Ben Cao Jing’, which is said to have originated from the emperor Chen Nung (2737 BCE) and was passed down orally until it was assembled (dated no later than 221 BCE). In Egypt, the medicinal use of Cannabis was first mentioned in ‘Papyrus Ramesseum III’ (1550 BCE), where Cannabis was ground and used to treat eye diseases [12]. In India, ‘Soma’ (potentially originating from Cannabis) was considered paradisiacal. The description of Soma in Rig Veda (1400 BCE), an ancient Indian literature, may be the earliest reference to the psychoactive activity of Cannabis [13]. Subsequently, Cannabis has been recorded in many important medical texts such as ‘Shi Liao Ben Cao’ and ‘Compendium of Materia Medica’ in China, providing detailed information on the medicinal effects of Cannabis inflorescence and seeds. Furthermore, classical medicinal recipes frequently included Cannabis, for example, Hua Tuo used Cannabis to make ‘Ma Fei San’ for anesthesia in 207 AD, demonstrating the widespread recognition and appreciation of the therapeutic properties of Cannabis in the ancient society [14]. A series of recent events on the medicinal properties of Cannabis have gradually unraveled its pharmacological mechanism. In 1798, Napoleon brought Cannabis back to France from Egypt and investigated it for its pain-relieving and sedative qualities [15]. In the 20th century, CBD and THC were successfully isolated and synthesized in succession [16–18]. Subsequently, the discovery of CB1 and CB2 receptors, the target of cannabinoids in the human body, and endocannabinoid system (ECS) has revealed the relationship between cannabinoids and human health in maintaining homeostasis and influencing various functions such as sleep, appetite, pain perception, inflammation, memory, mood, and reproduction [19, 20].
Historically, Cannabis use has encompassed various productive and religious purposes, playing a crucial role in diverse aspects of people’s lives. Originally, Cannabis was primarily cultivated to obtain seeds and fiber [21]. Approximately 3000 years ago, ‘The Book of Songs’ provided a detailed record of the entire process involved in hemp clothing production, likely making it the earliest written record. Moreover, the ‘Shuo Wen Jie Zi’ (121 AD) described that disorderly padded hemp fibers were used to keep warm during winter. These ancient written records highlight the significance of hemp as an important textile material at that time. Additionally, increasing archaeological evidence indicated the spiritual use of Cannabis smoke before 1260 BCE. It is believed to be the earliest discovery of the narcotic and hallucinogenic effects of Cannabis [22]. Cannabinol (CBN), an oxidation product of THC, was detected on the surface of ten wooden fire pots and cobblestones excavated from the Jirzankal cemetery in the Pamir plateau (500 BCE), suggesting the clear evidence of the burning of Cannabis [23].
The psychoactive properties of marijuana have led to restrictions on its usage in various countries. In recent times, however, some countries and regions have begun liberalizing the restrictions on Cannabis for its economic and pharmacological benefits. California was the first US state to legalize the drug marijuana, ushering in the era of store-bought, commercially made edibles in 1996 [24]. Colorado was the first state to legalize recreational marijuana, imposing a limit of 10 mg THC per serving for edibles in 2014 [25]. In the same year, Uruguay became the first country to legalize Cannabis. Subsequently, Thailand was the first Asian country to legalize Cannabis in 2022 [26]. The legalization has allowed the development of more diverse properties of Cannabis for industrial and medical usage.
Origin and domestication of Cannabis
The origin and domestication of Cannabis are debated. Previously, it was widely believed that Cannabis originated in Central Asia, supported by the presence of its pollen in India around 32 000 years ago and in Japan around 10 000 BCE [27–29]. Central and Southeast Asia have been proposed as potential regions for its natural origin and primary domestication, playing a significant role in its evolutionary history [30]. However, recent microfossil (fossil pollen) data suggest a center of origin in the northeastern Tibetan Plateau [7]. Further, studies examining the correlation between genetic and geographic distances based on the chloroplast genome propose that Cannabis likely originated in low-latitude regions [31]. The complex genetic makeup of Cannabis, characterized by high heterozygosity, has posed challenges in studying its domestication history [32]. According to achene fossils found in East Asia and Europe, early humans employed hemp as a fiber plant, whereas the ancient use of the drug marijuana dates back to at least 2700 years ago in Central Asia, as discovered via wooden pots containing THC, which were probably employed for ritualistic and medicinal purposes [23, 33, 34]. However, large-scale whole-genome resequencing studies involving 110 Cannabis resources from around the world suggest that Cannabis was domesticated in East Asia during the early Neogene [35]. Over millennia, both artificial domestication and wild cultivation led to the development of diverse Cannabis species, which are cultivated for their fiber and as drugs (Fig. S1, see online supplementary material).
Cannabinoids
Cannabinoids and human health
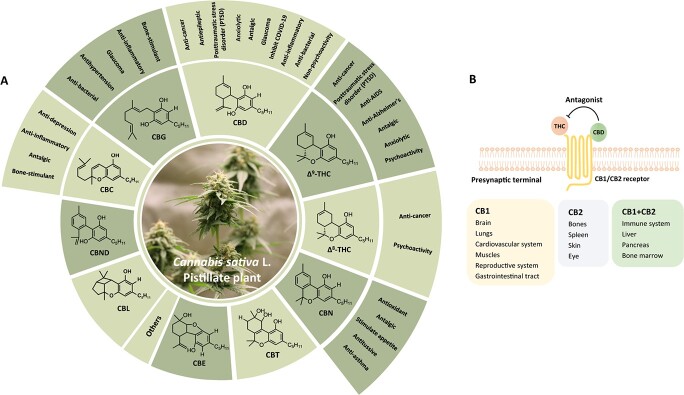
Cannabis contains numerous natural compounds [36] and over 560 compounds have been identified, such as cannabinoids, phenolics, terpenes, and alkaloids. Cannabinoids form the majority of compounds found in Cannabis, with more than 130 cannabinoids having been isolated. They can be classified into eleven subclasses based on their chemical structures, including ∆9-THC, ∆8-THC, CBG (cannabigerol), CBC (cannabichromene), CBD, CBND (dehydrocannabidiol), CBE (cannabielsoin), CBL (cannabicyclol), CBN, and CBT (dihydroxycannabinol) [37, 38] (Fig. 1A). Cannabinoids exist in two chemical forms: decarboxylated forms and carboxylated forms. In fresh Cannabis tissues, carboxylated form is the predominant form. However, through nonenzymatic reactions such as drying, aging, heating, or incineration, Cannabis and its extracts undergo decarboxylation of carboxylated forms to form decarboxylated forms [39].
Figure 1.
. Cannabinoids and human health. A Eleven subclasses of cannabinoids in Cannabis and their respective pharmacological activities. ∆9-THC (∆9-tetrahydrocannabinol), ∆8-THC (∆8-tetrahydrocannabinol), CBG (cannabigerol), CBC (cannabichromene), CBD (cannabidiol), CBND (dehydrocannabidiol), CBE (cannabielsoin), CBL (cannabicyclol), CBN (cannabinol), CBT (dihydroxycannabinol). B Cannabinoid receptors CB1/CB2 are distributed in the presynaptic terminal. THC activates the CB1/CB2 cannabinoid receptors, which leads to the release of neurotransmitters, the control of pain perception and memory learning, and the regulation of metabolic and cardiovascular systems. CBD can antagonize THC and reduce its side effects and addiction. Rectangle indicates the distribution of CB1/CB2 cannabinoid receptors in the human body.
THC and CBD are the most significant compounds in Cannabis, and their effects can be perceived as both positive and negative [40]. Clinical pharmacological studies have demonstrated that THC has potent hallucinogenic and addictive properties by activating both CB1 and CB2 receptors, whereas excessive consumption of exogenous cannabinoids can lead to the overstimulation of CB1 and CB2 receptors, causing reduced sensitivity and making it difficult for the body’s natural endocannabinoids to activate them [41, 42]. Thus, some individuals may be more prone to the effects of THC, although it also exhibits antiepileptic, antitumor, and antiemetic effects [43, 44]. In contrast, CBD, a nonpsychoactive component of Cannabis, acts as an antagonist to the association between THC and the receptors, effectively mitigating the hallucinogenic effects of THC on the body. CBD can be used in the restoration of the ECS, helping alleviate physical ailments and emotional distress [45, 46] (Fig. 1B). It has verified that CBD possesses diverse pharmacological effects, including antibacterial, anti-inflammatory, anxiolytic, and antiepileptic properties, and it is considered safe for humans [47–49].
Recent studies have revealed promising findings regarding the therapeutic properties of various cannabinoids [6, 50]. For instance, CBC is reported to exert anti-inflammatory, antitumor, antidepressant, and antifungal effects [51–54]. CBG shows potential as an antibiotic and has also been associated with antitumor, antidepressant, analgesic, and glaucoma-alleviating properties [51, 52, 55, 56]. Meanwhile, CBN stimulates appetite and possesses anti-asthma, tranquilizing, and pain-relieving properties [51, 52, 57]. However, some of the lesser-known cannabinoids may be scarce and difficult to extract, resulting in limited research on their medicinal functions and functional activities. Further exploration is needed to fully understand their potential benefits.
