Abstract
Blood vessel chips are bioengineered microdevices, consisting of biomaterials, human cells, and microstructures, which recapitulate essential vascular structure and physiology and allow a well-controlled microenvironment and spatial-temporal readouts. Blood vessel chips afford promising opportunities to understand molecular and cellular mechanisms underlying a range of vascular diseases. The physiological relevance is key to these blood vessel chips that rely on bioinspired strategies and bioengineering approaches to translate vascular physiology into artificial units. Here, we discuss several critical aspects of vascular physiology, including morphology, material composition, mechanical properties, flow dynamics, and mass transport, which provide essential guidelines and a valuable source of bioinspiration for the rational design of blood vessel chips. We also review state-of-art blood vessel chips that exhibit important physiological features of the vessel and reveal crucial insights into the biological processes and disease pathogenesis, including rare diseases, with notable implications for drug screening and clinical trials. We envision that the advances in biomaterials, biofabrication, and stem cells improve the physiological relevance of blood vessel chips, which, along with the close collaborations between clinicians and bioengineers, enable their widespread utility.
Keywords: disease modeling, hemodynamics, angiogenesis, drug screening, permeability, rare disease, COVID-19
Graphical Abstract
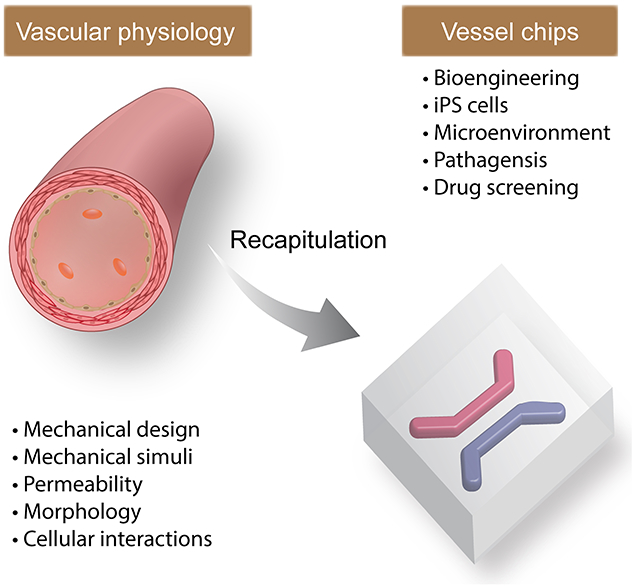
Blood vessel chips harness advanced bioengineering approaches, patients’ cells, and biomaterials to recapitulate essential features of vascular systems, including mechanical designs and stimuli, cellular compositions and interactions, and other microenvironmental cues. The physiological relevance is key to the capability of blood vessel chips, which requires an in-depth understanding of vascular physiology, continuous exploitation of biomaterials and microfabrication methods, and collaboration between clinicians and scientists.
1. Introduction
Tissue chips, or microphysiological systems (MPS),[1–3] are bioengineered microdevices that consist of microstructures, biomaterials (e.g., extracellular matrix (ECM) and other structural proteins, such as silk fibroin),[4–7] and often human cells.[8–10] Due to the bioengineering nature, tissue chips allow the manipulation of cellular composition [11] and a range of biophysical and biochemical environmental cues, including but not limited to cell-ECM interactions, dynamic flows, chemical species gradients, substrate stiffness,[12] and active mechanical stimuli.[13, 14] In particular, the on-chip adoption of patients’ cells, either primary or induced human pluripotent stem cells (hiPSC), makes a bespoke and patient-specific tissue model in line with the goal of precision medicine.[15] In contrast, small animal-based models exhibit notable genetic and cellular differences from human physiology, which may lead to biased results regarding disease pathogenesis and therapeutic effects.[16, 17]
Furthermore, tissue chips can precisely control and decouple multiple experimental factors, beneficial for exploiting the inherently complex human physiology, in contrast to other in vitro models, such as Petri dish-based ones, which are focused on maintaining cell proliferation in vitro. Thus, tissue chips, as a viable alternative to other tissue/disease-modeling approaches, are promising for in vitro recapitulation of the sophisticated human physiology and pathology at the tissue and organ level and are expected to transform the landscapes of fundamental biological research,[18–20] drug screening and toxicology,[21–24] and possibly, clinical trials.[25, 26]
Tissue chips can construct a broad range of tissue- and organ-analogs in vitro, including the vessel and vascular networks,[27–29] muscles,[30–33] liver,[34–36] lung,[37–39] brain,[40, 41] kidney,[42–46] gut,[47, 48] bone marrow,[49] corneal,[50, 51] tumor,[52–56] as well as the integrated multiple tissues/organs,[57–59] such as liver-kidney,[60–62] neuromuscular junction,[63] and a recirculating system with up to 13 organs.[64] Of note, these tissue chips may rely on the same set of technical approaches yet recapitulate the specific tissue (patho)physiology by considering tissue-specific cellular and matrix composition.
Among all tissues/organs modeled in the chips, the blood vessel is of particular interest, largely for the following reasons (Figure 1).[65, 66] i) The blood vessel exists in all tissues/organs throughout the body up to the epithelial layer covering surfaces. Thus, the on-chip reconstruction of the vessel or its analog is often a prerequisite for constructing a tissue analog. ii) The blood vessel is both the structural basis of circulating blood flow throughout the body and responsible for many physiological functions. The circulating blood flow enables the conventional and diffusive transport of nutrients, oxygen, metabolic wastes, and cells, which is important to maintain cells alive and immune responses.[67, 68] Also, the blood vessel is essential to gas-exchange, urine-enrichment, and digestion in the lung, kidney, and intestine, respectively. iii) The abnormal conditions of the blood vessel, such as intraluminal narrowing, obstruction, constriction, and stiffening, change the pulsatile blood flow patterns that both further change the vessel and contribute to the manifestations of severe disabilities and diseases, including heart attack and stroke (Figures 1 and 2). For example, one disease in the blood vessels, atherosclerosis, is the leading cause of mortality and morbidity in the U.S. and globally.[69] Thus, blood vessel chips that can construct in vitro physiologically relevant blood vessel analogs and provide tissue-level investigation would broadly benefit biomedicine.
Figure 1.
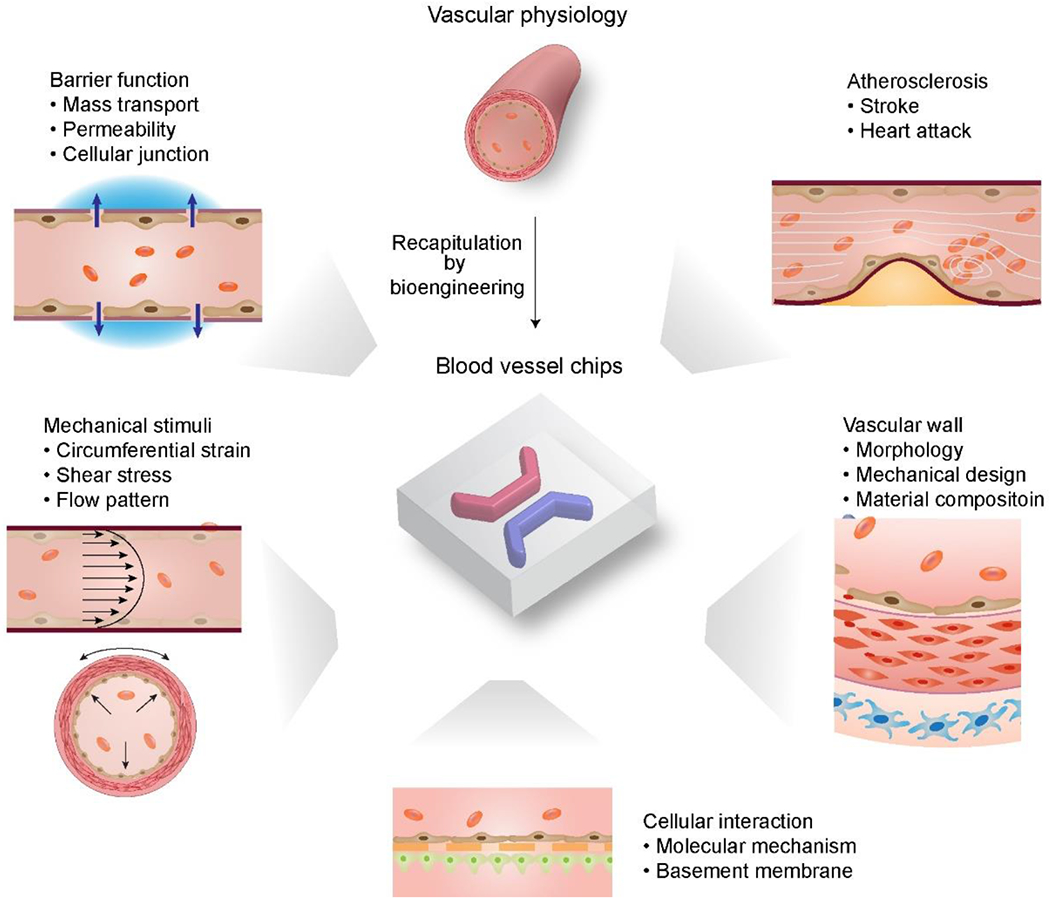
Overview of blood vessel chips recapitulating sophisticated vascular (patho)physiology, including morphology, mechanical properties and stimuli, mass transport, and compositions, laying a foundation for various biomedical research.
Figure 2.
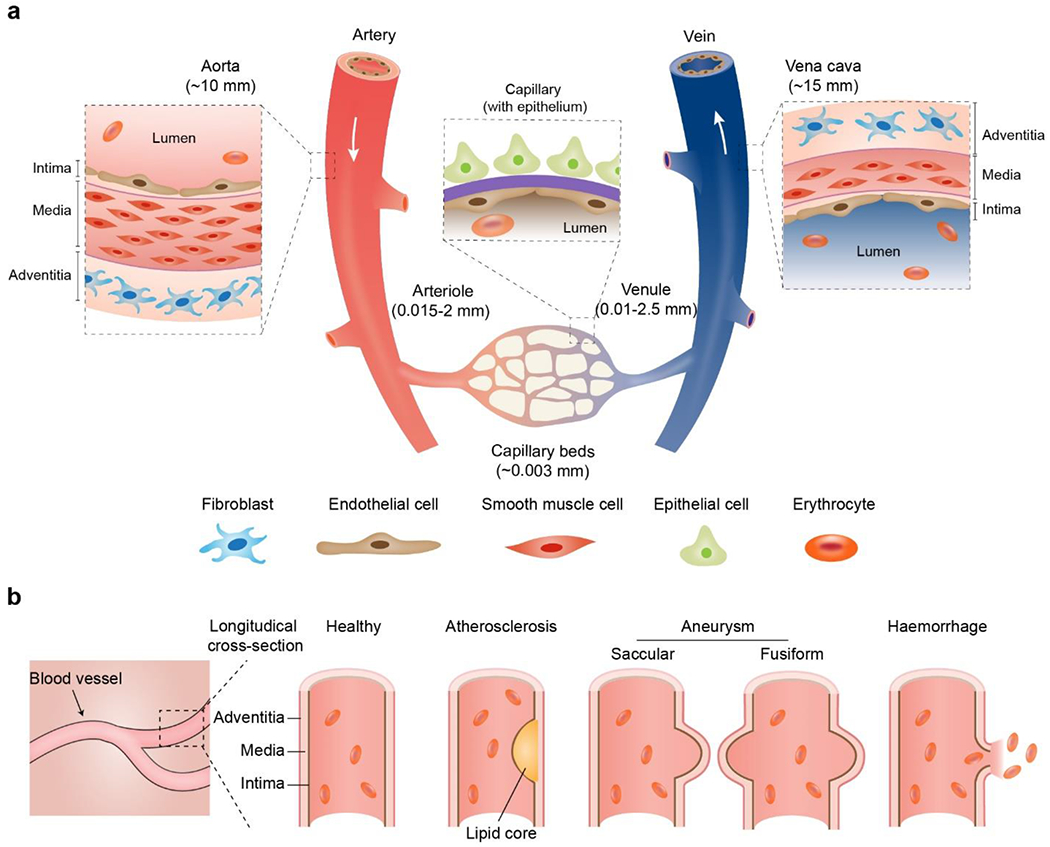
a) Schematic anatomy of blood vessels at hierarchical length scales. The vascular walls of the aorta and vena cava comprise three concentric layers termed the intima, media, and adventitia, respectively. The capillary wall usually contains a single layer of the intima. In some tissues, such as the lung alveoli and renal nephron, blood capillary is often accompanied by the epithelium. b) Schematic longitudinal cross-sections of healthy and pathological vascular walls, such as aneurysms (saccular and fusiform), atherosclerosis, and hemorrhage.
The most important feature of blood vessel chips is most likely the in vitro recapitulation of essential vascular (patho)physiology, as the physiological relevance of in vitro models underlies the accuracy of the outcome. The in vitro recapitulation is largely achieved by exploiting a variety of bioengineering approaches, including microfluidics[7, 66, 70–72] and, more recently, 3D printing and bioprinting.[73–79] These bioengineering approaches play a key role in organizing cells and biomaterials into physiology-relevant structures. For example, microfluidics is based on soft lithography/micromolding and has been widely used to construct vessel analogs, i.e., perfusable microchannels, on a 2D plane [36, 80] and 3D interconnected vasculature analogs.[81] In addition, 3D (bio)printing benefits from digital manufacturing and provides improved manufacturing flexibility.[78, 82–85] In particular, several strategies have been exploited to print ECM- and other structural protein-based inks,[86–89] as these protein materials are often not compatible with conventional thermal process-based 3D printing.[90] In addition, co-axial spinning or (bio)printing has proven useful for constructing vascular structures with unique manufacturing advantages, [91, 92][7, 93, 94] including diverse biomimetic sizes, layers, and shapes such as necklaces and knots[95][96] and meters-long hydrogel conduits with concave and convex topographies[97].
This review aims to elaborate on essential vascular physiology and to introduce state-of-art developments of blood vessel chips for biomedical research, with an emphasis on the correlation between native (patho)physiological features and the bioengineered ones. The vascular physiology, reviewed here, includes tissue morphology, mechanical properties and stimuli, flow dynamics, mass transport, and material composition, providing insight into the designing principles of blood vessel chips (Figure 1). We then review recent advances in blood vessel chips related to tissue-level function (i.e., intervascular mass transport), rare diseases, and atherosclerosis, corroborating the biomedical significance of blood vessel chips. In the end, we suggest future directions of blood vessel chips for better recapitulating vascular physiology, which is promising yet requires substantial effort and perhaps technical breakthroughs. This review article is expected to supplement other excellent ones in the field.[65, 66, 98–102]
2. Vascular physiology and on-chip recapitulation
Blood vessels, in a total length of over 60, 000 miles, constitute a continuous and hierarchical transport system for blood circulation, which primarily includes arteries, veins, and capillaries with distinct diameters, compositions, and mechanical properties (Figure 2 and Table 1). For example, the arteries are usually more elastic and stronger than others, which help sustain the surge of blood pressure and prevent wall-rupturing. According to size, the artery can be divided into conducting arteries (up to 12.5 mm) such as the aorta, distributing/media arteries (approximately 2 mm), and resistance arteries, such as arterioles (approximately 15 μm). Similarly, veins can be divided into several segments with different diameters, including the largest conducting veins (larger than 10 mm) such as vena cava, moderate veins (1-10 mm), and the smallest postcapillary veins (10-20 μm). Arteries and veins are interconnected at the tips via capillary beds that are small vessels (approximately 3 μm) organized into a web-like network (Figure 2a). At each level the blood vessels differ in the pulsatile waveform and pressure of blood flow. The pulsatile blood flow pattern also differs according to which tissue is being supplied. Blood vessels and endothelial cells can adopt tissue-specific morphology and composition to enable and maintain tissue-specific functions.[103] The in-depth understanding of the general and tissue-specific vascular physiology and the advances of blood vessel chips would mutually benefit each other.[104]
Table 1.
| Vessel | Average radius (mm) | Vessel number | Area (mm2) | Wall thickness (mm) | Wall shear stress (Pa) | Blood velocity (mm/s) |
|---|---|---|---|---|---|---|
| Aorta | 12.5 | 1 | 450 | 2 | 0.6-0.98 | 1200 |
| Distributing arteries | 2 | 159 | 2 × 103 | 1 | 1.1 | N/A |
| Arterioles | 1.5 × 10−2 | 5.7 × 107 | 4 × 104 | 0.2 | 5.1 | 15 |
| Capillaries | 3 × 10−3 | 1.6 × 1010 | 4.5 × 105 | 1 × 10−2 | 4.4 | 0.4 |
| Venules | 1 × 10−2 | 1.3 × 109 | 4 × 105 | 2 × 10−2 | 1.6 | 5 |
| Media veins | 2.5 | 200 | 4 × 103 | 0.5 | 0.71 | N/A |
| Vena cava | 15 | 1 | 180 | 1.5 | 0.42 | 80 |
This section will elaborate on five fundamental aspects of blood vessels, including hierarchical branching morphology, vascular wall morphology, mechanical design, permeability and mass transport, and mechanical forces on vessels. State-of-art blood vessel chips for recapitulating these essential vascular features are also discussed.
2.1. Hierarchical branching
The vascular system resembles a tree-like network, which starts from the largest conduits (Aorta and Vena cava) and continuously branches into smaller ones (capillaries) (Figure 2 and Table 1). The hierarchical branching structures of blood vessels play a critical role in maintaining optimal vascular functions and are precisely regulated by multiple biological and biophysical mechanisms. A large portion of the branched vascular network, as seen in the kidney, lung, mammary gland, as well as plants,[105] can be mathematically described by Murray’s law (Figure 3c),[106, 107] originated from the cardiovascular network, as shown below:
| (1) |
, where a0 is the radius of the parent vessel, and a1, a2, …, an indicate the radii of child vessels with n the hierarchical number. According to Murray’s law, the cube of the diameter of the parent vessel equals the sum of the cubes of the child vessels. Murray’s law indicates an optimal designing principle for branched transport systems that maximize hydraulic conductance with minimized material and energy costs. The mathematic relationship is useful for designing a hierarchically branched vascular network on chips.
Figure 3.
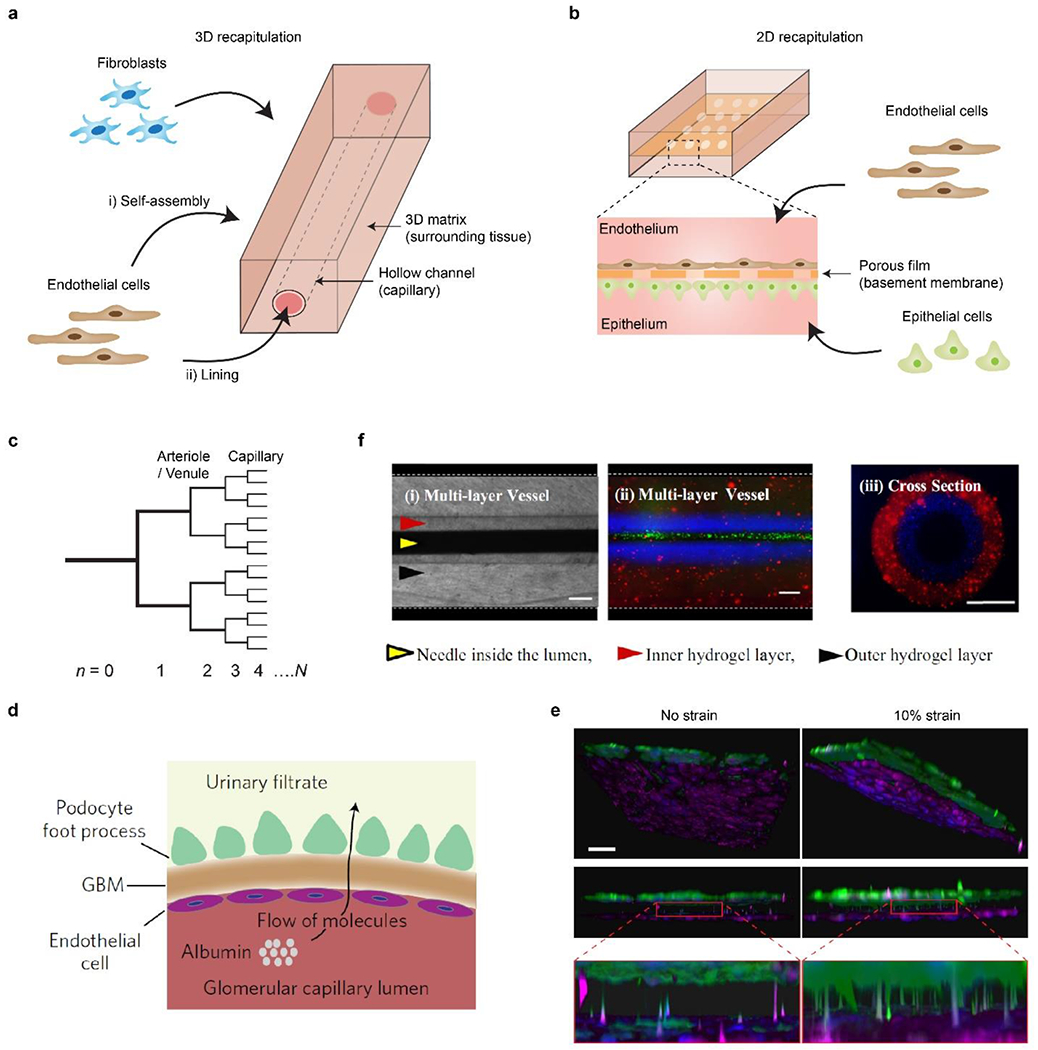
a) Schematic of 3D blood vessel chips. Microchannels are constructed within a 3D matrix. ECs and other cell types, e.g., fibroblasts, are seeded into the microchannel or the hydrogel matrix. b) Schematic of 2D blood vessel chips. Microchannels are separated by a porous membrane. ECs and other types of cells, often epithelial ones, are seeded onto the two sides of the membrane, respectively. c) Schematic of a bifurcated, hierarchical vascular network, where n is the hierarchical number. Reproduced with permission.[109] Copyright 2006, Royal Society of Chemistry. d) Schematic of glomerular cross-section reconstructed in tissue chips. EC-lined capillary is separated from the epithelial cell (podocyte)-lined urinary space by a glomerular basement membrane (GBM). Large molecules such as albumin are retained in capillaries, while small molecules like inulin are filtered into the urine. In the tissue chip, ECs and hiPSC-induced podocytes are cultured on the two sides of a porous elastic PDMS membrane, which mimic the urinary and capillary compartments and the GBM, respectively. The cells can be stretched by manipulating vacuum inside channels. e) 3D reconstruction of the on-chip interface between podocytes (green) and ECs (magenta). 10% cyclic strain enhanced the extension of podocytes through the porous membrane and insertion into the biomimicked glomerular endothelium. Scale bar, 100 μm. d and e) Reproduced with permission.[163] Copyright 2017, Springer Nature. f) Multi-layered vessel-like structure. (i) Top view, brightfield. (ii) Top view, fluorescence. Red and blue fluorescence indicate two layers of the vessel, respectively. Green fluorescence indicates flowing beads in the lumen. (iii) Cross-sectional view, fluorescence. Scale bars, 200 μm. Reproduced with permission.[165] Copyright 2015, Springer Nature.
