Abstract
Citrus is an important and widely cultivated fruit crop in South China. Although the species of fungal diseases of leaves and fruits have been extensively studied, the causal organisms of branch diseases remain poorly known in China. Species of Botryosphaeriaceae are known as important fungal pathogens causing branch diseases on citrus in the USA and Europe. To determine the diversity of Botryosphaeriaceae species associated with citrus branch diseases in China, surveys were conducted in the major citrus-producing areas from 2017 to 2020. Diseased tissues were collected from twigs, branches and trunks with a range of symptoms including cankers, cracking, dieback and gummosis. Based on morphological characteristics and phylogenetic comparison of the DNA sequences of the internal transcribed spacer region (ITS), the translation elongation factor 1-alpha gene (tef1), the β-tubulin gene (tub2) and the DNA-directed RNA polymerase II second largest subunit (rpb2), 111 isolates from nine provinces were identified as 18 species of Botryosphaeriaceae, including Botryosphaeria dothidea, B. fabicerciana, Diplodia seriata, Dothiorella alpina, Do. plurivora, Lasiodiplodia citricola, L. iraniensis, L. microconidia, L. pseudotheobromae, L. theobromae, Neodeightonia subglobosa, Neofusicoccum parvum, and six previously undescribed species, namely Do. citrimurcotticola, L. guilinensis, L. huangyanensis, L. linhaiensis, L. ponkanicola and Sphaeropsis linhaiensis spp. nov. Botryosphaeria dothidea (28.8 %) was the most abundant species, followed by L. pseudotheobromae (23.4 %), which was the most widely distributed species on citrus, occurring in six of the nine provinces sampled. Pathogenicity tests indicated that all 18 species of Botryosphaeriaceae obtained from diseased citrus tissues in this study were pathogenic to the tested Citrus reticulata shoots in vitro, while not all species are pathogenic to the tested Cocktail grapefruit (C. paradisi × C. reticulata) shoots in vivo. In addition, Lasiodiplodia was the most aggressive genus both in vitro and in vivo. This is the first study to identify Botryosphaeriaceae species related to citrus branch diseases in China and the results provide a theoretical basis for the implementation of prevention and control measures.
Citation: Xiao XE, Wang W, Crous PW, et al. 2021. Species of Botryosphaeriaceae associated with citrus branch diseases in China. Persoonia 47: 106–135. https://doi.org/10.3767/persoonia.2021.47.03.
Keywords: Botryosphaeria cankers, distribution, new taxa, pathogenicity, systematics
INTRODUCTION
The Botryosphaeriaceae was established by Theissen & Sydow (1918). The taxonomic status of Botryosphaeriaceae has been heavily debated and somewhat controversial until Schoch et al. (2006) proposed the Botryosphaeriales as a new order to accommodate the family (see Phillips et al. 2013). Presently, the Botryosphaeriaceae contains 23 genera and over 100 species that have been confirmed based on their DNA sequence data (Slippers et al. 2017, Yang et al. 2017, Zhang et al. 2021).
Species of Botryosphaeriaceae have a broad host range and cosmopolitan distribution (Slippers & Wingfield 2007, Phillips et al. 2013). Many species are important plant pathogens, especially for woody plant genera such as Citrus, causing bark rot, branch canker, gummosis, shoot blight, dieback and fruit rot, and even death of whole plants when conditions are conducive to disease development (Slippers & Wingfield 2007, Úrbez-Torres 2011). Citrus is one of the most important fruit crops globally. Citrus diseases caused by species in the Botryosphaeriaceae have been reported since the early 1900s when Fawcett & Burger (1911) isolated a Diplodia sp. from orange trees with gummosis, and from rotten grapefruits and oranges in Florida. The fungal agent was then considered to be Diplodia natalensis, which was regarded as the pathogen responsible for decay and gummosis in lemons and other citrus fruits in the USA and South Africa (Fawcett & Burger 1911, Adesemoye et al. 2014). Subsequent taxonomic revisions showed that D. natalensis represents as synonym of Lasiodiplodia theobromae (Alves et al. 2004). Further studies indicated that Diplodia stem-end rot caused by L. theobromae is one of the most important postharvest decays in warm, humid tropical and subtropical citrus-producing areas (Brown & Eckert 2000, Ismail & Zhang 2004, Zhang 2014). Several other species of Botryosphaeriaceae have subsequently been isolated from citrus with cankers, dieback, gummosis and fruit rot symptoms, including species of Botryosphaeria (Smith 1934, Adesemoye et al. 2011), Diplodia (Adesemoye et al. 2014, Berraf-Tebbal et al. 2020), Dothiorella (Adesemoye & Eskalen 2011, Abdollahzadeh et al. 2014, Berraf-Tebbal et al. 2020), Lasiodiplodia (Alves et al. 2008, Abdollahzadeh et al. 2010, Adesemoye et al. 2014, Linaldeddu et al. 2015, Coutinho et al. 2017, Guajardo et al. 2018, Bautista-Cruz et al. 2019, Berraf-Tebbal et al. 2020), Macrophomina (Azadeh et al. 2018), Neofusicoccum (Adesemoye & Eskalen 2011), Neoscytalidium (Polizzi et al. 2009, Adesemoye et al. 2014, Mayorquin et al. 2016) and Sphaeropsis (Phillips et al. 2013).
China has a history of more than 4 000 years of citrus cultivation (Deng et al. 2008, Shen 2019) and is the world’s largest producer of citrus, with 37.92 M tons in 2018 (FAO 2018). Branch diseases including twig blight, branch dieback, bark rot, canker, crack and gummosis are commonly observed on citrus, especially in regions where stress factors such as frost and sunburn often occur. Resin (gummosis) caused by Diaporthe citri has been recorded as the most important fungal branch disease (Cai et al. 2011, Huang et al. 2013b), followed by Alternaria brown spot (dieback) caused by Alternaria alternata pathotype tangerine (Huang et al. 2012, Qin et al. 2012), anthracnose (twig blight and branch dieback) caused by Colletotrichum gloeosporioides (Cai et al. 2011, Huang et al. 2013a), and foot root caused by Phytophthora spp. (Cheng et al. 2004, Cai et al. 2011, Zhu et al. 2011). Species in other genera such as Cytospora, Diplodia, Dothidea, Macrophoma, Phoma, Phyllosticta and Sphaeropsis, have also been associated with citrus branch diseases (Chinese Academy of Agricultural Sciences 1960, Tai 1979). However, all fungal identifications were based on morphology or simply based on the symptoms before the 1990s and pathogenicity tests were lacking for most species (Tai 1979).
During 2017–2020, several surveys of citrus branch diseases were conducted in the major citrus production regions in China. The objectives of this study were to:
– identify the species of Botryosphaeriaceae associated with citrus branch diseases in China based on morphological traits and phylogenetic analysis;
– identify the dominant species associated with citrus branch diseases; and
– determine their pathogenicity.
MATERIALS AND METHODS
Disease symptoms, sample collection and fungal isolations
From 2017 to 2020, citrus branch disease samples with symptoms of canker, gummosis, twig blight and branch dieback (Fig. 1) were collected from the main citrus-producing regions in nine provinces of China, namely Chongqing, Fujian, Guangdong, Guangxi, Hunan, Jiangxi, Shaanxi, Shanghai and Zhejiang. The citrus species investigated and the number of samples collected would depend on the incidence of branch diseases in the orchard and region.
Fig. 1.

Disease symptoms on citrus caused by Botryosphaeriaceae. a. Twig blight of Citrus reticulata; b. twig blight on Cocktail grapefruit; c. branch dieback of C. reticulata; d. death tree of C. reticulata; e. branch canker on C. reticulata; f. trunk canker of C. unshiu; g–h. gummosis on twig and trunk of Cocktail grapefruit; i. fungal fruitbody structures formed on dead branch of Cocktail grapefruit.
Fungal strains were isolated via two methods. Firstly, sporocarps visible on diseased tissue were transferred to a microtube containing sterile water to make a spore suspension. After dilution, 150 μL spore suspension was spread over the surface of water agar (WA) plates amended with 100 μg/mL ampicillin and 100 μg/mL streptomycin to suppress bacterial growth. After 24–36 h, germinating spores were retrieved and transferred onto potato dextrose agar plates (PDA, 200 g potatoes, 20 g glucose and 15 g agar/L water) with 100 μg/mL ampicillin and 100 μg/mL streptomycin (PDA-AS) and incubated at 25 °C. Axenic cultures were obtained by transferring a single colony onto PDA. Secondly, for samples lacking sporocarps, a tissue isolation method was used. A small section (about 3 × 3 mm) between the healthy and diseased tissue was aseptically cut and surface-sterilised in 70 % ethanol for 1 min, followed by 1 % NaClO solution for 1 min, and rinsed three times in sterile water. Tissue sections were dried on sterilised filter paper, placed on 1/2 PDA-AS plates and incubated at 25 °C. Axenic cultures were obtained by transferring single hyphal tips onto PDA. Specimens and isolates from this study were deposited in Zhejiang University, and ex-type cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC), Beijing, China.
DNA extraction, PCR amplification and sequencing
Isolates were grown on PDA plates and incubated at room temperature for 4–7 d. Surface mycelia were collected using a sterile scalpel blade and genomic DNA was extracted by the CTAB (Cetyl trimethylammonium bromide) method (Van Burik et al. 1998). Partial regions of four loci were amplified. The internal transcribed spacer region (ITS) was amplified with primers ITS1 and ITS4 (White et al. 1990). Part of the translation elongation factor 1-alpha gene (tef1) was amplified with primers EF1-688F (Alves et al. 2008) or EF1-728F and EF1-986R (Carbone & Kohn 1999). Part of the β-tubulin gene (tub2) was amplified with Bt2a and Bt2b (Glass & Donaldson 1995). Part of the DNA directed RNA polymerase II second largest subunit (rpb2) was amplified with RPB2-6F and fRPB2-7cR (Liu et al. 1999) or rpb2-lasF and rpb2-lasR (Cruywagen et al. 2017). All amplification reactions were performed in a total volume of 25 μL mixture consisted of 12.5 μL of 2 × Taq Master Mix (Dye Plus) (Vazyme), 9.5 μL ddH2O, 1 μL of each forward and reverse primer, and 1 μL DNA template. The amplification conditions consisted of an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 45 s, and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 5 min. The PCR products were separated by agarose gel electrophoresis and sent to Qingke Biotechnology (Hangzhou, China) for Sanger DNA sequencing. The nucleotide sequences were assembled and edited with MEGA v. 7.0.26 (Kumar et al. 2016). Sequences obtained in this study were deposited in GenBank nucleotide database (http://www.ncbi.nlm.nih.gov; Table 1).
Table 1.
Details of Botryosphaeriaceae isolates studied.
| Speciesa | Isolate | Location | Collector | Host | Associated symptom | GenBank Accession no.b |
|||
|---|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | rpb2 | ||||||
| Botryosphaeria dothidea | BE1 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MT772261 | MT775839 | MT775849 | MW884107 |
| (C. paradisi × C. reticulata) | |||||||||
| BE2 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MT772262 | MT775840 | MT775850 | MW884108 | |
| (C. paradisi × C. reticulata) | |||||||||
| BE60 | Chenggu, Shaanxi, China | H.Y. Li & X.E. Xiao | C. unshiu | Trunk canker | MW862113 | MW884017 | MW884086 | MW884109 | |
| BE61 | Changxing Island, Shanghai, China | X.E. Xiao | hybrid cv. Hongmeiren | Twig dieback | MW862116 | MW884020 | MW884087 | MW884110 | |
| BE62 | Chunan, Zhejiang, China | H.Y. Li & X.E. Xiao | C. unshiu | Twig dieback | MW862110 | MW884014 | – | – | |
| BE63 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig gummosis | MW862111 | MW884015 | – | – | |
| BE64 | Quzhou, Zhejiang, China | H.Y. Li & J.W. Lv | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862112 | MW884016 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE65 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862114 | MW884018 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE66 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862115 | MW884019 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE67 | Xiangshan, Zhejiang, China | H.Y. Li | C. unshiu | Trunk canker | MW862117 | MW884021 | – | – | |
| BE68 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862118 | MW884022 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE69 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig gummosis | MW862119 | MW884023 | – | – | |
| BE72 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig dieback | MW881452 | MW884024 | – | – | |
| BE73 | Chenggu, Shaanxi, China | H.Y. Li & X.E. Xiao | C. unshiu | Trunk gummosis | MW862120 | MW884025 | – | – | |
| BE75 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862121 | MW884026 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE76 | Hangzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. maxima | Branch dieback | MW862122 | MW884027 | – | – | |
| BE77 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Branch canker | MW862123 | MW884028 | – | – | |
| BE79 | Hangzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. maxima | Twig dieback | MW862124 | MW884029 | – | – | |
| BE81 | Hangzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. maxima | Branch dieback | MW862125 | MW884030 | – | – | |
| BE90 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW862126 | MW884031 | – | – | |
| BE91 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig gummosis | MW862127 | MW884032 | – | – | |
| BE92 | Shaoyang, Hunan, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW862128 | MW884033 | – | – | |
| BE93 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862129 | MW884034 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE94 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Branch gummosis | MW862130 | MW884035 | – | – | |
| BE95 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig gummosis | MW862131 | MW884036 | – | – | |
| BE96 | Chun’an, Zhejiang, China | H.Y. Li & X.E. Xiao | C. unshiu | Branch dieback | MW862132 | MW884037 | – | – | |
| BE97 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862133 | MW884038 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE98 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862134 | MW884039 | – | – | |
| (C. paradisi × C. reticulata) | |||||||||
| BE101 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW862135 | MW884040 | – | – | |
| BE103 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW862136 | MW884041 | – | – | |
| BE105 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW862137 | MW884042 | – | – | |
| BE106 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW862138 | MW884043 | – | – | |
| Botryosphaeria fabicerciana | BE3 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MT772263 | MT775841 | MT775851 | MW884111 |
| (C. paradisi × C. reticulata) | |||||||||
| BE78 | Hangzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. maxima | Twig dieback | MW862139 | MW884044 | MW884088 | MW884112 | |
| Botryosphaeria fabicerciana | BE85 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862140 | MW884045 | MW884089 | MW884113 |
| (cont.) | (C. paradisi × C. reticulata) | ||||||||
| BE86 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit | Twig gummosis | MW862141 | MW884046 | MW884090 | MW884114 | |
| (C. paradisi × C. reticulata) | |||||||||
| Diplodia seriata | BE4 | Chenggu, Shaanxi, China | H.Y. Li & X.E. Xiao | C. unshiu | Branch canker | MW862142 | MW884047 | MW884091 | MW884115 |
| Dothiorella alpina | BE17 | Shimen, Hunan, China | H.Y. Li & Y.T. Zeng | C. unshiu | Twig dieback | MW862143 | MW884048 | MW884092 | MW884116 |
| Do. citrimurcotticola | BE5 = CGMCC3.20392 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW880663 | MW884166 | MW884195 | MW884140 |
| BE6 | Huangyan, Zhejiang, China | H.K. Wang & X.E. Xiao | C. maxima | Twig dieback | MW880664 | MW884167 | MW884196 | MW884141 | |
| BE7 = CGMCC3.20393 | Quzhou, Zhejiang, China | H.Y. Li | C. maxima | Twig dieback | MW880665 | MW884168 | MW884197 | MW884142 | |
| BE8 * = CGMCC3.20394 | Wanzhou, Chongqing, China | H.Y. Li & X.E. Xiao | hybrid cv. Murcott (C. reticulata × C. sinensis) | Twig dieback | MW880661 | MW884164 | MW884193 | MW884138 | |
| BE9 = CGMCC3.20395 | Wanzhou, Chongqing, China | H.Y. Li & X.E. Xiao | hybrid cv. Murcott | Twig dieback | MW880662 | MW884165 | MW884194 | MW884139 | |
| (C. reticulata × C. sinensis) | |||||||||
| BE71 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW880666 | MW884169 | MW884198 | MW884143 | |
| Do. plurivora | BE16 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | C. reticulata cv. Ponkan | Twig dieback | MT772270 | MT775848 | MT775858 | MW884117 |
| BE74 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW862144 | MW884049 | MW884093 | MW884118 | |
| Lasiodiplodia citricola | BE13 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | hybrid cv. Cocktail grapefruit (C. paradisi × C. reticulata) | Branch gummosis | MT772267 | MT775845 | MT775855 | MW884119 |
| BE38 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit (C. paradisi × C. reticulata) | Twig gummosis | MW862145 | MW884050 | MW884094 | MW884120 | |
| BE45 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | C. unshiu | Twig dieback | MW862146 | MW884051 | MW884095 | MW884121 | |
| BE83 | Wanzhou, Chongqing, China | H.Y. Li & X.E. Xiao | hybrid cv. Hongmeiren | Twig dieback | MW862148 | MW884053 | MW884096 | MW884122 | |
| BE89 | Chenggu, Shaanxi, China | H.Y. Li & X.E. Xiao | C. unshiu | Trunk canker | MW862147 | MW884052 | – | – | |
| BE99 | Lishui, Zhejiang, China | X.E. Xiao | C. sinensis | Twig dieback | MW862149 | MW884054 | – | – | |
| BE102 | Xiangshan, Zhejiang, China | H.Y. Li & X.E. Xiao | C. unshiu | Branch canker | MW862150 | MW884055 | MW884097 | MW884123 | |
| BE104 | Lishui, Zhejiang, China | X.E. Xiao | hybrid | Branch canker | MW862151 | MW884056 | MW884098 | MW884124 | |
| L. guilinensis | BE31 * = CGMCC3.20378 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW880672 | MW884175 | MW884204 | MW884149 |
| BE59 = CGMCC3.20379 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Branch gummosis | MW880673 | MW884176 | MW884205 | MW884150 | |
| L. huangyanensis | BE33 * = CGMCC3.20380 | Huangyan, Zhejiang, China | X.E. Xiao & Q.B. Huang | C. reticulata | Twig dieback | MW880674 | MW884177 | MW884206 | MW884151 |
| BE50 = CGMCC3.20381 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Branch canker | MW880675 | MW884178 | MW884207 | MW884152 | |
| BE111 | Huangyan, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW880676 | MW884179 | MW884208 | MW884153 | |
| L. iranensis | BE27 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW880686 | MW884189 | MW884215 | MW884160 |
| BE30 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW880687 | MW884190 | MW884216 | MW884161 | |
| BE36 | Taizhou, Zhejiang, China | X.E. Xiao & Q.B. Huang | C. reticulata | Trunk canker | MW880688 | MW884191 | MW884217 | MW884162 | |
| BE41 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | C. maxima | Trunk canker | MW862152 | MW884057 | MW884099 | MW884125 | |
| BE42 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | C. maxima | Trunk canker | MW862153 | MW884058 | MW884100 | MW884126 | |
| BE100 | Lishui, Zhejiang, China | X.E. Xiao | C. maxima | Trunk gummosis | MW880684 | MW884187 | MW884213 | MW884158 | |
| L. linhaiensis | BE51 * = CGMCC3.20386 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Branch canker | MW880677 | MW884180 | MW884209 | MW884154 |
| BE28 = CGMCC3.20383 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW880678 | MW884181 | MW884210 | MW884155 | |
| BE34 = CGMCC3.20384 | Huangyan, Zhejiang, China | X.E. Xiao & Q.B. Huang | C. reticulata | Branch canker | MW880679 | MW884182 | MW884211 | MW884156 | |
| BE40 = CGMCC3.20385 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | hybrid cv. Cocktail grapefruit (C. reticulata × C. sinensis) | Twig dieback | MW880680 | MW884183 | MW884212 | MW884157 | |
| BE43 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | C. reticulata | Branch canker | MW880681 | MW884184 | – | – | |
| BE49 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Branch canker | MW880682 | MW884185 | – | – | |
| BE52 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Branch canker | MW880683 | MW884186 | – | – | |
| L. microconidia | BE32 | Huangyan, Zhejiang, China | H.K. Wang & X.E. Xiao | C. reticulata | Branch canker | MW880668 | MW884171 | MW884200 | MW884145 |
| BE35 | Huangyan, Zhejiang, China | X.E. Xiao & Q.B. Huang | C. reticulata | Branch canker | MW880669 | MW884172 | MW884201 | MW884146 | |
| BE80 | Chun’an, Zhejiang, China | H.Y. Li & X.E. Xiao | C. grandis | Trunk canker | MW880670 | MW884173 | MW884202 | MW884147 | |
| BE87 | Chun’an, Zhejiang, China | H.Y. Li & X.E. Xiao | C. unshiu | Trunk canker | MW880671 | MW884174 | MW884203 | MW884148 | |
| BE88 | Chun’an, Zhejiang, China | H.Y. Li & X.E. Xiao | C. grandis | Twig dieback | MW880667 | MW884170 | MW884199 | MW884144 | |
| L. ponkanicola | BE44 * = CGMCC3.20388 | Quzhou, Zhejiang, China | H.K. Wang & X.E. Xiao | C. reticulata | Trunk canker | MW880685 | MW884188 | MW884214 | MW884159 |
| L. pseudotheobromae | BE10 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig gummosis | MT772264 | MT775842 | MT775852 | MW884127 |
| BE11 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MT772265 | MT775843 | MT775853 | MW884128 | |
| BE12 | Fuzhou, Jiangxi, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig dieback | MT772266 | MT775844 | MT775854 | MW884129 | |
| BE19 | Guangzhou, Guangdong, China | H.Y. Li | C. reticulata | Twig dieback | MW862154 | MW884059 | MW884101 | MW884130 | |
| BE22 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW862158 | MW884063 | MW884102 | MW884131 | |
| BE23 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit (C. paradisi × C. reticulata) | Twig gummosis | MW862160 | MW884065 | MW884103 | MW884132 | |
| BE24 | Sihui, Guangdong, China | H.Y. Li | C. reticulata | Twig dieback | MW862155 | MW884060 | – | – | |
| BE26 | Sihui, Guangdong, China | H.Y. Li | C. reticulata | Twig dieback | MW862156 | MW884061 | – | – | |
| BE29 | Sihui, Guangdong, China | H.Y. Li | C. reticulata | Twig dieback | MW862157 | MW884062 | – | – | |
| BE37 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. sinensis | Twig dieback | MW862159 | MW884064 | – | – | |
| BE39 | Quzhou, Zhejiang, China | H.Y. Li & X.E. Xiao | hybrid cv. Cocktail grapefruit (C. paradisi × C. reticulata) | Twig gummosis | MW862161 | MW884066 | – | – | |
| BE46 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862162 | MW884067 | – | – | |
| BE47 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862163 | MW884068 | – | – | |
| BE48 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862164 | MW884069 | – | – | |
| BE53 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862165 | MW884070 | – | – | |
| BE54 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862166 | MW884071 | – | – | |
| BE55 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862167 | MW884072 | – | – | |
| BE56 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862168 | MW884073 | – | – | |
| BE57 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862169 | MW884074 | – | – | |
| BE58 | Linhai, Zhejiang, China | W.L. Li | C. unshiu | Trunk canker | MW862170 | MW884075 | – | – | |
| BE70 | Wanzhou, Chongqing, China | H.Y. Li & X.E. Xiao | C. limon | Twig dieback | MW862171 | MW884076 | – | – | |
| BE82 | Fuzhou, Jiangxi, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig dieback | MW862172 | MW884077 | – | – | |
| BE84 | Chun’an, Zhejiang, China | H.Y. Li & X.E. Xiao | C. unshiu | Trunk canker | MW862173 | MW884078 | – | – | |
| BE107 | Yongchun, Fujian, China | X.E. Xiao | C. reticulata | Twig dieback | MW862174 | MW884079 | – | – | |
| BE109 | Yongchun, Fujian, China | X.E. Xiao | C. sinensis | Twig dieback | MW862175 | MW884080 | – | – | |
| BE110 | Yongchun, Fujian, China | X.E. Xiao | C. reticulata | Twig dieback | MW862176 | MW884081 | – | – | |
| L. theobromae | BE20 | Sihui, Guangdong, China | H.Y. Li | C. reticulata | Twig dieback | MW862177 | MW884082 | MW884104 | MW884133 |
| BE21 | Sihui, Guangdong, China | H.Y. Li | C. reticulata | Twig dieback | MW862178 | MW884083 | MW884105 | MW884134 | |
| BE25 | Guilin, Guangxi, China | H.Y. Li & X.E. Xiao | C. reticulata | Twig dieback | MW862179 | MW884084 | MW884106 | MW884135 | |
| BE108 | Yongchun, Fujian, China | X.E. Xiao | C. sinensis | Twig dieback | MW862180 | MW884085 | – | – | |
| Neodeightonia subglobosa | BE14 | Xiangshan, Zhejiang, China | H.Y. Li & B. Liu | C. unshiu | Trunk gummosis | MT772268 | MT775846 | MT775856 | MW884136 |
| Neofusicoccum parvum | BE15 | Xiangshan, Zhejiang, China | X.E. Xiao | C. unshiu | Branch gummosis | MT772269 | MT775847 | MT775857 | MW884137 |
| Sphaeropsis linhaiensis | BE18 * = CGMCC3.20382 | Linhai, Zhejiang, China | H.Y. Li | C. unshiu | Twig dieback | MW880689 | MW884192 | MW884218 | MW884163 |
a Species names in bold represent new species described in this study.
