Abstract
During a 2-year period, 10 strains of Arthrobacter cumminsii were isolated in or received by a Swiss routine clinical bacteriology laboratory, and 5 further isolates were referred to a Swedish bacteriology reference center over a 5-year period, making A. cumminsii the most frequently encountered Arthrobacter species in these two laboratories. The present report outlines the clinical features of the 15 A. cumminsii strains and presents an extended biochemical characterization of this microorganism. A. cumminsii exhibits a unique cellular fatty acid pattern with the consistent presence of C14:0i and C14:0 fatty acids as well as relatively large amounts of C16:0i and C16:0 fatty acids usually not seen in other Arthrobacter spp. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was found to be a useful tool for confirmation of the identification of A. cumminsii. The MICs of 39 antimicrobial agents were determined, and it was demonstrated that aminoglycosides and quinolones had only weak activities against A. cumminsii strains, in contrast to their activities against most other coryneform bacteria. As a result of the extended characterization of A. cumminsii, an emended description of this species is presented. Due to the lack of A. cumminsii in established identification systems, it is most likely that this species is underdiagnosed in many routine clinical bacteriology laboratories.
In recent years, some genera (e.g., Aureobacterium, Cellulomonas, and Microbacterium) belonging to the coryneform bacteria (i.e., aerobically growing, asporogenous, irregular, non-partially acid-fast, gram-positive bacteria) were shown not only to be present in environmental specimens but also to be derived from human clinical material and to cause infections in selected patients (10). The genus Arthrobacter is another genus which is very widely distributed in the environment, especially in soil (12), and it has only been recently described as being isolated from human clinical specimens (2, 7, 11). It was demonstrated that there is an enormous heterogeneity within Arthrobacter strains isolated from humans, suggesting that many strains are representatives of separate individual yet undescribed species (7). However, 3 of 11 clinical Arthrobacter strains in the study by Funke et al. (7) were shown to be Arthrobacter cumminsii strains, whereas all the other Arthrobacter strains were single representatives of different taxa. This indicated for the first time that A. cumminsii might be the most frequently encountered Arthrobacter species in clinical specimens. The present report from two different European laboratories confirms this initial observation. In addition, an extended characterization of A. cumminsii comprising 15 new strains is presented. The extended characterization has led to a slightly emended description of A. cumminsii.
MATERIALS AND METHODS
Strains and culture conditions.
Workers in the Department of Medical Microbiology at the University of Zürich (DMMZ) isolated from patients’ material 9 of the 15 strains listed in Table 1 by established methods (3). The nine Swiss patients were all hospitalized in the Zürich metropolitan area. Only strain DMMZ 2279 was referred from abroad to DMMZ for identification. The other five strains listed in Table 1 were referred from various laboratories in Sweden to the Culture Collection of the University of Göteborg (CCUG) for identification. All strains were subcultured on Columbia agar (Difco, Detroit, Mich.) supplemented with 5% sheep blood (SBA) at 37°C in a 5% CO2 atmosphere before they were used for further testing.
TABLE 1.
Origins of the A. cumminsii strains studied
| Strain no. | Patient’s sex,a age (yr) | Material or clinical diagnosis | Other bacteria isolated |
|---|---|---|---|
| DMMZ 1490 (CCUG 38876) | f, 28 | Urinary tract infection | |
| DMMZ 2279 (CCUG 38877) | f, 66 | Urinary tract infection | Members of the family Enterobacteriaceae, enterococci |
| DMMZ 2358 (CCUG 38878) | f, 45 | Chronic otorrhea | Staphylococcus aureus |
| DMMZ 2554 (CCUG 38879) | m, 73 | Urinary tract infection | |
| DMMZ 2558 (CCUG 38880) | f, 17 | Infected amniotic fluid | |
| DMMZ 2591 (CCUG 38881) | f, 33 | Vaginal swab | Lactobacilli |
| DMMZ 3147 (CCUG 38882) | f, 40 | Calcaneus osteomyelitis | Corynebacterium striatum |
| DMMZ 3406 (CCUG 38883) | f, 23 | Urinary tract infection | |
| DMMZ 3419 (CCUG 38884) | m, 70 | External otitis | Miscellaneous gram-negative bacteria |
| DMMZ 3452 (CCUG 38885) | m, 61 | Deep tissue infection, upper leg | |
| CCUG 28647 | f, 36 | Chronic cervicitis | |
| CCUG 28802 | ND,b 70 | ND | |
| CCUG 33745 | m, 47 | Blood culture, urosepsis | |
| CCUG 35241 | m, 83 | Blood culture | |
| CCUG 35863 | f, 81 | Leg wound |
f, female; m, male.
