Abstract
The aim of the study was to investigate the frequency and types of mutations on the retinoblastoma gene (RB1 gene) in Turkish population. RB1 gene mutation analysis was performed in a total of 219 individuals (122 probands with retinoblastoma, 14 family members with retinoblastoma and 83 clinically healthy family members). All 27 exons and close intronic regions of the RB1 gene were sequenced for small deletions and insertions using both the Sanger sequencing or NGS methods, and the large deletions and duplications were investigated using the MLPA analysis and CNV algorithm. The bilateral/trilateral retinoblastoma rate was 66% in the study population. The general frequency of RB1 gene mutation in the germline of the patients with retinoblastoma was 41.9%. Approximately 51.5% of the patients were diagnosed earlier than 12 months old, and de novo mutation was found in 32.4% of the patients. Germline small genetic rearrangement mutations were detected in 78.9% of patients and LGRs were detected in 21.1% of patients. An association was detected between the eye color of the RB patients and RB1 mutations. 8 of the mutations detected in the RB1 gene were novel in the study.
Keywords: hereditary, mutation, RB1 gene, retinoblastoma, sporadic
1. Introduction
Retinoblastoma is the most common intra-ocular malignancy in children.[1] The incidence is 1 in 15,000 to 18,000 live births.[2] The incidence of retinoblastoma is reported to be higher in developing countries, such as in Central and South America[3] and survival rate is lower in resource limited countries.[4] Two-thirds of the cases are diagnosed before the age of 2 years and 95% are diagnosed by the age of 5 years.[5] The diagnosis of retinoblastoma in children aged 5 years and over is extremely rare.[6]
Important genetic factors are known to play a role in the pathogenesis of retinoblastoma.[7–9] Mutations are known to initiate the disease in the retinoblastoma gene (RB1). RB1 gene (Gene ID: 5925, OMIM 614041) produces a nuclear protein weighing 110 kd called pRB. This protein which normally regulates the cell proliferation and prevents uncontrolled cell division, acts as a tumor suppressor.[10] The effects of tumor suppressor genes on cancer were described first in retinoblastoma.[11] Except for the RB1 gene, other genetic alteration in retinoblastoma genetics is the MYCN- amplification.[12] Patients with hereditary retinoblastoma have congenital mono-allelic RB1 mutations in their germline DNA and develop tumors in their retinal cells due to a second hit in the RB1 gene. While patients with familial retinoblastoma inherit an unknown mutant gene allele, patients with sporadic hereditary retinoblastoma develop the disease with de novo mutations in RB1 or other genes. MYCN amplification causes small portion (2%) of non-hereditary retinoblastomas that do not carry RB1 gene mutation. In the last decade, McEvoy et al showed that chromothripsis formed on chromosome 13, disrupts the RB1 locus and is an alternative mechanism for RB1 inactivation.[13] This structural alteration/mutation may be responsible for disease occurrence in cases without RB1 gene mutation and promoter methylation.[14]
Bilateral disease manifestation is detected in 40%, however, unilateral manifestation is detected in 60% of the retinoblastoma patients.[15] In about 5% of cases, trilateral tumor manifestation can also be detected (associated with a midline brain tumor).[16–18] All bilateral tumors are associated with germline alterations. Not all are hereditary/familial retinoblastoma as some are de novo retinoblastoma. Thus, (25%–35%) of retinoblastomas are hereditary/familial and (65%–75%) are non-hereditary for RB1 gene mutations. The patients with a family history, bilateral retinoblastoma, or RB1 mutation are considered to have hereditary retinoblastoma. Hereditary retinoblastoma is an autosomal dominant disease with a germline mutation and 85% of hereditary types of tumors occur at an early stage of childhood.[19,20]
RB1 gene contains a wide range of mutation types including single nucleotide variations, small insertions and deletions, large deletions and duplications.[14] While evaluating the RB1 gene mutations, both the 27 exons and the neighboring intronic regions (at least 50 bases from the start and end bases of the exons) of the RB1 gene should be scanned and also large deletions and duplications should be investigated using the copy number variation (CNV) or multiplex ligation probe amplification (MLPA) method.
Retinoblastoma is a rare tumor and accounts for about 3% of all pediatric malignancies in the SEER data[21] and the rate is around 3% of all pediatric malignancies in Türkiye according to the Turkish Pediatric Oncology Group pediatric cancer registry.[22] Our center is a referral center for retinoblastoma constituting 8.6% of all pediatric malignancies.[23] Our clinic is the first in Türkiye to provide genetic counseling and gene screening services on cancer. It is the only center that provides counseling and raising funds for genetic counseling and gene screening especially for patients with retinoblastoma.
