Abstract
A quick genetic approach for the screening of influenza virus variants was developed in this laboratory (S. Zou, J. Clin. Microbiol. 35:2623–2627, 1997). It uses multiplex reverse transcription and multiplex PCR to amplify and differentiate the variable region of the hemagglutinin genes of different types and subtypes of influenza viruses. Variants within the same type or subtype are then identified by the heteroduplex mobility shift assay of the amplicons. The method was used to screen influenza virus isolates received from provincial laboratories during the 1996–1997 season and was able to identify new influenza B virus variants. Sequencing of the amplicons derived from the hemagglutinin gene of the identified variants and comparison with the vaccine strain B/Harbin/7/94 showed substitution rates of 2.26 to 2.55% at the nucleotide level and 4.26 to 4.68% at the amino acid level. The result further demonstrated that the approach provides a quick, sensitive, and reliable screening for influenza virus variants. It also suggested the necessity of close monitoring of influenza B virus isolates in the 1997–1998 season and critical evaluation of the reference strain for the type B influenza virus.
Influenza viruses contain a genome of single-stranded RNA segments, two of which encode two envelope proteins, hemagglutinin (HA) and neuraminidase (5). The two proteins are the major immunogens that induce protective antibodies (7). Because of the error-prone nature of the viral RNA polymerase and the immune selection pressure from the host, the proteins keep changing, giving rise to antigenic variants (antigenic drift) (6). As well, reassortment (recombination) of viral RNA segments from animal and human type A virus strains results in the generation of new subtypes (antigenic shift) (9).
To monitor the emergence of new variants or new subtypes of influenza viruses, isolates across the world are tested for changes in the HA antigen by either serological (hemagglutination inhibition [HI] assay) or genetic (PCR and sequence analysis) means (1, 3, 4). Some strains are also tested for changes in the neuraminidase antigen (11). However, the HI test is time-consuming, whereas sequencing of all isolates would be highly demanding, if not impossible. Recently, a quick genetic approach to screening for influenza virus variants was developed in this laboratory, and it was found to be highly sensitive on the basis of the testing of reference influenza virus strains (12). The method uses multiplex reverse transcription (RT) and multiplex PCR to amplify and differentiate the variable regions located in the HA1 portion of the HA genes of different types or subtypes of influenza viruses. Subsequently, variations within the same type or subtype are further characterized by heteroduplex mobility shift assay (HMA) of the amplicons. The approach was used to screen influenza virus isolates from the 1996–1997 season, and new type B influenza virus variants were identified. Further sequence analysis confirmed the divergence of these variants.
MATERIALS AND METHODS
Viruses.
Influenza virus isolates were received from Canadian provincial laboratories as frozen cell culture fluids. Upon arrival and thawing of the specimens, 50 μl was aliquoted into a 1.5-ml microcentrifuge tube and the rest was inoculated into cell culture (MDCK or MRC-5 cells). The virus amplified in cell culture was processed as described before (14) and was tested by the standard HI assay (8).
Multiplex RT-PCR.
RNA extraction, RT, and PCR were performed essentially as described before (12). Briefly, viral RNA was extracted from the 50-μl aliquot of cell culture fluid and was converted into cDNA with three primers, SZA+, SZB+, and SZC+, which are complementary to the conserved 3′ termini of all viral genome segments of influenza virus type A, type B, and type C, respectively. The cDNA was then amplified with five primers, primers SZAHA+, SZAH1-3, and SZAH3-5B (for HA genes of subtypes H1 and H3 of type A) and primers SZBHA+1 and SZBHA-4 (for HA genes of type B). The amplicons cover all or most of the variable regions located in the HA1 portion of the HA gene. The other two primers, SZCHE+1 and SZCHE-9, specific for type C viruses, were omitted from testing for the late-season isolates because none of the isolates had been found to be type C influenza viruses. Amplicons of different types and subtypes were differentiated according to the expected sizes: 1,117, 942, and 751 nucleotides for subtype H3 of type A, subtype H1 of type A, and type B, respectively.
HMA.
Amplicons were then subjected to HMA by the established protocol (12). Basically, the amplicon of an isolate was mixed with the amplicon of a reference strain, denatured, reannealled, and separated by electrophoresis. Variations in the amplicon sequences of the test strains from that of the reference strain generate mismatches and, therefore, heteroduplexes following denaturation and reannealling. Heteroduplexes will migrate more slowly than the homoduplex of the same size, and the degree of mobility shift correlates with the divergence of amplicons in the mixture (2).
