Abstract
The identification and speciation of strains of Legionella is often difficult, and even the more successful chromatographic classification techniques have struggled to discriminate newly described species. A sequence-based genotypic classification scheme is reported, targeting approximately 700 nucleotide bases of the mip gene and utilizing gene amplification and direct amplicon sequencing. With the exception of Legionella geestiana, for which an amplicon was not produced, the scheme clearly and unambiguously discriminated among the remaining 39 Legionella species and correctly grouped 26 additional serogroup and reference strains within those species. Additionally, the genotypic classification of approximately 150 wild strains from several continents was consistent with their phenotypic classification, with the exception of a few strains where serological cross-reactivity was complex, potentially confusing the latter classification. Strains thought to represent currently uncharacterized species were also found to be genotypically unique. The scheme is technically simple for a laboratory with even basic molecular capabilities and equipment, if access to a sequencing laboratory is available.
The genus Legionella comprises approximately 40 species, at least 7 of which have more than one serotype (3, 15, 31). Approximately half of the species have been associated with human disease (28). Legionella-like organisms isolated from clinical specimens, or from the environment during the course of an outbreak, need to be identified to elucidate the disease process and to identify the source. Legionellae have proved to be relatively unreactive when traditional biochemical tests are utilized, necessitating more complex identification methods (6, 7, 26, 41). Serologically based methods are widely used in clinical laboratories, but antigen cross-reactivity limits specificity and restricts their confident use to a few frequently isolated species (38). This is especially true for countries where legionellosis caused by species other than L. pneumophila is common (12). More complex classification schemes have been proposed (26, 38), the most successful being one based on the range and proportion of cellular fatty acids and ubiquinones (21, 22, 40, 43). As additional species have been characterized, this method has become less discriminating, since apparently unique patterns were proved to be shared by several species (43). The inclusion of hydroxylated fatty acids has improved discrimination, but it requires the analysis of both mono- and dihydroxylated fatty acids, and individual patterns are complex, making analysis difficult (21).
Gene sequence-based phylogenic (genotypic) schemes have become widely used for organisms which are difficult to classify, as more sequences have been determined and sequencing methods have become simpler, more widely available, and cost effective (11, 23, 24, 29, 32, 34). Genotypic schemes have the great advantage of being unaffected by colony age and growth conditions and, in contrast to chromatographic methods, are not subject to extraction and chromatographic conditions or constituent equipment. Additionally, because a gene sequence is essentially a long digital string, with each digit being one of only four nucleotides, genotypic schemes are less ambiguous and can utilize significantly more discriminatory data than phenotypic ones, and in a form that lends itself to widely available computer analysis software. Many genotypic schemes utilize variation in the 16S rRNA sequence (11, 23, 24, 32, 34), because of the ease with which regions can be amplified and sequenced with universal primers. The 16S rRNA sequences of Legionella species have been reported (18), as have the sequences of the mip gene (2, 12, 13, 31), which codes for an immunophilin of the FK506 binding protein (FKBP) class (14). This protein, which ranges in size from 232 to 251 amino acids, depending on the Legionella species (31), is an outer membrane protein important in the intracellular cycle of Legionella. While it is known to be involved with the survival of the bacterium immediately after uptake into phagocytic cells (9, 12, 28), its exact role is unclear. Additionally, analogs are found widely in both prokaryotes and eukaryotes and are likely to have a significant cellular role (14). Other gene sequences have been determined for Legionella (5, 17, 36), but only the rRNA sequences and the mip gene have been comprehensively determined for most species, an essential prerequisite for any gene to be the basis of a genotyping scheme. Ratcliff et al. (31) recently phylogenetically compared most Legionella species, using the species variation among both the 16S rRNA and mip genes, and found over twice the variation in the mip gene at the DNA level (56% of base sites) as in 16S rRNA (23% of base sites). A pairwise comparison of species reveals a mip gene variation of 3 to 31% (mean, 20%) between species pairs compared with 1 to 10% (mean, 6%) for 16S rRNA. For the mip gene, interspecies nucleotide variation occurred throughout the gene but especially within a hypervariable insert of up to 51 bases immediately adjacent to the region coding for the signal sequence, at redundant third codon sites, and in sequences coding for either single or small regions of variable amino acids interspersed among regions coding for total or near-total amino acid homology, especially toward the 3′ end of the gene (31). These last regions are known to encode the active portions of the protein’s enzymatic peptidylprolyl cis-trans-isomerase (PPIase) activity (31).
Additionally the mip gene appeared to be relatively stable genetically, with no evidence of homologous recombination, in that identical or near-identical sequences were not found for the mip genes from phenotypically divergent species. With respect to genetic stability, the mip gene may therefore behave like housekeeping genes, which are known to be more stable than other gene classes (1). Homologous recombination would severely compromise a sequence-based classification scheme (1), and it is a theoretical possibility at least for rRNA targets (37). Thus, the genetic stability and greater mutational variation of the mip gene suggest that it is an ideal target for a classification scheme, with results likely to be more discriminating in identifying species and more resilient to clonal variation within each species. It may even be possible to discriminate between serogroups where these are present or to demonstrate distinct intraspecies clonal groups.
The present study reports the use of the mip gene to develop a sequence-based classification scheme for Legionella, the first proposed for this genus. Further, it reports the comparison of sequences from species which have additional serogroups, to determine if serogroups can be discriminated. Similarly, it reports the comparison of sequences from wild strains isolated on several continents, for which there is confirmatory phenotypic or DNA hybridization identification data, to test the robustness of the scheme for variation within strains of the same species. Lastly, isolates which appear phenotypically or from DNA hybridization studies to be different from currently characterized species were tested to determine if a sequence-based classification scheme can clarify their identities. Some of these unusual isolates have been previously reported (43).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and DNA preparation.
