Abstract
Treatment of chronic prostatitis/chronic pelvic pain syndrome is often empirical because clinical culture methods fail to detect prostate-associated pathogens in >90% of patients. Previously, we tested a variety of specific-microorganism PCRs and began a DNA sequence study after we found that 77% of prostatitis patients were PCR positive for prokaryotic rRNA-encoding DNA sequences (rDNAs) despite negative cultures using optimal techniques. In the present study, 36 rDNA clones from 23 rDNA-positive patients were sequenced. This study represents more than twice the total rDNA sequence and more than twice the number of patients in the previous study. The increased number of patients and clones sequenced allowed enhanced phylogenetic analyses and refinements in our view of rDNA species inhabiting the prostate. A continuum of related rDNAs that might be arbitrarily described as two major groups of rDNAs and several minor groups was found. Sequences termed Pros A, identified in 8 (35%) of 23 rDNA-positive patients, grouped with Aeromonas spp. in phylogenetic studies. Sequences termed Pros B, identified in 17 (74%) of 23 rDNA-positive patients, were distinct from previously reported sequences, although all were >90% similar to known gram-negative bacteria. Of the nine patients for whom multiple rDNAs were sequenced, six had biopsy specimens containing rDNAs from more than one species. Four (17%) patients had rDNAs different from those of the Pros A and Pros B groups. Of these four, one patient had rDNA similar to that of Flavobacterium spp., another had rDNA similar to that of Pseudomonas testosteroni, and two patients had rDNAs <70% similar to known rDNAs. These findings suggest that the prostate can harbor bacteria undetectable by traditional approaches. Most of these diverse sequences are not reported in environments outside the prostate. The sequence similarities suggest adaptation of limited groups of bacteria to the microenvironment of the prostate. Further studies may elucidate the relationship of prostate-associated bacteria to chronic prostatitis/chronic pelvic pain syndrome.
Approximately half of all men suffer from prostatitis syndromes at some time in their lives (24). Patients characteristically experience perineal, suprapubic, genital, and ejaculatory pains; voiding; or sexual dysfunction (15). Many relate the onset of their condition to sexual activity, commonly to an episode of acute urethritis (4). Antimicrobial treatment is often transiently effective in relieving symptoms, but efforts to identify pathogens have been successful only in small groups of patients (13, 19, 24). Bacterial prostatitis that can be diagnosed by standard, clinical culture methods occurs in fewer than 10% of patients (15). The new National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases) consensus classification puts patients in the chronic prostatitis/chronic pelvic pain category if they cannot be diagnosed by traditional culture methods.
Chlamydia trachomatis and ureaplasmas were implicated in ≤20% of men with prostatitis syndromes (5–7, 29). Trichomonas vaginalis, genital mycoplasmas, herpesviruses 1 and 2, and cytomegalovirus have also been implicated (7, 9, 18, 27–29). These organisms have been identified by culture or antigen detection in samples obtained from sites distal to the prostate, including urine, urethral swabs, and expressed prostatic secretions.
Many microorganisms are unculturable under the most refined conditions (1). The limitations of standard clinical laboratories are undoubtedly greater (1) because to culture an organism its ecological requirements must be duplicated. Previous studies have shown that most environmental bacteria do not multiply on conventional media (10, 25, 30). Although a higher percentage of bacteria infecting or inhabiting human tissues is known, even this source contains a significant number of uncharacterized species (30).
One approach to identification of culture-resistant bacteria is to amplify and sequence their rRNA-encoding DNA sequences (rDNAs). rDNAs lend themselves to this purpose because various regions of the rDNA gene sequence have unequal rates of evolutionary change (11). In a previous study, we detected C. trachomatis, T. vaginalis, or Mycoplasma genitalium in prostate biopsy specimens of 8% of 135 men with chronic prostatitis/chronic pelvic pain syndrome. In contrast, 77% of the same patient population had bacterial rDNAs (16). Patients were selected carefully, based on negative clinical evaluations and negative urethral, urine, and expressed prostatic secretion cultures for recognized urogenital pathogens.
The present study extends these observations by much more extensive DNA sequencing, database comparisons, and phylogenetic studies of larger numbers of cloned DNA sequences, allowing a significantly clearer profile of rDNAs in prostate tissue. The previous study involved an initial sequence investigation involving 10 patients and a total of 6 kb of rDNA sequence consisting of complete and partial sequences of rDNA clones. The present study involves 23 patients and 36 completely sequenced rDNA clones from prostate biopsy specimens (34 clones) and skin tissue at the site of biopsy (2 clones). The advantage of this approach is that positives obtained from very broad-spectrum PCR assays can yield precise molecular information that may be used to narrow down and, in some instances, identify bacterial species found in the prostate. This information is important for understanding the microbial ecology of the prostate and the potential role of such organisms in chronic prostatitis and related disorders.
MATERIALS AND METHODS
Patient population and clinical evaluation.
