Abstract
In a survey of bloodstream infection (BSI) isolates across the continental United States, 162 Candida albicans isolates were fingerprinted with the species-specific probe Ca3 and the patterns were analyzed for relatedness with a computer-assisted system. The results demonstrate that particular BSI strains are more highly concentrated in particular geographic locales and that established BSI strains are endemic in some, but not all, hospitals in the study and undergo microevolution in hospital settings. The results, however, indicate no close genetic relationship among fluconazole-resistant BSI isolates in the collection, either from the same geographic locale or the same hospital. This study represents the first of three fingerprinting studies designed to analyze the origin, genetic relatedness, and drug resistance of Candida isolates responsible for BSI.
Nosocomial infections are defined as those acquired by a patient after residence is established in a hospital setting (6). Although initiated in a hospital setting, the origin of the etiologic agent is not always obvious, and this is most evident in infections involving organisms like Candida albicans, which can be caused either by a commensal strain carried by the patient into the hospital setting or by a strain acquired from the hospital setting. Each year in the United States alone, approximately 2 million patients acquire a nosocomial infection (4, 6, 26). Of these, 250,000 will be life-threatening bloodstream infections (BSIs), and approximately 10% of these will be due to Candida spp. and other fungi (4, 6, 7, 16, 17, 26). Nosocomial BSIs resulting from Candida spp. carry an attributable mortality of approximately 40 to 50% and a mean excess length of hospitalization of 30 days (36). With the added concern of the emergence of antimicrobial resistance in Candida spp. (14, 18, 19, 21), it is now crucial that the origins and possible specialization of nosocomial strains of Candida be investigated.
C. albicans accounts for 50 to 70% of all nosocomial BSIs resulting from Candida spp. (1, 4, 7, 17). C. albicans also represents the most prevalent commensal Candida spp. (16) and has been demonstrated to reside in one or more body locations in more than 70% of healthy women (34). The origin of nosocomial BSIs caused by C. albicans, therefore, is complicated by the fact that more than half of the patients at risk as well as more than half of the hospital staff carry a commensal strain. By fingerprinting C. albicans and a number of related species with species-specific fingerprinting probes that include moderately repetitive elements and analyzing strain relatedness with computer-assisted systems (8, 11, 20, 29), one can test whether nosocomial BSI isolates from a single hospital are genetically identical, highly related, moderately related, or unrelated, whether in particular geographical locales, such as a city or region of the United States (e.g., Northeast [NE], Southeast [SE], Midwest [MW], Northwest [NW], or Southwest [SW]), particular nosocomial BSI strains predominate, whether particular nosocomial BSI strains established in a particular hospital setting are undergoing microevolution (10, 12), and whether particular nosocomial BSI strains in a particular geographical locale or hospital are becoming resistant to drugs.
In the nosocomial BSI surveillance study conducted during a 15-month period between 1995 and 1996 as part of the Surveillance and Control of Pathogens of Epidemiological Importance (SCOPE) Program, 162 C. albicans BSI isolates were collected from hospitals throughout the continental United States. All C. albicans isolates were fingerprinted with the probe Ca3, and the fingerprint patterns were compared with a computer-assisted system in order to assess genetic relatedness. All C. albicans isolates were also tested for fluconazole susceptibility. By generating dendrograms and computing the average similarity coefficients (SABs) for select sets of strains, genetic relatedness was assessed for isolates from specific hospitals, isolates from specific geographical locales within the continental United States, and isolates that were fluconazole resistant. The results suggest that established BSI strains are endemic in some, but not all, hospitals in the study and that these strains are undergoing microevolution in the hospital setting. The results also suggest nosocomial strain specificity in particular geographical regions of the continental United States. The results, however, suggest no genetic relationship among fluconazole-resistant isolates, even from the same hospital.
MATERIALS AND METHODS
Collection of BSI isolates.
The SCOPE Program was established under the auspices of Wyeth Ayerst Pharmaceuticals (Pearl River, N.J.) to identify, measure the frequency of, and assess the antimicrobial susceptibility patterns of the predominant pathogens of nosocomial BSIs obtained from approximately 50 medical centers throughout the continental United States. Each participating hospital collected isolates during a 15-month period between April 1995 and June 1996. Fungal isolates (one per patient) were cultured on nutrient agar slants and were sent on a monthly basis to the Microbiology Laboratories at the University of Iowa Hospitals and Clinics, Iowa City, for storage and characterization.
Organism identification.
All fungal blood culture isolates were initially identified at each participating institution by the routine procedures of that institution. Upon receipt at the University of Iowa, fungal isolates were subcultured onto potato dextrose agar (Remel, Lenexa, Kans.) and Chromagar Candida agar (Hardy Diagnostics, Santa Maria, Calif.) to assess viability and strain homogeneity. Species were then identified with Vitek and API products (bioMerieux, St. Louis, Mo.) and by other conventional methods, as required. All Candida isolates were stored as water suspensions or on agar slants at ambient temperature.
DNA fingerprinting.
Fingerprinting with the moderately repetitive DNA fingerprinting probe Ca3 was used to assess the genetic relatedness of the C. albicans isolates (2, 10, 24, 29, 33). This method was selected because it was previously demonstrated to reflect the genetic distance between highly related, moderately related, and unrelated isolates with approximately the same resolving power as the randomly amplified polymorphic DNA method and the multilocus enzyme electrophoresis method but to be faster than these last two methods and highly amenable to computer-assisted analysis for large-scale studies and future retrospective studies (20). In brief, cells from individual storage slants were grown to the late logarithmic phase in YPD broth (2% glucose, 2% Bacto Peptone, 1% yeast extract). DNA from each isolate was then prepared by the method of Scherer and Stevens (27). DNA was measured in a Sequoia-Turner 45 fluorometer (Barnstead/Thermodyne, Dubuque, Iowa), digested with the restriction enzyme EcoRI, and separated in a 0.8% (wt/vol) agarose gel containing 16 lanes. In each case, test isolates were run in the inner 14 lanes and the reference strain 3153A was run in the outer 2 lanes. When the indicator dye bromophenyl blue had migrated to a point 16 cm from the origin of the gel, electrophoresis was terminated and the gel was stained with ethidium bromide to verify equal loading. The gel was then destained, transferred (25) to a Nitropure membrane (Micron Separations, Inc., Wesborough, Mass.), and hybridized with randomly primed 32P-labeled Ca3 probe by previously described methods (29). Hybridized membranes were then autoradiographed with XAR-S film (Eastman Kodak Co., Rochester, N.Y.) with a Cronex Lightning-Plus intensifying screen (Dupont Co., Wilmington, Del.).
To compare the DNA fingerprints of the 162 isolates, autoradiogram images were digitized into Dendron software, version 2.0 (Solltech Inc., Oakdale, Iowa), by using the transparency option of a Scanjet II scanner (Hewlett-Packard, Palo Alto, Calif.). Each pattern was processed for distortions, and lanes and bands were automatically identified. The SAB between the patterns for every pair of strains A and B was computed by the formula SAB = 2E/(2E + a + b), where E is the number of bands in the patterns for strains A and B sharing the same positions, a is the number of bands in the pattern for strain A with no positional correlates in the pattern for strain B, and b is the number of bands in the pattern for strain B with no positional correlates in the pattern for strain A. This computation is based upon band position alone rather than band position plus intensity (35) and has proven to be most effective in the analysis of moderately related isolates (8, 12, 20). An SAB of 0.00 indicates that the patterns for strains A and B share no common bands, an SAB of 1.00 indicates that all bands in the pattern for strain A are common to those in the pattern for strain B, and SABs ranging between 0.01 and 0.99 represent increasing levels of similarity. In a recent analysis of unrelated C. albicans strains with the Ca3 probe and this computation of SAB, the average SAB was 0.65 ± 0.11 (20). Histograms of SABs were generated by direct comparison of the patterns for every pair of isolates in a selected group. Dendrograms based on SAB values were generated by the unweighted pair group method (31). In cluster analyses, the selection of SAB thresholds (12) is explained later in the text. To assess the integrity and stability of clusters generated by the unweighted pair group method (3), the Test Dendrogram Stability option of the Dendron, version 2.0, software package was used. In this assessment, the system first randomly permutes the order of isolates in the genesis of the dendrogram. This has the consequence of changing the order of group pairing, which represents a test of the stability of clusters (3). The system then adds noise to the dendrogram matrix by randomly computing values in the matrix with ±2% or ±5%, respectively, and generates dendrograms accordingly, again testing the stability of clusters.
Statistical tests.
