Abstract
Current methods of DNA extraction from different fungal pathogens are often time-consuming and require the use of toxic chemicals. DNA isolation from some fungal organisms is difficult due to cell walls or capsules that are not readily susceptible to lysis. We therefore investigated a new and rapid DNA isolation method using high-speed cell disruption (HSCD) incorporating chaotropic reagents and lysing matrices in comparison to standard phenol-chloroform (PC) extraction protocols for isolation of DNA from three medically important yeasts (Candida albicans, Cryptococcus neoformans, and Trichosporon beigelii) and two filamentous fungi (Aspergillus fumigatus and Fusarium solani). Additional extractions by HSCD were performed on Saccharomyces cerevisiae, Pseudallescheria boydii, and Rhizopus arrhizus. Two different inocula (108 and 107 CFU) were compared for optimization of obtained yields. The entire extraction procedure was performed on as many as 12 samples within 1 h compared to 6 h for PC extraction. In comparison to the PC procedure, HSCD DNA extraction demonstrated significantly greater yields for 108 CFU of C. albicans, T. beigelii, A. fumigatus, and F. solani (P ≤ 0.005), 107 CFU of C. neoformans (P ≤ 0.05), and 107 CFU of A. fumigatus (P ≤ 0.01). Yields were within the same range for 108 CFU of C. neoformans and 107 CFU of C. albicans for both HSCD extraction and PC extraction. For 107 CFU of T. beigelii, PC extraction resulted in a greater yield than did HSCD (P ≤ 0.05). Yields obtained from 108 and 107 CFU were significantly greater for filamentous fungi than for yeasts by the HSCD extraction procedure (P < 0.0001). By the PC extraction procedure, differences were not significant. For all eight organisms, the rapid extraction procedure resulted in good yield, integrity, and quality of DNA as demonstrated by restriction fragment length polymorphism, PCR, and random amplified polymorphic DNA. We conclude that mechanical disruption of fungal cells by HSCD is a safe, rapid, and efficient procedure for extracting genomic DNA from medically important yeasts and especially from filamentous fungi.
The rapid availability of genomic DNA from medically important fungi is becoming increasingly important in reference clinical microbiology laboratories for accurate molecular epidemiologic subtyping and diagnostic PCR. Direct sequencing systems which will rely upon the rapid availability of genomic DNA are currently under development for clinical microbiological laboratories. Previously described methods for genomic DNA extraction from fungal pathogens require hours to days to complete and often incorporate toxic chemicals. Additionally, the release of DNA is often poor due to cell walls or capsules that are not readily susceptible to lysis.
High-speed cell disruption (HSCD) is a method of DNA extraction which permits rapid lysis of cells and recovery of nucleic acids (11). To our knowledge, such rapid extraction procedures have not been applied to medically important fungi. We therefore investigated a new rapid DNA isolation method using HSCD and incorporating chaotropic reagents and lysing matrices in comparison to standard phenol-chloroform (PC) extraction protocols for DNA isolation from three medically important yeasts (Candida albicans, Cryptococcus neoformans, and Trichosporon beigelii) and two filamentous fungi (Aspergillus fumigatus and Fusarium solani). Additional extractions were performed on Saccharomyces cerevisiae, Pseudallescheria boydii, and Rhizopus arrhizus. Two different inocula (108 and 107 CFU) were compared for optimization of obtained yields. The integrity and quality of the extracted genomic DNA were further validated by restriction fragment length polymorphism (RFLP), PCR, and random amplified polymorphic DNA (RAPD) analysis.
(This work was presented in part at the 97th General Meeting of the American Society for Microbiology [29].)
MATERIALS AND METHODS
Rapid extraction procedure. (i) Culture and harvesting of fungal strains.
Forty-five milliliters of 2% Sabouraud medium was inoculated with one to three colonies of either C. albicans 4740, S. cerevisiae 4740 or 5645, or T. beigelii TSAS-1. C. neoformans 9658 or 134476 was inoculated into 45 ml of yeast nitrogen broth plus 2% glucose, 20 μg of uracil per ml, and 20 μg of adenine per ml. Cultures of C. albicans, S. cerevisiae, and T. beigelii were grown for 18 h at 37°C in a reciprocal shaking water bath set at 80 oscillations/min. Cultures of C. neoformans were grown for 2 to 3 days at 27°C in a reciprocal shaking water bath set at 80 oscillations/min. The cultures were harvested by centrifugation for 10 min at 2,000 × g (Damon/IEC CRU-5000 centrifuge). The cells were washed with normal saline, repelleted, and resuspended in 10 ml of normal saline.