Microfluidics is one of the widely-used approaches to constructing filamentary, hierarchical, branching networks resembling native vasculature.[108] However, microfluidic channels are often large in size and exhibit rectangular or trapezoidal cross-sections, in contrast to the small and circular cross-section of blood vessels. The geometry difference may lead to a biased flow profile and cellular behaviors. Yet it is possible to design non-circular microfluidic channels to follow Murray’s law by considering shear stress distribution.[109] Other methods have been developed to fabricate hierarchical branching microfluidic networks with largely circular cross-sections and proportionally correlated local height with local width in one step, such as backside lithography[110] and leaf scaffolds.[111, 112] Leaf venation shows an overall structural resemblance to mammalian vasculature, including hierarchical branching, small diameters (<50 μm), and an almost circular cross-section. The leaf venation-based tissue chips have been used for studying biological transport [113] and organ-specific metastasis.[114]
2.2. Morphology of vascular walls
Vascular walls are the interfaces between the blood and interstitial/functional tissues, which is composed of cells and ECMs in distinct concentric layers (Figure 2a).[115] The material and cellular composition and interactions between cells, either within the same or in different layers, are associated with vascular hemostasis and pathogenesis.[116, 117] Common clinical manifestations of abnormal vascular walls include atherosclerosis (narrowed vessels) and arterial aneurysms (dilated vessels) (Figure 2b).
2.2.1. Physiological Morphology
The walls of the arteries and veins usually exhibit a three-layer structure, including the tunica intima, tunica media, and tunica adventitia, from the inside out (Figure 2a). The tunica intima is the inner-most layer of the vessel wall, which contains a monolayer of endothelial cells (ECs) on the top of a basement membrane. The tunica intima directly contacts with blood and can play a critical barrier role, related to the transport of cells and soluble substances as well as platelet adhesion. The manipulation of the barrier function of the tunica intima is thus of intense interest for blood vessel chips.[118, 119]
The tunica media is in the middle of blood vessel walls and is the primary contributor to the overall mechanical strength and elasticity of vessels (Figure 2a). Notable features of the tunica media include the presence of smooth muscle cells (SMCs) and elastin/collagen-constituted elasticity. If there is a loss of SMCs and ECM in the tunica media, it thins the blood vessel wall softens which is often a sign of either aging or pathogenesis.[120] SMCs are oriented in a circumferential manner, thus allowing a vascular contraction in the circumferential direction. For example, arterial smooth muscle can constrict to decrease the vessel diameter by 20% to 50%.[121] The thickness and stiffness of the tunica media differ by blood vessel type; moreover, dilation or constriction of the blood vessel occurs in response to both specific tissue needs and to neuroendocrine mediators, some being produced locally by the vessel itself.[122–124] For example, the vena cava exhibits a thinner tunica media, a less amount of SMCs, and thus a weaker mechanical performance than the aorta. In addition to the intima and media, the third and outmost layer of vessels is the tunica adventitia that primarily comprises collagen-based connective tissues, nerves, and fibroblasts.
Unlike the three-layered arteries and veins, the capillary has one layer of the tunica intima (Figure 2). The single-layer configuration allows the capillary to interact to a larger extent with epithelial cells and other cells for mass exchange in tissues, such as in the nephrons and the pulmonary alveoli. Capillaries can be categorized into three subgroups: continuous, fenestrated, and sinusoids. Continuous capillaries have intercellular clefts, usually 4-nm-wide, allowing small molecules to pass through.[125] However, continuous capillaries in the blood-brain barrier lack clefts, exhibit tight cellular junctions, and are tightly wrapped by another cell type, the pericytes. Fenestrated capillaries have fenestrations or filtration pores ranging from 20 nm to 100 nm in diameter, allowing the transmission of small proteins. Sinusoids are discontinuous and exhibit the highest permeability among all capillaries, which allows the exchange of large proteins and cells.[125]
Of note, the interactions between ECs and pericytes play critical roles in the angiogenesis and the homogenesis of microvasculature,[126] which should be important technical considerations in modeling the vasculature, especially in the context of disease pathogenesis. The EC-pericyte interactions were studied in a microfluidic chip by characterizing the morphology and cellular composition of a self-assembled hollow vascular network.[127] Without transforming growth factor beta (TGF-β)-inhibition, ECs formed a long, tubular structure that followed the branched contours of a Y-shape microchannel. The ECs also exhibited tight junctions of platelet-endothelial cell adhesion marker-1 (PECAM-1); pericytes were located around the EC monolayer. In contrast, TGF-β-inhibition resulted in disorganized, tortuous vascular morphology. This result was attributed to the role of TGF-β in recruiting and differentiating pericytes.[128, 129] Another microfluidic channel filled with 3D fibrin gel was used to characterize different mural cells on vascular formation.[130] All three mural cells, including human induced pluripotent stem cells-derived vascular smooth muscle cells (hiPSC-VSMCs), human brain vascular smooth muscle cells (HBVSMCs), and primary human brain vascular pericytes (HBVPs), gave rise to a similar vascular diameter of approximately 75 μm. This result indicated that stem cell-differentiated cells might be an alternative to primary cells, thus enabling the investigation of genetic vascular diseases. Tissue-specific pericytes have been found to exhibit tissue-specific functions to maintain vascular morphology and vessel constriction.[131]
2.2.2. On-chip recapitulation
There are largely two approaches for on-chip recapitulation of vascular walls, which can be roughly categorized as “2D” and “3D” ones according to the format of the involved biomaterials. In general, the 3D approach uses bulk hydrogels, such as fibrin and collagen,[132, 133] to construct perfusable vessel-like conduits, while the 2D approach employs a porous membrane to recapitulate the tissue interface of a vascular wall (Figures 3a, 3b).
The 3D approach enables either the encapsulation of ECs in the 3D bulk hydrogel for self-assembly into capillary networks, the lining of ECs on the inner wall of hydrogel microchannels, or both (Figure 3a).[100, 134, 135] For example, a perfusable, interconnected and branching capillary-like network has been formed in bovine fibrin gels filled in a microfluidic chamber.[136] A supraphysiological flow velocity (approximately 0.5 mm/min) across the microfluidic chamber, as compared to lower ones, promoted the anastomosis of the capillary-like network and two adjacent microfluidic channels. Also, the pressure was applied to each side of the microfluidic chamber alternatively, leading to the alternating direction of interstitial flow, which might be beneficial to the anastomosis, but requires more investigations.
The 3D approach also enables the encapsulation and recruitment of auxiliary and functional cells in the 3D bulk hydrogels, analogous to the native tissues (Figure 3a).[133] Various auxiliary cells can be used to explore tissue-specific functions, such as renal reabsorption,[137] blood-brain barrier,[138, 139] and placenta.[140] In particular, one vessel chip constructed a cylindrical lumen of a brain capillary vessel with the co-culture of human brain ECs and pericytes.[138] The pericytes improved the barrier function of ECs and led to an immune response that is physiologically relevant. Besides, the 3D approach allows studying assembly-involved angiogenesis,[133, 141–145] vessel-matrix interactions in terms of matrix stiffness [146] and niche effects, [147] anti-angiogenetic therapeutic drugs,[148] morphogenesis,[149] cellular interactions,[150] mass transport,[151] and tissue regeneration.[152]
Capillary vessels are small in diameter yet make up the major volume of the circulation system and play central roles in mass transport and cell infiltration. It is important yet usually challenging to engineer capillary and its analogs in the diameter of 7-15 μm. One approach to fabricating capillary vessels is the self-assembly of ECs in a 3D matrix.[153] The co-culture of supporting cells and ECs may help the maintenance and generation of capillaries. For example, the tri-culture of iPSC-ECs, pericytes, and astrocytes led to a diameter of approximately 45 μm for the vessels, which is much smaller than the ones formed with iPSC-ECs alone, at 105 μm.[154] The spatial patterns of the cells have also been suggested to influence the capillary diameter. The mixed and non-patterned cellular composition led to the formation of capillaries with a diameter smaller than 20 μm.[155] Another approach combined soft lithography-based microfabrication and the self-assembly of ECs in a pro-vascular poly(ethylene glycol) (PEG) hydrogel, which gave rise to microvascular networks in diameters ranging from 15 to 50 μm.[156] Furthermore, the combination of engineered macrovessels and self-assembled capillaries was proven useful in promoting the vascular integration of a 3D implant.[157] Finally, 3D printing has been widely used to fabricate blood vessels but it is usually challenging to fabricate microchannels with a similar diameter to capillaries.[158, 159] One exception is multi-photon-based 3D printing that could exhibit a circular microchannel with a diameter of 18 μm and a wall thickness of 3 μm.[160] Additional newer demonstrations such as post-printing shrinkage was shown to be able to produce 10-μm-sized capillary networks.[161]
Besides the 3D approach, a segmental cross-section of vascular walls can be recapitulated by seeding ECs and other cells, such as epithelial cells, at the opposite sides of a porous membrane, i.e., the 2D approach (Figure 3b). The porous membrane is pivotal to the 2D approach, which allows the transport of soluble substances and cells, underlying various cellular interactions and tissue functions. It also enables mechanical support and stimuli, e.g., cyclic stretching, to vascular cells. Compared with the 3D approach, the membrane-based 2D approach is advantageous to the construction and real-time observation of the endothelium-epithelium interface, which is central to the primary physiological functions in, for example, the lung,[37] gut,[162] and kidney.[163]
One eminent example of the 2D approach is the construction of the kidney glomerular capillary on chips, characterized by the use of hiPSC-induced podocytes (Figures 3d, e).[163] The podocyte is one of the main cellular components in the glomerular capillary for selective filtration, whose malfunctions are associated with kidney diseases and degeneration.[164] Tissue-specific ECM (laminin-511 E8 fragment) and a rationally designed set of soluble factors, including vascular endothelial growth factor (VEGF), retinoic acid (RA), CHIR99021, bone morphogenetic protein-7 (BMP-7), and activin A, are used to induce the hiPSC line (PGP1) to produce highly functional and differentiated human podocytes with high efficiency (over 90%).[163] The hiPSC-induced podocytes exhibit similar molecular and morphological features of mature human podocytes, including narrow membrane protrusions and the expressions of nephrin, Wilms’ tumor 1 (WT1), and podocin, and thus surpass the podocyte cell line. The on-chip co-culture of hiPSC-induced podocytes and glomerular capillary ECs recapitulated important cellular interactions and investigated glomerular filtration (Figure 3e). For example, physiologically relevant mechanical stimuli, such as flow stress (0.0007 dyn/cm2 for podocytes and 0.017 dyn/cm2 for ECs) and 10% cyclic strain, promoted on-chip albumin retention and inulin clearance. This vessel chip also recapitulated the characteristics of proteinuria, which was related to the loss of filtering functions due to the toxicity of a cancer drug, Adriamycin.[163]
Most blood vessel chips recapitulated just one or two layers of the vessel, which doesn’t match the three-layer configuration of the aorta, arteries, veins and vena cava. One 3D vessel chip recapitulated the whole three-layer arrangement of vessels (Figure 3f).[165] The tunica media and adventitia were constructed in gelatin methacryloyl (GelMA) hydrogel layers containing SMCs and fibroblasts, respectively. The two hydrogel layers were molded by two concentric and removable needles in different diameters. A layer of human umbilical vein ECs (HUVECs), lining the lumen of GelMA hydrogel microchannels, was analogous to the tunica intima and exhibited the barrier function for 10-kDa dextran labeled with Alex Fluor 488. The confluence of HUVECs was associated with GelMA concentration. For example, less than 12 wt% GelMA led to sparsely distributed HUVECs without confluence.[165] Another vessel chip exploited the three-layer configuration of vascular walls for studying the metabolic rate and glucose consumption of the cocultured ECs and SMCs.[166] Of note, the recapitulation of three-layered vessels is often challenging due to the complex and multiple-layer morphology and distinct mechanical strength and composition in each layer. However, the three-layer configuration is essential for the homeostasis and regeneration of vessels,[167] thus representing a rewarding direction for developing physiology-relevant blood vessel chips.
2.3. Mechanical design of vascular walls
The mechanical design and properties of the blood vessels underlie their physiological functions, such as supplying blood and modulating blood pressure. Arteries are largely distensible elastic conduits and exhibit non-linear elasticity as J-shaped stress-strain curves (Figure 4a).[168] The non-linear mechanical behavior refers to the elastic stiffness or modulus increasing with increased strain, due to the composite nature of blood vessel walls (Figure 4a).[168] The blood vessel wall consists of mechanically distinct proteins, which include rubbery elastin and stiff collagen. The modulus of elastin is often lower than collagen by more than two orders of magnitude. For example, the elastin and collagen moduli of the porcine abdominal aorta are approximately 2.66 MPa and 682 MPa, respectively (Table 2).[121] At low strains, elastin bears more force than collagen. At high strains, collagen fibers begin to sustain forces, thus leading to increased stiffness/modulus. In other words, soft elastin and stiff collagen contribute to the two segments of the J-shaped stress-strain curve at low and high strains, respectively (Figure 4a and Table 2).
Figure 4.
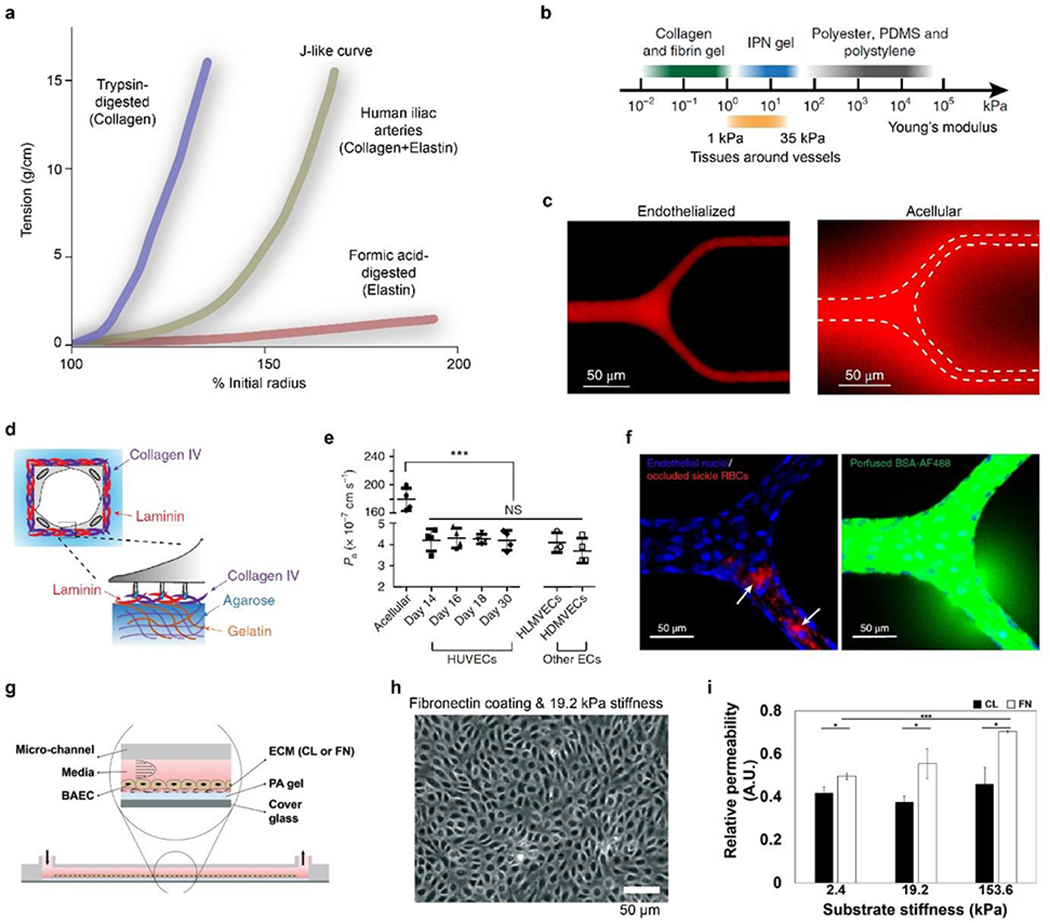
a) Composite components of elastin and collagen contribute to the J-like tension-length (stress-strain) behavior of human iliac arteries. Reproduced with permission.[168] Copyright 1999, Journal of Experimental Biology. b) Stiffness of IPN hydrogel covers a range between common soft hydrogels and stiff polymeric materials, agreeing well with physiological tissues surrounding vessels. c) ECs cultured in a branched IPH hydrogel channel form a tight monolayer and are impermeable to red-fluorescent Alexa Fluor 549-labeled bovine serum albumin after 15-min perfusion, compared to an acellular hydrogel channel with notable diffusion. White dash line indicates the boundary of microchannels. d) Schematic of the deposition of basement membrane components, such as laminin and collagen IV. e) Physiology-relevant permeability using HUVECs and other ECs can last up to 30 days in IPN microchannels. f) 3D confocal images of the endothelialized IPN microchannels perfused with sickle RBCs and Alexa 488-labeled bovine serum albumin. Sickle RBC occlusions, indicated by white arrows, lead to the local increase in permeability. b-f) Reproduced with permission.[146] Copyright 2018, Springer Nature. g) Schematic of a vessel mimicking microchannel constituted with PA hydrogels and ECM coatings. h) Phase-contrast image of an ECs monolayer on fibronectin (FN)-coated PA hydrogels with 19.2-kPa stiffness after one hour of shear stress. i) ECM coating influences cellular responses to substrate stiffness. Permeability of FN coating is higher than collagen (CL) coating and depends on stiffness. g-i) Reproduced with permission.[181] Copyright 2019, Royal Society of Chemistry.
Table 2.
Mechanics and composition of porcine vessels.[121]
| Vessel (direction) | Modulus (MPa) | Wet content (wt%) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Low strain | High strain | Collagen | Elastin | Smooth muscle | ||
| Aorta | (long) | 0.250 | 3.16 | 13.7 | 9.0 | 11.35 |
| (trans) | 0.0567 | 3.30 | ||||
| Carotid | (long) | 0.0345 | 0.876 | 12.2 | 9.3 | 8.94 |
| (trans) | 0.041 | 2.81 | ||||
| Vena cava | (long) | 0.319 | 2.31 | 18.3 | 2.67 | 8.97 |
| (trans) | 0.191 | 6.00 | ||||
The J-shaped stress-strain curve of the vessels is of physiological significance. Vascular dilation at the low strains smooths the pulsation of blood flow, and stiffness at the high strains helps maintain the integrity of blood vessels and prevents rupture. Of note, the mechanical behavior varies in transverse/radial and longitudinal directions and in the distinct regions of vessels, such as the aorta and vena cava, according to local mechanical requirements (Table 2). In particular, the transverse failure strain is approximately 0.6 for the vena cava and 0.9 for the aorta.[121] This mechanical difference is most likely due to the difference in wall morphology and material and cellular composition.