To compare the average SABs for defined collections, a two-sample t test for independent samples with unequal variances was used (23). The null hypothesis in this case was that no difference existed between tested pairs of average SABs. To compare proportions of isolates in clusters generated at a particular SAB threshold, a chi-square test was used (23). The null hypothesis in this case was that no difference existed between proportions. Fisher’s exact test (23) was used for small sample sizes. The null hypothesis in this case was that the results were due to randomness. In all cases, significance was considered to be a P value of ≤0.05.
Fluconazole susceptibility.
Fluconazole susceptibility was tested by the reference broth microdilution method described by the National Committee for Clinical Laboratory Standards (13). Quality control was performed with Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258. A fluconazole-resistant isolate was defined as an isolate requiring ≥64 μg of fluconazole per ml for growth inhibition, as described by Rex et al. (22) and by the National Committee for Clinical Laboratory Standards (13).
RESULTS
The general collection of BSI isolates.
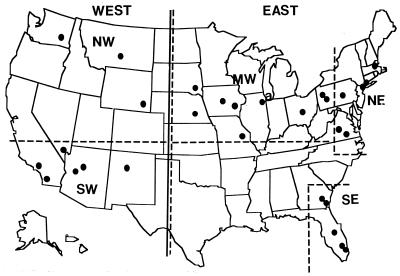
To test for geographical localization of BSI strains, the continental United States was arbitrarily separated into the NE, SE, MW, SW, and NW according to the dashed-line borders in Fig. 1. Although Pittsburgh, Pa., and Danville, Pa., are in a traditional NE state, they are geographically closer to the Ohio border, a traditional MW border, than to the remaining NE cities in this study. Therefore, an analysis of average SABs was performed. In that analysis the isolates from these two cities were included or excluded from the MW or NE collections, and an explanation for their inclusion in the MW locale is presented below. Similarly, although Norfolk, Va., and Richmond, Va., are in a traditional SE state, they are geographically closer to the NE cities than to the remaining SE cities in this study. Similarly, we performed an analysis of average SABs in which the isolates from these two cities were included or excluded from the NE and SE collections, and an explanation for their inclusion in the NE locale is presented below. The United States was further separated into East and West according to the solid vertical line bisecting North Dakota and the five states directly south of it. Hospitals which provided one or more BSI C. albicans isolates are noted by the filled circles in the general map (Fig. 1). Isolates were obtained from 44 patients in the NE, 41 patients in the SE, 37 patients in the MW, 33 patients in the SW, and 7 patients in the NW (Table 1). Two of the isolates that were originally typed as C. albicans by their sugar assimilation patterns did not generate a complex pattern when probed with the Ca3 probe (e.g., NYBX in Fig. 2B) and were later confirmed to be Candida dubliniensis (9). Within each geographical locale there was an uneven distribution of isolates between participating hospitals. For instance, in the NE collection two isolates were obtained from hospital PAL in Lancaster, Pa., while 14 isolates were obtained from hospital NYB in New York City, and in the SW collection two isolates were obtained from hospital CAF in Fullerton, Calif., while 22 isolates were obtained from hospital NVL in Las Vegas, Nev. (Table 1). In addition, only one-third as many isolates were obtained from the western division compared to the number obtained from the eastern division of the United States (Table 1). The effects of unequal distributions will therefore be considered in comparisons between individual geographical locales or East-West sectors.
FIG. 1.
Geographical separations and the locations of hospitals from which one or more BSI isolates of C. albicans were obtained. Dashed lines delineate the following geographical locales: NE, SE, MW, SW, and NW. The solid line separates East and West sections of the continental United States.
TABLE 1.
Geographical origin of C. albicans BSIs
| Region (total no. of isolates) | City, state | Code: Hospital and isolates |
|---|---|---|
| NE (n = 44) | Springfield, Mass. | MAS1 to MAS3 |
| New York, N.Y. | NYN1 to NYN13 | |
| New York, N.Y. | NYB1 to NYB14 | |
| Langhorne, Pa. | PAL1 and PAL2 | |
| Norfolk, Va. | VAN1 to PAL5 | |
| Richmond, Va. | VAR1 to PAL7 | |
| SE (n = 41) | Savannah, Ga. | GAS1 to GAS6 |
| Ft. Lauderdale, Fla. | FLF1 to FLF10 | |
| Savannah, Ga. | GAM1 to GAM7 | |
| Orlando, Fla. | FLO1 to FLO12 | |
| Miami, Fla. | FLM1 to FLM6 | |
| MW (n = 37) | Omaha, Nebr. | NEO1 and NEO2 |
| Pittsburgh, Pa. | PAP1 to PAP9 | |
| Danville, Pa. | PAD1 to PAD5 | |
| Des Moines, Iowa | IAD1 to IAD6 | |
| Iowa City, Iowa | IAI1 to IAI5 | |
| Park Ridge, Ill. | ILP1 and ILP2 | |
| Akron, Ohio | OHA1 | |
| Danville, Ill. | ILU1 | |
| Indianapolis, Ind. | INI1 | |
| Sioux Falls, S.D. | SDS1 to SDS5 | |
| NW (n = 7) | Billings, Mont. | MTB1 to MTB4 |
| Cheyenne, Wyo. | WYC1 | |
| Spokane, Wash. | WAS1 and WAS2 | |
| SW (n = 33) | Fullerton, Calif. | CAF1 and CAF2 |
| Bakersfield, Calif. | CAB1 and CAB2 | |
| Las Vegas, Nev. | NVL1 to NVL22 | |
| Sun City, Ariz. | AZS1 to AZS4 | |
| Albuquerque, N.M. | NMA1 to NMA3 |
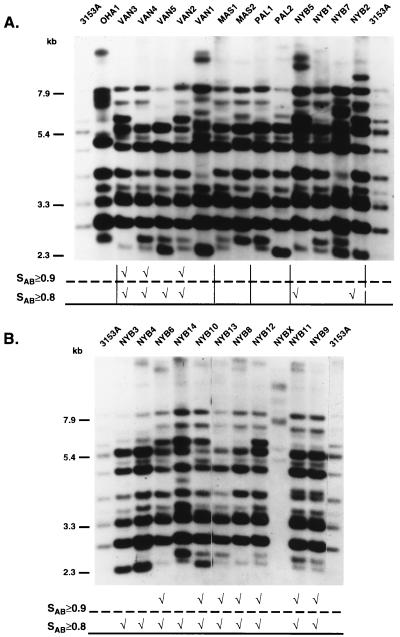
FIG. 2.
Examples of the Ca3 Southern blot hybridization patterns of the C. albicans BSI isolates tested in this study. (A) Example of one of the test blots in this study. Note that the standard strain 3153A is run in the first and last lanes to normalize the gel for comparison to the universal standard in the Dendron program. (B) Patterns of isolates from New York hospital NYB. At the bottom of each gel, comparison of the patterns at SAB thresholds of ≥0.90 and ≥0.80 are made. The checkmarks indicate the relatedness of the patterns at these thresholds. Molecular sizes (in kilobases) are given to the left of each gel.
Differences exist in the average SABs of the total collections from the different geographical locales of the continental United States.
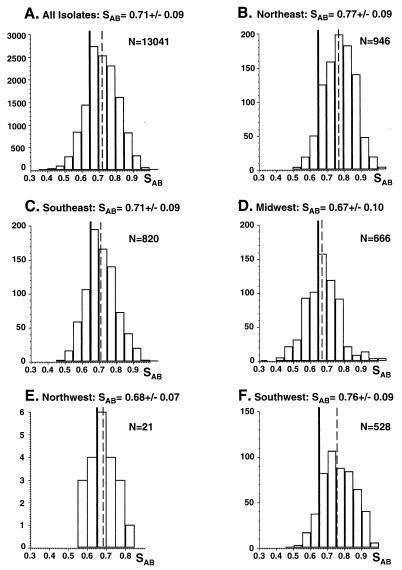
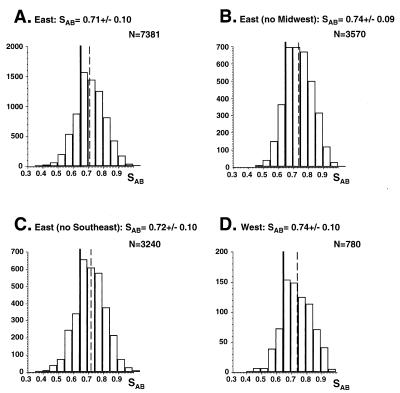
One hundred sixty-two BSI isolates exhibited complex Southern blot hybridization patterns with the Ca3 probe. In Fig. 2A, the patterns of isolates from Ohio, Virginia, Massachusetts, Pennsylvania, and New York are presented. In all cases, bands in the Ca3 pattern were discrete and amenable to automatic computer-assisted analysis. In all cases, the bands have been defined according to specific hybridization to endonuclease digestion fragments of the 11-kb Ca3 probe (2, 10). The average SAB for the entire collection of 162 C. albicans isolates was 0.71 ± 0.09 (Fig. 3A). The distribution of SABs was bell shaped and ranged from 0.35 to 1.00 (Fig. 3A). The average SAB for the entire collection was significantly higher than the average SAB value of 0.65 ± 0.11 previously computed by the same method (i.e., band position only) for 22 isolates originally selected for unrelatedness (20). The P value computed for the difference was 0.02. The average SABs for isolates from the East and West regions of the United States (Fig. 1) were 0.71 ± 0.10 and 0.74 ± 0.10, respectively (Fig. 4A and D, respectively). Again, the distributions of SABs were relatively bell shaped (Figure 4A and D). The average SABs for the total East and the total West collections were higher than the previously computed average SABs for unrelated isolates (20), with P values of <0.05 and 0.01, respectively. These results indicate that within the total collection and within the East or West collection there was a higher level of relatedness than in a random collection.