A. fumigatus 972 and 4215, F. solani 92-1484 and 93-1547, P. boydii 91-1216, and R. arrhizus 88-390 were inoculated onto potato dextrose agar slants, incubated for 24 h at 30°C, and then placed at room temperature until mature. Conidia were harvested by washing the slants with normal saline–0.0125% Tween 20 solution. The suspension was then filtered through a two-ply sterile gauze pad to remove any unwanted debris (hyphae and agar), resulting in a pure harvest of conidia. The cells were pelleted by centrifugation at 2,000 × g, washed once with normal saline, repelleted, and resuspended in 10 ml of normal saline. Cell counts were performed with a hemacytometer on each species, and volumes containing 107 and 108 cells were aliquoted and pelleted.
(ii) DNA isolation.
Lysis buffer solutions used in both HSCD and PC procedures were optimized for each organism. C. albicans, S. cerevisiae, T. beigelii, A. fumigatus, and P. boydii pellets were resuspended in 500 μl of lysis buffer (LB) (0.05 M EDTA [pH 8.0], 0.3% sodium dodecyl sulfate [SDS]). C. neoformans was resuspended in 500 μl of LB containing 0.5 M EDTA, 1 M Tris-HCl (pH 8), 1% Sarkosyl, and 0.2% proteinase K (19). F. solani was resuspended in 500 μl of LB containing 0.05 M EDTA, 0.1 M Tris-HCl (pH 8), and 0.5 M NaCl (12). Samples were transferred to a homogenization tube (Bio 101, Inc., Vista, Calif.) containing beads and immediately processed in the FP 120 FastPrep cell disrupter (Bio 101, Inc.) for 30 s. All samples were processed twice, except for C. neoformans, which was processed four times. Between processing, the tubes were cooled on ice for 10 min. The samples were then centrifuged for 5 min at 14,000 × g to pellet cell debris.
(iii) DNA binding and elution.
Supernatants were transferred to new tubes, and 300 μl of glass milk binding matrix (Bio 101, Inc.) was added. The supernatants and matrices were mixed by inversion for 2 to 3 min, followed by centrifugation for 3 min at 14,000 × g. Supernatants were discarded, and pellets were resuspended in 500 μl of 80% ethanol. The mixture was transferred to spin filters and centrifuged twice for 2 min at 14,000 × g to remove the last traces of ethanol. The spin filters containing the glass milk pellet were transferred to clean catch tubes. The 107-CFU pellets were gently resuspended in 100 μl of 0.01 M Tris-HCl–0.001 M EDTA (pH 8.0) (TE), and the 108-CFU pellets were resuspended in 200 μl of TE. Tubes were heated in a 70°C water bath for 10 min and centrifuged for 2 min at 14,000 × g. All samples were treated with RNase (RNase A; Sigma, St. Louis, Mo.) for 1 h at 37°C. Ten microliters of each sample was run on a 1% agarose gel. To control for potential interday and interuser variation, the extractions of each organism were performed on at least three different days by two different individuals.
PC extraction. (i) Culture and harvesting of fungal strains.
Culturing and harvesting of yeasts and filamentous fungi were performed as described above by a conventional PC extraction protocol (27).
(ii) Spheroplast formation.
Yeast spheroplasts for C. albicans, S. cerevisiae, and T. beigelii were prepared by resuspending cells in 1.5 ml of spheroplast buffer (1.0 M sorbitol, 0.05 M sodium phosphate [monobasic], 0.1% 2-mercaptoethanol, 100 μg of Lyticase [Sigma] per ml) (7). Cells from C. neoformans and filamentous fungi were resuspended in 1 ml of spheroplast buffer plus 75 μl of Novozyme (20 mg/ml; Sigma) (19, 23). Pellets were resuspended by vortexing and then incubated for 45 min at 30°C. Upon completion, spheroplasts were pelleted by centrifugation for 10 min at 2,000 × g.