The abnormal mechanical performance of blood vessels is a primary clinical manifestation of aging and vascular pathogenesis. For example, one impaired mechanical performance of vessels is the increased stiffness, possibly due to the breakdown of elastin fibers, accumulation of lipids, and formation of plaques (i.e., atherosclerosis) (Figure 2b).[169] In particular, the loss of elastin led to increased deposition of collagen fibers, thus leading to decreased vascular compliance and increased stiffness. Another mechanically abnormal vessel is characterized by mechanical weakening and geometric thinning due to the ECM degradation in the adventitia and the SMC loss in the media (Figure 2b).[170] The compromised mechanical strength led to the dilatation of a part of the circumference of the vessel, i.e., saccular aneurysms, or, more commonly, the whole circumference, i.e., fusiform aneurysms (Figure 2b). Both atherosclerosis and aneurysms may result in vessel rupture and hemorrhage (Figure 2b).
The stiffening of vascular walls is also associated with endothelial dysfunctions, for example, the abnormal cytoskeleton and morphology of ECs,[171] loose cell-cell junctions, and a leaky endothelium.[172] The molecular mechanism is the activation of Yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) pathway that causes dysregulated vascular metabolism, such as glutaminolysis and anaplerosis.[173] At the tissue level, vascular stiffening affects pulse wave velocity and increases the afterload seen by the left ventricle and decreases coronary perfusion, contributing to hypertension, angina, and other cardiovascular diseases.[174, 175]
Mechanical behavior is an important consideration in the design and development of vascular grafts [176–178] and blood vessel chips.[146] Several tissue chips have recapitulated the physiological mechanical properties of blood vessels. One vessel chip employed the agarose-gelatin interpenetrating polymer network (IPN) to provide a physiological substrate stiffness, ranging from 1 kPa to 35 kPa, which helps achieve month-long vascular barrier functions (Figures 4b–f).[146], outside the physiological range. The stiffness of the IPN hydrogel, containing 1% agarose and 1% gelatin, was around 20 kPa, within the physiological range, in contrast to other widely used materials, such as polydimethylsiloxane (PDMS) and polystyrene, ranging from 50 kPa to several MPa (Figure 4b). The IPN hydrogel vessels exhibited approximately a permeability of 4 × 10−7 cm s−1 (Figure 4c), similar to mammalian venules (1.5 × 10−7 cm s−1) in vivo[81] and lower than most previously reported results of in vitro models.[133, 179] Other stiffness, higher or lower than the physiological ones, may lead to increased permeability and compromised vascular barrier functions. For example, the IPN hydrogel with approximately 50 kPa stiffness resulted in a two-fold increase in permeability; the 5-kPa IPN hydrogel could not even support the formation of EC monolayers, resulting in minimal barrier function. Furthermore, substrate stiffness affects the deposition of the basement membrane. In 20-kPa IPN hydrogels, the on-chip cultured ECs deposited a continuous sheet of both collagen IV and laminin (Figure 4d), as well as enzymes to remodel the basement membrane, including matrix metalloproteinase-2 (MMP-2). In contrast, the continuous deposition of collagen IV and laminin was not seen for ECs cultured on PDMS substrates, probably due to the excessively high stiffness.[146]
Furthermore, the vessel chip implied that the physiologically relevant microenvironment, including substrate stiffness and flow conditions, helps elongate the physiology-relevant permeability of vessel chips, for example, over 4 weeks (Figure 4e).[146] This long-term maintenance of the vascular barrier function is rarely seen in previous results. It enables well-controlled spatial-temporal imaging and investigation of the chronic pathological processes of vessel-related diseases. For example, the vessel chip found the spatial correlation between the occlusion of sickle red blood cells (RBCs) and the increased permeability, offering insights into disease-pathogenesis and treatment-development (Figure 4f).
Another vessel chip exploited the impact of matrix stiffness, along with shear stress, on YAP mechanotransduction.[180] The percentage ratio of the PDMS crosslinkers to monomers was used to tune the substrate stiffness of the tissue chip. In particular, 5% and 10% crosslinker led to stiffnesses representing normal muscular coronary arteries and atherosclerotic vessels, respectively. The high stiffness increased the nuclear partitioning of YAP, compromising the protection of ECs from high shear stress. These results provided valuable insights into the correlation between mechanotransduction and vascular diseases, highlighting the benefits of incorporating multiple physiological conditions to exploit complicated atherosclerosis mechanisms.
Blood vessel chips also found that the ECM coating (collagen and fibronectin), as biochemical cues, interferes with the impact of substrate stiffness on vascular permeability (Figure 4g, 4h).[181] The stiffness of polyacrylamide (PA) hydrogels is controlled by the ratio between the monomer (acrylamide) and the crosslinker (bis-acrylamide). Three levels of PA hydrogel stiffness, including 2.4 kPa, 19.2 kPa, and 153.6 kPa, were examined in blood vessel chips; the first two are within the physiological range, while the latter is pathological. For collagen-coated PA gels, permeability is largely unvaried with the change of stiffness. In contrast, fibronectin coating resulted in a proportional relationship between permeability and stiffness (Figure 4i). The relative permeability was obtained by measuring the fluorescent intensity in the PA gel after incubating ECs with fluorescein isothiocyanate (FITC)-dextran for 10 min. These results may imply that the regulation of vascular permeability is subject to the combination of multiple factors, including chemical and mechanical cues.
2.4. Permeability
Permeability is an important measure of the barrier functions of blood vessels. As discussed in the previous sections, permeability is widely measured for on-chip vessels, quantifying the cellular responses to various stimuli and revealing insights into tissue regulation.[146, 182–185] Endothelial junctions are the primary mechanism for regulating vessel permeability and mass transport across vascular walls (Figure 5a). Vascular permeability can be regulated by a range of cytokines, including VEGF, tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), histamine, and serotonin,[186] and is associated with chronic inflammation and pathological conditions such as carcinogenesis, wounds, and rheumatoid arthritis. In particular, VEGF is widely used to alter vascular permeability; it decouples endothelial cell-cell junctions and induces the formation of fenestrations (small pores) and caveolae (small plasmalemmal invaginations), thus increasing permeability.[187] Also, hyperpermeability is often seen in tumor vasculature that exhibits defective endothelium and reduced basal lamina.[188] This section focuses on the on-chip measurements of permeability and permeability-related mass transport.
Figure 5. Permeability of vessels and blood vessel chips.
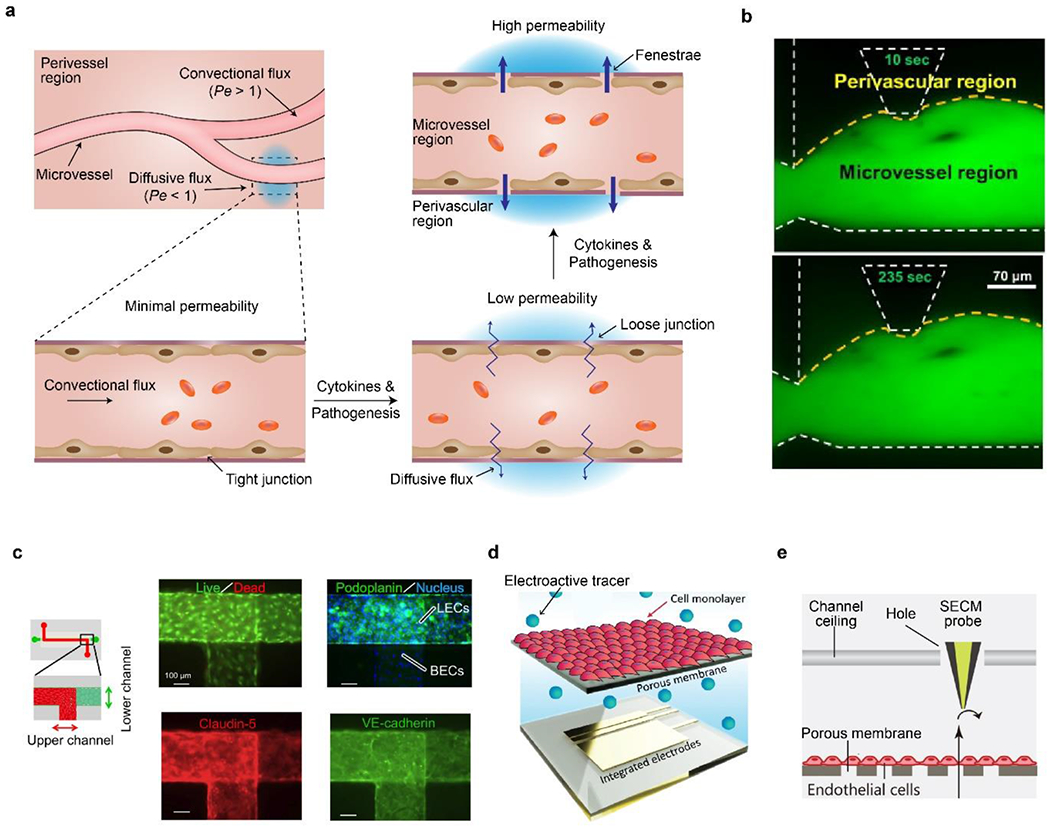
a) Schematic of vascular permeability related to endothelial junctions. Assumed flux and Pe number are also shown. b) On-chip measurement of vascular permeability by fluorescence diffusion from microvessel region to perivascular region at 10 and 253 seconds. Yellow dash line indicates boundaries of vessels. White dash line indicates on-chip microstructures. Reproduced with permission.[190] Copyright 2014, Elsevier. c) On-chip co-culture of blood vascular ECs (BECs, upper channel) and lymphatic ECs (LECs, lower channel) for investigating venom-indued hemorrhage and increased permeability. Both cells are confluent and express tight-junction proteins. Reproduced with permission.[182] Copyright 2015, Creative Commons Attribution License. d) Schematic of on-chip planar electrodes to detect electroactive tracers passing through an EC monolayer and a membrane. Reproduced with permission.[194] Copyright 2019, Royal Society of Chemistry. e) Schematic of an SECM probe for detecting permeability of EC monolayer. Reproduced with permission.[194] Copyright 2021, John Wiley and Sons.
2.4.1. Image-based measurements
The permeability of on-chip vessels is widely measured by fluorescence intensity,[189] which is often assumed to be linear to the concentration and barely influenced by the photo-bleaching effect. For example, the increase of fluorescence in the perivascular region over a certain amount of time was used to calculate the permeability of 3D microvessels (Figure 5b).[190] For vessels constructed in 3D hydrogels (Figure 3a), the permeability (P) can be determined using the following equation:[53, 146]
| (2) |
, where Is is the initial intensity, ΔI is the change of intensity during a period of time Δt, and a is the width/diameter of the channel. The intensity is measured from the fluorescence image covering the microchannel. A typical imaging field is 120-μm-wide and 160-μm-long for a microchannel of 20-μm-wide.[146] Alternatively, P for a 3D capillary can be calculated by either fitting the diffusion profile into a dynamic mass conservation equation[191–193] or another equation requiring 3D confocal images.[185]
For membrane-based 2D vascular walls (Figure 3b), the P of bioengineered vessels can be measured by analyzing centration in the two subchannels, analogous to vessels and epithelial tubes, respectively, using the following equation:[182, 194–197]
| (3) |
, where Cs is the initial concentration of the source channel, ΔCd is the concentration change of the drain channel during a certain time of Δt, Vd is the volume of the drain channel, and A is the area of the permeable membrane that separates the two channels. Concentration can be derived from fluorescence images and other measurable signals. P results from the combination of the EC monolayers (Pc) and the acellular structure (Pm), including the membrane and, if any, the coated hydrogel. Pc can be calculated using the following equation:[182, 198]
| (4) |
One vessel chip used fluorescence measurements and equations (3) and (4) to investigate snake venom-induced hemorrhage, i.e., increased permeability and reduced barrier function (Figure 5c).[182] The blood vessel chip could evaluate hemorrhagic damage within 30 minutes,[182] promising to evaluate anti-venom and anti-hemorrhagic drugs efficiently, in contrast to conventional examination that takes 24 hours.[199]
Blood vessel chips enable high-throughput screening of vascular permeability, for example, via multiple parallel on-chip structures.[198, 200] For example, an array of 96-blood vessels was constructed in one tissue chip that is in the format of a standard 384-well plate, compatible with the standard robotic liquid operation.[198] The chip with 96 vessels enabled the screening of multiple compounds with a range of concentrations, such as IL-8, interferon-γ (INF-γ), RA, TNF-α, VEGF, and IL-1β. Among these compounds, both TNF-α and IL-1β resulted in significantly increased permeability, which agrees well with in vitro [201] and in vivo results.[202] One interesting result was the biphasic response to VEGF.[198] VEGF at low (10 ng/mL) and high (100 ng/mL) concentrations decreased and increased permeability, respectively. The result at the low VEGF concentration was attributed to the generation of cyclic adenosine monophosphate (cAMP) that protects the barrier function of vessels.[203]
2.4.2. Electrical and electrochemical measurements
Besides fluorescence, electrical and electrochemical signals can be used to measure the permeability of on-chip vessels through transendothelial electrical resistance (TEER) measurement,[204–206] planar electrode measurement,[194, 195] and scanning electrochemical microscopy (SECM).[196] Electrodes are often easily incorporated into microfluidic chips and small enough compared with the dimension of microchannels, thus promising for real-time, long-term, and spatial-resolved detection. Planar electrodes can be fabricated using similar techniques for microfluidic chips. Unlike TEER, electrochemical detection relies on electroactive tracers, analogous to fluorescence tracers, including methylene blue, ruthenium chloride (RuHex),[194] and potassium ferricyanide.[196] Among these electroactive tracers, RuHex is reportedly well-qualified for measuring vascular permeability because of efficient electron transfer, electrochemical response over a wide concentration range, and cytocompatibility for ECs.[194]
One blood vessel chip with integrated planar electrodes investigated the impact of shear stress on vascular permeability (Figure 5d).[194] This blood vessel chip employed ethylene glycol-bis(β-aminoethyl)-N, N, N’, N’-tetraacetic acid (EGTA) and square wave voltammetry (SWV); SWV is advantageous in detection speed and sensitivity.[207] The on-chip electrochemical results showed that aortic flow with a high shear stress at 10 dyn/cm2 led to higher permeability 5.60 ± 0.55 × 10−5 cm/s, in comparison to a lower shear flow 0.01 dyn/cm2 led to lower permeability 2.80 ± 0.15 × 10−5 cm/s. This result is consistent with the fluorescence staining of VE-cadherin.
SECM is another useful tool to assess permeability in blood vessel chips. The probe of the SECM is small (tens of micron in diameter) and can be inserted into the microfluidic channel through a hole (Figure 5e).[196] The in situ probe eliminates the difficulty of imaging tissue chips which usually contain multiple layers and perhaps requiring an objective lens with a long working distance. Also, the inserted probe is promising to provide in situ and spatially resolved information of chemical flux and, thus, permeability at the length scale of single cells. A blood vessel chip equipped with the SECM was shown to be suitable for investigating vascular permeability of a mimicked intestinal vasculature by co-culturing intestinal epithelial cells, Caco-2, and ECs.[196] Such a co-culture of endothelium and epithelium underlies the recapitulation of complex vascular physiology, for example, tissue regeneration and inflammatory responses.[10, 208]
2.4.3. Mass transport
Mass transport is the primary physiological role of blood vessels, enabling the efficient transfer of oxygen and nutrients and the removal of wastes to meet the metabolic needs of every cell in the body, as well as the needs of other essential tissue-specific functions. The mass transport via the vascular network is mainly achieved by combining two distinct physical processes: convectional pulsatile flow and permeability-related diffusion (Figure 5a).[101] Convectional flow is driven by hydraulic pressure generated by the heart beating, dominating long-distance transport. Convective transport can be described by the Hagen-Poiseuille equation, as shown below:
| (5) |
, where Q is volumetric flow rate, Δp is the pressure difference, and R is the hydraulic resistance.
Diffusive transport is a spontaneous thermodynamic process, driven by the concentration gradient, and is effective over a short distance. Diffusive transport is described by Fick’s first law, as shown below:
| (6) |
, where J is the flux of transported species, De is the effective diffusion coefficient, and ∇c is the spatial gradients of concentration. De is associated with the size of solutes and the permeability of the endothelium.
Convective and diffusive transport occur simultaneously within and outside the vascular network. Given that C is a typical concentration, a typical diffusive flux can be described by DeC/L, and a typical convective flux is UC. Péclet number (Pe) is the ratio between convective and diffusive fluxes, allowing quantitative measurement of the competence between conductive and diffusive fluxes, as described below:
| (7) |
, where L is the typical length scale, and U is the flow velocity. A large Pe (>1) indicates the dominance of convective transport; a small Pe (<1) indicates the dominance of diffusive transport (Figure 5a). Normally, diffusion-based mass transfer is dominated in capillaries, compared with convection-dominated transport in large vessels (arteries, arterioles, veins, venules). Also, as indicated by Pe number, the flow velocity may be manipulated in the tissue chips to adjust mass transport.
2.5. Mechanical forces
Blood vessels, due to the pulsatile blood flow, consistently bear cyclic mechanical forces, including shear forces on the endothelium and normal and circumferential forces on the entire vessel wall (Figure 6a). These mechanical forces, just like soluble biochemical cues, play regulatory roles in vascular morphogenesis, remodeling, and pathogenesis, thus necessary to be considered in devising blood vessel chips.[209, 210] This section elaborates on the biomechanical roles of the shear and circumferential forces on vessels and the incorporation of these mechanical forces in blood vessel chips.
Figure 6.
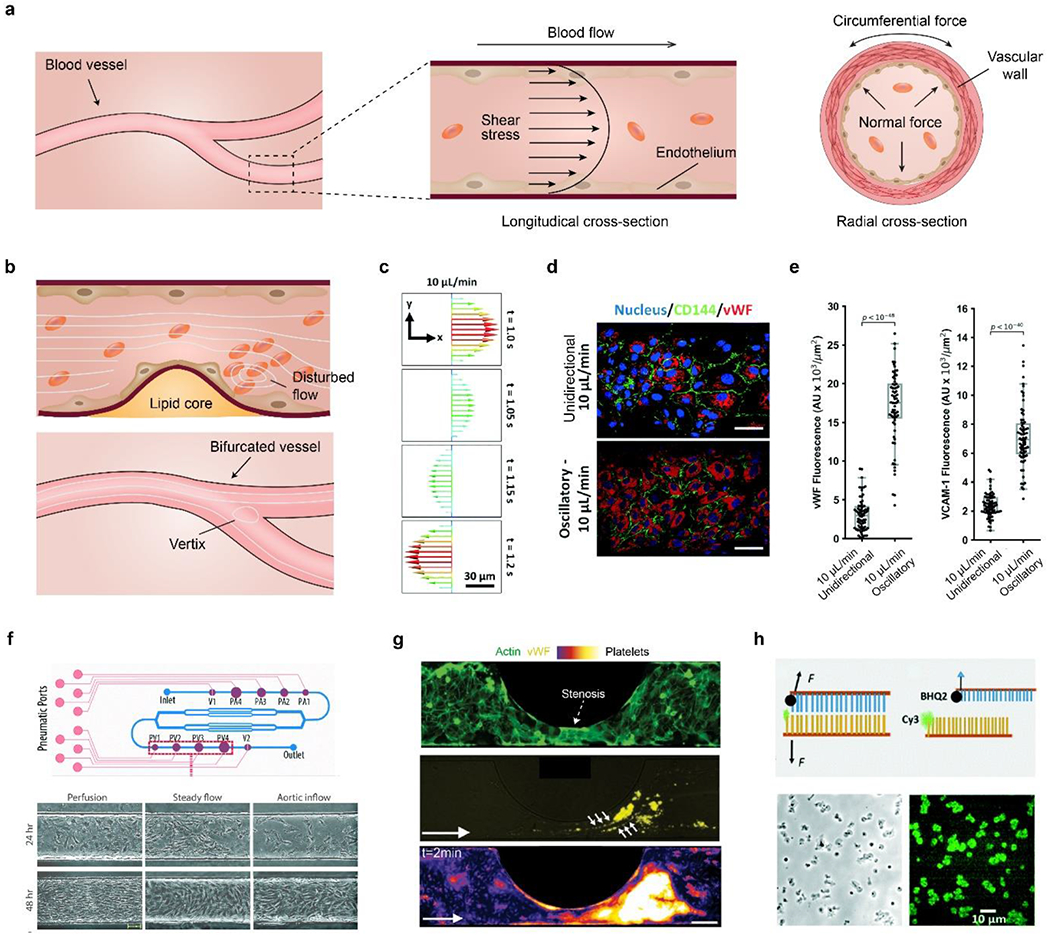
a) Schematic diagram of mechanical forces on vessels, including shear, normal and circumferential ones. b) Disturbed blood flow and vortices due to bifurcation and accumulated lipid-rich plaque. Platelets may aggregate at a post-stenosis site. c) Simulated velocity profile of an oscillatory flow at 10 μL/min and 0.3 Hz. d) Fluorescence images of ECs stained with the nucleus, cluster of differentiation (CD)-144, and vWF under unidirectional and oscillatory flows. Scale bar, 50 μm. e) Quantitative comparison of vWF and VCAM-1 under the two different flow conditions. c-e) Reproduced with permission.[180] Copyright 2021, Royal Society of Chemistry. f) Schematic of a vessel chip for generating a range of flow profiles. The frequency and magnitude of shear stress can be manipulated by on-chip pumping and valve size (P1-4, PV1-4). Primary valvular ECs exhibit different morphology and alignment under flow conditions. Scale bar, 50 μm. Reproduced with permission.[235] Copyright 2018, Royal Society of Chemistry. g) Platelet aggregation in a stenotic microfluidic channel lined with ECs. Dash arrow indicates the stenosis. Top, endothelial monolayer stained with F-actin (green); middle, vWF (yellow) expressed at post-stenotic sites, indicated by six small arrows; bottom, platelets, stained with DiOC6, are aggregated at the post-stenotic site after the whole blood perfusion. The platelet density increases as the color changes from black, purple, red, yellow to white. White arrow indicates the flow direction. Scale bar, 100 μm. Reproduced with permission.[180] h) Top, schematic of DNA-based integrin tension sensor. Black hole quencher 2 (BHQ2) quenches the fluorescence of the adjacent Cy3 probe. Upon removing the tope DNA chain by flow stress, the fluorescence of Cy3 becomes visible. Bottom, representative optical and force-mapping images (in green fluorescence) of platelets in the microfluidic channel. Reproduced with permission.[243] Copyright 2021, Royal Society of Chemistry.