b ITS, internal transcribed spacer region and intervening 5.8S nrRNA gene; tef1, translation elongation factor 1-alpha; tub2, β-tubulin; rpb2, DNA-directed RNA polymerase II second largest subunit.
* Isolates represent ex-type.
Phylogenetic analyses
Sequences of the ITS and tef1 locus for all the isolates obtained in this study were generated and blasted against the NCBIs GenBank nucleotide datasets (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to obtain an initial identification. Representative isolates were selected for sequencing of tub2 and rpb2 loci and further phylogenetic analyses. Sequences of ex-type strains closely related to the Botryosphaeriaceae isolates studied here were downloaded from NCBI and used for phylogenetic analyses (Table 2). Sequence alignments of each of the ITS, tef1, tub2 and rpb2 loci were initially aligned by using MAFFT v. 7 online service (https://mafft.cbrc.jp/alignment/server/index.html) (Katoh et al. 2019), with iterative refinement methods (FFT-NS-i), and then edited manually with MEGA v. 7.0.26 software. Aligned datasets and phylogenetic trees for the individual genes and combined alignments were deposited in TreeBASE (http://treebase.org; study number S28083).
Table 2.
Isolates from other studies used in the phylogenetic analyses.
| Species | Isolate numbersa | Host | Location | Collector | GenBank accession numbersb |
|||
|---|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | rpb2 | |||||
| Botryosphaeria agaves | CBS 133992 = MFLUCC11-0125 * | Agave sp. | Thailand | R. Phookamsak | JX646791 | JX646856 | JX646841 | – |
| MFLUCC 10-0051 | Agave sp. | Thailand | P. Chomnunti | JX646790 | JX646855 | JX646840 | – | |
| Botryosphaeria corticis | CBS 119047 * | Vaccinium corymbosum | USA | P.V. Oudemans | DQ299245 | EU017539 | – | – |
| ATCC 22927 | Vaccinium sp. | USA | R.D. Millholland | DQ299247 | EU673291 | EU673108 | – | |
| Botryosphaeria dothidea | CBS 115476 = CMW 8000 * | Prunus sp. | Switzerland | B. Slippers | AY236949 | AY236898 | AY236927 | EU339577 |
| CBS 110302 | Vitis vineifera | Portugal | A.J.L. Phillips | AY259092 | AY573218 | EU673106 | – | |
| CBS 145971 = CPC 29048 | Grevillea sp. | Australia | P.W. Crous | MT587332 | MT592034 | MT592470 | – | |
| Botryosphaeria fabicerciana | CBS 127194 = CMW 27094 * | Eucalyptus sp. | China | M.J. Wingfield | HQ332197 | HQ332213 | KF779068 | MF410137 |
| CERC 2948 | Eucalyptus sp. | China | M.J. Wingfield | KX277983 | KX278088 | KX278193 | MF410132 | |
| Botryosphaeria kuwatsukai | CBS 135219 = PG2 * | Malus domestica | China | C.S. Wang | KJ433388 | KJ433410 | – | – |
| LSP5 | Pyrus sp. | China | C.S. Wang | KJ433395 | KJ433417 | – | – | |
| Botryosphaeria qingyuanensis | CERC 2946 = CGMCC 3.18742 * | Eucalyptus hybrid | China | S.F. Chen & G.Q. Li | KX278000 | KX278105 | KX278209 | MF410151 |
| CERC 2947 = CGMCC 3.18744 | Eucalyptus hybrid | China | S.F. Chen & G.Q. Li | KX278001 | KX278106 | KX278210 | MF410152 | |
| Botryosphaeria ramosa | CBS 122069 = CMW 26167 * | Eucalyptus camaldulensis | Australia | T.I. Burgess | EU144055 | EU144070 | KF766132 | – |
| CGMCC 3.18006 | Myrtaceae | China | – | KX197072 | KX197092 | KX197099 | – | |
| Botryosphaeria scharifii | CBS 124703 = IRAN 1529C * | Mangifera indica | Iran | J. Abdollahzadeh | JQ772020 | JQ772057 | – | – |
| CBS 124702 = IRAN 1543C | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | JQ772019 | JQ772056 | – | – | |
| Diplodia africana | CBS 120835 = CPC 5908 * | Prunus persica | South Africa | U. Damm | KF766155 | KF766397 | KF766129 | – |
| STE-U 5946 | Prunus persica | South Africa | U. Damm | EF445344 | EF445383 | – | – | |
| Diplodia afrocarpi | CBS 131681 = CMW 35506 | Afrocarpus falcatus, healthy twigs | South Africa | E.M. Cruywagen | MT587333 | MT592035 | MT592471 | – |
| Diplodia agrifolia | CBS 124.30 | Ulmus sp. | USA | – | KX464087 | KX464557 | KX464783 | KX463953 |
| Diplodia allocellula | CBS 130408 = CMW 36468 * | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239397 | JQ239384 | JQ239378 | – |
| CMW 36470 | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239399 | JQ239386 | JQ239380 | – | |
| Diplodia arengae | MFLU 17-2769 = XTBG28 * | Arenga hookeriana | China | D.N. Wanasinghe | MG762771 | MG762774 | MG783039 | – |
| Diplodia bulgarica | CBS 124254 * | Malus sylvestris | Bulgaria | S.G. Bobev | GQ923853 | GQ923821 | – | – |
| CBS 124135 | Malus sylvestris | Bulgaria | S.G. Bobev | GQ923852 | GQ923820 | – | – | |
| Diplodia citricarpa | CBS 124715 = CJA 131 = IRAN 1578C * | Citrus sp., twigs | Iran | J. Abdollahzadeh & A. Javadi | KF890207 | KF890189 | KX464784 | – |
| Diplodia corticola | CBS 112549 = CAP 134 * | Quercus suber | Portugal | A. Alves | AY259100 | AY573227 | DQ458853 | – |
| CBS 112546 | Quercus ilex | Spain | – | AY259090 | EU673310 | EU673117 | KX463954 | |
| Diplodia crataegicola | MFLU 15-1311 * | Crataegus sp. | Italy | – | KT290244 | KT290248 | KT290246 | – |
| Diplodia cupressi | CBS 168.87 * | Cupressus sempervirens | Israel | Z. Solel | DQ458893 | DQ458878 | DQ458861 | – |
| CBS 261.85 | Cupressus sempervirens | Israel | Z. Solel | DQ458894 | DQ458879 | DQ458862 | – | |
| Diplodia eriobotryicola | CBS 140851 = BN-21 * | Eriobotrya japonica | Spain | E. Gonzalez-Domýnguez | KT240355 | KT240193 | MG015806 | – |
| Diplodia estuarina | CMW 41363 | Rhizophora mucronata | South Afric | J.A. Osorio & Jol. Roux. | KP860829 | KP860674 | KP860752 | – |
| CMW 41230 | Rhizophora mucronata | South Afric | J.A. Osorio & Jol. Roux. | KP860830 | KP860675 | KP860753 | – | |
| Diplodia fraxini | CBS 136010 * | Fraxinus angustifolia | Portugal | A. Deidda | KF307700 | KF318747 | MG015807 | – |
| CBS 136011 | Fraxinus angustifolia | Italy | B.T. Linaldeddu | KF307711 | KF318748 | MG015808 | – | |
| Diplodia galiicola | MFLU15-1310 * | Galium sp. | Italy | E. Camporesi | KT290245 | KT290249 | KT290247 | – |
| Diplodia gallae | CBS 211.25 | Quercus sp., fruit | – | – | KX464090 | KX464564 | KX464795 | – |
| CBS 212.25 | Quercus sp., gall | – | – | KX464091 | KX464565 | KX464796 | – | |
| Diplodia malorum | CBS 124130 * | Malus sylvestris | Portugal | A.J.L. Phillips | GQ923865 | GQ923833 | – | – |
| BN-37 | Eriobotrya japonica | Spain | – | KT240360 | KT240198 | – | – | |
| Diplodia mutila | CBS 112553 = CAP 062 * | Vitis vinifera | Portugal | A.J.L. Phillips | AY259093 | AY573219 | KY554743 | – |
| Diplodia neojuniperi | CPC 22753 = B0031 * | Juniperus chinensis | Thailand | T. Trakunyingcharoen | KM006431 | KM006462 | – | – |
| CPC 22754 = B0032 | Juniperus chinensis | Thailand | T. Trakunyingcharoen | KM006432 | KM006463 | – | – | |
| Diplodia olivarum | CBS 121887 = CAP 254 * | Olea europaea | Italy | C. Lazzizera | EU392302 | EU392279 | HQ660079 | – |
| IMI 390972 | Carob tree | Italy | – | HM028640 | HQ660078 | HQ660080 | – | |
| Diplodia pseudoseriata | CBS 124906 * | Blepharocalyx salicifolius | Uruguay | C. Pérez | EU080927 | EU863181 | MG015820 | – |
| Diplodia quercivora | CBS 133852 * | Quercus canariensis | Tunisia | B. T. Linaldeddu | JX894205 | JX894229 | MG015821 | – |
| MEAN 1017 | Quercus suber | Portugal | H. Braganca | KU311198 | KU311201 | – | – | |
| Diplodia rosulata | CBS 116470 * | Prunus africana | Ethiopia | A. Gure | EU430265 | EU430267 | EU673132 | – |
| CBS 116472 | Prunus africana | Ethiopia | A. Gure | EU430266 | EU430268 | EU673131 | – | |
| Diplodia sapinea | CBS 393.84 * | Pinus nigra | Netherlands | H.A. van der Aa | DQ458895 | DQ458880 | DQ458863 | – |
| CBS109726 = CMW 04880 | Pinus patula | South Africa | M.J. Wingfield | KX464094 | KX464568 | KX464800 | KX463956 | |
| Diplodia scrobiculata | CBS 118110 * | Pinus resinosa | USA | M.A. Palmer | AY253292 | AY624253 | AY624258 | KX463959 |
| Diplodia seriata | CBS 112555 = CAP 063 * | Vitis vinifera | Italy | A.J.L. Phillips | AY259094 | AY573220 | DQ458856 | – |
| CBS 117.82 | Rubus sp., dead stem | Italy | H.A. van der Aa | KX464108 | KX464598 | KX464834 | KX463964 | |
| Diplodiasp. 1 | CBS 678.88 | Quercus suber | Spain | J. Luque | AY259104 | GU799459 | GU799458 | – |
| UCD1275So | Grape vine | USA | – | GU799471 | GU799468 | GU799465 | – | |
| Diplodia subglobosa | CBS 124133 = JL 453 * | Lonicera nigra | Spain | J. Luque | GQ923856 | GQ923824 | MT592576 | – |
| CBS 124132 = JL 375 | Fraxinus excelsior | Spain | J. Luque | DQ458887 | DQ458871 | DQ458852 | – | |
| Diplodia tsugae | CBS 418.64 = IMI 197143 * | Tsuga heterophylla | Canada | A. Funk | DQ458888 | DQ458873 | DQ458855 | – |
| Dothiorella acacicola | CBS 141295 = CPC 26349 * | Acacia mearnsii | France | P.W. Crous & M.J. Wingfield | KX228269 | KX228376 | – | – |
| Dothiorella acericola | KUMCC 18-0137 * | Acer palmatum, dead hanging twigs | China | R. Phookamsak | MK359449 | MK361182 | – | – |
| HNXX032 | Ziziphus jujuba, branch | China | R. Zang | KY385661 | KY393212 | KY393178 | ||
| Dothiorella alpina | CGMCC 3.18001 * | Platycladus orientalis | China | W. He & J.R. Wu; det. Y. Zhang | KX499645 | KX499651 | – | – |
| Dothiorella brevicollis | CBS 130411 = CMW 36463 * | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239403 | JQ239390 | JQ239371 | – |
| CMW 36464 | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239404 | JQ239391 | JQ239372 | – | |
| Dothiorella capri-amissi | CBS 121763 = CMW 25403 * | Acacia erioloba | South Africa | F.J.J. van der Walt & G.J. Marais | EU101323 | EU101368 | KX464850 | – |
| CMW 25404 | Acacia erioloba | South Africa | F.J.J. van der Walt & G.J. Marais | EU101324 | EU101369 | – | – | |
| Dothiorella casuarini | CBS 120688 = CMW 4855 * | Casuarina sp. | Australia | M.J. Wingfield | DQ846773 | DQ875331 | – | KX463970 |
| CBS 120690 = CMW 4857 | Casuarinasp. | Australia | M.J. Wingfield | DQ846774 | DQ875333 | – | – | |
| Dothiorella citricola | CBS 124729 = ICMP 16828 * | Citrus sinensis | New Zealand | S.R. Pennycook, P.R. Johnston & B.C. Paulus | EU673323 | EU673290 | KX464853 | KX463971 |
| CBS 124728 = ICMP 16827 | Citrus sinensis | New Zealand | S.R. Pennycook, P.R. Johnston & B.C. Paulus | EU673322 | EU673289 | KX464852 | – | |
| Dothiorella diospyricola | CBS 145972 = CPC 34653 * | Diospyros mespiliformis | South Africa | P.W. Crous | MT587398 | MT592110 | MT592581 | |
| Dothiorella dulcispinae | CBS 130413 = CMW 36460 * | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239400 | JQ239387 | JQ239373 | – |
| CMW 36462 | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239402 | JQ239389 | JQ239375 | – | |
| Dothiorella eriobotryae | CBS 140852 = CPC 29679 = BN 81 * | Eriobotrya japonica, branch canker | Spain | E. Gonzalez-Domýnguez | KT240287 | KT240262 | MT592582 | – |
| Dothiorella heterophyllae | CMW46458 | Acacia heterophylla | La Réunion | M.J. Wingfield | MN103794 | MH548348 | MH548324 | – |
| Dothiorella iranica | CBS 124722 = IRAN 1587C * | Olea europea | Iran | A. Javadi | KC898231 | KC898214 | KX464856 | – |
| MFLUCC 15-0656 | Paliurus | Italy | E. Camporesi | KX765302 | KX765303 | – | – | |
| Dothiorella koae | CMW48017 | Acacia heterophylla | La Réunion | M.J. Wingfield | MH447652 | MH548338 | MH548327 | – |
| Dothiorella lampangensis | MFLUCC 18-0232 * | Rutaceae, fallen fruit pericarp | Thailand | S.C. Jayasiri | MK347758 | MK340869 | MK412874 | |
| Dothiorella longicollis | CBS 122068 = CMW 26166 * | Lysiphyllum cunninghamii | Australia | T.I. Burgess & M.J. Wingfield | EU144054 | EU144069 | KF766130 | KX463972 |
| CBS 122066 = CMW 26166 | Terminalia sp. | Australia | T.I. Burgess & M.J. Wingfield | EU144052 | EU144067 | KX464857 | – | |
| Dothiorella magnoliae | CFCC 51563 * | Magnolia grandiflora | China | C.J. You | KY111247 | KY213686 | – | |
| CFCC 51564 | Magnolia grandiflora | China | C.J. You | KY111248 | KY213687 | – | ||
| Dothiorella mangifericola | CBS 124727 = IRAN 1584C * | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | KC898221 | KC898204 | – | KX463973 |
| IRAN 1545C | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | KC898223 | KC898206 | – | – | |
| Dothiorella moneti | MUCC 505 = WAC 13154 * | Acacia rostellifera | Australia | K.M. Taylor | EF591920 | EF591971 | EF591954 | – |
| MUCC 507 | Acacia rostellifera | Australia | K.M. Taylor | EF591922 | EF591973 | EF591956 | – | |
| Dothiorella plurivora | CBS 124724 = IRAN 1557C * | Citrus sp. | Iran | J. Abdollahzadeh & A. Javadi | KC898225 | KC898208 | KX464874 | – |
| CBS 124725 | Prunus armeniaca | Iran | J. Abdollahzadeh & A. Javadi | KC898225 | KC898213 | KX464875 | – | |
| Dothiorella pretoriensis | CBS 130404 = CMW 36480 * | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239405 | JQ239392 | JQ239376 | – |
| CMW 36481 | Acacia karroo | South Africa | F. Jami & M. Gryzenhout | JQ239406 | JQ239393 | JQ239377 | – | |
| Dothiorella prunicola | CBS 124723 = CAP 187 * | Prunus dulcis | Portugal | E. Diogo | EU673313 | EU673280 | – | – |
| Dothiorella reunionis | CMW46457 * | Acacia heterophylla | La Réunion | M.J. Wingfield | MH447649 | MH548347 | – | – |
| Dothiorella santali | MUCC 509 = WAC 13155 * | Santalum acuminatum | Australia | K.M. Taylor | EF591924 | EF591975 | EF591958 | – |
| MUCC 508 | Santalum acuminatum | Australia | K.M. Taylor | EF591923 | EF591974 | EF591957 | – | |
| Dothiorella sarmentorum | IMI 63581b * | Ulmus sp. | England | E.A. Ellis | AY573212 | AY573235 | – | – |
| CBS 115038 | Malus pumila | Netherlands | A.J.L. Phillips | AY573206 | AY573223 | EU673101 | – | |
| Dothiorella sp. 1 | CBS 121783 = CMW 25432 = CAMS 1187 | Acacia mearnsii | South Africa | F.J.J. van der Walt & R.N. Heath | EU101333 | EU101378 | KX464859 | – |
| CBS 121784 = CMW 25430 = CAMS 1185 | Acacia mearnsii | South Africa | F.J.J. van der Walt & R.N. Heath | EU101331 | EU101376 | KX464860 | – | |
| Dothiorella striata | CBS 124731= ICMP 16824 * | Citrus sinensis | New Zealand | S.R. Pennycook, P.R. Johnston & B.C. Paulus | EU673321 | EU673288 | EU673143 | KX463976 |
| CBS 124730 = ICMP 16819 | Citrus sinensis | New Zealand | S.R. Pennycook, P.R. Johnston & B.C. Paulus | EU673320 | EU673287 | EU673142 | – | |
| Dothiorella tectonae | MFLUCC 12-0382 = MD-2014 * | Tectona grandis | Thailand | M. Doilom | KM396899 | KM409637 | KM510357 | – |
| Dothiorella thailandica | MFLUCC 11-0438 * | Bamboo culm | Thailand | D.Q. Dai | JX646796 | JX646861 | JX646844 | – |
| Dothiorella thripsita | CBS 125445 = BRIP 51876 * | Acacia harpophylla | Australia | D.J. Tree & C.E.C. Tree | FJ824738 | KJ573639 | KJ577550 | KX463977 |
| Dothiorella ulmacea | CBS 138855 = CPC 24416 * | Ulmus laevis | Germany | R.K. Schumacher | KR611881 | KR611910 | KR611909 | – |
| CPC 24945 | Ulmus laevis | Germany | R.K. Schumacher | KR611882 | KR857697 | – | – | |
| Dothiorella uruguayensis | CBS 124908 = CMW 26763 =UY672 * | Hexachlamis edulis | Uruguay | C.A. Pérez | EU080923 | EU863180 | KX464886 | – |
| Dothiorella vinea-gemmae | DAR 81012 = B116-3 * | Vitis vinifera | Australia | N. Wunderlich | KJ573644 | KJ573641 | – | – |
| Dothiorella viticola | CBS 117009 * | Vitis vinifera cv. Garnatxa Negra | Spain | J. Luque & S. Martos | AY905554 | AY905559 | EU673104 | DQ677985 |
| GAR09 | Vitis sp. | French | – | KT595694 | KX098285 | KT595695 | – | |
| Dothiorella yunnana | CGMCC 3.17999 * | Camellia sp. | China | W. He & J.R. Wu; det. Y. Zhang | KX499643 | KX499649 | – | – |
| CGMCC 3.18000 | Camellia sp. | China | W. He & J.R. Wu; det. Y. Zhang | KX499644 | KX499650 | – | – | |
| Lasiodiplodia acaciae | CBS 136434 = CPC 20820 * | Acacia sp., leaf spot | Indonesia | M.J. Wingfield | MT587421 | MT592133 | MT592613 | MT592307 |
| Lasiodiplodia americana | CERC 1961 = CFCC 50065 * | Pistachia vera | USA | T.J. Michailides | KP217059 | KP217067 | KP217075 | MF410161 |
| CERC 1960 = CFCC 50064 | Pistachia vera | USA | T.J. Michailides | KP217058 | KP217066 | KP217074 | MF410162 | |
| Lasiodiplodia aquilariae | CGMCC 3.18471 * | Aquilaria crassna | Laos | X. Sun | KY783442 | KY848600 | – | KY848562 |
| Lasiodiplodia avicenniae | CMW 41467 * | Avocennia marina | South Africa | J.A. Osorio & J. Roux | KP860835 | KP860680 | KP860758 | KU587878 |
| LAS 199 | Avocennia marina | South Africa | J.A. Osorio & J. Roux | KU587957 | KU587947 | KU587868 | KU587880 | |
| Lasiodiplodia brasiliense | CMM 4015 * | Mangifera indica | Brazil | M.W. Marques | JX464063 | JX464049 | – | – |
| IBL 344 | Adansonia madagascariensis | Madagascar | – | KT151808 | KT151802 | KT151805 | – | |
| Lasiodiplodia bruguierae | CMW 41470* | Bruguiera gymnorrhiza | South Africa | J.A. Osorio & J. Roux | KP860833 | KP860678 | KP860756 | KU587875 |
| CMW 41614 | Bruguiera gymnorrhiza | South Africa | J.A. Osorio & J. Roux | KP860834 | KP860679 | KP860757 | KU587877 | |
| Lasiodiplodia cinnamomi | CFCC 51997 * | Cinnamomum camphora | China | N. Jiang | MG866028 | MH236799 | MH236797 | MH236801 |
| CFCC 51998 | Cinnamomum camphora | China | N. Jiang | MG866029 | MH236800 | MH236798 | MH236802 | |
| Lasiodiplodia citricola | CBS 124707 = IRAN 1522C * | Citrus sp. | Iran | J. Abdollahzadeh & A. Javadi | GU945354 | GU945340 | KP872405 | KU696351 |
| CBS124706 = IRAN 1521C | Citrus sp. | Iran | A. Shekari | GU945353 | GU945339 | KP872406 | KU696350 | |
| Lasiodiplodia crassispora | CBS 118741 = WAC12533 * | Santalum album | Australia | T.I. Burgess & B. Dell | DQ103550 | EU673303 | KU887506 | KU696353 |
| CMM 4585 | – | – | – | MG954354 | MG979520 | MG979552 | MG979561 | |
| Lasiodiplodia euphorbicola | CMM 3609 * | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234543 | KF226689 | KF254926 | – |
| CMW 33350 | Adansonia digitata | Botswana | – | KU887149 | KU887026 | KU887455 | KU696346 | |
| Lasiodiplodia gilanensis | CBS 124704 = IRAN1523C= UCCE 940B * | Citrus sp., fallen twigs | Iran | J. Abdollahzadeh & A. Javadi | KX906851 | KX906853 | KX906849 | KU696357 |
| CBS 124705 = IRAN 1501C | Citrus sp., fallen twigs | Iran | J. Abdollahzadeh & A. Javadi | GU945352 | GU945341 | KP872412 | KU696356 | |