(iii) Cell lysis.
Each pellet was resuspended in 800 μl (yeast) and 500 μl (filamentous fungi) of LB. To the filamentous fungi samples, an additional 62.5 μl of 10% SDS and 0.25 g of glass beads were added, followed by 3 min of vortexing. Spheroplasts were then incubated for 30 min at 65°C. Tubes were once again centrifuged, and the lysate was transferred to a clean tube. Lysates were purified by PC-isoamyl alcohol extractions until a white interface was no longer present. A single chloroform extraction was performed to remove any residual phenol.
(iv) DNA precipitation.
Purified lysates were transferred to clean microcentrifuge tubes. To each tube, 32 μl of 5 M sodium chloride was added, followed by 2 volumes of 100% ethanol. DNA was precipitated for 1 h at −20°C and then pelleted at 4°C in a microcentrifuge at 14,000 × g for 30 min. The supernatant was decanted, and the pellets were gently rinsed with ice-cold 70% ethanol. Pellets were dried in a vacuum desiccator with P2O5 (Sigma).
Dried DNA pellets were resuspended overnight in TE as follows. DNA extracted from 107 CFU was resuspended in 50 μl of TE; DNA from 108 CFU was resuspended in 100 μl of TE. DNA samples were then treated with RNase as described above, and a 10-μl aliquot was run on a 1% agarose gel.
RFLP.
Restriction digests were performed with 15 μl of DNA extracted by HSCD from 108 cells of C. albicans, C. neoformans, S. cerevisiae, T. beigelii, A. fumigatus, F. solani, P. boydii, and R. arrhizus. The DNA was digested with 10 U of EcoRI (Boehringer Mannheim, Indianapolis, Ind.) for 2 h at 37°C. The digests were analyzed on a 0.7% agarose gel.
PCR.
DNA extracted by HSCD from 107 cells of C. albicans, C. neoformans, S. cerevisiae, T. beigelii, A. fumigatus, F. solani, P. boydii, and R. arrhizus was used for PCR. PCR samples were set up under standard reaction conditions (Qiagen, Santa Clarita, Calif.). For all organisms, oligonucleotides from 18S rRNA were used: FG-F, 5′-ATTGGAGGGCAAGTCTGGTG-3′, and FG-R, 5′-CCGATCCCTAGTCGGCATAG-3′. PCR was performed as follows: 94°C for 30 s, 62°C for 1 min, and 72°C for 2 min, for 35 cycles with a final extension of 7 min. A 0.53-kb product was amplified (15). Each PCR product (10 μl) was analyzed on a 1% agarose gel.
RAPD.
DNA extracted by HSCD from 107 cells of C. albicans, C. neoformans, S. cerevisiae, T. beigelii, A. fumigatus, F. solani, P. boydii, and R. arrhizus was used for RAPD analysis. The single oligonucleotide used was RP5, 5′-CGGTCACGCT-3′ (22). Amplification reactions were performed in 1.5 mM MgCl2–50 mM KCl–10 mM Tris-HCl–0.001% (wt/vol) gelatin, which contained 20 mM (each) dATP, dCTP, dGTP, and dUTP (Boehringer Mannheim); 10 μl of primer RP5 (diluted 1:100); 2.5 U of Taq DNA polymerase (Boehringer Mannheim); 2.5 U of AmpliTaq DNA polymerase Stoeffel fragment (Perkin-Elmer Cetus, Foster City, Calif.); 1 U of uracil DNA glycosylate (Boehringer Mannheim); and 1 μl (diluted 1:10) of DNA, in a 50-μl reaction mixture. Samples were incubated at 37°C for 15 min and then placed in a thermal cycler. PCR was performed as follows: 95°C for 7 min, 95°C for 1 min, 35°C for 1 min, and 72°C for 2 min for 10 cycles and 95°C for 30 s, 45°C for 30 s, and 72°C for 1 min for 36 cycles followed by a 7-min extension period at 72°C. Each RAPD product (25 μl) was run on a 2% agarose gel.
Image analysis for concentration of DNA.
