Abstract
CRISPR interference (CRISPRi) has been developed and widely used for gene repression in various hosts. Here we report an improved CRISPRi system in Pichia pastoris by fusing dCas9 with endogenous transcriptional repressor domains. The CRISPRi system shows strong repression of eGFP, with the highest efficiency of 85%. Repression of native genes is demonstrated by targeting AOX1 promoter. AOX1 is efficiently repressed and the mutant strains show much slower growth in methanol medium. Effects of gRNA expression and processing on CRISPRi efficiency is also investigated. It is found that gRNA processing by HH/HDV ribozymes or Csy4 endoribonuclease generating clean gRNA is critical to achieve strong repression, and Csy4 cleavage shows higher repression efficiency. However, gRNA expression using native tRNA transcription and processing systems results in relatively weaker repression of eGFP. By expression of two gRNAs targeting promoters of eGFP and AOX1 in an array together with Cys4 recognition sites, both genes can be repressed simultaneously. Cys4-mediated gRNA array processing is further applied to repress fatty acyl-CoA synthetase genes (FAA1 and FAA2). Both genes are efficiently repressed, demonstrating that Cys4 endoribonuclease has the ability to cleave gRNAs array and can be can be used for multiplexed gene repression in P. pastoris.
Keywords: Pichia pastoris, CRISPRi, HH/HDV, Csy4, gRNA array
Credit Author Statement
Shujing. Qiao: designed the experiments and wrote the manuscript. Lun Yao designed the experiments and wrote the manuscript. Shujing Qiao: performed the experiments. Fan Bai: designed and constructed the tetR-tetO regulated promoter. Peng Cai: provided the CRISPR/Cas9 gene editing plasmids and strains. Lun Yao: proposed the idea. Yongjin J. Zhou: proposed the idea.
1. Introduction
The methylotrophic yeast Pichia pastoris (P. pastoris, syn. Komagataella phaffii) has been widely used for protein production for decades. Recently, it also shows great potential for construction of cell factories for production of fuels and chemicals using methanol as carbon source [1,2]. Construction of robust microbial cell factories requires rewiring of cellular metabolism to promote the synthesis of target compounds and reduce the formation of by-products [[3], [4], [5]]. The development of synthetic biology tools [[6], [7], [48]], especially CRISPR/Cas9-based technology, has greatly facilitated metabolic engineering of P. pastoris [[8], [9], [10], [11], [12], [13]]. However, development of genetic tools in P. pastoris still lags far behind model organisms such as Saccharomyces cerevisiae (S. cerevisiae) [14,15]. Therefore, it is necessary to further expand synthetic biology toolbox to promote metabolic engineering and basic research in P. pastoris.
CRISPRi has been demonstrated to be an efficient tool for gene regulation [16], and has been successfully applied in bacteria [[17], [18], [19], [20]], mammalian cells [21,22], plant cells [23,24] and yeasts [[25], [26], [27]]. In CRISPRi system, a catalytically dead Cas9 (dCas9) is obtained by mutating the HNH and RuvC nuclease domains (D10A and H840A, respectively). dCas9 loses the ability of cutting target DNA, but still retains the activity of forming a complex with gRNA and binding to the target sequence [16]. By directing dCas9-gRNA complex to the promoter or coding region of a target gene, transcriptional repression (silencing) can be achieved by steric hindrance of RNA polymerase and/or transcription factors (Fig. 1a). Studies have shown that, unlike CRISPRi system in bacteria that dCas9 alone can play a strong inhibitory role, it is necessary to fuse a transcriptional repressor domain with dCas9 to achieve efficient repression in eukaryotic cells [28]. Several transcriptional repressor domains have been tested and used for gene repression in mammalian cells and yeasts [21,22,29]. Recently, Lian et al. screened native transcriptional repressor domains in S. cerevisiae and found that endogenous repressor domains resulted in stronger repression in S. cerevisiae than repressor domains from mammalian cells [25].
Fig. 1.
Construction of CRISPRi system by using endogenous transcriptional repression domains. a. Schematic illustration of CRISPRi system. dCas9 is fused with a transcriptional repression domain (RD), and forms a complex with gRNA. The dCas9-RD-gRNA complex binds to the promoter region and blocks transcription. b. Expression cassettes of dCas9 and gRNA. RD1 and RD2 are the endogenous repression domains of the general transcriptional repressor Tup1. RD1 and RD2 are fused to the C-terminal end of dCas9 using GS-linker. Term, terminator. c. Selection of gRNAs targeting Ogataea polymorpha promoter POpTEF1 to repress eGFP expression. Values show the position of targeting sites relative to the TSS of POpTEF1. d.eGFP repression of strains with different combinations of dCas9/dCas9-RD and gRNAs. Results are presented as mean ± standard deviation (SD) of at least three biological replicates.