| Lasiodiplodia gonubiensis | CBS 115812 = CMW 14077 * | Syzygium cordatum | South Africa | D. Pavlic | AY639595 | DQ103566 | DQ458860 | KU696359 |
| CMW 46621 = MTU 56 | Syzygium cordatum | South Africa | D. Pavlic | KY052944 | KY024623 | KY000126 | – | |
| Lasiodiplodia gravistriata | CMM 4564 * | Anacardium humile | Brazil | M.S.B. Netto | KT250949 | KT250950 | – | – |
| CMM 4565 | Anacardium humile | Brazil | M.S.B. Netto | KT250947 | KT266812 | – | – | |
| Lasiodiplodia hormozganensis | CBS 124709 = IRAN 1500C * | Olea sp. | Iran | J. Abdollahzadeh & A. Javadi | GU945355 | GU945343 | KP872413 | KU696361 |
| CBS 124708 = IRAN 1498C | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | GU945356 | GU945344 | KP872414 | KU696360 | |
| Lasiodiplodia indica | IBP 1 * | Angiospermous tree | India | I.B. Prasher & G. Singh | KM376151 | – | – | – |
| Lasiodiplodia iranensis | CBS 124710 = IRAN 1520C * | Salvadora persica | Iran | J. Abdollahzadeh & A. Javadi | GU945348 | GU945336 | KU887516 | KU696363 |
| CBS 124711 = IRAN 1502C = CMM 4603 | Juglans sp. | Iran | A. Javadi | GU945347 | GU945335 | MG979537 | – | |
| Lasiodiplodia laeliocattleyae | CBS 167.28 * | Laeliocattleya | Italy | C. Sibilia | KU507487 | KU507454 | – | – |
| CMM 4724 | Vitis vinifera | Brazil | – | MG954343 | MG979508 | MG979541 | – | |
| Lasiodiplodia lignicola | CBS 134112= MFLUCC 11-0435 * | Dead wood | Thailand | A.D. Ariyawansa | JX646797 | KU887003 | KT852958 | KU696364 |
| Lasiodiplodia macrospora | CMM 3833 * | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234557 | KF226718 | KF254941 | – |
| Lasiodiplodia mahajangana | CBS 124925 = CMW 27801 * | Terminalia catappa | Madagascar | J. Roux | FJ900595 | FJ900641 | FJ900630 | KU696365 |
| CMW 27818 | Terminalia catappa | Madagascar | J. Roux | FJ900596 | FJ900642 | FJ900631 | – | |
| Lasiodiplodia margaritacea | CBS 122519 = CMW 26162 * | Adansonia gibbosa | Australia | T.I. Burgess & M.J. Wingfield | EU144050 | EU144065 | KX464903 | KU696367 |
| CBS 122065 | Adansonia gibbosa | Australia | T.I. Burgess & M.J. Wingfield | EU144051 | EU144066 | – | – | |
| Lasiodiplodia mediterranea | CBS 137783 * | Holm oak | Italy | B.T. Linaldeddu | KJ638312 | KJ638331 | KU887521 | KU696368 |
| CBS137784 | Grapevine | Italy | S. Serra | KJ638311 | KJ638330 | KU887522 | KU696369 | |
| Lasiodiplodia microconidia | CGMCC 3.18485 * | Aquilaria crassna | Laos | X. Sun | KY783441 | KY848614 | – | KY848561 |
| Lasiodiplodia parva | CBS 456.78 * | Cassava-field soil | Colombia | O. Rangel | EF622083 | EF622063 | KU887523 | KU696372 |
| CBS 494.78 | Cassava-field soil | Colombia | O. Rangel | EF622084 | EF622064 | EU673114 | KU696373 | |
| Lasiodiplodia plurivora | CBS 120832 = STE-U5803 * | Prunus salicina | South Africa | F. Halleen | EF445362 | EF445395 | KP872421 | KU696374 |
| CBS 121103 = STE-U4583 | Vitis vinifera | South Africa | F. Halleen | AY343482 | EF445396 | KP872422 | KU696375 | |
| Lasiodiplodia pontae | CMW 1277 = IBL12 * | Spondias purpurea | Brazil | J.S. Lima & F.C.O. Freire | KT151794 | KT151791 | KT151797 | – |
| Lasiodiplodia pseudotheobromae | CBS 116459 * | Gmelina arborea | Costa Rica | J. Carranza & Velásquez | EF622077 | EF622057 | EU673111 | KU696376 |
| CMM 3887 | Jatropha curcas | Brazil | A.R. Machado | KF234559 | KF226722 | KF254943 | – | |
| Lasiodiplodia rubropurpurea | CBS 118740 = CMW 14700 = WAC 12535 * | Eucalyptus grandis | Australia | T.I. Burgess & G. Pegg | DQ103553 | DQ103571 | KU887529 | KU696380 |
| WAC 12536 = CMW 15207 | Eucalyptus grandis | Australia | T.I. Burgess & G. Pegg | DQ103554 | DQ103572 | KP872425 | KU696381 | |
| Lasiodiplodia subglobosa | CMM 3872 * | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234558 | KF226721 | KF254942 | – |
| CMM 4046 | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234560 | KF226723 | KF254944 | – | |
| Lasiodiplodia syzygii | MFLUCC 19-0219.1 = GUCC 9719.1 * | Syzygium samarangense | Thailand | Q. Zhang | MT990531 | MW016943 | MW014331 | – |
| GUCC 9719.3 | Syzygium samarangense | Thailand | Q. Zhang | MW081992 | MW087102 | MW087105 | – | |
| Lasiodiplodia thailandica | CPC 22795 * | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193637 | KJ193681 | – | – |
| BJFU DZP160123-13 | Albizia chinensis | China | Z.P. Dou & Z.C. Liu | KY676789 | KY676798 | KY751301 | KY751298 | |
| Lasiodiplodia theobromae | CBS 164.96 * | fruit along coral reef coast | Papua New Guinea | A. Aptroot | AY640255 | AY640258 | KU887532 | KU696383 |
| CBS 111530 | Leucospermum sp. | USA | J.E. Taylor | EF622074 | EF622054 | KU887531 | KU69638 | |
| CBS 124.13 | – | USA | J.J. Taubenhaus | DQ458890 | DQ458875 | DQ458858 | KY472887 | |
| Lasiodiplodia tropica | CGMCC 3.18477 * | Aquilaria crassna | Laos | X. Sun | KY783454 | KY848616 | KY848540 | KY848574 |
| Lasiodiplodia venezuelensis | CBS 118739 = CMW 13511 = WAC 12539 * | Acacia mangium | Venezuela | S. Mohali | DQ103547 | EU673305 | KU887533 | KU696384 |
| CBS 129757 | Acacia mangium | Venezuela | S. Mohali | JX545102 | JX545122 | JX545142 | – | |
| Lasiodiplodia viticola | CBS 128313 = UCD 2553AR * | Vitis vinifera | USA | R.D. Cartwright & W.D. Gubler | HQ288227 | HQ288269 | HQ288306 | KU696385 |
| CBS 128315 = UCD 2604MO | Vitis vinifera | USA | K. Striegler & W.D. Gubler | HQ288228 | HQ288270 | HQ288307 | KU696386 | |
| Lasiodiplodia vitis | CBS 124060 * | Vitis vinifera | Italy | S. Burruano | KX464148 | KX464642 | KX464917 | – |
| Neodeightonia licuriensis | COAD 1780 * | Syagrus coronata | Brazil | O.L. Pereira | KP165429 | KP165430 | KP165431 | – |
| Neodeightonia microspora | MFLUCC 11-0483 | bamboo | Thailand | D.Q. Dai | KU940110 | – | – | – |
| MFLUCC 11-0504 | bamboo | Thailand | D.Q. Dai | KU940111 | – | – | – | |
| Neodeightonia palmicola | MFLUCC10-0822 * | Arenga westerhoutii | Thailand | J.K. Liu | HQ199221 | – | – | – |
| FAFU 002 | Caryota mitis | China | – | MK203813 | – | MK208460 | – | |
| Neodeightonia phoenicum | CBS 122528 * | Phoenix sp. | Spain | F. Garcia | EU673340 | EU673309 | EU673116 | KX463999 |
| CBS 169.34 | Phoenix dactylifera | USA | H.S. Fawcett | EU673338 | EU673307 | EU673138 | – | |
| Neodeightonia planchoniae | MFLUCC 17-2427 | Planchonia sp. | Thailand | S.C. Jayasiri | MK347755 | – | – | – |
| Neodeightonia rattanica | MFLUCC 15-0712 * | Calamus sp. | Thailand | S. Konta | KX646357 | KX646360 | – | – |
| MFLUCC 15-0313 | Calamus sp. | Thailand | S. Konta | KX646358 | KX646361 | – | – | |
| Neodeightonia rattanicola | MFLUCC 15-0319 * | Calamus sp. | Thailand | S. Konta | KX646359 | KX646362 | – | – |
| Neodeightonia subglobosa | CBS 448.91 * | Bambusa arundinacea | Sierra Leone | F.C. Deighton | EU673337 | EU673306 | EU673137 | – |
| MFLUCC 11-0607 | bamboo | Thailand | D.Q. Dai | KU940113 | – | – | – | |
| Neofusicoccum algeriense | CBS 137504 = ALG1 * | Vitis vinifera | Algeria | A. Berraf-Tebbal | KJ657702 | KJ657715 | – | – |
| ALG9 | Vitis vinifera | Algeria | A. Berraf-Tebbal | KJ657704 | KJ657721 | – | – | |
| Neofusicoccum andinum | CBS 117453 = CMW 13455 * | Eucalyptus sp. | Venezuela | S. Mohali | AY693976 | AY693977 | KX464923 | KX464002 |
| CBS 117452 = CMW 13446 | Eucalyptus sp. | Venezuela | S. Mohali | DQ306263 | DQ306264 | KX464922 | KX464001 | |
| Neofusicoccum arbuti | CBS 116131 * | Arbutus menziesii | USA | A. Rossman | AY819720 | – | KF531793 | KX464003 |
| CBS 116575 | Arbutus menziesii | USA | M. Elliott | KX464155 | KX464650 | KX464927 | – | |
| Neofusicoccum australe | CMW 6837 * | Acacia sp. | Australia | M.J. Wingfield | AY339262 | AY339270 | AY339254 | EU339573 |
| C1.2 | Arctostphylos glauca | USA | L. Drake-Schultheis | MH777002 | MH754929 | – | – | |
| Neofusicoccum batangarum | CBS 124924 = CMW 28363 * | Terminalia catappa | Cameroon | D. Begoude & J. Roux | FJ900607 | FJ900653 | FJ900634 | FJ900615 |
| OB45 | Opuntia ficus-indica | Italy | – | MG609042 | MG609076 | MG609059 | – | |
| Neofusicoccum brasiliense | CMM 1338 * | Mangifera indica | Brazil | M.W. Marques | JX513630 | JX513610 | KC794031 | – |
| CMM 1269 | Mangifera indica | Brazil | M.W. Marques | JX513629 | JX513609 | KC794032 | – | |
| Neofusicoccum cordaticola | CBS 123634 = CMW 13992 * | Syzygium cordatum | South Africa | D. Pavlic | EU821898 | EU821868 | EU821838 | EU821928 |
| CBS123635 = CMW 14056 | Syzygium cordatum | South Africa | D. Pavlic | EU821903 | EU821873 | EU821843 | EU821933 | |
| Neofusicoccum cryptoaustrale | CBS 122813 = CMW 23785 * | Eucalyptus tree | South Africa | H.M. Maleme | FJ752742 | FJ752713 | FJ752756 | KX464014 |
| Neofusicoccum eucalypticola | CBS 115679 = CMW 6539 * | Eucalyptus sp. | Australia | M.J. Wingfield | AY615141 | AY615133 | AY615127 | – |
| CBS 6539 = CMW 6217 | Eucalyptus sp. | Australia | M.J. Wingfield | AY615143 | AY615135 | AY615125 | – | |
| Neofusicoccum eucalyptorum | CBS 115791 = CMW 10125 * | Eucalyptus grandis | South Africa | H. Smith | AF283686 | AY236891 | AY236920 | – |
| CAA 518 | Eucalyptus globulus | Portugal | – | KX871883 | KX871839 | KX871776 | – | |
| Neofusicoccum grevilleae | CBS 129518 = CPC 16999 * | Grevillea aurea | Australia | P.W. Crous & R.G. Shivas | JF951137 | – | – | – |
| Neofusicoccum hellenicum | CERC 1947 = CFCC 50067 * | Pistacia vera | Greece | T.J. Michailides | KP217053 | KP217061 | KP217069 | – |
| CERC 1948 = CFCC 50068 | Pistacia vera | Greece | T.J. Michailides | KP217054 | KP217062 | KP217070 | – | |
| Neofusicoccum kwambonambiense | CBS 123639 = CMW 14023 * | Syzygium cordatum | South Africa | D. Pavlic | EU821900 | EU821870 | EU821840 | EU821930 |
| CBS 123641 = CMW 14140 | Syzygium cordatum | South Africa | D. Pavlic | EU821919 | EU821889 | EU821859 | EU821949 | |
| Neofusicoccum lumnitzerae | CMW 41469 * | Lumnitzera racemosa | South Africa | J.A Osorio & Jol. Roux | KP860881 | KP860724 | KP860801 | KU587925 |
| CMW 41228 | Lumnitzera racemosa | South Africa | J.A Osorio & Jol. Roux | KP860882 | KP860725 | KP860803 | KU587926 | |
| Neofusicoccum luteum | CBS 562.92 = ATCC 58193* | Actinidia deliciosa | New Zealand | S.R. Pennycook | KX464170 | KX464690 | KX464968 | KX464020 |
| BRIP 5016 | Persea americana | USA | – | MH057191 | MH102254 | – | – | |
| Neofusicoccum macroclavatum | CBS 118223 = WAC 12444 * | Eucalyptus globulus | Australia | T.I. Burgess | DQ093196 | DQ093217 | DQ093206 | KX464022 |
| WAC 12446 | Eucalyptus globulus | Australia | T.I. Burgess | DQ093197 | DQ093218 | DQ093207 | – | |
| Neofusicoccum mangiferae | CBS 118531 = CMW 7024 * | Mangifera indica | Australia | G.I. Johnson | AY615185 | DQ093221 | AY615172 | – |
| CBS 118532 = CMW 7797 | Mangifera indica | Australia | G.I. Johnson | AY615186 | DQ093220 | AY615173 | KX464023 | |
| Neofusicoccum mangroviorum | CMW 41365 * | Avicennia marina | South Africa | J.A. Osorio | KP860859 | KP860702 | KP860779 | KU587905 |
| CMW 42481 | Bruguiera gymnorrhiza | South Afric | J.A. Osorio | KP860848 | KP860692 | KP860770 | KU587895 | |
| Neofusicoccum mediterraneum | CBS 12718 = PD 312 * | Eucalyptus sp. | Greece | P.W. Crous, M.J. Wingfield & A.J.L. Phillips | GU251176 | GU251308 | GU251836 | KX464024 |
| Neofusicoccum microconidium | CGMCC 3.18750 = CERC 3497 | Eucalyptus urophylla × E. grandis | China | S.F. Chen & G.Q. Li | KX278053 | KX278158 | KX278262 | MF410203 |
| Neofusicoccum nonquaesitum | CBS 126655 = PD 484 * | Umbellularia californica | USA | F.P. Trouillas | GU251163 | GU251295 | GU251823 | KX464025 |
| PD301 | Vaccinium corymbosum | Chile | E.X. Briceño, J.G. Espinoza & B. A.Latorre | GU251164 | GU251296 | GU251824 | – | |
| Neofusicoccum occulatum | CBS 128008 = MUCC 227 * | Eucalyptus grandis | Australia | T.I. Burgess | EU301030 | EU339509 | EU339472 | EU339558 |
| MUCC 286 = WAC 12395 | Eucalyptus pellita | Australia | T.I. Burgess | EU736947 | EU339511 | EU339474 | EU339560 | |
| Neofusicoccum parvum | CMW 9081 = ATCC 58191* | Populus nigra | New Zealand | S.R. Pennycook | AY236943 | AY236888 | AY236917 | EU821963 |
| CBS 110301 | Vitis vinifera | Portugal | A.J.L. Phillips | AY259098 | AY573221 | EU673095 | – | |
| Neofusicoccum pennatisporum | MUCC 510 = WAC 13153 * | Allocasuarina fraseriana | Australia | K.M. Taylor | EF591976 | EF591976 | EF591959 | – |
| Neofusicoccum pistaciae | CBS 595.76 | Pistacia vera | Greece | D.G. Zachos | KX464163 | KX464676 | KX464953 | KX464008 |
| Neofusicoccum pistaciarum | CBS113083 = CPC 5263 * | Pistacia vera | USA | T.J. Michailides | KX464186 | KX464712 | KX464998 | KX464027 |
| CBS113084 = CPC 5284 | Redwood | USA | T.J. Michailides | KX464187 | KX464713 | KX464999 | KX464028 | |
| Neofusicoccum protearum | CMW 39280 | Acacia karroo | Africa | – | KF270041 | KF270011 | – | – |
| CMW 39282 | Acacia karroo | Africa | – | KF270043 | KF270013 | – | – | |
| Neofusicoccum ribis | CBS 115475 = CMW 7772 * | Ribes sp. | USA | B. Slippers & G. Hudler | AY236935 | AY236877 | AY236906 | EU821958 |
| Neofusicoccum sinense | CGMCC 3.18315 | Unknown wood plant | China | J.J. Gan | KY350148 | KY817755 | KY350154 | – |
| Neofusicoccum stellenboschiana | CBS 110864 | Vitis vinifera | South Africa | F. Halleen | AY343407 | AY343348 | KX465047 | KX464042 |
| Neofusicoccum terminaliae | CBS 125263 = CMW26679 * | Terminalia sericea | South Africa | D. Begoude & J. Roux | GQ471802 | GQ471780 | KX465052 | KX464045 |
| CBS 125264 = CMW26683 | Terminalia sericea | South Africa | D. Begoude & J. Roux | GQ471804 | GQ471782 | KX465053 | KX464046 | |
| Neofusicoccum umdonicola | CBS 123645 = CMW 14058 * | Syzygium cordatum | South Africa | D. Pavlic | EU821904 | EU821874 | EU821844 | EU821934 |
| CBS 123646 = CMW 14060 | Syzygium cordatum | South Africa | D. Pavlic | EU821905 | EU821875 | EU821845 | EU821935 | |
| Neofusicoccum ursorum | CBS 122811 = CMW 24480 * | Eucalyptus sp. | South Africa | H.M. Maleme | FJ752746 | FJ752709 | KX465056 | KX464047 |
| CMW 23790 | Eucalyptus sp. | South Africa | H.M. Maleme | FJ752745 | FJ752708 | KX465057 | – | |
| Neofusicoccum viticlavatum | CBS 112878 = STE-U 5044 * | Vitis vinifera | South Africa | F. Halleen | AY343381 | AY343342 | KX465058 | KX464048 |
| CBS 112977 = STE-U 5041 | Vitis vinifera | South Africa | F. Halleen | AY343380 | AY343341 | KX465059 | – | |
| Neofusicoccum vitifusiforme | CBS 110887 = STE-U 5252 * | Vitis vinifera | South Africa | J.M. van Niekerk | AY343383 | AY343343 | KX465061 | KX464049 |
| CBS 110880 = STE-U 5050 | Vitis vinifera | South Africa | J.M. van Niekerk | AY343382 | AY343344 | – | – | |
| Sphaeropsis chromolaenicola | MFLUCC 17-1499 * | Chromolaena odorata | Thailand | A. Mapook | MT214366 | – | – | – |
| Sphaeropsis citrigena | ICMP 16812 * | Citrus sinensis | New Zealand | S.R. Pennycook, P.R. Johnston & B.C.Paulus | EU673328 | EU673294 | EU673140 | – |
| Sphaeropsis citrigena (cont.) | ICMP 16818 | Citrus sinensis | New Zealand | S.R. Pennycook, P.R. Johnston & B.C.Paulus | EU673329 | EU673295 | EU673141 | – |
| Sphaeropsis eucalypticola | MFLUCC 11-0579 * | Eucalyptus sp. | Thailand | M. Doilom | JX646802 | JX646867 | JX646850 | – |
| MFLUCC 11-0654 | Eucalyptus sp. | Thailand | M. Doilom | JX646803 | JX646868 | JX646851 | – | |
| Sphaeropsis porosa | CBS 110496 = STE-U 5132 * | Vitis vinifera | South Africa | J.M. van Niekerk | AY343379 | AY343340 | EU673130 | KX464076 |
| CBS 110574 = STE-U 5046 | Vitis vinifera | South Africa | J.M. van Niekerk | AY343378 | AY343339 | – | – | |
| Sphaeropsis ulmicola | CBS 174.63 | Ulmus glabra | Finland | – | MK134681 | – | – | – |
| PB-11f | Ulmus glabra | Poland | – | MK134682 | – | – | – | |
| Sphaeropsis visci | CBS 100163 * | Vitis vinifera | South Africa | J.M. van Niekerk | EU673324 | EU673292 | EU673127 | KX464077 |
| CBS 186.97 | Viscum album | Germany | T. Graefenhan | EU673325 | EU673293 | EU673128 | KX464080 | |
a ALG: Personal culture collection A. Berraf-Tebbal; ATCC: American Type Culture Collection, Virginia, USA; BL: Personal number of B.T. Linaldeddu; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; CAA: Personal culture collection Artur Alves, Universidade de Aveiro, Portugal; CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; CERC: Culture collection of China Eucalypt Research Centre, Chinese Academy of Forestry, ZhanJiang, GuangDong, China; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Microbiological Culture Collection Center, Beijing, China; CJA: Collection of J. Abdollahzadeh, Department of Plant Protection, Faculty of Agriculture, University of Kurdistan, Sanandaj, Iran; CMM: Culture Collection of Phytopathogenic Fungi ‘Prof. Maria Menezes, Universidade Federal Rural de Pernambuco, Recife, Brazil; CMW: Tree Pathology Co-operative Program, Forestry and Agricultural Biotechnology Institute, University of Pretoria, South Africa; CPC: Working collection of P.W. Crous, housed at CBS; GUCC: Guizhou University Culture Collection; GZCC: Guizhou Academy of Agricultural Sciences Culture Collection, GuiZhou, China; IBL: Personal culture collection, I.B.L. Coutinho; IBP: Personal culture collection, I.B. Prasher; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; IMI: Kew Royal Botanical Gardens, Kew, England; IRAN: Iranian Fungal Culture Collection, Iranian Research Institute of Plant Protection, Iran; JL: Personal culture collection of J. Luque, IRTA, Barcelona, Spain; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUCC: Culture collection of Murdoch University, Perth, Australia; PD: Culture Collection, University of California, Davis, USA; STE-U: Culture collection of the Department of Plant Pathology, University of Stellenbosch, South Africa; UCD: University of California, Davis, Plant Pathology Department Culture Collection; UCROK: Department of Plant Pathology and Microbiology, University of California, Riverside; UY: Department of Plant Pathology, University of Minnesota; WAC: Department of Agriculture, Western Australia Plant Pathogen Collection, South Perth, Western Australia; XTBG: Institutional Repository of Xishuangbanna Tropical Botanical Garden.