Sequencing of amplicons.
Amplicons of new virus variants identified by HMA were purified and sequenced as described earlier (14). The sequences for the HA1 region of B/Harbin/7/94 and B/Beijing/184/93 were kindly provided by the World Health Organization Collaborating Center for Influenza at the Centers for Disease Control and Prevention, Atlanta, Ga.
RESULTS
Identification of type B virus variants by HMA.
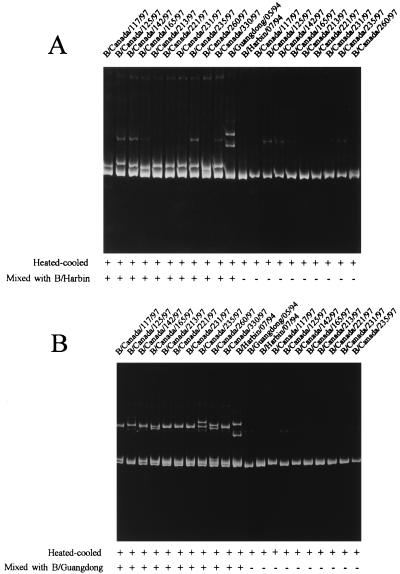
During the 1996–1997 season, no type A isolates of the H1 subtype were received from provincial laboratories, although a few isolations of the H1 subtype were reported in Canada (unpublished data). For a selected group of type A isolates of the H3 subtype, variations were not detected in the variable region of the HA gene by HMA. However, of 25 type B virus isolates received from provincial laboratories and tested by HMA, 10 isolates were clearly identified as new variants on the basis of noticeable mobility shifts of the DNA duplexes formed from amplicons of these isolates and the amplicons of the reference strains for the 1996–1997 season, B/Harbin/7/94 and B/Guangdong/5/94 (Fig. 1). In Fig. 1A, B/Harbin/7/94 was used as the reference, and in Fig. 1B, B/Guangdong/5/94 was used as the reference. It is obvious from the HMA gels that amplicons of these 10 type B isolates showed bands with shifted mobilities when they were mixed with the amplicon of the reference strain in comparison with the mobilities of the same amplicons when they were not mixed with that of the reference strain. Nevertheless, it is also clearly shown, from comparison of the degree of mobility shift between Fig. 1A and B, that these isolates were close to B/Harbin/7/94 but distant from B/Guangdong/5/94. The higher-molecular-weight bands observed in some lanes are probably nonspecific PCR products and should not affect the interpretation of results when the patterns of the amplicons with and without mixing with the amplicon of the reference strain are compared.
FIG. 1.
HMA of amplicons of influenza B virus isolates. (A) B/Harbin/7/94 was used as the reference strain; (B) B/Guangdong/5/94 was used as the reference strain. A total of 5 μl of amplicon of the reference strain was mixed with 5 μl of that of each of the 10 isolates, heated at 94°C for 2 min, chilled on wet ice immediately, and loaded onto a 6% polyacrylamide gel and run in 1× TBE (89 mM Tris-borate, 2 mM EDTA) at 240 V for 4 h. DNA bands were visualized by ethidium bromide staining. +, with the indicated treatment; −, without the treatment.
Sequencing of amplicons.
