Abstract
The complexity of the human brain gives the illusion that brain activity is intrinsically high-dimensional. Nonlinear dimensionality-reduction methods such as uniform manifold approximation and t-distributed stochastic neighbor embedding have been used for high-throughput biomedical data. However, they have not been used extensively for brain activity data such as those from functional magnetic resonance imaging (fMRI), primarily due to their inability to maintain dynamic structure. Here we introduce a nonlinear manifold learning method for time-series data—including those from fMRI—called temporal potential of heat-diffusion for affinity-based transition embedding (T-PHATE). In addition to recovering a low-dimensional intrinsic manifold geometry from time-series data, T-PHATE exploits the data’s autocorrelative structure to faithfully denoise and unveil dynamic trajectories. We empirically validate T-PHATE on three fMRI datasets, showing that it greatly improves data visualization, classification, and segmentation of the data relative to several other state-of-the-art dimensionality-reduction benchmarks. These improvements suggest many potential applications of T-PHATE to other high-dimensional datasets of temporally diffuse processes.
As we move through the world, the human brain performs innumerable operations to process, represent, and integrate information. How does a system composed of limited computational units represent so much while handling a constant barrage of incoming information? One possible explanation is that individual neural units operate in tandem to encode and update information in population codes, which exponentially expands the distinct ways in which information can be represented by a fixed number of units. Neural population codes better predict behavior than single neurons, particularly in how representations change over time in response to new information1–5. Although neural population codes exist in high ambient dimensions6, these dimensions are redundant7, and from them emerge dominant latent signals that code for behavior and information8,9.
The principles of neural population codes were defined using direct neural recordings, and were extended to non-invasive, indirect measurements of brain activity using functional magnetic resonance imaging (fMRI). Multivariate pattern analysis10,11 of fMRI activation has yielded valuable insights into the structure and content of cognitive representations, including low-12 and high-level11,13,14 sensory stimuli, as well as higher-order cognitive processes such as memory15, emotion16, narrative comprehension17, and theory-of-mind18. These insights have largely come from group-level analyses, requiring aggregation over subjects and time points to overcome the spatiotemporal noise inherent to fMRI. Like other high-throughput biomedical data, fMRI noise is pervasive at multiple levels, from subject movement to blood-oxygenation-level-dependent (BOLD) signal drift and physiological confounds19. fMRI has a lower temporal resolution than the cognitive processes many studies attempt to measure, with acquisitions every one to two seconds20. Finally, the BOLD signal is a slow, vascular proxy of neuronal activity, peaking approximately four to five seconds after stimulation before returning to the baseline.
In summary, many brain representations of interest are coded in high-dimensional patterns of activation2–4,10,11, which can be characterized by low-dimensional latent signals1,9,21. fMRI affords unique insight into the healthy, behaving human brain, but the data are noisy, sampled slowly, and blurred in time, giving rise to high signal autocorrelation20,22. Furthermore, complex forms of cognition and learning unfurl and integrate over varying intrinsic timescales along processing hierarchies in the brain23,24. Addressing these issues requires consideration of how time and dimensionality interact with the measured BOLD signal, with the goal of interpreting fMRI activity at the single-subject level.
Here we introduce temporal potential of heat-diffusion for affinity-based transition embedding (T-PHATE) as a nonlinear manifold learning algorithm designed for high-dimensional, temporally dynamic signals. We apply T-PHATE to fMRI data measured during cognitive tasks–a ripe testbed known for its high ambient dimensionality, multi-source noise, and temporal autocorrelation. Most previous studies exploring fMRI activity with dimensionality-reduction methods have relied on linear heuristics25–27. Nonlinear dimensionality-reduction algorithms have been used previously to characterize the geometry and underlying temporal dynamics of neural recordings6,8,21 by integrating local similarities among data points into a global representation9,28–31, but they remain underutilized in fMRI21. We build on PHATE28–a manifold learning algorithm designed for high-throughput, high-noise biomedical data–with a second, explicit model of the temporal properties of BOLD activity. This second view is learned from the data to capture the temporal autocorrelation of the signal and the dynamics specific to the stimulus (Fig. 1a).
Fig. 1 |. T-PHATE procedure.
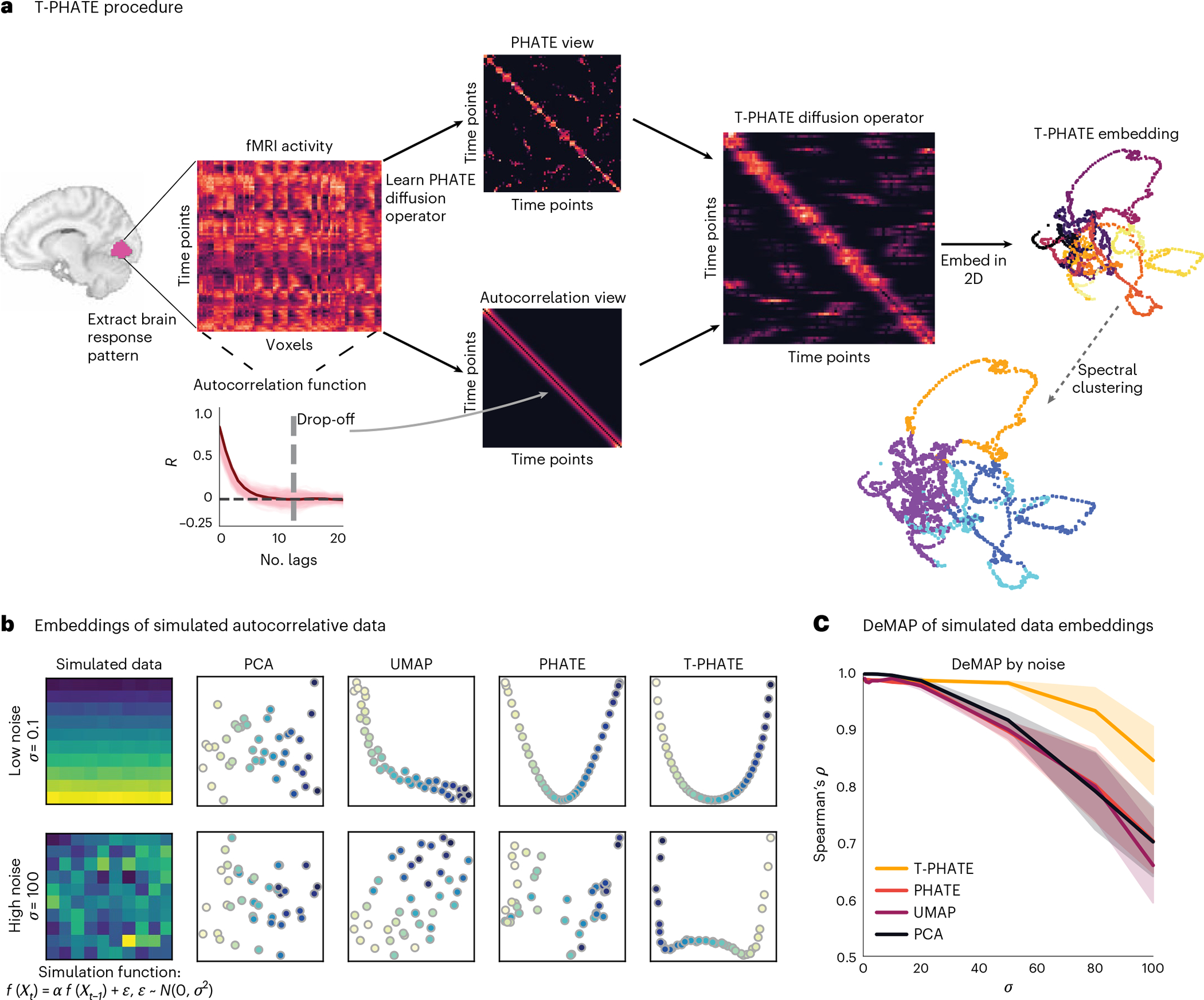
a, A multivariate pattern of fMRI activation over time (time points by voxels) is extracted from a region of interest (ROI). PHATE is then run to learn a PHATE-based affinity matrix between time points. The autocorrelation function is also estimated for the data separately to make an autocorrelation-based affinity matrix. These two views are combined into the T-PHATE diffusion operator and then embedded into lower dimensions. b,c, We tested this approach on simulated data with a known autocorrelative function. To do this, we took a matrix of pristine data (here, a simulated multivariate time-series in which and added noise sampled from a normal distribution to each sample of pristine data. We tested various values of from 0 to 100 (c, x-axis) and compared the fidelity of the manifold distances recovered by each embedding as a function of the amount of noise added to the input data using DeMAP28. We show two sample input matrices (b, far left) to the DeMAP analysis for noise sampled from a narrow distribution (top row, and noise sampled from the widest distribution (bottom row; . T-PHATE both visually better reconstructs the autocorrelative signal from the noise (b) and quantitatively better preserves manifold distances (c) compared with PCA, UMAP, and PHATE. The error bands in c represent the 90% confidence interval of the mean DeMAP score from 1,000 bootstrap iterations. See Supplementary Fig. 1a for more benchmarks.
Using the manifold preservation metric denoised manifold affinity preservation (DeMAP)28, we benchmark T-PHATE against principal component analysis (PCA) and uniform manifold approximation (UMAP)32–dimensionality-reduction methods that have been commonly applied to fMRI data25,26,33,34–as well as PHATE28, a reduced form of T-PHATE excluding the above-mentioned second, temporal view. In Supplementary Information, we compare T-PHATE with further benchmarks such as locally linear embeddings (LLE)30,35, isometric mapping29, and t-distributed stochastic neighbor embedding (t-SNE)36. We test T-PHATE on two movie-viewing fMRI datasets and find that it both denoises the data and affords enhanced access to brain-state trajectories relative to voxel data and other embeddings. By mitigating noise and voxel covariance, this subspace yields clearer access to regional dynamics of brain signals, which we then relate to time-dependent cognitive features. In all, T-PHATE reveals that information about brain dynamics during naturalistic stimuli lies in a low-dimensional latent space that is best modeled in nonlinear, temporal dimensions.
