Abstract
Many European countries have recently reported upsurges in invasive group A Streptococcus (iGAS) infections, mainly caused by emm1 Streptococcus pyogenes, specifically the toxigenic M1UK lineage. We present the epidemiology of emm1 causing iGAS in Belgium during 2018–August 2023, and describe an emergence of the toxigenic M1UK lineage in Belgium in mid-2022 that was observed as an increase in bloodstream infections caused by emm1 S. pyogenes that continued into 2023.
Keywords: Streptococcus pyogenes, emm1, invasive group A Streptococcus, bloodstream infections
Since mid-2022, invasive group A Streptococcus (iGAS) infections caused by Streptococcus pyogenes harbouring emm1, which encodes the M1 protein, have been increasingly reported across different European countries [1-4]. Specifically, this increase seems to be related to an increase in the proportion of the toxigenic M1UK lineage [5] compared to the original M1global lineage [6,7]. The M1UK lineage differs from the original by 27 defining single nucleotide polymorphisms (SNPs), which leads to an increased expression of the superantigen gene speA [8]. Since June 2023, another M1 sublineage was reported from Denmark (M1DK), which is characterised by the presence of speC and 15 defining SNPs [4]. In Belgium, a remarkable increase in iGAS infections was also observed from mid-2022, mostly caused by emm1 S. pyogenes. Here, we present the epidemiology, genetic characteristics and associated lineages of emm1 S. pyogenes causing iGAS infections in Belgium.
Invasive group A Streptococcus in Belgium in 2022–2023
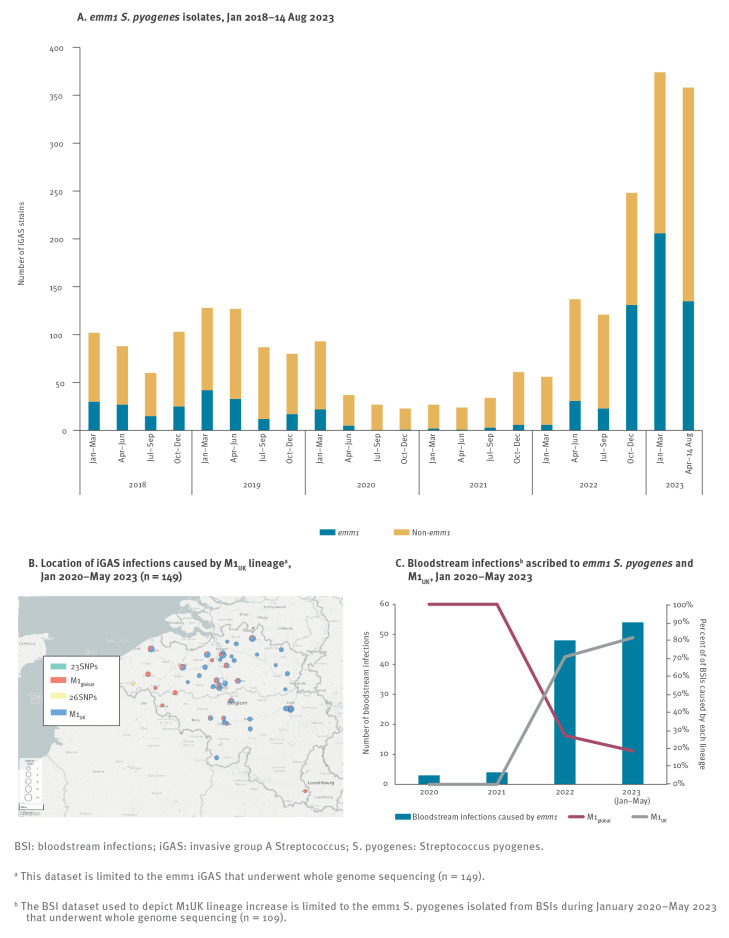
In Belgium, the total number of iGAS infections increased 1.3 to 1.6-fold in 2022 and 1.8- to 2-fold in 2023 compared to the pre-COVID-19 years (2018 and 2019, respectively) (Figure 1A). Of all iGAS infections, 45% (341/752) and 34% (191/566) were caused by emm1 S. pyogenes in 2023 (data available up to 14 August 2023) and in 2022, respectively, as compared to 25−27% in 2018 (97/354) and 2019 (104/422) (p ≤ 0.0056, Fisher’s exact test, Figure 1A and 2A). Although emm1 has been the predominant emm type among Belgian iGAS, emm1 comprises only 10%−25% of the yearly iGAS strains since the start of the surveillance in 2012 [9]. Age-based stratification of emm1 iGAS infections from May 2022 up to 14 August 2023 did not show a predilection for any age group (Table). Of the 518 emm1 iGAS infections reported during this period, 31% (n = 160) were identified in the 0–12 age group, 16% (n = 85) in the 18–40, and 25% and 26% in the 41–65 (n = 129) and 65 and older (n = 135) age groups, respectively (Table).