ND, no data.
Biochemical tests.
All isolates were initially screened for their biochemical reactions according to the system outlined by von Graevenitz and Funke (17). The commercial API Coryne system (bioMérieux, Marcy l’Etoile, France) was used according to the manufacturer’s instructions except that the strips were incubated and read after up to 7 days at 37°C in ambient air.
Chemotaxonomic investigations.
The diamino acid of the cell wall peptidoglycan was determined by thin-layer chromatography analysis (7). The cellular fatty acid (CFA) patterns were generated by using the Sherlock Microbial Identification System (Microbial ID, Inc., Newark, Del.) (18).
Susceptibility tests with antimicrobial agents.
The MICs (always given in micrograms per milliliter) of 39 antimicrobial agents were determined by a microdilution method with the Merlin MIC system (Merlin Diagnostics, Bornheim-Hersel, Germany) with H medium (Merlin Diagnostics), as outlined previously (5). β-Lactamase activity was tested for by using the chromogenic substrate nitrocefin (14). MICs were interpreted by using the criteria for staphylococci (which are not as strict as the criteria for streptococci) established by the National Committee for Clinical Laboratory Standards (13), although it must be emphasized that the National Committee for Clinical Laboratory Standards has not explicitly published guidelines for coryneform bacteria.
PAGE of whole-cell proteins.
The strains were grown on horse blood agar (Columbia base) at 37°C in the presence of 5% CO2. Polyacrylamide gel electrophoresis (PAGE) of whole-cell proteins was performed as described by Pot et al. (15). For densitometric analysis, normalization and interpretation of protein patterns, the Gelcompar (version 3.1) software package (Applied Maths, Kortrijk, Belgium) was used. The levels of similarity between pairs of traces were expressed by the Pearson product moment correlation coefficient converted for convenience to a percentage value.
RESULTS AND DISCUSSION
The 10 DMMZ A. cumminsii strains included in this study (Table 1) were isolated or received between January 1996 and December 1997, whereas the 5 CCUG A. cumminsii strains were referred to CCUG for identification between June 1991 and May 1996. Workers in DMMZ isolated or received as reference cultures seven other Arthrobacter strains during the 2-year period described above, and none of those was identical to any other strain or belonged to any of the other presently established Arthrobacter species. CCUG had received three other clinical Arthrobacter strains during the 5-year period mentioned above. Therefore, A. cumminsii was the most frequently encountered Arthrobacter species isolated from clinical specimens in these two European laboratories.
Of the 14 patients for whom data were available, 9 were female and 5 were male. Their mean age was 50.2 years (age range, 17 to 83 years). We did not observe multiple isolates from the same patients. None of the patients was epidemiologically linked. Additional clinical data were, unfortunately, not available for the patients from whom the five CCUG strains were isolated. We observed three patients with urinary tract infections, and A. cumminsii grew (104 to >105 CFU/ml) in pure cultures from samples from these patients. In a sample from one patient with a deep tissue infection, A. cumminsii grew in pure culture to approximately 103 to 104 CFU/ml; Gram staining showed a moderate leukocyte response, but no microorganisms were seen. Short, small gram-positive rods were seen in the direct Gram stain of a sample of infected amniotic fluid. A. cumminsii was recovered together with multiple other bacteria from two patients with external otitis. It was also not grown in pure culture from a sample from a patient with calcaneus osteomyelitis or in a pure culture from a vaginal swab. In summary, A. cumminsii seems to be a microorganism with a rather low level of pathogenicity since cases of severe, life-threatening infections were not observed in our series of patients. However, A. cumminsii might be the bacterial agent responsible for selected cases of urinary tract infections. In the initial study of Funke et al. (7) on A. cumminsii, two of the three strains also came from patients with urinary tract infections, and a third strain was isolated from a patient with a skin infection. It seems not unlikely that A. cumminsii is part of the normal human skin and mucous membrane flora, in particular, in the genitourinary tract, although these anatomical sites have not been systematically screened for A. cumminsii. Up to now, humans have not been described as reservoirs for Arthrobacter spp. (12).