Extracted DNA and DNA standards (Life Technologies, Gaithersburg, Md.) were run on a 1% agarose gel. A standard curve was generated by using the IS-1000 Digital System software program (version 2.03; Alpha Innotech Corp., San Leandro, Calif.). DNA was quantitated against the standard curve.
Statistical analysis.
Differences between means were analyzed by the Wilcoxon rank sum test. Values are expressed as means ± standard errors of the means (SEMs). All P values were two-sided, and a P value of ≤0.05 was considered to be significant.
RESULTS
The DNA yields of HSCD are presented in Tables 1 and 2. The entire extraction procedure was performed on as many as 12 samples within 1 h compared to 6 h for PC extraction. The extracted genomic DNA was of large molecular size (greater than 25 kb). Yields obtained from 108 CFU by HSCD were significantly higher than those obtained by PC extraction (P < 0.0001), but the difference was not significant for 107 CFU.
TABLE 1.
Yields obtained by HSCD versus PC DNA extractiona
| Organism and CFU | Mean ± SEM (μg/ml)
|
95% CI (μg/ml) (min, max)
|
Two-tailed P value | ||
|---|---|---|---|---|---|
| HSCD | PC | HSCD | PC | ||
| Yeasts | |||||
| C. albicans | |||||
| 108 | 19.02 ± 1.94 | 3.03 ± 0.06 | +14.54, +23.50 | +2.88, +3.18 | ≤0.005 |
| 107 | 0.76 ± 0.08 | 0.70 ± 0.05 | +0.57, +0.94 | +0.58, +0.83 | NS |
| T. beigelii | |||||
| 108 | 6.07 ± 0.98 | 1.71 ± 0.15 | +3.82, +8.33 | +1.37, +2.05 | ≤0.005 |
| 107 | 0.22 ± 0.02 | 0.39 ± 0.04 | +0.17, +0.27 | +0.29, +0.49 | ≤0.05 |
| C. neoformans | |||||
| 108 | 1.71 ± 0.43 | 2.32 ± 0.31 | +0.72, +2.69 | +1.62, +3.02 | NS |
| 107 | 0.99 ± 0.23 | 0.47 ± 0.11 | +0.46, +1.51 | +0.22, +0.72 | ≤0.05 |
| Filamentous fungi | |||||
| A. fumigatus | |||||
| 108 | 29.94 ± 1.88 | 10.22 ± 1.42 | +25.59, +34.28 | +6.95, +13.49 | ≤0.005 |
| 107 | 2.59 ± 0.73 | 1.22 ± 0.26 | +0.92, +4.27 | +0.63, +1.81 | ≤0.01 |
| F. solani | |||||
| 108 | 49.12 ± 5.50 | 0.90 ± 0.03 | +36.43, +61.81 | +0.82, +0.97 | ≤0.005 |
| 107 | 2.48 ± 0.22 | QNS | +1.96, +3.00 | QNS | |
CI, confidence interval; NS, not significant; QNS, quantity not sufficient. n is 9 for both procedures, all organisms, and all CFU values.
TABLE 2.
Yields obtained by HSCD DNA extractiona
| Organism and CFU | Mean ± SEM (μg/ml) | 95% CI (μg/ml) (min, max) | Two-tailed P value |
|---|---|---|---|
| Yeast | |||
| S. cerevisiae | |||
| 108 | 19.62 ± 1.65 | +15.83, +23.42 | ≤0.005 |
| 107 | 0.99 ± 0.14 | +0.67, +1.32 | |
| Filamentous fungi | |||
| P. boydii | |||
| 108 | 19.68 ± 2.59 | +13.71, +25.65 | ≤0.005 |
| 107 | 0.67 ± 0.05 | +0.56, +0.77 | |
| R. arrhizus | |||
| 108 | 27.70 ± 4.21 | +17.98, +37.42 | ≤0.005 |
| 107 | 4.05 ± 0.22 | +3.54, +4.56 |
n is 9 for all organisms and CFU values. CI, confidence interval.