Results
fMRI is a safe, powerful, and ubiquitous tool for studying how the healthy human brain generates the mind and behavior. However, fMRI data are highly noisy in both space and time. The measured BOLD signal is delayed and blurred by the hemodynamic response with respect to underlying neuronal activity. The naturalistic, dynamic stimuli (for example, movies) increasingly used to probe real-world cognition extend over multiple timescales (for example, conversations, plot lines). For these reasons, fMRI data are autocorrelated in time, and this varies across brain regions according to their functional role in cognition (for example, sensory processing versus narrative comprehension). We therefore designed T-PHATE as a variant of the PHATE algorithm that combines the robust manifold-geometry-preserving properties of PHATE with an explicit model of signal autocorrelation in a dual-diffusion operator.
Evaluating manifold quality on simulated data
We validated the quality of manifolds learned from data with a simulated high signal autocorrelation using DeMAP28. DeMAP takes pristine, noiseless data (here, a simulated multivariate time-series in which and computes the Spearman correlation between the geodesic distances on the noiseless data and the Euclidean distances of the embeddings learned from the noisy data. Noisy data were generated by adding an error term to the pristine data, where . We tested robustness to noise by varying . Higher DeMAP scores suggest that an embedding method performs effective denoising and preserves the geometric relationships in the original data (Fig. 1c; see Supplementary Fig. 1a for additional benchmarks). All of the methods achieve high DeMAP scores at low noise levels, although PCA visualizations do not seem to reflect much structure (Fig. 1b). With increasing noise, T-PHATE outperforms the other methods at denoising the simulated data, providing visualizations that most closely resemble lower noise conditions.
Temporal trajectories and static information in fMRI embeddings
Having validated that T-PHATE can learn meaningful manifolds from simulated data under noise, we next applied T-PHATE to real fMRI data. First, we embedded the movie-viewing data from the Sherlock and StudyForrest datasets with PCA, UMAP, PHATE, and T-PHATE in two dimensions to visually inspect the properties of the data highlighted by the manifold. Embeddings were computed separately per subject for four regions of interest (ROIs): early visual cortex (EV), high visual cortex (HV), early auditory cortex (EA), and posterior medial cortex (PMC). Visually, T-PHATE embeddings both denoise the time-series data and capture stimulus-related trajectory structure better than other methods, as shown in all brain regions for a sample subject (Fig. 2a). PCA visualizations show no apparent structure or clustering. UMAP shows slight clustering in HV and EA, but often creates small, shattered clusters with local structure placed through the latent space. PHATE yields slight improvements over UMAP, notably in HV and EA, with larger and less disjointed clusters of nearby time points. T-PHATE reveals trajectories through the latent space that are clearly reflective of temporal structure, and also shows a hub in the center of the space.
Fig. 2 |. Visual validation of T-PHATE embeddings.
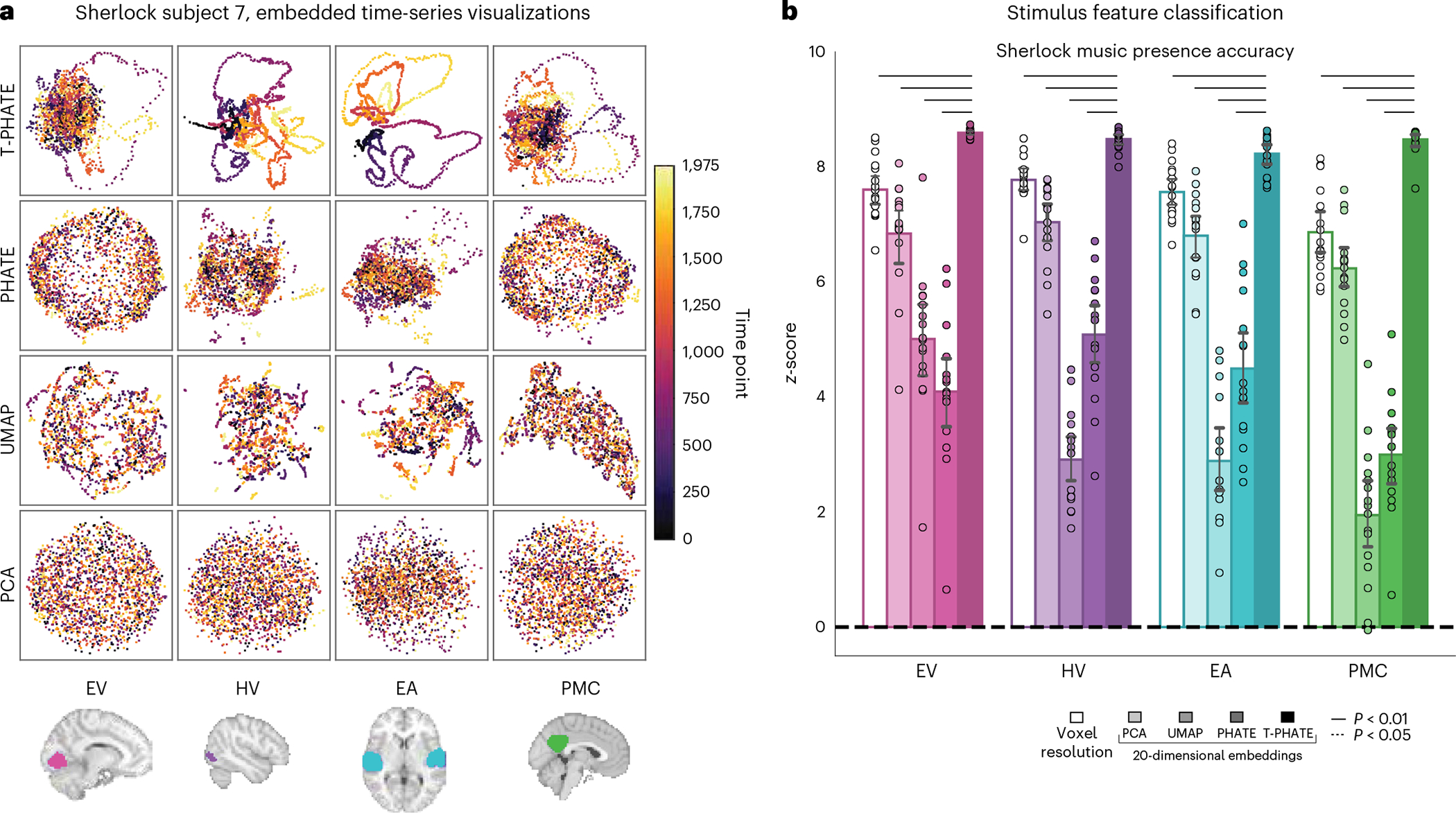
a, Manifold embeddings using PCA, UMAP, PHATE, and T-PHATE for the Sherlock dataset in an example subject and four ROIs. Scatter plot points are colored by time index. b, Support vector classification accuracy predicting the presence of music in the movie at the corresponding time point. Results are presented as the z-score of the true prediction accuracy relative to a null distribution of accuracies, obtained by shifting the music labels in time relative to the embedding data 1,000 times. Dots represent individual subjects (n = 16), bars represent the average z-score across subjects, and error bars represent the 95% confidence interval of the mean, estimated with 1,000 bootstrap iterations. The significance of differences between T-PHATE and other methods is evaluated with permutation tests (10,000 iterations) and corrected for multiple comparisons. The extended results are included in Supplementary Fig. 3.
T-PHATE manifolds also better reflect non-temporal movie features, as shown by classification of these features (Fig. 2b and Supplementary Fig. 3). We trained a support vector machine to predict whether music was playing in the movie from embeddings of neural data (and as a baseline, the original voxel-resolution data). The significance of these predictions was tested by shifting the labels in time with respect to the brain data–which breaks the correspondence between the labels and the data without breaking the internal temporal structure–and recalculating the classification accuracy at each shift to obtain a null distribution of accuracies. Results are presented as the z-score of the true prediction accuracy between brain data and movie labels normalized to the mean and standard deviation of the shifted null distribution.
Alternative representations of time in the brain and tasks
The clarity of the dynamic structure recovered by T-PHATE prompted two follow-up questions. First, does the performance of T-PHATE depend on autocorrelation to represent a temporal structure during manifold learning? We tested two alternative models. PHATE + Time adds the time index label for each sample as an additional feature vector to the voxel data before embedding with PHATE. PHATE + Time manifolds show the clearest temporal trajectory through the embedding space (Fig. 3a), unraveling the brain data in time due to the disproportionate weighting of time in the input data, with less of the hub structure revealed by T-PHATE. Smooth PHATE performs temporal smoothing along each voxel across a number of time points t, where t is learned as the width of the autocorrelation function of the input data before embedding with PHATE. Smooth PHATE yields comparable structure to PHATE or UMAP, with mild clustering and shattered trajectories.
Fig. 3 |. Alternative embeddings featuring time.