Compared with S. cerevisiae, much fewer studies on CRISPR/dCas9-based gene regulation have been reported in P. pastoris. Baumschabl et al. demonstrated the transcriptional activation in P. pastoris by fusing dCas9 with trans-activator domain (VP64) through RNA scaffold, and this system also showed repression effect for targets close to transcription start site (TSS) of the promoter [30]. Liao et al. constructed a CRISPR-ARE system which could achieve simultaneous gene activation, repression and editing, of which gene repression was realized by fusing dCas9 with heterologous transcriptional repressors (Mix1/RD1152) from S. cerevisiae [31]. The application of CRISPRi in P. pastoris is still limited by relatively low repression efficiency and lack of effective tools for multiplexed repression. It is thus necessary to screen more transcriptional repressor domains, especially native ones, to improve CRISPRi efficiency. Efficient gRNA expression and processing are also critical for achieving strong repression and multiplexed CRISPRi [32]. Pol-II promoters are commonly used for gRNA expression in P. pastoris due to their strong transcription level, however, ribozymes (such as HH and HDV) are required for gRNA processing in this case [9,33]. In addition to HH/HDV, native tRNA transcription and processing systems have also been characterized and harnessed for gRNA expression in P. pastoris [11,31]. crRNA processing proteins (e.g., Csy4 endoribonuclease) in native CRISPR systems can also be used for gRNA processing [34]. The Csy4 endoribonuclease from Pseudomonas aeruginosa has a high degree of substrate specificity toward a 28 nucleotides RNA stem-loop and cleaves at a fixed site [35], and has been successfully used for multiplexed gRNA expression in S. cerevisiae [34,36,37].
In this study, the endogenous repression domains (RD1 and RD2) of the general transcriptional repressor Tup1 were fused with dCas9 for construction of CRISPRi system in P. pastoris, and it achieved efficient eGFP repression. Targeting the promoter region of endogenous alcohol oxidase 1 (AOX1) efficiently repressed transcription of AOX1 and slowed cell growth when cultivated in methanol medium. Different gRNA expression and processing systems, including HH/HDV ribozymes, native tRNA expression and processing systems and Csy4 endoribonuclease were tested, and Csy4-mediated gRNA cleavage showed strongest repression of eGFP. Csy4 also showed the ability to process gRNAs array and could be used for multiplexed CRISPRi. Overall, this work provides an improved CRISPRi system, and it can be used for both applied and basic research in P. pastoris.
2. Materials and methods
2.1. Strains and cultivation
Escherichia coli (E. coli) DH5α was used for plasmid construction and propagation. P. pastoris GS115 was offered by Prof. Menghao Cai from East China University of Science and Technology. All strains constructed in this study were listed in Supplementary Table S1. E. coli DH5α was cultivated in LB medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) and antibiotics were added when necessary (ampicillin at 100 μg/mL, zeocin at 100 μg/mL, kanamycin at 50 μg/mL). For preparation of electrocompetent cells, P. pastoris were cultivated in YPD medium consisting of 10 g/L yeast extract, 20 g/L peptone and 20 g/L glucose. For solid plates, 20 g/L agar was added. P. pastoris shake flask fermentation was carried out in Delft minimal medium (pH5.6) containing 2.5 g/L (NH4)2SO4, 14.4 g/L KH2PO4, 0.5 g/L MgSO4•7H2O, 0.04 g/L His, 2 mL/L trace metals, 1 mL/L vitamin solution and one of the following carbon sources, 20 g/L glucose (Delft + G20) or 10 g/L methanol (Delft + M10). 200 μg/mL G418 was added to maintain gRNA plasmid. All strains were cultivated (37 °C for E. coli DH5α and 30 °C for P. pastoris) in shake incubators (Zhichu Shaker ZQZY-CS8).
2.2. DNA assembly and genetic manipulation of P. pastoris
Genes (dCas9, eGFP, Csy4 and tetR) were either chemically synthesized or amplified using existing templates in the lab (all sequences are listed in Table S2). Gene expression cassettes consisting upstream homologous arm (HA), promoter, gene, terminator and downstream HA, was assembled by one-pot overlap extension PCR. The assembled PCR products were purified and used as donor for chromosome integration. DNA integration into P. pastoris chromosome was performed using a CRISPR-Cas9 genome engineering technology established previously [9]. Donor DNA and Cas9/gRNA expression plasmid (500 ng donor DNA and 500 ng gRNA plasmid) were co-transformed into P. pastoris competent cells via electroporation and selected on YPD plates supplemented with zeocin (100 μg/mL). Clones were selected, and their genotype were verified by PCR and Sanger sequencing. Clones with correct integration were cultivated overnight in YPD liquid medium and then plated on YPD plates without antibiotics to remove Cas9/gRNA plasmid. All primers used in this study are listed in Supplementary Table S3.