Table 1 illustrates the comparison of yields obtained from C. albicans, T. beigelii, C. neoformans, A. fumigatus, and F. solani (107 to 108 CFU) by HSCD DNA extraction with yields obtained by PC DNA extraction. The HSCD DNA extraction protocol demonstrates significantly greater yields for 108 CFU of C. albicans, T. beigelii, A. fumigatus, and F. solani (P ≤ 0.005); 107 CFU of C. neoformans (P ≤ 0.05); and 107 CFU of A. fumigatus (P ≤ 0.01). Yields were within the same range for 108 CFU of C. neoformans and 107 CFU of C. albicans for both HSCD extraction and PC extraction. For 107 CFU of T. beigelii, the PC extraction resulted in a greater yield than did HSCD (P ≤ 0.05).
Table 2 demonstrates that the yield of DNA derived from S. cerevisiae is similar to that of C. albicans at 107 and 108 CFU. Moreover, the yield of DNA from P. boydii and R. arrhizus is comparable to that of A. fumigatus and F. solani. DNA yields for each organism were significantly greater at 108 than at 107 CFU.
Yields obtained from 108 and 107 CFU were significantly greater for the filamentous fungi than for yeasts by the HSCD extraction procedure (P < 0.0001). By the PC extraction procedure, the differences were not significant.
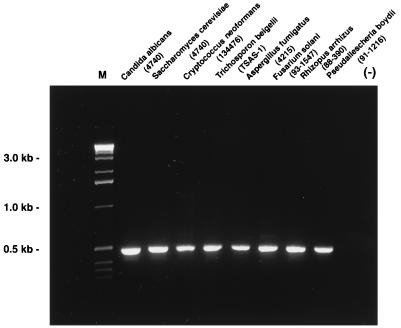
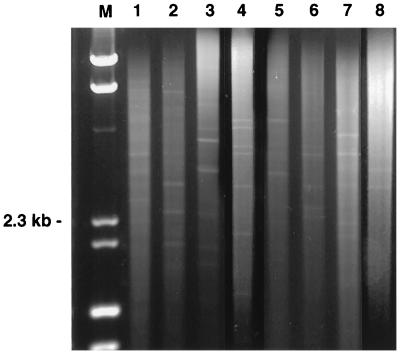
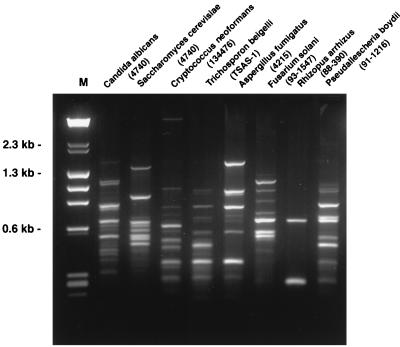
The integrity and quality of fungal genomic DNA obtained by the HSCD procedure were validated by RFLP, PCR, and RAPD. With DNA from 107 CFU of all eight organisms extracted by the HSCD procedure, single bands were obtained by PCR. The bands corresponded to the anticipated size of 0.53 kb (Fig. 1). The restriction digest fragments of genomic DNA were clearly delineated and produced distinct patterns for different species (Fig. 2). Further consistent with maintaining the quality of DNA during the rapid extraction procedure are the results of the RAPD analysis, which demonstrate distinct band patterns distinguishing the different organisms (Fig. 3).
FIG. 1.
PCR products from four yeasts and four filamentous fungi. Lane M, molecular size markers; lane (−), no organism.
FIG. 2.
RFLP patterns from four yeasts and four filamentous fungi. Lane M, molecular size markers; lane 1, C. albicans isolate 4740; lane 2, S. cerevisiae isolate 4740; lane 3, C. neoformans isolate 134476; lane 4, T. beigelii isolate TSAS-1; lane 5, A. fumigatus isolate 4215; lane 6, F. solani isolate 93-1547; lane 7, R. arrhizus isolate 88-390; lane 8, P. boydii isolate 91-1216.
FIG. 3.
RAPD patterns obtained from four yeasts and four filamentous fungi. Lane M, molecular size markers.
DISCUSSION
Filamentous fungi have strong cell walls which are often resistant to traditional DNA extraction procedures (26). Fungal nucleases, polysaccharides, and pigments also contribute to difficulties in isolating DNA from filamentous fungi. These difficulties have led to time-consuming (2) and expensive methods involving ultracentrifugation (22) or column chromatography (21). A variety of minipreparation procedures have been described elsewhere for the isolation of fungal genomic DNA (5, 20). Many of these methods are not suited for the clinical microbiology laboratory, where processing many samples simultaneously is necessary (17). Moreover, the use of toxic chemicals such as PC further limits the use of conventional DNA extraction procedures in clinical microbiology laboratories (5, 6, 18, 25). While enzymatic methods are generally employed for obtaining consistent release of fungal DNA, the yields are often low (25, 26).