The following type strains, with their corresponding American Type Culture Collection (ATCC) designations and GenBank, EMBL, and/or DDBJ accession numbers, were used to construct the classification scheme: L. adelaidensis (ATCC 49625; U91606); L. anisa (ATCC 35292; U91607); L. birminghamensis (ATCC 43702; U91608); L. bozemanii serogroup 1 (ATCC 33217; U91609); L. brunensis (ATCC 43878; U92227); L. cherrii (ATCC 35252; U91635); L. cincinnatiensis (ATCC 43753; U91636); L. dumoffii (ATCC 33279; U91637); L. erythra (ATCC 35303; U92203); L. fairfieldensis (ATCC 49588; U92204); L. feeleii serogroup 1 (ATCC 35072; U92205); L. geestiana (ATCC 49504); L. gormanii (ATCC 33297; U91638); L. gratiana (ATCC 49413; U92206); L. hackeliae serogroup 1 (ATCC 35250; U92207); L. israelensis (ATCC 43119; U92208); L. jamestowniensis (ATCC 35298; U92228); L. jordanis (ATCC 33623; U92209); L. lansingensis (ATCC 49751; U92210); L. londiniensis (ATCC 49505; U92229); L. longbeachae serogroup 1 (ATCC 33462; X83036); L. longbeachae serogroup 2 (ATCC 33484; AF000958); L. maceachernii (ATCC 35300; U92211); L. micdadei (ATCC 33218; S62141); L. moravica (ATCC 43877; U92212); L. nautarum (ATCC 49506; U92213); L. oakridgensis (ATCC 33761; U92214); L. parisiensis (ATCC 35299; U92215); L. pneumophila serogroup 1 Philadelphia-1 (ATCC 33152; S42595); L. quateirensis (ATCC 49507; U92216); L. quinlivanii (ATCC 43830; U92217); L. rubrilucens (ATCC 35304; U92218); L. sainthelensi serogroup 1 (ATCC 35248; U92219); L. santicrucis (ATCC 35301; U92220); L. shakespearei (ATCC 49655; U92221); L. spiritensis (ATCC 35249; U92222); L. steigerwaltii (ATCC 35302; U92223); L. tucsonensis (ATCC 49180; U92224); L. wadsworthii (ATCC 33877; U92225); and L. worsleiensis (ATCC 49508; U92226). Table 1 presents additional reference type strains and wild strains, used to test the robustness of the classification scheme, and the GenBank, EMBL, and DDBJ accession numbers of sequences determined from those strains during the study. All sequences which were not identical in the region tested to the type strain sequences listed above were submitted. Wild strains consisted of 102 isolates obtained from clinical or environmental specimens from a variety of laboratories within Australia over a period of several years and stored at −70°C and 52 isolates obtained from Europe, Israel, Singapore and the United States. For this latter group of strains, the identification determined by the originating laboratory was used in this study. For some strains, identification was based solely on colony morphology and serological agglutination or immunofluorescence with polyvalent antisera, although cross-reactivity caused some ambiguity. These isolates were still included in the study because they were the only wild strain of a species or the only isolate from a region or because they produced a variant sequence for a species. In-house storage numbers are used to identify strains from which unique sequences were obtained and submitted to GenBank, EMBL, and DDBJ.
TABLE 1.
Isolates used to test the robustness of the classification scheme
| Legionella isolatea | Accession no.b | Phenotypic identificationc | Sequence identification | No. of differencesd
|
|
|---|---|---|---|---|---|
| DNA | AA | ||||
| L. anisa | |||||
| 5 wild strains, LC3934 | NA | L. anisa | L. anisa | 0 | 0 |
| IMVS-946 | AF022312 | L. anisa | L. anisa | 18 | 1 |
| L. birminghamensis | |||||
| IMVS-C4D5 | NA | L. birminghamensis | L. birminghamensis | 0 | 0 |
| IMVS-K7H8 | AF047743 | L. birminghamensis | L. birminghamensis | 1 | 0 |
| LC2720 | AF047744 | L. birminghamensis | L. birminghamensis | 41 | 1 |
| L. bozemanii | |||||
| LC4348 | NA | L. bozemanii serogroup 1 | L. bozemanii | 0 | 0 |
| Serogroup 2 ATCC 35545, [IMVS-A5A1]c | AF022308 | NA, [L. bozemanii serogroup 2] | L. bozemanii | 3 | 1 |
| IMVS-A5F7, [IMVS-A8E7] | AF022309 | L. bozemanii serogroup 1, [serogroup 2] | L. bozemanii | 3 | 1 |
| IMVS-A5I7, -K7B3, [LC2763] | AF022310 | L. bozemanii serogroup 1 [or L. parisiensis] | L. bozemanii | 6 | 1 |
| IMVS-D2/7, -K7B4 | AF022311 | L. bozemanii serogroup 2 | L. bozemanii | 6 | 1 |
| L. brunensis species E (IMVS-594) | AF022350 | NC | L. brunensis | 6 | 0 |
| L. cherrii LC3664 | NA | L. cherrii | L. cherrii | 0 | 0 |
| L. cincinnatiensis | |||||
| IMVS-C4B3 | NA | L. cincinnatiensis | L. cincinnatiensis | 0 | 0 |
| IMVS-K5B7 | AF022358 | L. cincinnatiensis | L. cincinnatiensis | 3 | 0 |
| IMVS-K8D7 | AF022359 | L. cincinnatiensis | L. cincinnatiensis | 1 | 0 |
| IMVS-K8I2 | AF047745 | L. cincinnatiensis | L. cincinnatiensis | 10 | 3 |
| LC3936 | AF047746 | Cross-reactivity | L. cincinnatiensis | 1 | 0 |
| L. dumoffii IMVS-K7D6, -C7A3, LC0455c | AF022313 | L. dumoffii | L. dumoffii | 2 | 0 |
| L. erythra | |||||
| IMVS-926, LC0709, LC3719 | NA | L. erythra | L. erythra | 0 | 0 |
| LC1317c | AF047747 | L. erythra | L. erythra | 3 | 1 |
| L. feeleii | |||||
| Serogroup 2 ATCC 35849, LC4210 | AF022341 | NA | L. feeleii | 2 | 0 |
| IMVS-913 | AF023174 | L. feeleii | L. feeleii | 9 | 1 |
| IMVS-853, -865 | AF022340 | L. feeleii | L. feeleii | 11 | 1 |
| Species J (IMVS-933) | AF022354 | NC | L. feeleii | 12 | 1 |
| L. geestiana LC3644 | NA | L. geestiana | NA | −i | − |
| L. gormanii LC0777C | AF047748 | L. tucsonensis/L. feeleii serogroup 2 | L. gormanii | 26 | 4 |
| L. hackeliae serogroup 2 ATCC 35999 | NA | NA | L. hackeliae | 0 | 0 |
| L. jamestowniensis | |||||