Cannabinoid biosynthesis
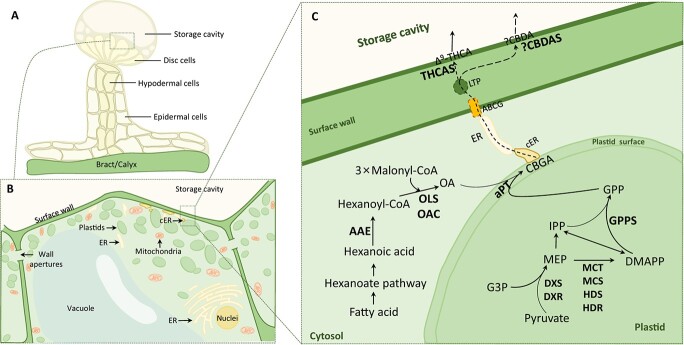
The secretory GTs are known as the ‘plant chemical factory’, secreting and storing multiple secondary metabolites (Fig. 2A). Cannabinoids are synthesized and accumulated in the Cannabis secretory GTs, and the content of cannabinoid is related to the secretion, type, and density of GTs [5]. Until now, the skeleton biosynthesis of cannabinoids has been elucidated, including two main biosynthetic pathways (Fig. 2B and C). The first pathway is polyketide synthesis occurring within the cytosol. It involves the conversion of hexanoic acid to thiolate hexanoyl coenzyme A by acyl-activating enzyme (AAE) [58]. Olivetol synthase (OLS) then catalyzes hexanoyl coenzyme A condensation with malonyl coenzyme A, leading to the formation of an intermediate tetraketide-CoA [59]. This intermediate is further cyclized by olivetolic acid cyclase (OAC) to produce olivetolic acid (OA), which serves as the polyketide nucleation component required for cannabinoid synthesis [60, 61]. The second pathway is the methylerythritol 4-phosphate (MEP) pathway, which generates isopentenyl diphosphate and dimethyl allyl diphosphate. These two compounds combine to form geranyl pyrophosphate (GPP), an isoprenoid compound that provides the monoterpene fraction needed for cannabinoid biosynthesis [40]. The GPP biosynthesis occurs in the plastid matrix, where it can freely move within the hydrophobic membrane [62]. Furthermore, on the plasma membrane, GPP and OA are catalyzed into cannabigerolic acid (CBGA) via the plastid envelope-localized aromatic prenyltransferase (aPT), known as CBGAS [63]. CBGA serves as a common precursor of partial oxidative cyclization to tetrahydrocannabinolic acid (THCA), cannabichromenic acid (CBCA), and cannabidiolic acid (CBDA), which is catalyzed by THCA synthase (THCAS), CBCA synthase, and CBDA synthase (CBDAS) [64–66]. The final step in cannabinoid biosynthesis occurs on the cell wall surface in the extracellular storage lumen [67]. The acidic forms of Δ9-THCA, CBCA, and CBDA are then converted to THC, CBC, and CBD, respectively, through a nonenzymatic decarboxylation process. Other cannabinoids can be synthesized through isomerization from THC, CBD, and CBC, and conversions between cannabinoids can also occur under specific conditions [34].
Figure 2.
. Biosynthesis, trafficking, and secretion of cannabinoids in Cannabis glandular trichomes (GTs). A Structural diagram of secretory GTs. B The distribution of plastids in secretory-stage GT disc cell. Plastids move between cells via apertures in the GT cell wall, aggregating below the surface wall and storage cavity. C The biosynthesis and secretion of THCA in GT disc cell. Cytosol-produced olivetolic acid (OA) and plastid-produced geranyl pyrophosphate (GPP) are converted by plastid envelope-localized aromatic prenyltransferase (aPT) into cannabigerolic acid (CBGA). CBGA likely partitions into the plastid membrane bilayer owing to its lipophilicity. Endoplasmic reticulum (ER) membrane contact between the ER and plastids may facilitate lipid metabolite transfer through lipid-binding proteins or transient hemifusions. CBGA may be exported by ATP-binding cassette G-family (ABCG) proteins and transported through the hydrophilic cell wall via lipid transfer proteins (LTPs). Cannabinoid biosynthesis occurs at the disk cell surface wall, where THCA synthase (THCAS) or CBDA synthase (CBDAS) catalyzes CBGA to produce THCA or CBDA, respectively. Cannabinoids form lipophilic metabolite droplets within the apical cell wall, creating storage cavity space. Abbreviations: DMAPP, dimethylallyl pyrophosphate; DXR, deoxyxylulose phosphate reductoisomerase; DXS, deoxyxylulose phosphate synthase; G3P, glyceraldehyde 3-phosphate; GPPS, geranylpyrophosphate synthase; HDR, hydroxymethylbutenyl diphosphate reductase; HDS, hydroxymethylbutenyl diphosphate synthase; IPP, isopentenyl diphosphate; MCS, methylerythritol cyclodiphosphate synthase; MCT, methylerythritol phosphate cytidylyltransferase; MEP, methylerythritol phosphate.
With advancements in sequencing technology, multiple versions of the Cannabis genome have been published, providing a deeper understanding of the synthesis mechanism of THC and CBD from CBGA at the genomic level. Grassa et al. sequenced the Cannabis genome and identified the specific location of THCAS and CBDAS on chromosome 9 [68]. Subsequent studies using updated genome versions further examined THCAS and CBDAS in two Cannabis strains (PK and FN) and revealed significant rearrangements of motifs related to THC/CBD synthesis [32, 69]. Further, CBDAS has a higher affinity for CBGA than that for THCAS. Consequently, when both enzymes are present, the production of CBDA is favored, suggesting why Cannabis plants typically contain higher CBD levels than THC levels [70]. Initially, THCAS and CBDAS were considered as an allele pair [71]. However, de Meijer et al. found that the THCA/CBDA ratio in medical marijuana F1 plants followed a Mendelian expectation of 1:2:1 [72]. This observation, along with the identification of major quantitative trait loci, suggested the presence of a single locus that exhibits codominant Mendelian inheritance patterns [73]. However, Kojoma et al. evidenced the presence of THCAS and CBDAS genes at two distinct loci on the genome that are linked together [74]. These findings have complicated our understanding of the genetic basis of THC and CBD synthesis in Cannabis.
Whole-genome resequencing of Cannabis revealed that nearly all drug Cannabis samples contained the complete coding sequence of THCAS and two CBDAS pseudogenes, while the majority of fiber Cannabis samples only had the full CBDAS-encoding gene [35], suggesting that both genes were initially present and functional in the ancestral state. Early domestication involved polymorphism and random loss of function in one of the genes, with THCAS and CBDAS losing their functions in hemp and in drug Cannabis, respectively, after strong artificial selection. The competitive relationship between these two genes in cannabinoid synthesis likely contributed to their loss of function, which was associated with the increase and decrease in THC content in drug Cannabis and in industrial hemp, respectively. Although the de novo sequencing and resequencing of the Cannabis genomes provided insights into the genetic basis and localization of genes involved in cannabinoid biosynthesis, further research is needed to explore the genomic evolution, biosynthesis, and regulation of target compounds. A deeper understanding at the chromosomal level as well as genome-wide association studies (GWAS) will contribute to a more comprehensive understanding of these aspects.
Regulatory network of cannabinoid biosynthesis
Manipulation of external environmental factors is a significant area for increasing the cannabinoid content like CBD and decreasing the content of THC. Exogenous hormone treatments, including gibberellic acid, methyl jasmonate, and salicylic acid, influence cannabinoid synthesis in Cannabis [75–79]. Additionally, the use of mevinolin can effectively reduce THC content by inhibiting the MEP and mevalonate pathways [75]. Polyploidization treatments have increased CBD content without altering THC accumulation [80, 81]. Furthermore, light conditions differently affect the synthesis of different cannabinoids [79, 82, 83]. These findings highlight the potential of manipulating external factors to modulate cannabinoid composition in Cannabis. Additionally, transcription factors (TFs) have been identified as key regulators that activate or inhibit the biosynthesis of plant natural products, effectively enhancing the synthesis of desired secondary metabolites. Numerous reports have highlighted the significance of TFs in regulating secondary metabolite synthesis [84–87]. To date, many TF gene families, such as WRKY, HD-ZIP, MYB, and bHLH, have been identified as being related to cannabinoid biosynthetic regulation based on genome-wide strategies in Cannabis [88–91]. However, the lack of a robust genetic transformation system hinders comprehensive studies on the mechanisms regulating cannabinoid biosynthesis.
Plant genetic transformation allows plants to acquire new traits, contributing significantly to variety improvement and gene function elucidation. However, for Cannabis as a regenerative and genetically recalcitrant plant, it is difficult to develop an efficient genetic transformation system, with low regeneration efficiency influenced by factors such as variety, tissue type, plant age, and growth regulator combination [92, 93]. Previous studies employed different methods, such as Agrobacterium-mediated transformation, virus-induced gene silencing, and nanomaterial translocation, to achieve transient expression in Cannabis leaves, albeit with limited efficiency [94–99]. Recently, efforts have been made to develop stable genetic transformation systems for Cannabis. Wahby et al. successfully induced transgenic hairy roots; however, cannabinoids were not detected, possibly owing to their accumulation primarily in GTs rather than that in roots [100]. Therefore, it is not feasible to study the cannabinoid biosynthesis regulation through transgenic hairy roots. Zhang et al. developed transgenic plants through Agrobacterium-mediated infection but with low redifferentiation efficiency (≤7.09%) and a high proportion of chimeras [101]. Further, Galán-Ávila et al. optimized the transgenic system and successfully induced regenerated seedlings directly from hypocotyls or cotyledons [102]. Nevertheless, the establishment of an efficient transgenic system, for both transient and stable transformation, remains a significant challenge in Cannabis research, particularly for investigating cannabinoid synthesis regulation and other important traits.
Glandular trichomes in Cannabis and their potential transcriptional regulatory network
GTs arise from the differentiation of epidermal cells and are prominent on the surface of numerous plant species [103]. GTs are important defense organs against environmental stress, while also offering significant economic and practical value through their specialized metabolites [104–106]. Secretory GTs are specifically involved in the synthesis, accumulation, and release of a wide range of metabolites, including organic acids, polysaccharides, polyphenols, flavonoids, alkaloids, and terpenoids [107–109]. In contrast, nonsecretory GTs primarily function as protective structures without chemical compound secretion [110].
Secretory GTs are abundantly present on the tissue surface of multiple plants such as Solanaceae, Labiatae and Asteraceae. Given their similar structure, the secretory GTs may have similar developmental events in different plants species [111]. Arabidopsis thaliana only possesses nonsecretory GTs [112, 113]. Consequently, although more comprehensive studies on the morphological aspects, density, and developmental processes of GTs have been conducted on Arabidopsis, the mechanisms underlying the development and specialized metabolite biosynthesis in secretory GTs may not be fully elucidated by using the model plant A. thaliana. On the contrary, the recently extensive studies in Artemisia annua and Solanum lycopersicum exemplify the transcription factors regulating secretory GTs development. Therefore, we summarized a transcriptional regulatory network of GTs development in these both species in the later part of this review to provide a reference for the regulation of GTs in Cannabis (Fig. 3).
Figure 3.
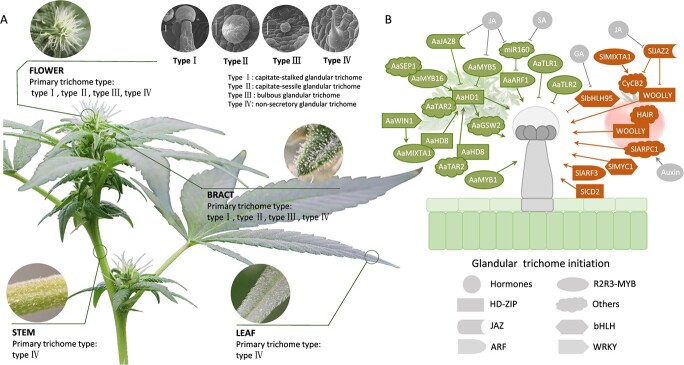
. GTs in Cannabis and their transcriptional regulatory network in Solanum lycopersicum and Artemisia annua. A Scanning electron microscopy analysis illustrating the different GT types on the surface of various tissues in Cannabis sativa. Type I, capitate-stalked GT; Type II, capitate-sessile GT; Type III, bulbous GT; Type IV, nonsecretory GT. Bars = 25 μm. B Summarized transcriptional regulatory network of GT initiation in S. lycopersicum and A. annua. Different shapes represent distinct regulatory factors. Arrow-headed lines indicate upregulation, whereas blunted lines indicate downregulation or inhibition.