b ITS, internal transcribed spacer region and intervening 5.8S nrRNA gene; tef1, translation elongation factor 1-alpha; tub2, β-tubulin; rpb2, DNA-directed RNA polymerase II second largest subunit.
* Isolates represent ex-type.
The maximum parsimony (MP) analyses were conducted using PAUP v. 4.0b10 (Swofford 2003), with gaps treated as a fifth character. The characters were unordered and of equal weight with 1 000 random addition replicates. The equally most parsimonious trees were generated using the heuristic search option with the tree bisection-reconnection (TBR) branch swapping. MAXTREES were set to 5 000 and zero-length branches were collapsed. To assess clade stability, a bootstrap analysis was conducted with 1 000 replicates. Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC) were recorded to evaluate the trees (Hillis & Bull 1993).
The maximum-likelihood (ML) analyses for each dataset were conducted using PhyML v. 3.0 (Guindon et al. 2010). The software package jModeltest v. 2.1.5 (Darriba et al. 2012) was used to determine the best nucleotide substitution model for each dataset. In PhyML, the retention of the maximum number of 1 000 trees was set and nodal support was determined by non-parametric bootstrapping with 1 000 replicates. For both the MP and ML analyses, the phylogenetic trees were viewed in MEGA v. 7.0.26 and FigTree v. 1.4.4 (http://tree.bio.ed.ac. uk/software/figtree).
Morphology
Representative isolates of Botryosphaeriaceae that were identified as new species based on DNA sequence analysis were selected for morphological study. Sporulation was induced on pine needle agar (PNA) (Smith et al. 1996) by incubating cultures at 25 °C in 12/12 h fluorescent light/dark cycle for 4–6 wk. Sporocarps were embedded in a Leica Biosystem Tissue Freezing Medium (Leica Biosystems Nussloch GmbH, Nussloch, Germany) and sectioned (8 μm thick) using a freezing microtome (CryoStar NX50 HOP, Thermo Fisher Scientific, Walldorf, Germany) at -20 °C (Chen et al. 2018). Conidia and other microstructures were examined with a compound microscope (Eclipse 80i, Nikon, Japan) and images were recorded with a Nikon digital camera (NIS-Elements F3.0, Nikon, Japan). Measurements were made with Fiji-ImageJ software (Schindelin et al. 2012). One hundred conidia were measured per isolate, and 30 measurements were taken of other morphological structures. Results are presented as (minimum–) (mean – standard deviation) – (mean + standard deviation) (–maximum). The average length/average width ratio (L/W) of the conidial measurements were also calculated.
Colony characters on PDA were noted and colony colours were determined according to the colour charts of Rayner (1970). To determine growth rates in culture, agar plugs (5 mm diam) were taken from the edge of actively growing cultures of each representative isolate and transferred onto the centre of 90 mm diam Petri dishes containing PDA. Cultures were incubated at five temperature intervals from 5–40 °C in the dark. Five replicate plates of each representative isolate were incubated at each temperature. Perpendicular colony diameters were measured daily until the fastest growing cultures reached the edge of the Petri dish.
Pathogenicity tests
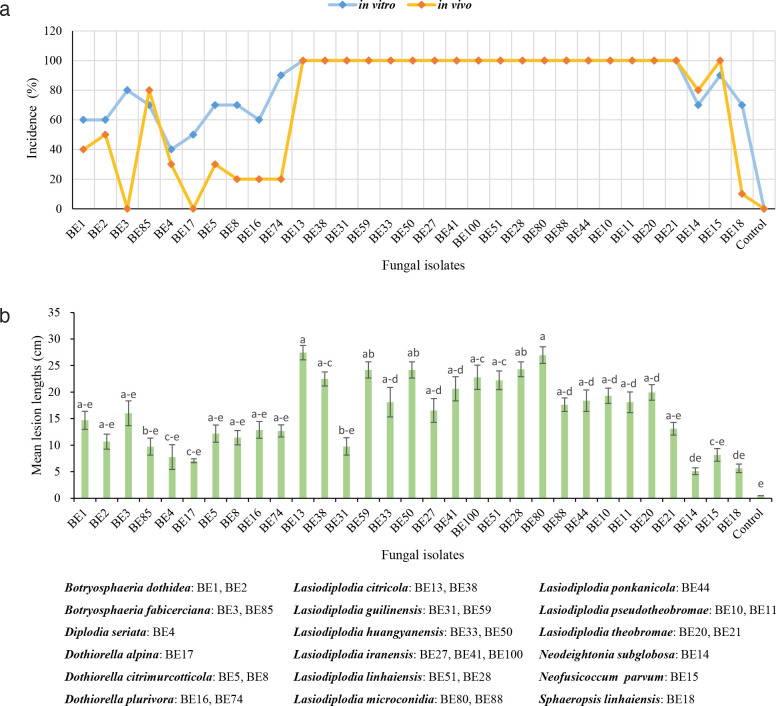
At least two representative isolates from each identified group, except for those with only one isolate, were selected for pathogenicity testing in this study. Inoculation tests were conducted both in vitro and in vivo. For in vitro inoculation, isolates were used to inoculate detached healthy green shoots (40 cm long, 0.6–1 cm diam) collected from Citrus reticulata trees and 10 shoots were inoculated with each isolate. One wound per shoot was made using a cork borer (5 mm diam) and a mycelial plug taken from the margins of colonies grown on PDA for 5 d in the dark was placed on the freshly wounded surface of each shoot, and the inoculated area was covered with Parafilm. The control treatment was inoculated with sterile PDA plugs. The inoculated shoots and controls were covered with liquid paraffin at their ends to prevent desiccation and incubated at 25 °C in moist chambers. Eight days after inoculation, the disease incidences were calculated and the internal lesions or wound lengths were measured. Data were analysed by one-way analysis of variance (ANOVA) using SPSS Statistics 20 software (SPSS 2011). To prove Koch’s postulates, fungi were re-isolated by cutting small pieces of necrotic tissue from the edges of each lesion and plating them in PDA plates at 25 °C. The species were confirmed based on morphology.
For in vivo inoculation, the pathogenicity test was conducted on 6-yr-old healthy plants of Cocktail grapefruit (C. paradisi × C. reticulata). The plants were grown in vinyl house of the Xielong Family Farm in Kecheng District, Quzhou City, Zhejiang Province from 24 June to 9 July 2021. During this time, the environmental temperature ranged from 20–38 °C. Each representative isolate, as well as the control, was inoculated onto 10 shoots. After 15 d, the symptoms and disease incidences were assessed. Re-isolation was also conducted in the same way to fulfil Koch’s postulates.
RESULTS
Isolates
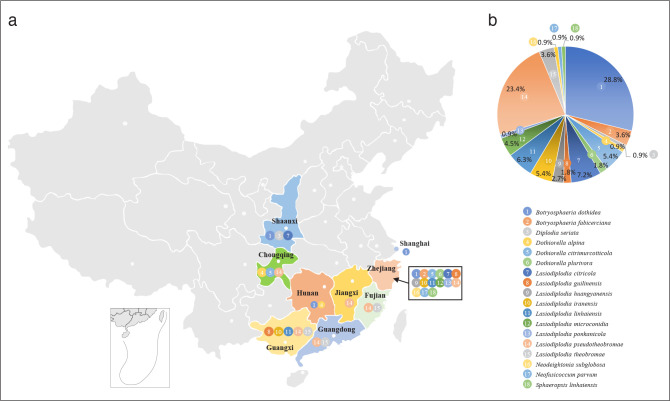
A total of 111 isolates from 88 collected citrus samples exhibited typical morphological characteristics of Botryosphaeriaceae. Eighty-one isolates were collected from Zhejiang, seven from Guangxi, six from Guangdong, four respectively from Chongqing, Fujian and Shaanxi, two respectively from Hunan and Jiangxi, and one from Shanghai. Among them, 52 isolates were obtained from twigs and branches with dieback, 31 were associated with branches and trunks with canker, and 28 from gummosis symptoms. In terms of Citrus species, 42 isolates were obtained from C. unshiu, 25 from C. reticulata, 10 from C. sinensis, nine from C. maxima, one from C. limon, and 24 from hybrids.
Phylogenetic analyses
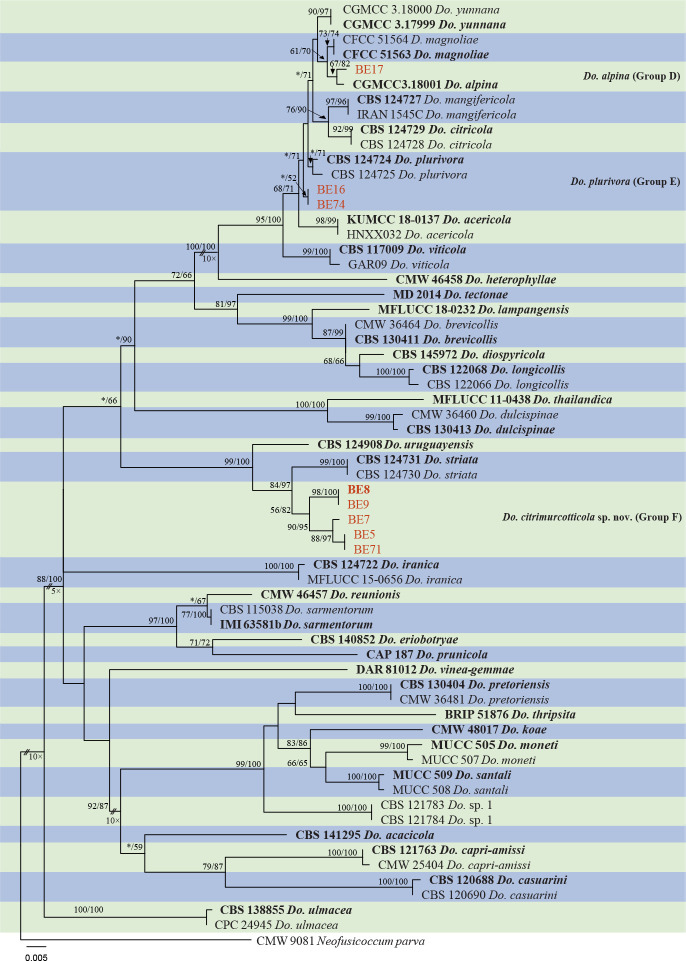
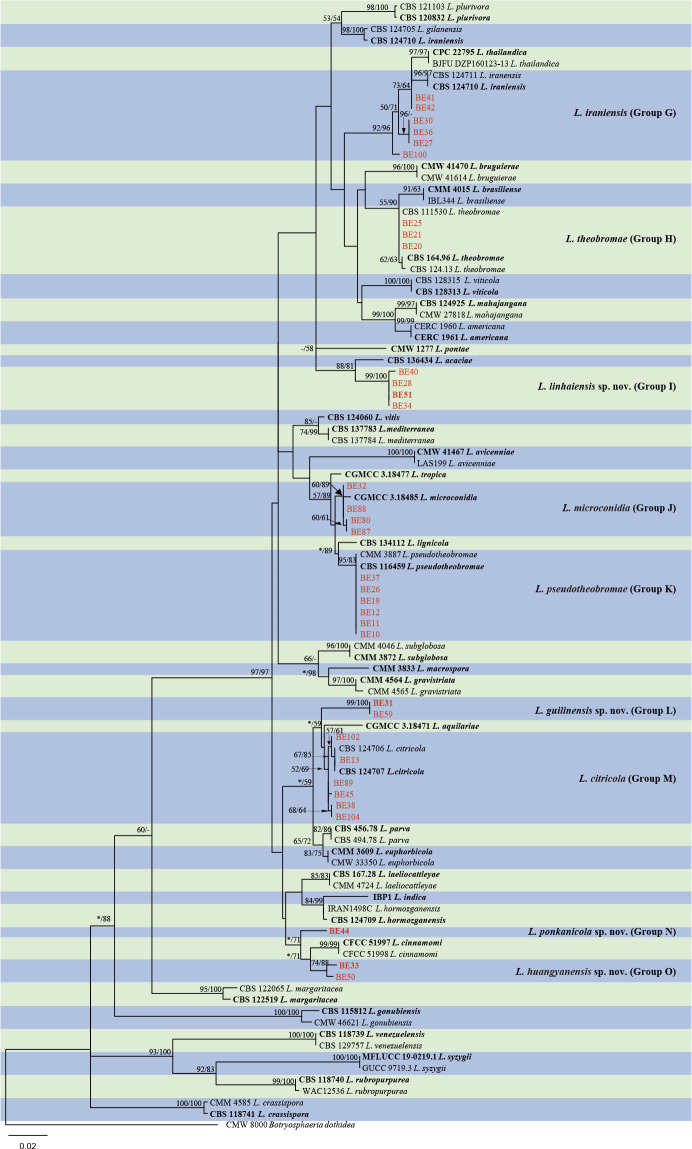
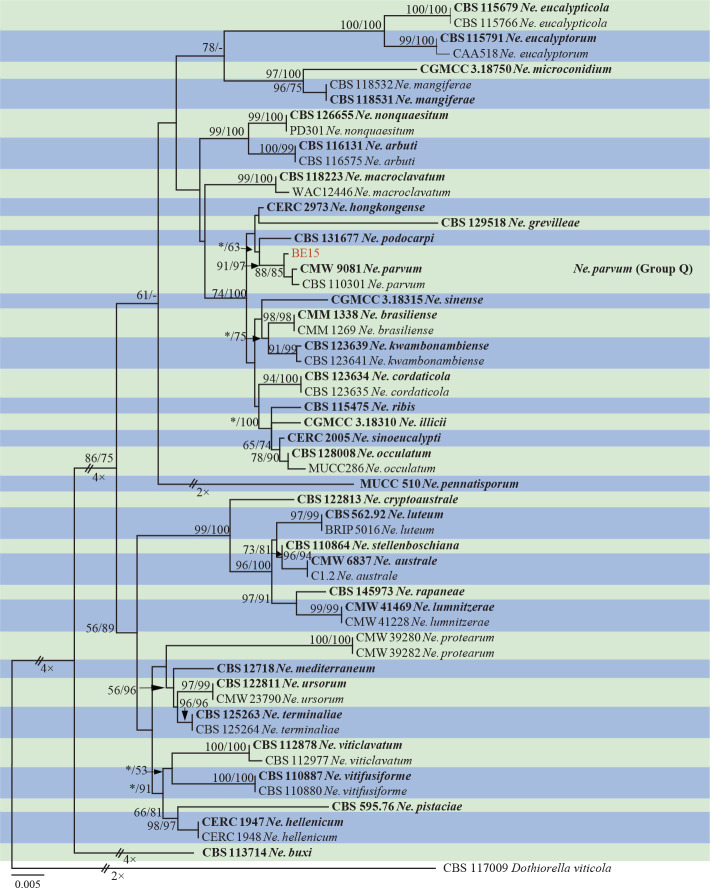
The ITS and tef1 sequences were amplified for all 111 isolates obtained in this study, and blast results indicated that these isolates resided in Botryosphaeria, Diplodia, Dothiorella, Lasiodiplodia, Neodeightonia, Neofusicoccum and Sphaeropsis. Fifty-seven representative isolates were subsequently selected to be sequenced for their tub2 and rpb2 loci (Table 1). Datasets for the seven genera, the parameters of the statistical values of the trees for the MP analyses and the best-fit substitution models for ML analyses are provided in Table 3. All sequences of Botryosphaeriaceae species obtained in this study were deposited in GenBank (Table 1).
Table 3.