A. cumminsii colonies are smaller than 2 mm in diameter after 24 h of incubation at 37°C either in a 5% CO2-enriched atmosphere or in ambient air. In contrast, all other presently defined Arthrobacter spp. which are able to grow at 37°C exhibit colonies which are larger than 2 mm in diameter after 24 h of incubation (4, 7). Colonies of A. cumminsii are more grayish than those of other Arthrobacter spp. and are also less convex. Like the other Arthrobacter spp., A. cumminsii does not give off the typical cheese-like smell which is observed in the biochemically closely related Brevibacterium spp. (6, 7). Gram stains of colonies from SBA cultures (24 h of incubation at 37°C) showed ordinary (i.e., not branching), relatively small (1 to 2 μm in length) coryneform bacteria. A typical rod-coccus growth cycle was not observed for A. cumminsii strains on SBA.
According to the biochemical typing system for coryneform bacteria developed by von Graevenitz and Funke (17), we observed the following characteristics for all 15 strains studied: catalase positive, oxidative metabolism, nonmotile, nitrate reduction negative, esculin hydrolysis negative, no acid formation in cystine Trypticase agar medium from glucose, maltose, sucrose, mannitol, and xylose, CAMP reaction negative, and no lipophilism. Lysine was detected as the cell wall diamino acid in all 15 strains. When extended morphological characterization as well as extended biochemical typing based on the API Coryne system were performed, we found some features which had not been observed for the three A. cumminsii strains included in the original description of this species (7): (i) 4 of 15 strains had a sticky consistency, (ii) two strains were urease positive (which was confirmed by the observation of urease activity in Christensen’s medium), (iii) all 15 strains produced acid from ribose after 7 days of incubation, and (iv) 2 of 15 strains also produced acid from glucose (but more weakly than from ribose).
Numerous API Coryne codes were observed: after 24 h, 0000004 (n = 2 isolates), 2101004 (n = 1), 4002004 (n = 2), 6000004 (n = 3), 6002004 (n = 5), 6101004 (n = 1), and 6102004 (n = 1); after 48 h, 0000004 (n = 1), 0002004 (n = 1), 2103004 (n = 1), 4002004 (n = 2), 6000004 (n = 1), 6002004 (n = 7), 6102004 (n = 1), and 6103004 (n = 1); and after 7 days, 0002204 (n = 2), 2103204 (n = 1), 4002204 (n = 1), 4002304 (n = 1), 6002204 (n = 7), 6002304 (n = 1), 6102204 (n = 1), and 6103204 (n = 1). It is noteworthy that A. cumminsii is not included in the database (version 2.0) of the API Coryne system but that Arthrobacter spp. are included (9). However, because up to eight different numerical profiles were observed for A. cumminsii strains at each time that the strips were read, it might be difficult to include this species in the system’s database. Eleven of the 15 strains exhibited pyrazinamidase activity and 12 of 15 exhibited pyrrolidonyl arylamidase activity. Weak alkaline phosphatase activity was present in 3 of 15 strains. Gelatinase activity was consistently observed in all strains after 7 days.
The CFA patterns of the A. cumminsii strains are given in Table 2. It is obvious that C15:0ai and C15:0i are not as dominant in A. cumminsii as they are in other Arthrobacter spp. In contrast to other Arthrobacter spp., C14:0i and C14:0 are always detectable in A. cumminsii. In addition, larger amounts of C16:0i and C16:0 are present in A. cumminsii than in most other Arthrobacter spp. In nearly all Brevibacterium spp., the amounts of C15:0ai and C17:0ai together are greater than 75% of the total CFAs (6), whereas these CFAs represent only 50 to 60% of the total CFAs in A. cumminsii strains. In contrast, significantly more C16:0 is found in A. cumminsii than in Brevibacterium spp. In summary, A. cumminsii strains exhibit a very unique CFA pattern which is not observed in any other coryneform bacterium (1, 18).
TABLE 2.