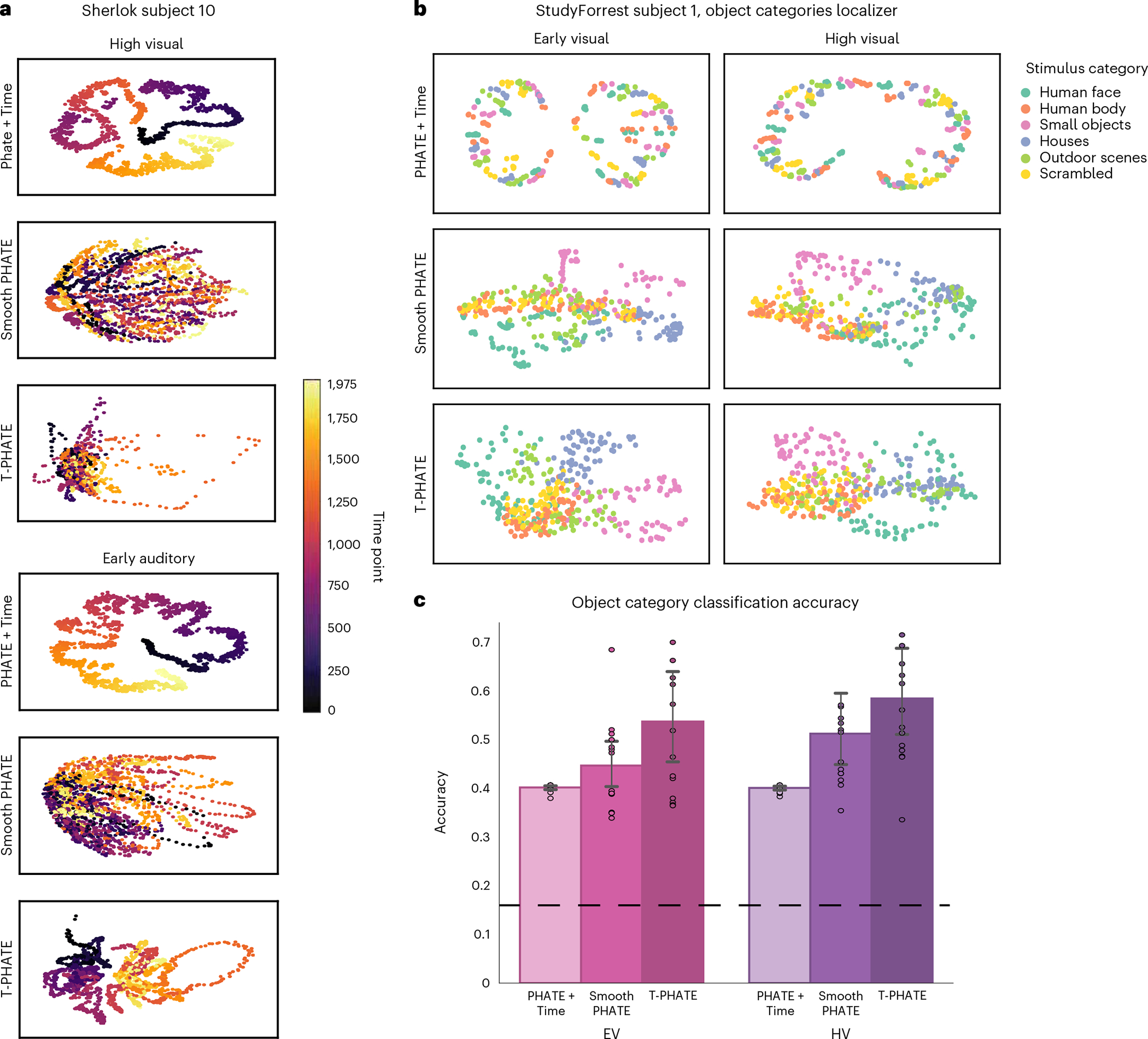
a, Manifold embedding of Sherlock data with PHATE+Time, Smooth PHATE, and T-PHATE, in HV and EA ROIs, colored by time index. b, StudyForrest localizer data embeddings using PHATE+Time, Smooth PHATE, and T-PHATE in EV and HV. Points are colored by the stimulus category presented at a given time point, which are presented in a random order. T-PHATE reveals the best clustering by stimulus category, showing that the manifold successfully denoises data to learn structure in the data also independent of time. c, Support vector classification of stimulus categories presented during the StudyForrest localizer task based on the embedding data. Dots represent individual subjects (n = 14); bars represent the average classification accuracy across subjects;error bars represent the 95% confidence interval of the mean, estimated with 1,000 bootstrap iterations; the dashed line represents chance classification (1/6). See Supplementary Fig. 1b for further benchmark results.
Second, are the benefits of T-PHATE for stimulus feature classification specific to fMRI tasks that are intrinsically linked to time, such as movie watching? We tested T-PHATE on an object category localizer task from StudyForrest with no temporal structure, during which static images of faces, bodies, objects, houses, scenes, and scrambled images were presented in a random order. Data were embedded with T-PHATE, PHATE + Time, and Smooth PHATE in EV and HV. Due to the lack of temporal structure in the task, a meaningful manifold of fMRI activity in visual regions would show a time-independent clustering of categories. T-PHATE manifolds showed the best clustering of stimulus categories in the embedding space (Fig. 3b) when compared with PHATE + Time and Smooth PHATE. Smooth PHATE showed some less-pronounced clustering, whereas PHATE + Time showed minimal clustering by category but retained temporal structure. We used a support vector machine with leave-one-run-out cross-validation to quantify the classification of object categories in the latent space (chance = 1/6; Fig. 3c). Classification accuracy on T-PHATE embeddings surpassed other dimensionality-reduction methods in both the EV and HV ROIs (Fig. 3c; see Supplementary Fig. 1b for further benchmarks). T-PHATE therefore captures the structure in brain activity via autocorrelative denoising, whereas adding time explicitly into the manifold does little to capture data geometry aside from unrolling the signal in time. Temporal smoothing provides an advantage over time-agnostic dimensionality reduction (PCA, PHATE, UMAP), but less of an advantage than the autocorrelation kernel in T-PHATE for both the dynamic movie task and the non-temporal localizer task.
Segmentation of brain-state trajectories into events
We designed T-PHATE to incorporate temporal dynamics (namely, autocorrelation) into a manifold learning algorithm for two reasons. First, as shown above, this helps to denoise fMRI data, given its spatiotemporal noise, and improve subsequent analyses such as feature classification. Second, this may help recover cognitive processes that are represented over time in fMRI data. Many cognitive processes operate on long timescales, including our ability to segment continuous sensory experience into discrete mental events37–39. That is, we neither perceive the world as transient from moment to moment nor as amorphous and undifferentiated, but rather as a series of coherent, hierarchically structured epochs40,41. For example, consider taking a flight: traveling to the airport, passing through security, boarding, flying, deplaning, and transiting to the destination. These mental events are reflected in stable states of the brain, which can be captured with a hidden Markov model (HMM) to identify the boundaries between events42–44. We hypothesized that an HMM would better segment events from fMRI data after embedding them into a manifold learned with T-PHATE. This would suggest that T-PHATE increases sensitivity to the neural dynamics associated with cognitive processing of the stimulus.
We started by learning the optimal number of events experienced by each ROI using a nested leave-one-subject-out cross-validation procedure on the voxel resolution data (Fig. 4a). We find a consistent pattern of results across datasets where the number of events K is higher for early sensory cortices (EV) than late (HV) and integrative cortices (PMC) (mean ± s.d. across subjects): Sherlock, EV = 46 ± 2, HV = 21 ± 1, EA = 18 ± 1, PMC = 24 ± 1; StudyForrest, EV = 121 ± 3, HV = 70 ± 2, EA = 69 ± 2, PMC = 110 ± 4. After fixing the K parameter for each ROI based on the voxel data, we then learned the number of manifold dimensions that would best reveal temporal structure for each region, subject, and embedding method with a leave-one-subject-out cross-validation procedure. The number of dimensions is low (between 3 and 6) with minor variance across subjects and methods (Supplementary Fig. 2), indicating substantial redundancy, covariance, and noise among voxels in the ROIs.
Fig. 4 |. HMM procedure and analysis.
