Abstract
A dominant epitope within the human herpesvirus 8 (HHV8) ORF 65-encoded protein was mapped to an 8-amino-acid (aa) sequence (RKPPSGKK [aa 162 to 169]) by an amino acid replacement method. Using a 14-aa peptide (P4) encompassing this epitope as the antigen, we developed an enzyme immunoassay for HHV8 antibodies. The presence of P4 antibodies in a panel of 61 human serum specimens was highly correlated with biopsy-confirmed Kaposi’s sarcoma. The homologous Epstein-Barr virus peptide derived from BFBR3-encoded protein did not interfere with the assay, suggesting that P4 is specific for HHV8.
A recently identified gammaherpesvirus, human herpesvirus 8 (HHV8), may be the causative agent of Kaposi’s sarcoma (KS), a tumor commonly associated with human immunodeficiency virus (HIV) infection (1, 4, 19, 21). The presence of HHV8 antibodies in sera from HIV-infected patients is highly correlated with the presence or likelihood of developing KS (7, 13, 14, 16, 17, 22). The occasional detection of HHV8 DNA sequences in semen samples from KS patients indicates that this agent may be sexually transmissible (9, 11, 12). Furthermore, frequent detection of HHV8 virions in saliva samples from KS patients suggests that salivary contact could contribute to HHV8 transmission (15). A rapid and sensitive test for HHV8 infection is needed for large-scale epidemiologic studies to determine the prevalence of HHV8 infection in the general population and to study its role in disease.
Several assays for HHV8 antibodies have been developed. They include immunofluorescence assay (7, 13, 14, 16, 18), immunoblotting (8, 17), and enzyme immunoassays (EIA) in a microtiter plate format (5, 22). The first two assays used antigens expressed in HHV8-carrying cell lines derived from primary effusion lymphomas. The last assay used either a recombinant protein (22) derived from the HHV8 ORF 65 gene or an 18-amino-acid (aa) peptide (5) of the HHV8 capsid protein (ORF 26) conjugated to bovine serum albumin as the antigen. EIA has features that would be useful for routine seroepidemiologic studies of HHV8 infection because they can be configured for high throughput. The use of synthetic peptide(s) or recombinant antigens may make this assay more specific than infected-cell-based assays. However, neither of these assays was 100% sensitive (60 to 80%) in detecting HHV8 antibodies in KS patients, and discordant results were observed when they were compared with other assays. In this study, we identified the dominant continuous epitope of the ORF 65-encoded protein and developed a peptide-based EIA for the detection of HHV8 antibodies in human sera.
MATERIALS AND METHODS
Serum panel.
All serum specimens (n = 61) were collected from the Atlanta metropolitan area as part of past Centers for Disease Control and Prevention studies, and were unlinked from personal identifiers prior to testing. One specimen was from a patient with classical KS (i.e., an elderly patient who was HIV seronegative), and the remaining 60 specimens were from three different groups of 20 individuals each. The first group (KS+ HIV+) consisted of HIV-infected homosexual men who had biopsy-confirmed KS (CD4+ T-cell counts ranged from 10 to 660/μl; mean, 269/μL). The second group (KS− HIV+) consisted of HIV-infected homosexual men who did not have KS (CD4+ T-cell counts ranged from 7 to 1,246/μl; mean, 255/μl). The third group (KS− HIV−) consisted of healthy HIV-negative blood donors (10 men and 10 women).
Synthetic peptides.
Peptides were synthesized according to the manufacturer’s protocol on an automatic synthesizer (model 432A; Applied Biosystems, Foster City, Calif.), partially purified by reverse-phase high-performance liquid chromatography (Bio-Rad, Richmond, Calif.), lyophilized, and stored desiccated at room temperature until use.
Three overlapping peptides of 31 to 34 residues (P1, aa 91 to 124; P2, aa 117 to 147; and P3, aa 140 to 170) encompassing the C-terminal 80-residue HHV8 ORF 65 protein were synthesized for initial antibody screening. A shorter version of P3 (P4, aa 157 to 170) corresponding to the last 14 aa of the protein, which was predicted to be highly immunogenic by a hydrophilicity-based algorithm of Hoop and Woods (10) (not shown), was also used. For epitope mapping, P4 and 11 of its analogs (P4.1 to P4.11) which differ from P4 by 1 aa were used (Table 1). For the competition study, the Epstein-Barr virus (EBV) homolog (QPHDTAPRGARKKQ) derived from the corresponding segment of the EBV BFRF3-encoded protein (2) was used as the competing peptide.