CFA patterns of A. cumminsii and phenotypically related taxa
| Taxon | % total CFAsa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13:0ai | 14:0i | 14:0 | 15:0i | 15:0ai | 16:0i | 16:0 | 17:0i | 17:0ai | 18:1ω9c | 18:1ω7t | 18:0 | Feature 6b | |
| A. cumminsiic | 1 ± 1 (0–2) | 3 ± 1 (1–5) | 3 ± 2 (1–7) | 3 ± 1 (2–5) | 37 ± 6 (26–49) | 13 ± 2 (9–19) | 13 ± 5 (6–26) | 1 ± 1 (1–2) | 18 ± 5 (11–28) | 2 ± 1 (1–4) | 1 ± 1 (0–2) | 3 ± 2 (1–7) | 1 ± 1 (0–2) |
| Other Arthrobacter spp.d | 0–4 | 0–3 | 3–29 | 34–75 | 3–15 | 2–10 | 0–11 | 6–25 | 0–3 | 0–4 | |||
| Brevibacterium spp.e | 4–11 | 36–72 | 2–13 | 1–5 | 0–4 | 17–47 | |||||||
The antimicrobial susceptibility patterns of the A. cumminsii strains are given in Table 3. With the exception of ceftibuten, the β-lactams showed good activity against A. cumminsii. Rifampin exhibited an excellent activity, with MICs of ≤0.01 μg/ml, and all A. cumminsii strains tested were susceptible to glycopeptides, as has been reported before (7). Only strains DMMZ 2358, CCUG 35241, and CCUG 35863 were resistant to tetracyclines, and no strain was resistant to macrolides. Aztreonam and fosfomycin MICs for all 15 strains were relatively high, but the MICs for A. cumminsii were slightly lower than those for other coryneform bacteria, especially true corynebacteria (4, 8). Surprisingly, for all isolates the MICs of the aminoglycosides, in particular, those of netilmicin and tobramycin, were relatively high. The MICs of the following other aminoglycosides were also elevated for A. cumminsii: kanamycin (MIC at which 50% of strains are inhibited [MIC50], 64 μg/ml; MIC90, 256 μg/ml), streptomycin (MIC50, 16 μg/ml; MIC90, 64 μg/ml), spectinomycin (MIC50, 32 μg/ml; MIC90, 64 μg/ml), apramycin (MIC50, 64 μg/ml; MIC90, >128 μg/ml), ribostamycin (MIC50, >128 μg/ml; MIC90, >128 μg/ml), and livitomycin (MIC50, >128 μg/ml; MIC90, >128 μg/ml). Surprisingly, high quinolone MICs were also observed, in particular, those of norfloxacin and ofloxacin. The MICs of the following other quinolones were also elevated: enoxacin (MIC50, >32 μg/ml; MIC90, >32 μg/ml), fleroxacin (MIC50, 16 μg/ml; MIC90, >32 μg/ml), and pefloxacin (MIC50, 32 μg/ml; MIC90, >32 μg/ml). Poor activities of aminoglycosides and quinolones combined with good activities of most beta-lactams as well as those of macrolides and tetracyclines have so far not been observed for any other taxon of coryneform bacteria (4, 8, 10, 16). β-Lactamase activity was not detected in any of the 15 strains tested. The better activity of amoxicillin-clavulanic acid compared to that of amoxicillin (Table 3) is probably due to an intrinsic activity of clavulanic acid against A. cumminsii strains. Whether the resistance against aminoglycosides and quinolones is acquired or intrinsic is not clear at present, but its presence in all strains tested makes an intrinsic mechanism (e.g., complicated or blocked transport through the bacterial cell wall) more likely.
TABLE 3.
Antimicrobial susceptibility pattern of A. cumminsii
| Antimicrobial agent | MIC (μg/ml)
|
||
|---|---|---|---|
| Range | 50% | 90% | |
| Amikacin | 4–64 | 16 | 64 |
| Amoxicillin | ≤0.06–2 | ≤0.06 | 1 |
| Amoxicillin-clavulanic acid | ≤0.06–0.25 | ≤0.06 | ≤0.06 |
| Azithromycin | 0.06–4 | 0.125 | 0.5 |
| Aztreonam | 1–>64 | 16 | >64 |
| Cefaclor | ≤0.06–0.5 | ≤0.06 | 0.125 |
| Ceftazidime | 0.5–8 | 1 | 2 |
| Ceftibuten | 1–>64 | 4 | >64 |
| Chloramphenicol | 1–64 | 2 | 32 |
| Ciprofloxacin | 0.5–>32 | 2 | 16 |
| Clarithromycin | ≤0.03–0.5 | ≤0.03 | 0.06 |
| Clindamycin | 0.03–1 | 0.125 | 1 |
| Doxycycline | 0.25–8 | 0.5 | 1 |
| Erythromycin | ≤0.03–1 | ≤0.03 | 0.125 |
| Fosfomycin | 16–>256 | 32 | >256 |
| Fusidic acid | 0.125–0.5 | 0.25 | 0.25 |
| Gentamicin | 2–>128 | 8 | 16 |
| Imipenem | ≤0.03–0.125 | 0.06 | 0.125 |
| Loracarbef | ≤0.125–0.5 | ≤0.125 | ≤0.125 |
| Netilmicin | 4–>128 | 64 | >128 |
| Norfloxacin | 8–>64 | 32 | >64 |
| Ofloxacin | 1–32 | 4 | 32 |
| Oxacillin | ≤0.03–4 | 0.125 | 2 |