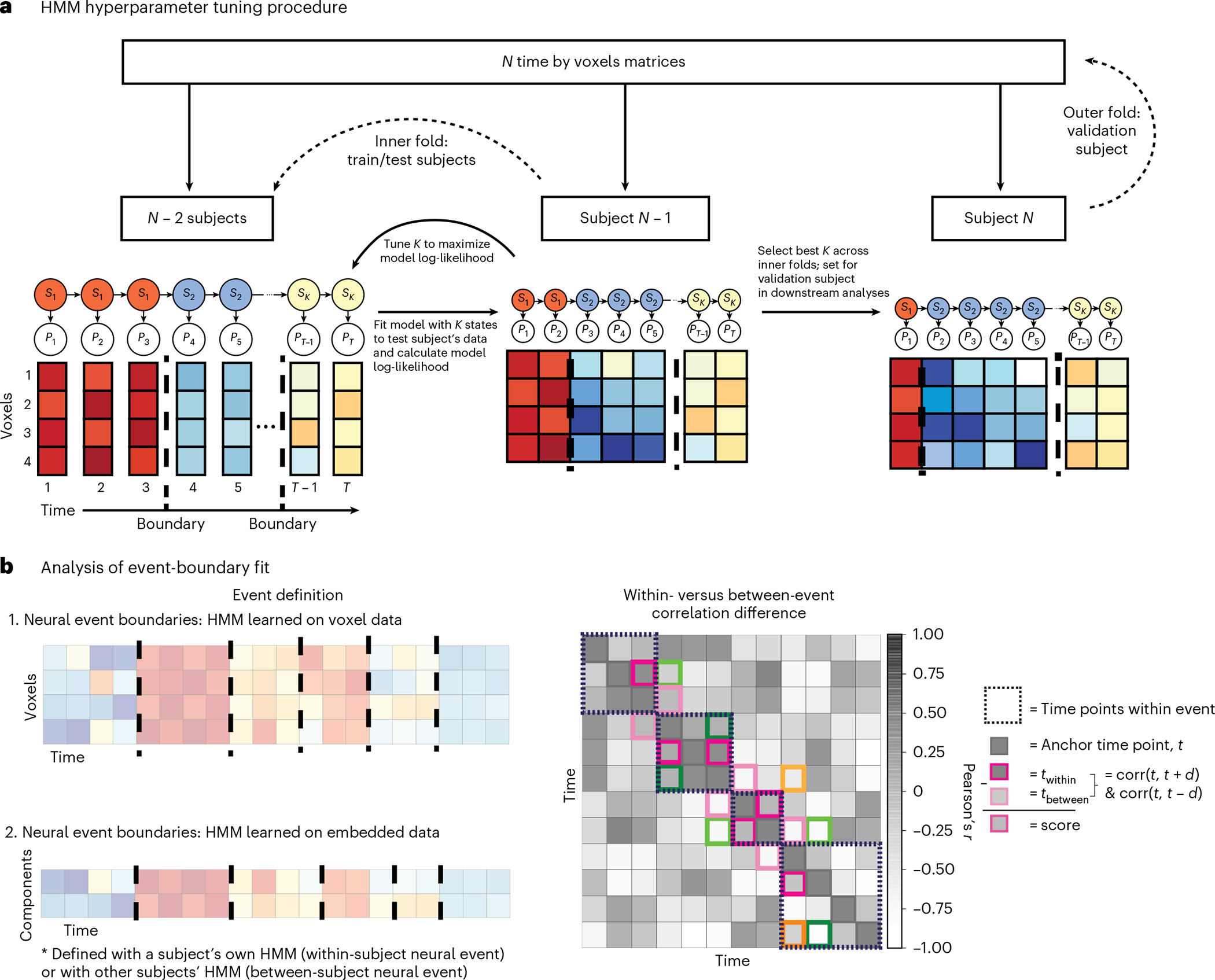
a, Nested cross-validation procedure for hyperparameter tuning. The HMM is trained on the average data for subjects to find stable patterns of voxel activation, where the voxel patterns for each time point belong to a given hidden state is tuned according to model fit on subject (folding over all of the iterations of train/test splits) and then the best value across the folds is applied to the validation subject . This procedure is repeated for all subjects (see the ‘Optimizing the number of neural events’ section in Methods for more information). Event boundaries (dashed lines) are identified by the model as time points at a transition between hidden states. For dimensionality-reduced approaches, is learned with leave-one-subject-out cross-validation, within embedding method and ROI (see the ‘Optimizing the manifold dimensionality’ section in Methods for more information). b. Left: event boundaries are learned both from the data (by applying the HMM on voxel resolution or embedded time-series data and applying those boundaries either within or across subjects) and from independent sources such as human behavioral ratings. Right: representation of dynamic information in a latent space is quantified by applying event boundaries to embedded data. Data emphasize brain-state transitions if the boundaries maximize the difference between brain activity at time points within- versus between-events. For each time point in the time-series, we correlated the activation pattern across features at with that of and such that is in the same event as (within a dashed box) and is in a different event. The within- versus between-event correlation difference score is the subtraction of from . To account for intrinsically higher correlations of proximal time points, we restricted comparisons to where . Here, is denoted by a darker colored box and its corresponding by a lighter shade, where both and are time-steps from . Pink, green, and orange boxes indicate time points when , and 3, repsectively.
A goal of T-PHATE is to recover dynamic cognitive processes that may have been obscured by redundancy and noise. If the T-PHATE latent space unveils dynamical structure during movie watching, we should see better HMM performance on data embedded with T-PHATE compared with other methods, or on the voxel resolution data. This can be framed as: (1) how well can an HMM fit the data and (2) how well do boundaries learned by the model capture structure of the neural data. If an HMM can fit the data well, then there is event-related structure in the data representation for the model to capture. If the event boundaries learned by the model capture this structure, we would further expect a distinction between time points within the same event label versus those spanning across event labels. For each subject, ROI, and embedding method, we fitted an HMM with K hidden states on a M-dimensional embedding (hyperparameter K was cross-validated at the subject level and within ROI; M was cross-validated within ROI and embedding method).
The model fit was quantified on a held-out subject after hyperparameter tuning on a training sample using log-likelihood, which captures the probability that the data input to the model were generated from the learned event structure, and Akaike’s information criterion (AIC45), which penalizes the log-likelihood of a model by the number of parameters of that model. The statistical significance of the difference in performance between T-PHATE and other methods was assessed within ROIs using a permutation test, correcting for the four multiple comparisons. The model fit for T-PHATE was much better for both the Sherlock and StudyForrest datasets in almost all regions (Supplementary Fig. 4). This indicates that an HMM trained on one representation of the data (the embedding for a given ROI and subject using one of the methods) was more confident in its learned event structure than a very similar (or, in the case of Supplementary Fig. 7, identical) HMM trained on a different data representation. This is a somewhat unorthodox approach to model fitting (comparing the fit of the same data between models that have a different number of parameters, architecture, or objective function), so we further evaluated the benefits of T-PHATE by examining event-related dynamics and additional outcomes in the brain and behavior.
Within- versus between-event stability of brain states
To validate that the HMM boundaries identified real transitions between brain states, we next tested whether patterns of fMRI activity from pairs of time points labeled as within the same event were more similar to each other than equidistant pairs of time points labeled as from two different events (Fig. 4b). We first compared the performance of voxel resolution data to T-PHATE embeddings, as this type of within- versus between-event correlation difference analysis has been applied successfully at the voxel level42,46. For all subjects, T-PHATE embeddings outperformed the voxel resolution data (Fig. 5a). We next asked whether this benefit was specific to T-PHATE or a generic effect of dimensionality reduction. We thus repeated this analysis on PCA, UMAP, and PHATE embeddings (Fig. 5b; see Supplementary Figs. 5a and 6a for additional benchmarks) and again found better performance for T-PHATE embeddings in all ROIs. In three out of four ROIs for both datasets, T-PHATE doubled the performance of the second-best method. Except for two instances, all of the dimensionality-reduction methods outperformed the voxel resolution data. Thus, although there is a generic advantage of lower dimensionality when revealing signal structure, T-PHATE captures dynamic signals beyond a temporally agnostic manifold.
Fig. 5 |. Evaluating event segmentation within-subject.
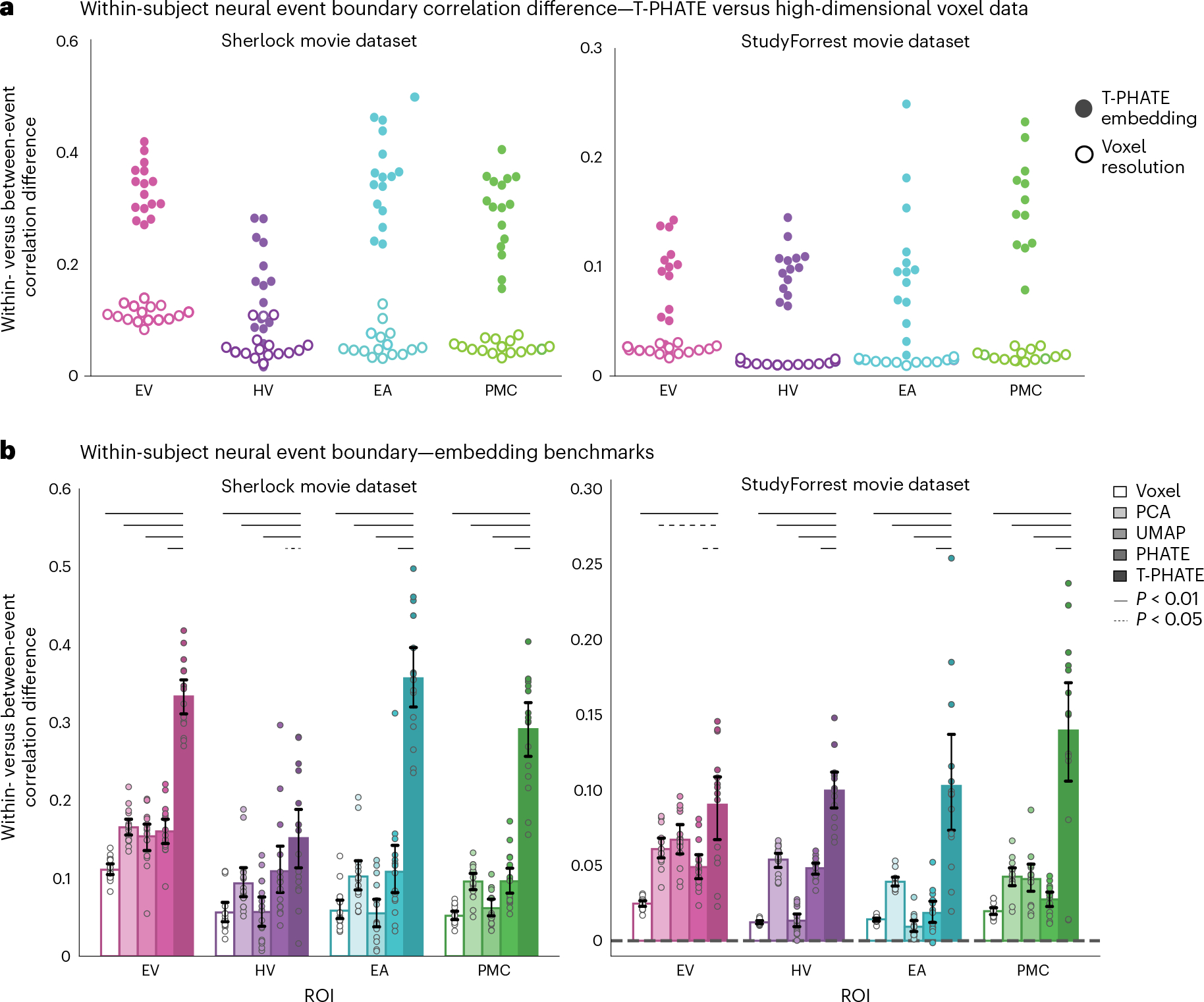
a, After hyperparameters K and M were tuned with cross-validation, a new HMM with these parameters was fit to each subject’s data to identify neural event boundaries and calculate the within- versus between-event score, shown as one point per subject. Embedding the data with T-PHATE greatly increased the within- versus between-event score over the voxel-resolution data. b, We expanded this analysis to embeddings performed with three additional dimensionality-reduction methods. Dots represent individual subjects (n = 16 for Sherlock, n = 14 for StudyForrest); bars represent the average within- versus between-event score across subjects; error bars represent the 95% confidence interval of the mean, estimated with 1,000 bootstrap iterations. The significance of the differences between T-PHATE and other methods was evaluated with permutation tests (10,000 iterations) and corrected for multiple comparisons. See Supplementary Figs. 5a and 6a for additional benchmark results.