FIG. 3.
Distributions, average SABs, and number of pairwise comparisons (N) of all isolates (A), NE isolates (B), SE isolates (C), MW isolates (D), NW isolates (E), and SW isolates (F). SABs are presented as average ± standard deviation. The solid vertical line notes the average SAB computed for unrelated isolates (20). Dashed vertical lines note the average SAB of each respective collection.
FIG. 4.
Distributions, average SABs, and number of comparisons (N) of complete East collection (A), East collection minus MW collection (B), East collection minus SE collection (C), and complete West collection (D). See legend to Fig. 3 for details.
There were also significant differences in average SABs between the five geographical locales, NE, SE, MW, NW, and SW (Fig. 3B through F, respectively). The average SABs for isolates from the MW and NW were 0.67 ± 0.10 and 0.68 ± 0.07, respectively (Fig. 3D and E, respectively), neither of which was significantly different from the average SAB of 0.65 ± 0.11 previously determined for unrelated isolates (20). Again, the distributions of SABs for the MW and NW collections were relatively bell shaped (Fig. 3D and E, respectively). The average SAB for isolates from the SE was 0.71 ± 0.09 (Fig. 3C), which was significantly higher than that previously determined for unrelated isolates (20), with a P value of <0.05. The average SABs for isolates from the NE and SW were 0.77 ± 0.09 and 0.76 ± 0.09 (Fig. 3B and F, respectively), both of which were significantly higher than that previously determined for unrelated isolates (20), with P values in both cases of 0.001. The average SABs for isolates from the SW or NE were significantly higher than those for isolates from the NW, SE, and MW, with P values ranging between <0.05 and 0.001. These results suggest that the individual collections of isolates from the NE and SW were each more intrarelated than the collections from the MW, NW, or SE.
Since Pittsburgh and Danville, Pa., are close to the traditional Pennsylvania-Ohio border of the MW locale, they could be considered in either the NE or the MW locale. With and without isolates from these two cities, the average SABs for the MW collection were 0.677 ± 0.099 and 0.672 ± 0.093, respectively; with and without these isolates, the average SABs for the NE collection were 0.749 ± 0.096 and 0.771 ± 0.089, respectively. Therefore, addition of the isolates to the NE collection lowered the average SAB slightly, while addition to the MW collection increased it only slightly. These results suggested that, on average, the Pittsburgh and Danville, Pa., isolates were more related to the MW collection than to the NE collection. For that reason, they were incorporated into the former collection. Since Norfolk and Richmond, Va., are traditionally considered SE cities but are close to the traditional NE-SE border and, in absolute distance, are closer to the average NE city than to the average SE city in this study, they could have been considered in the NE or SE. With and without isolates from these two cities, the average SABs for the NE collection were 0.771 ± 0.087 and 0.756 ± 0.096, respectively; with and without these isolates, the average SABs for the SE collection were 0.677 ± 0.099 and 0.672 ± 0.093, respectively. While addition of the isolates to either the NE or SE collections increased the SAB for each collection slightly, the effect was greater for the NE collection. These results suggest that the Norfolk and Richmond, Va., isolates are more related to the NE isolates than to the SE isolates. For that reason they were included in the NE collection.
The average SAB for the MW collection was significantly lower than that for the NE or the SE collection (Fig. 3). When the MW collection was removed from the East collection, the average SAB increased from 0.71 ± 0.10 to 0.74 ± 0.09 (Fig. 4A and B, respectively). The change was significant, with a P value of 0.001, supporting the suggestion that isolates of the MW collection were less interrelated than the isolates of the NE and SE collections. In contrast, if the SE collection was removed from the East collection, the average SAB increased insignificantly from 0.71 ± 0.10 to 0.72 ± 0.10 (Fig. 4A and C, respectively), reinforcing the conclusion that isolates of the NE and SE collections were more interrelated than they were with isolates of the MW collection.
The results obtained by analyzing the full collections from the five geographical locales could be interpreted to suggest that isolates from select geographical locales differ in their levels of interrelatedness. However, because the collections were not matched for the number of hospitals per geographical location or the number of isolates per hospital and because groups of highly related isolates from individual hospitals would skew the average SAB to higher values, this interpretation must be more carefully scrutinized. Therefore, the collection from each geographical locale was first individually tested for highly related sets of isolates from individual hospitals. In such cases, collections from geographical locales were refined so that only one isolate from any highly related cluster of isolates from the same hospital was used in the computation of the average SAB for each geographical locale.
NE collection.
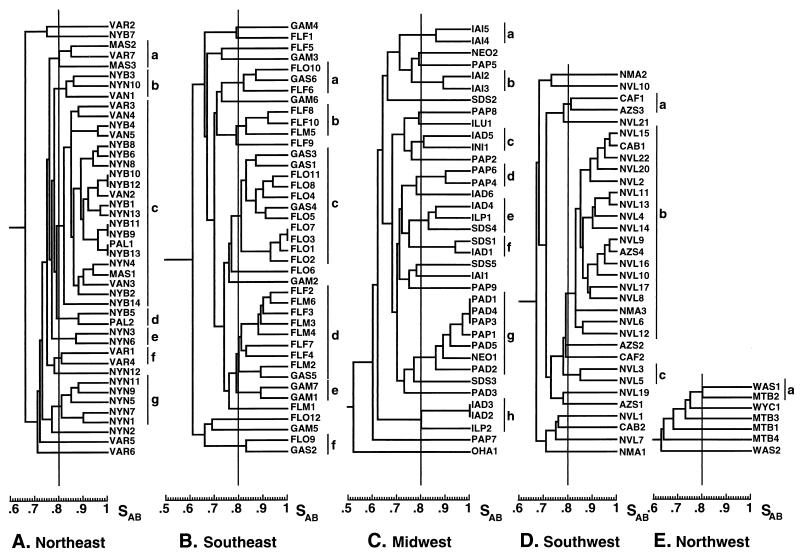
The collection of NE BSI isolates totaled 44 from six hospitals (Table 1). The isolates from New York City hospitals NYB and NYN represented 61% (27 of 44) of the total NE collection (Table 1). A dendrogram for all NE isolates based on SABs computed between all pairs is presented in Fig. 5A. The initial threshold used to discriminate clusters in this and all subsequent dendrograms was 0.80, which was approximately 45% the distance between an SAB of 0.65, the previous estimate of unrelatedness (20), and an SAB of 1.00, representing identicalness. Thirty-eight of the 44 isolates (86%) separated into seven clusters, clusters a through g (Fig. 5A). The distribution of isolates from individual hospitals in the clusters was nonrandom. For instance, the largest cluster, cluster c, contained 79% (11 of 14) of the NYB isolates. If the distribution of isolates among clusters were random, only 32% of the NYB isolates would have fallen within this cluster (i.e., the proportion of NYB isolates in the total NE collection). The concentration of NYB isolates in cluster c was significant, with a P value of 0.008. In addition, 7 of the 11 NYB isolates in this cluster formed subclusters of two or three for which SABs of 0.97 or greater. The high level of pattern similarity at SAB thresholds of ≥0.80 and ≥0.90 between the NYB isolates is demonstrated in Fig. 2A and B. The high level of relatedness of NYB isolates suggests that they originated from a single clone of C. albicans endemic to the NYB hospital setting. The small differences in the Ca3 patterns of highly related isolates with SABs of ≥0.90 (Fig. 2B) were due primarily to changes in the hypervariable C-fragment bands at molecular sizes greater than 7.9 kb (1, 10), which have been shown to be indicators of microevolution (10, 11).