Subjects over 18 years old were recruited from a special Prostatitis Clinic at the University of Washington Medical Center. Potential subjects were offered a standardized evaluation for infectious, inflammatory, structural, and functional conditions associated with prostatitis. Patients who had taken antibiotics within 6 weeks or who had documentation of bacteriuria were excluded. We obtained specimens for urethral Gram stain and cultures for Neisseria gonorrhoeae, C. trachomatis, genital mycoplasmas, and T. vaginalis (3, 14). Most patients visited the clinic four times for this study. As previously described (16), patients underwent lower urinary tract localization cultures, expressed prostatic secretion leukocyte count determination, uroflow studies, urethral cultures, seminal fluid analysis, and prostatic biopsies with control skin swabs or biopsies at the site of entry.
Of 260 men who met the entry criteria, 135 had urethral urine and expressed prostatic secretion evaluations and agreed to prostate biopsy. Of these 135 men, 17 rDNA-positive patients with the highest number of leukocytes in their expressed prostate secretions or in seminal fluid were selected for DNA sequencing. Of these 17 men, 13 were positive for inflammation in expressed prostate secretions and 4 were positive for seminal fluid inflammation while having either negative or undetermined inflammation in expressed prostate secretions. As a pilot control group (see Discussion), four men were selected because they had 0 leukocytes in their prostate secretions but were nevertheless positive for rDNA. Of the 4 patients who lacked leukocytes in prostatic secretions, one was later found to have elevated levels of seminal fluid leukocytes, defined as >104 leukocytes/ml and so was also included in the group of 17 patients with inflammation. Two of these four patients had undetermined seminal fluid inflammation, and the fourth patient was negative for seminal fluid inflammation. Cloned rDNAs were also sequenced from three additional patients whose inflammation status was undetermined for both expressed prostate secretions and seminal fluid. A total of 23 patients had their rDNAs sequenced. Biopsy tissue preparation and anticontamination procedures, including use of a perineal approach and a double-needle method to limit potential contamination, were as previously described (16).
Environmental and patient controls.
Our sampling protocol includes numerous control swabs of the procedure room, equipment, and clinical solutions evaluated by rDNA PCR to detect contamination (16). PCR reagents were treated with UV light to inactivate DNA contaminants. Skin biopsy specimens were taken from patients undergoing prostate biopsy, and positives were characterized by DNA sequencing.
Bacterial cultures.
Biopsy specimens were placed on sterile gauze moistened with saline and transported to the culture laboratory immediately. Upon receipt, the specimen was placed into a glass Dounce homogenizer with phosphate-buffered saline. The tissue was ground, and the solution was then inoculated onto culture medium for aerobic (Columbia sheep blood agar; PML Microbiologicals, Wilsonville, Conn.) and anaerobic (brucella agar; University of Washington Clinical Laboratories) bacteria as well as genital mycoplasmas (A7b; University of Washington Clinical Laboratories). One hundred microliters was inoculated on each plate as well as into broths used to detect low quantities of bacteria. Slides were prepared for Gram staining. Aerobe plates were incubated at 37°C in a 5% CO2 incubator. Anaerobic plates were incubated at 37°C in a Bactron anaerobic environmental chamber (Sheldon Manufacturing, Cornelius, Oreg.).
PCR amplification, cloning, and sequencing.
We PCR amplified rDNAs whose sequences constitute molecular signatures of prokaryotes (16, 22). The 475-bp PCR products, amplified with primers 91E and 13B, were cloned into the PCR II vector (Stratagene, Inc.; LaJolla, Calif.) and then sequenced with either Sequenase (United States Biochemical; Cleveland, Ohio) or Cycle sequencing (Perkin-Elmer Cetus, Norwalk, Conn.) as described previously (16).
To determine matches or similarities with known sequences, rDNAs homology searches were conducted with both the GenBank and EMBL databases on CD-ROM by using the Genepro (Bainbridge Island, Wash.) program. Searches were also done by using the BLAST feature at EMBL (16). When homology searches revealed >85% similarity to a known rDNA sequence, pairwise sequence alignments were performed, comparing prostatic rDNA with the similar sequence(s). Subsequently, multiple sequence alignments were performed with 5 to 14 prostatic rDNAs at a time and known rDNAs. Multiple sequence alignments were used as input into phylogenetic inference programs.
Sequence phylogenies.
Phylogenetic trees were constructed by using the neighbor-joining program of Clustal V. Positions with sequence gaps were excluded, and 1,000 bootstrap replicates were analyzed for each phylogenetic tree. In separate studies, the programs Seqboot, Fitch, Neighbor, and Consense (Phylip Phylogenetic Inference Package [8]) were used, in that order, to infer bootstrapped phylogenetic relationships. Phylogenies were computed with and without the removal of gaps and ambiguously aligned sequences. Randomized data inputs (J option) were employed. In independent studies, the program DNAML was used to infer unrooted phylogenies.
RESULTS
Cloned prostate PCR product sequences and database searches.