Datasets used and statistics resulting from phylogenetic analyses in the current study.
| Genus | Dataset | Maximum likelihood | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| Subst. model1 | NST2 | Rate matrix | Ti/Tv ratio3 | p-inv | Gamma | Rates | ||||||
| Botryosphaeria | ITS | TrN+I | 6 | 1.0000 | 1.4653 | 1.0000 | 1.0000 | 7.9294 | – | 0.8090 | – | equal |
| tef1 | TPM3uf+I | 6 | 0.3783 | 2.6378 | 1.0000 | 0.3783 | 2.6378 | – | 0.6000 | – | equal | |
| tub2 | TrN+I | 6 | 1.0000 | 6.0784 | 1.0000 | 1.0000 | 14.3581 | – | 0.7170 | – | equal | |
| rpb2 | TIM3+G | 6 | 9756.0724 | 15799.8965 | 1.0000 | 9756.0724 | 70110.9963 | – | – | 0.1480 | gamma | |
| ITS/tef1/tub2 | TrN+I | 6 | 1.0000 | 3.4860 | 1.0000 | 1.0000 | 7.2459 | – | 0.761 | – | equal | |
| Diplodia | ITS | TVMef+I+G | 6 | 6.1639 | 23.8085 | 4.6946 | 13.6614 | 23.8085 | – | 0.6580 | 0.6850 | gamma |
| tef1 | TrN+G | 6 | 1.0000 | 3.3131 | 1.0000 | 1.0000 | 5.5220 | – | – | 0.5530 | gamma | |
| tub2 | TrN+I | 6 | 1.0000 | 3.1072 | 1.0000 | 1.0000 | 5.7192 | – | 0.676 | – | equal | |
| rpb2 | TrN+G | 6 | 1.0000 | 5.1679 | 1.0000 | 1.0000 | 13.7446 | – | – | 0.228 | gamma | |
| ITS/tef1/tub2 | TrN+I+G | 6 | 1.0000 | 3.6215 | 1.0000 | 1.0000 | 5.0395 | – | 0.5940 | 1.3320 | gamma | |
| Dothiorella | ITS | TrN+I+G | 6 | 1.0000 | 1.4234 | 1.0000 | 1.0000 | 3.2819 | – | 0.4180 | 0.6840 | gamma |
| tef1 | TPM2uf+G | 6 | 2.0341 | 5.0971 | 2.0341 | 1.0000 | 5.0971 | – | – | 0.765 | gamma | |
| tub2 | HKY+I+G | 2 | – | – | – | – | – | 1.7295 | 0.5600 | 0.9450 | gamma | |
| rpb2 | TrN+I | 6 | 1.0000 | 3.8616 | 1.0000 | 1.0000 | 10.6645 | – | 0.6030 | – | equal | |
| ITS/tef1/tub2 | TIM2+G | 6 | 1.2403 | 2.7176 | 1.2403 | 1.0000 | 4.1029 | – | – | 0.2160 | gamma | |
| Lasiodiplodia | ITS | K80+I | 2 | – | – | – | – | – | 2.4444 | 0.7840 | – | equal |
| tef1 | K80+G | 2 | – | – | – | – | – | 1.7723 | – | 0.4130 | gamma | |
| tub2 | TrNef+I | 6 | 1.0000 | 1.6752 | 1.0000 | 1.0000 | 6.7164 | – | 0.6540 | – | equal | |
| rpb2 | TrNef+G | 6 | 1.0000 | 5.3397 | 1.0000 | 1.0000 | 12.4894 | – | – | 0.3110 | gamma | |
| ITS/tef1/tub2/rpb2 | TrN+I+G | 6 | 1.0000 | 3.8560 | 1.0000 | 1.0000 | 7.6054 | – | 0.5410 | 0.6090 | gamma | |
| Neodeightonia | ITS | TPM3uf+I+G | 6 | 3.2767 | 4.6927 | 1.0000 | 3.2767 | 4.6927 | – | 0.6640 | 0.5000 | gamma |
| tef1 | TIM1 | 6 | 1.0000 | 1.0118 | 0.2049 | 0.2049 | 2.8685 | – | – | – | equal | |
| tub2 | TIM1+I | 6 | 1.0000 | 1.2183 | 0.3558 | 0.3558 | 2.4674 | – | 0.6860 | – | equal | |
| ITS/tef1/tub2 | TIM1+G | 6 | 1.0000 | 1.2491 | 0.5067 | 0.5067 | 3.6475 | – | – | 0.1940 | gamma | |
| Neofusicoccum | ITS | TIM1ef+I+G | 6 | 1.0000 | 6.4363 | 2.1879 | 2.1879 | 17.3039 | – | 0.5780 | 0.6010 | gamma |
| tef1 | HKY+G | 2 | – | – | – | – | – | 2.5159 | – | 0.7750 | gamma | |
| tub2 | TrN+G | 6 | 1.0000 | 3.1372 | 1.0000 | 1.0000 | 6.7052 | – | – | 0.2270 | gamma | |
| rpb2 | TrN+G | 6 | 1.0000 | 5.5358 | 1.0000 | 1.0000 | 17.053 | – | – | 0.235 | gamma | |
| ITS/tef1/tub2/rpb2 | K80+G | 2 | – | – | – | – | – | 2.265 | – | – | gamma | |
| Sphaeropsis | ITS | HKY+I+G | 2 | – | – | – | – | – | 1.7294 | 0.5610 | 0.9480 | gamma |
| tef1 | HKY+I+G | 2 | – | – | – | – | – | 1.7294 | 0.5610 | 0.9480 | gamma | |
| tub2 | TIM2+G | 6 | 1.2403 | 2.7176 | 1.2403 | 1.0000 | 4.1029 | – | – | 0.2160 | gamma | |
| rpb2 | TIM2+G | 6 | 1.2403 | 2.7176 | 1.2403 | 1.0000 | 4.1029 | – | – | 0.2160 | gamma | |
| ITS/tef1/tub2/rpb2 | HKY+I+G | 2 | – | – | – | – | – | 1.7294 | – | 0.5610 | gamma | |
| Botryosphaeria | ITS | 26 | 504 | 25 | 1 | 39 | 0.7949 | 0.8769 | 0.6970 | 0.2051 | ||
| tef1 | 26 | 347 | 103 | 42 | 129 | 0.8992 | 0.9378 | 0.8433 | 0.1008 | |||
| tub2 | 21 | 406 | 16 | 1 | 21 | 0.9048 | 0.9444 | 0.8545 | 0.0952 | |||
| rpb2 | 14 | 659 | 12 | 3 | 18 | 0.8889 | 0.9583 | 0.8519 | 0.1111 | |||
| ITS/tef1/tub | 26 | 1257 | 144 | 3 | 193 | 0.8601 | 0.9129 | 0.7852 | 0.1399 | |||
| Diplodia | ITS | 47 | 535 | 91 | 1354 | 233 | 0.6652 | 0.8534 | 0.5677 | 0.3348 | ||
| tef1 | 47 | 287 | 117 | 5000 | 275 | 0.6291 | 0.9016 | 0.5672 | 0.3709 | |||
| tub | 39 | 388 | 46 | 1103 | 95 | 0.6105 | 0.8571 | 0.5233 | 0.3895 | |||
| rpb2 | 7 | 594 | 41 | 2 | 61 | 0.8033 | 0.7857 | 0.6311 | 0.1967 | |||
| ITS/tef1/tub2 | 47 | 1210 | 42 | 42 | 658 | 0.5881 | 0.8536 | 0.5020 | 0.4119 | |||
| Dothiorella | ITS | 63 | 496 | 97 | 384 | 287 | 0.5714 | 0.8955 | 0.5117 | 0.4286 | ||
| tef1 | 63 | 324 | 217 | 400 | 791 | 0.5310 | 0.8594 | 0.4563 | 0.4690 | |||
| tub2 | 45 | 387 | 95 | 185 | 203 | 0.6946 | 0.8758 | 0.6083 | 0.3054 | |||
| rpb2 | 16 | 594 | 83 | 2 | 145 | 0.6621 | 0.8333 | 0.5517 | 0.3379 | |||
| ITS/tef1/tub2 | 63 | 1540 | 409 | 6 | 1336 | 0.5427 | 0.8584 | 0.4658 | 0.4573 | |||
| Lasiodiplodia | ITS | 99 | 486 | 49 | 5000 | 89 | 0.6517 | 0.8905 | 0.5803 | 0.3483 | ||
| tef1 | 98 | 291 | 132 | 802 | 284 | 0.6092 | 0.8911 | 0.5428 | 0.3908 | |||
| tub2 | 90 | 345 | 34 | 10 | 54 | 0.7222 | 0.9227 | 0.6664 | 0.2778 | |||
| rpb2 | 77 | 521 | 86 | 201 | 166 | 0.6024 | 0.8981 | 0.5411 | 0.3976 | |||
| ITS/tef1/tub2/rpb2 | 99 | 1643 | 313 | 670 | 956 | 0.485356 | 0.8422 | 0.4087 | 0.5146 | |||
| Neodeightonia | ITS | 15 | 502 | 51 | 1 | 111 | 0.7840 | 0.8490 | 0.6650 | 0.2160 | ||
| tef1 | 9 | 229 | 14 | 1 | 19 | 0.8420 | 0.8750 | 0.7370 | 0.1580 | |||
| tub2 | 7 | 418 | 6 | 1 | 9 | 0.8890 | 0.8570 | 0.7620 | 0.1110 | |||
| ITS/tef1/tub2 | 15 | 1150 | 78 | 3 | 162 | 0.7346 | 0.8114 | 0.5960 | 0.2654 | |||
| Neofusicoccum | ITS | 57 | 494 | 54 | 28 | 146 | 0.5548 | 0.8516 | 0.4725 | 0.4452 | ||
| tef1 | 55 | 280 | 115 | 5000 | 273 | 0.6923 | 0.9012 | 0.6239 | 0.3077 | |||
| tub2 | 51 | 422 | 67 | 5000 | 130 | 0.6231 | 0.8345 | 0.5199 | 0.3769 | |||
| rpb2 | 33 | 560 | 75 | 68 | 143 | 0.6364 | 0.8267 | 0.5261 | 0.3636 | |||
| ITS/tef1/tub2/rpb2 | 57 | 1756 | 334 | 24 | 817 | 0.5704 | 0.8208 | 0.4682 | 0.4296 | |||
| Sphaeropsis | ITS | 10 | 518 | 54 | 1 | 19 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | ||
| tef1 | 10 | 297 | 77 | 1 | 117 | 0.8462 | 0.8583 | 0.7262 | 0.1538 | |||
| tub2 | 9 | 375 | 20 | 3 | 21 | 0.8095 | 0.8261 | 0.6687 | 0.1905 | |||
| rpb2 | 5 | 557 | 18 | 2 | 27 | 0.7410 | 0.6110 | 0.4530 | 0.2590 | |||
| ITS/tef1/tub2/rpb2 | 10 | 1747 | 130 | 2 | 188 | 0.8245 | 0.8245 | 0.6797 | 0.1755 | |||
1 Subst. model = best fit substitution model.
2 NST = number of substitution rate categories.
3 Ti/Tv ratio = transition/transversion ratio.
4 bp = base pairs.
5 PIC = number of parsimony informative characters.
6 CI = consistency index.
7 RI = retention index.
8 RC = rescaled consistency index.
9 HI = homoplasy index.
Species of Botryosphaeria
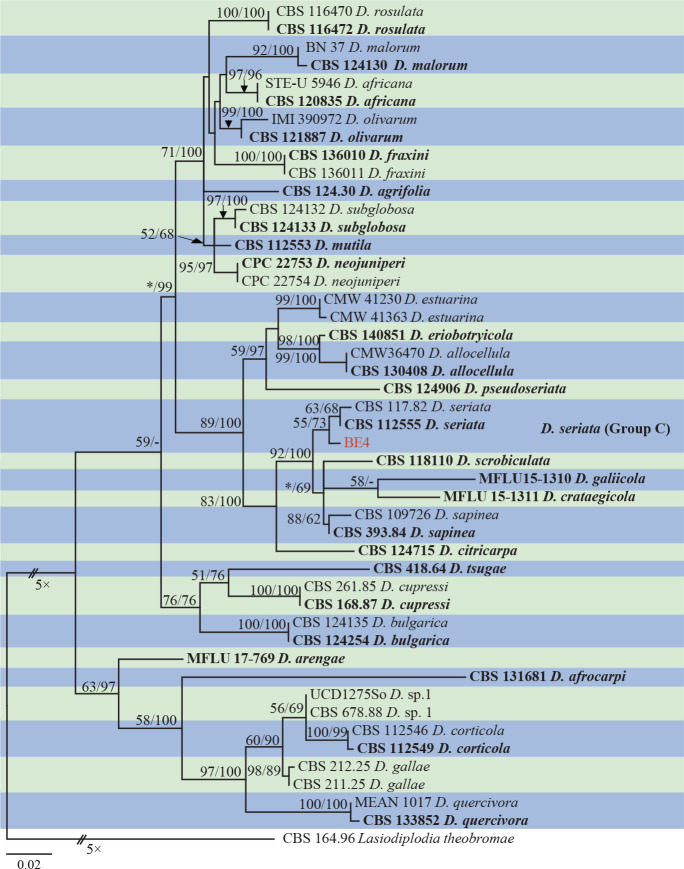
Isolates clustered into two phylogenetic groups (Group A and B) for the individual genes (ITS, tef1, tub2, rpb2), as well as the combined gene dataset (Fig. 2, S1a–d). For the ITS sequences, isolates BE3, BE78, BE85 and BE86 (Group A) grouped with several species, while isolates BE1, BE2, BE63 and BE66 (Group B) formed a clade distinct from the other species (Fig. S1a). For the tef1 and rpb2 and combined ITS/tef1/tub2/rpb2 datasets, isolates in Group A were closely related to B. fabicerciana, and isolates in Group B were most closely related to B. dothidea (Fig. 2, S1b, d). Therefore, isolates in Group A were identified as B. fabicerciana, and isolates in Group B were identified as B. dothidea.
Fig. 2.
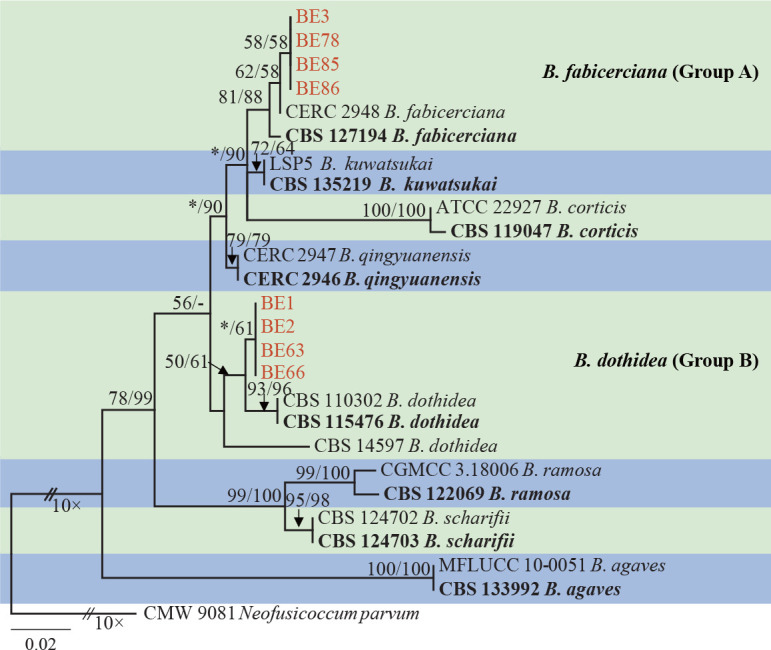
Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1 and tub2 sequence alignments of Botryosphaeria. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Neofusicoccum parvum (CMW 9081).
Species of Diplodia
Isolate BE4 (Group C) clustered more closely to D. seriata and D. galiicola in the ITS datasets (Fig. S2a), while more closely to D. seriata and D. sapinea in the tef1 datasets (Fig. S2b). For the tub2 and rpb2 sequences, isolate BE4 clustered with D. seriata (Fig. S2c–d). The analyses of the combined ITS, tef1, tub2 and rpb2 sequences demonstrated that isolate BE4 was most closely related to D. seriata (Fig. 3).
Fig. 3.
Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1 and tub2 sequence alignments of Diplodia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Lasiodiplodia theobromae (CBS 164. 96).
Species of Dothiorella
Eight isolates clustered in three clades (Group D–F). Isolate BE17 in Group D grouped with Do. alpina and Do. magnoliae based on the ITS sequences (Fig. S3a). For the tef1 sequences, isolate BE17 formed an independent lineage close to Do. alpina and Do. acericola (Fig. S3b). For the tub2 and rpb2 sequences, isolate BE17 formed an independent lineage (Fig. S3c, d). For the combined ITS, tef1 and tub2 sequences, isolate BE17 clustered with Do. alpina (Fig. 4). For Group E, the sequence analyses of ITS, tef1, tub2 and ITS/tef1/tub2 sequences showed that isolates BE16 and BE74 clustered in the same clade (ITS, tub2) or close (tef1, ITS/tef1/tub2) to Do. plurivora (rpb2 sequences are not available for Do. plurivora) (Fig. 4, S3a–c). Thus, isolates in Group D were identified as Do. alpina, while those in Group E were identified as Do. plurivora.
Fig. 4.
Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1 and tub2 sequence alignments of Dothiorella. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Neofusicoccum parva (CMW 9081).
Isolates in Group F (BE5, BE7, BE8, BE9, BE71) clustered close to Do. striata based on the ITS and tef1 datasets (Fig. S3a–b), but formed independent clades that were separated from Do. striata with high bootstrap values in the tub2, rpb2 and combined ITS/tef1/tub2 datasets (tub2, ML/MP = 92 % / 94 %; rpb2, ML/MP = 100 % / 100 %; ITS/tef1/tub2, ML/MP = 84 % / 97 %) (Fig. 4, S3c–d). Thus, isolates in Group F were considered as an undescribed species in Dothiorella.
Species of Lasiodiplodia
Isolates resided in nine groups (Group G–O) based on the tef1 and combined ITS/tef1/tub2/rpb2 datasets (Fig. 5, S4b). Isolates in Group G (BE27, BE30, BE36, BE41, BE42, BE100) were closest to L. iraniensis and various other species based on the ITS, tub2 and rpb2 datasets (Fig. S4a, c–d). For the tef1 and combined ITS/tef1/tub2/rpb2 sequences, the six isolates were closest to L. iraniensis (Fig. 5, S4b). Thus, the six isolates in Group G were identified as L. iraniensis.
Fig. 5.
Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1, tub2 and rpb2 sequence alignments of Lasiodiplodia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
For isolates in Group H (BE20, BE21, BE25) and Group J (BE32, BE80, BE87, BE88), the analyses of the ITS, tef1, tub2 and rpb2 sequences indicated that isolates in Group H clustered into the same (ITS, tub2, rpb2) clade or close (tef1) to L. theobromae, while isolates in Group J clustered into the same (rpb2) clade or close (ITS, tef1) to L. microconidia (Fig. S4a–d). The combined ITS/tef1/tub2/rpb2 datasets showed that isolates in Group H were more closely related to L. theobromae, while isolates in Group J clustered with L. microconidia (Fig. 5).
Isolates in Group I (BE28, BE34, BE40, BE51) and Group L (BE31, BE59) clustered with various Lasiodiplodia species based on ITS, tub2 and rpb2 datasets (Fig. S4a, c–d), but formed independent clades based on the tef1 and combined ITS/tef1/tub2/rpb2 trees, with high bootstrap values (Group I: tef1, ML/MP = 99 % / 100 %; ITS/tef1/tub2/rpb2, ML/MP = 99 % / 100 %; Group L: tef1, ML/MP = 98 % / 99 %; ITS/tef1/tub2/rpb2, ML/MP = 99 % / 100 %) (Fig. 5, S4b). Therefore, isolates in Group I and Group L were considered to represent two novel species.
Isolates in Group K (BE10, BE11, BE12, BE19, BE26, BE37) clustered with L. pseudotheobromae and various other species based on the ITS and tub2 datasets (Fig. S4a, c). For the analyses of tef1, rpb2 and the combined ITS/tef1/tub2/rpb2 datasets, the six isolates resided in the same clade with L. pseudotheobromae (Fig. 5, S4b, d). Therefore, the isolates in Group K were treated as L. pseudotheobromae. Isolates in Group M (BE13, BE38, BE45, BE89, BE102, BE104) clustered into the same (ITS, rpb2) clade or close (tef1, rpb2) to L. citricola (Fig. S4a–d). The combined ITS/tef1/tub2/rpb2 datasets showed that isolates in Group M clustered with L. citricola (Fig. 5).
Isolate BE44 in Group N resided in a clade with L. citricola based on the analyses of the ITS and tub2 datasets (Fig. S4a, c). For the tef1 datasets, BE44 formed an independent lineage phylogenetically close to L. aquilariae (Fig. S4b). For the rpb2 datasets, BE44 grouped with various other species (Fig. S4d). The analyses of the combined ITS/tef1/tub2/rpb2 datasets indicated that isolate BE44 formed an independent lineage that was distinguished from other known phylogenetically related species (Fig. 5). Isolates in Group O (BE33, BE50) grouped together with various other Lasiodiplodia species in the ITS, tub2 and rpb2 trees (Fig. S4a, c–d). For the tef1 datasets, the two isolates formed two independent clades but with low bootstrap support based on the tef1 in the ML analyses, while they clustered together with Group L in the MP analyses (data not shown). The analyses of the combined ITS/tef1/tub2/rpb2 datasets indicated that isolates in Group O formed an independent clade with high support bootstrap values (ML/MP = 74 % / 88 %) (Fig. 5). Consequently, isolates in Group N and Group O were identified as two new species of Lasiodiplodia.
Species of Neodeightonia
Isolate BE14 (Group P) grouped with N. subglobosa in one clade with high support value on the basis of the phylogenetic analyses for the ITS, tef1, tub2 and ITS/tef1/tub2 datasets (Fig. 6, S5a–d).
Fig. 6.
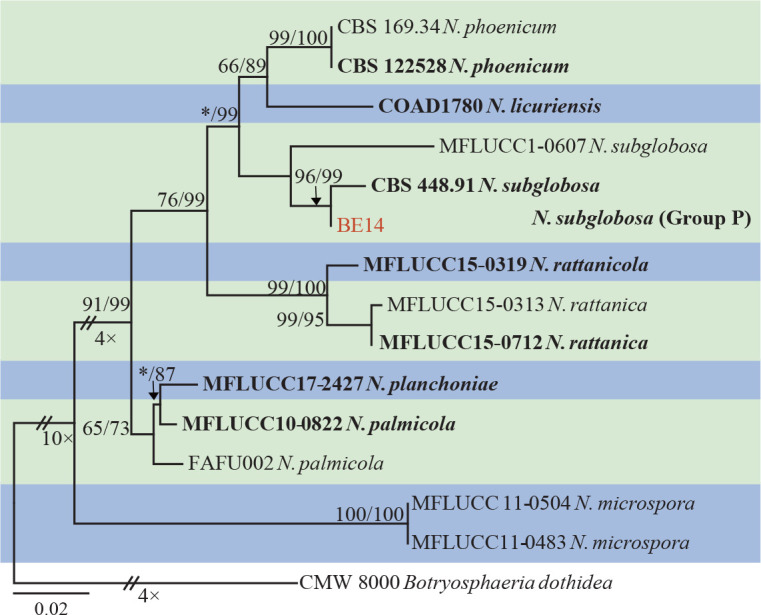
Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1 and tub2 sequence alignments of Neodeightonia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
Species of Neofusicoccum
Phylogenetic analyses of ITS and tef1 consistently indicated that isolate BE15 (Group Q) resided in one phylogenetic clade with Ne. parvum (Fig. S6a–b), and with Ne. pennatisporum based on tub2 (Fig. S6c). Isolate BE15 clustered with Ne. parvum, Ne. cryptoaustrale and Ne. mangiferae based on rpb2 (Fig. S6d). The phylogeny based on the combined ITS/tef1/tub2/rpb2 sequences indicated that isolate BE15 was closely related to Ne. parvum (Fig. 7).