Recently, an HSCD system incorporating chaotropic reagents and lysing matrices for disruption of cell membranes was developed (FP 120 FastPrep). The cell disruption is caused by the collision between the fungal cell wall and the beads within the reaction tube. The effectiveness of the cell disruption process depends on the rate of collision and the energy of impact, which are functions of the speed settings (range, 4.0 to 6.5 m/s), and the specific gravity of the bead material used. The rate of collision is proportional to the speed, while the energy of impact is proportional to the square of the speed.
The HSCD system has been successfully applied to extractions of DNA and RNA from bacterial pathogens and in agricultural studies. For example, the FastPrep cell disrupter in combination with the FastRNA kit has been used for total RNA extraction from Vibrio cholerae, followed by reverse transcriptase PCR to detect viable cells of V. cholerae (1). RNA extracted from Staphylococcus aureus, Streptococcus pyogenes, and Mycobacterium tuberculosis by similar methods was found to be suitable for Northern blotting (9, 10, 13, 14). RNA extraction based on homogenization of plant tissue by high-speed reciprocal shaking in the presence of a mixture of sand and glass beads was successfully performed on plant samples (14). HSCD DNA extraction from agricultural soil was five times faster than previous methods and resulted in very pure DNA suitable for restriction digestion, cloning, and PCR amplification. The method provided far less shearing of DNA (predominantly 9 to 23 kb) than did previous bead methods used to extract DNA from sediments (0.5 to 9 kb) (3). DNA extracted from soybean plants by the HSCD system was also suitable for PCR (8). To our knowledge, HSCD heretofore has not been applied to medically important fungi for potential use in clinical microbiology laboratories.
The use of different lysis buffers bears note. For DNA isolation, conidia from filamentous fungi and blastoconidia from yeasts were pelleted and each pellet was resuspended in lysis solution from traditional extraction procedures. Our experience with different lysis solutions from traditional extraction procedures was confirmed in this system. Lysis solutions containing EDTA and SDS gave good yields of DNA for C. albicans, S. cerevisiae, T. beigelii, A. fumigatus, P. boydii, and R. arrhizus, but for F. solani and C. neoformans, different lysis solutions were necessary to obtain optimal yields within the same range. Additional lysis solutions could be explored for other fungal organisms to be used in the HSCD system.
DNA extraction by this rapid mechanical method was achieved within 1 h for up to 12 samples and was superior to PC extraction, which requires about 6 h. All samples were processed twice, except for those from C. neoformans, which, due to the tenacious capsule, were processed four times. A glass milk binding matrix was added to the supernatant, and following centrifugation, the pellet was resuspended and transferred to a spin column. The DNA was eluted from the column and run on a native agarose gel for confirmation of integrity. Up to the stage of binding and elution of the DNA, this procedure was able to be completed within 1 h.
In this study, DNA yields were not quantified by nucleic acid absorbance. Several recent studies have indicated that measurement of nucleic acid absorbance ratios at 260 nm/280 nm was inaccurate for determining the quantity or purity of nucleic acid preparations (16, 18, 24). Our findings confirm these observations. In order to further quantitatively assess the yields of this rapid extraction procedure, the concentration of the extracted genomic DNA was measured in comparison to DNA standards on a 1% agarose gel. A standard curve was generated, and the DNA was quantitated against the standard curve.
In general, DNA yields obtained by mechanical breakage of the fungal cell wall by HSCD were greater than or equal to those obtained by PC extraction. Due to the fast breakage of the fungal cell wall within seconds by the high-speed cell disrupter, the whole extraction procedure is much faster than PC DNA extraction, which involves several lysing steps. For C. albicans, A. fumigatus, and difficult organisms such as T. beigelii and F. solani, HSCD resulted in significantly higher yields from 108 CFU, and for C. neoformans and A. fumigatus, yields from 107 CFU were higher than those obtained by the PC extraction procedure. For the yeasts C. albicans and C. neoformans, yields were within the same range for both methods from 107 CFU. Only from 107 CFU of T. beigelii did PC extraction result in a greater yield than that of HSCD.