TABLE 1.
C-terminal sequences of the HHV8 ORF 65 protein (P4) and its peptide analogs for fine mapping of the dominant epitope
| Peptide | Sequencea | Substitution |
|---|---|---|
| P4 (wild type) | AVADARKPPSGKKK | |
| P4.1 | –––G–––––––––– | D160G |
| P4.2 | ––––L––––––––– | A161L |
| P4.3 | –––––G–––––––– | R162G |
| P4.4 | ––––––G––––––– | K163G |
| P4.5 | –––––––G–––––– | P164G |
| P4.6 | ––––––––G––––– | P165G |
| P4.7 | –––––––––G–––– | S166G |
| P4.8 | ––––––––––L––– | G167L |
| P4.9 | –––––––––––G–– | K168G |
| P4.10 | ––––––––––––G– | K169G |
| P4.11 | –––––––––––––G | K170G |
The entire P4 (wild type) sequence is shown. For the peptide analogs, amino acids identical to those in P4 are represented by the dashes; only the residues that are different from those in P4 are shown.
Peptide EIA.
Published procedures for peptide EIA were followed (20). Briefly, peptides were dissolved in carbonate-bicarbonate buffer (0.1 M, pH 9.4) to a final concentration of 5 μg/ml, and 100 μl of this solution was used to coat microtiter wells by overnight incubation at 4°C. Peptide-coated wells were washed once in phosphate-buffered saline (PBS) (pH 7.4) containing 0.05% Tween 20, air dried, and stored desiccated at −20°C until use. Nonspecific binding sites of the peptide-coated wells were blocked with 5% nonfat dry milk (Nestle Food Co., Glendale, Calif.) in PBS containing 0.1% Tween 20 (milk buffer) for 30 min at 37°C just prior to the assay. Sera were diluted 1:100 in milk buffer, and allowed to react with peptide-coated wells for 1 h at 37°C. Bound antibodies were detected with peroxidase-conjugated goat anti-human immunoglobulin G (IgG) (heavy and light chains) (Bio-Rad) and tetramethyl-benzidine and hydrogen peroxide substrates (Kirkegaard and Perry Laboratories, Gaithersburg, Md.) after the plates were washed five times with PBS containing 0.05% Tween 20. The baseline-corrected optical density at 450 nm (OD450) was calculated as follows: A450 − A630. The mean corrected OD450 of the 20 KS− HIV− specimens plus 5 standard deviations was arbitrarily chosen as the assay cutoff for each peptide.
Epitope mapping.
Three serum specimens highly reactive to peptide P4 were tested on all 12 peptides listed in Table 1 on a single microtiter plate. The antibody reactivity of each peptide analog was then compared with that of P4.
Competition assay.
Six P4-reactive serum specimens (three highly and three moderately reactive) were chosen for this study. Diluted serum samples (1:100 in milk buffer) were first incubated with the competing EBV peptide or P4 itself ranging from 0.01 to 10.0 μg/100 μl at room temperature for 15 min. The peptide-serum mixtures were then added to the P4-coated plate, and the peptide EIA procedure as described above was then followed.
Detection of HHV8 antibodies by immunofluorescence assay.
An mouse monoclonal antibody-enhanced immunofluorescence assay (MIFA) as described by Lennette et al. (16) with a slight modification in slide preparation (3) was performed. Briefly, tetradecanoyl phorbol ester acetate (Sigma)-induced BCBL-1 cells were harvested on day 6 postinduction, washed once in PBS, and suspended in PBS to give a final concentration of 106 cells/ml. One drop of this suspension was applied to the slide, air dried, and fixed in cold acetone for 5 min at −20°C. Fixed cells were incubated first with diluted (1:10) serum specimens for 30 min, then with mouse monoclonal anti-human IgG (ATCC HF6508), and finally with fluorescein-conjugated anti-mouse IgG (Cappel, Durham, N.C.) at 1:100 dilution. The cells were then examined with a fluorescence microscope.
RESULTS
Antigenicity of overlapping peptides.