2.3. gRNA design, plasmid construction and transformation
The promoter region within 300-bp upstream of the TSS of either template or non-template strand was used for gRNA design for CRISPRi. 20 bp target sequences of gRNAs were designed using a web-based gRNA design toolbox (http://chopchop.cbu.uib.no). All gRNA spacer sequences are listed in Supplementary Table S4. gRNA plasmids were constructed by overlap-PCR using previously constructed gRNA plasmid as template according to the described method [9]. Transformation of gRNA plasmids into P. pastoris was performed by electroporation according to the described protocol [38]. About 50 ng plasmid was used for each transformation. Cells were plated on agar plates supplemented with G418 (200 μg/mL) after transformation, and incubated under 30 °C for two to three days to obtain single colonies.
2.4. Fluorescence measurement
For determination of eGFP fluorescent intensity, single clones were pre-cultured in 400 μL YPD containing G418 in 2 mL Eppendorf tubes for 12 h, then transferred into 4 mL YPD containing G418 in 15 mL centrifuge tubes for 12–16 h. Cells were collected by centrifugation, re-suspended with 20 mL Delft medium with initial OD600 of 0.1, and then cultivated for 20 h with Delft + G20 medium or 24 h with Delft + M10 medium. Cells were harvested by centrifugation at 500g for 4 min and washed with 1 mL PBS twice, and diluted to OD600 = 0.2–0.8. Then cell suspension was used for fluorescence measurement and absorbance determination in 96-well black-walled clear bottom plate in a Tecan SPARK microplate reader (Tecan, Switzerland). The excitation and emission wavelengths were set to 485 nm and 525 nm, respectively. All fluorescence intensity was normalized by OD600.
2.5. qPCR analysis
Total RNA was extracted using RNA simple Total RNA Kit (DP419, TIANGEN). No more than 1 μg total RNA of each sample was reverse-transcribed to cDNA using the PrimeScript™ RT reagent Kit with DNA Eraser (Perfect Real Time) (RR047A, TAKARA) according to the manufacturer's protocol. Actin gene ACT1 was used as the endogenous reference. A two-step PCR reaction was employed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (RR820A, Takara Bio Inc.). Data analysis was conducted using 2−ΔΔCT method. Primers used for qPCR were listed in Supplementary Table S3.
2.6. Quantification of free fatty acids (FFAs)
Quantification of FFAs was carried out following published protocol [39]. Cell cultures were collected after 5 days fermentation in Delft + M10 medium. 200 μL culture was used for extraction of FFAs. 0.1 mg/mL pentadecanoic acid was used as internal standard for FFAs quantification. FFAs was analyzed on gas chromatography (Focus GC, Thermo Fisher Scientific) equipped with a Zebron ZB-5MS GUARDIAN capillary column (30 m × 0.25 mm × 0.25 μm, Phenomenex).
3. Results
3.1. Construction of CRISPRi system using endogenous transcriptional repressor domains
In eukaryotic cells, efficient CRISPRi can be achieved by fusion expression of transcriptional repressor domains with dCas9 (Fig. 1a). Repressor domains of Tup1, Mig1 and Ume6 have been tested and showed strong repression when fused with dCas9 in S. cerevisiae [25]. However, these repressor domains had low repression efficiency in P. pastoris [31]. We hypothesized that using native repressor domains of P. pastoris might be able to improve the repression efficiency. Therefore, we selected two repression domains (RD1 and RD2) of the endogenous Tup1 (encoding a general transcriptional repressor) from P. pastoris and fused them to the C-terminal of dCas9 through a flexible GS linker (GGSGGS). Repressor domain of Tup1 from S. cerevisiae was also fused with dCas9 to compare its repression efficiency with that of using endogenous repressor domains. A PTEF1(tetO2) promoter with reduced activity by tetO/tetR system (Fig. S1) was used to express dCas9/dCas9-repressor to avoid the potential toxicity (Fig. 1b). dCas9/dCas9-repressor cassettes were integrated into NSI-9 site of PC110 strain [9]. eGFP expression driven by POpTEF1 of Ogataea polymorpha was integrated into NSI-6 site and used for evaluation of repression efficiency. Three strains with dCas9/dCas9-RD and eGFP expression cassettes QSJ037 (dCas9, eGFP), QSJ038 (dCas9-RD1, eGFP) and QSJ039 (dCas9-RD2, eGFP) were obtained.