Fig. 7.
Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1, tub2 and rpb2 sequence alignments of Neofusicoccum. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Dothiorella viticola (CBS 117009).
Species of Sphaeropsis
Isolate BE18 (Group R) formed an independent lineage that was distinct from any known species of Sphaeropsis based on the phylogenetic analyses for ITS, tef1, tub2, rpb2 and the combined four gene datasets. The bootstrap values associated with the other species were higher than 50 % in ITS, tef1, rpb2 and the combined datasets (ITS, ML/MP = 68 % / 64 %; tef1, MP = 93 %; rpb2, MP = 100 %; ITS/tef1/tub2/rpb2, ML/MP = 50 % / 55 %) (Fig. 8, S7a–b, d). Therefore, isolate BE18 was treated as a novel species of Sphaeropsis.
Fig. 8.

Phylogenetic tree generated by maximum likelihood analyses based on the combined ITS, tef1, tub2 and rpb2 sequence alignments of Sphaeropsis. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
Morphology and taxonomy
The selected isolates for morphological studies produced pycnidia on PNA within 4–6 wk. No sexual structures were observed in this study. Based on DNA sequences and morphology, 18 species belonging to seven genera were identified. Of these, Botryosphaeria dothidea, B. fabicerciana, Diplodia seriata, Dothiorella alpina, Do. plurivora, Lasiodiplodia citricola, L. iraniensis, L. microconidia, L. pseudotheobromae, L. theobromae, Neodeightonia subglobosa and Neofusicoccum parvum are known species. The remaining six species are described below.
Dothiorella citrimurcotticola X.E. Xiao, P.W. Crous & H.Y. Li, sp. nov. — MycoBank MB 840681; Fig. 9
Fig. 9.
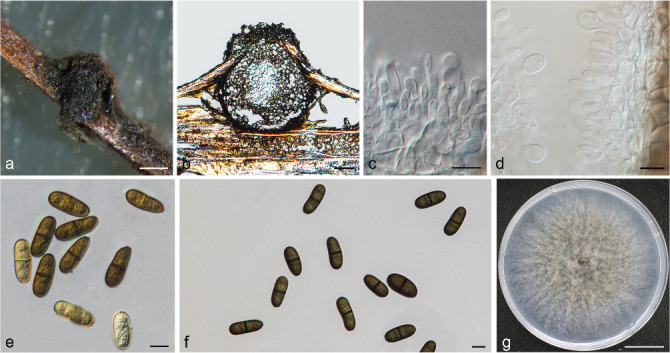
Dothiorella citrimurcotticola. a. Conidioma formed on PNA; b. section view of conidioma; c–d. conidiogenous cells and developing conidia; e–f. conidia; g. colony growing on PDA after 3 d. — Scale bars: a = 100 μm; b = 50 μm; c–f = 10 μm; g = 2.1 cm.
Etymology. Referring to the citrus host (Murcott) which it was isolated.
Typus. CHINA, Chongqing Municipality, Wanzhou City, from a twig of Murcott (C. reticulata × C. sinensis), 23 Mar. 2019, H.Y. Li & X.E. Xiao, conidiomata induced on PNA (holotype ZJUE H-0008, culture ex-type CGMCC 3.20394 = BE8).
Sexual morph unknown. Conidiomata pycnidial, produced on PNA within 2–4 wk, dark brown to black, up to 823 μm diam, globose, superficial or semi-immersed, unilocular, thick-walled. Conidiophores absent. Conidiogenous cells cylindrical to fusiform or lageniform, hyaline, thin-walled, smooth, 4.5–10.5 × 2–5 μm. Conidia subcylindrical to ellipsoid or ovoid, initially hyaline, thin-walled, aseptate, becoming brown, thick-walled, 1-septate, externally smooth, internally verrucose, apex rounded, base truncate or rounded, (21.5–)23–25.5(–27) × (8.5–)9.5–11(–14) μm (av. = 24.4 × 10.3 μm, n = 100; L/W ratio = 2.4) (Table 4).
Table 4.
Conidial measurements of Botryosphaeriaceae species.
| Species1 | Conidia |
Paraphyses |
Reference | |||
|---|---|---|---|---|---|---|
| Conidial size (μm) (L × W)2 | Mean (μm) (L × W)3 | L/W4 | long (μm)5 | wide (μm)6 | ||
| Do. citrimurcotticola | (21.5–)23–25.5(–27)× (8.5–)9.5–11(–14) | 24.4×10.3 | 2.4 | – | – | This study |
| Do. striata | (21–)23–26(–29.4) × (8.9–)9–12(–15.1) | 25.1 × 10.7 | 2.4 | – | – | Abdollahzadeh et al. (2014) |
| Do. uruguayensis | (17–)22–22.5(–26.5) × (7–)9–9.5(–12) | 22 × 9.25 | 2.4 | – | – | Pérez et al. (2010) |
| Lasiodiplodia acaciae | (21.5–)25–29.5(–31) × (11–)12–14(–15) | 27.3 × 12.9 | 2.1 | 69 | 2–5 | Zhang et al. (2021) |
| L. aquilariae | (23–)25–28(–29) × 12–16 | 26.9 × 14.1 | 1.8 | 100 | 3 | Wang et al. (2019) |
| L. cinnamomi | (17.5–)18.7–21.1(–22.4) × (11.5–)12.7–14.1(–15.5) | 19.9 × 13.4 | 1.5 | 106 | 3–4 | Jiang et al. (2018) |
| L. citricola | (20–)22–27(–31) × (10.9–)12–17(–19) | 24.5 × 15.4 | 1.6 | 125 | 3–4 | Abdollahzadeh et al. (2010) |
| L. guilinensis | (23–)28–31(–33.5)× (13.5–)15–16.5(–17) | 29.6 ×15.7 | 1.9 | 75 | 2–5 | This study |
| L. huangyanensis | (21–)28–32.5(–34) × (13–)14–16(–17) | 30.1× 15 | 2 | 82 | 3–4 | This study |
| L. linhaiensis | (24.5–)27–30(–32)× (12.5–)13.5–15(–16) | 28.5 × 14.2 | 2 | 80 | 2–6 | This study |
| L. microconidia | (18–)19–22(–23) × 10–15 | 20.8 × 13.2 | 1.5 | 90 | 3 | Wang et al. (2019) |
| L. ponkanicola | (16–)23.5–27.5(–28.5)× (11–)13–14.5(–15.5) | 25.4 × 13.7 | 1.9 | 87 | 2–5 | This study |
| Sphaeropsis citrigena | (27–)28–33(–34) × (14.5–)15–18.5(–19) | 30.5 × 16.8 | 1.8 | 25 | 3–5 | Phillips et al. (2008) |
| S. linhaiensis | (26.5–)28.5–35(–38)× (11.5–)14–18(–19.5) | 31.6 × 15.9 | 2 | 27 | 1–5 | This study |
1 Isolates and measurements in bold were examined in this study.
2 Minimum – (average – standard deviation) – (average + standard deviation) – maximum or minimum – maximum, L × W = length × width.
3 L × W = average length × average width.
4 L/W = average length/average width.
5 Maximum.
6 Maximum or minimum – maximum.
Culture characteristics — Colonies on PDA have abundant aerial mycelia, initially leaden grey in the centre, becoming pale mouse grey at the surface and greenish olivaceous to dull green at the reverse. Colonies cover the 90 mm plates after 3 d at the optimum temperature of 25 °C. No growth was observed at 40 °C. After 3 d, colonies at 5 °C, 10 °C, 15 °C, 20 °C, 30 °C and 35 °C reach 11 mm, 11 mm, 59 mm, 79 mm, 42 mm and 12 mm, respectively.
Additional materials examined. CHINA, Zhejiang Province, Yongquan Town, from a twig of C. unshiu, Aug. 2018, H.Y. Li, conidiomata induced on PNA (holotype ZJUE H-0005, culture ex-type CGMCC 3.20392 = BE5); Chongqing Municipality, Wanzhou City, from a twig of Murcott (C. reticulata × C. sinensis), 23 Mar. 2019, H.Y. Li & X.E. Xiao, conidiomata induced on PNA (ZJUE H-0009, culture CGMCC 3.20395 = BE9); Zhejiang Province, Quzhou City, from a twig of C. maxima, 27 Apr. 2019, H.Y. Li, conidiomata induced on PNA (ZJUE H-0007, culture CGMCC 3.20393 = BE7).
Notes — Phylogenetically, Do. citrimurcotticola is closely re-lated to Do. striata and Do. uruguayensis, but morphologically it can be distinguished based on their average conidial dimensions. Conidia of Do. citrimurcotticola (av. 24.4 × 10.3; L/W = 2.4) are larger than Do. uruguayensis (av. 22 × 9.25; L/W = 2.4) (Pérez et al. 2010) but smaller than Do. striata (av. 25.1 × 10.7; L/W = 2.4) (Abdollahzadeh et al. 2014) (Table 4). Moreover, Do. citrimurcotticola differs from these species based on nucleotide differences in ITS (Do. striata: 5 bp, Do. uruguayensis: 1 bp), tef1 (Do. striata: 5 bp, Do. uruguayensis: 15 bp and including three gaps), tub2 (Do. striata: 4 bp, Do. uruguayensis: 6 bp) and rpb2 loci (Do. striata: 8 bp).
Lasiodiplodia guilinensis X.E. Xiao, P.W. Crous & H.Y. Li, sp. nov. — MycoBank MB 840682; Fig. 10
Fig. 10.
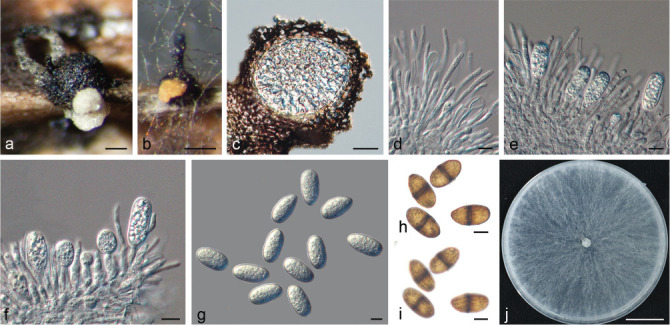
Lasiodiplodia guilinensis. a–b. Pale yellow to saffron yellow conidial mass released from conidiomata formed on PNA; c. section view of conidioma; d. paraphyses; e–f. conidia developing on conidiogenous cells between paraphyses; g. hyaline, aseptate conidia; h–i. dark-brown, 1-septate conidia at two different focal planes to show the longitudinal striations; j. colony growing on PDA after 2 d. — Scale bars: a–b = 200 μm; c = 50 μm; d–i = 10 μm; j = 2 cm.
Etymology. Referring to the city, Guilin, where it was collected.
Typus. CHINA, Guangxi Province, Guilin City, from a twig of C. sinensis cv. Valencia, 26 Mar. 2019, H.Y. Li & X.E. Xiao, conidiomata induced on PNA (holotype ZJUE H-0031, culture ex-type CGMCC 3.20378 = BE31).
Sexual morph unknown. Conidiomata stromatic, produced on PNA within 2–4 wk, superficial or semi-immersed, dark brown to black, up to 2 mm diam, solitary or aggregated, unilocular, covered by dense mycelium, globose, thick-walled, often releasing pale yellow to saffron yellow conidial tendrils or mass. Paraphyses hyaline, cylindrical, septate, unbranched, ends rounded, up to 75 μm long, 2–5 μm wide, formed among conidiogenous cells. Conidiophores absent. Conidiogenous cells holoblastic, hyaline, smooth, thin-walled, cylindrical, 8–54 × 3–9 μm. Conidia initially hyaline, aseptate, ellipsoid to ovoid, thin-walled with granular content, rounded at apex, base round or truncate, becoming dark brown, 1-septate with longitudinal striations, (23–)28–31(–33.5) × (13.5–)15–16.5(–17) μm (av. = 29.6 × 15.7 μm, n = 100; L/W ratio = 1.9) (Table 4).
Culture characteristics — Colonies on PDA with moderately dense aerial mycelium, initially white to smoke grey, turning grey olivaceous on the surface and greenish grey in reverse, becoming dark slate-blue with age. Colonies cover the 90 mm plates after 2 d in the dark at the optimum temperature of 25–30 °C. No growth was observed at 5 °C. After 2 d, colonies at 10 °C, 15 °C, 20 °C, 35 °C and 40 °C reach 11 mm, 32 mm, 64 mm, 72 mm and 12 mm, respectively.
Additional material examined. CHINA, Zhejiang Province, Yongquan Town, from the branch of C. unshiu, 26 Sept. 2017, H.Y. Li, conidiomata induced on PNA (ZJUE H-0059, culture CGMCC 3.20379 = BE59).
Notes — Phylogenetically, L. guilinensis is closely related to L. aquilariae and L. citricola, but morphologically it can be separated from these species based on average conidial dimensions and length of its paraphyses. Conidia of L. guilinensis (av. 29.6 × 15.7; L/W = 1.9) are larger than those of L. aquilariae (av. 26.9 × 14.1; L/W = 1.8) (Wang et al. 2019) and L. citricola (av. 24.5 × 15.4; L/W = 1.6) (Abdollahzadeh et al. 2010). In terms of paraphyses, those of L. guilinensis (up to 75 μm long) are shorter than L. aquilariae (up to 100 μm long) (Wang et al. 2019) and L. citricola (up to 125 μm long) (Abdollahzadeh et al. 2010) (Table 4). Furthermore, L. guilinensis differs from these species by nucleotide differences in ITS (L. aquilariae: 6 bp, L. citricola: 3 bp), tef1 (L. aquilariae: 16 bp, L. citricola: 14 bp), tub2 (L. citricola: 3 bp) and rpb2 loci (L. aquilariae: 3 bp).
Lasiodiplodia huangyanensis X.E. Xiao, P.W. Crous & H.Y. Li, sp. nov. — MycoBank MB 840683; Fig. 11
Fig. 11.
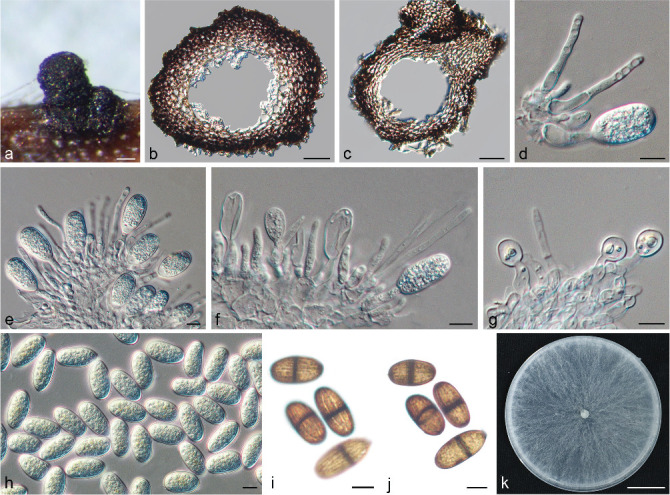
Lasiodiplodia huangyanensis. a. Conidioma formed on PNA; b–c. section view of conidiomata; d–g. conidia developing on conidiogenous cells between paraphyses; h. hyaline, aseptate conidia; i–j. dark-brown, 1-septate conidia at two different focal planes to show the longitudinal striations; k. colony growing on PDA after 2 d. — Scale bars: a = 200 μm; b–c = 50 μm; d–j = 10 μm; k = 2.1 cm.
Etymology. Referring to the district, Huangyan, where it was collected.
Typus. CHINA, Zhejiang Province, Huangyan District, from a twig of C. reti-culata cv. Succosa, 22 Jan. 2019, X.E. Xiao & Q.B. Huang, conidiomata induced on PNA (holotype ZJUE H-0033, culture ex-type CGMCC 3.20380 = BE33).
Sexual morph unknown. Conidiomata stromatic, formed on PNA within 2–4 wk, superficial or semi-immersed, dark brown to black, up to 1.5 mm diam, solitary or aggregated, unilocular, covered by dense mycelium, globose, thick-walled, often releasing in pale yellow to saffron yellow conidial tendrils or mass. Paraphyses hyaline, cylindrical, septate, unbranched, ends rounded, up to 82 μm long, 3–4 μm wide, formed among conidiogenous cells. Conidiophores absent. Conidiogenous cells holoblastic, hyaline, smooth, thin-walled, cylindrical, 8–35 × 3.5–7 μm. Conidia initially hyaline, aseptate, ellipsoid to ovoid, thin-walled with granular content, rounded at apex, base round or truncate, becoming dark brown, 1-septate with longitudinal striations, (21–)28–32.5(–34) × (13–)14–16(–17) μm (av. = 30.1 × 15 μm, n = 100; L/W ratio = 2) (Table 4).
Culture characteristics — Colonies on PDA with moderately dense aerial mycelium, initially white to smoke grey, turning grey olivaceous on the surface and greenish grey in reverse, becoming dark slate-blue with age. Colonies cover the 90 mm plates after 2 d in the dark at the optimum temperature of 25–30 °C. No growth was observed at 5 °C. After 2 d, colonies at 10 °C, 15 °C, 20 °C, 35 °C and 40 °C reach 10 mm, 24 mm, 53 mm, 28 mm and 12 mm, respectively.
Additional material examined. CHINA, Zhejiang Province, Linhai City, from the branch of C. unshiu, 13 Dec. 2018, W.L. Li, conidiomata induced on PNA (ZJUE H-0050, culture CGMCC 3.20381 = BE50).
Notes — Phylogenetically, L. huangyanensis is closely related to L. cinnamomi and L. ponkanicola. Morphologically, however, it can be distinguished based on the average conidial dimensions and length of its paraphyses. Conidia of L. huangyanensis (av. 30.1 × 15; L/W = 2) are larger than L. cinnamomi (av. 19.9 × 13.4; L/W = 1.5) (Jiang et al. 2018) and L. ponkanicola (av. 25.4 × 13.7; L/W = 1.9) (this study). Moreover, the paraphyses of L. huangyanensis (up to 82 μm long) are shorter than those of L. cinnamomi (up to 106 μm long) (Jiang et al. 2018) and L. ponkanicola (up to 87 μm long) (this study) (Table 4). Furthermore, L. huangyanensis differs from these species by nucleotide differences in ITS (L. ponkanicola: 3 bp), tef1 (L. cinnamomi: 7 bp, L. ponkanicola: 10 bp), tub2 (L. ponkanicola: 3 bp) and rpb2 loci (L. cinnamomi: 9 bp).
Lasiodiplodia linhaiensis X.E. Xiao, P.W. Crous & H.Y. Li, sp. nov. — MycoBank MB 840684; Fig. 12
Fig. 12.
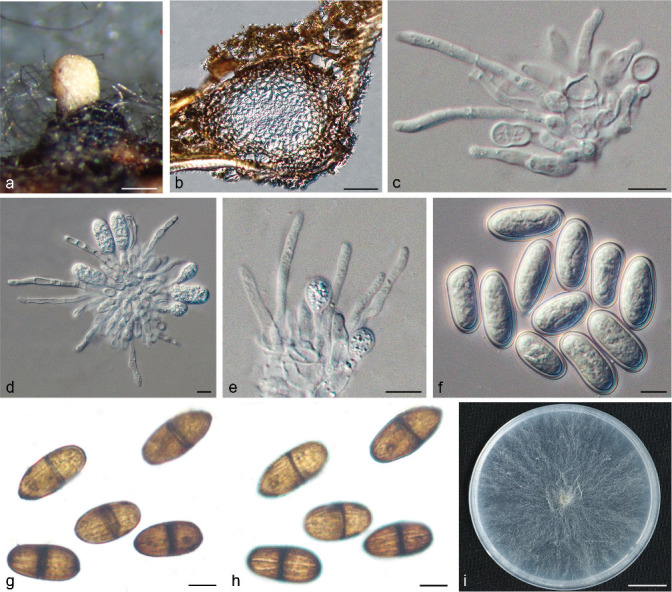
Lasiodiplodia linhaiensis. a. Pale yellow conidial mass released from conidioma formed on PNA; b. section view of conidioma; c–e. conidia developing on conidiogenous; f. hyaline, aseptate conidia; g–h. dark-brown, 1-septate conidia at two different focal planes to show the longitudinal striations; i. colony growing on PDA after 2 d. — Scale bars: a = 200 μm; b = 50 μm; c–h = 10 μm; i = 1.8 cm.
Etymology. Referring to the city, Linhai, where it was collected.
Typus. CHINA, Zhejiang Province, Linhai City, from the branch of C. unshiu, 14 Dec. 2018, W.L. Li, conidiomata induced on PNA (holotype ZJUE H-0051, culture ex-type CGMCC 3.20386 = BE51).
Sexual morph unknown. Conidiomata stromatic, produced on PNA within 2–4 wk, superficial or semi-immersed, dark brown to black, up to 950 μm diam, solitary or aggregated, unilocular, covered by dense mycelium, globose, thick-walled, often releasing in pale-yellow to saffron-yellow conidial tendrils or mass. Paraphyses hyaline, cylindrical, septate, unbranched, ends rounded, up to 80 μm long, 2–6 μm wide, formed among conidiogenous cells. Conidiophores absent. Conidiogenous cells holoblastic, hyaline, smooth, thin-walled, cylindrical, 7.5–22.5 × 3–5.5 μm. Conidia initially hyaline, aseptate, ellipsoid to ovoid, thin-walled with granular content, rounded at apex, base round or truncate, becoming dark brown, 1-septate with longitudinal striations, (24.5–)27–30(–32) × (12.5–)13.5–15(–16) μm (av. = 28.5 × 14.2 μm, n = 100; L/W ratio = 2) (Table 4).