FIG. 5.
Dendrograms of the C. albicans collections from the NE (A), SE (B), MW (C), SW (D), and NW (E). The vertical line within each diagram denotes the SAB threshold of 0.80. The lines to the right of each dendrogram delineate clusters based on an SAB threshold of ≥0.80.
Similar results were obtained with the isolates from hospital NYN in New York City. Five of the 13 isolates coclustered in cluster g (Fig. 5A). No isolates from other hospitals in the NE coclustered with the NYN isolates in cluster g (Fig. 5A). The concentration of NYN isolates in cluster g was significant, with a P value equal to 0.001. In addition, three NYN isolates coclustered with the majority of NYB isolates in cluster c and two NYN isolates clustered alone in cluster e (Fig. 5A). In contrast, of the seven isolates from the Virginia hospital VAR, only two coclustered in cluster f (Fig. 5A). The remaining five VAR isolates did not cocluster. Only one VAR isolate fell in the dominant cluster, cluster c, and one VAR isolate coclustered with two Massachusetts isolates in cluster a (Fig. 5A). The remaining three VAR isolates were unrelated to the rest of the isolates in the collection as well as to each other and separated at the top or bottom of the dendrogram (Fig. 5A). Finally, two of the three isolates from the Massachusetts hospital MAS coclustered in cluster a. These results suggest that while some hospitals, such as NYB and NYN, harbor endemic strains that have undergone microevolution and that are responsible for a significant portion of nosocomial BSI infections, other hospitals, such as VAR, do not.
SE collection.
The collection of SE BSI isolates totaled 41 (Table 1), and the average SAB for the collection was 0.71 ± 0.09 (Fig. 3C), which was significantly lower than that for the entire NE collection (Fig. 3A) (P = 0.01). Collections from the five individual hospitals in the SE contained between 6 and 12 isolates (Table 1). A dendrogram for all SE isolates is presented in Fig. 5B. Thirty of the 41 isolates (73%) separated into six different clusters, a smaller proportion than that of the total NE collection (Table 2). Again, the distribution of isolates in clusters defined by a SAB threshold of 0.80 was not random. For example, FLO isolates represented 72% (8 of 11) of the isolates in the largest cluster, cluster c (Fig. 5B). If the distribution were random, the proportion of FLO isolates in cluster c would have been 29%, which is the proportion of FLO isolates in the SE collection (Table 1). The concentration of FLO isolates in cluster c was significant, with a P value of 0.006. Similar nonrandom distributions were obtained for isolates from hospitals FLF and FLM. FLF and FLM isolates each accounted for 44% of cluster d (Fig. 5B) but for only 24 and 14%, respectively, of the SE collection (Table 1). In addition, two of the three isolates in cluster c were from hospital FLF and both isolates in cluster e were from hospital GAM (Fig. 5B).
TABLE 2.
Relatedness of isolates in refined collections from the different geographical localesa
| Region | Avg. SAB for total collection | Avg. SAB for refined collection | % Isolates in refined collection in clusters for which SABs were as follows:
|
|
|---|---|---|---|---|
| ≥0.80 | ≥0.90 | |||
| NE | 0.77 ± 0.09 | 0.74 ± 0.08 | 77 | 27 |
| SE | 0.71 ± 0.09 | 0.68 ± 0.08 | 65 | 8 |
| MW | 0.67 ± 0.10 | 0.67 ± 0.09 | 61 | 14 |
| SW | 0.76 ± 0.09 | 0.70 ± 0.08 | 39 | 22 |
In the refinement process all but one member of isolates from a single hospital in each cluster (defined by an SAB of ≥0.80) were removed from each collection.
MW collection.
The collection of MW BSI isolates totaled 37 (Table 1), and the average SAB for the collection was 0.67 ± 0.10 (Fig. 3D), which was close to the computed SAB for unrelated isolates (20). The number of isolates per hospital ranged between one and nine (Table 1). A dendrogram for all MW isolates is presented in Fig. 5C. Twenty-three of the 37 isolates (62%) separated into eight clusters (Fig. 5C). This represents a significantly lower proportion than those of the NE and SE collections, with P values of 0.025 and 0.05, respectively. The MW collection did not form any clusters as large as the largest cluster in either the NE or the SW collection (Fig. 5). In spite of the lower average SAB, the distribution of many of the isolates in the dendrogram for the MW collection was nonrandom. For instance, four of the seven isolates (57%) in the largest cluster, cluster g, were from hospital PAD (Fig. 5C). If the distribution were random, PAD isolates would have represented 14% of this cluster, the proportion of PAD isolates in the MW collection (Table 1). The concentration of PAD isolates in cluster g was significant, with a P value of 0.0025. The seven additional clusters contained between two and three isolates each, and four of these clusters each contained two isolates from the same hospital (Fig. 5C).
SW collection.
The collection of SW BSI isolates totaled 33 (Table 1), and the average SAB for the collection was 0.76 ± 0.09 (Fig. 3F). The collection was dominated by 22 isolates (67%) from a single hospital, hospital NVL (Table 1). A dendrogram for all SW isolates is presented in Fig. 5D. Twenty-two of the 33 isolates (67%) separated into three clusters. This represents a lower proportion than those for the NE or the SE collection (Table 2). The differences were significant, with P values of <0.05. A single cluster, cluster b, contained 18 isolates. Fifteen of these 18 isolates, or 83% of the isolates in cluster b, were from hospital NVL (Fig. 5D). If the distribution were random, only 67% of the isolates in cluster b would have been from NVL. Two additional NVL isolates coclustered, but none of the isolates from the remaining four SW hospitals coclustered.
NW collection.
The collection of NW BSI isolates was small, consisting of seven isolates from three hospitals (Table 1), and the average SAB for the collection was 0.68 ± 0.07 (Fig. 3E), a value close to that for previously analyzed unrelated isolates (20). A dendrogram for NW isolates is presented in Fig. 5E. Only one cluster containing two isolates formed, and it formed right at the threshold of 0.80. The isolates were from different hospitals. Although this collection was too small for meaningful comparisons with the collections from the other geographical regions, it should be noted that the collection of four isolates from hospital MTB did not cocluster.
The NE harbors the most related BSI isolates.
To obtain a meaningful comparison of the levels of interrelatedness of isolates from the different geographical locales, the SABs were computed for each collection after a refinement process in which all but one member of each cluster (defined by an SAB of ≥0.80) from a single hospital were removed from each collection (Table 2). The region with the highest average SAB for the refined collection was the NE. The average SAB dropped from 0.77 ± 0.09 for the entire NE collection to 0.74 ± 0.08 for the refined NE collection (Table 2). The latter value was still well above the measure of unrelatedness (0.65 ± 0.11) determined previously (20). The average SAB for the refined collection from the SE represented the next highest SAB (Table 2). The average SAB in this case dropped from 0.76 ± 0.09 for the entire SE collection to 0.70 ± 0.08 for the refined SE collection (Table 2). The latter was significantly lower than that for the refined collection from the NE, with a P value of 0.001. The average SAB for the refined collections from the MW and SW were even lower than that for the refined collection from the SE. The average SAB for the MW remained at 0.67 when the collection was refined (Table 2), demonstrating that the isolates in the collection were relatively unrelated to begin with. The average SAB for the SW collection dropped from 0.71 ± 0.09 for the entire collection to 0.68 ± 0.08 for the refined collection (Table 2).
The high level of relatedness between isolates in the refined NE collection was reflected in higher proportions of isolates in clusters defined by an SAB threshold of 0.80 and an SAB threshold of 0.90 (Table 2).
Testing the stability of clustering.