| IMVS-724, -935 | AF022339 | L. jamestowniensis | L. jamestowniensis | 2 | 0 |
| IMVS-707, -708, -871, -945 | AF022337 | L. jamestowniensis | L. jamestowniensis | 24 | 2 |
| IMVS-857 | AF022338 | L. jamestowniensis | L. jamestowniensis | 27 | 4 |
| L. jordanis LC3940 | NA | L. jordanis | L. jordanis | 0 | 0 |
| L. londiniensis | |||||
| Species C (IMVS-449), IMVS-755, -914, -967, LC4049 | NA | L. londiniensis | L. londiniensis | 0 | 0 |
| IMVS-912 | AF022346 | L. londiniensis | L. londiniensis | 1 | 0 |
| L. longbeachae | |||||
| 26 wild strainse | NA | L. longbeachae serogroup 1 | L. longbeachae | 0 | 0 |
| Serogroup 2 ATCC 33484, [IMVS-C4E7] | NA, [L. longbeachae serogroup 2] | L. longbeachae | 2 | 0 | |
| L. maceachernii | |||||
| IMVS-984 and IMVS-A4H7, LC4349 | AF022315 | L. maceachernii | L. maceachernii | 7 | 0 |
| IMVS-910, -943, -962 | AF022314 | L. maceachernii | L. maceachernii | 6 | 0 |
| L. micdadei | |||||
| 10 wild strains, D4307, D4310, M097-017C1, LC0858 | AF023175 | L. micdadei | L. micdadei | 1 | 0 |
| D4309, D4363, D4534 | AF047749 | L. micdadei | L. micdadei | 2 | 0 |
| IMVS-K5D3 | AF023176 | L. micdadei | L. micdadei | 2 | 0 |
| L. oakridgensis LC3780 | NA | L. oakridgensis | L. oakridgensis | 0 | 0 |
| L. parisiensis IMVS-916 | NA | L. parisiensis | L. parisiensis | 0 | 0 |
| L. pneumophila | |||||
| Serogroup 1 Bellingham-1 ATCC 43111 | AF022329 | NA | L. pneumophila | 9 | 1 |
| Serogroup 1 Knoxville-1 ATCC 33153 | AF022332 | NA | L. pneumophila | 2 | 0 |
| Serogroup 1 OLDA ATCC 43109 | AF022335 | NA | L. pneumophila | 0 | 0 |
| Serogroup 1 Allentown ATCC 43106 | AF022330 | NA | L. pneumophila | 2 | 0 |
| Serogroup 1 Benidom 030 E ATCC 43108 | AF022336 | NA | L. pneumophila | 9 | 1 |
| Serogroup 1 Camperdown-1 ATCC 43113 | AF022331 | NA | L. pneumophila | 0 | 0 |
| Serogroup 1 Heysham 1 ATCC 43107 | AF022334 | NA | L. pneumophila | 0 | 0 |
| Serogroup 1 Oxford 4032 E ATCC 43110 | AF022333 | NA | L. pneumophila | 0 | 0 |
| Serogroup 1 Wadsworth 130b | Reference 13 | NA | L. pneumophila | 9 | 1 |
| Serogroup 2 ATCC 33154 | AF022316 | NA | L. pneumophila | 11 | 1 |
| Serogroup 3 ATCC 33155 | AF022317 | NA | L. pneumophila | 12 | 1 |
| Serogroup 4 ATCC 33156 | AF022318 | NA | L. pneumophila | 11 | 1 |
| Serogroup 5 ATCC 33216 | AF022319 | NA | L. pneumophila | 30 | 5 |
| Serogroup 6 ATCC 33215 | AF022320 | NA | L. pneumophila | 9 | 1 |
| Serogroup 7 ATCC 33823 | AF022321 | NA | L. pneumophila | 3 | 0 |
| Serogroup 8 ATCC 35096 | AF022322 | NA | L. pneumophila | 6 | 0 |
| Serogroup 9 ATCC 35289 | AF022323 | NA | L. pneumophila | 8 | 1 |
| Serogroup 10 ATCC 43283 | AF022324 | NA | L. pneumophila | 8 | 1 |
| Serogroup 11 ATCC 43130 | AF022325 | NA | L. pneumophila | 2 | 0 |
| Serogroup 12 ATCC 43290 | AF022326 | NA | L. pneumophila | 8 | 1 |
| Serogroup 13 ATCC 43736 | AF022327 | NA | L. pneumophila | 2 | 0 |
| Serogroup 14 ATCC 43703 | AF022328 | NA | L. pneumophila | 2 | 0 |
| IMVS-C7C3, -C7D1 | AF023173 | L. pneumophila serogroup ? | L. pneumophila | 31 | 3 |
| IMVS-C6A1, -876 | NA | L. pneumophila serogroup 1 | L. pneumophila | 0 | 0 |
| IMVS-C5E1, -C5E3, -A4C7, -D2/21, [IMVS-D1/77] | NAf | L. pneumophila serogroup 1 [serogroup 13] | L. pneumophila | 2 | 0 |
| IMVS-C6A2 | NAg | L. pneumophila serogroup 1 | L. pneumophila | 9 | 1 |
| LC1329 | AF047750 | L. pneumophila serogroup 16 | L. pneumophila | 6 | 1 |
| L. quinlivanii | |||||
| LC0870 | NA | L. quinlivanii | L. quinlivanii | 0 | 0 |
| IMVS-C1/1007 | AF022347 | L. quinlivanii | L. quinlivanii | 19 | 3 |
| IMVS-731a | AF022348 | L. quinlivanii | L. quinlivanii | 17 | 3 |
| IMVS-941 | AF022349 | L. quinlivanii | L. quinlivanii | 23 | 3 |
| Genomospecies ATCC 51913 | AF022356 | NA | L. quinlivanii | 20 | 3 |
| L. rubrilucens | |||||
| IMVS-901, -906, -939, LC0704, LC0805Hc, LC1092c, LC4557 | NA | L. rubrilucens | L. rubrilucens | 0 | 0 |
| IMVS-896, -915, [LC4042] | AF022342 | L. rubrilucens, [? L. spiritensis] | L. rubrilucens | 25 | 2 |
| L. sainthelensi serogroup 2 ATCC 49322, IMVS-K7B9, [LC4261] | AF022357 | L. sainthelensi [serogroup 2 or L. santicrucis] | L. sainthelensi | 14 | 0 |
| L. santicrucis IMVS-K5E4 | NA | L. santicrucis | L. santicrucis | 0 | 0 |
| L. spiritensis | |||||
| IMVS-C4E5 | AF047751 | L. spiritensis | L. spiritensis | 1 | 0 |
| IMVS-C4E1, ML0076 | AF047752 | L. spiritensis | L. spiritensis | 2 | 0 |
| L. waltersii ATCC 51914, species D (IMVS-500, -532) | AF022343 | NC | L. waltersii | 0 | 0 |
| Species not characterized | |||||
| Species A (IMVS-36) | AF022344 | NC | NC | 160h | 33h |
| Species B (IMVS-86) | AF022345 | NC | NC | 95h | 21h |
| Species G (IMVS-823, -895) | AF022351 | NC | NC | 33h | 3h |
| LC3043, LC3044 | NC/? L. shakespearei | Species G | 0 | 0 | |
| Species H (IMVS-911, -960) | AF022352 | NC | NC | 144h | 46h |
| Species I (IMVS-959) | AF022353 | NC | NC | 146h | 57h |
| Species K (IMVS-K5G3) | AF047753 | NC | NC | 81h | 17h |
| Species L (LC1863) | AF047754 | NC | NC | 123h | 32h |
| Species M (LC4046c) | AF047756 | NC | NC | 151h | 54h |
| Species N (LC4048c) | AF047755 | NC | NC | 143h | 43h |
| LC4381 | NA | NC | NA | − | − |
Wild strains were isolated within Australia, unless otherwise indicated. Strains prefixed LC were isolated within Europe, with the exception of LC1863 (Kenya) and strains LC0455, LC2720, LC3780, LC3940, and LC4210, which were isolated in Singapore. Strains D4309, D4310, D4363, D4534, and M097-017C1 were isolated in the United States, and D4307 was isolated in Israel.