Characteristics of glandular trichomes in Cannabis
Cannabis GTs are distinctive secretory structures that are crucial in the production and reservation of cannabinoids, primarily found on the bracts and flowers of female Cannabis plants (Fig. 2A) [114]. In Cannabis, the density of GTs per unit area is generally higher in female plants than that in male ones [115]. Female plants exhibit a greater abundance of GTs on bracts, flowers, and other plant parts (Fig. 3A). GTs in Cannabis can be classified into two main types: secretory GTs and nonsecretory GTs [116]. Secretory GTs are further categorized into stalked GTs, sessile GTs, and bulbous GTs [5]. Stalked GTs consist of a stalked base with 12–16 secretory disc cells; sessile GTs possess a short-stalked base with eight secretory disc cells; and bulbous GTs are the smallest in size and consist of a short-stalk base with a bulbous head. Additionally, Cannabis plants possess two types of nonsecretory GTs, which primarily serve as a protective barrier in the plant’s epidermis, shielding it from external damage [116, 117]. The type and density of secretory GTs exhibit variations during development. In the early developmental stages in bract and flower, all GT types are present, whereas in the later stages stalked GTs are predominant, indicating that sessile GTs may represent an early stage in stalked GT development [5].
The synthesis and storage of cannabinoids primarily occur in secretory GTs [118]. Through the isolation and analysis of Cannabis GTs, Happyana et al. confirmed that both bulbous GTs and stalked GTs can produce CBGA and THCA. Additionally, CBC and CBN were specifically detected in bulbous stalked GTs [119]. Autofluorescence studies further supported these findings and revealed the higher accumulation of monoterpenes in the stalked GTs [5]. The analysis of Cannabis GT wall components revealed the presence of loosely bound xyloglucan and pectin polysaccharides. Within the cell wall, the interaction between polysaccharide emulsions and metabolite droplets creates a crucial microenvironment where the boundary between metabolites and polysaccharides acts as a hydrophobic–hydrophilic interface [120]. Cannabinoid synthases have greater metabolic efficiency in hydrophobic environments than that in aqueous ones [118]. This metabolite–polysaccharide wall boundary may be a favorable microenvironment that is well suited for THCA biosynthesis, which could explain the abundant synthesis and secretion of cannabinoids in GTs [120]. Despite their small size, the ability of these GTs to produce and secrete significant quantities of secondary metabolites remains a mystery. Unraveling the mechanisms behind their metabolite synthesis should provide valuable insights for improving the production capacity of natural products in microorganisms and plant chassis.
Furthermore, ultra-rapid cryofixation, quantitative electron microscopy, and immuno-gold labeling of enzymes involved in the cannabinoid pathway revealed that metabolically active cells in Cannabis GTs form a specialized ‘supercell’ [67, 121]. These supercells exhibit extensive cytoplasmic bridges across their cell walls and a polarized distribution of organelles adjacent to the apical surface, which is responsible for metabolite secretion. These supercells are organized as polarized cell syncytia, representing a synchronized fusion of multiple cells with no discernible cytoplasmic connection to the underlying tissue. This polarized cell syncytium configuration presents a potential mechanism for preventing the backflow of metabolites from the trichomes by physically isolating the syncytium from the surrounding tissue.
Transcriptional regulation in the development of secretory GTs
The regulatory mechanisms governing the GT development in Cannabis are not yet fully understood. However, the MIXTA gene, an MYB family transcription factor, has been identified as a regulator of multicellular epidermal hairs and conical cells in the leaves of Antirrhinum majus [122, 123]. Attempts to overexpress the MIXTA1 homolog from Cannabis in tobacco led to an increase in GTs [124]. To date, the regulatory mechanisms underlying the development of secretory GTs in S. lycopersicum and A. annua have been extensively studied. Herein, we summarize the progress in research on TFs and noncoding RNAs that regulate the development of secretory GTs.
Molecular mechanism of secretory GT development in S. lycopersicum
Several TFs involved in the development and morphology of GTs in tomatoes have been identified. Chang et al. discovered a C2H2 zinc finger protein gene, Hair, associated with the absence of GTs using GWAS analysis [125]. Transgenic experiments showed that the hair-absent phenotype was caused by deletion of the entire coding region of Hair. Additionally, Hair was found to interact with WOOLLY to promote GT development [126]. Further, Chun et al. explored the transcriptome differences between hairless mutants and the wild type and discovered that downregulation of the actin-related protein component 1 (SlARPC1) led to the severe curvature of GTs in hairless mutants [127]. SlCycB2, encoding a B-type cell cycle protein, reduces the secretory GT density. Further study revealed that SlCycB2 expression was increased in plants overexpressing WOOLLY and SlMIXTA1, whereas it was reduced in woolly mutant plants [128]. Yu et al. discovered that SlJAZ2 inhibited GT development by suppressing WOOLLY and SlCycB2 activity [129]. An ARF family transcription factor, SlARF3, highly expressed in GTs, positively regulates GT initiation [130]. SlMYC1 is also involved in the initiation and morphogenesis of secretory GTs. Knockdown of SlMYC1 led to smaller and less dense trichomes, whereas its knockout resulted in the disappearance of secretory GTs. Furthermore, Chen et al. discovered that SlbHLH95 suppressed GT development by repressing the expression of key genes (SlGA20ox2 and SlKS5) involved in endogenous gibberellic acid synthesis [131]. Overall, these tomato TFs related to GT development have provided valuable insights into the genetic mechanisms underlying the development and regulation of GTs in Cannabis (Fig. 3B).
Molecular mechanism of GT development in A. annua
The World Health Organization recommends artemisinin as an essential component of standard antimalarial therapy, with A. annua being its unique natural plant source. Artemisinin is specifically distributed in the secretory GTs in A. annua, and the developmental transcriptional regulatory networks of these trichomes have been extensively investigated. AaHD1, an HD-ZIP family transcription factor, as a key regulator of GT initiation in A. annua, has functions in increasing trichome density and artemisinin production [132]. AaJAZ8 antagonizes AaHD1 expression and suppress GT development. Another HD-ZIP family TF, AaHD8, promotes AaHD1 expression and trichome initiation. The overexpression of AaGSW2, a gene specifically expressed in GTs and coexpressed with AaHD1, increased trichome density [132]. AaMIXTA1, a homologous gene of the positive regulator MIXTA, is also involved in regulating GT development by binding to the AaHD8 promoter [133]. The AP2/ERF family TF AaWIN1 interacts with AaMIXTA1 to induce AaGSW2 and promote trichome development [134]. AaMYB16, another homolog of AaMIXTA-like2, is bound to the L1-box on the AaHD1 promoter to enhance AaHD1 expression, whereas AaMYB5 is competitively bound to the AaHD1 promoter by binding to AaJAZ8, exerting a repressive effect [135]. The MADS-box family TF AaSEPALLATA1 (AaSEP1) enhances the regulatory effect of AaHD1 on AaGSW2 by binding to AaMYB16, thereby regulating trichome development [136]. These identified TFs indirectly or directly regulate AaHD1 expression, forming an AaHD1-centered regulatory network that serves as a reference for studies on GT initiation. Other MYB family TFs like MYB1, TrichomeLess Regulator 1 (TLR1), and TLR2 also regulate GT initiation [137, 138].
MicroRNAs (miRNAs) are crucial in regulating terpenoid biosynthesis by targeting key enzymes and TFs involved in secondary metabolite synthesis. Overexpression of miR160 leads to significant inhibition of GT initiation and artemisinin biosynthesis [139]. Conversely, inhibition of miR160 expression had the opposite effect, indicating that miR160 negatively regulates the GT initiation and artemisinin biosynthesis. This regulatory mechanism is mediated by miR160’s cleavage of AaARF1, providing valuable insights into the transcriptional regulatory network governing GT synthesis in A. annua (Fig. 3B).
Based on the progress in research on the TF-mediated regulation of secretory GTs in tomato and A. annua, the homologous genes in Cannabis and/or the trichome-specific TFs could be identified as a potential transcriptional regulatory network associated with GT development in Cannabis. Furthermore, the regulatory mechanisms of candidate TFs related to cannabinoid biosynthesis and secretory GT initiation need to be further elucidated by in vivo and in vitro biochemical strategies. Furthermore, single-cell transcriptome sequencing technology could be used to identify genes specifically expressed in Cannabis secretory GTs, providing an important reference for precise mining of regulatory genes related to Cannabis secretory GT development and cannabinoid biosynthesis [140, 141].
Advances in metabolic engineering of cannabinoids
At present, Δ9-THC and CBD, the primary compounds of interest, are extracted directly from Cannabis. However, the study and medicinal use of cannabinoids have faced challenges owing to legal restrictions on Cannabis and the low natural abundance of most cannabinoids. In addition, the complex structures of cannabinoids hinder their large-scale chemical synthesis. Green bioproduction of cannabinoids based on the theory of synthetic biology could efficiently overcome these limitations by providing a comprehensive understanding of the cannabinoid synthesis pathway, synthesizing and assembling the essential components in microbial cells, and optimizing the fermentation production of target cannabinoids.
OA production
CBGA, formed through the condensation of OA and GPP, is the primary precursor molecule in cannabinoid biosynthesis. Therefore, achieving high levels of OA production is crucial for effective metabolic engineering of cannabinoids.
Escherichia coli is widely recognized as an ideal host for microbial production owing to its straightforward genetic background and rapid reproduction [142]. Tan et al. successfully achieved OA synthesis in E. coli by coexpressing OLS and OAC with the required modules of a β-oxidation reversal for hexanoyl-CoA generation. Furthermore, the integration of supplementary enzymes to enhance the production of hexanoyl-CoA and malonyl-CoA, along with the evaluation of varying fermentation conditions, enabled the synthesis of 80 mg/L OA, marking a significant milestone as the first reported production of OA in E. coli [143].
By introducing a group of fungal tandem polyketide synthases in the model fungus Aspergillus globulus, OA was produced at high yields of up to 80 mg/L. This finding presents a novel and alternative method for the microbial production of cannabinoid precursors, bypassing the need for the OLS and OAC enzymes derived from Cannabis [144].