| Penicillin | ≤0.01–0.5 | ≤0.01 | 0.5 |
| Piperacillin | ≤0.125–4 | ≤0.125 | 4 |
| Rifampin | ≤0.01 | ≤0.01 | ≤0.01 |
| Teicoplanin | 0.125–0.5 | 0.5 | 0.5 |
| Tetracycline | 1–64 | 1 | 16 |
| Tobramycin | 16–>128 | 64 | >128 |
| Vancomycin | 0.5–1 | 1 | 1 |
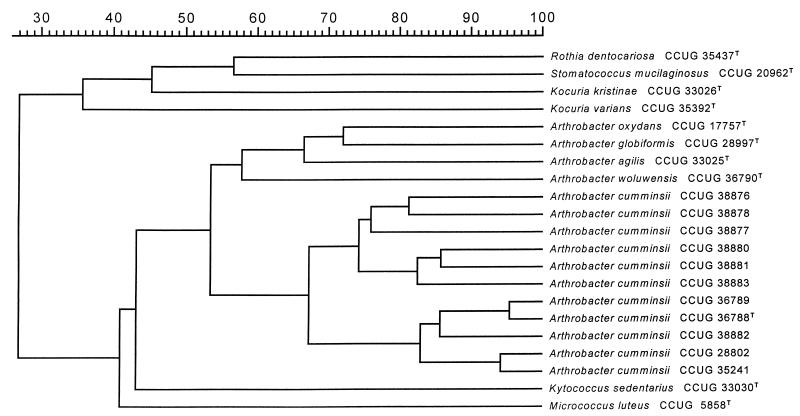
The whole-cell protein profiles of 10 randomly chosen strains included in this study were examined by sodium dodecyl sulfate (SDS)-PAGE. A dendrogram derived from a numerical analysis of the protein profiles is presented in Fig. 1. All 10 strains grouped together with the type strain of A. cumminsii, strain CCUG 36788 (DSM 10493). In addition, they formed a distinct branch with a within-group correlation level of >65%. This indicated that the strains examined represent a homogeneous group. It has been established that comparative quantitative whole-cell protein profiles and quantitative DNA-DNA hybridizations correlate very well (15). It is further evident from the SDS-PAGE analysis that the clinical strains were distinct from other Arthrobacter spp. and phylogenetically related taxa examined. We acknowledge that the use of SDS-PAGE for the identification of species of coryneform bacteria is reserved for a reference laboratory, especially since the laboratory must at first create a large database for later comparative studies and since the technique is labor-intensive. However, once this has been established, SDS-PAGE is a useful tool for the identification of coryneform bacteria to the species level, in particular, for clustering analysis.
FIG. 1.
Similarity dendrogram based on whole-cell protein patterns of A. cumminsii and phylogenetically related taxa. Levels of correlation were expressed as percentages of similarity for convenience.
Although the pathogenic potential of A. cumminsii seems to be rather low compared to those of other gram-positive bacteria, we believe that the present report will make clinical bacteriologists more aware of A. cumminsii strains. For the routine clinical laboratory we recommend that nonfermenting, short, gram-positive rods without a typical corynebacterial Gram staining result (10) but with whitish-grayish colonies of <2 mm in diameter should raise the suspicion of A. cumminsii. On the basis of the data presented above, an emended description of A. cumminsii is given.
Emended description of Arthrobacter cumminsii Funke, Hutson, Bernard, Pfyffer, Wauters, and Collins 1996.
The following description is based on the studies of 18 strains (3 strains were from the study of Funke et al. [7] and 15 are from the present study).
The cells are coryneform bacteria without irregular branching. No spores are formed. The cells are nonmotile. The organism is obligately aerobic. Colonies are whitish-grayish, smooth, slightly convex, and either of creamy or of sticky consistency and are of less than 2 mm in diameter after 24 h of incubation at 37°C in 5% CO2 on SBA. Catalase activity is detected, but nitrate reductase activity is not. Urease activity is variable. Esculinase activity is not detected. DNase and gelatinase activities are observed within 10 days. Acid is produced from ribose but not from maltose, sucrose, mannitol, xylose, lactose, or glycogen. Acid production from glucose is variable. The CAMP reaction is negative. The following other enzyme activities are detected: esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, and phosphoamidase. The activities of pyrazinamidase, pyrrolidonyl arylamidase, alkaline phosphatase, and cystine arylamidase are variable. Chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not present. The peptidoglycan type is l-lysine-l-serine-l-glutamic acid. The DNA base composition ranges from 60 to 62 mol% G+C. The organism is isolated from human clinical specimens. The type strain is strain DSM 10493 (CCUG 36788), which has been deposited in the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany, and in the Culture Collection of the University of Göteborg, Göteborg, Sweden.