After verification by two investigators, sequences were used to search an rDNA sublibrary derived from GenBank and EMBL sequences. Homology searches revealed few prostate rDNA sequence matches with known rDNAs. There was recurring similarity (>85% identical nucleotides) with a limited number of gram-negative organisms including Proteus vulgaris, Escherichia coli, Vibrio furnisii, and a variety of Aeromonas spp. (Table 1) as well as two patients with novel sequences, defined as rDNAs <70% similar to any DNA sequences in GenBank. While consistent with our earlier results (16), the present study revealed greater diversity of rDNA sequences.
TABLE 1.
rDNA clones from idiopathic-prostatitis patients
| Patient | Inflammationa
|
Cloneb | 16S rDNA (% similarity)c | |
|---|---|---|---|---|
| Expressed prostatic secretion | Seminal fluid | |||
| 1521 | E2 | Pros A (95) | ||
| E4 | Pros B (89) | |||
| 0236 | + | + | A3 | Novel |
| B2 | Pros B (95) | |||
| B3 | Pros B (94) | |||
| B4 | Pros B (96) | |||
| B5 | Novel | |||
| 7576 | + | + | F3 | Pros A (97) |
| 6550 | 5725d | Pros B (96) | ||
| 5735d | Pros A (98) | |||
| 5744e | S. epidermidis (98)f | |||
| 6527 | 5751 | Pros B (97) | ||
| 5752 | Pros B (97) | |||
| 5754e | B. subtilis (97) | |||
| 5682 | + | 9035 | Pros B (95) | |
| 9036 | Pros B (95) | |||
| 2512 | + | 9041 | Pros A (100) | |
| 9046 | Pros B (97) | |||
| 9269 | + | + | 9051 | Pros B (97) |
| 9052 | Pros A (97) | |||
| 2652 | + | 9061 | Pros B (95) | |
| 9068 | Pros B (95) | |||
| 0537 | − | 9075 | Pros B (95) | |
| 9720 | + | 90123 | Novel | |
| 9632 | + | + | 90211 | Pros B (95) |
| 90212 | Pros B (95) | |||
| 2586 | + | 9251 | Pros A (100) | |
| 3263 | Pros B (98) | |||
| 5751 | + | 9284 | Flavobacterium (95) | |
| 9532 | + | 3251 | Pros A (100) | |
| 5732 | − | 3272 | Pros B (92) | |
| 5632 | − | + | 3284 | P. testosteroni (93) |
| 5747 | + | 4114 | Pros B (95) | |
| 5300 | + | 4122 | Pros B (96) | |
| 1348 | + | 4131 | Pros B (96) | |
| 0211 | + | + | 32312 | Pros B (94) |
| 9393 | + | 3243 | Pros A (99) | |
| 0355 | − | − | 4143 | Pros B (95) |
Analyzed by leukocyte counts and by histology. +, >1 × 104 leukocytes/ml.
16S rDNA PCR products were cloned into the plasmid PCR II. Clones are listed sequentially; multiple clones were sequenced from some patients.
Pros A and Pros B are groups of rDNA sequence species closely related to one another as determined by phylogenetic analysis. Pros A includes members with 100% similarity to A. hydrophila rDNA within the region of 16S rDNA spanned by primers 91E13B; percent similarity to A. hydrophila is indicated in parentheses. Members of the Pros B group were not identical to known 16S rDNA sequences, although all had >90% similarity to 16S rDNAs of known gram-negative organisms; percent similarity to E. coli is indicated in parentheses. 16S rDNA sequences with <70% similarity to any previously reported rDNA in GenBank and EMBL were categorized as novel; these sequences were also unrelated to one another phylogenetically.
In reference 16, clones 5725 and 5735 were incorrectly referred to as coming from separate patients; they are from the same patient.
Isolated from skin.
Also 97% similar to S. aureus. The sequence of this clone was published previously (16).
Of 23 patients whose cloned prostate rDNAs were selected for sequencing, 8 (35%) had rDNAs related to those of gram-negative bacteria of the genus Aeromonas (23). Three separate patients had rDNAs 100% similar to that of Aeromonas hydrophila, a freshwater species that is known to cause gastroenteritis, septicemia, and flesh wound infections (2, 20, 26). Four other patients had rDNAs ≥95% similar to that of A. hydrophila. Since A. hydrophila inhabits freshwater, additional controls sampling all aqueous solutions in the procedure rooms and PCR laboratory were performed, with negative findings of A. hydrophila contamination. Despite the similarities, there were clear sequence differences among these rDNA clones, suggesting a family of closely related but diverse microorganisms (see Fig. 1 and 3). This family of sequences was arbitrarily termed Pros A, defined in terms of percent similarity to A. hydrophila, the species with the strongest rDNA similarity to this group of patient sequences (Table 1; Fig. 1 to 3). The rDNA for A. hydrophila was entered in the GenBank database since our previous report. Similarity of several of our prostate rDNAs to this species contributed to our decision to make A. hydrophila, rather than Vibrio spp., the comparison standard for the Pros A group (see Discussion).
FIG. 1.
Clustal V multiple sequence alignment and contrast map of six cloned 16S rDNA PCR products from six prostatitis patients. Ahydro, A. hydrophila. Differences are highlighted. Gap penalties were set at 10 (fixed) and 10 (floating). The weighted option of the transition toggle was selected. I5735, previously reported DNA sequence of clone INV5735 (16).