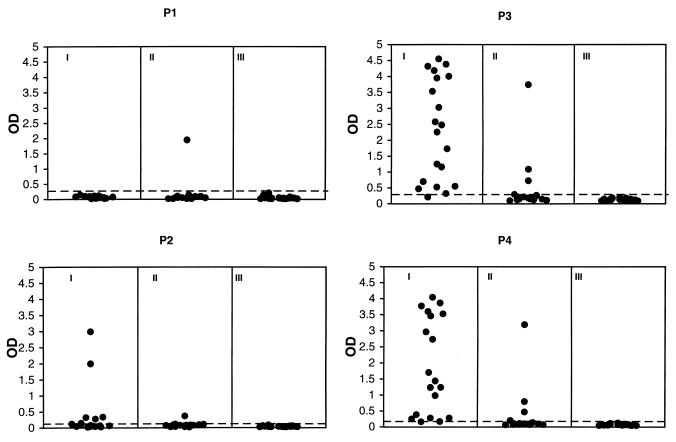
The seroreactivity patterns of the 60 human serum specimens determined by the peptide EIA with overlapping peptides P1, P2, P3, and P4 are shown in Fig. 1. Eighteen (90%) of the 20 KS+ HIV+, 4 (20%) of the 20 KS− HIV+, and none of the KS− HIV− specimens reacted with P4. Virtually identical reactivity patterns were observed with P3. However, no KS+ HIV+ and only one KS− HIV+ (5%) specimen reacted with P1. Five KS+ HIV+ (25%) specimens and one KS− HIV+ (5%) specimen reacted with P2. The specimen from the patient with classical KS was tested only on P3 and P4 and was positive for both peptides (data not shown).
FIG. 1.
Seroreactivities of specimens from 20 KS+ HIV+ (I), 20 KS− HIV+ (II), and 20 KS− HIV− (III) persons with overlapping peptides P1, P2, P3, and P4 derived from the C-terminal half of the HHV8 ORF 65 gene product. The broken lines indicate the cutoff values, as defined by the mean baseline-corrected OD450 of the 20 KS− HIV− serum specimens plus 5 standard deviations.
Fine epitope mapping.
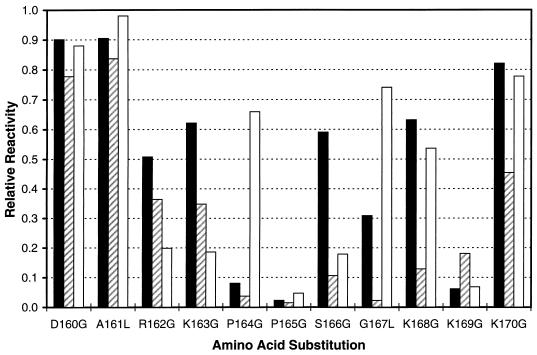
Figure 2 depicts the reactivity patterns of three HHV8-positive serum specimens with P4 analogs. Although the effect of amino acid substitutions varied from specimen to specimen, residues 165 (P) and 169 (K) appeared to be the most important for antibody recognition in all three specimens examined. On the basis of the overall lower reactivities of these three specimens with peptide analogs having substitutions at residues 162 to 169, we deduced that the sequence RKPPSGKK comprises the immunodominant domain of the C-terminal region of the ORF 65 gene product.
FIG. 2.
Seroreactivities of three HHV8 antibody-positive serum specimens with P4 peptide analogs which differ from P4 by one residue. Substitutions and residue number corresponding to the HHV8 ORF 65 protein are shown in the x axis. A relative reactivity of 1.0 was assigned to peptide P4.
Competition study with EBV peptide analog.
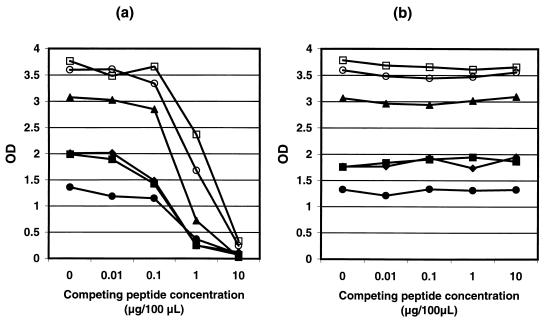
No inhibition of P4 reactivity by the homologous EBV peptide (up to 10 μg/100 μl) was observed by peptide EIA (Fig. 3b), while the autologous peptide greatly diminished the assay signals at 0.1 to 1.0 μg/100 μl with the six serum specimens examined (Fig. 3a).
FIG. 3.
Seroreactivities of P4 with serum specimens (n = 6) preincubated with 0 to 10 μg/100 μl of competing peptides. P4 itself (a) and the P4 homolog derived from the C terminus of BFRF3-encoded EBV protein (b) were used as the competing peptide.