Figure 1.
Overview of invasive group A Streptococcus infections caused by Streptococcus pyogenes, Belgium, 2018–August 2023
BSI: bloodstream infections; iGAS: invasive group A Streptococcus; S. pyogenes: Streptococcus pyogenes.
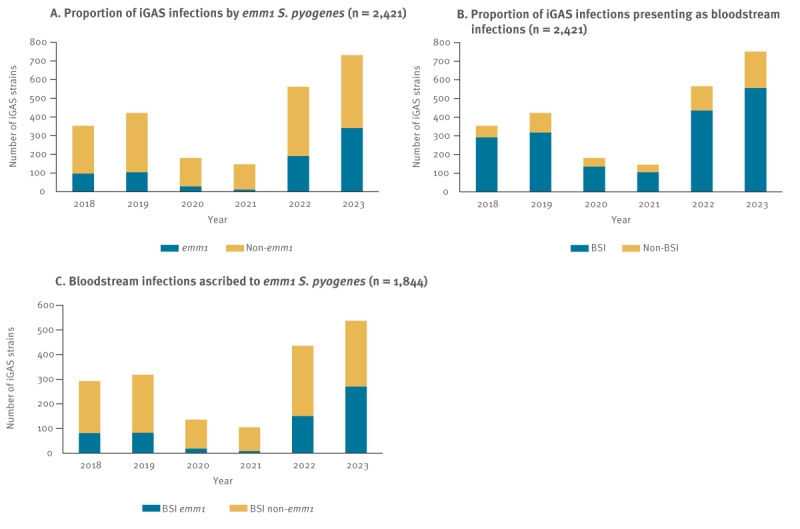
Figure 2.
Distribution of emm1 invasive group A Streptococcus and bloodstream infections caused by the M1UK lineage, Belgium, January 2018–August 2023
BSI: bloodstream infections; iGAS: invasive group A Streptococcus; S. pyogenes: Streptococcus pyogenes.
a This dataset is limited to the emm1 iGAS that underwent whole genome sequencing (n = 149).
b The BSI dataset used to depict M1UK lineage increase is limited to the emm1 S. pyogenes isolated from BSIs during January 2020–May 2023 that underwent whole genome sequencing (n = 109).
Table. Number of emm1 invasive group A Streptococcus (iGAS) infections, 1 May 2022–14 August 2023 (n = 518), and number of M1UK lineage among sequenced emm1 S. pyogenes causing iGAS infections, May 2022–May 2023 (n = 130) by age, Belgium.
| Case characteristics | emm1 iGAS infectionsa | Sequenced emm1 iGASb | M1 UK in iGAS infectionsd | M1 global in iGAS infectionsd | M1 UK causing BSIse | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age range (years) | Total | Sexb,c | May−Dec 2022 | Jan−Aug 2023 | |||||||||||||
| Males | Females | ||||||||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| 0–12 | 160 | 86 | 54 | 74 | 46 | 64 | 36 | 96 | 28 | 40 | 25 | 27 | 68 | 12 | 30 | 19 | 70 |
| 13–17 | 4 | 2 | 50 | 2 | 50 | 1 | 1 | 3 | 1 | 1 | 25 | 1 | 100 | 0 | 0 | 1 | 100 |
| 18–40 | 85 | 36 | 42 | 49 | 58 | 26 | 15 | 59 | 17 | 34 | 40 | 26 | 76 | 8 | 24 | 19 | 73 |
| 41–65 | 129 | 76 | 59 | 53 | 41 | 46 | 26 | 83 | 24 | 31 | 24 | 27 | 87 | 4 | 13 | 22 | 81 |
| > 65 | 135 | 65 | 48 | 69 | 51 | 38 | 21 | 97 | 28 | 22 | 16 | 20 | 91 | 2 | 9 | 16 | 80 |
| Unknown | 5 | 3 | 60 | 2 | 40 | 2 | 1 | 3 | 1 | 2 | 40 | 1 | 50 | 1 | 50 | 1 | 100 |
| Total | 518 | 268 | 52 | 249 | 48 | 177 | 100 | 341 | 100 | 130 | 25 | 102 | 78 | 27 | 21 | 78 | 76 |
BSIs: bloodstream infections; iGAS: invasive group A Streptococcus.
a For emm1 iGAS infections: % based on total isolates in that period.
b For sex and sequenced isolates: % based on total number of isolates per age group.
c Sex is unknown for one case in the > 65 age group.
d For M1 isolates: % based on sequenced isolates. The dataset for lineage assignment is limited to the emm1 iGAS isolated during May 2022–May 2023 that underwent whole genome sequencing (n = 130).
e For M1UK causing BSI: % based on M1UK causing iGAS.