Culture characteristics — Colonies on PDA with slightly dense aerial mycelium, initially white to smoke grey, turning grey olivaceous on the surface and greenish grey in reverse, becoming dark slate-blue with age. Colonies cover the 90 mm plates after 2 d in the dark at the optimum temperature of 25–30 °C. No growth was observed at 5 °C. After 2 d, colonies at 10 °C, 15 °C, 20 °C, 35 °C and 40 °C reach 12 mm, 18 mm, 42 mm, 22 mm and 11 mm, respectively. Isolates produced a pink pigment in PDA cultures at 35 °C.
Additional materials examined. CHINA, Guangxi Province, Guilin City, from the dieback of C. sinensis cv. Valencia, 26 Mar. 2019, H.Y. Li & X.E. Xiao, conidiomata induced on PNA (ZJUE H-0028, culture CGMCC 3.20383 = BE 28); Zhejiang Province, Taizhou City, from the branch of C. reticulata cv. Succosa, 22 Jan. 2019, X.E. Xiao & Q.B. Huang, conidiomata induced on PNA (ZJUE H-0034, culture CGMCC 3.20384 = BE34); Zhejiang Province, Quzhou City, from the trunk of C. reticulata vs Ponkan, 23 Mar. 2018, H.K. Wang & X.E. Xiao, conidiomata induced on PNA (ZJUE H-0040, culture CGMCC3.20385 = BE40).
Notes — Phylogenetically, L. linhaiensis is closely related to L. acaciae, but can be separated from that species based on the length of its paraphyses. Paraphyses of L. linhaiensis (up to 80 μm long) are longer than those of L. acaciae (up to 69 μm long) (Zhang et al. 2021) (Table 4). Moreover, L. linhaiensis differs from L. acaciae by nucleotide differences in ITS (1 bp), tef1 (5 bp) and rpb2 loci (4 bp).
Lasiodiplodia ponkanicola X.E. Xiao, P.W. Crous & H.Y. Li, sp. nov. — MycoBank MB 840685; Fig. 13
Fig. 13.
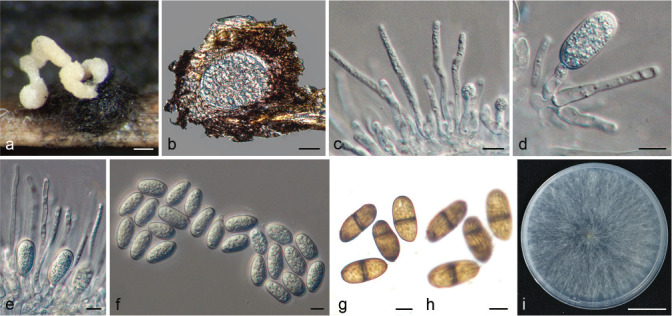
Lasiodiplodia ponkanicola. a. Pale yellow conidial mass oozing from conidioma formed on PNA; b. section view of conidioma; c–e. conidia developing on conidiogenous; f. hyaline, aseptate conidia; g–h. dark-brown, 1-septate conidia at two different focal planes to show the longitudinal striations; i. colony growing on PDA after 7 d. — Scale bars: a = 200 μm; b = 50 μm; c–h = 10 μm; i = 2.3 cm.
Etymology. Referring to the host variety (Ponkan) from which the fungus was isolated.
Typus. CHINA, Zhejiang Province, Quzhou City, from the trunk of C. reticulata cv. Ponkan, 23 Mar. 2018, H.K. Wang & X.E. Xiao, conidiomata induced on PNA (holotype ZJUE H-0044, culture ex-type CGMCC 3.20388 = BE44).
Sexual morph unknown. Conidiomata stromatic, produced on PNA within 2–4 wk, superficial or semi-immersed, dark brown to black, up to 1 mm diam, solitary or aggregated, unilocular, covered by mycelium, globose, thick-walled, often releasing pale yellow to saffron yellow conidial tendrils or mass. Paraphyses hyaline, cylindrical, septate, not branched, ends rounded, up to 87 μm long, 2–5 μm wide, formed among conidiogenous cells. Conidiophores absent. Conidiogenous cells holoblastic, hyaline, smooth, thin-walled, cylindrical, 8.5–40 × 2.5–9 μm. Conidia initially hyaline, aseptate, ellipsoid to ovoid, thin-walled with granular content, rounded at apex, base round or truncate, becoming pigmented, 1-septate with longitudinal striations, (16–)23.5–27.5(–28.5) × (11)–13–14.5(–15.5) μm (av. = 25.4 × 13.7 μm, n = 100; L/W ratio = 1.9) (Table 4).
Culture characteristics — Colonies on PDA with moderately dense aerial mycelium, initially white to smoke grey, turning grey olivaceous on the surface and greenish grey in reverse, becoming dark slate-blue with age. Colonies cover the 90 mm plates after 2 d in the dark at the optimum temperature of 25–30 °C. No growth was observed at 5 °C. After 2 d, colonies at 10 °C, 15 °C, 20 °C, 35 °C and 40 °C reach 9 mm, 19 mm, 43 mm, 71 mm and 26 mm, respectively.
Notes — Phylogenetically, L. ponkanicola is closely related to L. aquilariae (based on tef1), L. citricola (based on ITS and tub2), L. cinnamomic and L. huangyanensis (based on ITS/tef1/tub2/rpb2), but morphologically they can be separated on their average conidial dimensions and length of their paraphyses. Conidia of L. ponkanicola (av. 25.4 × 13.7; L/W = 1.9) are longer than L. cinnamomi (av. 19.9 × 13.4; L/W = 1.5) (Jiang et al. 2018) and L. citricola (av. 24.5 × 15.4; L/W = 1.6) (Abdollahzadeh et al. 2010), but shorter and narrower than L. aquilariae (av. 26.9 × 14.1; L/W = 1.8) (Wang et al. 2019) and L. huangyanensis (av. 30.1 × 15; L/W = 2) (this study). Moreover, the paraphyses of L. ponkanicola (up to 87 μm long) are longer than L. huangyanensis (up to 82 μm long) (this study) but shorter than L. aquilariae (up to 100 μm long) (Wang et al. 2019), L. cinnamomi (up to 106 μm long) (Jiang et al. 2018) and L. citricola (up to 125 μm long) (Abdollahzadeh et al. 2010) (Table 4). Furthermore, L. ponkanicola differs from these species by nucleotide differences in ITS (L. aquilariae: 3 bp, L. cinnamomi: 3 bp, L. huangyanensis: 3 bp), tef1 (L. aquilariae: 2 bp, L. cinnamomi: 6 bp, L. citricola: 7 bp, L. huangyanensis: 10 bp), tub2 (L. cinnamomi: 3 bp, L. huangyanensis: 3 bp) and rpb2 loci (L. aquilariae: 15 bp, L. cinnamomi: 9 bp, L. citricola: 15 bp). In view of the fact that isolate BE44 clustered with different species at different loci, it was considered that it may be a hybrid, which requires further research.
Sphaeropsis linhaiensis X.E. Xiao, P.W. Crous & H.Y. Li, sp. nov. — MycoBank MB 840686; Fig. 14
Fig. 14.
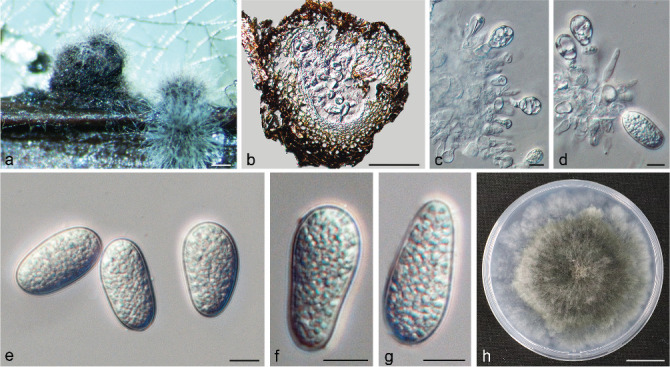
Sphaeropsis linhaiensis. a. Conidiomata formed on PNA; b. section view of conidioma; c–d. conidia developing on conidiogenous cells; e–g. conidia; h. colony growing on PDA after 7 d. — Scale bars: a = 200 μm; b = 40 μm; c–g = 10 μm; h = 1.9 cm.
Etymology. Named after the Linhai City where it was isolated for the first time.
Typus. CHINA, Zhejiang Province, Linhai City, from a twig of C. unshiu, 2 June 2018, H.Y. Li, conidiomata induced on PNA (holotype ZJUE H-0018, culture ex-type CGMCC 3.20382 = BE18).
Sexual morph unknown. Conidiomata pycnidial, produced on PNA within 2–4 wk, dark brown to black, unilocular, up to 880 μm diam, immersed in the needle tissue, globose to subglobose, ostiolate, wall composed of several layers of dark brown textura angularis. Paraphyses hyaline, aseptate, up to 27 μm long, 1–5 μm wide. Conidiogenous cells hyaline, discrete, proliferating internally to form periclinal thickenings, 4.5–11 × 3–8 μm. Poor sporulation, conidia hyaline, aseptate, guttulate, oval to broadly ellipsoid, apex obtuse, base obtuse or truncate, moderately thick-walled, (26.5–)28.5–35(–38) × (11.5–)14–18(–19.5) μm (av. = 31.6 × 15.9 μm, n = 30; L/W ratio = 2) (Table 4).
Culture characteristics — Colonies on PDA initially white, turning olivaceous grey gradually on the surface and leaden grey at the reverse. Colonies cover the 90 mm plates after 6 d at the optimum temperature of 25 °C. No growth was observed at 5 °C, 35 °C and 40 °C. After 6 d, colonies at 10 °C, 15 °C, 20 °C and 30 °C reach 31 mm, 64 mm, 80 mm and 19 mm, respectively.
Notes — Phylogenetically, S. linhaiensis is closely related to S. citrigena, but can be distinguished from S. citrigena based on its average conidial dimensions. Conidia of S. linhaiensis (av. 31.6 × 15.9; L/W = 2) are longer than those of S. citrigena (av. 30.5 × 16.8; L/W = 1.8) (Phillips et al. 2008). Moreover, S. linhaiensis differs from S. citrigena by nucleotide differences in ITS (6 bp), tef1 (10 bp) and tub2 (7 bp).
Prevalence of Botryosphaeriaceae species
In total, 18 species of Botryosphaeriaceae were identified from citrus branch diseases in the Chongqing, Fujian, Guangdong, Guangxi, Hunan, Jiangxi, Shaanxi, Shanghai and Zhejiang provinces of China (Fig. 15a). These species include Botryosphaeria dothidea (32 isolates, 28.8 %), B. fabicerciana (4 isolates, 3.6 %), Diplodia seriata (1 isolate, 0.9 %), Dothiorella alpina (1 isolate, 0.9 %), Do. citrimurcotticola (6 isolates, 5.4 %), Do. plurivora (2 isolates, 1.8 %), Lasiodiplodia citricola (8 isolates, 7.2 %), L. guilinensis (2 isolates, 1.8 %), L. huangyanensis (3 isolates, 2.7 %), L. iraniensis (6 isolates, 5.4 %), L. linhaiensis (7 isolates, 6.3 %), L. microconidia (5 isolates, 4.5 %), L. ponkanicola (1 isolate, 0.9 %), L. pseudotheobromae (26 isolates, 23.4 %), L. theobromae (4 isolates, 3.6 %), Neodeightonia subglobosa (1 isolate, 0.9 %), Neofusicoccum parvum (1 isolate, 0.9 %) and Sphaeropsis linhaiensis (1 isolate, 0.9 %) (Fig. 15b). Of these 18 species, B. dothidea (28.8 %) was most commonly isolated, followed by L. pseudotheobromae (23.4 %), L. citricola (8 isolates, 7.2 %) and L. linhaiensis (7 isolates, 6.3 %) (Fig. 15a). In terms of the source of isolates, Zhejiang Province has the most isolates and the most diversity in species, most likely because of the more intensive sampling in this province (Fig. S8). In addition, L. pseudotheobromae was the most widely distributed species found in six provinces, including Chongqing, Fujian, Guangdong, Guangxi, Shaanxi and Zhejiang (Fig. 15b, S8).
Fig. 15.
The prevalence of Botryosphaeriaceae species isolated from citrus. a. Distribution of Botryosphaeriaceae species in China; b. overall isolation rate (%) of Botryosphaeriaceae species. Different species are represented by numbers with different colours.
Pathogenicity tests
For in vitro inoculation, all 31 isolates inoculated were pathogenic to C. reticulata shoots with visible lesions. No lesions were produced on the shoots inoculated with PDA plugs (Fig. S9). Specifically, isolates of Lasiodiplodia spp. and Ne. parvum caused symptoms on all inoculated shoots (with incidence of 100 %), the isolates of D. seriata and Do. alpina caused symptoms on less than 60 % of the shoots, and the remaining isolates caused symptoms on shoots ranging from 60 to 100 % (Fig. 16a). Most isolates of Lasiodiplodia produced relatively longer lesions (mean lesion length > 15 cm) than the isolates from the other six genera. In contrast, isolates in N. subglobosa (5.09 cm), S. linhaiensis (5.64 cm), Do. alpina (7.06 cm) and D. seriata (7.75 cm) produced the shortest mean lesion lengths (Fig. 16b). In consideration of both disease incidence and lesion length, we concluded that species of Lasiodiplodia were more virulent to citrus than the other genera encountered.
Fig. 16.
Pathogenicity tests results of inoculated isolates in Botryosphaeriaceae. a. Incidences of shoots inoculated in vitro and in vivo, the blue diamond represents the incidence of shoots inoculated in vitro, while the orange diamond represents the incidence of shoots inoculated in vivo; b. mean lesion lengths on C. reticulata shoots inoculated in vitro after 8 d. Bars represent standard errors. Columns with different letters indicate significant differences according to LSD test with confidence level α = 0.05. Control: PDA plugs.
For in vivo inoculation, all shoots of Cocktail grapefruit inoculated with the isolates listed in Fig. 16 produced lesions after 15 d inoculation, except for the isolates BE3 (B. fabicerciana) and BE17 (Do. alpina). Consistent with the results of in vitro inoculation, 100 % of the shoots inoculated with isolates of Lasiodiplodia spp. produced symptoms. The symptoms were similar to those observed in the field (Fig. S10a–g). However, the remaining isolates caused symptoms on less than 60 % of the shoots, except for isolates BE15 (100 %) and BE85 (80 %) (Fig. 16a). On the contrary, no necrosis symptoms were observed on the control shoots inspected 15 days after inoculation (Fig. 17h). All isolates inoculated in vitro and in vivo were re-isolated successfully from these lesions. As expected, no isolates of Botryosphaeriaceae were isolated from the control inoculations.
DISCUSSION
This study represents the first comprehensive characterisation of species in Botryosphaeriaceae isolated from citrus trees with twig and branch dieback, cankers and gummosis in China. Based on phylogenetic analyses and morphological characteristics, 18 species belonging to seven genera of Botryosphaeriaceae were identified. These species include Botryosphaeria dothidea, B. fabicerciana, Diplodia seriata, Dothiorella alpina, Do. plurivora, Lasiodiplodia citricola, L. iraniensis, L. microconidia, L. pseudotheobromae, L. theobromae, Neodeightonia subglobosa, Neofusicoccum parvum, and the new species described here, namely Do. citrimurcotticola, L. guilinensis, L. huangyanensis, L. linhaiensis, L. ponkanicola and Sphaeropsis linhaiensis. Of the 12 known species reported here, B. fabicerciana, Do. alpina, L. microconidia and N. subglobosa are reported on citrus in China for the first time.
Results of the pathogenicity tests indicate that all Lasiodiplodia species obtained in this study are pathogenic to the tested C. reticulata shoots in vitro and Cocktail grapefruit in vivo, with disease incidences of 100 %. Most of the remaining taxa were not as aggressive however, especially in the in vivo inoculation, which may be due to the higher ambient temperatures observed in the field (Zhang et al. 2016, 2021, this study). Moreover, significant differences in aggressiveness were observed among species. Most species of Lasiodiplodia were strongly aggressive. Conversely, N. subglobosa, S. linhaiensis, Do. alpina and D. seriata were weakly aggressive. In general, Lasiodiplodia was the most aggressive genus in the present study.
Botryosphaeria dothidea is the type species of Botryosphaeria and was considered as one of the most common and important pathogens of woody plants (Phillips et al. 2013, Marsberg et al. 2017). In this study, B. dothidea was the most commonly isolated species. Results of pathogenicity tests indicate that isolates of B. dothidea are moderately aggressive on C. reticulata shoots, which is similar to observations on Pistacia vera in California (Chen et al. 2014). Botryosphaeria fabicerciana, the other species of Botryosphaeria isolated in this study, was first reported on Eucalyptus and was weakly aggressive to this host (Chen et al. 2011). This is the first report of B. fabicerciana on citrus, and on C. reticulata it appeared to be mild to highly aggressive, while on Cocktail grapefruit it appeared to be weakly aggressive.
Diplodia seriata is regarded as a pathogen of citrus, causing branch canker and dieback in Algeria and the USA (Adesemoye et al. 2014, Berraf-Tebbal et al. 2020). Similarly, we isolated a single strain of D. seriata from C. sinensis with branch canker. This finding contrasts with that of Linaldeddu et al. (2015), who reported D. seriata to be the dominant species on Vitis in Italy. Pathogenicity tests indicate that D. seriata is less aggressive than most species of Botryosphaeriaceae obtained in this study.
Dothiorella gummosis refers to the occurrence of branch or trunk cankers on citrus caused by species of Botryosphaeriaceae. The pathogen was long believed to be Do. gregaria, the asexual morph of Botryosphaeria ribis (Adesemoye et al. 2011). However, other species that resided in the Botryosphaeriaceae were also found causing Dothiorella gummosis in citrus (Adesemoye et al. 2011). Therefore, the term ‘Dothiorella gummosis’ was no longer suitable for describing such symptoms, while ‘Botryosphaeria gummosis’ (Adesemoye et al. 2011) and ‘Bot gummosis’ (Adesemoye et al. 2014) were proposed as alternative. In this study, one undescribed Dothiorella species (Do. citrimurcotticola) and two previously reported species (Do. alpina and Do. plurivora) were isolated from branch dieback of Citrus spp. Dothiorella alpina was described from a dead tree of Platycladus orientalis in China (Zhang et al. 2016) and has not been reported from other hosts. Thus, this study represents the first report of this fungus on citrus. Dothiorella plurivora was first reported by Abdollahzadeh et al. (2014) in Iran and Spain, named for its broad host range, including twigs of Casuarina sp., Citrus sp., Cupressus sempervirens, Eucalyptus sp., Juglans regia, Malus domestica, Prunus armeniaca and Vitis vinifera. In this study, the isolates of Do. plurivora were collected from C. reticulata and C. unshiu with twig dieback. To our knowledge, this is the first report of Do. plurivora occurring on citrus in China. Pathogenicity tests demonstrated that the three species of Dothiorella were pathogenic to C. reticulata shoots, and that Do. alpina was less aggressive than the other two species. However, isolates of Do. citrimurcotticola and Do. plurivora were weakly aggressive on Cocktail grapefruit shoots, with incidences lower than 50 %, while Do. alpina was non-pathogenic on Cocktail grapefruit shoots.
We obtained nine species of Lasiodiplodia from citrus diseased branches, accounting for 55.9 % of the total number of isolates, making Lasiodiplodia the most prevalent genus with the highest number of species encountered in this study. This finding is consistent with previous reports that Lasiodiplodia is common on citrus (Abdollahzadeh et al. 2010, Adesemoye et al. 2014, Coutinho et al. 2017, Guajardo et al. 2018, Bautista-Cruz et al. 2019, Berraf-Tebbal et al. 2020). Probable reasons why species of Lasiodiplodia are dominant on citrus include the following: Firstly, it was observed that species of Lasiodiplodia are fast growing, covering 90 mm plates in only 2 d, while species of other genera of Botryosphaeriaceae have slower growth rates. Species of Lasiodiplodia also proved to be more aggressive to citrus compared to other genera tested. Secondly, the optimum temperature of the other genera in this study is 25 °C, while the optimum growth temperature of Lasiodiplodia spp. ranges from 25 °C to 30 °C, thereby giving it an advantage over other species at higher temperatures, and this is probably the reason why Lasiodiplodia species are mostly found in tropical or sub-tropical regions, and rare in regions with temperate climates.
Lasiodiplodia pseudotheobromae can infect citrus, causing gummosis, trunk canker, and twig blight (Abdollahzadeh et al. 2010, Bautista-Cruz et al. 2019, Ahmed et al. 2020). In the present study, L. pseudotheobromae was the second most abundant species isolated from citrus with symptoms of gummosis, dieback and canker. Furthermore, the results of pathogenicity tests showed that L. pseudotheobromae is one of the most aggressive species on citrus shoots. Stem-end rot is a common and economically important postharvest disease of citrus fruits worldwide, and it is usually thought to be caused by L. theobromae (Zhang 2014). Sultana et al. (2018) found that L. pseudotheobromae was also associated with citrus stem-end rot in Bangladesh. Stem-end rot is also an important postharvest disease on citrus in China (Cai et al. 2011). Since L. pseudotheobromae is similar to L. theobromae and is common on citrus according to the isolation results obtained in this study, it is possible that several reports of L. theobromae could have in fact been L. pseudotheobromae. In summary, L. pseudotheobromae is widely distributed on citrus, highly aggressive and can cause stem-end rot and branch diseases. Therefore, we consider L. pseudotheobromae to be an important pathogen on citrus in China, urgently requiring further research to elucidate its impact on this crop.
Neodeightonia subglobosa was initially reported on dead culms of Bambusa arundinacea in Sierra Leone (Punithalingam 1969). Furthermore, N. subglobosa was also found to cause keratomycosis in human eyes (Phillips et al. 2008). There are few reports about N. subglobosa, and to our knowledge, this study is the first to report N. subglobosa associated with gummosis on citrus. However, pathogenicity tests indicated that N. subglobosa was only weakly aggressive on citrus.