Yarrowia lipolytica, a safe and lipid-rich yeast, has been successfully engineered to produce various valuable natural products [145, 146]. Introduction of LvaE from Pseudomonas sp., which encodes a short-chain acyl-CoA synthetase, enables efficient conversion of hexanoic acid to hexanoyl-CoA in Y. lipolytica via coexpressing acetyl-CoA carboxylase, the pyruvate dehydrogenase bypass, the NADPH-generating malic enzyme, as well as activation of the peroxisomal β-oxidation pathway and ATP export pathway. These modifications effectively redirect the carbon flux toward OA production. The optimized protocols led to a remarkable 83-fold increase in OA production, with a titer of 9.18 mg/L, providing an excellent microbial chassis for high-throughput production of cannabinoids [147].
In contrast to fungi and plants, amoebae represent a unique group of eukaryotes that express terpene synthases, indicating the presence of an active GPP biosynthesis pathway [148]. Reimer et al. utilized the amoeba Dictyostelium discoideum as a host organism to successfully produce phlorocaprophenone, methyl-olivetol, resveratrol, and OA by sequentially expressing native cannabinoid polyketide synthase. To facilitate OA synthesis, an amoeba/plant inter-kingdom hybrid enzyme was further engineered to produce OA from primary metabolites by only two enzymes, thus providing a shortcut to the synthetic cannabinoid pathway using the D. discoideum chassis [149]. Subsequently, the optimization of reaction conditions in D. discoideum led to OA production with productivity of 0.04 μg/L/h and a concentration of 4.8 μg/L by scaling up the bioreactor to an industrial scale of 300 L [150].
De novo cannabinoid synthesis
De novo and high-yield production of cannabinoids in microorganisms or heterogenous plants have been gaining attention for its global demand. Luo et al. first achieved de novo bioproduction of cannabinoids in yeast via the introduction of OA biosynthetic modules, optimization of the mevalonate pathway, and the identification of novel prenyltransferase and different cannabinoid synthases [63]. The exogenous hexanoyl-CoA enzymes including RebktB, CnpaaH1, Cacrt, and Tdter [151], combined with Cannabis CsTKS and CsOAC, improved the yield of OA to 1.6 mg/L. Further, site-direction mutations of ERG20F96W/N127W for increasing GPP accumulation, and the identification and introduction of novel Cannabis CsaPT4 in yeast led to the successful synthesis of 7.2 mg/L and 1.4 mg/L CBGA by adding galactose and 1 mM hexanoic acid and by adding galactose, respectively. Furthermore, the replacement of N-terminal secretory signal peptides of THCAS and CBDAS with a vacuolar localization tag led to yields of THCA and CBDA of 2.3 mg/L and 4.2 μg/L, respectively. Finally, by increasing the copy numbers of the key enzymes CsTKS, CsOAC, CsaPT4, and THCAS, the THCA yield reached 8 mg/L. Although this yield is only at the milligram level, it demonstrates the potential of Saccharomyces cerevisiae as a microbial chassis for cannabinoid synthesis. Qiu et al. further de novo-synthesized CBD in S. cerevisiae by integrating the CBD biosynthetic gene modules. In addition, the transporter protein BPT1 could effectively transfer CBGA from cytoplasm to vacuole, and the combination of BPT1 in engineered yeast improved the CBD content to 6.92 mg/L [152].
NphB, a prenyltransferase from Streptomyces sp., has been ever served as an enzyme to catalyze the production of CBGA [153], and the nonspecific regioselectivity of NphB greatly limits cannabinoid synthesis. Therefore, Lim et al. modified and optimized the NphBV49W/Y288P variants, leading to a 13.6-fold improvement in CBGA yield [154]. The cannabinoid synthesis titer in yeast was mainly restricted by the enzymatic activities related to the OA pathway; there is thus a need for further exploitation of the modification and optimization of these enzymes.
Nicotiana benthamiana is a potential plant chassis to efficiently synthesize cannabinoids. Thies et al. introduced 12 aPTs into N. benthamiana and found that CsaPT4 can convert GPP and OA into GBGA. However, OA and its glycosylation products were detected owing to the presence of endogenous glycosyltransferase in engineered N. benthamiana [155]. Reddy et al. also engineered N. benthamiana to synthesize cannabinoid precursors via the stable expression of AAE and OLS, and transient expression of OAC genes. Owing to the endogenous modifications of cannabinoid intermediates in tobacco, the chassis may be further directly engineered, especially the gene silencing or knockdown of modified enzymes, to make it applicable for cannabinoid bioproduction [156].
Cell-free system for cannabinoid engineering
Microbial production offers an alternative to natural extraction for prenylated cannabinoids but has challenges such as carbon flux diversion, product toxicity, and GPP essentiality, which can hinder large-scale production. Valliere et al. developed a cell-free platform to enhance the prenylation of natural products, with a focus on cannabinoid production [159]. Through the introduction of the pyruvate dehydrogenase bypass and optimization of cofactors, enzymes, and environmental factors, the cell-free system achieved CBGA titers of 132 mg/L. Further advancements were made by redesigning the NphB prenyltransferase, resulting in a CBGA titer of 600 mg/L. To enhance enzyme stability, a flow system for CBGA capture was implemented, leading to a titer of 1.25 g/L, representing a 140-fold improvement. The cell-free system was further optimized and designed, optimizing the number of enzymes to only 12 enzymes to produce CBGA, and using the relatively inexpensive substrate acetyl-CoA. The final optimized system could produce 570 ± 60 mg/L CBGA, which was lower than that of the previous system, but the cost was substantially reduced; with the subsequent optimization, the yield could be further increased [160]. These solutions were rapidly achieved through iterative design-build-test cycles, surpassing previously reported results using living cells. However, the large-scale construction of highly complex systems involving numerous enzymes, cofactors, and metabolites using a cell-free system remains a challenge.
The utilization of metabolic engineering to produce cannabinoids offers unparalleled advantages compared with both plant extracts and chemical synthesis. Although significant efforts have been made to produce cannabinoids using various chassis cells such as E. coli, yeast, and tobacco, the resulting yields remain considerably low and fall short of the requirements for industrial production (Table 1). Considerable efforts are still required to enhance synthetic efficiency, including the improvement of metabolic flux into the cannabinoid biosynthesis in the chassis, exploration and modification of efficient synthetic enzymes, and optimization of fermentation processes. With the global changes in the legalization of Cannabis, further expansion in the market for green cannabinoid bioproduction is expected.
Table 1.
Biosynthesis of OA and cannabinoids using metabolic engineering
| Organism | Methods | Products | Yield | Reference |
|---|---|---|---|---|
| Yarrowia lipolytica | Used a short-chain acyl-CoA synthetase co-expressed with the acetyl-CoA carboxylase | Olivetolic acid (OA) | 9.18 mg/L | Ma et al. (2022) [147] |
| Escherichia coli | Co-expressed olivetol synthase (OLS) and olivetolic acid cyclase (OAC), with the module required for β-oxidation to reverse hexanoyl-CoA, hexanoyl-CoA and malonyl-CoA | OA | 80 mg/L | Tan et al. (2018) [143] |
| Aspergillus globulus | Discovered a set of fungal tandem polyketide synthases without relying on the OLS and OAC found in Cannabis | OA | 80 mg/L | Okorafor et al. (2021) [144] |
| Dictyostelium discoideum | Sequentially expressed native cannabinoid polyketide synthase and introduced an amoeba/plant inter-kingdom hybrid enzyme | OA | 4.5 μg/L | Reimer et al. (2022) [149] |
| Pichia pastoris | Introduced tetrahydrocannabinolic acid synthase (THCAS) in Pichia pastoris | Tetrahydrocannabinolic acid (THCA) | Zirpel et al. (2015) [157] | |
| K. phaffii | Used NphB replace the function of THCAS, | THCA | 82 ± 4.6 pmol/L | Zirpel et al. (2017) [153] |
| Saccharomyces cerevisiae | Introduced and modified more than 15 genes from different species into yeast | THCA, Tetrahydrocannabivarinic acid (THCVA) | 8 mg/L, 4.8 mg/L | Luo et al. (2019) [63] |
| S. cerevisiae | Integrated the CBD biosynthetic gene modules and the transporter protein BPT1 | Cannabidiol (CBD) | 6.92 mg/L | Qiu et al. (2022) [152] |
| Nicotiana benthamiana | Introduced acyl-activating enzyme and OLS | Cannabinoid derivatives | Reddy et al. (2022) [156] | |
| N. benthamiana | Transient Expression of CsaPT4, SrUGT71E1 and OsUGT5 | Cannabigerolic acid (CBGA), olivetolic acid glucoside and cannabigerolic acid glucoside | Gulck et al. (2020) [155] | |
| Obtained more active enzyme OACF24I and TKSL190G | different acyl groups on C-3 of OA | Lee et al. (2022) [158] | ||
| Obtained more active enzyme NphBV49W/Y288P | CBGA | Lim et al. (2022) [154] | ||
| Cell-free | Constructed a 23-enzyme reaction system | CBGA | 1.25 g/L | Valliere et al. (2019) [159] |
| Cell-free | Constructed a 12-enzyme reaction system | CBGA | 570 ± 60 mg/L | Valliere et al. (2020) [160] |
Conclusion and future perspectives
Cannabis, an economically important crop, is restricted or banned for cultivation in many countries owing to it containing the psychoactive compound THC. However, the significant medicinal value of various cannabinoids, such as CBD, found in Cannabis has led to increased demand for cannabinoids, accelerating the legalization of industrial Cannabis cultivation globally. Nevertheless, the lack of comprehensive research on Cannabis GT development and cannabinoid biosynthesis regulation poses major challenges for high CBD and low THC Cannabis varieties breeding and efficient green bioproduction of cannabinoids. To address these issues and provide crucial support for the globalization of the Cannabis industry, there is an urgent need for innovative interdisciplinary and technological research on Cannabis. Recently, metabolic engineering of cannabinoids and the cultivation of new medicinal Cannabis varieties with high CBD and low THC content have gained importance.
Understanding the regulatory mechanism of cannabinoid biosynthesis and secretory GTs
TFs, noncoding RNAs, and epigenetic modifications have been identified as key players in plant development and responses to abiotic/biotic stresses, and the biosynthesis of valuable natural products [85, 161]. Cannabinoid biosynthetic genes have been fully elucidated, and they present trichome-specific accumulation. However, the mechanisms regulating cannabinoid biosynthesis and secretory GTs remain largely unclear. Although the regulation of secretory GT development in tomato and A. annua could provide homologous evidence on the Cannabis trichome initiation, further exploration of Cannabis-specific cannabinoid biosynthesis and trichome development are necessary. With the advancements in sequencing technologies, multi-omics data for Cannabis, including the T2T genome, 3D genome architecture and epigenomes for flower or trichomes, single-cell transcriptomes, and transcriptomes and metabolomes of different tissues, will provide important genetic resources for the selection of candidate TFs and noncoding RNAs related to cannabinoid biosynthesis regulation and secretory GT initiation [162, 163].