In the previous analysis, each HMM was trained and tested within the same subject. To ensure that these results were not driven by overfitting, we performed between-subject cross-validation by applying the event boundaries learned from N – 1 subjects to held-out data from subject N. The premise for this analysis is that the event structure learned from voxel data can be shared across subjects42,46. We asked whether the same holds true in lower dimensions with embeddings performed at the subject level. For each of N subjects (per ROI and embedding method), we repeated the analysis N – 1 times using each of the other subject’s HMM-identified boundaries. We again found better performance for T-PHATE relative to other embedding methods overall (Fig. 6a), though with a smaller advantage than when the analysis was performed within-subject. Although T-PHATE outperformed the voxel resolution data in all cases, the voxel resolution data outperformed some of the embedding methods (Sherlock dataset: EV voxel > PCA, PHATE, UMAP; HV voxel > UMAP; PMC voxel > UMAP. StudyForrest dataset: EV voxel > PCA, PHATE; HV voxel > UMAP; EA voxel > UMAP, PHATE; PMC voxel > PCA, UMAP, PHATE). This was not true for the within-subject analysis, which suggests that embedding data in lower, subject-specific dimensions highlights idiosyncratic aspects of event structure. Voxel resolution data are assumed to be in the same anatomical template space for all subjects, which would lend itself better to generalization across subjects. T-PHATE still outperforms other methods in most cases on this task, suggesting that the event structure highlighted in subject-specific T-PHATE embeddings is at least partly generalizable to other subjects.
Fig. 6 |. Out-of-sample event boundary fit.
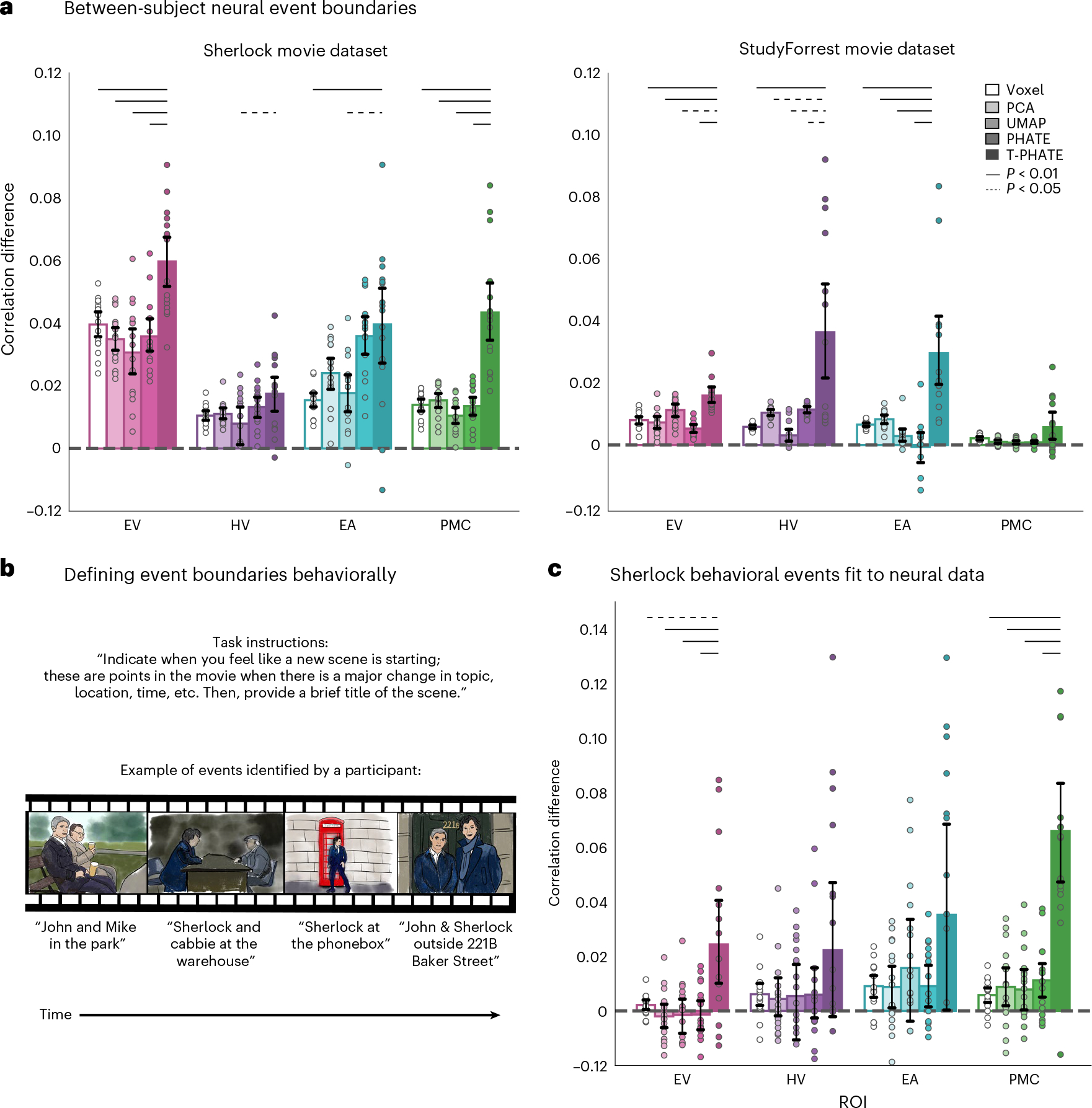
a, Event boundaries were rotated between subjects by testing subject N iteratively with the event boundaries learned from HMMs applied to each of the remaining N – 1 subjects (within ROI and embedding type). Within- versus between-event correlation difference was evaluated as in Fig. 4b. T-PHATE embeddings revealed robust event-related structure that generalized across subjects. See Supplementary Figs. 5b and 6b for additional benchmark results for this analysis. b, Event boundaries were also identified in behavioral ratings of the Sherlock movie by other subjects. These behavioral boundaries were then applied to the neural data and the within- versus between-event correlation difference was calculated as in Fig. 4b. Images in the figure are drawn for copyright reasons; movies were shown at high resolution in the experiment. c, T-PHATE revealed event structure in PMC (and to a weaker extent EV) that mirrored the behavioral boundaries. Dots represent individual subjects (n = 16 for Sherlock; n = 14 for StudyForrest); bars represent the average within- versus between-event correlation difference across subjects; error bars represent the 95% confidence interval of the mean, estimated with 1,000 bootstrap iterations. The significance of differences between T-PHATE and other methods was evaluated with permutation tests (10,000 iterations) and corrected for multiple comparisons.
Relationship between neural and behavioral event boundaries
Identifying neural event boundaries with an HMM is a data-driven approach to understanding how the brain makes sense of continuous experience. This can be compared with boundaries obtained from other tasks, such as having subjects watch the same movie and indicate behaviorally when they believe “something changed”37–39,47 (Fig. 6b). Such behavioral boundaries have been used to model event structure in the brain previously47–50, incuding in PMC42,51. We used behavioral boundaries to test the generalization of neural event structure to the conscious experience of an out-of-sample cohort. We applied behavioral boundaries from this cohort to the neural embedding data and analyzed their fit as in Fig. 4b. We found much higher fMRI pattern similarity within-versus-between behavioral boundaries in the T-PHATE embedding than in the other methods, at least for EV and PMC (Fig. 6c). Critically, T-PHATE improved on voxel performance in PMC by an order of magnitude, a particularly striking gain as the behavioral data being predicted were generated by other subjects performing a distinct behavioral task.
Discussion
Many complex aspects of cognition, including attention, memory, language, thinking, navigation, and decision-making, play out over long timescales52. These cognitive processes mutually drive our conscious experience in the moment, structuring continuous input into meaningful events37–39,42,46,53. To understand the dynamics of how this integration occurs seamlessly, it can be helpful to consider the geometry of brain states. The human fMRI literature suggests that flexible population codes underlie complex cognitive processing13,18,42,51,54. Linear manifolds have so far been used to track discrete, task-related brain states25,26, and nonlinear manifolds have been used to model dynamics across tasks21. These approaches aggregated whole-brain data across dozens of subjects when defining a manifold, which resulted in coarse, population-average representations that could limit utility for understanding individual brains.
With the proper choice of dimensionality-reduction algorithm6,9, we accounted for these challenges to unveil the dynamics and contents of naturalistic cognition. We investigated the latent space of brain activity from movie-viewing fMRI data. T-PHATE manifolds revealed brain-state trajectories obscured in high-dimensional, noisy, and redundant neural population activity by modeling its latent spatiotemporal geometry. The advantages of T-PHATE representations for capturing temporal dynamics in continuous tasks did not come at the expense of decoding discrete stimuli without temporal structure. Thus, although time-considerate and time-agnostic manifolds can encode static information (Figs. 2b and 3c, and Supplementary Figs. 1b and 3), only the former additionally unveil dynamic information.
There are other multi-view variants of dimensionality-reduction methods, such as Multi-SNE55 or Multi-LLE56, which capture multiple views of data by summarizing different types of measurements from the same samples. Whether alternative dimensionality-reduction methods, when combined with an autocorrelation kernel, could yield similar insights remains an open question; however, given that vanilla PHATE yields better performance on most tasks here than benchmarked methods, we predict that T-PHATE benefits from the combination of superior manifold learning of fMRI by PHATE plus modeling temporal signals with the autocorrelation view.