2.5.1. Shear stress
Intravascular wall shear stress (τw) originates from the viscous drag force of the blood flow per unit area, and is associated with the development, regeneration, and morphology of ECs, and vascular homeostasis and functions.[211–213]. ECs sense shear stress via a complex biophysical mechanism that involves glycocalyx, a thin layer of proteoglycans, glycoproteins, and glycosaminoglycans at the apical surface of cells.[214, 215] The glycocalyx transduces the external shear stress to the endothelial cytoskeleton.[214] The physiological range of τw is roughly from 0.1 to 7 Pa, while the τw in a stenotic vessel can be as high as 150 Pa (Table 1).[216] Within the physiological range of shear stress, the high end is beneficial to preventing atherosclerosis, while the low end, especially with the disturbed flow, promotes atherogenesis.[217] The pathological shear stress, above or below the physiological range, can lead to vascular dysfunction, including thrombus formation via platelet aggregation [218] and atherosclerosis.[211]
In a straight cylindrical vessel under laminar flow, τw can be defined as below:
| (8) |
, where μ is the viscosity of the blood, Q is the blood flow rate, and d is the diameter of the blood vessel. Also, for bioengineered vessels with a rectangular cross-section, τw can be calculated using the following equation:
| (9) |
, where w and h are the width and height of the microchannel, respectively. According to equations (8) and (9), τw is proportional to flow rate and inversely proportional to the vessel diameter/size. Thus, the magnitude of τw can be tuned by manipulating the geometry of microchannels and the flow rate.
Of note, equations (8) and (9) for calculating τw are based on several assumptions of vessels and blood flow, such as the straight geometry and the laminar flow condition. However, vessels may have complex shapes, such as bifurcation and stenosis; at these locations, blood may exhibit turbulence and vortices (Figure 6b), leading to challenges in estimating shear stress by equations (8) and (9). The spatial distribution and magnitude of the shear stress in blood vessels with complex shapes have been estimated by finite element analysis-based simulation.[219–221]
Blood vessel chips have been employed to investigating the impact of τw on vascular morphology and functions.[222] Compared with conventional approaches, including cone and plate systems[223] and parallel flow chamber,[224] blood vessel chips are advantageous in incorporating an array of biomimetic microenvironments,[222] including complex and tissue-specific flow profiles,[225, 226] 3D culturing matrices,[198, 227] and structured scaffold materials,[228] which support long-term cell culture and are associated with cellular response to the shear stress.
One vessel chip incorporated electrospun fibrous scaffolds that enhanced the flow-induced alignment of ECs.[228] Three strategies were employed in the vessel chip to change the flow rate from static conditions to artery flow-related ones, including gradual, immediate, and intermittent schemes. The gradual increase in flow rate was better than the other two for reducing the shedding of ECs. This result agrees with the previous understanding that the changing pattern of stress, in addition to the absolute magnitude, is associated with vascular homeostasis and remodeling.[217] This strategy of the gradually increased flow rate can be useful for screening culture parameters and investigating vascular biology.[228]
Another vessel chip allows the well-controlled manipulation of a disturbed, oscillatory flow profile for studying YAP mechanobiology.[180] YAP is sensitive to both matrix stiffness[229] and shear stress,[230] and has important implications for atherosclerotic progression.[231, 232] In particular, an oscillatory flow (10 μL/min, 0.3 Hz, 11.2 dyn/cm2) was applied to HUVEC cultured in the blood vessel chip, leading to significantly improved YAP nuclear partition (the ratio of nuclear YAP to total YAP) and cell circularity, compared with the unidirectional flow at the same flow rate (Figure 6c).[180] The activation of YAP was also confirmed by the upregulation of two downstream targets of YAP, including connective tissue growth factor (CTGF) and ankyrin repeat domain 1 (ANKRD1). Furthermore, the YAP activation led to the elevation of two atherosclerosis markers, vascular cell adhesion molecule 1 (VCAM-1), and von Willebrand factor (vWF), suggesting a pathological role of the disturbed, oscillatory flow via the activation of YAP (Figure 6d, e).[233, 234]
On-chip shear stress magnitudes and pulsatile frequencies can be manipulated using pneumatically actuated micropumps and size-controlled microvalves (Figure 6f).[235] Three different flow profiles, including perfusion (0.47 dyn/cm2), static (8.9 dyn/cm2), and aortic inflow (5.9 dyn/cm2, 1.2 Hz), were applied to valvular ECs, which exhibit influence on cellular alignment, elongation, and α-smooth muscle actin (α-SMA) expression (Figure 6f).[236] In particular, the aortic inflow increased the expression of α-SMA by 1.9 and 1.5 times compared with the perfusion and static flow profiles, respectively, implying the need for positive feedback to enhance cellular contraction.[235]
Platelet adhesion and thrombus formation are associated with life-threatening cardiovascular diseases [237, 238] and are regulated by complex biochemical and mechanical cues,[239, 240] which are usually challenging to recapitulate in vitro by conventional approaches. A blood vessel chip allows the manipulation of channel width, thus constructing a stenotic vessel analog to investigate the biochemical factor of platelet aggregation (Figure 6g).[241] The stenosis diameter and geometry of the microchannel can be tuned to generate a series of shear stress gradients, benefiting the quantitative, in vitro investigation of shear-dependent thrombus formation. vWF expression increases at the post-stenosis site, probably due to increased share stress, and is spatially correlated with the platelet-aggregation, suggesting the role of vWF in the stenosis-dependent platelet aggregation. Furthermore, another blood vessel chip containing a network of stenosed arterial vessels was developed for real-time, quantitative evaluation of blood clotting, featuring a small sample volume, real-time observation, and parallel measurements.[242]
Platelet adhesion can be monitored in blood vessel chips using DNA-based integrin tension sensors (Figure 6h).[243] The integrin tension sensor is fluorescence-labeled double-strand DNA, coated on the bottom surface of microchannels, which translates the detachment of platelets into fluorescent signals, allowing in situ and real-time imaging. This blood vessel chip investigated the effect of a Rho-associated kinase (ROCK) inhibitor, Y-27632,[244] and shear stress on platelet adhesion. Notably, the vessel chip employed a classic Christmas tree-like microchannel network to generate a series of Y-27632 concentrations, thus facilitating high throughput screening.
2.5.2. Circumferential force
The entire vascular wall experiences circumferential forces and, thus, various strains, resulting from pulsatile blood flow and mechanical motions of tissues, such as breathing (Figure 6a). In a physiological circulation between systolic and diastolic pressures, the circumferential strain of the vascular wall could vary between 5% to 10% at the heat rate (around 1 Hz).[245] The strain of 16%-20% and above has been suggested to be pathological and associated with hypertension.[245, 246]
The circumferential force has profound implications for the physiology and pathology of vascular cells.[247, 248] For example, the physiological circumferential strain (6% at 1 Hz) was found to prevent the apoptosis of ECs, probably through the activation of heme oxygenase-1 (HO-1), an inhibitor for the apoptosis of ECs.[249] Also, the physiological circumferential force regulates proliferation, migration, focal adhesions, and matrix remodeling of vascular cells,[250, 251] and promotes secretion of pro-angiogenetic cytokines,[248, 252] including VEGF, platelet-derived growth factor-ββ (PDGF-ββ), and angiopoietin 2 (Ang-2).
Compared with conventional methods of mechanical stimuli, such as FlexCell,[253] blood vessel chips are advantageous in parallel operations, high-throughput, and a small volume of culture media.[210, 254–256] A vessel chip allowed semi-cylindrical circumferential strain in an array of microfluidic channels spanning from 20 to 500 μm in diameter, analogous to vascular capillaries and small arteries (Figure 7a).[254] The circumferential strain, ranging from 0% to 8%, was controlled by the hydrostatic pressure under the elastic membrane. The vessel chip investigated several signaling proteins of mesenchymal stem cells (MSCs) under the circumferential strain, including Smad family proteins, β-catenin, and glycogen synthase kinase (GSK)-3β. For instance, the nuclear accumulation of β-catenin under 8.0% strain was significantly higher than in 0 and 1.1 % strains (Figure 7b). β-catenin is key to the Wnt signaling pathway that involves the proliferation and differentiation of MSCs.
Figure 7.
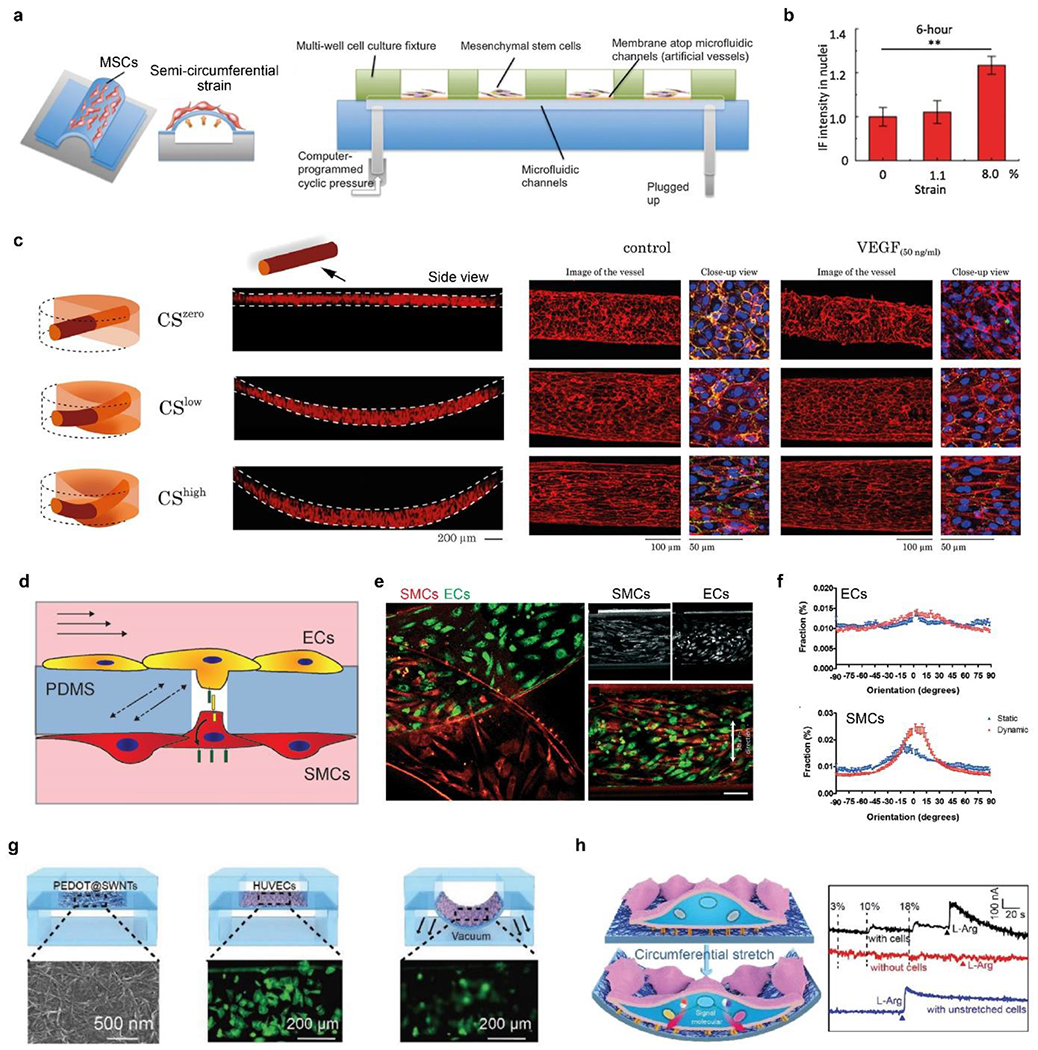
a) Schematic of microfluidic chips with semi-circumferential deformation. b) quantitative comparison of the nucleus accumulation of β-catenin under circumferential strains. a-b) Reproduced with permission.[254] Copyright 2012, Oxford University Press. c) Schematic and fluorescence images (side view) of the 3D vessels under zero, low, and high circumferential strains (CS). CS increases cellular alignment and counteracts VEGF to stabilize vessels. F-actin (red), platelet endothelial cell adhesion molecule-1 (PECAM-1, green), and DAPI (blue). Reproduced with permission.[257] Copyright 2021, Creative Commons Attribution license. d) Schematic of ECs and SMCs cocultured on a porous PDMS membrane in a vessel chip and exposed to shear stress and circumferential strain. e) Fluorescence image of ECs and SMCs after 4-day of culturing under mechanical stimuli, i.e., dynamic culture. Scale bar, 100 μm. f) Quantitative analyses of cellular alignment under static and dynamic culturing conditions. d-f) Reproduced with permission.[258] Copyright 2018, Creative Commons Attribution license. g) Schematic of in situ, real-time monitoring of mechanical stimuli using a flexible electrochemical sensor. HUVECs (in green) become blurred when they stretched and were out of focus. H) Right, schematic of cellular mechanotransduction to release NO. Left, electrochemical curves of cells responding to 0%, 10%, and 18% strains, and L-arginine. G-h) Reproduced with permission.[262] Copyright 2019, John Wiley and Sons.
Another vessel chip applied the circumferential strain to 3D perfusable vessels (Figure 7c).[257] The 3D vessel is made by seeding ECs in a needle-molded channel (200 μm in diameter) within a layer of fibrin hydrogel (500-μm-thick). The gel was supported by PDMS microchannels for medium perfusion and was under three circumferential strains, including zero strain (CSzero), low strain (CSlow) of 8-12% and high strain (CShigh) of 17-22%. The increased circumferential strain increases cell surface area, length, alignment, permeability, and the expression of adhesion molecules, such as PECAM-1 (Figure 7c).[257] Also, the circumferential strain can help stabilize the 3D vessels, suppress sprouting, and decrease permeability, thus seeming to counteract VEGF. These particular roles of high circumferential strain in the 3D vascular model might be different from some 2D results that the mechanical strains may disrupt endothelial barriers, thus implying the importance of a 3D vessel model and requiring further investigation.[257]
Blood vessel chips co-culture vascular cells, thus offering insights into the role of cell-cell interactions in sensing mechanical forces and vascular pathogenesis.[258, 259] For example, the cellular interactions between ECs and SMCs play an important role in sensing hemodynamic shear and circumferential forces for vascular homeostasis and pathogenesis, including atherosclerosis and hypertension.[117, 260] In one vessel chip, ECs and vascular SMCs were co-cultured on the two sides of a porous PDMS membrane, allowing cell-cell contacts through pores (10 μm in diameter) (Figure 7d).[258] The two cells were exposed to physiological mechanical stimuli, including both shear stress (1-1.5 Pa) and circumferential strain (5-8%). The mechanical stimuli didn’t align ECs but made SMCs more elongated and aligned (Figures 7e, f).[258] Of note, the alignment of SMC was parallel to the flow direction but perpendicular to the strain direction, which is not similar to the native SMCs in the tunica media. This result indicates the complex regulation of cellular alignment that may involve environmental cues other than mere mechanical stain and shear stress.[258] Furthermore, the on-chip EC-SMC interactions allowed further understanding of the Notch signaling pathway, which is crucial for vascular development, maturation, and homeostasis.[261] In particular, when cultured on a porous membrane and interacted with ECs, SMCs exhibited an increased expression of a component of the Notch pathway, HEY1, compared to ones cultured on non-porous membranes and without cellular interactions.[258]
Blood vessel chips can incorporate on-chip sensors for in situ, real-time monitoring of cellular responses to mechanical stimuli.[3, 262, 263] One vessel chip integrated a stretchable film electrode of poly(3, 4-ethylenedioxythiophene)-coated single-walled carbon nanotubes (SWNTs@ PEDOT) beneath cells, which detected mechanical stimuli-related cellular signals, including reactive oxygen species (ROS) and nitric oxide (NO) (Figure 7g).[262] The detection limits for ROS and NO were found at 1.6 × 10−9 M and 1 × 10−6 M, respectively. Of note, the NO signal was proportional to the strain magnitude and increased upon the addition of a precursor for the synthesis of NO, l-arginine (L-Arg) (Figure 7h). In contrast, the absence of cells in the chip led to no NO signal under the same conditions of applied strain and added L-Arg. Also, a NO-inhibitor, NG-nitro-l-arginine methyl ester hydrochloride (l-NAME) eliminated the NO signal, verifying the utility of the vessel chip for measuring the NO signal.[262]
3. Biomedical applications
As discussed in the above sections, blood vessel chips have recapitulated several essential aspects of vascular physiology and established useful in vitro models, which are promising for gaining insights into a range of vessel-related pathological conditions,[264] such as physical injury,[265] malaria infection,[146, 266] cancer metastasis,[267–269] aneurysm,[270, 271] thrombosis,[272, 273] and atherosclerosis.[181, 204, 241, 274, 275] In particular, blood vessel chips feature relatively short turnaround times compared with animal models, thus enabling early and rapid investigations of emerging diseases, such as the outbreak of the coronavirus disease 2019 (COVID-19).[39, 276, 277]
This section reviews the utility of blood vessel chips with more details for more focused aspects of the promising biomedical application, including intravascular mass transport, rare diseases, and atherosclerosis.
3.1. Intravascular mass transport
Intravascular mass transport, or controlled barrier function, is the mass transport between blood vessels and other tubular structures, and is instrumental to a broad range of tissue-level physiological functions, such as nutrient-absorption in the intestine, gas-exchange in the lung, and reabsorption in the kidney (Figures 8a–c). In particular, intensive mass transport occurs in the kidney, which processes blood up to 180 L daily.[278] The mass transport in the kidney is via passive diffusion and active reabsorption, leading to a concentration of drugs different from that in the blood and, thus, unexpected renal toxicity. Kidney toxicity is one major hurdle in drug development and often remains undetectable until phase 3 of the clinical trials,[279, 280] thus demanding improved in vitro testing models.[119]
Figure 8. Intravascular mass transport and kidney reabsorption.