Although the unweighted pair group method (31) provides a rapid method for approximating clusters, it does not represent in all cases the individual SABs obtained by direct pairwise SAB computation. Therefore, mistakes can be made by this method in the genesis of higher-order clusters (3). One way to test the stability of a dendrogram generated by this method is to randomize the order of isolates chosen in the genesis of the dendrogram, an option of the software system used in this study. In addition, a second way to test the stability of the generated dendrogram is to add randomly noise of ±2% and ±5% to SABs. These two methods were combined to test the stability of the representative cluster c of the NE dendrogram (Fig. 5A). For each noise level (0, 2, or 5%), 10 random permutations of the order used to generate the dendrogram were performed. At 0% noise, randomization in no case resulted in the loss of the original 21 isolates of the c cluster and in three cases led to the addition of two to four isolates to the c cluster (Table 3). At a noise level of ±2%, the c cluster in 9 of the 10 dendrograms generated contained all 21 isolates of the original c cluster; only 1 isolate was dropped from the c cluster of one dendrogram generated through randomization at ±2% noise (Table 3). In five of the dendrograms, one to two new isolates were added to the cluster, and in all cases, the SAB between the added isolate and the c cluster in the original dendrogram was 0.78 or 0.79 (Table 3). At a noise level of 5%, the c clusters of 6 of the 10 dendrograms contained all of the original 21 isolates of the original c cluster, 3 contained 20 of the 21 isolates, and 1 contained 19 of the 21 isolates. Again, additions to the c cluster occurred, this time in a majority of the dendrograms, and the SABs between the cluster with the additional isolates and the c cluster in the original dendrogram ranged between 0.73 and 0.79. Together, these results demonstrate that the c cluster is relatively stable and suggest that use of the Ca3 probe to generate fingerprint patterns through the unweighted pair group method provides relatively stable clusters at a threshold SAB of 0.80.
TABLE 3.
Testing of the integrity of clustering by randomizing the initial pair and adding noise in the genesis of dendrogramsa
| Noise level (%) | % Original c isolates in new cluster | No. of new isolates penetrating c cluster | Original SAB(s) for added isolate and the original c cluster |
|---|---|---|---|
| 0 | 100 (21) | 0 | |
| 100 (21) | 0 | ||
| 100 (21) | 0 | ||
| 100 (21) | 0 | ||
| 100 (21) | 0 | ||
| 100 (21) | 0 | ||
| 100 (21) | 2 | 0.78, 0.78 | |
| 100 (21) | 2 | 0.78, 0.78 | |
| 100 (21) | 0 | ||
| 100 (21) | 4 | 0.79, 0.79, 0.78, 0.78 | |
| 2 | 100 (21) | 0 | |
| 100 (21) | 0 | ||
| 100 (21) | 2 | 0.79, 0.79 | |
| 100 (21) | 1 | 0.78 | |
| 100 (21) | 2 | 0.79, 0.79 | |
| 100 (21) | 2 | 0.79, 0.79 | |
| 100 (21) | 0 | ||
| 95 (20) | 1 | 0.78 | |
| 100 (21) | 0 | ||
| 100 (21) | 0 | ||
| 5 | 100 (21) | 1 | 0.78 |
| 100 (21) | 1 | 0.78 | |
| 100 (21) | 0 | ||
| 95 (20) | 1 | 0.79 | |
| 95 (20) | 1 | 0.73 | |
| 90 (19) | 4 | 0.78, 0.78, 0.73, 0.73 | |
| 100 (21) | 3 | 0.79, 0.76, 0.79 | |
| 100 (21) | 3 | 0.78, 0.78, 0.79 | |
| 95 (20) | 3 | 0.73, 0.73, 0.78 | |
| 100 (21) | 0 |
The integrity of the c cluster of the NE dendrogram (Fig. 5A) was assessed in the analysis. The original c cluster contained 21 isolates. Ten random starts were analyzed at each noise level.
Individual hospitals harbor endemic nosocomial BSI strains.
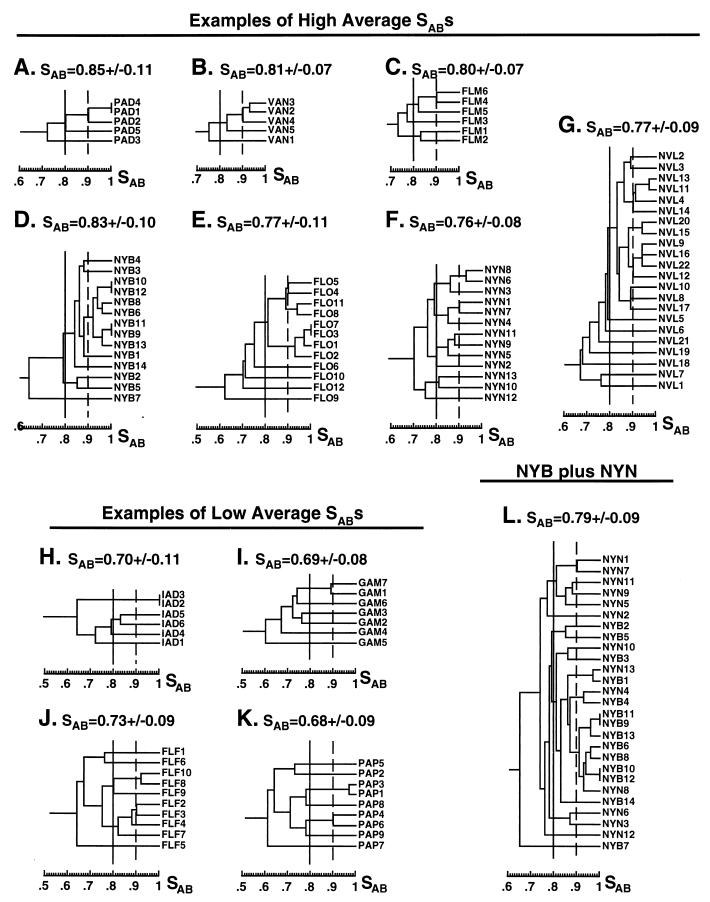
In the dendrograms generated from the total collections of each geographical locale, it was demonstrated that isolates from several of the individual hospitals coclustered in a nonrandom fashion (Figure 5), suggesting the existence of hospital-entrenched or endemic strains. To examine this point further, the average SAB was computed and dendrograms were generated for select individual hospital collections (Fig. 6). Of the 24 hospitals from which two or more isolates were obtained, 7 produced collections for which average SABs were ≥0.75 and 4 produced collections for which average SABs were ≥0.80 (Table 4). The NE contained the greatest proportion of individual hospital collections for which average SABs were ≥0.75, while the SW contained the lowest proportion (Table 4) (the NW collection was too small to be included in this comparison).
FIG. 6.
Dendrograms of collections from individual hospitals. Vertical solid and dashed lines denote SAB thresholds of 0.80 and 0.90, respectively. The average ± standard deviation SAB for each collection is noted at the top of each panel.
TABLE 4.
Isolates from the same hospital are in many cases highly related
| Region | Hospital | No. of isolates | Average SAB | % Isolates in clusters for which SABs are as follows
|
|
|---|---|---|---|---|---|
| ≥0.80 | ≥0.90 | ||||
| Unrelateda | 22 | 0.65 ± 0.11 | 59 | 27 | |
| NE | NYB | 14 | 0.83 ± 0.10b | 93 | 50 |
| NYN | 13 | 0.76 ± 0.08 | 92 | 31 | |
| VAN | 6 | 0.81 ± 0.07 | 80 | 60 | |
| VAR | 7 | 0.71 ± 0.07 | 29 | 0 | |
| NYB + NYN | 27 | 0.79 ± 0.09 | 89 | 52 | |
| SE | FLF | 10 | 0.73 ± 0.09 | 70 | 40 |
| FLM | 6 | 0.80 ± 0.07 | 83 | 33 | |
| FLO | 12 | 0.77 ± 0.11 | 67 | 67 | |
| GAM | 7 | 0.69 ± 0.08 | 29 | 29 | |
| GAS | 6 | 0.72 ± 0.10 | 50 | 0 | |
| MW | IAD | 6 | 0.70 ± 0.11 | 67 | 33 |
| IAI | 5 | 0.74 ± 0.08 | 80 | 0 | |
| PAD | 5 | 0.85 ± 0.10 | 80 | 60 | |
| PAP | 9 | 0.68 ± 0.09 | 44 | 44 | |
| SDS | 5 | 0.67 ± 0.08 | 40 | 0 | |
| SW | AZS | 4 | 0.72 ± 0.06 | 50 | 0 |
| NVL | 22 | 0.77 ± 0.09 | 68 | 68 | |
| NW | MTB | 4 | 0.69 ± 0.05 | 0 | 0 |
This set represents a collection of C. albicans isolates which was considered unrelated in a previous study (20) and includes the following isolates: FC-12, FC-15, FC-5, FC-27, FC-2, FC-21, FC-25, FC-6, FC-18, FC-24, FC-22, FC-4, FC-13, FC-8, FC-26, FC-9, FC-10, FC-20, FC-28, FC-29, FC-7, and FC-16.