Four gRNAs targeting promoter POpTEF1 were designed to repress eGFP expression (Fig. 1c). gRNA0 without targeting any site was designed and used as control. These gRNA plasmids were transformed into QSJ037 (dCas9, eGFP), QSJ038 (dCas9-RD1, eGFP) and QSJ039 (dCas9-RD2, eGFP) to evaluate their repression efficiency. Fluorescence analysis showed that gRNA1(−157 to −138 bp) had no repression of eGFP in all three strains (Fig. 1d). gRNA 2, 3 and 4 showed various repression effects, among which gRNA3(−106 to −187 bp) achieved the strongest repression. Repression of eGFP showed the same pattern with previous report that repression effect was stronger for targets closer to TATA box region of the promoter than other binding sites [16,40]. Fused expression of dCas9 and RD1 resulted in strongest repression of eGFP in all cases, and achieved 85% repression of eGFP for gRNA3. Repression of eGFP using native RD1 was also more efficient than using RD1 of Tup1 from S. cerevisiae (Fig. S2). Therefore, the strain with fusion expression of dCas9 and RD1 was used for the following experiment.
3.2. Repression of endogenous gene in P. pastoris
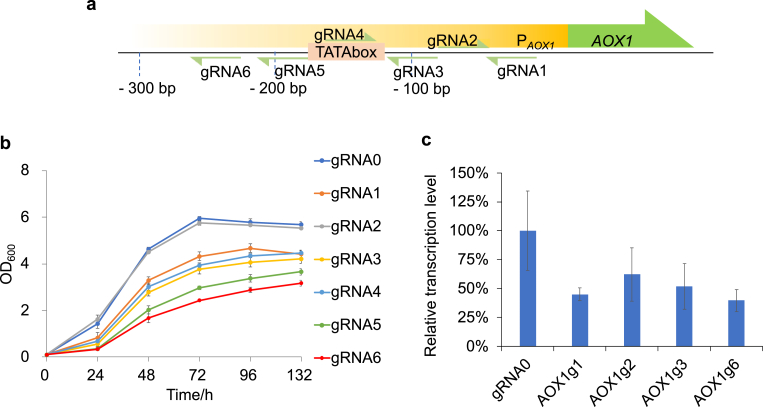
To evaluate the repression efficiency of the CRISPRi system on endogenous gene, six gRNAs targeting promoter region of alcohol oxidase 1 (PAOX1) were designed (Fig. 2a). AOX1-mediated conversion of methanol to formaldehyde is the first step of methanol metabolism and PAOX1 is a strong promoter induced by methanol [41]. According to previous report, AOX1 knockout (MutS strain) drastically reduced methanol consumption and growth rate in methanol medium, but did not affect cell growth in glucose medium [42,43]. As expected, gRNAs targeting different regions within PAOX1 did not affect cell growth in glucose medium (Fig. S3), but slowed cell growth in methanol medium (except gRNA2) (Fig. 2b), while gRNA0 did not show any effect on cell growth in either glucose or methanol medium. Relative transcription level of AOX1 was analyzed by qPCR, and repression of AOX1 was observed for all tested gRNAs (Fig. 2c). gRNA6 targeting −254 to −235 bp of PAOX1 showed the strongest repression effect on both cell growth and transcription among all gRNAs.
Fig. 2.
Repression endogenous gene alcohol oxidase 1 (AOX1). a. Schematic map of gRNAs targeting endogenous promoter PAOX1. Numbers show the relative position of gRNAs to the start codon (ATG) of AOX1. b. Growth curve of strains harboring different gRNAs targeting PAOX1 cultivated in Delft + M10 medium. gRNA0 (g0) binding no site in the genome was used as negative control. c. Relative transcription level of AOX1 repression strains harboring different gRNAs measured by qPCR. Data are presented as mean ± s. d. of three biological replicates.
3.3. Optimization of gRNA expression and processing
Efficient gRNA expression and processing are also critical to achieve efficient CRISPR/dCas9 mediated gene regulation. Pol-II promoters are widely used for gRNA expression, since they generally produce sufficient amount of gRNA and are easy to be regulated. We first tested the commonly used PHTX1 promoter for gRNA expression. The hammerhead ribozyme (HH) and the hepatitis delta virus ribozyme (HDV) were added to the left and right side of each pre-gRNA respectively for gRNA processing (Fig. 3a). Additionally, the 5′-end of HH (6 bp) was variable and reverse complementary to the 5′-end of the spacer region to generate a clean gRNA. However, the 5′ variable region of HH makes the construction of gRNA plasmid require a pair of long primers, which increases the cost and difficulty in gRNA plasmid construction. To simplify gRNA construction, we also fixed the 6-bp variable sequence of HH ribozyme [9], and constructed gRNAs with variable (gRNA2 and gRNA3) and fixed (gRNA2′ and gRNA3′) 6-bp HH ribozymes. These gRNA plasmids were transformed into QSJ038 (dCas9-RD1, eGFP) and the obtained repression strains were cultivated in glucose (Delft + G20) and methanol (Delft + M10) medium. The eGFP repression efficiency of the simplified gRNAs (gRNA2′ and gRNA3′) was about 10%–15% lower than the gRNAs with perfect matched spacer (gRNA2 and gRNA3) in both glucose (Fig. 3b) and methanol medium (Fig. S4). The decreased repression efficiency of the simplified gRNAs is probably due to the 6-bp mismatched 5′ extension of base-pairing region [16], indicating that it is critical of generating clean gRNA to achieve strong repression.