FIG. 3.
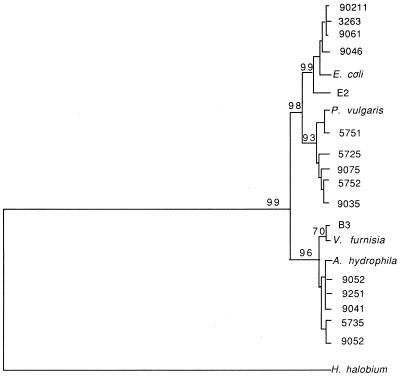
Unrooted dendrogram using the Phylip phylogenetic inference program. Sequences aligned by Clustal V were entered into the Seqboot program of Phylip to generate 1,000 bootstrap replicates consisting of 1,000 sequence alignments each with randomly selected portions of the sequences rendered inactive. The numbers to the left of the nodes indicate the percentage of resulting phylogenetic trees with the nodal topologies shown. These topologies also agreed with the best tree found by using the maximum likelihood program DNAML. H. holobium was entered as the outgroup species, although the phylogeny is unrooted. Horizontal line lengths are proportional to inferred evolutionary distances. Sequences identified by species name are 16S rRNA sequences referenced in GenBank); other entries represent patient prostatic sequences.
Of the 23 patients, 17 (74%) had rDNAs that were >94% similar to either P. vulgaris or E. coli, two gram-negative bacteria that, although members of distinct genera, are strongly similar (93%) in the region of rDNA spanned by primers 91E and 13B. This group of rDNAs was termed Pros B, defined in terms of percent similarity to E. coli (Table 1). In one case (clone 5751), P. vulgaris was a closer match in pairwise alignments. In a two-by-two matrix comparing 21 cloned Pros B type sequences with one another, the group was heterogeneous, with a range of similarity of one Pros B sequence to another of 89 to 98% (not shown). Conceivably, sequences more similar to P. vulgaris than E. coli could be categorized separately. For simplicity, we limited groupings to Pros A and Pros B, with the caveat that subgroupings within both Pros A and Pros B are possible. Also, some members of the Pros A group are as much as 86% similar to Pros B sequences. This reflects the fact that the phylogenetic analysis relies on more than simple homologies such that similar sequences based on homology (clones 5725 and 5735) are not always found in the same phylogenetic group.
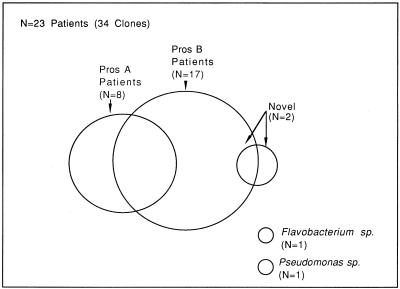
Some rDNAs clearly fell outside the groupings defined by Pros A and Pros B. Three rDNA clones, from two patients, exhibited sequences with low similarity (<70%) to any previously reported rDNAs. These rDNAs were classified as novel. One patient had sequences 95% similar to rDNA of Flavobacterium spp. with no other similarities to known sequences, and another patient had sequences 93% similar to that of Pseudomonas testosteroni without other similarities. Neither of the latter-patient rDNAs could be classified as Pros A or Pros B. The rDNA groups found in prostatitis patients are illustrated in Fig. 2.
FIG. 2.
Venn diagram illustrating the major and minor groups of rDNAs found in prostate biopsy specimens of prostatitis patients. Of the patients whose rDNAs were sequenced, 35% had Pros A type sequences, 74% had Pros B type sequences, and 22% had both Pros A and Pros B sequences (overlap). One patient (4%) had both Pros B and another sequence classified as novel due to lack of similarity with database entries. One patient had rDNA similar to that of Pseudomonas spp., and one patient had rDNA similar to that of Flavobacterium spp., neither of which could be classified as Pros A or Pros B. Two clones from skin at the site of biopsy are not included in the diagram.
Of the 23 prostate biopsy specimens evaluated, 6 had rDNAs of more than one type, including Pros A plus Pros B in 5 patients, and Pros B plus two separate, novel rDNAs in another patient. Although 17 (74%) of the biopsy specimens had a single type of rDNA, this number may be skewed due to the fact that a single rDNA clone was sequenced from a number of these specimens.
Analysis of E. coli rDNAs from men with acute urinary tract infections.
Since few perfect matches with GenBank database entries were found, we were concerned that natural genetic variation in gram-negative organisms such as P. vulgaris and E. coli might obscure the identities of such microorganisms when rDNAs isolated from individual patients were sequenced. In theory, genetic drift of a common known species could lead to similarities of <100% with the database entries. To help assess genetic drift, cloned rDNA PCR products were sequenced from two urine cultures (from a separate population of men with acute urinary tract infection) identified as E. coli at the University of Washington Clinical Microbiology Laboratories (17). These sequences were 100% similar to the GenBank E. coli sequence, indicating that urogenital tract E. coli, identical with the database entry, can easily be found, although such E. coli rDNA was absent in all 36 prostate rDNA clones. This does not rule out the possibility that some members of the Pros B group may be close relatives of E. coli.