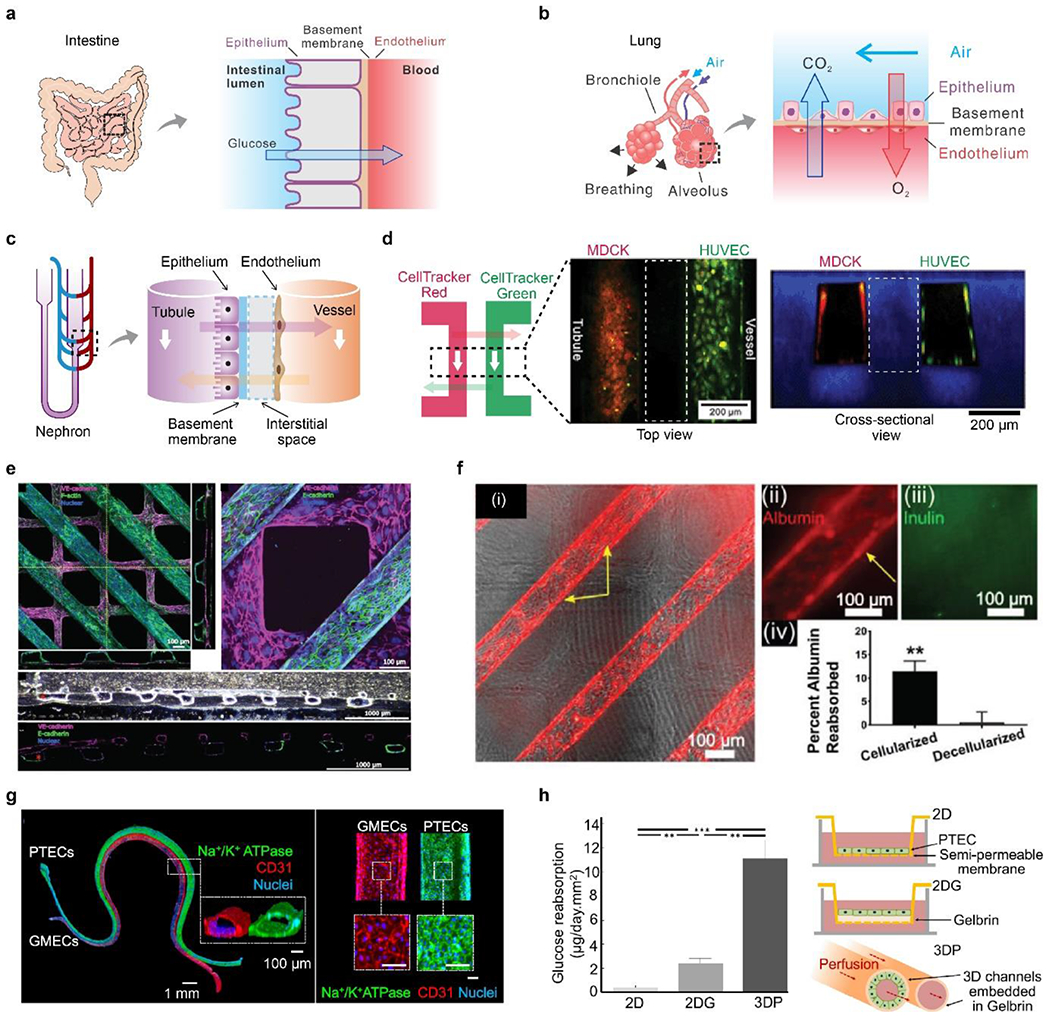
A) Schematic of intervascular mass transport in the intestine. b) Schematic of intervascular mass transport in the lung. C) Schematic of intervascular mass transport in the nephron. d) A biomimetic nephron constructed in a monolithic hydrogel vessel chip. Two channels, lined by Madin-Darby canine kidney cells (MDCKs) and HUVECs, represented the tubule and vessel, respectively. Two fluorescent dyes, CellTraker red and green, were perfused into one channel and allowed to diffuse into the other channel. White dash boxes indicate hydrogel located between two channels. Reproduced with permission.[42] Copyright 2013, Royal Society of Chemistry. e) Cellularized tubules (in purple) and vessels (in green) were constructed in collagen hydrogels, mechanically supported by polymer scaffolds. f) Albumin-reabsorption in the tubule, compared with inulin and decellularized devices. e-f) Reproduced with permission. [293] Copyright 2018, John Wiley and Sons. g) PTECs and glomerular microvascular ECs (GMECs) for confluent monolayers in hydrogel channels to mimic renal tubules and vessels. Scale bar, 100 μm. h) Schematic and quantitative glucose-reabsorption in the 3D hydrogel channel, compared to two 2D models. G-h) Reproduced with permission.[137] Copyright 2019, Creative Commons Attribution license.
Blood vessel chips with either 2D or 3D approaches have been exploited for intravascular mass transport (Figure 3a, 3b).[53, 281–284] Compared to the 2D approach, the 3D approach may be more flexible in constructing multiple distinct vessels/tubules, recruiting tissue-specific auxiliary cells, and mimicking the 3D water-containing, cell-remodelable ECM.[42, 285, 286] We briefly review several 3D hydrogel-enabled blood vessel chips for recapitulating the functional tubule unit and the solute transport in the kidney.
One monolithic hydrogel tissue chip has two microchannels, which are lined by epithelial cells and ECs to recapitulate renal tubules and vessels, respectively(Figures 8c, d).[42] The mass transport between the hydrogel channels, as exemplified by using small molecule dyes, is able to mimick the passive diffusion in the nephron (Figure 8d).[42] Passive diffusion, driven by the concentration gradient, is the main mechanism in the kidney for transporting soluble organic molecules, such as anti-gout probenecid analogs[287] and antioxidant coenzyme Q10.[288]
The unique fabrication method of the hydrogel tissue chip is key to the recapitulation of renal mass transport and is based on the synergy of collagen fibrillogenesis and a technique termed “liquid molding”. Collagen fibrillogenesis refers to the formation and extension of water-insoluble collagen nanofibrils and has been used to bond bulk hydrogels together for counteracting cell-induced hydrogel contraction.[289] As a primary component of ECM, collagen also promotes cytocompatibility and cellular attachment. “Liquid molding” refers to adopting a liquid or saline solution as a mold for casting shapes and structures in a sol-gel transition.[290] The liquid mold protects the space of microchannels from hydrogel precursor solution. After gelation, the liquid can be easily removed, leaving perfusable microchannels in the hydrogels. The synergy of collagen fibrillogenesis and “liquid molding” for constructing hydrogel channels offers high cytocompatibility, manufacturing flexibility, and simplicity.[42] In contrast, other methods often involve cell-harmful materials and conditions, such as chaotropic agents, ultraviolet (UV) light, organic solvents, and high temperatures.[7, 291, 292]
Another blood vessel chip is made entirely of collagen hydrogels for constructing human renal vascular-tubular units (hRVTU) (Figures 8e, 8f).[293] Two collagen channels and a collagen membrane were assembled and supported by external acrylic scaffolds. The thickness of the collagen membrane decreased from 31.7 μm to around 3.1 μm after 14 days of culture, suggesting collagen remodeled by cells, a notable difference from the synthetic polymer-made membranes.[293] Human renal epithelial cells and ECs were seeded in the top (parallel lines) and bottom (grids) channels, respectively, exhibiting tight connections (Figure 8e). Human fetal kidney pericytes were also incorporated in the bulk collagen hydrogel to grow along the endothelial and epithelial channels, potentially regulating barrier functions. The on-chip mass transport was examined by the reabsorption of three representatives, including albumin, inulin, and glucose (Figure 8f). The cellularized channels exhibited approximately 11.1% albumin-reabsorption, compared to nearly 0% of decellularized ones, implying the active role of cells (Figure 8f). This tissue chip also demonstrated that the albumin in the effluent from the vascular channel was not increased, which is suggested to be consistent with in vivo albumin-reabsorption that is via receptor-mediated endocytosis and may not elevate albumin concentration in the blood.[294]
3D printing with fugitive inks is suitable for fabricating 3D serpentine, perfusable hydrogel channels that allow mass transport (Figures 8g, 8h).[295, 296] The fugitive ink consists of Pluronic F-127 that could be liquefied and removed upon cooling. The hydrogel channels were made in Gelbrin, a mixture of gelatin and fibrin.[297] Proximal tubule ECs (PTECs) and glomerular microvascular ECs (GMECs) formed confluent monolayers within the two hydrogel channels, mimicking the enal tubule and blood vessel, respectively. Glucose-reabsorption in the 3D hydrogel channels was significantly higher than in two Transwell-based 2D models by 5-fold and 10-fold, respectively (Figure 8h). This result was attributed to perfusion-induced shear stress and improved cellular morphology (cell height and brush density and length) in 3D culture.[295, 296]
3.2. Rare diseases
Rare diseases are caused by single gene mutations and are characterized by a limited number of patients, fewer than 2,000 (defined by Europe) or 1,250 (by the USA).[298] However, rare diseases include a large number of genetic disorders with severe consequences on lifespan and physical and mental abilities. Rare diseases have been increasingly recognized as important opportunities to understand fundamental physiological and pathological processes and as a testbed for developing therapeutic strategies for other diseases.[299] Due to the scarcity of patients, rare diseases are often challenging to obtain sufficient clinical samples for statistically significant clinical trials.[300]
Tissue chips, along with other in vitro models,[301–303] are particularly compelling for investigating rare diseases [304–307] and replacing traditional clinical trials.[308] It is primarily because the tissue chip can work with a small number of human cells, including hiPSC and primary cells from limited patients,[309] as well as can exhibit tissue-level physiology related to clinical manifestations. Several rare diseases related to vessels, including Marfan’s syndrome and Hutchinson-Gilford progeria syndrome (HGPS), are and have been investigated using tissue chips,[32, 127, 246, 310, 311] as summarized in Table 3.
Table 3.
Brief summary of blood vessel chips for rare and uncommon diseases.
| Disease | Mutated gene | Targeted tissues/organs | Tissue-level functions | Refs |
|---|---|---|---|---|
| Barth syndrome (BTHS) | TAFAZZIN | Heart | Contraction stress | [32] |
| Hutchinson-Gilford progeria syndrome (HGPS) | LMNA | Vessel | Vasoactivity | [310] |
| Inflammation factors | [246] | |||
| Dilated cardiomyopathy (DCM) | TTN | Heart | Contractile function | [311] |
| Hereditary hemorrhagic telangiectasia (HHT) | ENG | Vessel | Morphology | [127] |
HGPS is caused by mutations and abnormal splicing of a gene, LMNA, that encodes an inner nuclear membrane protein, lamin A/C (Figure 9a).[299, 312, 313] The mutated lamin A/C, permanently farnesylated and uncleaved, is called progerin, which accumulates in the inner nuclear envelope,[299] leading to multiple nuclear defects and affecting downstream cellular pathways and organismic behaviors, especially related to vthe ascular system (Figure 9a).[314] Thus, HGPS patients exhibit premature aging and the corresponding vascular diseases, including progressive cardiovascular atherosclerosis, calcific aortic stenosis, and strokes.[314]
Figure 9.
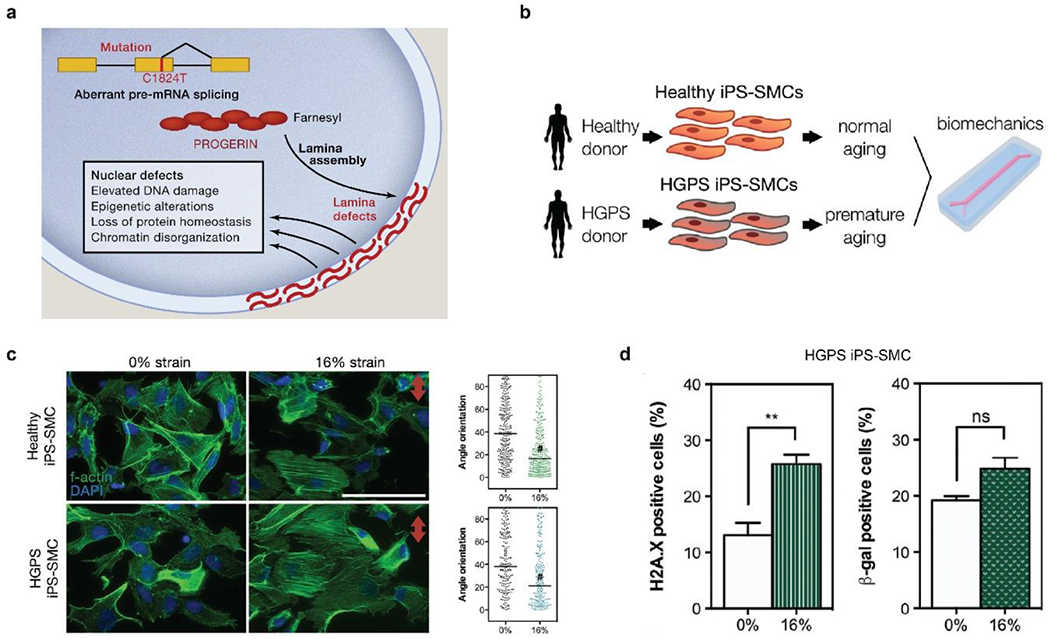
a) Schematic of HGPS pathogenesis. Mutations of the LMNA gene, such as C1824T, lead to aberrant mRNA splicing and permanent farnesylation of lamin A, i.e., progerin, that accumulates at the nuclear periphery. Reproduced with permission.[299] Copyright 2014, Elsevier. b) Schematic of blood vessel chips to investigate HGPS hiPSC-SMCs under mechanical stimuli. c) Both healthy and HGPS hiPSC-SMCs exhibit improved alignment under 16% strain. #, P < 0.0001. Red arrow indicates stretching direction. Scale bar, 50 μm. d) 16% mechanical strain leads to DNA double-strand breaks (H2A.X immunostaining) and a slight increase of senescence (β-galactosidase-induced fluorescence) of HGPS hiPSC-SMC. b-d) Reproduced with permission.[246] Copyright 2017, John Wiley and Sons.
Early and pervasive stiffness of vascular walls is one important clinical manifestation of HGPS patients. The stiffened blood vessels of HGPS patients have been linked to abnormally dense, fibrotic, and thickened vascular adventitia, the depletion of SMCs from the tunica media, and the over-deposition of replacement proteoglycans and collagen fibers.[315] Pulse wave velocity is a standard clinical measure for vessel stiffness.[316–320] The carotid-femoral pulse wave velocity of HGPS patients with a mean age of 7.4±3.4 years is around 13 m/s,[124] similar to normal adults over age 60.[321] Furthermore, the vascular wall density can be measured by the pixel intensity of ultrasonography.[322] The pixel intensity of the intima-media and adventitia of HGPS patients is significantly higher than the control group.[124] These clinical results and approaches would be useful in designing blood vessel chips to investigate HGPS and aging-associated vascular diseases.
One vessel chip investigated the inflammatory response of HGPS hiPSC-SMCs under mechanical strains (Figure 9b).[246] hiPSC-SMCs from healthy and HGPS donors were cultured on an elastic PDMS membrane inside a microchannel, representing the media layer (Figure 9b). Two strains, 9 and 16%, were used to mimic physiological and pathological conditions, respectively. The in vivo strain level of HGPS patients’ blood vessels remains unknown,[246] yet some studies have shown the mechanical sensitivity of progeria fibroblasts.[323] The pathological 16% stain alignment of both healthy and HGPS iPSC-SMCs, perpendicular to the strain direction (Figure 9c).[246] For HGPS hiPSC -SMCs, the pathological 16% strain also worsened DNA damage and increased cellular senescence, which are characterized by H2A.X staining and β-galactosidase (β-gal) activity, respectively (Figure 9d). Both DNA damage and senescence are markers of aging.[324] Furthermore, the 16% strain elevated mRNA expressions of HGPS hiPS-SMCs, including several pro-inflammatory factors, such as Caveolin 1 (CAV1), IL-6, Jun, and IL-1β, implying the role of inflammation in HGPS pathogenesis. The pro-inflammation response of HGPS hiPS-SMCs could be reversed by two pharmaceutical compounds, lovastatin and lonafarnib, suggesting potential therapeutic strategies.[246] Another work employed HGPS hiPSC-ECs to construct tissue-engineered blood vessels,[310] which exhibited many cellular differences from healthy ECs, including reduced vasoactivity, increased medial thickness, augmented calcification, and elevated apoptosis rate. These in vitro cellular behaviors help understand the cellular pathogenesis of HGPS and may be incorporated in blood vessel chips.
3.3. Atherosclerosis
Atherosclerosis refers to the buildup of plaques in vascular walls and the narrowing of vessels (Figures 2b, 6b), which leads to a broad range of cardiovascular diseases, including stroke and myocardial infarction. The pathology of atherosclerosis is associated with aberrant mechanical and solvent environments, including chronic inflammation,[325, 326] disturbed flow,[327] increased substrate stiffness, increased permeability,[327] and imbalanced lipid metabolism.[328, 329] Blood vessel chips, as an emerging alternative to mouse and other animal-based models,[330] have shed light on the pathogenesis of atherosclerosis, which is achieved largely by precisely controlling the atherosclerosis-related cellular microenvironment, including mechanical properties, soluble cues, and cell-cell interactions.
One 3D blood vessel chip has helped discover a non-canonical Notch mechanism for regulating vascular permeability and endothelial junctions (Figure 10a–b).[193] In particular, the blood vessel chip took the advantage of convenient permeability measurement and the precise control of flow, which are largely inaccessible to animal models. In particular, the blood vessel chip revealed that an inhibitor of Notch transcription signaling, N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), significantly increased the vascular permeability within 15-50 minutes.[193] This result was too rapid for a transcription-dependent regulation, the canonical Notch signaling pathway.[331] Then, the transmembrane domain (TMD) of NOTCH1, after cleavage of the intracellular domain, was found to form a complex with multiple signaling molecules, including vascular endothelial cadherin, the transmembrane protein tyrosine phosphatase, leukocyte common antigen-related protein (LAR), and triple functional domain protein (TRIO) (Figure 10b). The TMD-NOTCH1 complex activates Rac Family Small GTPase 1 (RAC1) to drive the assembly of adherent junctions for rescuing barrier functions (Figure 10b). This non-conical Notch mechanism is largely consistent with a complex-mediated mechanism for diverse biological events.[332]
Figure 10.
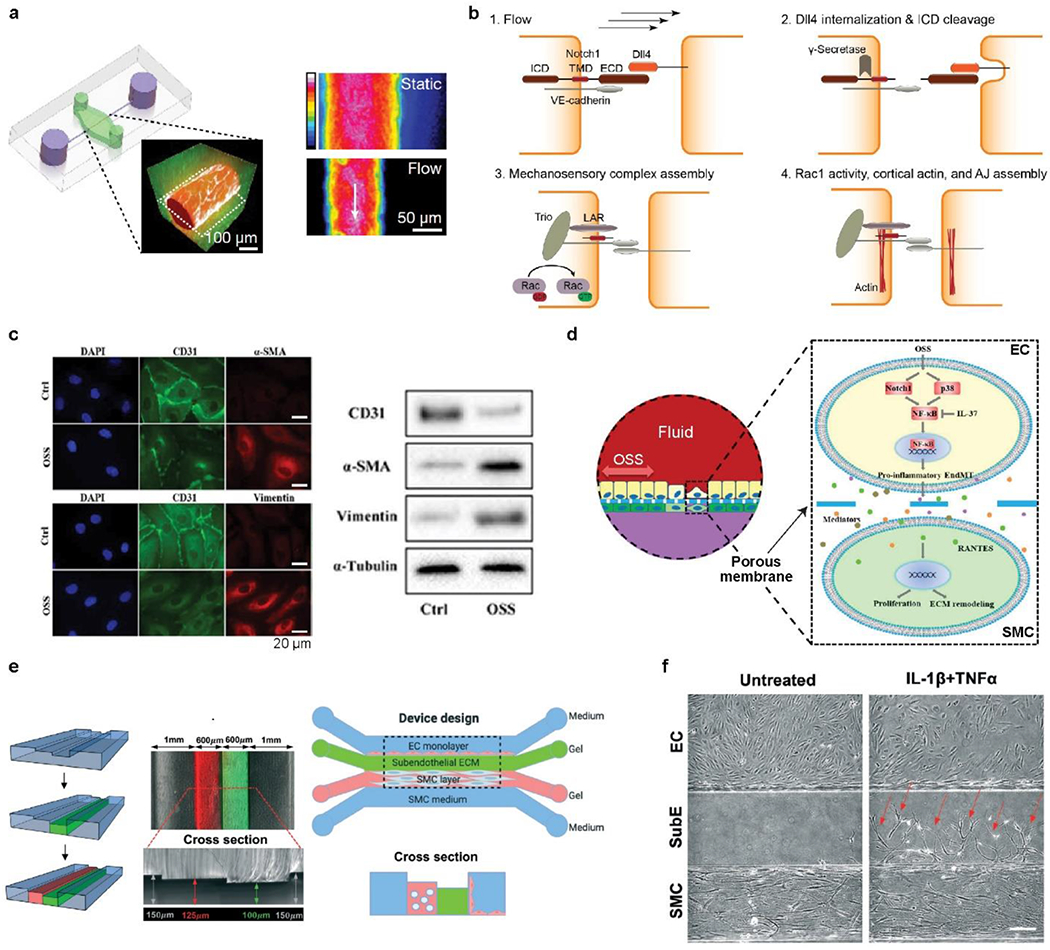
a) Schematic, fluorescence image and heat map of a vessel chip, consisting of human ECs (red) and ECM (green). Flow-induced shear stress, around 5 dyn/cm2, led to low vascular permeability, compared with static conditions. b) Schematic of NOTCH1 mechanosensory complex, encompassing LAR and TRIO, for stabilizing cellular junctions via the activation of RAC1. a-b) Reproduced with permission.[193] Copyright 2017, Springer Nature. c) Fluorescence images and western blots indicating the reduced expression of cluster of CD31, and the increased level of vimentin and α-SMA upon exposure to oscillatory flow. d) Schematic of the underlying mechanism of interactions between ECs and SMCs, involving paracrine communication and RANTES activation. c-d) Reproduced with permission.[335] Copyright 2021, Creative Commons Attribution NonCommercial License. e) Schematic of constructing intima-media interface using varying channel heights and CBV effect. f) Phase-contrast images of migrated SMC into the subendothelial layer in response to the treatment of inflammatory cytokines, including IL-1β and TNF-α. e-f) Reproduced with permission.[336] Copyright 2021, Royal Society of Chemistry.