Multi-omics-guided breeding and cultivation of novel Cannabis varieties
Cannabis varieties cultured mainly by conventional breeding such as hybridization and systematic selection, and some varieties with high CBD content have been cultured by hybridization, such as ACDC, Harle-Tsu, and Corazón (https://www.leafly.com/news/strains-products/high-cbd-marijuana-strains-according-tolab-data). However, conventional breeding often involves lengthy selection cycles and is less efficient [71]. In contrast, molecular breeding offers a more efficient and precise approach, thereby accelerating the breeding process [164]. Herbgenomics lays the foundation for molecular genetics of medicinal plants. The integration of multi-omics technology has provided additional genetic information for molecular breeding in Cannabis. Combining multiple published versions of the Cannabis genomes with multi-omics approaches with the aid of artificial intelligence-based data analysis tools has significantly improved the efficiency with which the genetic and molecular mechanisms regulating cannabinoid biosynthesis can be uncovered. Particularly, the relationship between the expression of key enzymes involved in cannabinoid biosynthesis and the THC:CBD ratio and cannabinoid content can be better understood at the genetic and molecular levels. Other breeding strategies such as GWAS use natural populations to identify molecular markers associated with important traits in Cannabis [165]. Moreover, GWAS enables the discovery of molecular markers and facilitates the identification of loci and candidate genes related to crucial agronomic traits such as CBD and THC biosynthesis, sexual development, and cellulose quality [166, 167]. Constructing a genetic linkage map of Cannabis using molecular marker technology, leveraging the genetic information from diverse genetic resources, and developing molecular markers for desirable traits will further advance the process of Cannabis breeding [168, 169].
Efficient green bioproduction of cannabinoids
Although significant progress has been made in the de novo synthesis of cannabinoids in yeast, achieving high yields of cannabinoids is still a major challenge. The rate-limiting steps and metabolic bottlenecks in cannabinoid biosynthetic pathways within chassis cells need to be further addressed and optimized to improve the overall efficiency and yield of cannabinoid production. Site-directed mutations of crucial rate-limiting enzymes in cannabinoid biosynthesis could efficiently improve the yields in chassis cells. Cannabinoid accumulation via parallel evolution has been discovered in Helichrysum umbraculigerum, providing a set of alternative enzymes for the synthetic biology of cannabinoids [170, 171]. Further studies can explore the construction of fusion proteins from different plant sources to enhance catalytic efficiency. In addition to microbial and plant chassis, OA synthesis has also been reported in D. discoideum. Amoebas offer a wider and larger gene repertoire for polyketide and terpenoid biosynthesis, presenting a new approach for the synthetic chassis of cannabinoids [172].
Robust model research system for medicinal plants
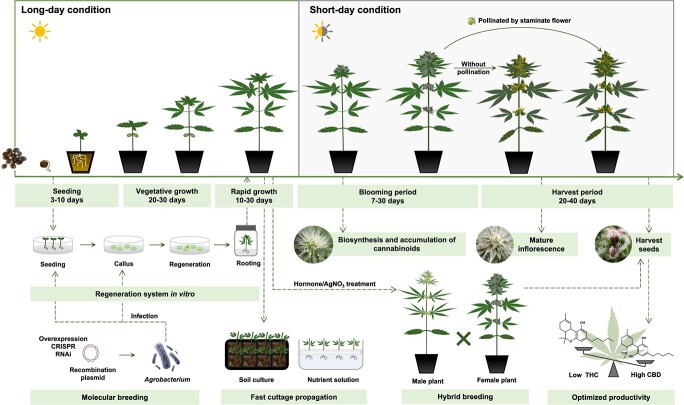
Since the late Northern Wei Dynasty, detailed techniques for hemp cultivation have been recorded in ‘Qi Min Yao Shu’. In recent times, research on efficient Cannabis cultivation has gained significant attention owing to the substantial market demand. Dioecious Cannabis offers several advantages as a robust model research system for understanding the development of secretory GTs. Cannabis has a relatively short growth cycle of approximately three months, and short-day conditions can induce Cannabis flowering initiation. The generation time can be shortened within nine weeks by controlling light condition [173]. Here, we construct a schematic to describe the Cannabis growth cycle and corresponding biotechnologically experimental timeline (Fig. 4). Briefly, multiple methods developed and exploited for the genetic transformation of Cannabis generally occur at or before seedlings. Fast cutting propagation usually takes place in the rapid growth stage before inflorescence [174]. To avoid genetic heterozygosity introduced by hybrid breeding, as Cannabis is dioecious, female plants can be transformed to produce male flowers by treating with hormones or AgNO3 [175]. The establishment of a stable genetic transformation system in Cannabis deepens our understanding of regulatory mechanisms and developmental biology and is critical in establishing Cannabis as a model plant. Genetic improvement to obtain new Cannabis varieties with superior traits serves as a valuable resource for the medicinal and health utilization of cannabinoids.
Figure 4.
Cannabis growth and experimental timelines. Cannabis exhibits a relatively short growth cycle, lasting 2–5 months. The seedling stage takes approximately 3–10 days after germination. Subsequently, it enters the vegetative growth stage characterized by slow growth and nutrient absorption focused on root development, lasting for approximately 20–30 days. The subsequently rapid growth of the aerial part is maintained for 10–30 days. Preflowering shoots as cuttings are suitable for asexual propagation. Callus induction and genetic transformation can be performed by taking explants from the hypocotyledonary axis, cotyledon, and tender leaf. The flowering initiation in Cannabis can be induced by subjecting the plants to short-day conditions. After approximately a week, flowers begin to appear at the top of the branches. The flowering phase typically lasts for a month. When treated with hormones or AgNO3, female plants can be transformed into male plants. The harvesting of inflorescences for cannabinoid extraction can be performed once the pistils have withered. If female flowers are pollinated during the flowering phase, seeds can be collected after approximately a month. New varieties with high CBD yield can be obtained through hybridization breeding or molecular breeding techniques.
In conclusion, Cannabis, an ancient medicinal plant with a longstanding history of global usage over millennia, can make a substantial transformative effect on human health in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82204579), the Fundamental Research Funds for the Central Universities (2572022DX06), the Scientific and Technological Innovation Project of China Academy of Chinese Medical Science (CI2021A04113), and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Author contributions
Z.X., W.S., and S.C. designed and coordinated the review. Z.X. structured, drafted, and revised the manuscript. Y.M, L.K., and M.G. revised the manuscript. S.C., W.C., and X.M. contributed to the critical revision of the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Supplementary Material
Contributor Information
Ziyan Xie, Key Laboratory of Saline-alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China.
Yaolei Mi, Key Laboratory of Saline-alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China; Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China.
Lingzhe Kong, Key Laboratory of Saline-alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China.
Maolun Gao, Key Laboratory of Saline-alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China.
Shanshan Chen, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China.
Weiqiang Chen, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China.
Xiangxiao Meng, Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China.
Wei Sun, College of Life Science, Northeast Forestry University, Harbin 150040, China; Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China.
Shilin Chen, College of Life Science, Northeast Forestry University, Harbin 150040, China; Institute of Herbgenomics, Chengdu University of Traditional Chinese Medicine, Chengdu 611137, China.
Zhichao Xu, Key Laboratory of Saline-alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education, Harbin 150040, China; College of Life Science, Northeast Forestry University, Harbin 150040, China.