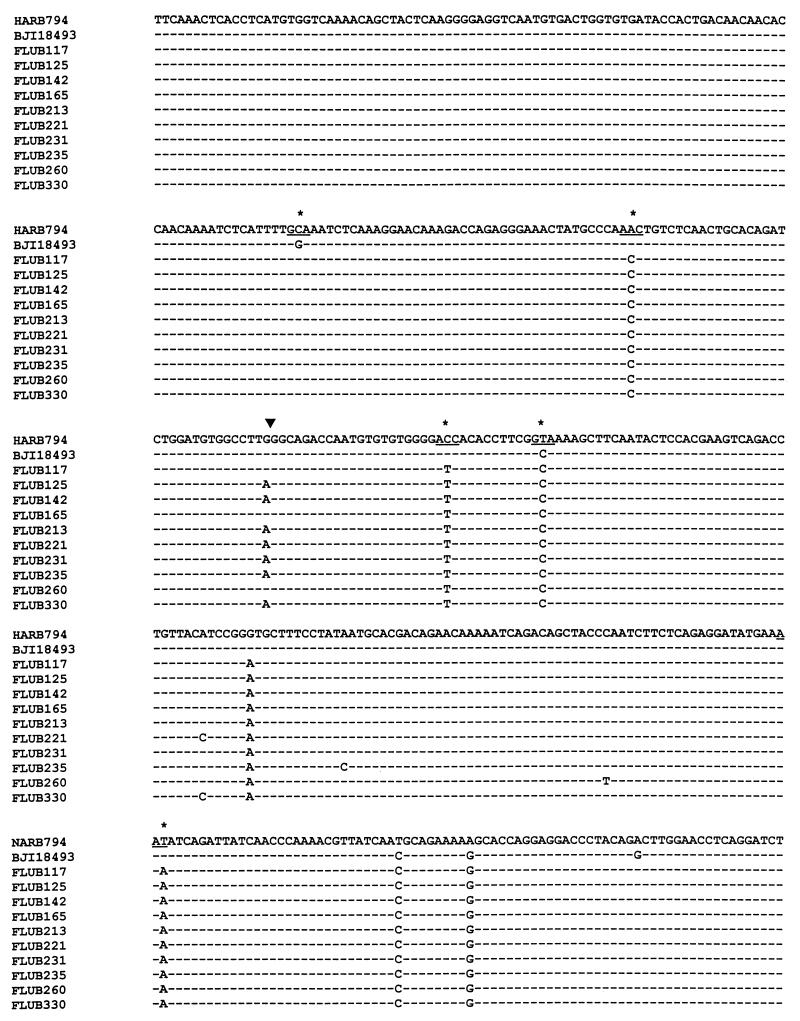
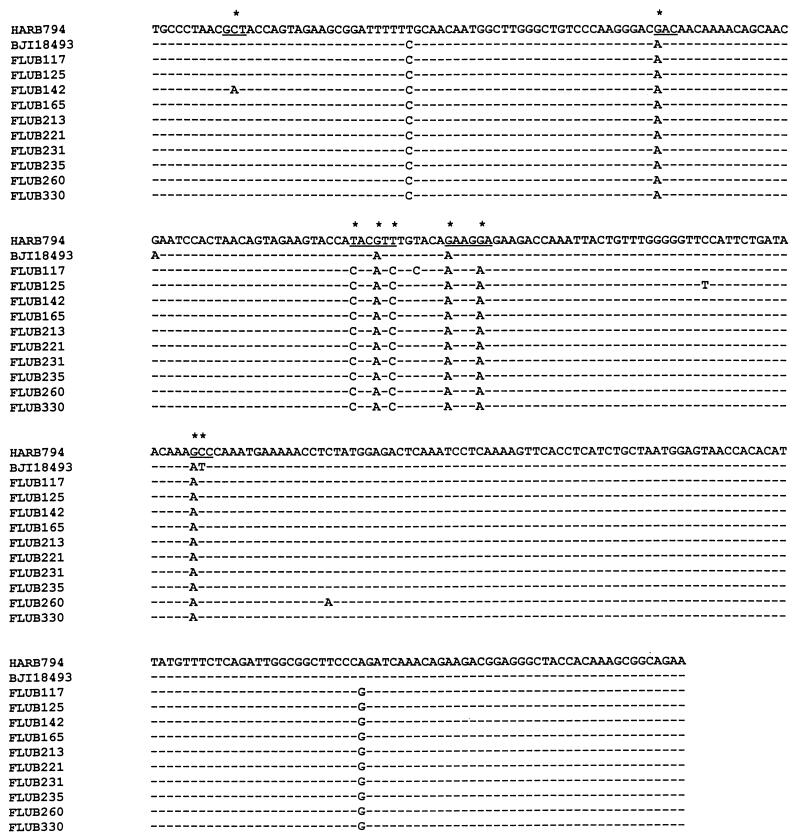
Because the region amplified covers most of the HA1 region where antigenic determinants are located, amplicons of variants identified by HMA were sequenced directly to characterize the genetic changes responsible for the variations. The nucleotide and the deduced amino acid sequences of the amplicons of all 10 type B variants were then compared to the sequences of B/Harbin/7/94, the type B component of the trivalent influenza vaccine, and another reference strain, B/Beijing/184/93, the recommended vaccine strain (10) (Fig. 2). It was found that these 10 strains showed substitution rates of 2.26 to 2.55% at the nucleotide level and 4.26 to 4.68% at the amino acid level compared to the sequence of B/Harbin/7/94 and substitution rates of 1.70 to 1.98% and 2.98 to 3.40%, respectively, compared to the sequence of B/Beijing/184/93 (Table 1). Sixteen nucleotide substitutions were consistent among the 10 isolates, although two mutations occurred only in some isolates and five other mutations appeared in just 1 isolate (Fig. 2). All but one of the amino acid substitutions were conserved among the 10 isolates (Fig. 2).