Underscores represent SABs significantly different from a random collection.
In the seven hospital collections for which average SABs were above 0.75, the proportion of isolates in clusters defined by a threshold of 0.80 ranged between 93 and 67% (Table 4). In all cases, this was above 59%, the level obtained with an unrelated collection (Table 4) (20). In six of these seven hospital collections, the proportion of isolates in clusters defined by a threshold of 0.90 ranged between 33 and 68%, all higher than the 27% value obtained with an unrelated collection (Table 4). In the dendrograms of the NYB, FLO, and NVL collections, clusters defined by a threshold of 0.90 contained four or more isolates (Fig. 6D, E, and G, respectively). In particular, one cluster in the NYB dendrogram contained 7 of the 14 isolates (50%) of this particular hospital collection (Fig. 6D). Since several NYB and NYN isolates coclustered in the dendrogram of the entire NE collection (Fig. 5A), a combined dendrogram was generated for the two hospital collections (Fig. 6L). One NYN isolate, NYN8, entered the highly related NYB cluster defined by an SAB threshold of 0.90. At a threshold of 0.80, 15 of the 27 isolates (56%) from the combined collection generated a single mixed cluster (Fig. 6L).
In one hospital collection, FLF, for which the average SAB was 0.73 ± 0.09, SABs for two pairs of isolates were ≥0.90 (Fig. 6J), representing 40% of the isolates (Table 4), and in another collection, PAP, for which the average SAB was 0.68 ± 0.09, SABs for two pairs of isolates were ≥0.90 (Fig. 6K), representing 44% of the isolates (Table 4). Together, these results demonstrate that while the dendrograms of some hospital collections contain significant clusters of moderately (threshold SAB, ≥0.80) and highly (threshold SAB, ≥0.90) related isolates, suggesting that they harbor endemic BSI strains, the dendrograms of other hospital collections do not contain similar clusters, suggesting the absence, in the latter cases, of endemic strains.
Geographical specificities of select BSI isolates.
To test whether highly related isolates from a single hospital also exhibited geographical specificity, we compared the proportion of isolates in collections from specific geographical locales and the proportion of isolates in the total collection for which SABs with select hospital isolates were ≥0.80 or ≥0.90 (Table 5). To compute these proportions, we refined the collection by removing all but one of the isolates from the same hospital which clustered at an SAB threshold of ≥0.80. Several of the isolates selected from hospital-enriched clusters exhibited geographical specificity. For instance, for isolate NYB10, which originated from a cluster containing seven BSI isolates from hospital NYB (Fig. 6D), the SAB with 38 isolates in the total collection, 16 (42%) of which were from the NE, was ≥0.80 (Table 5). If relationships were random, the latter value would have been 22%, the proportion of isolates in the total collection for which SABs with NYB10 were ≥0.80. The difference was significant, with a P value of 0.002. For NYB10 the SAB with 16 isolates of the total collection, 9 (56%) of which were from the NE, was ≥0.85 (Table 5). If the relationships were random, the latter value would have been 10%, the proportion of isolates in the total collection for which SABs with NYB10 were ≥0.85. Again, the difference was significant, with a P value of 0.002. Isolate NYB9 showed even stronger geographical specificity. The SAB for NYB9 with 11 isolates in the total collection, eight (73%) of which were from the NE, was ≥0.80 and the SAB with two isolates in the total collection, both (100%) of which were from the NE, was ≥0.85 (Table 5). Again, if random, the latter proportions in each case would have been 7 and 10%, respectively, the proportions of isolates in the total collection for which SABs with NYB9 were ≥0.80 and ≥0.85, respectively.
TABLE 5.
Geographical specificities of select isolates from clusters
| Hospital collection isolate | Geo- graphical locale | East or West | SAB threshold | No. of isolates specific to geographical locale/total no. of isolates (%) | No. of isolates specific to East or West/total no. of isolates (%) |
|---|---|---|---|---|---|
| FL03 | SE | E | 0.80 | 2/6 (33) | 5/6 (83) |
| 0.85 | 1/3 (33) | 3/3 (100) | |||
| 0.90 | 1/2 (50) | 2/2 (100) | |||
| FL08 | SE | E | 0.80 | 1/7 (14) | 7/7 (100) |
| 0.85 | |||||
| FLF10 | SE | E | 0.80 | 2/5 (40) | 4/5 (80) |
| 0.85 | |||||
| IAD2 | MW | E | 0.80 | ||
| PAP3 | MW | E | 0.80 | 0/11 (0) | 6/11 (55) |
| 0.85 | 0/3 (0) | 3/3 (100) | |||
| NYB10 | NE | E | 0.80 | 16/38 (42) | 28/38 (74) |
| 0.85 | 9/16 (56) | 13/16 (81) | |||
| 0.90 | 0/1 (0) | 1/1 (100) | |||
| NYB9 | NE | E | 0.80 | 8/11 (73) | 11/11 (100) |
| 0.85 | 2/2 (100) | 2/2 (100) | |||
| NYN6 | NE | E | 0.80 | 1/5 (20) | 5/5 (100) |
| 0.85 | 0/2 (0) | 2/2 (100) | |||
| NVL9 | SW | W | 0.80 | 6/17 (35) | 7/17 (41) |
| 0.85 | |||||
| NVL11 | SW | W | 0.80 | 4/22 (18) | 4/22 (18) |
| 0.85 | 0/3 (0) | 0/3 (0) |
In the case of isolates FLO3, FLO8, NYN6, and PAP3, there appeared to be marginal or no specificity to geographical locales (i.e., NE, SE, etc.). In the case of NVL, there was neither specificity to the geographical locale nor specificity to the general region (Table 5). These results suggest that while some isolates are relatively specific to a geographical locale, others are not.
The genetic relatedness of fluconazole-resistant isolates.
All isolates were tested for fluconazole resistance according to the criteria set forth in Materials and Methods. Fifteen such isolates (isolates FLO10, FLF6, FLM5, NYB13, NYB3, FLM2, NVL8, VAR1, SDS2, GAS2, IAI3, NYN5, IAI5, GAM5, and MTB4) were identified, including representatives from all five geographical locales. While the NE and SW were underrepresented according to their proportions in the entire collection, the SE was overrepresented. The average SAB for fluconazole-resistant isolates was 0.67 ± 0.09, which is very close to the average SAB of 0.65 ± 0.09 for unrelated isolates (20). At an SAB threshold of 0.80, the two NYB isolates, isolates NYB3 and NYB13, represented the only fluconazole-resistant isolates from the same hospital which coclustered, but even in this case the SAB was less than 0.85 (dendrogram not shown). Several of the fluconazole-resistant isolates were members of hospital-specific clusters but were the sole representatives in the fluconazole-resistant collection of isolates. For instance, NYB13 was a member of a cluster of seven NYB isolates for which SABs were ≥0.90, yet it was the only member of that cluster which was fluconazole resistant. These results suggest that the fluconazole-resistant isolates in the present collection were not highly related across the continental United States or even in a particular geographical locale.
DISCUSSION
In order to control nosocomial infections, it is imperative that the origins of the infecting organisms be identified. In the case of C. albicans and related species, this problem is complicated by commensalism. Since C. albicans is an opportunistic pathogen capable of living benignly in the oral cavity, gastrointestinal tract, and genitalia of healthy individuals (15), the origin of a nosocomial infection may be the microflora of the patient, the attending physicians and medical staff, other neighboring patients, visiting family and friends, or the physical hospital environment. If the infecting pathogen originates in the established microflora of the patient, there should be no hospital-associated specificity. In other words, nosocomial isolates should exhibit the same genetic diversity as isolates collected randomly. If infecting pathogens originate from other hospitalized patients, the hospital staff, or the hospital environment, genetic diversity should be restricted. The latter prediction is based upon a number of recent observations. First, it has been demonstrated that specific strains of C. albicans are highly adapted to different anatomical locations since different strains may be carried by the same healthy individual at different body locations (34). These strains have been demonstrated to be maintained over relatively long periods of time (32). Therefore, the restricted set of strains carried by a resident medical staff should not vary significantly over a time period of 15 months, the period of collection in this study. Second, it has been demonstrated for both C. albicans (10, 11, 30) and Candida glabrata (12) that the major scenario in recurrent vaginal infections is strain maintenance over time, with or without microevolution. In addition, if C. albicans can adapt to an anatomical niche and establish and maintain itself in that niche over long periods of time, then select strains may also establish and maintain themselves over long periods of time in a hospital setting. Nosocomial isolates from such a hospital setting would be highly related, but not identical, since such a hospital-entrenched strain would undergo microevolution (10–12). If specific strains adapt or specialize in a hospital setting, they may also establish themselves in a number of hospitals within the same geographical setting, such as the NE.