Significantly higher yields were obtained from filamentous fungi than from yeasts by HSCD. In comparison to yeast cells, conidia from most filamentous fungi are often resistant to enzymatic lysis procedures. HSCD may be superior to PC for extracting DNA from conidia by virtue of the high-energy mechanical disruptive forces. The DNA yield of F. solani was notably higher than that of all other filamentous fungi. The macroconidia of F. solani are cylindrical structures, in contrast to the sphere-like structures of the conidia of other filamentous fungi, such as A. fumigatus. The differences in DNA yields obtained may be related to the differences in high-energy particles impacting on a sphere versus a cylinder. A spherical shape is inherently stiffer than a cylinder of similar size because the cylinder behaves like a flat plate when viewed along its longitudinal axis. Moreover, the energy of a high-speed particle impacting on a sphere is dissipated parallel to the walls and normally in regard to the point of impact. Both energy waves are dissipated along the surfaces of the sphere, while the normal component imparts momentum to the overall cell. On the other hand, a high-energy particle striking a cylinder (analogous to a Fusarium macroconidium) has a more damaging effect. At the point of impact, there are again two energy vectors: a parallel component and a normal component. The parallel component of the energy wave is dissipated along the wall of the cylinder. However, the normal component of the energy wave impacts upon the opposite side of the wall. This results in local stretching of the cell wall under increasing tensile stress. The net result is a fracture of the cylinder (4).
An HSCD system has the potential for shearing DNA and thereby damaging its integrity for subsequent studies. The quality of the extracted genomic DNA was therefore investigated under standard conditions for RFLP from 108 CFU and for PCR and RAPD analysis from 107 CFU. The quality and concentration of DNA were sufficient to obtain PCR products and distinguishable DNA patterns from RFLP and RAPD analysis. The rapidly extracted genomic DNA was of high molecular weight, and the integrity of DNA was equal to that obtained by a traditional PC extraction.
Further corroborating the effectiveness of DNA extraction by mechanical disruption of medically important filamentous fungi, van Burik et al. compared five nonenzymatic methodologies with an enzymatic method for greatest yield of high-quality DNA from A. fumigatus hyphae (26). The nonenzymatic methods were (i) glass bead pulverization with vortexing, (ii) grinding with mortar and pestle followed by glass bead pulverization with gentle rocking, (iii) use of 1% hydroxyacetyltrimethylammonium bromide buffer followed by glass bead pulverization in a water bath sonicator, (iv) use of 1% hydroxyacetyltrimethylammonium bromide buffer followed by water bath sonication alone, and (v) grinding with mortar and pestle in liquid nitrogen, and the enzymatic method was Lyticase (Sigma) enzymatic fungal cell lysis. Genomic DNA yields were measured by visual reading of 2% agarose gels and spectrophotometry, with shearing assessed by the appearance on the gel. Fungal DNA yields were highest for the first method, followed by the fifth method; the other four methods yielded 10-fold-smaller amounts than did the first method. These results from A. fumigatus hyphae support our findings that high-speed vortexing with glass beads provides a rapid alternative for high-yield extraction of fungal DNA, especially from filamentous fungi.
ACKNOWLEDGMENT
F.-M. Müller was supported by a grant (Mu 1289/1-1) from the Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Bej A K, Ng W Y, Morgan S, Jones D D, Mahbubani M H. Detection of viable Vibrio cholerae by reverse-transcriptase polymerase chain reaction (RT-PCR) Mol Biotechnol. 1996;5:1–10. doi: 10.1007/BF02762407. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard M M, Nowotny V. High-throughput rapid yeast DNA extraction. Genet Anal Tech Appl. 1994;11:7–11. doi: 10.1016/1050-3862(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 3.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, L. B. (Brown Optronics Corp., Cochranville, Pa.). Personal communication.