Comparison of Pros A and Pros B group standards.
Pairwise sequence alignments indicated that A. hydrophila, the standard for our Pros A group had rDNA 87% similar to E. coli rDNA, the standard for our Pros B group. Thus, the Pros A and Pros B groupings are somewhat arbitrary but useful working classifications that indicate which type of rDNA a particular clone is most similar to. In extensive database homology searches with each prostatic rDNA compared with all recorded rDNAs, A. hydrophila and E. coli alternated as sources of rDNAs most similar to the majority of prostatic rDNAs, further supporting the use of these standards for comparison.
Phylogenetic studies.
The neighbor-joining method of phylogenetic inference involving 1,000 bootstrap replicates suggested the prostatic rDNA sequence groupings shown in Fig. 3. Inferred evolutionary distances are proportional to the horizontal lines of the dendrogram of Fig. 3. As shown, the majority of prostatic rDNAs appeared closely interrelated. In contrast, the selected outgroup species, Halobacterium halobium is a clearly unrelated prokaryote, as are many other known prokaryotic species. Phylogenies inferred by maximum likelihood, using the DNAML program, had similar topologies. As was suspected based on two-by-two sequence comparisons, two or possibly three (if Pros B sequences are split into two categories) groups of closely related sequences were inferred. Pros B sequences appear at the top of Fig. 3, while Pros A sequences appear at the bottom, from clone B3COM through 9052. The novel sequences were not included in Fig. 3.
Some individual patients shared nearly identical rDNA sequences. For example, clones 90211, 3263, and 9061 (top of Fig. 3), from different patients, were indistinguishable in phylogenetic studies, yet these sequences were divergent from other patient sequences and from E. coli, the closest known relative of this group. Sequences that are indistinguishable in the dendrogram are likely to be very similar but not necessarily identical in the pairwise alignments in Table 1. Clones 9251 and 9041, from different patients, were almost indistinguishable in phylogenetic studies and 99 and 100% similar, respectively, to A. hydrophila in pairwise alignments. Twenty cloned sequences not shown in Fig. 3 grouped with either Pros A type sequences (3 clones), Pros B type sequences (10 clones), or novel sequences (3 clones) or were similar to assorted bacteria that were implicated only once each in this study (e.g., Pseudomonas testosteroni, Flavobacterium spp., and the skin-associated clones).
Culture studies.
The majority of patients had parallel biopsy specimens sent to PCR and culture laboratories. The PCR and culture laboratories were blinded to one another’s results. The culture laboratory was instructed to perform routine clinical culture analysis of the samples. Of the 23 patients that were positive for bacterial rDNA PCR, 14 (61%) were scored as “no growth” by the culture laboratory. Of the same 23 patients, 4 were not processed for culture and 5 were both PCR and culture positive for bacteria. Four of the five patients that were both PCR and culture positive had multiple bacterial species by culture. Bacterial species identified in these four patients included Diphtheroides sp., E. coli, and coagulase-negative Staphylococcus spp.
DISCUSSION
Since our patient population was selected to be negative for microorganisms by traditional and advanced culture techniques (16), molecular methods were used document or refute that these men might harbor microorganisms in their prostate glands. We previously showed that there was a strong correlation between inflammation in expressed prostatic secretions and detection of rDNAs in prostatic tissue (16). This study extends and expands the earlier study that included fewer and, in some cases, partial DNA sequences. The initial study represented mostly specific PCRs and a beginning of our rDNA sequence investigation consisting of 6 kb of partial and some complete rDNAs sequenced from 10 patients. The present study represents 23 patients and 36 separate clones of PCR products. The present study involves more than twice the number of patients of the previous study, much more extensive phylogenetic analysis, and confirmation that rDNAs of the Vibrio family, specifically Aeromonas spp., appear in multiple patients.
The 36 clones were obtained from 23 patients—13 with inflammation in their expressed prostatic secretions; 4 without inflammation in expressed prostatic secretions; and 6 whose expressed prostatic secretion status was undetermined, although 3 had seminal fluid inflammation (see Materials and Methods). Although we reported a correlation between inflammation and rDNA positivity in the previous study, we chose four rDNA-positive, expressed prostatic secretion- and inflammation-negative biopsy specimens for comparison with rDNA-positive, inflammation-positive men in the present study. The four patients with rDNA-positive but inflammation-negative expressed prostatic secretions exhibited either Pros B rDNA or Pseudomonas testosteroni rDNA, but none exhibited Pros A rDNAs. Further studies will be required to determine if this is a consistent finding.