Sequences determined and deposited during this study. NA, not applicable (the isolate is an ATCC reference strain or a wild strain with a sequence identical to that of a type or reference strain).
Phenotypic identification included colony morphology and autofluorescence, and slide agglutination or immunofluorescence with polyvalent antisera for all isolates. Australian strains were additionally identified by chromatographic analysis of whole-cell fatty acids and ubiquinones (43). Strains LC0455, LC1317, LC0805H, LC1092, LC4046, and LC4048 were additionally identified by ribotyping and, with the exception of LC0455, DNA hybridization. Identifications enclosed in brackets relate solely to the similarly marked isolates. NC, not characterized (identification does not conform to currently characterized species).
Number of nucleotide (DNA) or amino acid (AA) differences from the type strain. Sequence differences for L. pneumophila isolates are with respect to Philadelphia-1.
Includes 11 isolates from the United States and 1 from Israel.
Sequence identical to L. pneumophila serogroup 1 (Knoxville-1) and other strains.
Sequence identical to L. pneumophila serogroup 1 (Wadsworth) and other strains.
Number of differences from closest characterized species.
−, no amplicon.
Cultures were grown on charcoal yeast extract agar base (CM665; Oxoid, Basingstoke, Hampshire, United Kingdom) with α-ketoglutarate and l-cysteine supplement (code SR110; Oxoid) and 1% bovine serum albumin in a humidified incubator at 35°C for 4 to 7 days. Chromosomal DNA was extracted either by the method of Manning et al. (25) or by the rapid method of Saunders et al. (35) from a subculture of a single colony checked visually for purity. A heat lysis method was also tested, where a moderately turbid suspension of organisms was made in 500 μl of sterile water and either microwaved on full power (650 W) for 2 min or boiled or steamed for 5 min. Two microliters of this crude extract was used in the amplification reaction.
Sequencing strategy.
From an alignment of the mip sequences of 40 type strains, forward and reverse primers were chosen to target the ribosomal binding site or open reading frame region and the PPIase site, respectively, of the mip gene to amplify a fragment of 661 to 715 bases, depending on the presence and size of the hypervariable region, which is species dependent (31). This equates to nearly 90% of the gene from the 5′ end. While the inferred amino acid sequence is highly conserved in the targeted regions, the primers required redundancies to account for variation at the third codon site.
The primer sequences are as follows (with parentheses indicating a mixed-base site): forward primer (Legmip_f) (27-mer), 5′-GGG(AG)ATT(ACG)TTTATGAAGATGA(AG)A(CT)TGG-3′; reverse primer (Legmip_r) (23-mer), 5′-TC(AG)TT(ATCG)GG(ATG)CC(ATG)AT(ATCG)GG(ATCG)CC(ATG)CC-3′. The primers were constructed with a model 392 DNA/RNA synthesizer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). For gene amplification (33), the reagent mixture (50 μl) consisted of 200 μM (each) deoxynucleoside triphosphate, 200 ng of each primer, PCR buffer (Applied Biosystems), 1.5 mM magnesium chloride (Applied Biosystems), 2.5 U of Taq polymerase (Applied Biosystems), chromosomal DNA (approximately 1 μg), and purified water. Gene amplification consisted of 35 cycles of denaturation at 94°C for 1 min, annealing for 2 min at 58°C, and extension for 2 min at 72°C. Products were visualized by gel electrophoresis, using SPP1 (EcoRI digest of bacteriophage SSP-1 DNA) (Bresatec, Adelaide, Australia) as a marker. Products to be sequenced were purified with the QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions.
Sequences were determined by dye terminator chemistry (Applied Biosystems), according to the manufacturer’s instructions, with 100 to 150 ng of purified amplicon and 40 ng of Legmip_f primer or 60 ng of Legmip_r primer per reaction. Legmip_f proved to be unreliable in priming the sequencing reaction, producing “noisy” sequence for the first 50 to 200 bases for some species. It is believed the primer also binds to a second indeterminable site, producing overlapping reaction products for these species. A forward sequencing primer was designed to overlap Legmip_f but stepped into the amplicon by 6 bases at the 3′ end. This primer, used only for sequencing and at the same concentration as Legmip_f, was designated Legmip_fs, and its sequence is as follows: 5′-TTTATGAAGATGA(AG)A(CT)TGGTC(AG)CTGC-3′. Electropherograms were determined with an Applied Biosystems model 377 DNA sequencer. Sequences were compared with the type strains by using the GeneCompar program (Applied Maths, Kortrijk, Belgium), using the homology search option to find related sequences and the cluster analysis module for the unweighted pair group method with averages (UPGMA) phylogeny.
Nucleotide sequence accession numbers.
The GenBank, EMBL, and DDBJ accession numbers of sequences determined during this study are listed in Table 1.
RESULTS
With Legmip_f and Legmip_r, all type strains gave a single amplification product of the predicted size (661 to 715 bp, depending on the species and the presence of the hypervariable insert), with the exception of L. geestiana, which failed to produce any product. The sequence of each amplicon was determined for the complete length of the fragment, with both Legmip_r and Legmip_fs reliably priming the sequencing reaction to produce unambiguous sequence after approximately 15 bases (to the end of the amplicon). The combination of Legmip_f, as the forward amplifying primer, with Legmip_r to produce the amplicon and Legmip_fs as the forward sequencing primer consistently produced the best results. During the course of this study all amplicons were sequenced in both forward and reverse directions. Amplicon sequences from the type strains were identical to the published sequences (2, 12, 13, 31) and contained sufficient variation (3.6 to 30.5%) to uniquely identify each species.