ACKNOWLEDGMENTS
A. von Graevenitz is acknowledged for careful review of the manuscript. We thank P. Riegel, Institut de Bactériologie Strasbourg, Strasbourg, France, for kindly referring strain DMMZ 2279.
This study was funded in part by the Swiss National Science Foundation (contract 3100-050648 97/1). G.F. is a recipient of an ESCMID research fellowship.
REFERENCES
- 1.Bernard K A, Bellefeuille M, Ewan E P. Cellular fatty acid composition as an adjunct to the identification of asporogenous, aerobic gram-positive rods. J Clin Microbiol. 1991;29:83–89. doi: 10.1128/jcm.29.1.83-89.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteban J, Bueno J, Perez-Santonja J, Soriano F. Endophthalmitis involving an Arthrobacter-like organism following intraocular lens implantation. Clin Infect Dis. 1996;23:1180–1181. doi: 10.1093/clinids/23.5.1180. [DOI] [PubMed] [Google Scholar]
- 3.Forbes B A, Granato P A. Processing specimens for bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 265–281. [Google Scholar]
- 4.Funke, G. Unpublished data.
- 5.Funke G, Alvarez N, Pascual C, Falsen E, Akervall E, Sabbe L, Schouls L, Weiss N, Collins M D. Actinomyces europaeus sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:687–692. doi: 10.1099/00207713-47-3-687. [DOI] [PubMed] [Google Scholar]
- 6.Funke G, Carlotti A. Differentiation of Brevibacterium spp. encountered in clinical specimens. J Clin Microbiol. 1994;32:1729–1732. doi: 10.1128/jcm.32.7.1729-1732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke G, Hutson R A, Bernard K A, Pfyffer G E, Wauters G, Collins M D. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J Clin Microbiol. 1996;34:2356–2363. doi: 10.1128/jcm.34.10.2356-2363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funke G, Pünter V, von Graevenitz A. Antimicrobial susceptibility patterns of some recently defined coryneform bacteria. Antimicrob Agents Chemother. 1996;40:2874–2878. doi: 10.1128/aac.40.12.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funke G, Renaud F N R, Freney J, Riegel P. Multicenter evaluation of the updated and extended API (RAPID) Coryne database 2.0. J Clin Microbiol. 1997;35:3122–3126. doi: 10.1128/jcm.35.12.3122-3126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funke G, von Graevenitz A, Clarridge III J E, Bernard K A. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, C. L., L. Y. Shih, H. S. Leu, C. L. Wu, and G. Funke.Arthrobacter sp. septicemia in a neutropenic patient with acute lymphoblastic leukemia. Submitted for publication. [DOI] [PubMed]
- 12.Keddie R M, Collins M D, Jones D. Genus Arthrobacter. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1288–1301. [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Minimum inhibitory concentration (MIC) interpretive standards (μg/ml) for organisms other than Haemophilus spp., Neisseria gonorrhoeae, and Streptococcus spp. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.O’Callaghan C H, Morris A, Kirby S M, Shingler A H. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprints. In: Goodfellow M, O’Donnell A G, editors. Modern microbial methods. Chemical methods in prokaryotic systematics. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1994. pp. 493–521. [Google Scholar]
- 16.Soriano F, Zapardiel J, Nieto E. Antimicrobial susceptibilities of Corynebacterium species and other non-spore-forming gram-positive bacilli to 18 antimicrobial agents. Antimicrob Agents Chemother. 1995;39:208–214. doi: 10.1128/aac.39.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Graevenitz A, Funke G. An identification scheme for rapidly and aerobically growing gram-positive rods. Zentrbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1996;284:246–254. doi: 10.1016/s0934-8840(96)80100-9. [DOI] [PubMed] [Google Scholar]
- 18.von Graevenitz A, Osterhout G, Dick J. Grouping of some clinically relevant gram-positive rods by automated fatty acid analysis. APMIS. 1991;99:147–154. doi: 10.1111/j.1699-0463.1991.tb05132.x. [DOI] [PubMed] [Google Scholar]