Although we chose prostatitis patients who had no inflammation in their expressed prostatic secretions as a control group for patients with inflammation, we have also explored several other potential control groups. For example, a recently obtained sample of prostate tissue removed during radical prostatectomy for prostate cancer was subjected to 16S rDNA PCR and was found to be positive for rDNA. The meaning of this is uncertain since it is possible that infectious microorganisms might contribute to the etiology of prostate cancer. Prostate cancer patients have a median age 20 years more than that of our prostatitis group. Benign prostatic hypertrophy patients also tend to be considerably older than prostatitis patients, and microorganism involvement is possible in this population as well. In selecting a control population, we were also concerned that prostatitis and the presence of 16S rDNA might be unrecognized risk factors for the development of benign prostatic hypertrophy or of prostate cancer in later decades.
In another pilot control study, we evaluated paid volunteer control subjects with no reported prostatic symptoms. Biopsy specimens from volunteers had a substantially lower rate of 16S rDNA positives than those from the prostatitis patients (data not shown). However, preliminary studies indicate that even in the volunteer group, 16S rDNA-positive PCRs that were found were associated with inflammation in prostatic secretions. Histologically, it is possible that some of the 16S rDNA-positive volunteers may be considered to have prostatitis although they lack pain symptoms. Thus, it is unclear at this time whether 16S rDNA that may be associated with prostatic inflammation is unique to patients with chronic prostatitis-associated pain, the focus of the present study. It is possible that 16S rDNA is more associated with inflammation than with pain, but this remains to be established. Although it is impossible to determine cause and effect by documenting microorganism DNA alone, our studies suggest that precise molecular techniques may prove fruitful for investigating the etiology of chronic prostatitis syndromes.
The group of prostatic rDNA sequences we termed Pros A includes members identical to the rDNA of A. hydrophila, gram-negative rods of the family Vibrionaceae. In the previous study, members of the Vibrio family were also implicated (16). Since A. hydrophila inhabits freshwater, additional PCR controls sampling all aqueous solutions in operating and PCR areas were performed. No evidence of bacterial DNA was found in these solutions in three separate control investigations performed at three different times during this study.
A. hydrophila is known to cause wound infections, eye infections, lung abscesses, and diarrhea (2, 20, 26). A. hydrophila is not a common commensal organism. Two previous studies of large population samples showed that Aeromonas spp. most likely do not inhabit the healthy human body. Specifically, Aeromonas spp. were isolated from 1.4% of subjects with diarrhea (n, 978) in one study (12) and 1.9% of subjects (n, 13,027) in another study (21). In 343 asymptomatic subjects in the latter study, no Aeromonas sp. was found. Thus, the microorganism was found infrequently in asymptomatic patients.
In the present study of prostate biopsy specimens, 3 (13%) of 23 patients had Pros A rDNAs identical to that of A. hydrophila, and 5 additional patients had rDNAs ≥95% similar to that of A. hydrophila (Fig. 1). The 5% sequence variation within the Pros A group is consistent with more than one species of Aeromonas based on the low amount of sequence variation (<5%) in comparisons of A. hydrophila and A. allosaccharophila with six other Aeromonas spp. (not shown) in the GenBank database. Based on these observations, the Pros A sequence variants in our patients with chronic prostatitis may represent variants within a single species such as A. hydrophila or separate but closely related species. We are currently unable to distinguish between these possibilities. None of the four patients lacking inflammation in their expressed prostatic secretions had Aeromonas sp. rDNAs by our techniques. Sequencing of larger numbers of samples from men with and without inflammation will be needed to determine if this is a consistent finding.
Observation of 100% similarity of rDNAs with that of A. hydrophila should not at present be taken as identification of a species, since further sequencing may reveal differences. The prokaryotes responsible for the Pros A rDNAs in our patient population may or may not have physical properties and nutritional requirements similar to those of other Aeromonas spp. In studies to be reported elsewhere, cultures of prostate biopsy specimens sampled in parallel to those studied by PCR, as reported here, yielded occasional bacterial colonies, but none of these were identified as Aeromonas sp. It is possible that prostatic bacteria of our selected urine and urethral culture-negative patient population are culture resistant. Studies involving media specific for Aeromonas spp. are in progress.
Despite these caveats, it is likely that the Pros A phylogenetic group detected by PCR is closely related to Aeromonas spp. based on the significant association (99% confidence) in phylogenetic studies involving 1,000 bootstrap replicates. The finding that 3 of 36 clones were 100% similar to a known human pathogen rarely present in healthy individuals indicates the need for further investigation of A. hydrophila and related organisms as potential genitourinary tract pathogens.
The Pros B group of sequences found in our prostatitis patients appears to represent a second group of closely related rDNA species. In two-by-two comparisons, members of the Pros B group were >89% related to one another. One clone was 98% similar to E. coli rDNA. It is possible that this and other Pros B clones originated from the intestine or skin via an ascending urethral route. Alternatively, the Pros B group may have arisen from contamination of the biopsy specimens in the operating room or PCR laboratory. Contamination seems unlikely due to the extensive anticontamination procedures, skin clone sequence composition, and negative environmental controls. In addition, none of these Pros B sequences actually matched E. coli, while isolates from men with acute lower urinary tract infections from another population showed 100% similarity to E. coli sequences in GenBank.