Comparison between peptide EIA and MIFA.
MIFA identified 19 of 20 (95%) KS+ HIV+, 9 of 20 (45%) KS− HIV+, and 5 of 20 (25%) KS− HIV− specimens positive for HHV8 antibodies. The results were consistent with previously published results in these categories (16). The specimen from the patient with classical KS also tested positive by the MIFA.
Nearly all KS+ HIV+ specimens (18 of 19) which scored positive by the MIFA were also positive by the peptide EIA. Only one of the 20 KS+ HIV+ specimens was negative by both assays. For both assays, an intermediate number of positive results were found in the KS− HIV+ set and the fewest number was found in the KS− HIV− set. Less agreement between the two assays was observed in these two sets of specimens. For the KS− HIV+ set, 3 of the 9 MIFA-positive specimens were also positive by the peptide EIA, and only 1 of the 11 MIFA-negative specimens was positive by the peptide EIA.
DISCUSSION
Peptide EIA results indicated that P3 (the last 31 residues of ORF 65) and P4 (the last 14 residues of ORF 65) were equally antigenic across the panel of 60 serum specimens tested. Only sporadic reactivity was observed with either P1 or P2. These results were consistent with the observation that a dominant epitope was located within the C-terminal 14 residues of the HHV8 ORF 65 gene product. Using short peptide analogs of P4, in which amino acids were sequentially replaced one at a time, we further mapped the epitope to an 8-aa sequence (aa 162 to 169) of the ORF 65 gene product. This epitope was tested for its utility in detecting HHV8 antibody.
In our preliminary study, we detected anti-P4 antibodies in 19 of 21 (90%) of known KS-positive individuals. This sensitivity was comparable to that reported from a similar assay (81%) using a recombinant ORF 65 gene product as the antigen (22) and appeared better than the 60% sensitivity of an 18-aa peptide derived from the minor capsid protein of HHV8 (5).
A 95% concordance was observed between the peptide EIA and a published MIFA with the KS+ HIV+ specimens. Lower concordances of 65 and 75% were observed for the KS− HIV+ and KS− HIV− specimens, respectively. All but 1 of the 13 discordant specimens were scored positive by the MIFA and negative by the peptide EIA. This discordance may indicate that MIFA is more sensitive than peptide EIA because of the multiple antigens present in the BCBL-1 cells and the use of a 10-fold-higher sample concentration in the MIFA. Alternatively, MIFA may be less specific than peptide EIA due to the presence of cross-reactive antibodies against other human herpesviruses, such as EBV. Although no strong correlation was observed between EBV and HHV8 antibodies when tested by the MIFA in specimens with high EBV IgG antibody titers, low-level cross-reaction is still possible (16). The use of short synthetic peptides as antigens will reduce the chance of cross-reactions. We compared P4 to the homologous EBV peptide and found only three identical residues (D160, K168, and K169), suggesting a low probability of cross-reaction. A competition study revealed no inhibition of P4 reactivity by this EBV peptide analog, indicating that P4 was specific for HHV8 antibody.
The conservatively high choice for the assay cutoff (5 standard deviations above the mean OD of the 20 KS− HIV− specimens) was made to ensure the specificity of the results, in the absence of “gold standard” negative-control specimens. Further work will be required to determine whether any high negative specimens are true positive.
Although sequence variation among different HHV8 isolates is generally low (0.1% in 2,500 bp), unique amino acid substitutions within ORF 25 or 26 and ORF 75 gene products have been reported (6, 23). However, ORF 65 sequences of various HHV8 isolates were not available. Therefore, sequencing the HHV8 ORF 65 of geographically divergent isolates is needed to ensure that P4 is capable of detecting HHV8 infections in various geographic regions and population groups.
Despite the high correlation of P4 antibodies and KS, this assay may not be able to detect latent HHV8 infections, since the ORF 65 gene product may not be highly expressed during latent infection. A highly sensitive assay for HHV8 infection may require a cocktail of peptides derived from latent- and lytic-cycle proteins. Therefore, identification of dominant epitopes within the latent antigens is critical for the development of better diagnostics for HHV8 infection. In addition, independent methods such as virus culture or PCR are needed to validate serological methods.