The latent dimensions learned by T-PHATE with this combination of nonlinear dimensionality reduction and temporal diffusion outperforms methods lacking either of those components. This suggests that the relation between the dimensions of individual voxels and the dominant signals that emerge from them are tied by nonlinear relations. By modeling a subject’s BOLD time-series in these dimensions, we can gauge the trajectory of their brain states throughout the course of the experiment, and we can relate this trajectory to theoretical frameworks such as event segmentation theory that facilitate higher-order cognitive processes in the brain. As this is performed at the level of individual subjects, future investigations could use this approach to probe individual differences, developmental trajectories, and/or clinical disorders in the native latent space.
Beyond the applications in cognitive neuroscience explored here, T-PHATE has promise for other forms of high-dimensional, noisy data. PHATE and variants have already been applied successfully to high-throughput biomedical data types including spatial transcriptomics, single-cell RNA sequencing, and blood sample data from SARSCoV-2 data28,57,58. As T-PHATE converges with PHATE when there is no temporal relation among samples, we foresee T-PHATE having similar advantages in these data types. We expect T-PHATE to afford particular insight into data with temporal structure, given the multi-view approach to integrating temporal dynamics in learning the embedding. Such data types could include, for example, calcium imaging, developmental trajectories, longitudinal health data, disease progression, climate change, economic trends, language evolution, and more. In all, we present a method with promising application to various big-data challenges.
Methods
Manifold embeddings
Different dimensionality-reduction algorithms focus on different aspects of a dataset, and thus amplify distinct signals depending on the structure targeted by the algorithm. We hypothesized that a nonlinear manifold learning algorithm designed to handle noisy, high-dimensional biological data would be best suited for the signal and intrinsic noise of fMRI data. PHATE28 is a diffusion-based manifold learning method that models local and global structures simultaneously in nonlinear dimensions58. Brain activity measured with fMRI is highly noisy in both space and time, with the BOLD signal canonically peaking 4–5 s after stimulus onset before slowly returning to baseline. With a temporally dependent stimulus such as a movie, where conversations and plot lines play out over different timescales, the autocorrelation of nearby time points will likely extend beyond the curve of the BOLD signal and vary by ROI along the hierarchy of temporal integration in the brain59. We estimate an autocorrelation kernel for each ROI by correlating each voxel time-series with lagged versions of itself until the correlation drops and stays below zero, then averaging the functions across voxels to get a regional autocorrelation function. This function is then expanded to calculate the transitional probability between all pairs of time points based solely on their estimated autocorrelation and then combined with the PHATE-based transitional probability matrix and embedded in lower dimensions (Fig. 1a).
PHATE
Given a dataset of voxel time-series data, , where at time is a -dimensional vector and is the number of voxels. Construction of the PHATE28 diffusion geometry has five main steps:
- The Euclidean distance matrix is computed between data pairs, where:
(1) - is converted from a distance matrix into a local affinity matrix using an adaptive bandwidth Gaussian kernel, to capture local neighborhoods in the data:
(2) - is then row-normalized to define transition probabilities into the row stochastic matrix, :
(3) -
The probabilities are then used for the Markovian random-walk diffusion process. The PHATE diffusion timescale, , is then computed, which specifies the number of steps taken in the random-walk process. This parameter provides a tradeoff between encoding local and global information in the embedding, where a larger corresponds to more steps than a smaller is computed automatically using the spectral or von Neumann entropy of the diffusion operator. is raised to the power of to perform the the -step random walk over . Based on the representation of , PHATE then computes the diffusion potential distance between the distribution at the ith row and the distribution at the jth row (both of which are distributions as is Markovian):
(4) The log scaling in the diffusion potential distance calculation acts as a damping factor which makes faraway points similarly equal to nearby points in terms of diffusion probabilities, giving PHATE the ability to maintain global context.
The potential distance matrix is finally embedded with metric MDS (a distance embedding method, using stochastic gradient descent as a solver) as a final step to derive an M-dimensional embedding (or 2–3 dimensions for visualization). For more details about the PHATE algorithm, we refer readers to ref. 28.
T-PHATE
We designed T-PHATE as a variant of PHATE that uses a dual-view diffusion operator to embed time-series data in a low-dimensional space. The first view of the T-PHATE diffusion operator is identical to the PHATE matrix, , defined above. The second view is based on an affinity matrix that summarizes the autocorrelation function of the data60. Computing the T-PHATE diffusion operator has the following steps:
For each voxel time-series vector where is the number of time points, calculate its autocovariance using lags, resulting in a matrix where is the number of voxels. Then, average across the N voxels to obtain a single vector c of autocorrelation at each lag.
- Smooth with a rolling average over time points, where is set by the user:
serves as a damping tool to account for possible jittering around where . Here we use , the default, as we did not find much instability in this dataset where . In other data types, this parameter may be more useful.(5) - Find the first lag () where . This defines the maximum width of smoothing for the temporal affinity matrix , which is calculated as:
(6) Convert the autocorrelation matrix into the transition probability matrix by row-normalizing the affinity matrix and powering it to , as in equation (3).
- Combine with the result of step 4 from the PHATE algorithm via alternating diffusion:
(7) Embed with metric MDS into dimensions (where for visualization or higher for downstream analysis).
This dual-view diffusion step allows T-PHATE to learn data geometry and latent signals that represent cognitive processes that play out over longer temporal windows. We compare T-PHATE’s performance at learning dynamic neural manifolds with common dimensionality-reduction algorithms that are agnostic to time including PCA, UMAP32, vanilla PHATE58, LLE30,35, isometric mapping29, and t-SNE36. To test whether our autocorrelation kernel was the best approach to modeling temporal dynamics in this manifold, we tested two additional versions of PHATE: incorporating time labels as a feature vector in the input data (PHATE + Time) and smoothing the data temporally over the data’s autocorrelation function as learned by T-PHATE (Smooth PHATE), to test whether T-PHATE’s effects can be accomplished by incorporating time into the PHATE embeddings without the additional kernel. fMRI data were extracted from ROIs and z-scored before embedding.
Event segmentation modeling
Human experience of real life is continuous, yet perception and conception of this experience is typically divided into discrete moments or events. Event segmentation theory explains that humans automatically divide continuous streams of information into discrete events37,39 to form, organize, and recollect memories, make decisions, and predict the future61. Participants show high consistency in explicitly segmenting continuous stimuli38 and also in how their brains represent these event boundaries42,46. Event boundaries can be represented–both behaviorally and in brain activity–at different timescales depending on the information being used to draw event boundaries53. In the brain, event boundaries are reflected by shifts in the stability of regional activity patterns, and we can learn the structure of events with a variant of an HMM42. During a continuous stimulus such as a movie, different brain regions represent events along different timescales, which reflect the dynamics of the information being represented by a given region. For example, early sensory regions represent low-level information about a stimulus. In most movie stimuli, low-level sensory features shift quickly as cameras change angles or characters speak, and so early sensory regions show more frequent event shifts42,44. Later sensory regions represent longer timescale events, such as conversations and physical environments that change less frequently than features such as camera angles. Regions associated with memory or narrative processing represent events on longer timescales, and these boundaries best correspond with scene changes marked by human raters43.
We used the HMM variant presented in ref. 42 and implemented in BrainIAK62 to learn from different representations of BOLD time-series data where a brain region experiences event boundaries, or transitions between stable states of activation. See ref. 42 for more details about the formulation of this model. Given an activation time-series of a brain region during a continuous stimulus, the HMM identifies stable activity patterns or “events” that are divided by boundaries, where the time-series transitions between two stable patterns of activation. This is done iteratively and the log-likelihood of the model fit is evaluated at each iteration, and model fitting stops when the log-likelihood begins to decrease. The first step of this process is to use the HMM to learn from the data the number of events a brain region represents for a given stimulus. Past studies have used HMMs for event segmentation on multivoxel activity patterns and have validated this approach against behavioral segmentation42,43,46. This shows that voxel resolution data reflect meaningful event segmentation, so we chose to estimate the optimal number of events (K) for each brain region using the voxel resolution data (Fig. 4), which also prevents overfitting to the embedding data.
To run an HMM for event segmentation on our manifold embedding data, we needed to tune two parameters for each subject: K, or the number of hidden states through which a brain region passes during the stimulus, and M, the dimensionality of the latent space that captures this signal.
Optimizing the number of neural events.
We estimated a hyperparameter K for each ROI and subject from the voxel resolution data and held this constant for a subject and ROI across all embedding methods. This was performed with a nested cross-validation procedure. In the outer loop, a subject N was held out as a validation subject, and the parameter identified by the inner loop was applied to this subject. In the inner loop, another subject N – 1 was held out as a test subject, and the BOLD data were averaged over the remaining N – 2 training subjects to get an average time-series matrix for this region. Importantly, as voxel space is assumed to be the same across subjects (as they have all been aligned to a standard template brain based on common anatomy), we can assume correspondence between the features of these time-series matrices and can thus average them together (the same assumption cannot be made for embedding data, as those spaces are learned within-subject and are thus subject-specific). We define K as the number of hidden states in an HMM, and we optimize the K parameter by fitting a model with K states to the training data from the N – 2 subjects and then applying that model to subject N – 1, where the model is scored using the log-likelihood. This is repeated for all K values, testing K from 2 to 200 for Sherlock and 2 to 400 for StudyForrest; the test subject N – 1 is then folded over for each subject in the training set, resulting in N – 1 sets of scores for each validation subject, which are then averaged over the N – 1 subjects. The K value that maximizes the log-likelihood after averaging over the inner-fold is set as subject N’s K value for this ROI. After definition on the voxel resolution data, K for each subject and ROI was held constant for all embedding types. For subsequent analyses, a new HMM was fitted with K states to each subject’s data.
Optimizing the manifold dimensionality.