Some sequences of the Pros B group were as much as 86% similar to some Pros A group sequences. While the distinction between these two groups based on sequence similarity appears arbitrary, the more sophisticated phylogenetic studies that examine maximum likelihoods of sequence changes, weighted transitions, and other factors indicate the existence of two main groups, with possible subgroups within each.
Classification of some rDNAs in this study as novel must be tentative at this time because of sequence database limitations. For example, previous database searches revealed similarity of patient clones 9041 and 3251 to the rDNA of the gram-negative species A. allosaccharophila but no absolute matches to this or any other species in the database. A more recent on-line search revealed 100% matches of these clones to the recently entered rDNA sequence of A. hydrophila. Thus, conclusions about prokaryotic rDNAs in prostatitis patients, or in any study where less than perfect matches are found, may depend on the database size and content. Some bacteria that are well characterized at the biochemical level are still absent from the rDNA database, while others that lack culture and biochemical characterization are present.
Most (61%) of the 16S rDNA PCR-positive patients were negative for cultured microorganisms. Several interpretations are possible. First, the patients were selected to be culture negative, which may have selected for culture-resistant microorganisms. Also, prostatitis patients frequently have had extensive treatment with antibiotics prior to coming to our clinic. Our results are consistent with PCR detection, in some cases, of defective or otherwise culture-resistant microorganisms. The biopsy specimens sent to the culture and PCR laboratories were nonidentical, although they were from the same patients. If focal infections exist in these patients, this may also explain some differences between culture and PCR results. For the five patients who were both PCR and culture positive, the culture lab found multiple microorganisms, including Diphtheroides sp., E. coli, and coagulase-negative Staphylococcus spp., in four of them. Since the PCR laboratory characterized an average of only 2.5 clones per patient, the PCR results are limited in the diversity that would have been seen. The culture laboratory reported no Aeromonas sp. The biopsy specimens were destructively processed for PCR, so we were unable to culture biopsy specimens that were PCR positive for microorganisms such as Aeromonas sp. A more detailed culture study is planned, specifically attempting to culture Aeromonas spp. from nondestructively stored portions of prostate biopsy specimens.
We have yet to attempt 16S rDNA PCR on expressed prostatic secretions. This source may be expected to present with urethral contaminants, and the nonspecific rDNA PCR would likely amplify these readily. However, expressed prostatic secretion sampling would be less invasive and preferable to prostate biopsies, and for these reasons design of an expressed prostatic secretion assay is in progress. Sequence data such as that presented here may be used to design PCR primers specific to prostatic species if indeed such specificity can be demonstrated in further studies. Such specific PCR primers might be used on expressed prostatic secretion samples in the future.
In summary, rDNA sequencing led to detection of diverse and related 16S rDNAs previously unsuspected by culture-based methods. Further rDNA sequencing and sequence comparisons may lead to categorization of prostatic bacteria and facilitate design of customized primers for more rapid identification of prostatic species. PCRs using such customized primers could be used to monitor the effects of drug treatment. Directed molecular techniques may prove useful for detecting prostate-localized microorganisms shed in urine or expressed prostatic secretions. Specific molecular tools may emerge from sequencing studies such as this. These tools might be used to begin determination of what role, if any, prostatic bacteria may have in the etiology of chronic prostatitis/chronic pelvic pain syndrome.
ACKNOWLEDGMENTS
This research was supported in part by NIH grant RO1 DK38955 and by a grant from the Allen Foundation, Seattle, Wash.
We thank Sue Ross, Ivan Rothman, and Roberta Jacobs for assistance and Kathy Agnew for providing bacterial culture results.