FIG. 2.
Comparison of amplicon sequences of the 10 type B virus isolates with the reference strains, B/Harbin/7/94 (HARB794) and B/Beijing/184/93 (BJI18493). The region is located in the HA1 portion of the HA gene from nucleotides 27 to 733 from the start of the HA1-coding region. Nucleotide substitutions leading to amino acid changes (underscores) are marked by stars. The arrowhead indicates the single substitution which is a silent mutation but which may be associated with noticeable mobility shift.
TABLE 1.
Divergence of new type B variants from B/Harbin/7/94 and B/Beijing/184/93a
| Isolate | No. (%) Nucleotide changes
|
No. (%) Amino acid changes
|
||
|---|---|---|---|---|
| B/Harbin | B/Beijing | B/Harbin | B/Beijing | |
| 117 | 17 (2.40) | 13 (1.84) | 10 (4.26) | 7 (2.98) |
| 125 | 18 (2.55) | 14 (1.98) | 10 (4.26) | 7 (2.98) |
| 142 | 18 (2.55) | 14 (1.98) | 11 (4.68) | 8 (3.40) |
| 165 | 16 (2.26) | 12 (1.70) | 10 (4.26) | 7 (2.98) |
| 213 | 17 (2.40) | 13 (1.84) | 10 (4.26) | 7 (2.98) |
| 221 | 18 (2.55) | 14 (1.98) | 10 (4.26) | 7 (2.98) |
| 231 | 17 (2.40) | 13 (1.84) | 10 (4.26) | 7 (2.98) |
| 235 | 18 (2.55) | 14 (1.98) | 10 (4.26) | 7 (2.98) |
| 260 | 18 (2.55) | 14 (1.98) | 10 (4.26) | 7 (2.98) |
| 330 | 18 (2.55) | 14 (1.98) | 10 (4.26) | 7 (2.98) |
The region sequenced has 707 nucleotides and 235 amino acids.
DISCUSSION
Epidemics caused by influenza viruses have been a major public health challenge. Although not responsible for pandemics, influenza B virus infections contribute substantially to the disease burden each year and account for as much as half of all infections in some seasons (15). In the 1996–1997 season, influenza B virus caused more than one-third of the laboratory-confirmed infections in Canada according to data from the National Disease Surveillance (11a) and the Canadian Virus Reporting data (13). This was reminiscent of the high level of activity which occurred in Canada during the 1985–1986 season, when the epidemic strains were significantly different from the influenza B virus component in the vaccine (14). A 3.44% substitution rate at the nucleotide level and a 3.33% substitution rate at the amino acid level were found in the complete HA gene between one of the type B strains isolated in that season, B/Canada/3/85, and the vaccine component strain for that season, B/USSR/100/83. For the HA1 region that is covered by the RT-PCR–HMA and sequenced in this study, the substitution rates between these two strains (B/Canada/3/85 and B/USSR/100/83) are 5.09 and 6.38%, respectively. To investigate whether the high level of activity in the 1996–1997 season was caused by emergence of new type B variants, amplicons of HMA-identified isolates were sequenced and compared to the sequence of the actual vaccine component strain, B/Harbin/7/94, and the recommended vaccine component strain, B/Beijing/184/93, even though the HI test showed that these isolates were similar to each other and to the reference strains (data not shown). It was observed that these isolates were indeed divergent from the reference strains both at the nucleotide level (2.26 to 2.55%) and at the amino acid level (4.26 to 4.68%). The result is consistent with what was observed at the World Health Organization Collaborating Center for Influenza at the Centers for Disease Control and Prevention. According to Regnery (8a), the majority of isolates that were collected since October 1996 and tested there by restriction fragment length polymorphism analysis belong to the same group of type B viruses. The sequences of these 10 isolates were further compared with those of 84 other influenza B virus strains for which the corresponding sequences are available in GenBank. Phylogenetic analysis showed that these 10 isolates form a distinct group within the B/Beijing/184/93-like sublineage of the B/Yamagata/16/88 lineage. The results indicate the necessity of closer monitoring of influenza B virus strains that will be circulating in the coming seasons and critical evaluation of the type B component in the trivalent vaccine.