The ratio of bloodstream infections (BSI) and non-BSI remained fairly constant, with BSI comprising 74−82% of the iGAS infections from 2018−23 (Figure 1B). However, the proportion of BSIs caused by emm1 S. pyogenes, which was 26−28% in 2018−19, dropped to 9%−14% during the COVID-19 pandemic (2020–21). In 2022, the proportion increased to 35%, and in 2023 constituted almost 50% of all BSIs in Belgium (p ≤ 0.0105 when comparing 2023 with the other years, Fisher’s exact test, Figure 1C). Patients with BSIs included those where emm1 S. pyogenes was isolated from blood cultures, or those with a primary differential diagnosis of septicaemia or meningitis, with or without other conditions, where emm1 S. pyogenes was isolated from another sample type, e.g. cerebrospinal fluid, pleural fluid.
Whole genome sequencing
An at-random selection of S. pyogenes isolated from patients with suspected iGAS infections, which were submitted to the National Reference Centre for invasive β-haemolytic streptococci between January 2020 and May 2023 and typed as emm1, were subjected to whole genome sequencing (n = 149, llumina, MiSeq). Approximately, 20–45% of all Belgian emm1 iGAS strains between January 2020 and May 2023 were sequenced (2020: 9/28, 2021: 5/11, 2022: 66/191 and 2023: 69/341. Strains were isolated from blood (90/149, 60%), wounds (13/149, 9%), pleural fluid (12/149, 8%), biopsies (8/149, 5%) and other invasive sample types (26/149, 17%) with a clinical presentation of septicaemia, streptococcal toxic shock syndrome (STSS), erysipelas, cellulitis, necrotising fasciitis, necrotising pneumonia, empyema, osteomyelitis and meningitis.
After initial quality assessment and trimming with FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and TrimGalore v0.6.7 (https://wiki.rc.usf.edu/index.php/TrimGalore), a whole genome alignment against the emm1 reference MGAS5005 was generated with Snippy v4.6.0 (https://github.com/tseemann/snippy), and the SNP distance matrix was extracted from the whole genome alignment using snp-dists v0.7.0 (https://github.com/tseemann/snp-dists). Isolates were assigned to the M1UK or the M1DK lineages based on the presence of the 27 or 15 defining SNPs in the whole genome alignment, as described previously [4,5]. Genome assemblies were generated with SPAdes v3.15.0 [10] and were annotated with prokka v1.14.5 [11]. Annotated assemblies were further analysed with BacPipe [12] for detection of acquired antimicrobial resistance genes based on the CARD database. Metadata was visualised using Microreact online interface.
Remarkably, BSI isolates constituted 73% (109/149) of the sequenced emm1 S. pyogenes in this study. The emm1-associated BSIs were detected in patients of all ages, ranging from 0−92 years. Sequencing identified the rapid increase in the proportion of the M1UK lineage among iGAS isolates in Belgium during 2022–23 (Figure 2B and C). This toxigenic lineage was already detected in Belgium in one BSI sample as early as January 2020. No isolate was identified that presented the 15 SNPs defining the recently described M1DK lineage, although six (two M1global and four M1UK) presented the speC superantigen gene. From May 2022 to May 2023, of 130 emm1 isolates sequenced, 102 (78%) were M1UK, and one presented 23 of the 27 defining SNPs, making M1UK the currently dominant emm1 lineage in Belgium (Figure 2B and C). This lineage also constituted 72% (78/109) of the BSI-associated emm1 isolates sequenced in this study. However, disease outcomes did not vary notably between patients developing infections caused by M1UK or M1global. Of the 13 of 149 patients who died, eight were infected with M1UK and were aged 7, 40, 56, 65 and over 65 (n = 4) years. Five patients who died were infected with M1global (ages 0, 1 (n = 2), 33 and 34 years). Of these 13 patients, twelve had been diagnosed with BSI (septicaemia, with or without pneumonia, fasciitis or STSS), while one presented with pneumonia. In our dataset, neither severity of disease nor disease presentation varied between the two lineages.