References
- 1. McPartland JM, Small E. A classification of endangered high-THC cannabis (Cannabis sativa subsp. indica) domesticates and their wild relatives. PhytoKeys. 2020;144:81–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schluttenhofer C, Yuan L. Challenges towards revitalizing hemp: a multifaceted crop. Trends Plant Sci. 2017;22:917–29 [DOI] [PubMed] [Google Scholar]
- 3. Callaway JC. Hempseed as a nutritional resource: an overview. Euphytica. 2004;140:65–72 [Google Scholar]
- 4. Ryz NR, Remillard DJ, Russo EB. Cannabis roots: a traditional therapy with future potential for treating inflammation and pain. Cannabis Cannabinoid Res. 2017;2:210–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Livingston SJ, Quilichini TD, Booth JKet al. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 2020;101:37–56 [DOI] [PubMed] [Google Scholar]
- 6. Datta S, Ramamurthy PC, Anand Uet al. Wonder or evil?: multifaceted health hazards and health benefits of Cannabis sativa and its phytochemicals. Saudi J Biol Sci. 2021;28:7290–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McPartland JM. Cannabis systematics at the levels of family, genus, and species. Cannabis Cannabinoid Res. 2018;3:203–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li QS, Meng Y, Chen SL. A new cannabis germplasm classification system and research strategies of non-psychoactive medicinal cannabis. Zhongguo Zhong Yao Za Zhi. 2019;44:4309–16 [DOI] [PubMed] [Google Scholar]
- 9. Carvalho A, Hansen EH, Kayser Oet al. Designing microorganisms for heterologous biosynthesis of cannabinoids. FEMS Yeast Res. 2017;17:fox037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Charitos IA, Gagliano-Candela R, Santacroce Let al. The cannabis spread throughout the continents and its therapeutic use in history. Endocr Metab Immune Disord Drug Targets. 2021;21:407–17 [DOI] [PubMed] [Google Scholar]
- 11. Kohek M, Aviles CS, Romani Oet al. Ancient psychoactive plants in a global village: the ritual use of cannabis in a in Catalonia. Int J Drug Policy. 2021;98:103390 [DOI] [PubMed] [Google Scholar]
- 12. Tomida I, Azuara-Blanco A, House Het al. Effect of sublingual application of cannabinoids on intraocular pressure: a pilot study. J Glaucoma. 2006;15:349–53 [DOI] [PubMed] [Google Scholar]
- 13. Bai YJ, Zhou XY, Yuan Yet al. Origin of medicinal Cannabis sativa and its early spread. Chinese Traditional and Herbal Drugs. 2019;50:5071–79 [Google Scholar]
- 14. Brand EJ, Zhao Z. Cannabis in Chinese medicine: are some traditional indications referenced in ancient literature related to cannabinoids? Front Pharmacol. 2017;8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kabelik J. Hemp (Cannabis sativa); antibiotic drug. I. Hemp in the old & popular medicine. Die Pharmazie. 1957;12:439–43 [PubMed] [Google Scholar]
- 16. Degenhardt F, Stehle F, Kayser O. Ch. 2 - The Biosynthesis of Cannabinoids. In: Preedy VR, ed. Handbook of Cannabis and Related Pathologies. San Diego: Academic Press, 2017 [Google Scholar]
- 17. Adams R, Hunt M, Clark JH. Structure of cannabidiol, a product isolated from the marihuana extract of Minnesota wild hemp. I. J Am Chem Soc. 1940;62:196–200 [Google Scholar]
- 18. Mechoulam R, Gaoni Y. A total synthesis of dl-Δ1-tetrahydrocannabinol, the active constituent of Hashish1. J Am Chem Soc. 1965;87:3273–5 [DOI] [PubMed] [Google Scholar]
- 19. McPartland JM, Duncan M, Di Marzo Vet al. Are cannabidiol and Delta(9) -tetrahydrocannabivarin negative modulators of the endocannabinoid system? A systematic review. Br J Pharmacol. 2015;172:737–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu HC, Mackie K. An introduction to the endogenous cannabinoid system. Biol Psychiatry. 2016;79:516–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. John MJ, Thomas S. Biofibres and biocomposites. Carbohydr Polym. 2008;71:343–64 [Google Scholar]
- 22. Zhao MY, Jiang HG, Grassa CJ. Archaeobotanical studies of the Yanghai cemetery in Turpan, Xinjiang. China. Archaeol Anthropol Sci. 2019;11:1143–53 [Google Scholar]
- 23. Ren M, Tang Z, Wu Xet al. The origins of cannabis smoking: chemical residue evidence from the first millennium BCE in the Pamirs. Sci Adv. 2019;5:eaaw1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borodovsky JT, Budney AJ. Legal cannabis laws, home cultivation, and use of edible cannabis products: a growing, relationship? Int J Drug Policy. 2017;50:102–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livingston MD, Barnett TE, Delcher Cet al. Recreational cannabis legalization and opioid-related deaths in Colorado, 2000–2015. Am J Public Health. 2017;107:1827–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sommano SR, Tangpao T, Pankasemsuk Tet al. Growing ganja permission: a real gate-way for Thailand's promising industrial crop? J Cannabis Res. 2022;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Russo EB, Jiang HE, Li Xet al. Phytochemical and genetic analyses of ancient cannabis from Central Asia. J Exp Bot. 2008;59:4171–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McPartland JM, Hegman W, Long TW. Cannabis in Asia: its center of origin and early cultivation, based on a synthesis of subfossil pollen and archaeobotanical studies. Veg Hist Archaeobotany. 2019;28:691–702 [Google Scholar]
- 29. Crocq MA. History of cannabis and the endocannabinoid system. Dialogues Clin Neurosci. 2020;22:223–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonini SA, Premoli M, Tambaro Set al. Cannabis sativa: a comprehensive ethnopharmacological review of a medicinal plant with a long history. J Ethnopharmacol. 2018;227:300–15 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Q, Chen X, Guo Het al. Latitudinal adaptation and genetic insights into the origins of Cannabis sativa L. Front Plant Sci. 2018;9:1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laverty KU, Stout JM, Sullivan MJet al. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 2019;29:146–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang HE, Wang L, Merlin MDet al. Ancient cannabis burial shroud in a central Eurasian cemetery. Econ Bot. 2016;70:213–21 [Google Scholar]
- 34. Kovalchuk I, Pellino M, Rigault Pet al. The genomics of cannabis and its close relatives. Annu Rev Plant Biol. 2020;71:713–39 [DOI] [PubMed] [Google Scholar]
- 35. Ren G, Zhang X, Li Yet al. Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa. Sci Adv. 2021;7:eabg2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bautista JL, Yu S, Tian L. Flavonoids in Cannabis sativa: biosynthesis, bioactivities, and biotechnology. ACS Omega. 2021;6:5119–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanus LO, Meyer SM, Munoz Eet al. Phytocannabinoids: a unified critical inventory. Nat Prod Rep. 2016;33:1357–92 [DOI] [PubMed] [Google Scholar]
- 38. Welling MT, Deseo MA, Bacic Aet al. Biosynthetic origins of unusual cannabimimetic phytocannabinoids in Cannabis sativa L: a review. Phytochemistry. 2022;201:113282 [DOI] [PubMed] [Google Scholar]
- 39. Deguchi M, Kane S, Potlakayala Set al. Metabolic engineering strategies of industrial hemp (Cannabis sativa L.): a brief review of the advances and challenges. Front. Plant Sci. 2020;11:580621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gulck T, Moller BL. Phytocannabinoids: origins and biosynthesis. Trends Plant Sci. 2020;25:985–1004 [DOI] [PubMed] [Google Scholar]
- 41. Rezende B, Alencar AKN, Bem GFet al. Endocannabinoid system: chemical characteristics and biological activity. Pharmaceuticals (Basel). 2023;16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Breivogel CS, Childers SR, Deadwyler SAet al. Chronic delta9-tetrahydrocannabinol treatment produces a time-dependent loss of cannabinoid receptors and cannabinoid receptor-activated G proteins in rat brain. J Neurochem. 1999;73:2447–59 [DOI] [PubMed] [Google Scholar]
- 43. Wang GS, Bourne DWA, Klawitter Jet al. Disposition of oral delta-9 tetrahydrocannabinol (THC) in children receiving cannabis extracts for epilepsy. Clin Toxicol. 2020;58:124–8 [DOI] [PubMed] [Google Scholar]
- 44. Nagarkatti P, Pandey R, Rieder SAet al. Cannabinoids as novel anti-inflammatory drugs. Future Med Chem. 2009;1:1333–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Russo E, Guy GW. A tale of two cannabinoids: the therapeutic rationale for combining tetrahydrocannabinol and cannabidiol. Med Hypotheses. 2006;66:234–46 [DOI] [PubMed] [Google Scholar]
- 46. Campos AC, Ortega Z, Palazuelos Jet al. The anxiolytic effect of cannabidiol on chronically stressed mice depends on hippocampal neurogenesis: involvement of the endocannabinoid system. Int J Neuropsychopharmacol. 2013;16:1407–19 [DOI] [PubMed] [Google Scholar]
- 47. Keimpema E, Marzo VD, Harkany T. Biological basis of cannabinoid medicines. Science. 2021;374:1449–50 [DOI] [PubMed] [Google Scholar]
- 48. Blessing EM, Steenkamp MM, Manzanares Jet al. Cannabidiol as a potential treatment for anxiety disorders. Neurotherapeutics. 2015;12:825–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Atalay S, Jarocka-Karpowicz I, Skrzydlewska E. Antioxidative and anti-inflammatory properties of cannabidiol. Antioxidants. 2020;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grof CPL. Cannabis, from plant to pill. Br J Clin Pharmacol. 2018;84:2463–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. El-Alfy AT, Ivey K, Robinson Ket al. Antidepressant-like effect of Delta(9)-tetrahydrocannabinol and other cannabinoids isolated from Cannabis sativa L. Pharmacol Biochem Behav. 2010;95:434–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Petrocellis L, Ligresti A, Moriello ASet al. Effects of cannabinoids and cannabinoid-enriched cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br J Pharmacol. 2011;163:1479–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Udoh M, Santiago M, Devenish Set al. Cannabichromene is a cannabinoid CB2 receptor agonist. Br J Pharmacol. 2019;176:4537–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zagozen M, Cerenak A, Kreft S. Cannabigerol and cannabichromene in Cannabis sativa L. Acta Pharma. 2021;71:355–64 [DOI] [PubMed] [Google Scholar]
- 55. Anil SM, Shalev N, Vinayaka ACet al. Cannabis compounds exhibit anti-inflammatory activity in vitro in COVID-19-related inflammation in lung epithelial cells and pro-inflammatory activity in macrophages. Sci Rep. 2021;11:1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nachnani R, Raup-Konsavage WM, Vrana KE. The pharmacological case for Cannabigerol. J Pharmacol Exp Ther. 2021;376:204–12 [DOI] [PubMed] [Google Scholar]
- 57. Izzo AA, Borrelli F, Capasso Ret al. Non-psychotropic plant cannabinoids: new therapeutic opportunities from an ancient herb. Trends Pharmacol Sci. 2009;30:515–27 [DOI] [PubMed] [Google Scholar]
- 58. Stout JM, Boubakir Z, Ambrose SJet al. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J. 2012;71:353–65 [DOI] [PubMed] [Google Scholar]
- 59. Taura F, Tanaka S, Taguchi Cet al. Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 2009;583:2061–6 [DOI] [PubMed] [Google Scholar]