We used a leave-one-subject-out cross-validation scheme to select the dimensionality of manifold embeddings. Holding out each subject one at a time, we embedded the data for each of N – 1 training subjects into M dimensions, testing values of M ranging from 2 to 10. As each training subject’s data were embedded into a distinct feature space, we fit an HMM to each embedding separately, and scored the model fit to each M-dimensional embedding using the within- versus between-event correlation difference (presented in Fig. 4b). We then averaged the correlation difference scores across the N – 1 training subjects to obtain one average score per M-value tested. We chose the M-value that maximized the within-versus-between score in the training set and fixed that as the M-value for the held-out subject N in all subsequent analyses. This procedure was performed separately for each ROI and embedding method.
To control for possible effects of embedding in different numbers of dimensions across different methods, we repeated all analyses without the M optimization step (Supplementary Fig. 7). We instead tested all embedding methods in M = 3 dimensions (chosen because three dimensions is the maximum embedding dimensionality for t-SNE). Our control and optimized M analyses were highly correlated (within- versus between-event correlation difference, Pearson’s r = 0.930; log-likelihood of model fit, r = 0.993; AIC of model fit, r = 0.998).
Evaluating the fit of event boundaries.
To quantify how well a learned manifold embedding amplifies dynamic structure in the data, we calculated difference in correlation across latent dimensions for pairs of time points within and across event boundaries. For this calculation, we restricted the time points that went into the calculation as follows. We calculated the length of the longest event, and only considered time points that were less than that temporal distance apart. We anchored a comparison on each time point t in the time-series, and for each temporal distance n ranging from one time point to the length of the longest event; we considered a pair of time points t − d and t + d if one time point fell in the same event as the anchor and the other a different event. We then took the correlation between the anchor and each time point, binning them according to whether they were a within-event comparison or a between-event comparison. This process assures that there are equal numbers of time points going into each comparison type (Fig. 4b).
One issue of defining and testing event boundaries within-subject is that the dynamics captured are subject-specific. Studies using voxel-resolution data to perform this HMM event segmentation analysis have shown that there is shared structure in voxel representations of event dynamics42,44,46. We therefore asked whether boundaries identified on one subject generalized across all other subjects. To test this, we cross-validated the event boundaries across subjects with a nested cross-validation procedure. In the outer loop, we held out one subject’s data for testing. In the inner loop, we performed leave-one-subject-out cross-validation to learn the best number of events (K) for the HMM and the optimal dimensionality (M) for embedding methods. After learning this K parameter, we fit an HMM with K states on each of the training subjects’ data to identify their event boundaries. These boundaries were then applied to the test subject one by one, scored with the within- versus between-event correlation difference, and then averaged over the training segmentations. This procedure was iterated with all subjects serving as test subject.
To assess the behavioral relevance of the neural event segmentation, we used a set of event boundaries identified by a separate cohort of Sherlock study participants. These participants watched the Sherlock episode outside of the scanner and were asked to indicate where they believed a new event began63 (Fig. 6b). We applied these human-labeled boundaries to the neural data and measured the fit of the boundaries (as outlined above) to gauge how the embeddings not only highlight neural dynamics but how those neural dynamics relate to the conscious, real-world experience of events.
Statistical tests
All results present the following pairwise statistical testing comparing T-PHATE with each other method. For each pair of methods, within ROI and dataset, we calculated the difference in scores across the two methods, within subjects to preserve subject-wise random effects. We then generated a null distribution of difference scores across the two methods by randomly permuting the method label of a subject’s score 10,000 times and recomputing the mean difference between the methods. We then tested whether T-PHATE outperformed the other method by calculating a one-tailed P-value of the true difference relative to the null distribution, and corrected for multiple comparisons using the Bonferroni method. All figures show bootstrapped 95% confidence intervals of the mean performance across subjects within method, tested with 1,000 iterations of bootstrap resampling.
Data
Sherlock dataset.
See the original publication of ref. 51 for full details on the Sherlock data. Here we used data from the sixteen participants who viewed a 48 min clip of the BBC television series ‘Sherlock’. Data were collected in two fMRI runs of 946 and 1,030 time points (repetition time; TRs) and was downloaded from the DataSpace public repository (http://arks.princeton.edu/ark:/88435/dsp01nz8062179). This experiment was approved by the Princeton University Institutional Review Board. The data were collected on a Siemens Skyra 3 T scanner with a 20-channel head coil. Functional images were acquired with a T2*-weighted echo-planar imaging sequence (TE, 28 ms; TR, 1.5 s; 64° flip angle, whole brain coverage with 27 slices of 4 mm thickness and 3 × 3 mm2 in-plane resolution, 192 × 192 mm2 FOV). Anatomical images were acquired with a T1-weighted MPRAGE pulse sequence with 0.89 mm3 resolution. Slice-time correction, motion correction, linear detrending, high-pass filtering (140 s cut-off), and co-registration and affine transformation of functional volumes to the Montreal Neurological Institute template were all performed with fMRI Software Library (FSL). Functional images were then resampled from native resolution to 3 mm isotropic voxels for all analyses, z-scored across time at every voxel and smoothed with a 6 mm kernel.
StudyForrest dataset.
For full details on the StudyForrest data, see the original publication of the movie dataset64 and the localizer extension dataset65. Here we included data from fourteen participants who completed both tasks. All participants were native German speakers. This experiment was approved by the Ethics Committee of the Otto-von-Guericke University. In the movie-viewing task, participants watched a 2 h version of the movie ‘Forrest Gump.’ These data were collected in eight fMRI runs resulting in a full time-series of 3,599 TRs (451, 441, 438, 488, 462, 439, 542 and 338 per run). Movie data were collected on a 3 T Philips Achieva dStream scanner with a 32-channel headcoil. Functional images were acquired with a T2*-weighted echo-planar imaging sequence (TE, 30 ms; TR, 2 s; whole brain coverage with 35 slices of 3 mm thickness and 3 × 3 mm2 in-plane resolution, 240 mm FOV). In the localizer task, the same fifteen participants viewed 24 unique grayscale images from six categories (human faces, human bodies without heads, houses, small objects, outdoor scenes and scrambled images) in four (156 TR) block-design runs with two 16 s blocks per stimulus category per run. Localizer data were collected on a 3 T Philips Achieva scanner with a 32-channel headcoil. Functional images for the localizer task were acquired with a T2*-weighted echoplanar imaging sequence (TE, 30 ms; TR, 2 s; 90° flip angle, 35 slices of 3 mm thickness and 3 × 3 mm2 in-plane resolution, 240 mm FOV). Structural images were acquired with a 3 T Philips Achieva using a 32-channel headcoil. T1-weighted anatomical images were acquired with a 3D turbo field echo sequence with 0.67 mm isotropic resolution. Slice-time correction, co-registration and affine transformation to Montreal Neurological Institute template were performed with FSL where functional images were resampled to 3 mm isotropic voxels. Additional preprocessing included linear detrending, high-pass filtering (100 s cut-off), spatial smoothing with a 6 mm kernel, and nuisance regression (including six motion parameters, global signal, white matter and cerebrospinal fluid) and z-scoring within voxel to match the Sherlock preprocessing.
ROI selection
We selected four ROIs based on the original publication of the Sherlock dataset51, subsequent publications42 and regions known to have reliably strong signals in response to audiovisual movie stimuli66. The EV, EA and HV region masks were based on a functional atlas defined with resting-state connectivity67. As in the original Sherlock publication, we defined a PMC ROI as the posterior medial cluster of voxels within the dorsal default mode network51. In the Sherlock data, the dimensionality of voxel-resolution ROIs are as follows: EV = 307, HV = 571, EA = 1,018, PMC = 481. In the StudyForrest data, the dimensionality of voxel-resolution ROIs are as follows: EV = 166, HV = 456, EA = 657, PMC = 309.
Supplementary Material
Acknowledgements
We thank Ariadne Letrou for reviewing the code and the helpful discussions. E.L.B. was supported by a NSF Graduate Research Fellowship (award no. 2139841). G.L. was supported by Canada CIFAR AI Chair and Canada Research Chair in Neural Computations and Interfacing. G.W. was supported by Canada CIFAR AI Chair and IVADO Professor research funds. S.K. was supported by the NIH (grant nos. R01GM135929 and R01GM130847), an NSF Career Grant (grant no. 2047856) and a Sloan Fellowship (grant no. FG-2021-15883). N.B.T-B. was supported by an NSF Grant CCF (grant no. 1839308) and CIFAR.
Footnotes
Competing interests
S.K. is a visiting professor at META Fundamental AI Research (FAIR). The other authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Data analysis code is written as custom Python scripts (v.3.6.13) based on sci-kit learn v.0.23.2 (https://scikit-learn.org/), nilearn v.0.9.2 (https://nilearn.github.io), nibabel v.4.0.1 (https://github.com/nipy/nibabel), PHATE v.1.0.7 (https://phate.readthedocs.io/en/stable/)28 and Brainiak v.0.11 (https://brainiak.org/)62. T-PHATE is available as a Python package at: https://github.com/KrishnaswamyLab/TPHATE (ref. 70). The pipeline to replicate all of the analyses presented here is available at: https://github.com/ericabusch/tphate_analysis_capsule (ref. 69).
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s43588-023-00419-0.
Peer review information Nature Computational Science thanks the anonymous reviewers for their contribution to the peer review of this work. Handling editor: Ananya Rastogi, in collaboration with the Nature Computational Science team.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The Sherlock dataset was downloaded from the Dataspace Public Repository at the following link: http://arks.princeton.edu/ark:/88435/dsp01nz8062179. The StudyForrest dataset was accessed via DataLad68 from: https://github.com/psychoinformatics-de/studyforrest-data. Steps to reproduce our preprocessing pipeline and ROI extraction are available here: https://github.com/ericabusch/tphate_analysis_capsule (ref. 69). Source Data are provided with this paper.