To test the specificities of the primers, DNA extracted from the following organisms was tested: Staphylococcus aureus, Streptococcus pneumoniae, Escherichia coli, Haemophilus influenzae, Salmonella (multiple serovars), Shigella flexneri, Proteus mirabilis, Aeromonas hydrophila, Pseudomonas aeruginosa, Yersinia enterocolitica, Campylobacter (multiple species), Helicobacter pylori, Corynebacterium diphtheriae, Mycobacterium abscessus, Vibrio parahaemolyticus, and Nocardia asteroides. No amplification was detected, despite the presence of genes coding for Mip-like analogs in some species (19, 20, 30, 44).
Heat lysis proved adequate for extracting DNA for amplification, although boiling or steaming gave slightly more consistent results than microwave heating. Prolonged boiling or steaming for 15 min did not appear to degrade the DNA, as measured by the amount of amplicon produced by subsequent amplification.
Table 1 presents the results for additional reference strains, as well as the comparison of results from the wild strains. This includes both those strains which conform to type strains and those for which speciation cannot be determined, which may represent new species. For a few of these latter strains, amplification produced only limited product which sequenced poorly. Lowering the annealing temperature to 50°C produced more product, which in turn produced longer unambiguous sequence. Sequencing was also attempted after amplification at a below-optimal annealing temperature to produce multiple products of different sizes, and excellent sequence was still obtained with Legmip_fs but not with Legmip_r, as would be predicted. Isolate LC4381 was not able to be amplified, even with the annealing temperature reduced to 40°C.
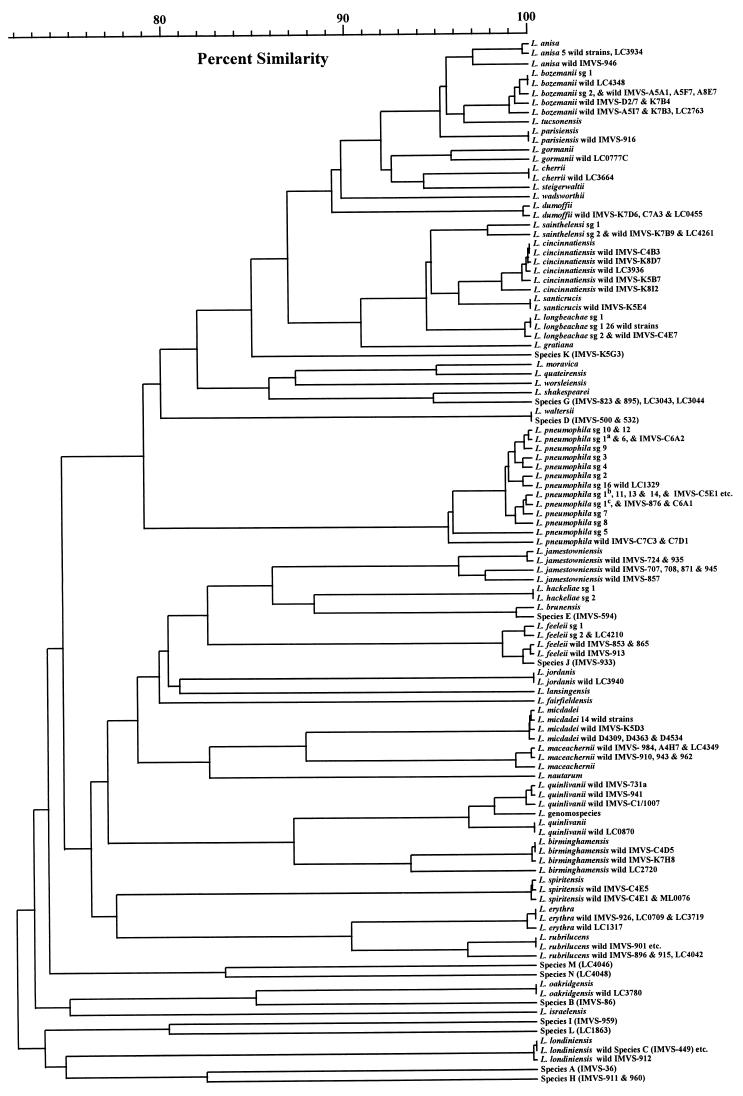
Figure 1 is a UPGMA phylogenic dendrogram of the inter- and intraspecies similarities.
FIG. 1.
UPGMA phylogenetic dendrogram of sequence similarities found among type strains and wild strains of Legionella. The vertical bar joining two isolates or clusters indicates the level of similarity. sg, serogroup; a, Wadsworth 130b, Benidom 030 E, and Bellingham-1 strains; b, Knoxville-1 and Allentown strains; c, Philadelphia-1, Camperdown-1, Heysham 1, OLDA, and Oxford 4032 E strains.
DISCUSSION
For a sequence-based classification scheme to be successful, the gene amplification needs to be specific for the genus but universal for the individual species and the subsequent interspecies sequence variation needs to be sufficient to discriminate clearly between species, even after allowing for the intraspecies sequence variation found among wild strains.
The primers reported here produced single amplicons of the correct size, and in sufficient quantity to enable accurate sequence determination, for all isolates tested, whether they were type strains or wild strains, with the exception of L. geestiana and one unusual, serologically nonreactive, nonidentifiable European wild strain, LC4381. Neither this last isolate nor the type strain or a European wild strain of L. geestiana produced an amplicon of the correct size, even after the annealing temperature was lowered to 40°C. The mip gene sequence for L. geestiana has not been published, and no sequence was determined with the various combinations of primers used by Ratcliff et al. to amplify and sequence this gene in 35 other Legionella species (31). However, a Mip analog has been detected in L. geestiana by using Mip-specific monoclonal antibody (16). Consequently, the lack of amplification for L. geestiana, and LC4381, is likely to be due to sequence variation in the sites targeted by the amplification primers. This was no impediment in our hands, as no locally derived wild strain tested failed to amplify. Similarly, other laboratories should have little problem, as such isolates are likely to be very rare.