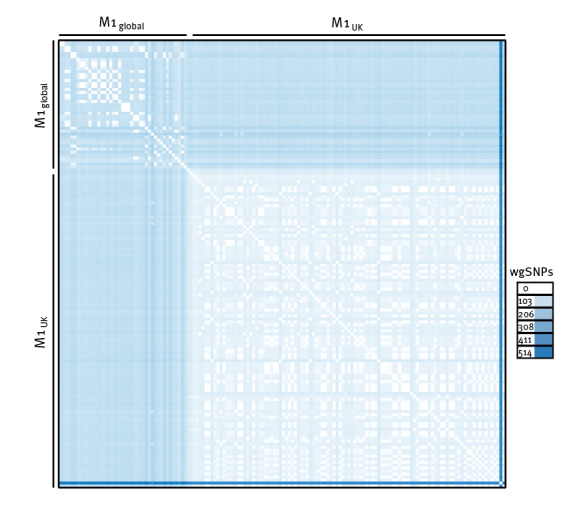
Within the M1UK isolates studied here, we identified on average 41 whole genome SNPs, indicating higher genome stability, in comparison to the M1global isolates, which were isolated during the same timeframe, and presented on average 85 whole genome SNPs within the lineage (Figure 3). However, one M1UK isolate presented a remarkably higher number of SNPs compared to the rest of isolates (452 whole genome SNPs on average), despite showing the same emm type and ST, and will require further investigation. Remarkably, M1UK isolates showed on average 105 whole genome SNPs when compared to the M1global isolates.
Figure 3.
Single nucleotide polymorphism (SNP) distance matrix representing number of whole genome SNPs between all included isolates, Belgium, Jan 2020–May 2023 (n = 149)
wgSNPs: whole genome single nucleotide polymorphisms.
Most of the isolates (139/149, 93%) belonged to sequence type (ST)28, one M1global isolate was ST785 and seven M1UK isolates were ST1357. Screening for antimicrobial resistance genes showed that mefE, which encodes a macrolide resistance-conferring efflux pump, was present in eight isolates (seven M1UK and one M1global), tetracycline resistance-encoding genes tetU and tetM were identified in four isolates (three M1UK and one M1global), and one isolate (M1UK) presented with the aminoglycoside resistance determinant, aad [6]. Although prevalence of antimicrobial resistance genes in the M1UK lineage was rather low, this was higher than among M1global isolates. The most commonly observed gene was mefE, as also reported in the original description of the M1UK lineage [5].
Discussion
The increase in emm1 iGAS observed during mid-2022 until August 2023 in Belgium was primarily because of an upsurge in BSIs caused by emm1 S. pyogenes. Remarkably, this coincided with the lifting of mandatory use of protective face masks in Belgium. Other countries have observed a similar increase in emm1 iGAS infections with different clinical presentations after the removal of COVID-19-related non-pharmaceutical protective measures. A rise in pleural empyema was reported in Scotland and England [1,13], meningitis in the Netherlands [6], and general infections in the Netherlands [2], France [3], Denmark [4] and England [7].
Our observations were similar to previous studies reporting that the M1UK lineage does not lead to more severe infections, although the emm1 genotype itself has been linked to higher virulence and requirement for intensive care [3,4]. In Belgium, the emm1-associated BSIs or those caused by the M1UK lineage did not show a proclivity for any age group, in contrast to previous findings that M1UK infections are more common among the paediatric population [7]. The success of the M1UK lineage has been hypothesised to be derived from the accumulation of SNPs in the genome that provide a fitness advantage in colonising the host [8], which caused this lineage to become predominant before the pandemic in the United Kingdom [5]. Genome stability, measured by accumulation of new SNPs in a defined timeframe, was higher among the M1UK isolates than the M1global studied here. These data support the hypothesis that the SNPs accumulated by the M1UK lineage are sufficient to provide a fitness advantage, which – coupled with a potentially lowered herd immunity to S. pyogenes because of decreased exposure during the pandemic and the increase in other respiratory viral infections – might explain the upsurge in iGAS infections reported in the 2022/23 winter season in many European countries.
Our investigation had some limitations. We studied a selection of the emm1 S. pyogenes that were all isolated from invasive infections, which did not allow a contextual analysis of other prevalent genotypes or of the prevalence of M1UK among non-invasive GAS infections. Despite the limitations, analysis of emm1 iGAS from years both pre- and post-COVID-19 pandemic nonetheless facilitated a clear picture of emm1 iGAS dynamics in Belgium.
Conclusion
The toxigenic M1UK lineage emerged in Belgium in mid-2022 and was observed as an increase in bloodstream infections caused by emm1 S. pyogenes that has continued until August 2023. These data call for increased vigilance and a sustained real-time monitoring of iGAS infections in Europe.