DNA fingerprinting provides a means for approaching these fundamental questions. Southern blot hybridization with the complex probe Ca3, multilocus enzyme electrophoresis, and randomly amplified polymorphic DNA analysis were recently compared for their effectiveness in assessing genetic distance in a collection of C. albicans isolates that contained identical, highly related, moderately related, and unrelated isolates (20). The results demonstrated relative equivalence in their capacities to discriminate and cluster isolates in the different categories, although Southern blot hybridization with the Ca3 probe had an advantage in discriminating between the most highly related isolates. Here we have used Southern blot hybridization with Ca3 not only because of its effectiveness in assessing relationships between moderately and highly related isolates and in identifying microevolution but also because of its amenability to computer-assisted analysis. In particular, we have taken advantage of the last characteristic to assess the relatedness of a large number of nosocomial isolates from patients with BSIs collected across the continental United States during a time period of 15 months. This represents the first of three studies. It will be followed by a second survey of isolates from the same hospitals 2 years after the first study to test the maintenance of endemic strains in select hospitals suggested in the present study. In addition, a detailed longitudinal study of BSI isolates from hospitals with established nosocomial strains will be performed to assess microevolution and to test the emergence of drug-resistant strains.
Use of SABs and dendrograms to assess genetic distance.
We have used in this analysis the SAB based on the position of bands in the Ca3 pattern to compare relatedness and to generate dendrograms. To compute average SABs, we have used the individual SABs between pairs computed directly from pairwise computations. Since dendrograms based on SABs generated by the unweighted pair group method are somewhat flawed because the branch points are influenced by the order of pairing (3), we have not used the dendrograms other than to consider general clustering patterns. We have tested the rigorousness of the clusters in the dendrograms by two methods, first by randomizing the order of the isolates scanned in the genesis of dendrograms and second by introducing noise to the SAB values in the genesis of the dendrograms. In both cases, we have demonstrated the relative stability of clusters formed at thresholds of ≥0.80. We have used this threshold because of the rigorousness of the clusters that it defines and because it is close to the halfway point between the SAB of 0.65 previously computed for unrelatedness (20) and the SAB of 1.00 representing identicalness.
Geographical specificities of BSI isolates.
Although the collections from different geographical locales were not matched, they provided us with enough information to suggest geographical differences. By using only one isolate in a cluster from each individual hospital, we have found that the average SAB for the refined NE collection was significantly higher than the average SABs for the refined collections of the remaining three geographical locales (i.e., SE, MW, and SW), from which more than 10 BSI isolates were obtained. The average SABs for the four locales were 0.74 ± 0.08, 0.68 ± 0.08, 0.67 ± 0.09 and 0.70 ± 0.08, respectively. In the case of the NE and the SW the average SABs for the refined collections were significantly higher than that previously computed for 22 unrelated isolates of C. albicans (20). These results suggest that the BSI isolates from a single geographical locale can be more highly related than a random set of isolates and that isolates from one geographical locale (in this case, the NE) can be, on average, more highly related than isolates from other geographical locales.
By selecting individual isolates from different geographical locales and computing the number of isolates in the general collection related to them at SABs of ≥0.80, 0.85, or 0.90, we have also demonstrated the geographical specificities of select isolates. Several BSI isolates showed geographical specificities not only in their restricted geographical locales but also in the East or West portion of the United States. These results support previous demonstrations of the geographical specificities of some strains (5, 28).
Hospital-entrenched strains.
Our results also suggest that while in some cases the SABs for BSI isolates from a hospital are close to those for a random group of isolates (e.g., the nine isolates from hospital PAP), for isolates from other hospitals (e.g., the 14 isolates from hospital NYB), the average SABs are remarkably high. In the former case, for 44% of the isolates SABs were ≥0.80, while in the latter case, for 93% of the isolates SABs were ≥0.80. This result suggests that while the BSI isolates from some hospitals reflect random origins, consistent with the heterogeneity of commensal organisms carried into the hospital by patients, the BSI isolates from other hospitals show restricted diversity, consistent with a hospital origin. The high degree of relatedness of the latter is consistent with ongoing transmission of C. albicans within the hospital setting. The reasons for the potential differences in transmission between different hospitals are under investigation but could reflect the patient population at the different hospitals, underlying diseases, duration of hospitalization, or differences in infection control practices. In the case of select hospitals in this study, bacterial BSI as well as fungal BSI isolates exhibited a high degree of relatedness (18a), suggesting that deficiencies in general infection control practices may be the reason for the endemic BSI strains of C. albicans identified. The possibility that such nosocomial transmission is mediated by carriage of the infecting strain on the hands of health care workers is under investigation. The detection of endemic strains of C. albicans does not prove that patients hospitalized in these settings are at greater risk for a C. albicans BSI, although it is a possibility that can now be tested.
Fluconazole-resistant isolates.
Our results suggest that in some hospitals, a single strain of C. albicans has established itself, and the fact that unrelated patients in the same hospital are infected with highly related but nonidentical isolates suggests that hospital-entrenched strains are diversifying through microevolution. If a hospital-entrenched strain became drug resistant, there arises the possibility that a significant proportion of highly related isolates from BSIs in a single hospital would be drug resistant. In the collection analyzed here, this would have been apparent as a cluster in a dendrogram of fluconazole-resistant isolates from the entire collection. In fact, we found that for fluconazole-resistant isolates in the collection SABs were low, and these isolates did not generate clusters for which the SAB threshold was 0.85. Furthermore, single isolates from highly related hospital clusters were fluconazole resistant, while the remaining members of the cluster were not. This result suggests that drug-resistant BSI strains may not dominate in a hospital setting; rather, drug-resistant isolates emerge from among the endemic strains. In other words, fluconazole-resistant strains of C. albicans may not predominate in a single geographical locale or a single hospital setting. For instance, NYB13, a fluconazole-resistant isolate, was a member of the very large c cluster of the NYB dendrogram containing 11 of the 14 NYB isolates (Fig. 4A). No other member of that cluster was fluconazole resistant. The only other NYB isolate that was fluconazole resistant was NYB3, and that isolate was one of the three NYB isolates not in the c cluster (i.e., NYB3 and NYB13 were not highly related). However, the number of fluconazole-resistant isolates in the present BSI collection was relatively low, and therefore, this does not exclude the possibility that fluconazole-resistant C. albicans strains can become endemic in some hospital settings, a possibility with negative ramifications for compromised patients.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grants AI2392 and DE1058 from the National Institutes of Health (to D.R.S.), by training grant AG00214 from the National Institutes of Health (to S.R.L.), and a grant from the Wyeth-Ayerst Research (Pearl River, N.J.).