- 60. Gagne SJ, Stout JM, Liu Eet al. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc Natl Acad Sci U S A. 2012;109:12811–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Flores-Sanchez IJ, Verpoorte R. PKS activities and biosynthesis of cannabinoids and flavonoids in Cannabis sativa L. plants. Plant Cell Physiol. 2008;49:1767–82 [DOI] [PubMed] [Google Scholar]
- 62. Vranova E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700 [DOI] [PubMed] [Google Scholar]
- 63. Luo X, Reiter MA, d'Espaux Let al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019;567:123–6 [DOI] [PubMed] [Google Scholar]
- 64. Marks MD, Tian L, Wenger JPet al. Identification of candidate genes affecting delta(9)-tetrahydrocannabinol biosynthesis in Cannabis sativa. J Exp Bot. 2009;60:3715–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Taura F, Morimoto S, Shoyama Y. Purification and characterization of cannabidiolic-acid synthase from Cannabis sativa L. Biochemical analysis of a novel enzyme that catalyzes the oxidocyclization of cannabigerolic acid to cannabidiolic acid. J Biol Chem. 1996;271:17411–6 [DOI] [PubMed] [Google Scholar]
- 66. Morimoto S, Komatsu K, Taura Fet al. Purification and characterization of cannabichromenic acid synthase from Cannabis sativa. Phytochemistry. 1998;49:1525–9 [DOI] [PubMed] [Google Scholar]
- 67. Livingston SJ, Rensing KH, Page JEet al. A polarized supercell produces specialized metabolites in cannabis trichomes. Curr Biol. 2022;32:4040–4047.e4 [DOI] [PubMed] [Google Scholar]
- 68. Grassa CJ, Weiblen GD, Wenger JPet al. A new cannabis genome assembly associates elevated cannabidiol (CBD) with hemp introgressed into marijuana. New Phytol. 2021;230:1665–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bakel SJM, Cote AGet al. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011;12:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weiblen GD, Wenger JP, Craft KJet al. Gene duplication and divergence affecting drug content in Cannabis sativa. New Phytol. 2015;208:1241–50 [DOI] [PubMed] [Google Scholar]
- 71. Meijer EPM, Hammond KM, Sutton A. The inheritance of chemical phenotype in Cannabis sativa L. (IV): cannabinoid-free plants. Euphytica. 2009;168:95–112 [Google Scholar]
- 72. Meijer EPM, Bagatta M, Carboni Aet al. The inheritance of chemical phenotype in Cannabis sativa L. Genetics. 2003;163:335–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Taura F, Sirikantaramas S, Shoyama Yet al. Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett. 2007;581:2929–34 [DOI] [PubMed] [Google Scholar]
- 74. Kojoma M, Seki H, Yoshida Set al. DNA polymorphisms in the tetrahydrocannabinolic acid (THCA) synthase gene in "drug-type" and "fiber-type" Cannabis sativa L. Forensic Sci Int. 2006;159:132–40 [DOI] [PubMed] [Google Scholar]
- 75. Mansouri H, Salari F. Influence of mevinolin on chloroplast terpenoids in Cannabis sativa. Physiol Mol Biol Plants. 2014;20:273–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Garrido J, Rico S, Corral Cet al. Exogenous application of stress-related signaling molecules affect growth and cannabinoid accumulation in medical cannabis (Cannabis sativa L.). Front. Plant Sci. 2022;13:1082554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mansouri H, Asrar Z, Amarowicz R. The response of terpenoids to exogenous gibberellic acid in Cannabis sativa L. at vegetative stage. Acta Physiol Plant. 2011;33:1085–91 [Google Scholar]
- 78. Apicella PV, Sands LB, Ma Yet al. Delineating genetic regulation of cannabinoid biosynthesis during female flower development in Cannabis sativa. Plant Direct. 2022;6:e412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Babaei M, Ajdanian L. Screening of different Iranian ecotypes of cannabis under water deficit stress. Sci Hortic. 2020;260:108904 [Google Scholar]
- 80. Parsons JL, Martin SL, James Tet al. Polyploidization for the genetic improvement of Cannabis sativa. Front Plant Sci. 2019;10:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Galan-Avila A, Garcia-Fortea E, Prohens Jet al. Development of a direct in vitro plant regeneration protocol from Cannabis sativa L. seedling explants: developmental morphology of shoot regeneration and ploidy level of regenerated plants. Front. Plant Sci. 2020;11:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Islam MJ, Ryu BR, Azad MOKet al. Cannabinoids accumulation in hemp (Cannabis sativa L.) plants under LED light spectra and their discrete role as a stress marker. Biology (Basel). 2021;10:710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Murovec J, Erzen JJ, Flajsman Met al. Analysis of morphological traits, cannabinoid profiles, THCAS gene sequences, and photosynthesis in wide and narrow leaflet high-cannabidiol breeding populations of medical cannabis. Front Plant Sci. 2022;13:786161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li C, Yu W, Xu Jet al. Anthocyanin biosynthesis induced by MYB transcription factors in plants. Int J Mol Sci. 2022;23:11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jan R, Asaf S, Numan Met al. Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy-Basel. 2021;11:968 [Google Scholar]
- 86. Feng K, Hou X-L, Xing G-Met al. Advances in AP2/ERF super-family transcription factors in plant. Crit Rev Biotechnol. 2020;40:750–76 [DOI] [PubMed] [Google Scholar]
- 87. Dubos C, Stracke R, Grotewold Eet al. MYB transcription factors in Arabidopsis. Trends in Plant Sci. 2010;15:573–81 [DOI] [PubMed] [Google Scholar]
- 88. Ma G, Zelman AK, Apicella PVet al. Genome-wide identification and expression analysis of homeodomain leucine zipper subfamily IV (HD-ZIP IV) gene family in Cannabis sativa L. Plants (Basel). 2022;11:1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sipahi H, Whyte TD, Ma Get al. Genome-wide identification and expression analysis of wall-associated kinase (WAK) gene family in Cannabis sativa L. Plants (Basel). 2022;11:2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhu X, Mi Y, Meng Xet al. Genome-wide identification of key enzyme-encoding genes and the catalytic roles of two 2-oxoglutarate-dependent dioxygenase involved in flavonoid biosynthesis in Cannabis sativa L. Microb Cell Factories. 2022;21:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang J, Zhang C, Huang Set al. Key cannabis salt-responsive genes and pathways revealed by comparative transcriptome and physiological analyses of contrasting varieties. Agronomy. 2021;11:2338 [Google Scholar]
- 92. Monthony AS, Page SR, Hesami Met al. The past, present and future of Cannabis sativa tissue culture. Plants (Basel). 2021;10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hesami M, Baiton A, Alizadeh Met al. Advances and perspectives in tissue culture and genetic engineering of cannabis. Int J Mol Sci. 2021;22:5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Deguchi M, Bogush D, Weeden Het al. Establishment and optimization of a hemp (Cannabis sativa L.) agroinfiltration system for gene expression and silencing studies. Sci Rep. 2020;10:3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schachtsiek J, Hussain T, Azzouhri Ket al. Virus-induced gene silencing (VIGS) in Cannabis sativa L. Plant Methods. 2019;15:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Matchett-Oates L, Mohamaden E, Spangenberg GCet al. Development of a robust transient expression screening system in protoplasts of cannabis. In Vitro Cell Dev Biol Plant. 2021;57:1040–50 [Google Scholar]
- 97. Alter H, Peer R, Dombrovsky Aet al. Tobacco rattle virus as a tool for rapid reverse-genetics screens and analysis of gene function in Cannabis sativa L. Plants (Basel). 2022;11:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sorokin A, Yadav NS, Gaudet Det al. Transient expression of the β-glucuronidase gene in Cannabis sativa varieties. Plant Signal Behav. 2020;15:1780037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ahmed S, Gao XF, Jahan MAet al. Nanoparticle-based genetic transformation of Cannabis sativa. J Biotechnol. 2021;326:48–51 [DOI] [PubMed] [Google Scholar]
- 100. Wahby I, Caba JM, Ligero F. Cannabis sativa infection of hemp (Cannabis sativa L.): establishment of hairy root cultures. J Plant Interact. 2013;8:312–20 [Google Scholar]
- 101. Zhang X, Xu G, Cheng Cet al. Establishment of an Cannabis sativa-mediated genetic transformation and CRISPR/Cas9-mediated targeted mutagenesis in hemp (Cannabis sativa L.). Plant Biotechnol J. 2021;19:1979–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Galán-Ávila A, Gramazio P, Ron Met al. A novel and rapid method for Cannabis sativa-mediated production of stably transformed Cannabis sativa L. plants. Ind Crop. Prod. 2021;170:113691 [Google Scholar]
- 103. Han G, Li Y, Yang Zet al. Molecular mechanisms of plant trichome development. Front Plant Sci. 2022;13:910228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Huchelmann A, Boutry M, Hachez C. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol. 2017;175:6–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li H, Wang G. Specialized metabolism in plant glandular trichomes. Scientia Sinica Vitae. 2015;45:557–68 [Google Scholar]
- 106. Chen H, Guo M, Dong Set al. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity. Plant Commun. 2023;4:100516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sun M, Zhang Y, Zhu Let al. Chromosome-level assembly and analysis of the thymus genome provide insights into glandular secretory trichome formation and monoterpenoid biosynthesis in thyme. Plant Commun. 2022;3:100413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang G. Recent progress in secondary metabolism of plant glandular trichomes. Plant Biotechnology. 2014;31:353–61 [Google Scholar]
- 109. Tissier A. Glandular trichomes: what comes after expressed sequence tags? Plant J. 2012;70:51–68 [DOI] [PubMed] [Google Scholar]
- 110. Schuurink R, Tissier A. Glandular trichomes: micro-organs with model status? New Phytol. 2020;225:2251–66 [DOI] [PubMed] [Google Scholar]
- 111. Chalvin C, Drevensek S, Dron Met al. Genetic control of glandular trichome development. Trends Plant Sci. 2020;25:477–87 [DOI] [PubMed] [Google Scholar]
- 112. Wang Z, Yang ZR, Li FG. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol J. 2019;17:1706–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Szymanski DB, Lloyd AM, Marks MD. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 2000;5:214–9 [DOI] [PubMed] [Google Scholar]
- 114. Aizpurua-Olaizola O, Soydaner U, Ozturk Eet al. Evolution of the cannabinoid and terpene content during the growth of Cannabis sativa plants from different chemotypes. J Nat Prod. 2016;79:324–31 [DOI] [PubMed] [Google Scholar]
- 115. Jin D, Dai K, Xie Zet al. Secondary metabolites profiled in cannabis inflorescences, leaves, stem barks, and roots for medicinal purposes. Sci Rep. 2020;10:3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dayanandan P, Kaufman PB. Trichomes of Cannabis sativa L. (Cannabaceae). Am J Bot. 1976;63:578–91 [Google Scholar]
- 117. López Carretero P, Pekas A, Stubsgaard Let al. Glandular trichomes affect mobility and predatory behavior of two aphid predators on medicinal cannabis. Biol Control. 2022;170:104932 [Google Scholar]
- 118. Rodziewicz P, Loroch S, Marczak Let al. Cannabinoid synthases and osmoprotective metabolites accumulate in the exudates of Cannabis sativa L. glandular trichomes. Plant Sci. 2019;284:108–16 [DOI] [PubMed] [Google Scholar]