Fig. 3.
gRNA expression and processing affect repression efficiency. a. Structure of gRNA expression and processing systems. gRNA cleavage sites are showed by triangle. b. Repression of eGFP by gRNAs generated by HH/HDV ribozymes cleavage system. g2, g2′, g3 and g3′ are gRNAs targeting promoter POpTEF1 for eGFP repression. g2 and g3 contain HH ribozymes with variable 5′ end (6 bp), while g2′ and g3′ contain HH ribozyme with fixed 5′ end. g0 targeting nowhere in the genome is used as control. Plasmid gRNA0 targeting no site transformed into strain PC110 was also used as negative control to show the background fluorescence intensity. All strains were cultivated in Delft + G20. c. Repression of eGFP by gRNA3 (g3) produced by different promoters and cleavage systems. g3 targeting POpTEF1 was expressed by different tRNA promoters and cleavage by native ribozymes. For PHTX1 driven gRNAs, HH/HDV ribozymes were used for gRNA cleavage. Plasmid gRNA0 targeting no site transformed into strain QSJ038 was used as control, gRNA0 was also transformed into strain PC110 and used to show the background fluorescence. d. Comparison of eGFP repression by g3 generated by HH/HDV and Csy4 processing systems. All results were presented as mean ± standard deviation (SD) of at least three biological replicates.
RNA Pol-III promoters have been widely used for gRNA expression due to their smaller size and high transcriptional activity of small RNAs [[44], [45], [46]]. The native tRNA promoters, which belong to type II RNA Pol-III promoter, have been characterized and harnessed for gRNA expression in P. pastoris, and result in efficient CRISPR/Cas9-based gene editing, as well as CRISPR/dCas9-based gene regulation [11,31]. We thus tried to use these well characterized endogenous tRNA promoters for gRNA3 expression and compared them with RNA Pol-II promoter (PHTX1). Results show that RNA Pol-III promoter-driven gRNAs achieve eGFP repression by about 35–60%, which is much lower than the RNA Pol-II promoter-driven gRNAs (Fig. 3c).
Another strategy for gRNA processing is using the CRISPR-associated ribonuclease Csy4 that recognizes a 28 bp stem–loop RNA sequence and cuts after the 20th nucleotide [47]. The codon optimized Csy4 gene was expressed using PADH2 and integrated into NSI-6 site in strain QSJ058 (dCas9-RD1, eGFP) and obtained strain QSJ079 (dCas9-RD1, eGFP, Csy4). gRNA3 plasmid containing Csy4 recognition sequence was used for eGFP repression and compared to gRNAs with HH/HDV ribozymes. It showed that gRNAs using Csy4 processing system resulted in stronger gene repression (Fig. 3d).
3.4. Processing of gRNA array using Csy4 endoribonuclease
To further test if Csy4 processing system can be used for construction of multiplexed CRISPRi, we constructed a gRNA array containing two gRNAs (OpTEF1g3 targeting POpTEF1 for eGFP repression, and AOX1g6 targeting AOX1 promoter) flanked by Csy4 recognition sites (Fig. 4a). The constructed gRNA array expression plasmid was transformed into strain QSJ079 (dCas9-RD1, eGFP, Csy4). Plasmids harboring single gRNA (OpTEF1g3 or AOX1g6), and a control gRNA (g0) without targeting any site were transformed into QSJ058 (dCas9-RD1, eGFP) and used as control. Fluorescence measurement showed that eGFP repression in the gRNA array strain was comparable to the strain with single gRNA construct (OpTEF1g3 with HH/HDV ribozymes sequence) (Fig. 4b). gRNAs array processed by Csy4 also achieved a similar effect on cell growth compared to the single gRNA construct (AOX1g6 with HH/HDV ribozymes sequence) (Fig. 4c). These results show that Csy4 is able to cleave gRNAs array and generate functional individual gRNA.
Fig. 4.