Atherosclerosis is associated with disturbed flow profiles that can be recapitulated by tissue chips. In one blood vessel chip, the oscillatory and low shear flow (<4 dyn/cm2) would lead to several alternations of ECs, including reduced junctional proteins, such as β-catenin, compromised barrier function, and increased inflammation.[333, 334] Another blood vessel chip found that disturbed flow could substantially change gene expressions characterized by single-cell RNA-seq.[335] In particular, disturbed flow resulted in the upregulation of 2,002 genes and the downregulation of 1,486 genes, the changes of which are associated with pro-inflammatory endothelial-to-mesenchymal transition (EndMT). Also, disturbed flow altered the protein expression of ECs, including reduced differentiation 31 (CD31) and increased α-SMA and vimentin (Figure 10c). Furthermore, this blood vessel chip demonstrated that the co-culture of ECs and SMCs promoted the atherogenic activity of SMCs, including proliferation and excessive deposition of ECM, most likely via a RANTES mediated paracrine mechanism (Figure 10d).[335] The vessel chip used an anti-inflammatory approach of IL-37 to mitigate the flow-induced vascular pathobiology, showing the potential for screening drug candidates.
Atherosclerosis is also associated with soluble environmental cues, such as inflammation, hyperglycemia, and hyperlipidemia.[274, 336] Atherosclerosis has also been reconstituted on chips. One blood vessel chip established early-stage atherosclerosis and found that the addition of glucose and cholesterol increased ROS, a proinflammatory factor related to atherosclerosis. However, the cholesterol-induced ROS was not found in Petri dish-based models, suggesting the improved and desired physiological relevance of blood vessel chips. Another 3D vessel chip with co-cultured ECs and SMCs examined the critical role of inflammatory cytokines and oxidized low-density lipoprotein (oxLDL) in promoting early inflammation-related atherogenic events, such as elevated intercellular adhesion molecule-1 (ICAM-1), increased SMCs migration, and increased monocyte-EC-adhesion (Figures 10e, 10f).[336] In particular, the treatment of IL-1β and TNF-α led to a 3.6-fold increase in SMC migration from 8.75 ± 2.36 to 31.5 ± 2.65 (Figure 10f). The vessel chip also tested two drugs, vitamin D and metformin, for mitigating the cytokine-induced inflammation responses of ECs. In addition, the vessel chip was characterized by using the capillary burst valve (CBV) effect and varying channel heights to construct pillar-free, full-contact, well-confined, and independent channels for accommodating ECMs, ECs, and SMCs (Figure 10e).
4. Conclusion and perspectives
This review article attempted to provide a glimpse of state-of-art blood vessel chips that exhibit huge promise for a broad range of vascular biomedical research. The crux of blood vessel chips is the ex vivo recapitulation of vascular physiology, including morphological, structural, compositional, mechanical, and cellular features of vessels, through exploiting the synergy of bioengineering approaches, human cells, and biomaterials. It is believed that the physiological relevance of in vitro models allows improved accuracy and increased value of outcomes. Blood vessel chips have demonstrated almost unprecedented physiological relevance compared with previous in vitro models by incorporating important and delicate physiological features, such as cellular interactions and mechanical and biochemical cues. Furthermore, blood vessel chips enable well-controlled experimental conditions and decouple multiple factors, facilitating the exploitation of sophisticated human physiology. Due to the conceptual and technical merits, blood vessel chips represent an emerging alternative to conventional in vitro models for identifying key elements of disease pathogenesis at cellular and tissue levels and for the personalized screening and efficacy verification of drug candidates.
Vascular physiology, especially structural features and material composition, provides a general guideline and valuable inspiration for devising blood vessel chips. In particular, the recapitulation of the basement membrane is a promising research theme. The basement membrane is a primarily proteinaceous structural unit separating endothelium and epithelium (or other layers of vascular walls), which is thin (<1 μm), proteinaceous (collagen and proteoglycan), and nanofibrous.[337, 338] These structural and compositional features enable cellular remodeling and intravascular mass transport. In contrast, these features of the basement membrane are largely inaccessible to the widely used PDMS membranes in the blood vessel chips. The PDMS membrane is not compatible with cell remodeling, is much thicker (>10 μm), and it can be haunted by bulk absorption issues.[339, 340] To construct more physiologically relevant basement membranes in blood vessel chips, a range of new materials and fabrication methods are emerging,[341–343] such as the poly(lactic-co-glycolic acid) (PLGA) electrospinning nanofiber membrane[344] and the gold mesh-supported composite membrane (collagen and elastin).[345] Notably, the improvement of physiological relevance requires a continuous and substantial input of efforts through collaboration between multiple disciplines.
Besides, human iPSC-derived vascular cells, including ECs [346, 347] and SMCs [348, 349] have been increasingly used in blood vessel chips. The iPSCs are an almost unlimited source of cells, but also characterized by human-specific features. Notably, several approaches have emerged to eliminate the heterogeneity of differentiated functional cells [350] and finetune the tissue specificity.[351] These advances in hiPSC thus help blood vessel chips recapitulate a particular segment of the vascular system and tissue-specific vessels.
Finally, we would like to join many other colleagues to emphasize the importance of the multidisciplinary collaboration between clinicians and bioengineers, [352–354] which is particularly important for advancing blood vessel chips. Clinical findings inspire and guide the design of blood vessel chips, while the insights and results from blood vessel chips would beneficially promote clinical treatment.
Acknowledgments
The authors acknowledge support from the National Institutes of Health (UG3TR003274, UH3TR003274), the National Science Foundation (CISE-IIS-2225698), and the Brigham Research Institute.
Biographies
Xuan Mu is an assistant professor in the Roy J. Carver Department of Biomedical Engineering, College of Engineering, University of Iowa. Before joining the current position, Dr. Mu worked at Brigham Women’s Hospital/Harvard Medical School, Tufts University, and Peking Union Medical Collage. Dr. Mu is interested in exploiting a set of bioinspired strategies, silk proteins, and bioengineering approaches to construct in vitro tissue/disease models and tissue engineering products, which help advance the healthcare for patients with respiratory and pulmonary diseases.

Dr. Marie Denise Gerhard-Herman received her MD from Northeastern College of Medicine (1987) and her MMsc from Harvard Medical School (2003). She is a cardiovascular physician at Brigham and Women’s Hospital and Associate Professor of Medicine at Harvard Medical School. Her research utilizes vascular imaging to understand disease, with a particular focus on understanding pathophysiology in accelerated atherosclerosis.

Dr. Y. Shrike Zhang received a B.Eng. in Biomedical Engineering from Southeast University (2008), a M.S. in Biomedical Engineering from Washington University in St. Louis (2011), and a Ph.D. in Biomedical Engineering at Georgia Institute of Technology/Emory University (2013). Dr. Zhang is currently an Assistant Professor at Harvard Medical School and Associate Bioengineer at Brigham and Women’s Hospital. Dr. Zhang’s research is focused on innovating medical engineering technologies, including bioprinting, organs-on-chips, microfluidics, and bioanalysis, to recreate functional tissues and their biomimetic models towards applications in precision medicine.

Footnotes
Conflicts of Interest
YSZ sits on the scientific advisory board of Allevi by 3D Systems and Xellar, which however, did not participate in or bias the work.
Contributor Information
Xuan Mu, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA; Roy J. Carver Department of Biomedical Engineering, College of Engineering, University of Iowa, Iowa City, IA 52242, USA.
Marie Denise Gerhard-Herman, Division of Cardiovascular Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02114, USA.
Yu Shrike Zhang, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Cambridge, MA 02139, USA.
References
- [1].Zhang B, Korolj A, Lai BFL, Radisic M, Nat. Rev. Mater 2018, 3, 257. [Google Scholar]
- [2].Bhatia SN, Ingber DE, Nat. Biotechnol 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- [3].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, Massa S, Riahi R, Chae S, Hu N, Proc. Natl. Acad. Sci 2017, 114, E2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ren K, Zhou J, Wu H, Accounts of Chemical Research 2013, 46, 2396. [DOI] [PubMed] [Google Scholar]
- [5].Ren K, Chen Y, Wu H, Current Opinion in Biotechnology 2014, 25, 78. [DOI] [PubMed] [Google Scholar]
- [6].Ahadian S, Civitarese R, Bannerman D, Mohammadi MH, Lu R, Wang E, Davenport-Huyer L, Lai B, Zhang B, Zhao Y, Adv. Healthcare Mater 2018, 7, 1700506. [DOI] [PubMed] [Google Scholar]
- [7].Xie R, Zheng W, Guan L, Ai Y, Liang Q, Small 2020, 16, 1902838. [DOI] [PubMed] [Google Scholar]
- [8].Wnorowski A, Yang H, Wu JC, Adv. Drug Delivery Rev 2019, 140, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mu X, Zheng W, Sun J, Zhang W, Jiang X, Small 2013, 9, 9. [DOI] [PubMed] [Google Scholar]
- [10].Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Sci. Rep 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sakthivel K, O’Brien A, Kim K, Hoorfar M, TrAC, Trends Anal. Chem 2019, 117, 166. [Google Scholar]
- [12].Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, Li S, Biomaterials 2011, 32, 3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ergir E, Bachmann B, Redl H, Forte G, Ertl P, Frontiers in physiology 2018, 9, 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhao Q, Cole T, Zhang Y, Tang S-Y, Micromachines 2021, 12, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Van Den Berg A, Mummery CL, Passier R, Van der Meer AD, Lab Chip 2019, 19, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Perlman RL, Evolution, medicine, and public health 2016, 2016, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rogers CS, Current Protocols in Human Genetics 2016, 90, 15.9. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vunjak-Novakovic G, Ronaldson-Bouchard K, Radisic M, Cell 2021, 184, 4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ingber DE, Development 2018, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mu X, He W, Rivera VAM, De Alba RAD, Newman DJ, Zhang YS, Life Sciences in Space Research 2022, 35, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ma C, Peng Y, Li H, Chen W, Trends in pharmacological sciences 2021, 42, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ching T, Toh Y-C, Hashimoto M, Zhang YS, Trends in Pharmacological Sciences 2021, 42, 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Caplin JD, Granados NG, James MR, Montazami R, Hashemi N, Adv. Healthcare Mater 2015, 4, 1426. [DOI] [PubMed] [Google Scholar]
- [24].Savoji H, Mohammadi MH, Rafatian N, Toroghi MK, Wang EY, Zhao Y, Korolj A, Ahadian S, Radisic M, Biomaterials 2019, 198, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shuler ML, Lab Chip 2019, 19, 9. [DOI] [PubMed] [Google Scholar]
- [26].Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA, Nature Reviews Drug Discovery 2021, 20, 345. [DOI] [PubMed] [Google Scholar]
- [27].Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A, Lab Chip 2014, 14, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bersini S, Yazdi IK, Talò G, Shin SR, Moretti M, Khademhosseini A, Biotechnology advances 2016, 34, 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sato K, Sato K, Anal. Sci 2018, 34, 755. [DOI] [PubMed] [Google Scholar]
- [30].Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, Aggarwal P, Zhang B, Conant G, Ronaldson-Bouchard K, Pahnke A, Protze S, Lee JH, Davenport Huyer L, Jekic D, Wickeler A, Naguib HE, Keller GM, Vunjak-Novakovic G, Broeckel U, Backx PH, Radisic M, Cell 2019, 176, 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Nature 2018, 556, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang G, McCain ML, Yang LH, He AB, Pasqualini FS, Agarwal A, Yuan HY, Jiang DW, Zhang DH, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen JH, Wang DZ, Li K, Wang JW, Wanders RJA, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT, Nat. Med 2014, 20, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Grosberg A, Alford PW, McCain ML, Parker KK, Lab Chip 2011, 11, 4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Zhang YS, Shin SR, Calzone G, Biofabrication 2016, 8, 014101. [DOI] [PubMed] [Google Scholar]
- [35].Hassan S, Sebastian S, Maharjan S, Lesha A, Carpenter AM, Liu X, Xie X, Livermore C, Zhang YS, Zarrinpar A, Hepatology 2020, 71, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ma L-D, Wang Y-T, Wang J-R, Wu J-L, Meng X-S, Hu P, Mu X, Liang Q-L, Luo G-A, Lab Chip 2018, 18, 2547. [DOI] [PubMed] [Google Scholar]
- [37].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, Moller R, Hoagland D, Oishi K, Horiuchi S, Uhl S, Blanco-Melo D, Albrecht RA, Liu W-C, Jordan T, Nilsson-Payant BE, Golynker I, Frere J, Logue J, Haupt R, McGrath M, Weston S, Zhang T, Plebani R, Soong M, Nurani A, Kim SM, Zhu DY, Benam KH, Goyal G, Gilpin SE, Prantil-Baun R, Gygi SP, Powers RK, Carlson KE, Frieman M, tenOever BR, Ingber DE, Nat. Biomed. Eng 2021, 5, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang D, Liu T, Liao J, Maharjan S, Xie X, Pérez M, Anaya I, Wang S, Mayer AT, Kang Z, Proc. Natl. Acad. Sci 2021, 118, e2016146118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bang S, Jeong S, Choi N, Kim HN, Biomicrofluidics 2019, 13, 051301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Harberts J, Fendler C, Teuber J, Siegmund M, Silva A, Rieck N, Wolpert M, Zierold R, Blick RH, ACS Nano 2020, 14, 13091. [DOI] [PubMed] [Google Scholar]
- [42].Mu X, Zheng W, Xiao L, Zhang W, Jiang X, Lab Chip 2013, 13, 1612. [DOI] [PubMed] [Google Scholar]
- [43].DesRochers TM, Palma E, Kaplan DL, Adv. Drug Delivery Rev 2014, 69, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Qu Y, An F, Luo Y, Lu Y, Liu T, Zhao W, Lin B, Biomaterials 2018, 155, 41. [DOI] [PubMed] [Google Scholar]
- [45].Ashammakhi N, Wesseling-Perry K, Hasan A, Elkhammas E, Zhang YS, Kidney international 2018, 94, 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lebedenko CG, Banerjee IA, Biomimetics 2021, 6, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ashammakhi N, Nasiri R, Barros N. R. d., Tebon P, Thakor J, Goudie M, Shamloo A, Martin MG, Khademhosseini A, Biomaterials 2020, 255, 120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shin W, Kim HJ, Proc. Natl. Acad. Sci 2018, 115, E10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tozzi L, Laurent P-A, Di Buduo CA, Mu X, Massaro A, Bretherton R, Stoppel W, Kaplan DL, Balduini A, Biomaterials 2018, 178, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bai J, Fu H, Bazinet L, Birsner AE, D’amato RJ, Frontiers in pharmacology 2020, 11, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Abdalkader R, Kamei K.-i., Lab Chip 2020, 20, 1410. [DOI] [PubMed] [Google Scholar]
- [52].Lai BFL, Lu RXZ, Hu Y, Davenport Huyer L, Dou W, Wang EY, Radulovich N, Tsao MS, Sun Y, Radisic M, Adv. Funct. Mater 2020, 30, 2000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cao X, Ashfaq R, Cheng F, Maharjan S, Li J, Ying G, Hassan S, Xiao H, Yue K, Zhang YS, Adv. Funct. Mater 2019, 29, 1807173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Carvalho M, Barata D, Teixeira L, Giselbrecht S, Reis R, Oliveira J, Truckenmüller R, Habibovic P, Sci. Adv 2019, 5, eaaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sontheimer-Phelps A, Hassell BA, Ingber DE, Nature Reviews Cancer 2019, 19, 65. [DOI] [PubMed] [Google Scholar]
- [56].Mu X, Zhang YS, in Engineering Technologies and Clinical Translation, Elsevier; 2022, p. 155. [Google Scholar]
- [57].Sasserath T, Rumsey JW, McAleer CW, Bridges LR, Long CJ, Elbrecht D, Schuler F, Roth A, Bertinetti-LaPatki C, Shuler ML, Hickman JJ, Adv. Sci 2020, 7, 2000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sung JH, Wang YI, Narasimhan Sriram N, Jackson M, Long C, Hickman JJ, Shuler ML, Analytical chemistry 2018, 91, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A, Maoz BM, Jeanty SSF, Somayaji MR, Burt M, Calamari E, Chalkiadaki A, Cho A, Choe Y, Chou DB, Cronce M, Dauth S, Divic T, Fernandez-Alcon J, Ferrante T, Ferrier J, FitzGerald EA, Fleming R, Jalili-Firoozinezhad S, Grevesse T, Goss JA, Hamkins-Indik T, Henry O, Hinojosa C, Huffstater T, Jang K-J, Kujala V, Leng L, Mannix R, Milton Y, Nawroth J, Nestor BA, Ng CF, O’Connor B, Park T-E, Sanchez H, Sliz J, Sontheimer-Phelps A, Swenor B, Thompson G, Touloumes GJ, Tranchemontagne Z, Wen N, Yadid M, Bahinski A, Hamilton GA, Levner D, Levy O, Przekwas A, Prantil-Baun R, Parker KK, Ingber DE, Nat. Biomed. Eng 2020, 4, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee SH, Sung JH, Adv. Healthcare Mater 2018, 7, 1700419. [Google Scholar]
- [61].Theobald J, Ghanem A, Wallisch P, Banaeiyan AA, Andrade-Navarro MA, Tasškova K, Haltmeier M, Kurtz A, Becker H, Reuter S, ACS Biomater. Sci. Eng 2018, 4, 78. [DOI] [PubMed] [Google Scholar]
- [62].Chang S-Y, Weber EJ, Sidorenko VS, Chapron A, Yeung CK, Gao C, Mao Q, Shen D, Wang J, Rosenquist TA, JCI insight 2017, 2, e95978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Osaki T, Uzel SG, Kamm RD, Nat. Protoc 2020, 15, 421. [DOI] [PubMed] [Google Scholar]
- [64].Miller PG, Shuler ML, Biotechnology and bioengineering 2016, 113, 2213. [DOI] [PubMed] [Google Scholar]
- [65].Lee H, Chung M, Jeon NL, MRS bulletin 2014, 39, 51. [Google Scholar]
- [66].Hu C, Chen Y, Tan MJA, Ren K, Wu H, Analyst 2019, 144, 4461. [DOI] [PubMed] [Google Scholar]
- [67].Pober JS, Tellides G, Trends in immunology 2012, 33, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR, Nature reviews Clinical oncology 2019, 16, 469. [DOI] [PubMed] [Google Scholar]
- [69].Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, Journal of the American College of Cardiology 2020, 76, 2982.33309175 [Google Scholar]
- [70].Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang Y-S, Dokmeci MR, Khademhosseini A, Biomaterials 2014, 35, 7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Van Duinen V, Trietsch SJ, Joore J, Vulto P, Hankemeier T, Current opinion in biotechnology 2015, 35, 118. [DOI] [PubMed] [Google Scholar]
- [72].Nie J, Gao Q, Wang Y, Zeng J, Zhao H, Sun Y, Shen J, Ramezani H, Fu Z, Liu Z, Small 2018, 14, 1802368. [DOI] [PubMed] [Google Scholar]
- [73].Thakare K, Jerpseth L, Pei Z, Elwany A, Quek F, Qin H, Journal of Manufacturing and Materials Processing 2021, 5, 91. [Google Scholar]