- 5.Cenis J L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Challen M P, Moore A J, Martinez-Carrera D. Facile extraction and purification of filamentous fungal DNA. BioTechniques. 1995;18:975–976. [PubMed] [Google Scholar]
- 7.Chanda V B, editor. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 13.13.1–13.3.9. [Google Scholar]
- 8.Chen W, Gray L E, Grau C R. Molecular differentiation of fungi associated with brown stem rot and detection of Phialophora gregata in resistant and susceptible soybean cultivars. Phytopathology. 1996;86:1140–1148. [Google Scholar]
- 9.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 10.Cheung A L, Wolz C, Yeaman M R, Bayer A S. Insertional inactivation of a chromosomal locus that modulates expression of potential virulence determinants in Staphylococcus aureus. J Bacteriol. 1995;177:3220–3226. doi: 10.1128/jb.177.11.3220-3226.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarkson A I, Lefevre P, Titchener-Hooker N J. A study of process interactions between cell disruption and debris clarification stages in the recovery of yeast intracellular products. Biotechnol Prog. 1993;9:462–467. doi: 10.1021/bp00023a004. [DOI] [PubMed] [Google Scholar]
- 12.Crowhurst R N, Hawthorne B T, Rikkerink E H, Templeton M D. Differentiation of Fusarium solani f. sp. cucurbitae races 1 and 2 by random amplification of polymorphic DNA. Curr Genet. 1991;20:391–396. doi: 10.1007/BF00317067. [DOI] [PubMed] [Google Scholar]
- 13.Dsna R C, Saghbiru M, Lippman D, Cheung A L. Isolating RNA using the new FastPrep system. J NIH Res. 1995;7:61. [Google Scholar]
- 14.Eggermont K, Goderis I J, Broekaert W F. High-throughput RNA extraction from plant samples based on homogenisation by reciprocal shaking in the presence of a mixture of sand and glass beads. Plant Mol Biol Rep. 1996;14:273–279. [Google Scholar]
- 15.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glasel J A. Validity of nucleic acid purities monitored by 260 nm/280 nm absorbance ratios. BioTechniques. 1995;18:62–63. [PubMed] [Google Scholar]
- 17.Glee P M, Russell P J, Welsch J A, Pratt J C, Cutler J E. Methods for DNA extraction from Candida albicans. Anal Biochem. 1987;164:207–213. doi: 10.1016/0003-2697(87)90387-3. [DOI] [PubMed] [Google Scholar]
- 18.Huberman J A. Importance of measuring nucleic acid absorbance at 240 nm as well as at 260 nm. BioTechniques. 1995;18:62–63. [PubMed] [Google Scholar]
- 19.Kwon-Chung K J, Varma A. Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol. 1991;29:810–812. doi: 10.1128/jcm.29.4.810-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 21.Saunders G, Rogers M E, Adlard M W, Holt G. Chromatographic resolution of nucleic acids extracted from Penicillium chrysogenum. Mol Gen Genet. 1984;194:343–345. [Google Scholar]
- 22.Specht C A, DiRusso C C, Novotny P C, Ullrich R C. A method for extracting high-molecular-weight deoxyribonucleic acid from fungi. Anal Biochem. 1982;119:158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- 23.Stevens D A, Denning D W, Clemons K V, Hanson L H. Restriction endonuclease analysis of total cellular DNA of Aspergillus fumigatus isolates of geographically and epidemiologically diverse origin. J Infect Dis. 1990;162:1151–1158. doi: 10.1093/infdis/162.5.1151. [DOI] [PubMed] [Google Scholar]
- 24.Subden R E, Krizus A. Correction factors for the diphenylamine test for deoxyribonucleic acid in yeasts. Microbios. 1985;43:233–243. [PubMed] [Google Scholar]
- 25.Tsai Y L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Burik J H, Schreckhise R W, Myerson D, White T C, Bowden R A. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Comparison of six extraction techniques for isolation of DNA from filamentous fungi, abstr. F-12; p. 262. [Google Scholar]
- 27.Walsh T J, Francesconi A, Kasai M, Chanock S J. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J Clin Microbiol. 1995;33:3216–3220. doi: 10.1128/jcm.33.12.3216-3220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir, S. Personal communication.
- 29.Werner K, Mueller F M, Kasai M, Francesconi A, Chanock S J, Walsh T J. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Rapid DNA isolation from five different fungal organisms using FastPrep system, abstr. C-239; p. 162. [Google Scholar]