Processing of synthetic gRNA array using Cys4 for efficient gene repression. a. Structure of the synthetic gRNA array and Csy4 recognition sites transcribed by one promoter. b.eGFP fluorescence of strains with single gRNA (OpTEF1g3, AOX1g6) or double-gRNA array (OpTEF1g3- AOX1g6). OpTEF1g3 targets POpTEF1 for eGFP repression and AOX1g6 targets AOX1 promoter. Strain with gRNA0 (g0) targeting nowhere in the genome was used as control. HH/HDV ribozymes were used for single gRNA cleavage. All gRNAs were transformed into strain with dCas9-RD1 and eGFP expression cassette. c. Growth curves of the same strains from b harboring single-gRNA plasmid or double-gRNA plasmid cultivated in Delft + M10 medium. Data is presented as mean ± standard deviation (SD) of at least three biological replicates.
We further applied the gRNA array system to repress fatty acyl-CoA synthetase genes (FAA1 and FAA2). It was previously reported that free fatty acids activation was completely abolished by deletion of FAA1 and FAA2, and the mutant strain could be used for overproduction of free fatty acids [2]. However, cell growth in methanol medium was affected by double deletion of FAA1 and FAA2 [2]. Therefore, we anticipated that repression of FAA using CRISPRi could be used to balance cell growth and fatty acid production. Five gRNAs were designed for each FAA and expressed individually using PHTX1 promoter. FAA1 and FAA2 was repressed by up to 53% (FAA1g1) and 68% (FAA2g2) respectively (Fig. S5). Comparable repression efficiency was achieved for both FAA1 and FAA2 when FAA1g1 and FAA2g2 were expressed in an array and processed by Csy4 (Fig. S5), which further demonstrated that Csy4 endoribonuclease could be used for multiplexed CRISPRi in P. pastoris. However, we did not detect accumulation of free fatty acids in the double-repression strain (data not shown). One of the reasons could be that FAA1 and FAA2 were only partially repressed, and the remain expression of FAA1 and FAA2 was enough for efficient activation of free fatty acids. Therefore, stronger repression of FAA is necessary to achieve accumulation of FFAs, which could be realized by using double or multiple gRNAs expressed in an array for both FAA1 and FAA2 repression.
4. Discussion
CRISPRi system has been demonstrated to be a robust tool for targeted gene repression. Transcriptional repressor domains are usually fused with dCas9 to increase repression efficiency. Repressor domains from S. cerevisiae has been used for construction of CRISPRi system in P. pastoris, however, the repression efficiency is relatively lower compared to CRISPRi system using the same repressor domains in S. cerevisiae. We selected repression domains of the native general transcriptional repressor Tup1 to construct CRISPRi system in P. pastoris. Fusion expression of dCas9 with repressor domain RD1 resulted in strong repression of eGFP, with the highest repression efficiency of 85%, which was higher than using repressor domains from S. cerevisiae. We anticipate that CRISPRi efficiency could be further improved by testing more endogenous repressor domains in P. pastoris. gRNAs targeting alcohol oxidase promoter PAOX1 were designed to verify the repression of native genes in P. pastoris. As expected, targeting PAOX1 significantly slowed cell growth in methanol medium, but did not affect cell growth in glucose medium (Fig. 2a, Fig. S3). Repression was further confirmed by analysis of relative transcription level of AOX1.
To simplify gRNA plasmid construction and achieve strong repression, different gRNA expression and processing systems were constructed and compared. Strong repression of eGFP could be achieved using Pol-II promoter for gRNA transcription and HH/HDV ribozymes for gRNA processing. However, fixing the variable 6-bp region of HH ribozyme reduced eGFP repression efficiency, indicating that generation of clean gRNA without mismatched 5’ extension is necessary for efficient repression. Among the tested gRNA processing systems, CRISPR-associated endoribonuclease Csy4 showed the strongest repression of eGFP, indicating its efficient processing of gRNA. We also demonstrated that Csy4 was functional for gRNA array cleavage and thus can be used for multiplexed gene regulation, which should be helpful for metabolic regulation of multiple pathways in P. pastoris. Though several RNA Pol-III promoters succeed in gene editing [31], the native RNA Pol-III promoter had much lower repression efficiency in our CRISPRi system. We hypothesize that it is a different situation for CRISPRi as it requires constant binding of dCas9/gRNA complex on target sites. Therefore, constant expression and efficient processing of gRNA is required for strong repression, while genome editing only requires temporal expression of gRNA to cut the target DNA.
To construct inducible CRISPRi and avoid the possible side effects of continuous expression of dCas9 and gRNA, weak tetracycline inducible promoters were tried for dCas9 or gRNA expression. However, neither strategy was successful, and the CRISPRi system could repress eGFP even without inducer (Fig. S6a). We propose that it is due to the leakiness of the tetR-tetO regulated promoters, therefore, further efforts are needed to construct tightly regulated inducible promoters. Additionally, we found that the promoter-less gRNA containing HH/HDV ribozymes also resulted in eGFP repression (Fig. S6b). The same phenomenon was also reported recently in S. cerevisiae, and it was proposed that the promoter-less gRNA could be transcribed by the base-pairing region, which was obtained from the target promoter sequence [26]. However, no repression of eGFP was observed for the same promoter-less gRNA without HH/HDV ribozymes (Fig. S6c), which further proved the requirement of gRNA processing for CRISPRi.