References
- 1.Averbeck BB, Latham PE & Pouget A Neural correlations, population coding and computation. Nat. Rev. Neurosci. 7, 358–366 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Laurent G Olfactory network dynamics and the coding of multidimensional signals. Nat. Rev. Neurosci. 3, 884–895 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Churchland MM, Cunningham JP, Kaufman MT, Ryu SI & Shenoy KV Cortical preparatory activity: representation of movement or first cog in a dynamical machine? Neuron 68, 387–400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang L & Tsao DY The code for facial identity in the primate brain. Cell 169, 1013–1028.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freiwald WA & Tsao DY Functional compartmentalization and viewpoint generalization within the macaque face-processing system. Science 330, 845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jazayeri M & Ostojic S Interpreting neural computations by examining intrinsic and embedding dimensionality of neural activity. Curr. Opinion Neurobiol. 70, 113–120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennig JA et al. Constraints on neural redundancy. eLife 7, e36774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieh EH et al. Geometry of abstract learned knowledge in the hippocampus. Nature 595, 80–84 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham JP & Yu BM Dimensionality reduction for large-scale neural recordings. Nat. Neurosci. 17, 1500–1509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman KA, Polyn SM, Detre GJ & Haxby JV Beyond mind-reading: multi-voxel pattern analysis of fmri data. Trends Cogn. Sci. 10, 424–430 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Cox DD & Savoy RL Functional magnetic resonance imaging (fMRI) ‘brain reading’: detecting and classifying distributed patterns of fMRI activity in human visual cortex. NeuroImage 19, 261–270 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Kamitani Y & Tong F Decoding the visual and subjective contents of the human brain. Nat. Neurosci. 8, 679–685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haxby JV Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293, 2425–2430 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Haynes J-D & Rees G Decoding mental states from brain activity in humans. Nat. Rev. Neurosci. 7, 523–534 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Polyn SM, Natu VS, Cohen JD & Norman KA Category-specific cortical activity precedes retrieval during memory search. Science 310, 1963–1966 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Peelen MV, Atkinson AP & Vuilleumier P Supramodal representations of perceived emotions in the human brain. J. Neurosci. 30, 10127–10134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeshurun Y, Nguyen M & Hasson U Amplification of local changes along the timescale processing hierarchy. Proc. Natl Acad. Sci. USA 114, 9475–9480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davatzikos C et al. Classifying spatial patterns of brain activity with machine learning methods: application to lie detection. NeuroImage 28, 663–668 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Birn RM, Smith MA, Jones TB & Bandettini PA The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. NeuroImage 40, 644–654 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turk-Browne NB Functional interactions as big data in the human brain. Science 342, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao S, Mishne G & Scheinost D Nonlinear manifold learning in functional magnetic resonance imaging uncovers a low-dimensional space of brain dynamics. Hum. Brain Mapp. 42, 4510–4524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olszowy W, Aston J, Rua C & Williams GB Accurate autocorrelation modeling substantially improves fMRI reliability. Nat. Commun. 10, 1220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golesorkhi M et al. The brain and its time: intrinsic neural timescales are key for input processing. Commun. Biol. 4, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Hearne LJ & Cole MW A cortical hierarchy of localized and distributed processes revealed via dissociation of task activations, connectivity changes, and intrinsic timescales. NeuroImage 221, 117141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shine JM et al. The dynamics of functional brain networks: integrated network states during cognitive task performance. Neuron 92, 544–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shine JM et al. Human cognition involves the dynamic integration of neural activity and neuromodulatory systems. Nat. Neurosci. 22, 289–296 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Allen EA et al. Tracking whole-brain connectivity dynamics in the resting state. Cerebral Cortex 24, 663–676 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon KR et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenenbaum JB, De Silva V & Langford JC A global geometric framework for nonlinear dimensionality reduction. Science 290, 2319–2323 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Roweis ST & Saul LK Nonlinear dimensionality reduction by locally linear embedding. Science 290, 2323–2326 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Salhov M, Bermanis A, Wolf G & Averbuch A Approximately-isometric diffusion maps. Appl. Comput. Harmon. Anal. 38, 399–419 (2015). [Google Scholar]
- 32.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Gotts SJ, Gilmore AW & Martin A Brain networks, dimensionality, and global signal averaging in resting-state fMRI: hierarchical network structure results in low-dimensional spatiotemporal dynamics. NeuroImage 205, 116289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casanova R et al. Embedding functional brain networks in low dimensional spaces using manifold learning techniques. Front. Neuroinform. 15, 740143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannfolk P, Wirestam R, Nilsson M, Ståhlberg F & Olsrud J Dimensionality reduction of fMRI time series data using locally linear embedding. Magn. Res. Mater. Phy. 23, 327–338 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Van der Maaten L & Hinton G Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 37.Radvansky GA & Zacks JM Event boundaries in memory and cognition. Curr. Opin. Behav. Sci. 17, 133–140 (2017). Memory in time and space. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zacks JM, Speer NK, Swallow KM & Maley CJ The brain’s cutting-room floor: segmentation of narrative cinema. Front. Hum. Neurosci. 4, 168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zacks JM, Speer NK, Swallow KM, Braver TS & Reynolds JR Event perception: a mind-brain perspective. Psychol. Bull. 133, 273–293 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zacks JM, Tversky B & Iyer G Perceiving, remembering, and communicating structure in events. J. Exp. Psychol. Gen. 130, 29–58 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Kurby CA & Zacks JM Segmentation in the perception and memory of events. Trends Cogn. Sci. 12, 72–79 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldassano C et al. Discovering event structure in continuous narrative perception and memory. Neuron 95, 709–721.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CS, Aly M & Baldassano C Anticipation of temporally structured events in the brain. eLife 10, e64972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldassano C, Hasson U & Norman KA Representation of real-world event schemas during narrative perception. J. Neurosci. 38, 9689–9699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacDonald IL & Zucchini W Hidden Markov and Other Models for Discrete-Valued Time Series Vol. 110 (CRC, 1997). [Google Scholar]
- 46.Yates TS et al. Neural event segmentation of continuous experience in human infants. Proc. Natl Acad. Sci. USA 119, e2200257119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speer NK, Zacks JM & Reynolds JR Human brain activity time-locked to narrative event boundaries. Psychol.Sci. 18, 449–455 (2007). [DOI] [PubMed] [Google Scholar]
- 48.DuBrow S & Davachi L The influence of context boundaries on memory for the sequential order of events. J. Exp. Psychol. Gen. 142, 1277–1286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DuBrow S & Davachi L Temporal binding within and across events. Neurobiol. Learn. Mem. 134, 107–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ezzyat Y & Davachi L Similarity breeds proximity: pattern similarity within and across contexts is related to later mnemonic judgments of temporal proximity. Neuron 81, 1179–1189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J et al. Shared memories reveal shared structure in neural activity across individuals. Nat. Neurosci. 20, 115–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasson U, Yang E, Vallines I, Heeger DJ & Rubin N A hierarchy of temporal receptive windows in human cortex. J. Neurosci. 28, 2539–2550 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zacks JM et al. Human brain activity time-locked to perceptual event boundaries. Nat. Neurosci. 4, 651–655 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Haxby JV, Connolly AC & Guntupalli JS Decoding neural representational spaces using multivariate pattern analysis. Ann. Rev. Neurosci. 37, 435–456 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Rodosthenous T, Shahrezaei V & Evangelou M S-multi-SNE: semi-supervised classification and visualisation of multi-view data. Preprint at https://arxiv.org/abs/2111.03519 (2021). [Google Scholar]
- 56.Rodosthenous T, Shahrezaei V & Evangelou M Multi-view data visualisation via manifold learning. Preprint at https://arxiv.org/abs/2101.06763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuchroo M et al. Multiscale phate identifies multimodal signatures of covid-19. Nat. Biotechnol. 40, 681–691 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moon KR et al. Manifold learning-based methods for analyzing single-cell RNA-sequencing data. Curr. Opin. Syst. Biol. 7, 36–46 (2018). [Google Scholar]
- 59.Himberger KD, Chien H-Y & Honey CJ Principles of temporal processing across the cortical hierarchy. Neuroscience 389, 161–174 (2018). Sensory Sequence Processing in the Brain. [DOI] [PubMed] [Google Scholar]
- 60.Brockwell PJ & Davis RA Introduction to Time Series and Forecasting (Springer, 2002). [Google Scholar]
- 61.Shin YS & DuBrow S Structuring memory through inference-based event segmentation. Topics Cogn. Sci. 13, 106–127 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Kumar M et al. Brainiak: The Brain Imaging Analysis Kit (BrainIAK, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vodrahalli K et al. Mapping between fMRI responses to movies and their natural language annotations. NeuroImage 180, 223–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanke M et al. A studyforrest extension, simultaneous fMRI and eye gaze recordings during prolonged natural stimulation. Sci. Data 3, 160092 (2016).. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sengupta A et al. A studyforrest extension, retinotopic mapping and localization of higher visual areas. Sci. Data 3, 160093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haxby JV, Guntupalli JS, Nastase SA & Feilong M Hyperalignment: modeling shared information encoded in idiosyncratic cortical topographies. eLife 9, e56601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirer WR, Ryali S, Rykhlevskaia E, Menon V & Greicius MD Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex 22, 158–165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halchenko YO et al. Datalad: distributed system for joint management of code, data, and their relationship. J. Open Source Softw. 6, 3262 (2021). [Google Scholar]
- 69.Busch E ericabusch/tphate_analysis_capsule Version 2 release (Zenoodo, 2023); 10.5281/zenodo.7626543 [DOI] [Google Scholar]
- 70.Busch E Krishnaswamylab/tphate Initial release (Zenodo, 2023); 10.5281/zenodo.7637523 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Sherlock dataset was downloaded from the Dataspace Public Repository at the following link: http://arks.princeton.edu/ark:/88435/dsp01nz8062179. The StudyForrest dataset was accessed via DataLad68 from: https://github.com/psychoinformatics-de/studyforrest-data. Steps to reproduce our preprocessing pipeline and ROI extraction are available here: https://github.com/ericabusch/tphate_analysis_capsule (ref. 69). Source Data are provided with this paper.