- [74].Singh NK, Han W, Nam SA, Kim JW, Kim JY, Kim YK, Cho D-W, Biomaterials 2020, 232, 119734. [DOI] [PubMed] [Google Scholar]
- [75].Polley C, Kussauer S, David R, Barkow P, Mau R, Seitz H, Current Directions in Biomedical Engineering 2020, 6, 469. [Google Scholar]
- [76].Heinrich MA, Liu W, Jimenez A, Yang J, Akpek A, Liu X, Pi Q, Mu X, Hu N, Schiffelers RM, Small 2019, 15, 1970126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Abudupataer M, Chen N, Yan S, Alam F, Shi Y, Wang L, Lai H, Li J, Zhu K, Wang C, Biomedical microdevices 2020, 22, 1. [DOI] [PubMed] [Google Scholar]
- [78].Mu X, Gonzalez-Obeso C, Xia Z, Sahoo JK, Li G, Cebe P, Zhang YS, Kaplan DL, Molecules 2022, 27, 2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang Y, Yu Y, Ozbolat IT, Journal of nanotechnology in engineering and medicine 2013, 4. [Google Scholar]
- [80].Korolj A, Laschinger C, James C, Hu E, Velikonja C, Smith N, Gu I, Ahadian S, Willette R, Radisic M, Lab Chip 2018, 18, 3112. [DOI] [PubMed] [Google Scholar]
- [81].Zhang B, Montgomery M, Chamberlain MD, Ogawa S, Korolj A, Pahnke A, Wells LA, Massé S, Kim J, Reis L, Nat. Mater 2016, 15, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lee A, Hudson A, Shiwarski D, Tashman J, Hinton T, Yerneni S, Bliley J, Campbell P, Feinberg A, Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- [84].Mu X, Agostinacchio F, Ning X, Pei Y, Guo C, Khan Y, Cebe P, Motta A, Kaplan DL, Prog. Polym. Sci 2021, 115, 101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Agostinacchio F, Mu X, Dirè S, Motta A, Kaplan DL, Trends Biotechnol. 2021, 39, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mu X, Wang Y, Guo C, Li Y, Ling S, Huang W, Cebe P, Hsu HH, De Ferrari F, Jiang X, Xu Q, Balduini A, Omenetto F, Kaplan DL, Macromol. Biosci 2020, 20, 1900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mu X, Fitzpatrick V, Kaplan DL, Adv. Healthcare Mater 2020, 9, 1901552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ouyang L, Highley CB, Sun W, Burdick JA, Adv. Mater 2017, 29, 1604983. [DOI] [PubMed] [Google Scholar]
- [89].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Włodarczyk-Biegun MK, del Campo A, Biomaterials 2017, 134, 180. [DOI] [PubMed] [Google Scholar]
- [91].Wang D, Maharjan S, Kuang X, Wang Z, Mille LS, Tao M, Yu P, Cao X, Lian L, Lv L, Sci. Adv 2022, 8, eabq6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Pi Q, Maharjan S, Yan X, Liu X, Singh B, van Genderen AM, Robledo-Padilla F, Parra-Saldivar R, Hu N, Jia W, Adv. Mater 2018, 30, 1706913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Tian Y, Wang Z, Wang L, Chemical Communications 2021, 57, 9166. [DOI] [PubMed] [Google Scholar]
- [94].Jun Y, Kang E, Chae S, Lee S-H, Lab Chip 2014, 14, 2145. [DOI] [PubMed] [Google Scholar]
- [95].Xie R, Xu P, Liu Y, Li L, Luo G, Ding M, Liang Q, Adv. Mater 2018, 30, 1705082. [DOI] [PubMed] [Google Scholar]
- [96].Xie R, Liang Z, Ai Y, Zheng W, Xiong J, Xu P, Liu Y, Ding M, Gao J, Wang J, Nat. Protoc 2021, 16, 937. [DOI] [PubMed] [Google Scholar]
- [97].Xie R, Korolj A, Liu C, Song X, Lu RXZ, Zhang B, Ramachandran A, Liang Q, Radisic M, ACS central science 2020, 6, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mandrycky CJ, Howard CC, Rayner SG, Shin YJ, Zheng Y, Journal of Molecular and Cellular Cardiology 2021, 159, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Caballero D, Blackburn SM, De Pablo M, Samitier J, Albertazzi L, Lab Chip 2017, 17, 3760. [DOI] [PubMed] [Google Scholar]
- [100].Lee S, Ko J, Park D, Lee S-R, Chung M, Lee Y, Jeon NL, Lab Chip 2018, 18, 2686. [DOI] [PubMed] [Google Scholar]
- [101].Moses SR, Adorno JJ, Palmer AF, Song JW, American Journal of Physiology-Cell Physiology 2021, 320, C92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Li Y, Zhu K, Liu X, Zhang YS, Current drug metabolism 2018, 19, 100. [DOI] [PubMed] [Google Scholar]
- [103].Rafii S, Butler JM, Ding B-S, Nature 2016, 529, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lin DS, Guo F, Zhang B, Nanotechnology 2018, 30, 024002. [DOI] [PubMed] [Google Scholar]
- [105].McCulloh KA, Sperry JS, Adler FR, Nature 2003, 421, 939. [DOI] [PubMed] [Google Scholar]
- [106].Murray CD, Proc. Natl. Acad. Sci. U. S. A 1926, 12, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Sherman TF, The Journal of general physiology 1981, 78, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Virumbrales-Muñoz M, Ayuso JM, Gong MM, Humayun M, Livingston MK, Lugo-Cintrón KM, McMinn P, Álvarez-García YR, Beebe DJ, Chemical Society Reviews 2020, 49, 6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Emerson DR, Cieślicki K, Gu X, Barber RW, Lab Chip 2006, 6, 447. [DOI] [PubMed] [Google Scholar]
- [110].Fenech M, Girod V, Claveria V, Meance S, Abkarian M, Charlot B, Lab Chip 2019, 19, 2096. [DOI] [PubMed] [Google Scholar]
- [111].Wu W, Guijt RM, Silina YE, Koch M, Manz A, RSC Adv. 2016, 6, 22469. [Google Scholar]
- [112].Wu W, Hansen CJ, Aragón AM, Geubelle PH, White SR, Lewis JA, Soft Matter 2010, 6, 739. [Google Scholar]
- [113].Miali ME, Colasuonno M, Surdo S, Palomba R, Pereira R, Rondanina E, Diaspro A, Pascazio G, Decuzzi P, ACS Appl. Mater. Interfaces 2019, 11, 31627. [DOI] [PubMed] [Google Scholar]
- [114].Mao M, Bei HP, Lam CH, Chen P, Wang S, Chen Y, He J, Zhao X, Small 2020, 16, 2000546. [DOI] [PubMed] [Google Scholar]
- [115].Kenneth SS, Anatomy & physiology: The unity of form and function, McGraw-Hill, 2017. [Google Scholar]
- [116].Bazzoni G, Dejana E, Physiological reviews 2004, 84, 869. [DOI] [PubMed] [Google Scholar]
- [117].Li M, Qian M, Kyler K, Xu J, Frontiers in Cardiovascular Medicine 2018, 5, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Arık YB, Helm M. W. v. d., Odijk M, Segerink LI, Passier R, Berg A. v. d., Meer A. D. v. d., Biomicrofluidics 2018, 12, 042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sakolish CM, Esch MB, Hickman JJ, Shuler ML, Mahler GJ, EBioMedicine 2016, 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Mistriotis P, Andreadis ST, Ageing Research Reviews 2017, 37, 94. [DOI] [PubMed] [Google Scholar]
- [121].Silver FH, Snowhill PB, Foran DJ, Ann. Biomed. Eng 2003, 31, 793. [DOI] [PubMed] [Google Scholar]
- [122].Miura K, PloS one 2020, 15, e0234759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Johnson C, Baugh R, Wilson C, Burns J, Journal of clinical pathology 2001, 54, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Gerhard-Herman M, Smoot LB, Wake N, Kieran MW, Kleinman ME, Miller DT, Schwartzman A, Giobbie-Hurder A, Neuberg D, Gordon LB, Hypertension 2012, 59, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Kenneth S D. Saladin, Anatomy & Physiology: The Unity of Form and Function, McGraw-Hill Education, 2017. [Google Scholar]
- [126].Gerhardt H, Betsholtz C, Cell and Tissue Research 2003, 314, 15. [DOI] [PubMed] [Google Scholar]
- [127].van der Meer AD, Orlova VV, ten Dijke P, van den Berg A, Mummery CL, Lab Chip 2013, 13, 3562. [DOI] [PubMed] [Google Scholar]
- [128].Goumans M-J, Ten Dijke P, Cold Spring Harbor perspectives in biology 2018, 10, a022210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Darland D, D’amore P, Angiogenesis 2001, 4, 11. [DOI] [PubMed] [Google Scholar]
- [130].Cuenca MV, Cochrane A, van den Hil FE, de Vries AA, Oberstein SAL, Mummery CL, Orlova VV, Stem cell reports 2021, 16, 2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kim S, Lee S, Lim J, Choi H, Kang H, Jeon NL, Son Y, Biomaterials 2021, 279, 121210. [DOI] [PubMed] [Google Scholar]
- [132].Polacheck WJ, Kutys ML, Tefft JB, Chen CS, Nat. Protoc 2019, 14, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD, Proc. Natl. Acad. Sci 2012, DOI: 10.1073/pnas.1201240109 doi:10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Yue T, Nakajima M, Takeuchi M, Hu C, Huang Q, Fukuda T, Lab Chip 2014, 14, 1151. [DOI] [PubMed] [Google Scholar]
- [135].Chen MB, Whisler JA, Fröse J, Yu C, Shin Y, Kamm RD, Nat. Protoc 2017, 12, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Moya ML, Hsu Y-H, Lee AP, Hughes CC, George SC, Tissue Eng., Part C 2013, 19, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lin NY, Homan KA, Robinson SS, Kolesky DB, Duarte N, Moisan A, Lewis JA, Proc. Natl. Acad. Sci 2019, 116, 5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Herland A, van der Meer AD, FitzGerald EA, Park T-E, Sleeboom JJ, Ingber DE, PLoS One 2016, 11, e0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Lee SWL, Campisi M, Osaki T, Possenti L, Mattu C, Adriani G, Kamm RD, Chiono V, Adv. Healthcare Mater 2020, 9, 1901486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Haase K, Gillrie MR, Hajal C, Kamm RD, Adv. Sci 2019, 6, 1900878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yeon JH, Ryu HR, Chung M, Hu QP, Jeon NL, Lab Chip 2012, 12, 2815. [DOI] [PubMed] [Google Scholar]
- [142].Phan DT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LY, George SC, Lee AP, Hughes CC, Lab Chip 2017, 17, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Song JW, Bazou D, Munn LL, Integr. Biol 2012, 4, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ko J, Ahn J, Kim S, Lee Y, Lee J, Park D, Jeon NL, Lab Chip 2019, 19, 2822. [DOI] [PubMed] [Google Scholar]
- [145].Bai J, Haase K, Roberts JJ, Hoffmann J, Nguyen HT, Wan Z, Zhang S, Sarker B, Friedman N, Ristić-Lehmann Č, Biomaterials 2021, 268, 120592. [DOI] [PubMed] [Google Scholar]
- [146].Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF, Lam WA, Nat. Biomed. Eng 2018, 2, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Marturano-Kruik A, Nava MM, Yeager K, Chramiec A, Hao L, Robinson S, Guo E, Raimondi MT, Vunjak-Novakovic G, Proc. Natl. Acad. Sci 2018, 115, 1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kim C, Kasuya J, Jeon J, Chung S, Kamm RD, Lab Chip 2015, 15, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Margolis EA, Cleveland DS, Kong YP, Beamish JA, Wang WY, Baker BM, Putnam AJ, Lab Chip 2021, 21, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].van Dijk CG, Brandt MM, Poulis N, Anten J, van der Moolen M, Kramer L, Homburg EF, Louzao-Martinez L, Pei J, Krebber MM, Lab Chip 2020, 20, 1827. [DOI] [PubMed] [Google Scholar]
- [151].Hsu Y-H, Moya ML, Abiri P, Hughes CCW, George SC, Lee AP, Lab Chip 2013, 13, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Paek J, Park SE, Lu Q, Park K-T, Cho M, Oh JM, Kwon KW, Yi Y.-s., Song JW, Edelstein HI, Ishibashi J, Yang W, Myerson JW, Kiseleva RY, Aprelev P, Hood ED, Stambolian D, Seale P, Muzykantov VR, Huh D, ACS Nano 2019, 13, 7627. [DOI] [PubMed] [Google Scholar]
- [153].Fritschen A, Blaeser A, Biomaterials 2021, 268, 120556. [DOI] [PubMed] [Google Scholar]
- [154].Campisi M, Shin Y, Osaki T, Hajal C, Chiono V, Kamm RD, Biomaterials 2018, 180, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Jang J, Park H-J, Kim S-W, Kim H, Park JY, Na SJ, Kim HJ, Park MN, Choi SH, Park SH, Biomaterials 2017, 112, 264. [DOI] [PubMed] [Google Scholar]
- [156].Cuchiara MP, Gould DJ, McHale MK, Dickinson ME, West JL, Adv. Funct. Mater 2012, 22, 4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Debbi L, Zohar B, Shuhmaher M, Shandalov Y, Goldfracht I, Levenberg S, Biomaterials 2022, 280, 121286. [DOI] [PubMed] [Google Scholar]
- [158].Kinstlinger IS, Miller JS, Lab Chip 2016, 16, 2025. [DOI] [PubMed] [Google Scholar]
- [159].Hann SY, Cui H, Esworthy T, Miao S, Zhou X, Lee S.-j., Fisher JP, Zhang LG, Translational Research 2019, 211, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Meyer W, Engelhardt S, Novosel E, Elling B, Wegener M, Krüger H, Journal of functional biomaterials 2012, 3, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Gong J, Schuurmans CC, van Genderen AM, Cao X, Li W, Cheng F, He JJ, López A, Huerta V, Manríquez J, Nat. Commun 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Kim HJ, Ingber DE, Integr. Biol 2013, 5, 1130. [DOI] [PubMed] [Google Scholar]
- [163].Musah S, Mammoto A, Ferrante TC, Jeanty SS, Hirano-Kobayashi M, Mammoto T, Roberts K, Chung S, Novak R, Ingram M, Nat. Biomed. Eng 2017, 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Reiser J, Sever S, Annual review of medicine 2013, 64, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hasan A, Paul A, Memic A, Khademhosseini A, Biomedical microdevices 2015, 17, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tan A, Fujisawa K, Yukawa Y, Matsunaga Y, Biomater. Sci 2016, 4, 1503. [DOI] [PubMed] [Google Scholar]
- [167].Yuan B, Jin Y, Sun Y, Wang D, Sun JS, Wang Z, Zhang W, Jiang XY, Adv. Mater 2012, 24, 890. [DOI] [PubMed] [Google Scholar]
- [168].Shadwick RE, Journal of Experimental Biology 1999, 202, 3305. [DOI] [PubMed] [Google Scholar]
- [169].Duca L, Blaise S, Romier B, Laffargue M, Gayral S, El Btaouri H, Kawecki C, Guillot A, Martiny L, Debelle L, Cardiovascular research 2016, 110, 298. [DOI] [PubMed] [Google Scholar]
- [170].Sakalihasan N, Michel J-B, Katsargyris A, Kuivaniemi H, Defraigne J-O, Nchimi A, Powell JT, Yoshimura K, Hultgren R, Nature reviews Disease primers 2018, 4, 1. [DOI] [PubMed] [Google Scholar]
- [171].Byfield FJ, Reen RK, Shentu T-P, Levitan I, Gooch KJ, Journal of Biomechanics 2009, 42, 1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Huynh J, Nishimura N, Rana K, Peloquin JM, Califano JP, Montague CR, King MR, Schaffer CB, Reinhart-King CA, Sci. Transl. Med 2011, 3, 112ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, Zhao J, Tai Y, Tang Y, Zhang Y-Y, The Journal of clinical investigation 2016, 126, 3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].DeLoach SS, Townsend RR, Clinical Journal of the American Society of Nephrology 2008, 3, 184. [DOI] [PubMed] [Google Scholar]
- [175].Safar ME, Nature Reviews Cardiology 2018, 15, 97. [DOI] [PubMed] [Google Scholar]
- [176].Akentjew TL, Terraza C, Suazo C, Maksimcuka J, Wilkens CA, Vargas F, Zavala G, Ocaña M, Enrione J, García-Herrera CM, Valenzuela LM, Blaker JJ, Khoury M, Acevedo JP, Nat. Commun 2019, 10, 3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lovett M, Cannizzaro C, Daheron L, Messmer B, Vunjak-Novakovic G, Kaplan DL, Biomaterials 2007, 28, 5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Lovett M, Lee K, Edwards A, Kaplan DL, Tissue Engineering Part B-Reviews 2009, 15, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Yuan W, Lv Y, Zeng M, Fu BM, Microvascular research 2009, 77, 166. [DOI] [PubMed] [Google Scholar]
- [180].Walther BK, Pandian NKR, Gold KA, Kiliç ES, Sama V, Gu J, Gaharwar AK, Guiseppi-Elie A, Cooke JP, Jain A, Lab Chip 2021, 21, 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Shin Y, Lim S, Kim J, Jeon JS, Yoo H, Gweon B, Lab Chip 2019, 19, 3664. [DOI] [PubMed] [Google Scholar]
- [182].Sato M, Sasaki N, Ato M, Hirakawa S, Sato K, Sato K, PloS one 2015, 10, e0137301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Ramón-Lozano C, Dessalles C, Babataheri A, Barakat A, Computer Methods in Biomechanics and Biomedical Engineering 2020, 23, S250. [Google Scholar]
- [184].Zeinali S, Bichsel CA, Hobi N, Funke M, Marti TM, Schmid RA, Guenat OT, Geiser T, Angiogenesis 2018, 21, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Hajal C, Offeddu GS, Shin Y, Zhang S, Morozova O, Hickman D, Knutson CG, Kamm RD, Nat. Protoc 2022, 1. [DOI] [PubMed] [Google Scholar]
- [186].Nagy JA, Benjamin L, Zeng H, Dvorak AM, Dvorak HF, Angiogenesis 2008, 11, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Weis SM, Cheresh DA, Nature 2005, 437, 497. [DOI] [PubMed] [Google Scholar]
- [188].McDonald DM, Baluk P, Cancer Res. 2002, 62, 5381. [PubMed] [Google Scholar]
- [189].Yuan F, Leunig M, Berk DA, Jain RK, Microvascular research 1993, 45, 269. [DOI] [PubMed] [Google Scholar]
- [190].Lee H, Kim S, Chung M, Kim JH, Jeon NL, Microvascular research 2014, 91, 90. [DOI] [PubMed] [Google Scholar]
- [191].Alimperti S, Mirabella T, Bajaj V, Polacheck W, Pirone DM, Duffield J, Eyckmans J, Assoian RK, Chen CS, Proc. Natl. Acad. Sci 2017, 114, 8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].ADAMSON RH, LENZ JF, CURRY FE, Microcirculation 1994, 1, 251. [DOI] [PubMed] [Google Scholar]
- [193].Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, Hirschi KK, Chen CS, Nature 2017, 552, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Wong JF, Simmons CA, Lab Chip 2019, 19, 1060. [DOI] [PubMed] [Google Scholar]
- [195].Wong JF, Mohan MD, Young EW, Simmons CA, Biosens. Bioelectron 2020, 147, 111757. [DOI] [PubMed] [Google Scholar]
- [196].Nashimoto Y, Abe M, Fujii R, Taira N, Ida H, Takahashi Y, Ino K, Ramon-Azcon J, Shiku H, Adv. Healthcare Mater 2021, 2101186. [DOI] [PubMed] [Google Scholar]
- [197].Funamoto K, Yoshino D, Matsubara K, Zervantonakis IK, Funamoto K, Nakayama M, Masamune J, Kimura Y, Kamm RD, Integr. Biol 2017, 9, 529. [DOI] [PubMed] [Google Scholar]
- [198].Van Duinen V, Van Den Heuvel A, Trietsch S, Lanz H, Van Gils J, Van Zonneveld A, Vulto P, Hankemeier T, Sci. Rep 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Kondo H, KONDO S, IKEZAWA H, MURATA R, OHSAKA A, Japanese Journal of Medical Science and Biology 1960, 13, 43. [DOI] [PubMed] [Google Scholar]
- [200].Probst C, Schneider S, Loskill P, Cur. Opini. Biomed. Eng 2018, 6, 33. [Google Scholar]
- [201].Friedl J, Puhlmann M, Bartlett DL, Libutti SK, Turner EN, Gnant MF, Alexander HR, Blood, The Journal of the American Society of Hematology 2002, 100, 1334. [PubMed] [Google Scholar]
- [202].Worrall NK, Chang K, Lejeune WS, Misko TP, Sullivan PM, Ferguson TB Jr, Williamson JR, American Journal of Physiology-Heart and Circulatory Physiology 1997, 273, H2565. [DOI] [PubMed] [Google Scholar]
- [203].Mirzapoiazova T, Kolosova I, Usatyuk PV, Natarajan V, Verin AD, American Journal of Physiology-Lung Cellular and Molecular Physiology 2006, 291, L718. [DOI] [PubMed] [Google Scholar]
- [204].Kim Y, Lobatto ME, Kawahara T, Chung BL, Mieszawska AJ, Sanchez-Gaytan BL, Fay F, Senders ML, Calcagno C, Becraft J, Proc. Natl. Acad. Sci 2014, 111, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Park JY, Ryu H, Lee B, Ha D-H, Ahn M, Kim S, Kim JY, Jeon NL, Cho D-W, Biofabrication 2018, 11, 015002. [DOI] [PubMed] [Google Scholar]
- [206].Maoz BM, Herland A, Henry OY, Leineweber WD, Yadid M, Doyle J, Mannix R, Kujala VJ, FitzGerald EA, Parker KK, Lab Chip 2017, 17, 2294. [DOI] [PubMed] [Google Scholar]
- [207].Mirceski V, Gulaboski R, Lovric M, Bogeski I, Kappl R, Hoth M, Electroanalysis 2013, 25, 2411. [Google Scholar]
- [208].Kim HJ, Li H, Collins JJ, Ingber DE, Proc. Natl. Acad. Sci 2016, 113, E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Kaarj K, Yoon J-Y, Micromachines 2019, 10, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Polacheck WJ, Li R, Uzel SG, Kamm RD, Lab Chip 2013, 13, 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Souilhol C, Serbanovic-Canic J, Fragiadaki M, Chico TJ, Ridger V, Roddie H, Evans PC, Nature Reviews Cardiology 2020, 17, 52. [DOI] [PubMed] [Google Scholar]
- [212].Lu D, Kassab GS, Soc JR, Interface 2011, 8, 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Tarbell JM, Cardiovascular research 2010, 87, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Weinbaum S, Tarbell JM, Damiano ER, Annu. Rev. Biomed. Eng 2007, 9, 121. [DOI] [PubMed] [Google Scholar]
- [215].Tarbell JM, Simon SI, Curry F-RE, Annu. Rev. Biomed. Eng 2014, 16, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Malek AM, Alper SL, Izumo S, Jama 1999, 282, 2035. [DOI] [PubMed] [Google Scholar]