In summary, this study provides an improved CRISPRi system using endogenous repressor domains in P. pastoris. Efficient gRNA processing can be achieved using Csy4 endoribonuclease. The CRISPRi system can be used for not only single gene knockdown but also multiplexed gene repression, which will greatly facilitate metabolic engineering and basic research in P. pastoris.
Declaration of competing interest
Yongjin J. Zhou is an associate editor for Synthetic and Systems Biotechnology and was not involved in the editorial review or the decision to publish this article. All authors declare that there are no competing interests.
Acknowledgments
This work was supported by National Key Research and Development Program of China (2021YFC2103500) and Dalian Institute of Chemical Physics Innovation Program (DICP I202111).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2023.06.008.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Peña D.A., Gasser B., Zanghellini J., Steiger M.G., Mattanovich D. Metabolic engineering of Pichia pastoris. Metab Eng. 2018;50:2–15. doi: 10.1016/j.ymben.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Cai P., Wu X., Deng J., Gao L., Shen Y., Yao L., Zhou Y.J. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris. Proc Natl Acad Sci U S A. 2022;119(29) doi: 10.1073/pnas.2201711119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamil O.K., Cravens A., Payne J.T., Kim C.Y., Smolke C.D. Biosynthesis of tetrahydropapaverine and semisynthesis of papaverine in yeast. Proc Natl Acad Sci U S A. 2022;119(33) doi: 10.1073/pnas.2205848119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu T., Liu Q., Wang X., Liu X., Chen Y., Nielsen J. Metabolic reconfiguration enables synthetic reductive metabolism in yeast. Nature Metabo. 2022;(4):1551–1559. doi: 10.1038/s42255-022-00654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou R., Gao L., Liu J., Liang Z., Zhou Y.J., Zhang L., Zhang Y. Comparative proteomics analysis of Pichia pastoris cultivating in glucose and methanol. Synth Syst Biotechnol. 2022;7(3):862–868. doi: 10.1016/j.synbio.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartner F.S., Ruth C., Langenegger D., Johnson S.N., Hyka P., Lin-Cereghino G.P., et al. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 2008;36(12):e76. doi: 10.1093/nar/gkn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogl T., Ruth C., Pitzer J., Kickenweiz T., Glieder A. Synthetic core promoters for Pichia pastoris. ACS Synth Biol. 2014;3(3):188–191. doi: 10.1021/sb400091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weninger A., Hatzl A.M., Schmid C., Vogl T., Glieder A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. 2016;235:139–149. doi: 10.1016/j.jbiotec.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Cai P., Duan X., Wu X., Gao L., Ye M., Zhou Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021;49(13):7791–7805. doi: 10.1093/nar/gkab535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogl T., Kickenweiz T., Pitzer J., Sturmberger L., Weninger A., Biggs B.W., et al. Engineered bidirectional promoters enable rapid multi-gene co-expression optimization. Nat Commun. 2018;9(1):3589. doi: 10.1038/s41467-018-05915-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalvie N.C., Leal J., Whittaker C.A., Yang Y., Brady J.R., Love K.R., Love J.C. Host-informed expression of CRISPR guide RNA for genomic engineering in Komagataella phaffii. ACS Synth Biol. 2020;9(1):26–35. doi: 10.1021/acssynbio.9b00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J., Ye C., Cheng J., Jiang L., Yuan X., Lian J. Enhancing homologous recombination efficiency in Pichia pastoris for multiplex genome integration using short homology arms. ACS Synth Biol. 2022;11(2):547–553. doi: 10.1021/acssynbio.1c00366. [DOI] [PubMed] [Google Scholar]
- 13.Weninger A., Fischer J.E., Raschmanova H., Kniely C., Vogl T., Glieder A. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J Cell Biochem. 2018;119(4):3183–3198. doi: 10.1002/jcb.26474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai P., Gao J., Zhou Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb Cell Factories. 2019;18(1):63. doi: 10.1186/s12934-019-1112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J., Jiang L., Lian J. vol. 6. 2021. pp. 110–119. (Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T., Guan C., Guo J., Liu B., Wu Y., Xie Z., et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang L., Fan J., Luo S., Chen Y., Wang C., Cao Y., Song H. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids. Nat Commun. 2021;12(1):4976. doi: 10.1038/s41467-021-25243-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao L., Shabestary K., Bjork S.M., Asplund-Samuelsson J., Joensson H.N., Jahn M., Hudson E.P. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat Commun. 2020;11(1):1666. doi: 10.1038/s41467-020-15491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao M., Li Y., Wang F., Ren Y., Wei D. A CRISPRi mediated self-inducible system for dynamic regulation of TCA cycle and improvement of itaconic acid production in Escherichia coli. Synth Syst Biotechnol. 2022;7(3):982–988. doi: 10.1016/j.synbio.2022.