REFERENCES
- 1.Aluigi M G, Albini A, Carlone S, Repetto L, De Marchi R, Icardi A, Moro M, Noonan D, Benelli R. KSHV sequences in biopsies and cultured spindle cells of epidemic, iatrogenic and Mediterranean forms of Kaposi’s sarcoma. Res Virol. 1996;147:267–275. doi: 10.1016/0923-2516(96)82285-0. [DOI] [PubMed] [Google Scholar]
- 2.Baer R J, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G F, Hudson G S, Satchwell S C, Sequin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Black J B, Sanderlin K C, Goldsmith C S, Gary H E, Lopez C, Pellett P E. Growth properties of human herpesvirus-6 strain Z29. J Virol Methods. 1989;26:133–146. doi: 10.1016/0166-0934(89)90143-2. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y, Moore P S. Kaposi’s sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5:215–222. [PubMed] [Google Scholar]
- 5.Davis D A, Humphrey R W, Newcomb F M, O’Brien T R, Goedert J J, Straus S E, Yarchoan R. Detection of serum antibodies to a Kaposi’s sarcoma-associated herpevirus-specific peptide. J Infect Dis. 1997;175:1071–1079. doi: 10.1086/516444. [DOI] [PubMed] [Google Scholar]
- 6.Di Alberti L, Ngui S L, Porter S R, Speight P M, Scully C M, Zakrewska J M, Williams I G, Artese L, Piattelli A, Teo C G. Presence of human herpesvirus 8 variants in the oral tissues of human immunodeficiency virus-infected persons. J Infect Dis. 1997;175:703–707. doi: 10.1093/infdis/175.3.703. [DOI] [PubMed] [Google Scholar]
- 7.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italian and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 8.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 9.Gupta P, Singh M K, Rinaldo C, Ding M, Farzadegan H, Saah A, Hoover D, Moore P, Kingsley L. Detection of Kaposi’s sarcoma herpesvirus DNA in semen of homosexual men with Kaposi’s sarcoma. AIDS. 1996;10:1596–1598. doi: 10.1097/00002030-199611000-00022. [DOI] [PubMed] [Google Scholar]
- 10.Hopp T P, Woods K R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard M R, Whitby D, Bahadur G, Suggett F, Boshoff C, Tenant-Flowers M, Schulz T F, Kirk S, Matthews S, Weller I V, Tedder R S, Weiss R A. Detection of human herpesvirus 8 DNA in semen from HIV-infected individuals but not healthy semen donors. AIDS. 1997;11:F15–F19. doi: 10.1097/00002030-199702000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y Q, Li J J, Poiesz B J, Kaplan M H, Friedman-Kien A E. Detection of the herpesvirus-like DNA sequences in matched specimens of semen and blood from patients with AIDS-related Kaposi’s sarcoma by polymerase chain reaction in situ hybridization. Am J Pathol. 1997;150:147–153. [PMC free article] [PubMed] [Google Scholar]
- 13.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus): distribution of infection in KS risk group and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 14.Kedes D H, Ganem D, Ameli N, Bacchetti P, Greenblatt R. The prevalence of serum antibody to human herpesvirus 8 (Kaposi sarcoma-associated herpesvirus) among HIV-seropositive and high-risk HIV-seronegative women. JAMA. 1997;277:478–481. [PubMed] [Google Scholar]
- 15.Koelle D M, Huang M L, Chandran B, Vieira J, Piekorn M, Corey L. Frequent detection of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in saliva of human immunodeficiency virus-infected man: clinical and immunologic correlates. J Infect Dis. 1997;176:94–102. doi: 10.1086/514045. [DOI] [PubMed] [Google Scholar]
- 16.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi’s sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 17.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 18.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Offermann M K. HHV8: a new herpesvirus associated with Kaposi’s sarcoma. Trends Microbiol. 1996;4:383–386. doi: 10.1016/0966-842X(96)10060-3. [DOI] [PubMed] [Google Scholar]
- 20.Pau C P, Lee-Thomas S, Auwanit W, George J R, Ou C Y, Parekh B S, Granade T C, Holloman D L, Phillips S, Schochetman G, Young N, Takebe Y, Gayle H D, Weniger B G. Highly specific V3 peptide enzyme immunoassay for serotyping HIV-1 specimens from Thailand. AIDS. 1993;7:337–340. doi: 10.1097/00002030-199303000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Russo J J, Bohenzky R A, Chien M C, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V, Weiss R A, Moore P S. Prevalence of Kaposi’s sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 23.Zong J C, Metroka C, Reitz M S, Nicholas J, Hayward G S. Strain variability among Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. J Virol. 1997;71:2505–2511. doi: 10.1128/jvi.71.3.2505-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]