Figure 1 shows that all of the remaining species could be easily discriminated and that the wild-strain isolates grouped within the same species as determined by serological, chromatographical, or molecular identification techniques, except for a few where the phenotypic identification was based solely on serology and cross-reactivity prevented unambiguous speciation. Many of these strains had been retained and stored at −70°C because of these nontypical reactions. For instance, LC2763, believed to be a L. bozemanii isolate, also showed strong cross-reactivity with L. parisiensis-specific antiserum. LC3936 reacted equally well with antisera specific for L. cincinnatiensis, L. santicrucis, and both serogroups of L. sainthelensi and L. longbeachae. LC4042 reacted weakly with only L. spiritensis-specific antiserum. However, this isolate was red autofluorescent, suggesting the isolate is likely to be related to either L. erythra or L. rubrilucens, the only red-autofluorescent species so far described. These results demonstrate how ambiguous serological identification can be for some species. The genotypic identification of other strains from the same laboratory, where the serological identification had been confirmed by ribotyping and/or DNA hybridization, was identical in every case.
The Legionella genomospecies clustered with L. quinlivanii, with which it has 69% DNA homology (4). With a difference of 3.6%, L. bozemanii and L. tucsonensis showed the least interspecies sequence variation. However, the four L. bozemanii wild strains and L. bozemanii serogroup 2, with an intraspecies variation of 1.3% or less, were easily discriminated from L. tucsonensis. Similarly, L. cincinnatiensis wild strains (maximum intraspecies variation, 1.6%) were easily discriminated from L. santicrucis strains (interspecies variation, 4.1%). Some species, such as L. anisa, L. longbeachae, L. micdadei, and L. londiniensis, for which several wild strains were tested, showed very little sequence difference, suggesting a single homogeneous clonal population, compared to L. jamestowniensis, L. rubrilucens, and L. quinlivanii, which appeared to demonstrate multiple divergent clonal populations. This is especially interesting given that the L. longbeachae strains were isolated on several continents whereas all but one of the L. quinlivanii isolates, including the type strain, came from one region within Australia. The monophyletic nature of L. longbeachae has been demonstrated by other workers (8). L. jamestowniensis, L. rubrilucens, and L. quinlivanii were generally quite divergent from their nearest neighbors, so the greater sequence variation among wild strains did not compromise the resolution of their speciation. This contrasts with chromatographic identification schemes, where ubiquinone and fatty acid profiles do not discriminate easily between some species, for example, L. erythra and L. rubrilucens (43). In fact, it was this lack of discrimination with such methods which was the motivation for this study.
LC0777C is a very interesting European strain. It reacted poorly serologically, and while there was moderate homology with L. gormanii, with which it was genotyped, there was also moderate homology with L. tucsonensis and L. feeleii serogroup 2. With 4.2% sequence variation from L. gormanii, LC0777C demonstrates more genotypic variation from L. gormanii than is typical for wild-strain variation in sister species. This is consistent with its reduced serological reactivity, and it may represent an additional serogroup, a new genotype clone, or even a new species.
However, the multiple serogroups of species where they have been determined could not be confidently discriminated by this scheme in every circumstance. For example, the sequences for L. hackeliae serogroups 1 and 2 are identical. Similarly, many of the serogroups of L. pneumophila are identical to each other, and as a species seem to mainly cluster into two closely related clonal populations, with serogroup 1 strains represented in both. L. pneumophila serogroup 5 and two nonserogrouped wild strains fall outside these two clonal populations. These two strains may represent a new serogroup, since they are both serologically and genotypically different from other L. pneumophila strains. IMVS-D1/77, a serogroup 13 strain, grouped with the serogroup 13 type strain. For L. bozemanii, IMVS-A8E7, a serogroup 2 isolate, and IMVS-A5F7, a serogroup 1 isolate, both grouped with the serogroup 2 type strain. Two other serogroup 2 isolates, IMVS-D2/7 and IMVS-K7B4, produced similar sequences, but they were different from that of the serogroup 2 type strain. Similarly, some serogroup 1 strains clustered uniquely together. Consequently, while these latter genotype groups may be indicative of a particular serogroup, as may some of the unique L. pneumophila serogroup sequences, the differences are small, and it is possible that such differences may be within the normal clonal variation found in the wild-strain population independent of a specific serogroup. The testing of more strains of these species would be necessary to determine if these few differences are characteristic of specific serogroups.
In contrast, the differences in sequences from the two serogroups of L. sainthelensi do appear discriminatory, with two wild strains producing sequences identical to that of the serogroup 2 type strain. Strain LC4261 has been serologically confirmed as L. sainthelensi serogroup 2, although it also demonstrated serological cross-reactivity with L. santicrusis and, to a lesser extent, L. cincinnatiensis and L. longbeachae. Similarly, although the L. longbeachae serogroup 2 type strain differs from the serogroup 1 type strain by only 2 bases, the 26 serogroup 1 strains and one serogroup 2 strain all produced sequences identical to those of their type strains. Therefore, further testing of additional serogroup 2 wild strains may prove this small difference between the two serogroups to be definitive.
Wilkinson et al. (43) reported the existence of strains representing six uncharacterized species, designated species A to F. These strains were amplified, and the products were sequenced to determine how well the genotyping scheme would identify them. Species F is the type strain of L. adelaidensis (3). Species A and B give unique sequences, supporting the ubiquinone and fatty acid profile data indicating that they are indeed new species. Species C, D, and E cluster closely with L. londiniensis, L. waltersii, and L. brunensis, respectively. However, either these three species were not characterized or isolates were not available at the time of testing (4, 10, 42). A comparison of the ubiquinone and fatty acid profiles for species C, D, and E and L. londiniensis, L. waltersii, and L. brunensis confirmed that the scheme had grouped them correctly. The speciation of species C and E has been independently confirmed with DNA-DNA hybridization (2a). In addition to these uncharacterized species, other isolates, stored because they possessed unusual or unique ubiquinone and fatty acid profiles, were also tested. Isolates IMVS-911 and IMVS-960, designated species H, produced sequences identical to each other but unique from all characterized species. Similarly, IMVS-959, designated species I, produced a unique sequence. These three results are in complete agreement with the ubiquinone and fatty acid profiles, and these strains are likely to represent two new undescribed species. Species G isolates (IMVS-823 and IMVS-895) produced identical sequences but grouped moderately close to L. shakespearei (5.4% variation). These two isolates produce ubiquinone and fatty acid profiles similar to those of L. shakespearei, a species which had not been described at the time of their storage (39). The degree of difference from L. shakespearei, however, is greater than the intraspecies variation found in other species, so their true identity needs to be further elucidated. Species J (isolate IMVS-933) has grouped with L. feeleii. While the isolate produces a ubiquinone profile similar to that of L. feeleii (27), the fatty acid profile shows some differences. The significance of this result is unclear, and the identity of this isolate also needs to be further elucidated. Five European wild strains, similarly uncharacterized, were also tested. LC3043 and LC3044, serologically similar to each other, were found to also have sequences identical to each other and identical to those of the two species G strains. Interestingly they demonstrated weak serological homology only with L. shakespearei. The remaining three isolates, designated species L (LC1863), species M (LC4046), and species N (LC4048), were all found to have unique sequences, consistent with the phenotypic classification.