The reason that these type B variants were not identified by the HI test in this laboratory may well be due to the restricted range of antisera that were used in the assay. With limited available resources, it is impossible for investigators to test all isolates received each season with a large panel of antisera, as in the international reference laboratories. This is the very reason that sensitive but quick testing methods are needed for the initial screening of isolates. The RT-PCR–HMA approach offers a solution to this problem in the influenza surveillance effort. All isolates from the community could be screened initially by this method, and the new variants that are identified could then be subjected to the HI test with a broad range of reference antisera. Sequencing of the new variants could then be done if it is considered necessary. The high sensitivity of HMA for the detection of less than 1% nucleotide changes (12) will ensure the identification of all new variants.
It should be pointed out that the variations identified by HMA may not necessarily be epidemiologically significant variations because some mutations are silent, resulting in no amino acid changes, and some mutations are not immunologically important. In addition, some mutations may negatively affect the survival or transmission of the virus so that viruses bearing these mutations cannot compete with other variants and will not become epidemic strains. Comparison of the mobility shift pattern in Fig. 1 with the genetic changes in Fig. 2 showed that the nucleotide substitution from a G in B/Harbin/7/94, isolates 117, 165, and 260, to an A in other isolates (indicated by an arrow in Fig. 2) was probably associated with a noticeable mobility shift. However, the substitution is silent and does not cause an amino acid change. Therefore, it is important to further characterize the variants identified by the HMA method by the HI assay or sequencing analysis, or both.
ACKNOWLEDGMENTS
We are grateful to the DNA Core Facility, Laboratory Centre for Disease Control, Ottawa, Ontario, Canada, for the sequencing work and to Carla Osiowy, Helen Regnery, Nancy Cox, and John Spika for valuable comments and support.
REFERENCES
- 1.Cox N J, Brammer T L, Regnery H L. Influenza: global surveillance for epidemic and pandemic variants. Eur J Epidemiol. 1994;10:467–470. doi: 10.1007/BF01719678. [DOI] [PubMed] [Google Scholar]
- 2.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 3.Hay A J, Douglas A R, Sparrow D B, Cameron K R, Skehel J J. Antigenic and genetic characterization of current influenza strains. Eur J Epidemiol. 1994;10:465–466. doi: 10.1007/BF01719677. [DOI] [PubMed] [Google Scholar]
- 4.Ikonen N, Kinnunen L, Forsten T, Pyhala R. Recent influenza B viruses in Europe: a phylogenetic analysis. Clin Diagn Virol. 1996;6:63–71. doi: 10.1016/0928-0197(96)00201-2. [DOI] [PubMed] [Google Scholar]
- 5.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, Fraenkel-Conrat H, Wagner R R, editors. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–88. [Google Scholar]
- 6.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1353–1395. [Google Scholar]
- 7.Murphy B R, Webster R G. Orthomyxoviruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1397–1445. [Google Scholar]
- 8.Noble G R, Kaye H S, Yarbrough W B, Fiedler B K, Reed C J, Felker M B, Kendal A P, Dowdle W R. Measurement of hemagglutination-inhibiting antibody to influenza virus in the 1976 influenza vaccine program: methods and test reproductivity. J Infect Dis. 1977;136:S429–S434. doi: 10.1093/infdis/136.supplement_3.s429. [DOI] [PubMed] [Google Scholar]
- 8a.Regnery, H. Personal communication.
- 9.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. Recommended composition of influenza virus vaccines for use in the 1996–1997 season. Weekly Epidemiol Rec. 1996;71:57–61. [PubMed] [Google Scholar]
- 11.Xu X, Cox N J, Bender C A, Regnery H L, Shaw M W. Genetic variation in neuraminidase genes of influenza A (H3N2) viruses. Virology. 1996;224:175–183. doi: 10.1006/viro.1996.0519. [DOI] [PubMed] [Google Scholar]
- 11a.Zabchuck, P. Personal communication.
- 12.Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou S. 1996–1997 influenza season: Canadian laboratory diagnosis and strain characterization. Can Communicable Dis Rep. 1997;23:137–141. [PubMed] [Google Scholar]
- 14.Zou S, Prud’homme I, Weber J M. Genetic characterization of the hemagglutinin gene of influenza B viruses that predominated in Canada in the 1985/86 season. Can J Infect Dis. 1997;8:265–269. doi: 10.1155/1997/187859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou S, Prud’homme I, Weber J M. Evolution of the hemagglutinin gene of influenza B virus was driven by both positive and negative selection pressure. Virus Genes. 1997;14:181–185. doi: 10.1023/a:1007927725332. [DOI] [PubMed] [Google Scholar]