We acknowledge the excellent cooperation and participation of all member institutions of the SCOPE Program. Participating institutions contributing data or isolates to the present study include the following: Chandler Hospital, Savannah, Ga. (A. Davis, L. Formby); St. Joseph Hospital, Omaha, Nebr. (S. Cavalieri, A. Lorenzen); St. Jude Medical Center, Fullerton, Calif. (D. Koga, P. Wardell); St. Alexus Medical Center, Bismark, N.D. (R. Baltzer, S. Ziemann); St. Joseph’s Hospital and Health Center, Dickinson, N.D. (D. Splichal); Parkview Episcopal Medical Center, Pueblo, Colo. (L. Fairbaks); St. Mary’s Hospital, Enid, Okla. (C. Williams, J. Word); Western Pennsylvania Hospital, Pittsburgh, Pa. (K Gartner, T. Montgomery); St. Mary’s Medical Center, Langhorne, Pa. (P. Arsdale, H. Kroh); Geisinger Medical Center, Danville, Pa. (P. Bourbeau, M. Dahlman); Mt. Sinai Medical Center, Miami, Fla. (J. Moore, S. Sharp); Boward General Hospital, Ft. Lauderdale, Fla. (P. Johnson, J. Stone); Memorial Medical Center, Savannah, Ga. (M. McNally, M. Shapiro); Veterans Affairs Medical Center, Portland, Oreg. (R. Tjolker, D. Sewell); Mercy Hospital Medical Center, Des Moines, Iowa (M. L. Davenport, C. Grout); Sioux Valley Hospital, Sioux Falls, S.D. (L. Docken, D. Ohrt); Sacred Heart Medical Center, Spokane, Wash. (D. Anderson, D. Leong); Immanuel Medical Center, Omaha, Nebr. (V. Oczki, G Pullen); Maricopa Medical Center, Phoenix, Ariz. (J. Chapman, S. Gamble); Columbia Presbyterian Hospital, New York, N.Y. (P. Della-Latta, M. Fracaro); Florida Hospital and Medical Center, Orlando, Fla. (H. Ferwerda, T. Otal, S. Hernandez); University of Iowa Hospitals and Clinics, Iowa City (L. Herwaldt, R. N. Jones, M. A. Pfaller); St. John’s Mercy Medical Center, St. Louis, Mo. (J. Block, L. Meyer); Lexington Veterans Affairs Medical Center, Lexington, Ky. (G. Fuller, T. Overman); St. Vincent Hospital and Health Center, Billings, Mont. (L. Temme, S. Skates); University Medical Center of Southern Nevada, Las Vegas (J. Bingham, V. Leslie); Central Maine Medical Center, Lewiston (M. A. Johnson, P. Noddin); United Medical Center, Cheyenne, Wyo. (S. Garner, C. Halverson); Walter O. Boswell Hospital, Sun City, Ariz. (J. Theis, V. Verhoeren); Lutheran General Hospital, Park Ridge, Ill. (N. Bharani, C. Galaviz); Akron General Medical Center, Akron, Ohio (D. Sailsbury, P. Steckel); Bronson Methodist Hospital, Kalamazoo, Mich. (R. Van Enk, K. Hanson); Sentara Norfolk General Hospital, Norfolk, Va. (B. Greene, L. Howell); University of Alabama Hospital, Birmingham (M. Long); Veterans Affairs Medical Center, Little Rock, Ark. (L. Illing); San Bernadino County Medical Center, San Bernadino, Calif. (M. Tomasulo); Veterans Affairs Medical Center, Palo Alto, Calif. (C. Valdon, G. Vigionese, C. Vyeda); Mercy Hospital, Bakersfield, Calif. (S. Eyherabide, S. Langenfeld); St. Luke’s Hospital, Houston, Tex. (V. Kennedy); Baystate Medical Center, Springfield, Mass. (M. Gardner, M. Schulte); Youville Hospital, Cambridge, Mass. (B. Mac Arthur, P. Souza); Robert Wood Johnson University Hospital, New Brunswick, N.J. (A. Potts, M. P. Weinstein); Beth Israel Medical Center, New York, N.Y. (W. McKinley, M. Motyl); United Samaritans Medical Center, Danville, Ill. (J. Allen, K. DeBoer); Parkview Regional Medical Center, Vicksburg, Miss. (L. Bane, G. Pierce, N. Taylor); Community Hospitals of Indianapolis, Indianapolis, Ind. (P. Gielerak); Ball Memorial Hospital, Muncie, Ind. (M. Langona, C. Risley); Medical College of Virginia, Richmond (M. Edmund, S. Wallace, R. Wenzel); 89th Medical Group, Andrews Air Force Base, Md. (A. Jarlijen); Suburban Hospital, Bethesda, Md. (Katy Serves); and Presbyterian Health Services, Albuquerque, N.M. (J. Ferranti).
REFERENCES
- 1.Abi-Said D, Anaissie E, Uzun O, Raad I, Pinzcowski H, Vartivarian S. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin Infect Dis. 1997;24:1122–1128. doi: 10.1086/513663. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J, Srikantha T, Morrow B, Miyasaki S, White T, Agabian N, Schmid J, Soll D. Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J Clin Microbiol. 1993;31:1472–1480. doi: 10.1128/jcm.31.6.1472-1480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backeljau T, De Bruyn L, De Wolf H, Jordaens K, Van Dongen S, Winnepenninckx B. Multiple UPGMA and neighbor-joining trees and the performance of some computer packages. Mol Biol Evol. 1996;13:309–313. [Google Scholar]
- 4.Banerjee S, Emori T, Culver D, Gaynes R, Jarvis W, Horan T, Edwards J, Tolson J, Henderson T, Martone W. Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1989. National Nosocomial Infections Surveillance System. Am J Med. 1991;91:86S–89S. doi: 10.1016/0002-9343(91)90349-3. [DOI] [PubMed] [Google Scholar]
- 5.Clemons K, Feroze F, Holmberg K, Stevens D. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;35:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emori T, Gaynes R. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fridkin S, Jarvis W. Epidemiology of nosocomial fungal. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joly S, Pujol C, Schroppel K, Soll D. Development of two species-specific fingerprinting probes for broad computer-assisted epidemiological studies of Candida tropicalis. J Clin Microbiol. 1996;34:3063–3071. doi: 10.1128/jcm.34.12.3063-3071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joly, S., M. Pfaller, F. Odds and D. Soll. Unpublished data.
- 10.Lockhart S, Fritch J, Meier A, Schroppel K, Srikantha S, Galask R, Soll D. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockhart S, Reed B, Pierson C, Soll D. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstrated by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J Clin Microbiol. 1996;34:767–777. doi: 10.1128/jcm.34.4.767-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockhart S, Joly S, Pujol C, Sobel J, Pfaller M, Soll D. Development and verification of fingerprinting probes for Candida glabrata. Microbiology. 1997;143:3733–3746. doi: 10.1099/00221287-143-12-3733. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Nguyen M, Peacock J, Morris A, Tanner D, Nguyen M, Snydman D R, Wagener M, Rinaldi M, Yu V. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 15.Odds F. Candida and candidosis: a review and bibliography. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 16.Pfaller, M. 1995. Epidemiology of candidiasis. J. Hosp. Infect. 30(Suppl.):329–338. [DOI] [PubMed]
- 17.Pfaller, M. 1996. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin. Infect. Dis. 22(Suppl.):589–594. [DOI] [PubMed]
- 18.Pfaller M, Rex J, Rinaldi M. Antifungal susceptibility testing: technical advances and potential clinical applications. Clin Infect Dis. 1997;24:776–784. doi: 10.1093/clinids/24.5.776. [DOI] [PubMed] [Google Scholar]
- 18a.Pfaller, M. A., S. A. Messer, M. B. Edmond, R. N. Jones, and R. P. Pfaller. Unpublished observations.
- 19.Price M, LaRocco M, Gentry L. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob Agents Chemother. 1994;38:1422–1427. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pujol C, Joly S, Lockhart S, Noel S, Tibayrenc M, Soll D. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rex J, Rinaldi M, Pfaller M. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rex J, Pfaller M, Galgiani J, Bartlett M, Espinel I, Ghannoum M, Lancaster M, Odds F, Rinaldi M, Walsh T, Barry A. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Subcommittee on Antifungal Susceptibility Testing of the National Committee for Clinical Laboratory Standards. Clin Infect Dis. 1997;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 23.Rosner B. Fundamentals of biostatistics. Belmont, Calif: Duxbury Press; 1995. [Google Scholar]
- 24.Sadhu C, McEachern M, Rustchenko B, Schmid J, Soll D, Hicks J. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J Bacteriol. 1991;173:842–850. doi: 10.1128/jb.173.2.842-850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schaberg D, Culver D, Gaynes R. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91:72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 27.Scherer S, Stevens D. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid J, Rotman M, Reed B, Pierson C, Soll D. Genetic similarity of Candida albicans isolates from vaginitis patients and their partners. J Clin Microbiol. 1993;31:39–46. doi: 10.1128/jcm.31.1.39-46.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid J, Voss E, Soll D. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroeppel K, Rotman M, Galask R, Mac K, Soll D. Evolution and replacement of Candida albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J Clin Microbiol. 1995;32:2646–2654. doi: 10.1128/jcm.32.11.2646-2654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sneath P, Sokal R. Numerical taxonomy. The principles and practice of numerical classification. San Francisco, Calif: W. H. Freeman & Co.; 1973. [Google Scholar]
- 32.Soll, D., and R. Galask. Unpublished observations.
- 33.Soll D, Langtimm C, McDowell J, Hicks J, Galask R. High-frequency switching in Candida strains isolated from vaginitis patients. J Clin Microbiol. 1987;25:1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soll D, Galask R, Schmid J, Hanna C, Mac K, Morrow B. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J Clin Microbiol. 1991;29:1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soll D R. DNA fingerprinting of Candida albicans. J Mycol Med. 1993;3:37–44. [Google Scholar]
- 36.Wey S, Mori M, Pfaller M, Woolson R, Wenzel R. Risk factors for hospital-acquired candidemia. A matched case-control study. Arch Intern Med. 1989;149:2349–2353. [PubMed] [Google Scholar]