In conclusion, the scheme was able to unambiguously discriminate among 39 of 40 species and correctly group 26 additional serogroups or reference strains within those species. Additionally, 102 wild strains isolated within Australia and 50 wild strains from Europe, the United States, Singapore, Israel, and Kenya, including strains which are thought to represent currently uncharacterized species, were grouped consistently with their phenotypic identification. Two isolates grouped genotypically differently from their phenotypic classification, but it is probable that DNA hybridization would support the genotypic classification. There were no regional clonal variations detected which would indicate that laboratories in other countries would be troubled with ambiguous classification. The scheme is technically simple for a laboratory with even basic molecular capabilities and equipment, if access to a sequencing laboratory is available, especially given that heat lysis is quite adequate for extracting DNA suitable for amplification. Although all wild-strain amplification products were sequenced in both directions during this study, routinely only one direction would be necessary. While both sequencing primers performed extremely well, Legmip_fs would be the preferred sequencing primer, as it proved to be unaffected by nonspecific amplification.
There was no evidence of genetic recombination horizontally across species in the sequences from the 220 strains used in this study, and while they are still theoretically possible, such events would be unlikely to affect the classification of wild strains in practice. However, unusual results or critical isolates could be confirmed by other phenotypic methods or rRNA gene sequences.
ACKNOWLEDGMENTS
We thank Robert Benson and Tim Harrison for the provision of wild strains from the United States and Israel and from Europe, Singapore, and Kenya, respectively, and Norma Sangster for the provision of the remaining isolates used in this study. We also thank Mark Achtman for succinct manuscript advice and the Clive and Vera Ramaciotti Foundation for generous assistance in the purchase of equipment.
REFERENCES
- 1.Achtman, M. A phylogenetic perspective on molecular epidemiology. In M. Sussman (ed.), Molecular medical microbiology, in press. Academic Press, Inc., London, England.
- 2.Bangsborg J M, Cianciotto N P, Hindersson P. Nucleotide sequence analysis of the Legionella micdadei mip gene, encoding a 30-kilodalton analog of the Legionella pneumophila Mip protein. Infect Immun. 1991;59:3836–3840. doi: 10.1128/iai.59.10.3836-3840.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Benson, R. (Centers for Disease Control and Prevention). 1997. Personal communication.
- 3.Benson R F, Thacker W L, Lanser J A, Sangster N, Mayberry W R, Brenner D J. Legionella adelaidensis, a new species isolated from cooling tower water. J Clin Microbiol. 1991;29:1004–1006. doi: 10.1128/jcm.29.5.1004-1006.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson R F, Thacker W L, Daneshvar M I, Brenner D J. Legionella waltersii sp. nov. and an unnamed Legionella genomospecies isolated from water in Australia. Int J Syst Bacteriol. 1996;46:631–634. doi: 10.1099/00207713-46-3-631. [DOI] [PubMed] [Google Scholar]
- 5.Black W J, Quinn F D, Tompkins L S. Legionella pneumophila zinc metalloprotease is structurally and functionally homologous to Pseudomonas aeruginosa elastase. J Bacteriol. 1990;172:2608–2613. doi: 10.1128/jb.172.5.2608-2613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner D J. Classification of Legionellae. Semin Respir Infect. 1987;2:190–205. [PubMed] [Google Scholar]
- 7.Brenner D J, Steigerwalt A G, Gorman G W, Wilkinson H W, Bibb W F, Hackel M, Tyndall R L, Campbell J, Feeley J C, Thacker W L, Skaliy P, Martin W T, Brake B J, Fields B S, McEachern H V, Corcoran L K. Ten new species of Legionella. Int J Syst Bacteriol. 1985;35:50–59. [Google Scholar]
- 8.Bull J Z, Nimmo G R, Giffard P. Genetic diversity and clonal population structure of Legionella longbeachae serogroup 1 in Australia. Microbiol Aust. 1997;18:A120. [Google Scholar]
- 9.Cianciotto N P, Eisenstein B I, Mody C H, Toews G B, Engleberg N C. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis P J, Brenner D J, Thacker W L, Wait R, Vesey G, Steigerwalt A G, Benson R F. Five new Legionella species isolated from water. Int J Syst Bacteriol. 1993;43:329–347. doi: 10.1099/00207713-43-2-329. [DOI] [PubMed] [Google Scholar]
- 11.Dewhirst F E, Paster B J, Olsen I, Fraser G J. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J Bacteriol. 1992;174:2002–2013. doi: 10.1128/jb.174.6.2002-2013.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle R M, Steele T W, McLennan A M, Parkinson I H, Manning P A, Heuzenroeder M W. Sequence analysis of the mip gene of the soilborne pathogen Legionella longbeachae. Infect Immun. 1998;66:1492–1499. doi: 10.1128/iai.66.4.1492-1499.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engleberg N C, Carter C, Webber D R, Cianciotto N P, Eisenstein B I. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect Immun. 1989;57:1263–1270. doi: 10.1128/iai.57.4.1263-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker J, Fischer G. Immunophilins: structure-function relationship and possible role in microbial pathogenicity. Mol Microbiol. 1993;10:445–456. doi: 10.1111/j.1365-2958.1993.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 15.Harrison T G, Saunders N A. Taxonomy and typing of Legionellae. Rev Med Microbiol. 1994;5:79–90. [Google Scholar]
- 16.Helbig J H, Ludwig B, Lück P C, Groh A, Witzleb W, Hacker J. Monoclonal antibodies to Legionella Mip proteins recognize genus- and species-specific epitopes. Clin Diagn Lab Immunol. 1995;2:160–165. doi: 10.1128/cdli.2.2.160-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman P S, Ripley M, Weeratna R. Cloning and nucleotide sequence of a gene (ompS) encoding the major outer membrane protein of Legionella pneumophila. J Bacteriol. 1992;174:914–920. doi: 10.1128/jb.174.3.914-920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hookey J V, Saunders N A, Fry N K, Birtles R J, Harrison T G. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int J Syst Bacteriol. 1996;46:526–531. [Google Scholar]