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radzisheuskaya A., Shlyueva D., Muller I., Helin K. Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucleic Acids Res. 2016;44(18):e141. doi: 10.1093/nar/gkw583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowder L.G., Zhang D., Baltes N.J., Paul J.W., 3rd, Tang X., Zheng X., et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169(2):971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piatek A., Ali Z., Baazim H., Li L., Abulfaraj A., Al-Shareef S., et al. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13(4):578–589. doi: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- 25.Lian J., HamediRad M., Hu S., Zhao H. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat Commun. 2017;8(1):1688. doi: 10.1038/s41467-017-01695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw W.M., Studena L., Roy K., Hapeta P., McCarty N.S., Graham A.E., et al. Inducible expression of large gRNA arrays for multiplexed CRISPRai applications. Nat Commun. 2022;13(1):4984. doi: 10.1038/s41467-022-32603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zalatan J.G., Lee M.E., Almeida R., Gilbert L.A., Whitehead E.H., La Russa M., et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1–2):339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amir M.R., Panos O., Maxwell Z., Tavazoie S. An inducible CRISPR interference library for genetic interrogation of Saccharomyces cerevisiae biology. COMMUN BIOL. 2020;3(723) doi: 10.1038/s42003-020-01452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumschabl M., Prielhofer R., Mattanovich D., Steiger M.G. Fine-tuning of transcription in Pichia pastoris using dCas9 and RNA scaffolds. ACS Synth Biol. 2020;9(12):3202–3209. doi: 10.1021/acssynbio.0c00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao X., Li L., Jameel A., Xing X.H., Zhang C. A versatile toolbox for CRISPR-based genome engineering in Pichia pastoris. Appl Microbiol Biotechnol. 2021;105(24):9211–9218. doi: 10.1007/s00253-021-11688-y. [DOI] [PubMed] [Google Scholar]
- 32.McCarty N.S., Graham A.E., Studena L., Ledesma A.R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat Commun. 2020;11(1):1281. doi: 10.1038/s41467-020-15053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y., Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56(4):343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 34.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54(4):698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haurwitz R.E., Sternberg S.H., Doudna J.A. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31(12):2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira R., Skrekas C., Nielsen J., David F. Multiplexed CRISPR/Cas9 genome editing and gene regulation using Csy4 in Saccharomyces cerevisiae. ACS Synth Biol. 2018;7(1):10–15. doi: 10.1021/acssynbio.7b00259. [DOI] [PubMed] [Google Scholar]
- 37.Kurata M., Wolf N.K., Lahr W.S., Weg M.T., Kluesner M.G., Lee S., et al. Highly multiplexed genome engineering using CRISPR/Cas9 gRNA arrays. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0198714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joan L.C., William W.W., See X., William G., Linda T.L., Jane V., et al. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. Biotechniques. 2005;38(1) doi: 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J., Li Y., Yu W., Zhou Y.J. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat Metab. 2022;4(7):932–943. doi: 10.1038/s42255-022-00601-0. [DOI] [PubMed] [Google Scholar]
- 40.Javier S.M., Yolanda S. CRISPR-based gene expression control for synthetic gene circuits. Biochem Soc Trans. 2020;48(5):1979–1993. doi: 10.1042/BST20200020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z., Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol Adv. 2018;36(1):182–195. doi: 10.1016/j.biotechadv.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Theron C.W., Berrios J., Steels S., Telek S., Lecler R., Rodriguez C., Fickers P. Expression of recombinant enhanced green fluorescent protein provides insight into foreign gene-expression differences between Mut+ and MutS strains of Pichia pastoris. Yeast. 2019;36(5):285–296. doi: 10.1002/yea.3388. [DOI] [PubMed] [Google Scholar]
- 43.Naatsaari L., Mistlberger B., Ruth C., Hajek T., Hartner F.S., Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie K., Minkenberg B., Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015;112(11):6. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi W., Zhu T., Tian Z., Li C., Zhang W., Song R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016;16(1):58. doi: 10.1186/s12896-016-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz C.M., Hussain M.S., Blenner M., Wheeldon I. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth Biol. 2016;5(4):356–359. doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- 47.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329(5997):1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X., Cai P., Yao L., Zhou Y.J. Genetic tools for metabolic engineering of Pichia pastoris. Eng. Microbiol. 2023;3(4) doi: 10.1016/j.engmic.2023.100094. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.