- [217].Chiu J-J, Chien S, Physiological reviews 2011, 91, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Nesbitt WS, Westein E, Tovar-Lopez FJ, Tolouei E, Mitchell A, Fu J, Carberry J, Fouras A, Jackson SP, Nat. Med 2009, 15, 665. [DOI] [PubMed] [Google Scholar]
- [219].LaDisa JF, Guler I, Olson LE, Hettrick DA, Kersten JR, Warltier DC, Pagel PS, Ann. Biomed. Eng 2003, 31, 972. [DOI] [PubMed] [Google Scholar]
- [220].Mu X, Kaplan DL, presented at Comsol Conference, Boston: 2019. [Google Scholar]
- [221].Jeong S, Seo J-H, Garud KS, Park SW, Lee M-Y, Biosens. Bioelectron 2021, 183, 113197. [DOI] [PubMed] [Google Scholar]
- [222].Chen H, Yu Z, Bai S, Lu H, Xu D, Chen C, Liu D, Zhu Y, TrAC, Trends Anal. Chem 2019, 117, 186. [Google Scholar]
- [223].Blackman BR, García-Cardeña G, Gimbrone MA Jr, J. Biomech. Eng 2002, 124, 397. [DOI] [PubMed] [Google Scholar]
- [224].Usami S, Chen H-H, Zhao Y, Chien S, Skalak R, Ann. Biomed. Eng 1993, 21, 77. [DOI] [PubMed] [Google Scholar]
- [225].Liu XF, Yu JQ, Dalan R, Liu AQ, Luo KQ, Integr. Biol 2014, 6, 511. [DOI] [PubMed] [Google Scholar]
- [226].Sei YJ, Ahn SI, Virtue T, Kim T, Kim Y, Sci. Rep 2017, 7, 10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Kim S, Lee H, Chung M, Jeon NL, Lab Chip 2013, 13, 1489. [DOI] [PubMed] [Google Scholar]
- [228].Zhang X, Wang Z, Zhang YS, Yan S, Hou C, Gong Y, Qiu J, Chen M, Li Q, Biotechnology and Bioengineering 2021, 118, 963. [DOI] [PubMed] [Google Scholar]
- [229].Totaro A, Panciera T, Piccolo S, Nature Cell Biology 2018, 20, 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Wang K-C, Yeh Y-T, Nguyen P, Limqueco E, Lopez J, Thorossian S, Guan K-L, Li Y-SJ, Chien S, Proc. Natl. Acad. Sci 2016, 113, 11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Panciera T, Azzolin L, Cordenonsi M, Piccolo S, Nat. Rev. Mol. Cell Biol 2017, 18, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Wang L, Luo J-Y, Li B, Tian XY, Chen L-J, Huang Y, Liu J, Deng D, Lau CW, Wan S, Ai D, Mak K-LK, Tong KK, Kwan KM, Wang N, Chiu J-J, Zhu Y, Huang Y, Nature 2016, 540, 579. [DOI] [PubMed] [Google Scholar]
- [233].Doddapattar P, Dhanesha N, Chorawala MR, Tinsman C, Jain M, Nayak MK, Staber JM, Chauhan AK, Arteriosclerosis, thrombosis, and vascular biology 2018, 38, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Cybulsky MI, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos J-C, Connelly PW, Milstone DS, The Journal of clinical investigation 2001, 107, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Lee J, Estlack Z, Somaweera H, Wang X, Lacerda CM, Kim J, Lab Chip 2018, 18, 2946. [DOI] [PubMed] [Google Scholar]
- [236].Farrar EJ, Butcher JT, Ann. Biomed. Eng 2014, 42, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Koupenova M, Clancy L, Corkrey HA, Freedman JE, Circulation research 2018, 122, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Koupenova M, Kehrel BE, Corkrey HA, Freedman JE, European Heart Journal 2016, 38, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Fogelson AL, Neeves KB, Annual review of fluid mechanics 2015, 47, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Xu XR, Carrim N, Neves MAD, McKeown T, Stratton TW, Coelho RMP, Lei X, Chen P, Xu J, Dai X, Thrombosis journal 2016, 14, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Westein E, van der Meer AD, Kuijpers MJ, Frimat J-P, van den Berg A, Heemskerk JW, Proc. Natl. Acad. Sci 2013, 110, 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Jain A, Graveline A, Waterhouse A, Vernet A, Flaumenhaft R, Ingber DE, Nat. Commun 2016, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Mao S, Sarkar A, Wang Y, Song C, LeVine DN, Wang X, Que L, Lab Chip 2021, 21, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Narumiya S, Ishizaki T, Ufhata M, Methods in enzymology 2000, 325, 273. [DOI] [PubMed] [Google Scholar]
- [245].Anwar M, Shalhoub J, Lim C, Gohel M, Davies A, Journal of vascular research 2012, 49, 463. [DOI] [PubMed] [Google Scholar]
- [246].Ribas J, Zhang YS, Pitrez PR, Leijten J, Miscuglio M, Rouwkema J, Dokmeci MR, Nissan X, Ferreira L, Khademhosseini A, Small 2017, 13, 1603737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Jufri NF, Mohamedali A, Avolio A, Baker MS, Vascular cell 2015, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Rosenfeld D, Landau S, Shandalov Y, Raindel N, Freiman A, Shor E, Blinder Y, Vandenburgh HH, Mooney DJ, Levenberg S, Proc. Natl. Acad. Sci 2016, 113, 3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Liu X.-m., Peyton KJ, Durante W, American Journal of Physiology-Heart and Circulatory Physiology 2013, 304, H1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Haga JH, Li Y-SJ, Chien S, Journal of Biomechanics 2007, 40, 947. [DOI] [PubMed] [Google Scholar]
- [251].Kwak BR, Bäck M, Bochaton-Piallat M-L, Caligiuri G, Daemen MJ, Davies PF, Hoefer IE, Holvoet P, Jo H, Krams R, European heart journal 2014, 35, 3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Yung YC, Chae J, Buehler MJ, Hunter CP, Mooney DJ, Proc. Natl. Acad. Sci 2009, 106, 15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Maul TM, Chew DW, Nieponice A, Vorp DA, Biomechanics and modeling in mechanobiology 2011, 10, 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Zhou J, Niklason LE, Integr. Biol 2012, 4, 1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Zhang W, Zhang YS, Bakht SM, Aleman J, Shin SR, Yue K, Sica M, Ribas J, Duchamp M, Ju J, Banan Sadeghian R, Kim D, Dokmeci MR, Atala AK, Ali, Lab Chip 2016, 16, 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Davis CA, Zambrano S, Anumolu P, Allen AC, Sonoqui L, Moreno MR, Journal of biomechanical engineering 2015, 137, 040801. [DOI] [PubMed] [Google Scholar]
- [257].Zeinali S, Thompson EK, Gerhardt H, Geiser T, Guenat OT, APL bioengineering 2021, 5, 026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].van Engeland NC, Pollet AM, den Toonder JM, Bouten CV, Stassen OM, Sahlgren CM, Lab Chip 2018, 18, 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Humayun M, Chow C-W, Young EW, Lab Chip 2018, 18, 1298. [DOI] [PubMed] [Google Scholar]
- [260].Dai Z, Zhu MM, Peng Y, Jin H, Machireddy N, Qian Z, Zhang X, Zhao Y-Y, Am. J. Respir. Crit. Care Med 2018, 198, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Loerakker S, Stassen OM, Ter Huurne FM, Boareto M, Bouten CV, Sahlgren CM, Proc. Natl. Acad. Sci 2018, 115, E3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Jin Z-H, Liu Y-L, Fan W-T, Huang W-H, Small 2020, 16, 1903204. [DOI] [PubMed] [Google Scholar]
- [263].Clarke GA, Hartse BX, Niaraki Asli AE, Taghavimehr M, Hashemi N, Abbasi Shirsavar M, Montazami R, Alimoradi N, Nasirian V, Ouedraogo LJ, Sensors 2021, 21, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Kim S, Kim W, Lim S, Jeon JS, Bioengineering 2017, 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Poventud-Fuentes I, Kwon KW, Seo J, Tomaiuolo M, Stalker TJ, Brass LF, Huh D, Small 2021, 17, 2004889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Arakawa C, Gunnarsson C, Howard C, Bernabeu M, Phong K, Yang E, DeForest CA, Smith JD, Zheng Y, Sci. Adv 2020, 6, eaay7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Wang X-Y, Pei Y, Xie M, Jin Z-H, Xiao Y-S, Wang Y, Zhang L-N, Li Y, Huang W-H, Lab Chip 2015, 15, 1178. [DOI] [PubMed] [Google Scholar]
- [268].Chen MB, Whisler JA, Jeon JS, Kamm RD, Integr. Biol 2013, 5, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD, Proc. Natl. Acad. Sci 2015, 112, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Abudupataer M, Zhu S, Yan S, Xu K, Zhang J, Luo S, Ma W, Alam MF, Tang Y, Huang H, eLife 2021, 10, e69310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Cai S, Li H, Zheng F, Kong F, Dao M, Karniadakis GE, Suresh S, Proc. Natl. Acad. Sci 2021, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Pandian NK, Mannino RG, Lam WA, Jain A, Cur. Opini. Biomed. Eng 2018, 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Mathur T, Singh KA, Pandian NK, Tsai S-H, Hein TW, Gaharwar AK, Flanagan JM, Jain A, Lab Chip 2019, 19, 2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Zheng W, Huang R, Jiang B, Zhao Y, Zhang W, Jiang X, Small 2016, 12, 2022. [DOI] [PubMed] [Google Scholar]
- [275].Venugopal Menon N, Tay HM, Pang KT, Dalan R, Wong SC, Wang X, Li KHH, Hou HW, APL bioengineering 2018, 2, 016103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Zhang M, Wang P, Luo R, Wang Y, Li Z, Guo Y, Yao Y, Li M, Tao T, Chen W, Adv. Sci 2021, 8, 2002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Ramezankhani R, Solhi R, Chai YC, Vosough M, Verfaillie C, Drug Discovery Today 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Valverde MG, Mille LS, Figler KP, Cervantes E, Li VY, Bonventre JV, Masereeuw R, Zhang YS, Nature Reviews Nephrology 2022, 10.1038/s41581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Bonventre JV, Vaidya VS, Schmouder R, Feig P, Dieterle F, Nat. Biotechnol 2010, 28, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Barnett LMA, Cummings BS, Seminars in nephrology 2019, 39, 141. [DOI] [PubMed] [Google Scholar]
- [281].Fathi P, Holland G, Pan D, Esch MB, ACS Applied Bio Materials 2020, 3, 6697. [DOI] [PubMed] [Google Scholar]
- [282].Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung SY, Suh KY, Ingber DE, Integr. Biol 2013, 5, 1119. [DOI] [PubMed] [Google Scholar]
- [283].Wang YI, Abaci HE, Shuler ML, Biotechnology and bioengineering 2017, 114, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Bossink EG, Zakharova M, De Bruijn DS, Odijk M, Segerink LI, Lab Chip 2021, 21, 2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Nguyen D-HT, Lee E, Alimperti S, Norgard RJ, Wong A, Lee JJ-K, Eyckmans J, Stanger BZ, Chen CS, Sci. Adv 2019, 5, eaav6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Maharjan S, Bonilla D, Zhang YS, Cur. Opini. Biomed. Eng 2022, 21, 100362. [Google Scholar]
- [287].Blanchard KC, Maroske D, May DG, Weiner IM, J Pharmacol Exp Ther February 1972, 180, 397. [PubMed] [Google Scholar]
- [288].Lippa S, Colacicco L, Bondanini F, Callà C, Gozzo ML, Ciccariello M, Angelitti AG, Clinica Chimica Acta 2000, 292, 81. [DOI] [PubMed] [Google Scholar]
- [289].Gillette BM, Jensen JA, Tang BX, Yang GJ, Bazargan-Lari A, Zhong M, Sia SK, Nat. Mater 2008, 7, 636. [DOI] [PubMed] [Google Scholar]
- [290].Liu X, Wang Q, Qin JH, Lin BC, Lab Chip 2009, 9, 1200. [DOI] [PubMed] [Google Scholar]
- [291].Price GM, Chu KK, Truslow JG, Tang-Schomer MD, Golden AP, Mertz J, Tien J, Journal of the American Chemical Society 2008, 130, 6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Ling Y, Rubin J, Deng Y, Huang C, Demirci U, Karp JM, Khademhosseini A, Lab Chip 2007, 7, 756. [DOI] [PubMed] [Google Scholar]
- [293].Rayner SG, Phong KT, Xue J, Lih D, Shankland SJ, Kelly EJ, Himmelfarb J, Zheng Y, Adv. Healthcare Mater 2018, 7, 1801120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Birn H, Christensen EI, Kidney International 2006, 69, 440. [DOI] [PubMed] [Google Scholar]
- [295].Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA, Sci. Rep 2016, 6, 34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proc. Natl. Acad. Sci 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Nat. Methods 2019, 16, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Schieppati A, Henter J-I, Daina E, Aperia A, The Lancet 2008, 371, 2039. [DOI] [PubMed] [Google Scholar]
- [299].Gordon LB, Rothman FG, López-Otín C, Misteli T, Cell 2014, 156, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Manfredi C, Tindall JM, Hong JS, Sorscher EJ, Science 2019, 365, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Varga R, Eriksson M, Erdos MR, Olive M, Harten I, Kolodgie F, Capell BC, Cheng J, Faddah D, Perkins S, Proc. Natl. Acad. Sci 2006, 103, 3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong Z-M, Tavarez UL, Davison LM, Gete YG, Mao X, Newby GA, Doherty SP, Narisu N, Sheng Q, Krilow C, Lin CY, Gordon LB, Cao K, Collins FS, Brown JD, Liu DR, Nature 2021, 589, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Erdos MR, Cabral WA, Tavarez UL, Cao K, Gvozdenovic-Jeremic J, Narisu N, Zerfas PM, Crumley S, Boku Y, Hanson G, Nat. Med 2021, 27, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Blumenrath SH, Lee BY, Low L, Prithviraj R, Tagle D, Experimental Biology and Medicine 2020, 245, 1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].de Mello CPP, Rumsey J, Slaughter V, Hickman JJ, Drug Discovery Today 2019, 24, 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Low LA, Tagle DA, Rare Diseases Epidemiology: Update and Overview 2017, 405. [Google Scholar]
- [307].Low LA, Tagle DA, Expert opinion on orphan drugs 2016, 4, 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Durmowicz AG, Lim R, Rogers H, Rosebraugh CJ, Chowdhury BA, Annals of the American Thoracic Society 2018, 15, 1. [DOI] [PubMed] [Google Scholar]
- [309].Liu G-H, Barkho BZ, Ruiz S, Diep D, Qu J, Yang S-L, Panopoulos AD, Suzuki K, Kurian L, Walsh C, Nature 2011, 472, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Atchison L, Zhang H, Cao K, Truskey GA, Sci. Rep 2017, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Science 2015, 349, 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Nature 2003, 423, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Gordon LB, Brown WT, Collins FS, in GeneReviews® [Internet], University of Washington, Seattle: 2019. [Google Scholar]
- [314].Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Engl N. J. Med 2008, 358, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Havelka GE, Kibbe MR, Vascular and endovascular surgery 2011, 45, 381. [DOI] [PubMed] [Google Scholar]
- [316].Laurent S, Boutouyrie P, Asmar R, Gautier I, Laloux B, Guize L, Ducimetiere P, Benetos A, Hypertension 2001, 37, 1236. [DOI] [PubMed] [Google Scholar]
- [317].Kim H-L, Kim S-H, Frontiers in cardiovascular medicine 2019, 6, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, Proc. Natl. Acad. Sci 2012, 109, 16666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Gordon LB, Kleinman ME, Massaro J, D’Agostino RB Sr, Shappell H, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland RH, Nazarian A, Circulation 2016, 134, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Corretti MC, Anderson TJ, Benjamin EJ, Celermajer D, Charbonneau F, Creager MA, Deanfield J, Drexler H, Gerhard-Herman M, Herrington D, Journal of the American College of Cardiology 2002, 39, 257. [DOI] [PubMed] [Google Scholar]
- [321].Redheuil A, Yu W-C, Wu CO, Mousseaux E, De Cesare A, Yan R, Kachenoura N, Bluemke D, Lima JA, Hypertension 2010, 55, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Tabel GM, Whittaker P, Vlachonassios K, Sonawala M, Chandraratna PA, Echocardiography: A Journal of Cardiovascular Ultrasound and Allied Techniques 2006, 23, 103. [DOI] [PubMed] [Google Scholar]
- [323].Verstraeten VL, Ji JY, Cummings KS, Lee RT, Lammerding J, Aging cell 2008, 7, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Cau P, Navarro C, Harhouri K, Roll P, Sigaudy S, Kaspi E, Perrin S, De Sandre-Giovannoli A, Lévy N, Seminars in cell & developmental biology 2014, 29, 125. [DOI] [PubMed] [Google Scholar]
- [325].Wolf D, Ley K, Circulation research 2019, 124, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Libby P, Ridker PM, Maseri A, Circulation 2002, 105, 1135. [DOI] [PubMed] [Google Scholar]
- [327].Hahn C, Schwartz MA, Nat. Rev. Mol. Cell Biol 2009, 10, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Lusis AJ, Nature 2000, 407, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Weber C, Noels H, Nat. Med 2011, 17, 1410. [DOI] [PubMed] [Google Scholar]
- [330].Getz GS, Reardon CA, Arteriosclerosis, thrombosis, and vascular biology 2012, 32, 1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Bray SJ, Nat. Rev. Mol. Cell Biol 2016, 17, 722. [DOI] [PubMed] [Google Scholar]
- [332].Coon BG, Baeyens N, Han J, Budatha M, Ross TD, Fang JS, Yun S, Thomas J-L, Schwartz MA, Journal of Cell Biology 2015, 208, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Estrada R, Giridharan GA, Nguyen M-D, Prabhu SD, Sethu P, Biomicrofluidics 2011, 5, 032006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Estrada R, Giridharan GA, Nguyen M-D, Roussel TJ, Shakeri M, Parichehreh V, Prabhu SD, Sethu P, Analytical chemistry 2011, 83, 3170. [DOI] [PubMed] [Google Scholar]
- [335].Zhao P, Yao Q, Zhang P-J, The E, Zhai Y, Ao L, Jarrett MJ, Dinarello CA, Fullerton DA, Meng X, Sci. Adv 2021, 7, eabg1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [336].Su C, Menon NV, Xu X, Teo YR, Cao H, Dalan R, Tay CY, Hou HW, Lab Chip 2021, 21, 2359. [DOI] [PubMed] [Google Scholar]
- [337].Vaccaro CA, Brody JS, Journal of Cell Biology 1981, 91, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Welling LW, Zupka MT, Welling DJ, Physiology 1995, 10, 30. [Google Scholar]
- [339].Grant J, Özkan A, Oh C, Mahajan G, Prantil-Baun R, Ingber DE, Lab Chip 2021, 21, 3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Toepke MW, Beebe DJ, Lab Chip 2006, 6, 1484. [DOI] [PubMed] [Google Scholar]
- [341].Mu X, Yuen JS, Choi J, Zhang Y, Cebe P, Jiang X, Zhang YS, Kaplan DL, Proc. Natl. Acad. Sci 2022, 119, e2115523119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Wang M, Li W, Hao J, Gonzales A, Zhao Z, Flores RS, Kuang X, Mu X, Ching T, Tang G, Nat. Commun 2022, 13, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Pei Y, Jordan KE, Xiang N, Parker RN, Mu X, Zhang L, Feng Z, Chen Y, Li C, Guo C, ACS Appl. Mater. Interfaces 2021, 13, 3186. [DOI] [PubMed] [Google Scholar]
- [344].Yang X, Li K, Zhang X, Liu C, Guo B, Wen W, Gao X, Lab Chip 2018, 18, 486. [DOI] [PubMed] [Google Scholar]
- [345].Zamprogno P, Wüthrich S, Achenbach S, Thoma G, Stucki JD, Hobi N, Schneider-Daum N, Lehr C-M, Huwer H, Geiser T, Communications biology 2021, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Gu M, Current protocols in human genetics 2018, 98, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Williams IM, Wu JC, Arteriosclerosis, thrombosis, and vascular biology 2019, 39, 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [348].Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O’Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, Nature cell biology 2015, 17, 994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Cheung C, Bernardo AS, Pedersen RA, Sinha S, Nat. Protoc 2014, 9, 929. [DOI] [PubMed] [Google Scholar]
- [350].Halaidych V, Cochrane A, van den Hil FE, Mummery CL, Orlova VV, Stem Cell Reports 2019, 12, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Cheung C, Bernardo AS, Trotter MW, Pedersen RA, Sinha S, Nat. Biotechnol 2012, 30, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Aboab J, Celi LA, Charlton P, Feng M, Ghassemi M, Marshall DC, Mayaud L, Naumann T, McCague N, Paik KE, Sci. Transl. Med 2016, 8, 333ps8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Fong EL, Watson BM, Kasper FK, Mikos AG, Adv. Mater 2012, 24, 4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [354].Chandra A, Journal of Vascular Surgery 2013, 57, 576. [DOI] [PubMed] [Google Scholar]
- [355].LaBarbera M, Science 1990, 249, 992. [DOI] [PubMed] [Google Scholar]
- [356].Papaioannou TG, Stefanadis C, Hellenic J Cardiol 2005, 46, 9. [PubMed] [Google Scholar]