Ethical statement
Genomic and epidemiological data presented in this study were obtained as part of national surveillance efforts.
Funding statement
The Belgian Reference Centre for invasive β-haemolytic streptococci is supported by the Belgian Ministry of Social Affairs through a fund within the Health Insurance System.
Data availability
The datasets generated and analysed during the current study are available at European Nucleotide Archive under bioproject number PRJEB64886 and at NCBI with bioproject number PRJNA1003449.
Acknowledgements
We would like to thank all Belgian clinical laboratories for sharing invasive β-haemolytic streptococcal isolates with the Belgian Reference Centre, and for sharing basic patient clinical information.
Conflict of interest: None declared.
Authors’ contributions: Conceptualisation: SM-K. Sequencing and data collection: VM, SKK, CL. Analysis: JPRR, QL. Original draft preparation: JPRR, SM-K. Writing, review and editing: SM-K, VM, QL, PRS. All authors read, gave input and approved the final manuscript.
References
- 1. Holdstock V, Twynam-Perkins J, Bradnock T, Dickson EM, Harvey-Wood K, Kalima P, et al. National case series of group A streptococcus pleural empyema in children: clinical and microbiological features. Lancet Infect Dis. 2023;23(2):154-6. 10.1016/S1473-3099(23)00008-7 [DOI] [PubMed] [Google Scholar]
- 2. van Kempen EB, Bruijning-Verhagen PCJ, Borensztajn D, Vermont CL, Quaak MSW, Janson JA, et al. Increase in invasive group a streptococcal infections in children in the Netherlands, a survey among 7 hospitals in 2022. Pediatr Infect Dis J. 2023;42(4):e122-4. 10.1097/INF.0000000000003810 [DOI] [PubMed] [Google Scholar]
- 3. Lassoued Y, Assad Z, Ouldali N, Caseris M, Mariani P, Birgy A, et al. Unexpected increase in invasive group A streptococcal infections in children after respiratory viruses outbreak in France: A 15-Year Time-Series Analysis. Open Forum Infect Dis. 2023;10(5):ofad188. 10.1093/ofid/ofad188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johannesen TB, Munkstrup C, Edslev SM, Baig S, Nielsen S, Funk T, et al. Increase in invasive group A streptococcal infections and emergence of novel, rapidly expanding sub-lineage of the virulent Streptococcus pyogenes M1 clone, Denmark, 2023. Euro Surveill. 2023;28(26):2300291. 10.2807/1560-7917.ES.2023.28.26.2300291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, Mosavie M, et al. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19(11):1209-18. 10.1016/S1473-3099(19)30446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van der Putten BCL, Vlaminckx BJM, de Gier B, Freudenburg-de Graaf W, van Sorge NM. Group A streptococcal meningitis with the M1UK variant in the Netherlands. JAMA. 2023;329(20):1791-2. 10.1001/jama.2023.5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alcolea-Medina A, Snell LB, Alder C, Charalampous T, Williams TGS, Tan MKI, et al. The ongoing Streptococcus pyogenes (Group A Streptococcus) outbreak in London, United Kingdom, in December 2022: a molecular epidemiology study. Clin Microbiol Infect. 2023;29(7):887-90. 10.1016/j.cmi.2023.03.001 [DOI] [PubMed] [Google Scholar]
- 8. Li HK, Zhi X, Vieira A, Whitwell HJ, Schricker A, Jauneikaite E, et al. Characterization of emergent toxigenic M1UK Streptococcus pyogenes and associated sublineages. Microb Genom. 2023;9(4):mgen000994. 10.1099/mgen.0.000994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.University of Antwerp and Université Libre de Bruxelles. Report 2021. National reference centre for invasive β-hemolytic streptococci non group B. Edegem: University of Antwerp and Université Libre de Bruxelles; 2021. Available from: https://www.sciensano.be/sites/default/files/annual_report_2021_streptococci.pdf
- 10. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 2020;70(1):e102. 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 11. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068-9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 12. Xavier BB, Mysara M, Bolzan M, Ribeiro-Gonçalves B, Alako BTF, Harrison P, et al. BacPipe: a rapid, user-friendly whole-genome sequencing pipeline for clinical diagnostic bacteriology. iScience. 2020;23(1):100769. 10.1016/j.isci.2019.100769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guy R, Henderson KL, Coelho J, Hughes H, Mason EL, Gerver SM, et al. Increase in invasive group A streptococcal infection notifications, England, 2022. Euro Surveill. 2023;28(1):2200942. 10.2807/1560-7917.ES.2023.28.1.2200942 [DOI] [PMC free article] [PubMed] [Google Scholar]