REFERENCES
- 1.Amann R, Springer N, Ludwig W, Gortz H, Schleifer K. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- 2.Archer B J, Holm R P. Treatment of Aeromonas hydrophila infection in a deep tissue wound. S D J Med. 1995;48:405–407. [PubMed] [Google Scholar]
- 3.Berger R E, Krieger J N, Kessler D, Ireton R C, Close C, Holmes K K, Roberts P L. Case-control study of men with suspected chronic idiopathic prostatitis. J Urol. 1989;141:328–331. doi: 10.1016/s0022-5347(17)40757-9. [DOI] [PubMed] [Google Scholar]
- 4.Bowie W. Urethritis in males. New York, N.Y: McGraw-Hill; 1990. p. 356. [Google Scholar]
- 5.Bruce A W, Chadwick P, Willett W S, O’Shaughnessy M. The role of chlamydiae in genitourinary disease. J Urol. 1981;126:625–629. doi: 10.1016/s0022-5347(17)54660-1. [DOI] [PubMed] [Google Scholar]
- 6.Bruce A W, Reid G. Prostatitis associated with Chlamydia trachomatis in 6 patients. J Urol. 1989;142:1006–1007. doi: 10.1016/s0022-5347(17)38970-x. [DOI] [PubMed] [Google Scholar]
- 7.Brunner H, Weidner W, Schiefer H G. Studies on the role of Ureaplasma urealyticum and Mycoplasma hominis in prostatitis. J Infect Dis. 1983;147:807–813. doi: 10.1093/infdis/147.5.807. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. Phylip—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 9.Gardner W A, Jr, Culberson D E, Bennett B D. Trichomonas vaginalis in the prostate gland. Arch Pathol Lab Med. 1986;110:430–432. [PubMed] [Google Scholar]
- 10.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 11.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 12.Hanninen M L, Salmi S, Mattila L, Taipalinen R, Siitonen A. Association of Aeromonas spp. with travellers’ diarrhoea in Finland. J Med Microbiol. 1995;42:26–31. doi: 10.1099/00222615-42-1-26. [DOI] [PubMed] [Google Scholar]
- 13.Krieger J. Evaluation and treatment of unconventional genitourinary tract infections. Semin Urol. 1985;3:193–199. [PubMed] [Google Scholar]
- 14.Krieger J N, Egan K J. Comprehensive evaluation and treatment of 75 men referred to chronic prostatitis clinic. Urology. 1991;38:11–19. doi: 10.1016/0090-4295(91)80004-q. [DOI] [PubMed] [Google Scholar]
- 15.Krieger J N, Egan K J, Ross S O, Jacobs R, Berger R E. Chronic pelvic pains represent the most prominent urogenital symptoms of “chronic prostatitis.”. Urology. 1996;48:715–721. doi: 10.1016/S0090-4295(96)00421-9. [DOI] [PubMed] [Google Scholar]
- 16.Krieger J N, Riley D E, Roberts M C, Berger R E. Prokaryotic DNA sequences in patients with chronic idiopathic prostatitis. J Clin Microbiol. 1996;34:3120–3128. doi: 10.1128/jcm.34.12.3120-3128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krieger J N, Ross S O, Simonsen J M. Urinary tract infections in healthy university men. J Urol. 1993;149:1046–1048. doi: 10.1016/s0022-5347(17)36292-4. [DOI] [PubMed] [Google Scholar]
- 18.Kurnatowska A, Kurnatowski A, Mazurek L, Wedzikowski P. Rare cases of prostatitis caused by invasion of Trichomonas vaginalis with Candida albicans. Wiad Parazytol. 1990;36:229–236. [PubMed] [Google Scholar]
- 19.Meares E M. Prostatis syndromes: new perspectives about old woes. J Urol. 1980;123:141–147. doi: 10.1016/s0022-5347(17)55828-0. [DOI] [PubMed] [Google Scholar]
- 20.Merino S, Rubires X, Knochel S, Tomas J M. Emerging pathogens: Aeromonas spp. Int J Food Microbiol. 1995;28:157–168. doi: 10.1016/0168-1605(95)00054-2. [DOI] [PubMed] [Google Scholar]
- 21.Rautelin H, Sivonen A, Kuikka A, Renkonen O V, Valtonen V, Lehti H, Kahanpaa A, Kosunen T U. Role of aeromonas isolated from feces of Finnish patients. Scand J Infect Dis. 1995;27:207–210. doi: 10.3109/00365549509019010. [DOI] [PubMed] [Google Scholar]
- 22.Relman D A, Falkow S. Identification of uncultured microorganisms: expanding the spectrum of characterized microbial pathogens. Infect Agents Dis. 1992;1:245–253. [PubMed] [Google Scholar]
- 23.Singh D V, Sanyal S C. Relationship between enterotoxicity and multiple drug resistance in Aeromonas spp. J Diarrhoeal Dis Res. 1995;13:172–175. [PubMed] [Google Scholar]
- 24.Stamey T. Pathogenesis and treatment of urinary tract infections. Baltimore, Md: Williams and Wilkins; 1980. pp. 342–349. [Google Scholar]
- 25.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 26.Weber C A, Wertheimer S J, Ognjan A. Aeromonas hydrophila—its implications in freshwater injuries. J Foot Ankle Surg. 1995;34:442–446. doi: 10.1016/S1067-2516(09)80019-7. [DOI] [PubMed] [Google Scholar]
- 27.Weidner W, Brunner H, Krause W. Quantitative culture of Ureaplasma urealyticum in patients with chronic prostatitis or prostatosis. J Urol. 1980;124:622–625. doi: 10.1016/s0022-5347(17)55586-x. [DOI] [PubMed] [Google Scholar]
- 28.Weidner W, Krause W, Schiefer H G, Brunner H, Friedrich H J. Ureaplasmal infections of the male urogenital tract, in particular prostatitis, and semen quality. Urol Int. 1985;40:5–9. doi: 10.1159/000281023. [DOI] [PubMed] [Google Scholar]
- 29.Weidner W, Schiefer H G, Krauss H. Role of Chlamydia trachomatis and mycoplasmas in chronic prostatitis: a review. Urol Int. 1988;43:167–173. doi: 10.1159/000281331. [DOI] [PubMed] [Google Scholar]
- 30.Wilson K H. Detection of culture-resistant bacterial pathogens by amplification and sequencing of ribosomal DNA. Clin Infect Dis. 1994;18:958–962. doi: 10.1093/clinids/18.6.958. [DOI] [PubMed] [Google Scholar]