- 19.Horne S M, Young K D. Escherichia coli and other species of the Enterobacteriaceae encode a protein similar to the family of Mip-like FK506-binding proteins. Arch Microbiol. 1995;163:357–365. doi: 10.1007/BF00404209. [DOI] [PubMed] [Google Scholar]
- 20.Horne S M, Kottom T J, Nolan L K, Young K D. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium copenhagen. Infect Immun. 1997;65:806–810. doi: 10.1128/iai.65.2.806-810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jantzen E, Sonesson A, Tangen T, Eng J. Hydroxy-fatty acid profiles of Legionella species: diagnostic usefulness assessed by principal component analysis. J Clin Microbiol. 1993;31:1413–1419. doi: 10.1128/jcm.31.6.1413-1419.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert M A, Moss C W. Cellular fatty acid compositions and isoprenoid quinone contents of 23 Legionella species. J Clin Microbiol. 1989;27:465–473. doi: 10.1128/jcm.27.3.465-473.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson P A, Llop-Perez P, Hutson R A, Hippe H, Collins M D. Towards a phylogeny of the clostridia based on 16S rRNA sequences. FEMS Microbiol Lett. 1993;113:87–92. doi: 10.1111/j.1574-6968.1993.tb06493.x. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig W, Schleifer K H. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 25.Manning P M, Heuzenroeder M W, Yeadon J, Leavesley D I, Reeves P R, Rowley D. Molecular cloning and expression in Escherichia coli K-12 of the O antigens of the Inaba and Ogawa serotypes of the Vibrio cholerae O1 lipopolysaccharides and their potential for vaccine development. Infect Immun. 1986;53:272–277. doi: 10.1128/iai.53.2.272-277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauchline W S, Keevil C W. Development of the BIOLOG substrate utilization system for identification of Legionella spp. Appl Environ Microbiol. 1991;57:3345–3349. doi: 10.1128/aem.57.11.3345-3349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moss C W, Bibb W F, Karr D E, Guerrant G O, Lambert M A. Cellular fatty acid composition and ubiquinone content of Legionella feeleii sp. nov. J Clin Microbiol. 1983;18:917–919. doi: 10.1128/jcm.18.4.917-919.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connell W A, Dhand L, Cianciotto N P. Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect Immun. 1996;64:4381–4384. doi: 10.1128/iai.64.10.4381-4384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahfeld J U, Rucknagel K P, Stoller G, Horne S M, Schierhorn A, Young K D, Fischer G. Isolation and amino acid sequence of a new 22-kDa FKBP-like peptidyl-prolyl cis/trans-isomerase of Escherichia coli—similarity to Mip-like proteins of pathogenic bacteria. J Biol Chem. 1996;271:22130–22138. doi: 10.1074/jbc.271.36.22130. [DOI] [PubMed] [Google Scholar]
- 31.Ratcliff R M, Donnellan S C, Lanser J A, Manning P A, Heuzenroeder M W. Interspecies sequence differences in the Mip protein from the genus Legionella; implications for function and evolutionary relatedness. Mol Microbiol. 1997;25:1149–1158. doi: 10.1046/j.1365-2958.1997.5471908.x. [DOI] [PubMed] [Google Scholar]
- 32.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christen R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 33.Saiki R K, Gelfand D H, Stoffel S, Schark S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 34.Sallen B, Rajoharison A, Desvarenne S, Quinn F, Mabilat C. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int J Syst Bacteriol. 1996;46:669–674. doi: 10.1099/00207713-46-3-669. [DOI] [PubMed] [Google Scholar]
- 35.Saunders N A, Harrison T G, Haththotuwa A, Kachwalla N, Taylor A G. A method for typing strains of Legionella pneumophila serogroup 1 by analysis of restriction fragment length polymorphisms. J Med Microbiol. 1990;31:45–55. doi: 10.1099/00222615-31-1-45. [DOI] [PubMed] [Google Scholar]
- 36.Segal G, Shuman H A. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun. 1997;65:5057–5066. doi: 10.1128/iai.65.12.5057-5066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strätz M, Mau M, Timmis K N. System to study horizontal gene exchange among microorganisms without cultivation of recipients. Mol Microbiol. 1996;22:207–215. doi: 10.1046/j.1365-2958.1996.00099.x. [DOI] [PubMed] [Google Scholar]
- 38.Veríssimo A, Morais P V, Diogo A, Gomes C, da Costa M S. Characterization of Legionella species by numerical analysis of whole-cell protein electrophoresis. Int J Syst Bacteriol. 1996;46:41–49. doi: 10.1099/00207713-46-1-41. [DOI] [PubMed] [Google Scholar]
- 39.Verma U K, Brenner D J, Thacker W L, Benson R F, Vesey G, Kurtz J B, Dennis P J L, Steigerwalt A G, Robinson J S, Moss C W. Legionella shakespearei sp. nov., isolated from cooling tower water. Int J Syst Bacteriol. 1992;42:404–407. doi: 10.1099/00207713-42-3-404. [DOI] [PubMed] [Google Scholar]
- 40.Wait R. Confirmation of the identity of Legionellae by whole cell fatty-acid and isoprenoid quinone profiles. In: Harrison T G, Taylor A G, editors. A laboratory manual for Legionella. London, England: John Wiley & Sons Ltd.; 1988. pp. 69–101. [Google Scholar]
- 41.Wilkinson H W. Reactions of Legionella species on biochemical tests. In: Wilkinson H W, editor. Hospital-laboratory diagnosis of Legionella infections. 2nd ed. Atlanta, Ga: Centers for Disease Control; 1988. p. 28. [Google Scholar]
- 42.Wilkinson H W, Drasar V, Thacker W L, Benson R F, Schindler J, Potuznikova B, Mayberry W R, Brenner D J. Legionella moravica sp. nov. and Legionella brunensis sp. nov. isolated from cooling-tower water. Ann Inst Pasteur Microbiol. 1988;139:393–402. doi: 10.1016/0769-2609(88)90102-0. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson I J, Sangster N, Ratcliff R M, Mugg P A, Davos D E, Lanser J A. Problems associated with identification of Legionella species from the environment and isolation of six possible new species. Appl Environ Microbiol. 1990;56:796–802. doi: 10.1128/aem.56.3.796-802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong C Y, Heuzenroeder M W, Quinn D M, Flower R L. Cloning and characterization of two immunophilin-like genes, ilpA and fkpA, on a single 3.9-kilobase fragment of Aeromonas hydrophila genomic DNA. J Bacteriol. 1997;179:3397–3403. doi: 10.1128/jb.179.11.3397-3403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]