- 119. Happyana N, Agnolet S, Muntendam Ret al. Analysis of cannabinoids in laser-microdissected trichomes of medicinal Cannabis sativa using LCMS and cryogenic NMR. Phytochemistry. 2013;87:51–9 [DOI] [PubMed] [Google Scholar]
- 120. Livingston SJ, Bae EJ, Unda Fet al. Cannabis glandular trichome cell walls undergo remodeling to store specialized metabolites. Plant Cell Physiol. 2021;62:1944–62 [DOI] [PubMed] [Google Scholar]
- 121. Livingston SJ, Chou EY, Quilichini TDet al. Overcoming the challenges of preserving lipid-rich Cannabis sativa L. glandular trichomes for transmission electron microscopy. J Microsc. 2022;291:119–27 [DOI] [PubMed] [Google Scholar]
- 122. Perez-Rodriguez M, Jaffe FW, Butelli Eet al. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development. 2005;132:359–70 [DOI] [PubMed] [Google Scholar]
- 123. Jaffe FW, Tattersall A, Glover BJ. A truncated MYB transcription factor from Antirrhinum majus regulates epidermal cell outgrowth. J Exp Bot. 2007;58:1515–24 [DOI] [PubMed] [Google Scholar]
- 124. Haiden SR, Apicella PV, Ma Yet al. Overexpression of CsMIXTA, a transcription factor from Cannabis sativa, increases glandular trichome density in tobacco leaves. Plants (Basel). 2022;11:1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chang J, Yu T, Yang QHet al. Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018;96:90–102 [DOI] [PubMed] [Google Scholar]
- 126. Chun JI, Kim SM, Kim Het al. SlHair2 regulates the initiation and elongation of type I trichomes on tomato leaves and stems. Plant Cell Physiol. 2021;62:1446–59 [DOI] [PubMed] [Google Scholar]
- 127. Chun JI, Kim SM, Jeong NRet al. Tomato ARPC1 regulates trichome morphology and density and terpene biosynthesis. Planta. 2022;256:38. [DOI] [PubMed] [Google Scholar]
- 128. Gao SH, Gao YN, Xiong Cet al. The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 2017;262:103–14 [DOI] [PubMed] [Google Scholar]
- 129. Yu XH, Chen GP, Tang BYet al. The jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2018;267:65–73 [DOI] [PubMed] [Google Scholar]
- 130. Zhang XL, Yan F, Tang YWet al. Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant Cell Physiol. 2015;56:2110–24 [DOI] [PubMed] [Google Scholar]
- 131. Chen Y, Su D, Li Jet al. Overexpression of bHLH95, a basic helix-loop-helix transcription factor family member, impacts trichome formation via regulating gibberellin biosynthesis in tomato. J Exp Bot. 2020;71:3450–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Yan T, Chen M, Shen Qet al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2017;213:1145–55 [DOI] [PubMed] [Google Scholar]
- 133. Shi P, Fu X, Shen Qet al. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua. New Phytol. 2018;217:261–76 [DOI] [PubMed] [Google Scholar]
- 134. Wang C, Chen T, Li Yet al. AaWIN1, an AP2/ERF protein, positively regulates glandular secretory trichome initiation in Artemisia annua. Plant Sci. 2023;329:111602 [DOI] [PubMed] [Google Scholar]
- 135. Xie L, Yan T, Li Let al. An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua. New Phytol. 2021;231:2050–64 [DOI] [PubMed] [Google Scholar]
- 136. Chen TT, Liu H, Li YPet al. AaSEPALLATA1 integrates jasmonate and light-regulated glandular secretory trichome initiation in Artemisia annua. Plant Physiol. 2023;192:1483–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lv Z, Li J, Qiu Set al. The transcription factors TLR1 and TLR2 negatively regulate trichome density and artemisinin levels in Artemisia annua. J Integr Plant Biol. 2022;64:1212–28 [DOI] [PubMed] [Google Scholar]
- 138. Matias-Hernandez L, Jiang W, Yang Ket al. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 2017;90:520–34 [DOI] [PubMed] [Google Scholar]
- 139. Guo Z, Hao K, Lv Zet al. Profiling of phytohormone-specific microRNAs and characterization of the miR160-ARF1 module involved in glandular trichome development and artemisinin biosynthesis in Artemisia annua. Plant Biotechnol J. 2023;21:591–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Liu ZX, Yu XL, Qin AZet al. Research strategies for single-cell transcriptome analysis in plant leaves. Plant J. 2022;112:27–37 [DOI] [PubMed] [Google Scholar]
- 141. Misra BB, Assmann SM, Chen SX. Plant single-cell and single-cell-type metabolomics. Trends Plant Sci. 2014;19:637–46 [DOI] [PubMed] [Google Scholar]
- 142. Yang D, Park SY, Park YSet al. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020;38:745–65 [DOI] [PubMed] [Google Scholar]
- 143. Tan Z, Clomburg JM, Gonzalez R. Synthetic pathway for the production of olivetolic acid in Escherichia coli. ACS Synth Biol. 2018;7:1886–96 [DOI] [PubMed] [Google Scholar]
- 144. Okorafor IC, Chen M, Tang Y. High-titer production of olivetolic acid and analogs in engineered fungal host using a nonplant biosynthetic pathway. ACS Synth Biol. 2021;10:2159–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ma Y, Li J, Huang Set al. Targeting pathway expression to subcellular organelles improves astaxanthin synthesis in Yarrowia lipolytica. Metab Eng. 2021;68:152–61 [DOI] [PubMed] [Google Scholar]
- 146. Luo Z, Liu N, Lazar Zet al. Enhancing isoprenoid synthesis in Yarrowia lipolytica by expressing the isopentenol utilization pathway and modulating intracellular hydrophobicity. Metab Eng. 2020;61:344–51 [DOI] [PubMed] [Google Scholar]
- 147. Ma J, Gu Y, Xu P. Biosynthesis of cannabinoid precursor olivetolic acid in genetically engineered Yarrowia lipolytica. Commun Biol. 2022;5:1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Chen X, Kollner TG, Jia Qet al. Terpene synthase genes in eukaryotes beyond plants and fungi: occurrence in social amoebae. Proc Natl Acad Sci U S A. 2016;113:12132–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Reimer C, Kufs JE, Rautschek Jet al. Engineering the amoeba Dictyostelium discoideum for biosynthesis of a cannabinoid precursor and other polyketides. Nat Biotechnol. 2022;40:751–8 [DOI] [PubMed] [Google Scholar]
- 150. Kufs JE, Reimer C, Steyer Eet al. Scale-up of an amoeba-based process for the production of the cannabinoid precursor olivetolic acid. Microb Cell Factories. 2022;21:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Dekishima Y, Lan EI, Shen CRet al. Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J Am Chem Soc. 2011;133:11399–401 [DOI] [PubMed] [Google Scholar]
- 152. Qiu J, Hou K, Li Qet al. Boosting the cannabidiol production in engineered Saccharomyces cerevisiae by harnessing the vacuolar transporter BPT1. J Agric Food Chem. 2022;70:12055–64 [DOI] [PubMed] [Google Scholar]
- 153. Zirpel B, Degenhardt F, Martin Cet al. Engineering yeasts as platform organisms for cannabinoid biosynthesis. J Biotechnol. 2017;259:204–12 [DOI] [PubMed] [Google Scholar]
- 154. Lim KJH, Hartono YD, Xue Bet al. Structure-guided engineering of prenyltransferase NphB for high-yield and regioselective cannabinoid production. ACS Catal. 2022;12:4628–39 [Google Scholar]
- 155. Gulck T, Booth JK, Carvalho Aet al. Synthetic biology of cannabinoids and cannabinoid glucosides in Nicotiana benthamiana and Saccharomyces cerevisiae. J Nat Prod. 2020;83:2877–93 [DOI] [PubMed] [Google Scholar]
- 156. Reddy VA, Leong SH, Jang ICet al. Metabolic engineering of Nicotiana benthamiana to produce cannabinoid precursors and their analogues. Meta. 2022;12:1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Zirpel B, Stehle F, Kayser O. Production of Δ9-tetrahydrocannabinolic acid from cannabigerolic acid by whole cells of Pichia (Komagataella) pastoris expressing Δ9-tetrahydrocannabinolic acid synthase from Cannabis sativa l. Biotechnol Lett. 2015;37:1869–75 [DOI] [PubMed] [Google Scholar]
- 158. Lee YE, Nakashima Y, Kodama Tet al. Dual engineering of olivetolic acid cyclase and tetraketide synthase to generate longer alkyl-chain olivetolic acid analogs. Org Lett. 2022;24:410–4 [DOI] [PubMed] [Google Scholar]
- 159. Valliere MA, Korman TP, Woodall NBet al. A cell-free platform for the prenylation of natural products and application to cannabinoid production. Nat Commun. 2019;10:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Valliere MA, Korman TP, Arbing MAet al. A bio-inspired cell-free system for cannabinoid production from inexpensive inputs. Nat Chem Biol. 2020;16:1427–33 [DOI] [PubMed] [Google Scholar]
- 161. Hassani D, Fu X, Shen Qet al. Parallel transcriptional regulation of artemisinin and flavonoid biosynthesis. Trends Plant Sci. 2020;25:466–76 [DOI] [PubMed] [Google Scholar]
- 162. Sun S, Shen X, Li Yet al. Single-cell RNA sequencing provides a high-resolution roadmap for understanding the multicellular compartmentation of specialized metabolism. Nat Plants. 2023;9:179–90 [DOI] [PubMed] [Google Scholar]
- 163. Sun W, Xu Z, Song Cet al. Herbgenomics: decipher molecular genetics of medicinal plants. Innovation. 2022;3:100322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Chen S, Yin X, Han Jet al. DNA barcoding in herbal medicine: retrospective and prospective. J Pharm Anal. 2023;13:431–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Welling MT, Liu L, Kretzschmar Tet al. An extreme-phenotype genome-wide association study identifies candidate cannabinoid pathway genes in cannabis. Sci Rep. 2020;10:18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Cascini F, Farcomeni A, Migliorini Det al. Highly predictive genetic markers distinguish drug-type from fiber-type Cannabis sativa L. Plants (Basel). 2019;8:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Watts S, McElroy M, Migicovsky Zet al. Cannabis labelling is associated with genetic variation in terpene synthase genes. Nat Plants. 2021;7:1330–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hurgobin B, Tamiru-Oli M, Welling MTet al. Recent advances in Cannabis sativa genomics research. New Phytol. 2021;230:73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Romero P, Peris A, Vergara Ket al. Comprehending and improving cannabis specialized metabolism in the systems biology era. Plant Sci. 2020;298:110571 [DOI] [PubMed] [Google Scholar]
- 170. Sommano SR, Sunanta P, Leksawasdi Net al. Mass spectrometry-based metabolomics of phytocannabinoids from non-cannabis plant origins. Molecules. 2022;27:3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Berman P, Haro LA, Jozwiak Aet al. Parallel evolution of cannabinoid biosynthesis. Nat Plants. 2023;9:817–31 [DOI] [PubMed] [Google Scholar]
- 172. Kufs JE, Reimer C, Stallforth Pet al. The potential of amoeba-based processes for natural product syntheses. Curr Opin Biotechnol. 2022;77:102766 [DOI] [PubMed] [Google Scholar]
- 173. Schilling S, Melzer R, Dowling CAet al. A protocol for rapid generation cycling (speed breeding) of hemp (Cannabis sativa) for research and agriculture. Plant J. 2023;113:437–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Kodym A, Leeb CJ. Back to the roots: protocol for the photoautotrophic micropropagation of medicinal cannabis. Plant Cell Tissue Organ Cult. 2019;138:399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Mohan Ram HY, Sett R. Induction of fertile male flowers in genetically female Cannabis sativa plants by silver nitrate and silver thiosulphate anionic complex. Theoret Appl Genetics. 1982;62:369–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.