Neofusicoccum parvum has a broad host range and distribution, and has been reported from 90 hosts across six continents and 29 countries (Sakalidis et al. 2013, Batista et al. 2021). The lack of host specificity, combined with both a sexual and an asexual cycle, and the ability to live as a latent pathogen are conducive to the infection and spread of Ne. parvum (Sakalidis et al. 2013). In China, Ne. parvum was found associated with canker and dieback on Cupressus funebris (Li et al. 2010), Eucalyptus (Chen et al. 2011), Juglans regia (Yu et al. 2015), Prunus (Li et al. 2019, Zhang et al. 2019), Rhododendron (Yang et al. 2015) and Vitis heyneana (Wu et al. 2015). In this study, we found that Ne. parvum also caused dieback and gummosis on C. unshiu in China. Based on the pathogenicity tests, Ne. parvum was less aggressive than most of the species in this study, but disease incidences in vitro and in vivo were high, second only to isolates in Lasiodiplodia.
Previous research revealed Sphaeropsis citrigena to occur on dead bark of citrus (Phillips et al. 2013). In the present study, another species of Sphaeropsis, namely S. linhaiensis, was found that was associated with twig dieback on C. unshiu. Pathogenicity tests, however, indicated that S. linhaiensis is weakly aggressiveness on citrus.
Branch diseases of citrus are a persistent and frequent problem in the main citrus production areas of China. Many fungal pathogens can induce branch diseases on citrus, including species within the Botryosphaeriaceae, Diatrypaceae and genera such as Colletotrichum and Diaporthe (Chinese Academy of Agricultural Sciences 1960, Tai 1979, Cai et al. 2011). Because branch diseases are complex, and symptoms may be induced by multiple pathogens at the same time, there are few effective measures to control such diseases. Previous research has shown that wounds caused by pruning, mechanical injury, frost and sunburn damage have become the entry point for Botryosphaeriaceae on woody hosts (Savocchia et al. 2007, Úrbez-Torres & Gubler 2009, Eskalen et al. 2013). Furthermore, fungal pathogens release the greatest number of spores during and after rainfall events (Eskalen & Gubler 2001, Amponsah et al. 2009, Eskalen et al. 2013). Therefore, to prevent and reduce the occurrence of citrus branch diseases, pruning should avoid rainy days, decaying or dead branches and twigs should be removed from orchards, wounds protected with sealant or fungicides, frost damage avoided where possible and trees protected from the sun in hot weather. Furthermore, because species of Botryosphaeriaceae are latent pathogens, they can become pathogenic when the trees are under stress or in weak vigour (Slippers & Wingfield 2007). Hence, the cultivation of good tree vigour is conducive to enhance disease resistance.
In conclusion, results of this study present the first detailed research of 18 species of Botryosphaeriaceae causing branch diseases on citrus in nine major citrus-producing provinces of China. Overall, Lasiodiplodia was found to be the most prevalent and aggressive genus in this study, which indicates that Lasiodiplodia is one of the most important genera causing citrus branch diseases. Besides, L. pseudotheobromae is one of the most abundant and most prevalent species, which together with its aggressiveness in branches and fruits, makes L. pseudotheobromae an economically important pathogen on citrus. To better prevent and control L. pseudotheobromae, further research is needed to study the relationship between L. pseudotheobromae in different regions and different citrus varieties, and its ability to cause stem-end rot. In the current study, several species were obtained as single isolates, but most of the isolates were from Zhejiang Province, so subsequent sampling needs to be expanded to investigate the prevalence of these species in other citrus-producing areas in China. Because species of Botryosphaeriaceae can be latent pathogens in woody host plants, and jump from hosts planted nearby (Damm et al. 2007, Slippers & Wingfield 2007, Begoude et al. 2012, James et al. 2017), collecting plant hosts adjacent to citrus is also useful for studying the diversity of the Botryosphaeriaceae species that could have an impact on citrus cultivation.
Acknowledgments
This research was supported by the Key Research and Development Program of Zhejiang Province (2019C02022), the National Key Research and Development Program (2017YFD0202000) and the China Agriculture Research System (CARS-26). We thank the China Eucalypt Research Centre (CERC) for technical guidance in the process of species identification, including phylogenetic analysis, morphology description and pathogenicity tests. We thank Mr. Qu Wu, Siqing Zhao, Dekuan Ding, Qianbin Huang, Weilong Li and Ms. Lan Cheng for their assistance during sample collection. We thank Mr. Xielong Yu for providing the citrus shoots for pathogenicity tests.
Supplementary material
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Botryosphaeria. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Neofusicoccum parvum (CMW 9081).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Diplodia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Lasiodiplodia theobromae (CBS 164. 96).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Dothiorella. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Neofusicoccum parva (CMW 9081).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Lasiodiplodia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and type species are in red bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1 and tub2 (a–c) sequence alignments of Neodeightonia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CBS 115476).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Neofusicoccum. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Dothiorella viticola (CBS 117009).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Sphaeropsis. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
The distribution of Botryosphaeriaceae species obtained from nine provinces. Provinces are represented by different colours.
Symptoms developed in the detached shoots of C. reticulata inoculated with isolates in Botryosphaeriaceae 8 d after inoculation. B. dothidea: BE1, BE2; B. fabicerciana: BE3, BE85; D. seriata: BE4; Do. alpina: BE17; Do. citrimurcotticola: BE5, BE8; Do. plurivora: BE16, BE74; L. citricola: BE13, BE38; L. guilinensis: BE31, BE59; L. huangyanensis: BE33, BE50; L. iranensis: BE27, BE41, BE100; L. linhaiensis: BE51, BE28; L. microconidia: BE80, BE88; L. ponkanicola: BE44; L. pseudotheobromae: BE10, BE11; L. theobromae: BE20, BE21; N. subglobosa: BE14; Ne. parvum: BE15; S. linhaiensis: BE18.
Symptoms developed in shoots of Cocktail grapefruit plants inoculated with isolates of Botryosphaeriaceae 15 d after inoculation. a–b. Shoots inoculated with B. fabicerciana (BE85) producing gum exudate; c. gummosis caused by Ne. parvum (BE15); d. shoot showing symptoms of dieback and gummosis after inoculation with L. guilinensis (BE59); e. dieback with a large amount of gummosis caused by L. pseudotheobromae (BE10); f. dieback with gummosis caused by L. huangyanensis (BE50); g. conidiomata formed on the inoculated shoots with dieback; h. shoots inoculated with sterile PDA plugs. Red arrows indicate the inoculated positions; orange arrows indicate gum exudate; white arrow indicates a conidioma.
REFERENCES
- Abdollahzadeh J, Javadi A, Mohammadi Goltapeh E, et al. 2010. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 25: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahzadeh J, Javadi A, Zare R, et al. 2014. Phylogenetic study of Dothiorella and Spencermartinsia species associated with woody plants in Iran, New Zealand, Portugal and Spain. Persoonia 32: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesemoye AO, Eskalen A. 2011. First report of Spencermartinsia viticola, Neofusicoccum australe, and N. parvum causing branch canker of citrus in California. Plant Disease 95: 770. [DOI] [PubMed] [Google Scholar]
- Adesemoye AO, Eskalen A, Faber B, et al. 2011. Multiple Botryosphaeria species causing ‘Dothiorella’ gummosis in citrus. Citrograph 32–34. [Google Scholar]
- Adesemoye AO, Mayorquin JS, Wang DH, et al. 2014. Identification of species of Botryosphaeriaceae causing bot gummosis in citrus in California. Plant Disease 98: 55–61. [DOI] [PubMed] [Google Scholar]
- Ahmed MZ, Shafique MS, Anwaar HMA, et al. 2020. First report of Lasiodiplodia pseudotheobromae causing trunk cankers in Citrus reticulata blanco (Kinnow) in Pakistan. Plant Disease 104: 2522. [Google Scholar]
- Alves A, Correia A, Luque J, et al. 2004. Botryosphaeria corticola, sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia 96: 598–613. [PubMed] [Google Scholar]
- Alves A, Crous PW, Correia A, et al. 2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13. [Google Scholar]
- Amponsah NT, Jones EE, Ridgway HJ, et al. 2009. Rainwater dispersal of Botryosphaeria conidia from infected grapevines. New Zealand Plant Protection 62: 228–233. [Google Scholar]
- Azadeh G, Abdoolnabi B, Majeed AS, et al. 2018. Citrus × aurantiifolia, a new host report of Macrophomina phaseolina in Iran. Australasian Plant Disease Notes 13: 15. [Google Scholar]
- Batista E, Lopes A, Alves A. 2021. What do we know about Botryosphaeriaceae? An overview of a worldwide cured dataset. Forests 12: 313–330. [Google Scholar]
- Bautista-Cruz MA, Almaguer-Vargas G, Leyva-Mir SG, et al. 2019. Phylogeny, distribution, and pathogenicity of Lasiodiplodia species associated with cankers and dieback symptoms of Persian lime in Mexico. Plant Disease 103: 1156–1165. [DOI] [PubMed] [Google Scholar]
- Begoude BAD, Slippers B, Perez G, et al. 2012. High gene flow and outcrossing within populations of two cryptic fungal pathogens on a native and non-native host in Cameroon. Fungal Biology 116: 343–353. [DOI] [PubMed] [Google Scholar]
- Berraf-Tebbal A, Mahamedi AE, Aigoun-Mouhous W, et al. 2020. Lasiodiplodia mitidjana sp. nov. and other Botryosphaeriaceae species causing branch canker and dieback of Citrus sinensis in Algeria. PLOS ONE 15: e0232448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GE, Eckert JW. 2000. Diplodia stem-end rot. In: Timmer LW, Gernsey SM, Graham JH. (eds), Compendium of citrus diseases, second ed: 43–44. APS Press, St Paul. [Google Scholar]
- Cai MD, Yi GJ, Peng CJ. 2011. An illustrated book of primary colours of citrus diseases and pests. China Agricultural Press. [In Chinese.] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Chen SF, Liu QL, Li GQ, et al. 2018. A new genus of Cryphonectriaceae isolated from Lagerstroemia speciosa in southern China. Plant Pathology 67: 107–123. [Google Scholar]
- Chen SF, Morgan DP, Michailides TJ. 2014. Botryosphaeriaceae and Diaporthaceae associated with panicle and shoot blight of pistachio in California, USA. Fungal Diversity 67: 157–179. [Google Scholar]
- Chen SF, Pavlic D, Roux J, et al. 2011. Characterization of Botryosphaeriaceae from plantation-grown Eucalyptus species in South China. Plant Pathology 60: 739–751. [Google Scholar]
- Cheng J, Wei X, Fan H. 2004. Phytophthora species infecting citrus in Guangdong Province. Journal of South China Agricultural University (Natural Science Edition) 25: 31–33. [In Chinese.] [Google Scholar]
- Chinese Academy of Agricultural Sciences. 1960. Chinese journal of fruit diseases and insects. Agricultural Press. [In Chinese.] [Google Scholar]
- Coutinho IBL, Freire FCO, Lima CS, et al. 2017. Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in Northeastern Brazil. Plant Pathology 66: 90–104. [Google Scholar]
- Cruywagen EM, Slippers B, Roux J, et al. 2017. Phylogenetic species recognition and hybridisation in Lasiodiplodia: a case study on species from baobabs. Fungal Biology 121: 420–436. [DOI] [PubMed] [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2007. Botryosphaeriaceae as potential pathogens of Prunus in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia 99: 664–680. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XX, Peng CJ, Chen ZS, et al. 2008. Chinese citrus varieties. China Agricultural Press. [In Chinese.] [Google Scholar]
- Eskalen A, Faber B, Bianchi M. 2013. Spore trapping and pathogenicity of fungi in the Botryosphaeriaceae and Diaporthaceae associated with avocado branch canker in California. Plant Disease 97: 329–332. [DOI] [PubMed] [Google Scholar]
- Eskalen A, Gubler W. 2001. Association of spores of Phaeomoniella chlamydospora, Phaeoacremonium inflatipes, and Pm. aleophilum with grapevine cordons in California. Phytopathologia Mediterranea 40: 429–432. [Google Scholar]
- FAO. 2018. Food and Agriculture Organization of the United Nations, Statistical Databases. http://www.fao.org. [Google Scholar]
- Fawcett HS, Burger OF. 1911. A gum-inducing Diplodia of peach and orange. Mycologia 3: 151–153. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guajardo J, Riquelme N, Tapia L, et al. 2018. First Report of Lasiodiplodia theobromae causing bot gummosis in citrus limon in Chile. Plant Disease 102: 818. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Huang F, Chen GQ, Hou X, et al. 2013a. Colletotrichum species associated with cultivated citrus in China. Fungal Diversity 61: 61–74. [Google Scholar]
- Huang F, Hou X, Dewdney M, et al. 2013b. Diaporthe species occurring on citrus in China. Fungal Diversity 61: 237–250. [Google Scholar]
- Huang F, Zhu L, Hou X, et al. 2012. Identification of the pathogenic fungus causing brown spot on Ougan. Journal of Zhejiang Agricultural Sciences 1281–1282. [In Chinese.] [Google Scholar]
- Ismail MA, Zhang JX. 2004. Post-harvest citrus diseases and their control. Outlooks Pest Management 15: 29–35. [Google Scholar]
- James WMM, Bernard S, Jolanda R, et al. 2017. Overlap of latent pathogens in the Botryosphaeriaceae on a native and agricultural host. Fungal Biology 121: 405–419. [DOI] [PubMed] [Google Scholar]
- Jiang N, Wang XE, Liang YM, et al. 2018. Lasiodiplodia cinnamomi sp. nov. from Cinnamomum camphora in China. Mycotaxon 133: 249–259. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Tang LH, Sun WX, et al. 2019. First report of stem canker and dieback caused by Neofusicoccum parvum on Plum in Guangxi, Southern China. Plant Disease 103: 2952–2952. [Google Scholar]
- Li SB, Li JZ, Lu ZH, et al. 2010. First report of Neofusicoccum parvum causing dieback disease of Chinese Weeping Cypress in China. Plant Disease 94: 641–641. [DOI] [PubMed] [Google Scholar]
- Linaldeddu BT, Deidda A, Scanu B, et al. 2015. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Diversity 71: 201–214. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Marsberg A, Kemler M, Jami F, et al. 2017. Botryosphaeria dothidea: a latent pathogen of global importance to woody plant health. Molecular Plant Pathology 18: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorquin JS, Wang DH, Twizeyimana M, et al. 2016. Identification, distribution, and pathogenicity of Diatrypaceae and Botryosphaeriaceae associated with citrus branch canker in the Southern California desert. Plant Disease 100: 2402–2413. [DOI] [PubMed] [Google Scholar]
- Pérez CA, Wingfield MJ, Slippers B, et al. 2010. Endophytic and canker-associated Botryosphaeriaceae occurring on non-native Eucalyptus and native Myrtaceae trees in Uruguay. Fungal Diversity 41: 53–69. [Google Scholar]
- Phillips AJL, Alves A, Abdollahzadeh J, et al. 2013. The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology 76: 51–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Pennycook SR, et al. 2008. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 21: 29–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi G, Aiello A, Vitale A, et al. 2009. First report of shoot blight, canker, and gummosis caused by Neoscytalidium dimidiatum on citrus in Italy. Plant Disease 93: 1215. [DOI] [PubMed] [Google Scholar]
- Punithalingam E. 1969. Studies on Sphaeropsidales in culture. Mycological Papers 119: 1–24. [Google Scholar]
- Qin X, Deng M, Yang T, et al. 2012. Pathogen identification of black rot of Gong’an. South China Fruits 41: 10–13. [In Chinese.] [Google Scholar]
- Rayner RW. 1970. A Mycological Colour Chart. Kew, Commonwealth Mycological Institute. [Google Scholar]
- Sakalidis ML, Slippers B, Wingfield BD, et al. 2013. The challenge of understanding the origin, pathways and extent of fungal invasions: global populations of the Neofusicoccum parvum-N. ribis species complex. Diversity and Distributions 19: 873–883. [Google Scholar]
- Savocchia S, Steel CC, Stodart BJ, et al. 2007. Pathogenicity of Botryosphaeria species isolated from declining grapevines in subtropical regions of Eastern Australia. Vitis 46: 27–32. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, et al. 2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Shen ZM. 2019. Present status and prospects of citrus production in China. Scientific Breeding 9: 5–10. [In Chinese.] [Google Scholar]
- Slippers B, Crous PW, Jami F, et al. 2017. Diversity in the Botryosphaeriales: Looking back, looking forward. Fungal Biology 121: 307–321. [DOI] [PubMed] [Google Scholar]
- Slippers B, Wingfield MJ. 2007. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biology Reviews 21: 90–106. [Google Scholar]
- Smith CO. 1934. Inoculations showing the wide host range of Botryosphaeria ribis. Journal of Agricultural Research 49: 467–476. [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, et al. 1996. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- SPSS. 2011. IBM SPSS Statistics Base 20. IBM Corporation, Armonk, NY. [Google Scholar]
- Sultana R, Islam MS, Rahman H, et al. 2018. Characterization of Lasiodiplodia pseudotheobromae associated with citrus stem-end rot disease in Bangladesh. International Journal of Biosciences 13: 252–262. [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tai FL. 1979. Sylloge Fungorum Sinicorum. Beijing, Science Press. [In Chinese.] [Google Scholar]
- Theissen F, Sydow H. 1918. Vorentwürfe zu den Pseudosphaeriales. Annales Mycologici 16: 1–34. [Google Scholar]
- Úrbez-Torres JR. 2011. The status of Botryosphaeriaceae species infecting grapevines. Phytopathologia Mediterranea 50: S5–S45. [Google Scholar]
- Úrbez-Torres JR, Gubler WD. 2009. Pathogenicity of Botryosphaeriaceae species isolated from grapevine cankers in California. Plant Disease 93: 584–592. [DOI] [PubMed] [Google Scholar]
- Van Burik JAH, Schreckhise RW, White TC, et al. 1998. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Medical Mycology 36: 299–303. [PubMed] [Google Scholar]
- Wang Y, Lin S, Zhao L, et al. 2019. Lasiodiplodia spp. associated with Aquilaria crassna in Laos. Mycological Progress 18: 683–701. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, CA, USA. [Google Scholar]
- Wu DD, Fu G, Ye YF, et al. 2015. First report of Neofusicoccum parvum causing panicle blight and leaf spot on Vitis heyneana in China. Plant Disease 99: 417–417. [DOI] [PubMed] [Google Scholar]
- Yang T, Groenewald JZ, Cheewangkoon R, et al. 2017. Families, genera, and species of Botryosphaeriales. Fungal Biology 2016: 322–346. [DOI] [PubMed] [Google Scholar]
- Yang XM, Wang JH, Zhang Y, et al. 2015. First report of Neofusicoccum parvum causing stem canker and dieback in Rhododendron in China. Plant Disease 99: 1179–1180. [Google Scholar]
- Yu ZD, Tang GH, Peng SB, et al. 2015. Neofusicoccum parvum causing canker of seedlings of Juglans regia in China. Journal of Forestry Research 26: 1019–1024. [Google Scholar]
- Zhang JX. 2014. Lasiodiplodia theobromae in citrus fruit (Diplodia stem-end rot). In: Bautista-Baños S, Postharvest decay: 309–335. Academic Press. [Google Scholar]
- Zhang LQ, Li XW, Su MS, et al. 2019. First report of Neofusicoccum parvum associated with shoot cankers of peach (Prunus persica) in Shanghai, China. Journal of Plant Pathology 101: 1257. [Google Scholar]
- Zhang M, He W, Wu JR, et al. 2016. Two new species of Spencermartinsia (Botryosphaeriaceae, Botryosphaeriales) from China. Mycosphere 7: 942–949. [Google Scholar]
- Zhang W, Groenewald JZ, Lombard L, et al. 2021. Evaluating species in Botryosphaeriales. Persoonia 46: 63–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhao XL, Wu RC, et al. 2011. Identification of the pathogen causing foot rot of Fortunella margarita (Lour.) Swingle. Acta Phytopathologica Sinica 41: 631–634. [In Chinese.] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Botryosphaeria. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Neofusicoccum parvum (CMW 9081).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Diplodia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Lasiodiplodia theobromae (CBS 164. 96).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Dothiorella. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Neofusicoccum parva (CMW 9081).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Lasiodiplodia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and type species are in red bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1 and tub2 (a–c) sequence alignments of Neodeightonia. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CBS 115476).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Neofusicoccum. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Dothiorella viticola (CBS 117009).
Phylogenetic trees generated by maximum likelihood analyses based on the individual ITS, tef1, tub2 and rpb2 (a–d) sequence alignments of Sphaeropsis. Bootstrap support values ≥ 50 % for ML and MP are presented above branches as follows: ML/MP bootstrap values < 50 % are marked with *, and absent are marked with -. Newly generated sequences are in red and ex-type strains are in bold. The tree was rooted to Botryosphaeria dothidea (CMW 8000).
The distribution of Botryosphaeriaceae species obtained from nine provinces. Provinces are represented by different colours.
Symptoms developed in the detached shoots of C. reticulata inoculated with isolates in Botryosphaeriaceae 8 d after inoculation. B. dothidea: BE1, BE2; B. fabicerciana: BE3, BE85; D. seriata: BE4; Do. alpina: BE17; Do. citrimurcotticola: BE5, BE8; Do. plurivora: BE16, BE74; L. citricola: BE13, BE38; L. guilinensis: BE31, BE59; L. huangyanensis: BE33, BE50; L. iranensis: BE27, BE41, BE100; L. linhaiensis: BE51, BE28; L. microconidia: BE80, BE88; L. ponkanicola: BE44; L. pseudotheobromae: BE10, BE11; L. theobromae: BE20, BE21; N. subglobosa: BE14; Ne. parvum: BE15; S. linhaiensis: BE18.
Symptoms developed in shoots of Cocktail grapefruit plants inoculated with isolates of Botryosphaeriaceae 15 d after inoculation. a–b. Shoots inoculated with B. fabicerciana (BE85) producing gum exudate; c. gummosis caused by Ne. parvum (BE15); d. shoot showing symptoms of dieback and gummosis after inoculation with L. guilinensis (BE59); e. dieback with a large amount of gummosis caused by L. pseudotheobromae (BE10); f. dieback with gummosis caused by L. huangyanensis (BE50); g. conidiomata formed on the inoculated shoots with dieback; h. shoots inoculated with sterile PDA plugs. Red arrows indicate the inoculated positions; orange arrows indicate gum exudate; white arrow indicates a conidioma.