The frequencies and types of RB1 gene mutation are unknown in the Turkish population. In our study, a total of 136 patients with retinoblastoma, 122 of whom were probands and 14 were family members (at least 3 generations) were analyzed for gene mutations of RB1. In addition, 83 clinically healthy family members of RB patients in whom a mutation was found were evaluated for RB1 mutation. Within the scope of the study, the RB1 gene was sequenced for the entire exon, exon-intron junction regions, and also large deletions or duplications. The aim of the present study was to assess the frequency of RB1 gene mutations and to identify mutation sites on the RB1 gene sequence, in a large cohort of Turkish patients with retinoblastoma and the healthy family members of RB patients with a mutation.
2. Materials and methods
2.1. Patient selection
A total of 122 pediatric patients with retinoblastoma who presented to Istanbul University, Istanbul Faculty of Medicine, Department of Ophthalmology and Istanbul University, Institute of Oncology, Division of Pediatric Hematology-Oncology between 2013 and 2021 and 14 family members (10 diagnosed with retinoblastoma in childhood, and 4 diagnosed after detailed fundus examination after the mutation was detected in the probands) were included in the study. In addition, 83 clinically healthy family members (at least 3 generations) with no diagnosis of retinoblastoma, of 47 probands with RB1 gene mutations were screened for known RB1 gene mutation. All patients with a diagnosis of RB presenting to our institute for treatment and their family members who were suspected to have the risk of carrying mutations were included in the study. Individuals who refused to give consent for genetic testing were excluded from the study. A detailed family trees of all families was created as shown in Figure 1. In this study, the germline RB1 gene mutations, rather than the somatic mutations of RB patients were examined and evaluated. The study was approved by the Istanbul University, Clinical Research Ethics Committee (Ethics approval no: 2013/252). Written consent was obtained from all parents.
Figure 1.
An example of a detailed family tree of the presenting family.
2.2. Mutation screening methods
Within the scope of the study, RB1 gene mutation screening was performed in a total of 219 people (122 probands, 14 family members with retinoblastoma and 83 clinically healthy family members). Peripheral blood samples of these individuals were collected and RB1 gene mutation was investigated. DNA isolation was performed using the QIAcube (Qiagen, Germany) in patients who were admitted to the clinic between 2013 and 2021. The measurement of genomic DNA was performed with a Qubit fluorimeter (ThermoFisher Scientific, Paisley PA4 9RF, UK).
2.3. Sanger sequencing
Sanger sequencing was used between 2013 and 2018. Sanger sequencing was used in 102 (70%) of 122 probands, mutation screenings in family members (14 family members with retinoblastoma and 83 clinically healthy family members) of patients with mutations. In Sanger sequencing method, all exon, exon and intron junction regions (±150–200 bp) and 5’UTR region of the RB1 gene were examined. MLPA analysis was performed for large deletions and duplications in cases sequenced with Sanger Sequencing. In the Sanger sequencing method, all coding exons of the RB1 gene and adjacent intronic binding sites were divided into 27 different fragments ranging in length from 197 to 823 base pairs and screened for RB1 mutations. MLPA analysis for the RB1 gene was performed using the Salsa MLPA Probemix P047 kit from MRC Holland (https://www.mrcholland.com/product/P047/911).
2.4. Next Generation Sequencing (NGS)
Next Generation Sequencing (NGS) was used between 2019 and 2021. NGS was used in 20 (30%) of 122 probands. All exon, exon and intron junction regions (±10 bp) of the RB1 gene were examined. The NGS process was performed on the MiSeq Platform. According to the panel protocol, DNA was fragmented after DNA quality was determined. Adapters were connected to this fragment with the ligation process to the DNA. The nonspecific DNA was removed by purification to provide an optimum level of sequencing quality. To obtain the amplified DNA, the marked library was replicated by a 10-cycle PCR process. The enriched library was loaded into the “Flow Cell” before placing on the device, and the “Flow Cell” loaded with the sample was placed in the MiSeq device for the sequencing process. Large deletions and duplications with CNV were investigated in patients screened with NGS. CNV status was confirmed by MLPA analysis in individuals with changes in CNV data.
2.5. Data analysis and interpretation of the results
Sanger Sequencing analysis was read using the SnapGene Viewer 5.0.6 software program. SOPHIA DDM platform was used for NGS data analysis in the study.[24] After the sequencing, the BCL format data obtained from the Illumina MiSeq device was first converted to the VCF file format and these files were uploaded to the SOPHIA DDM software program (Version 5.10.19.1). As a result, the descriptions of the variants specified in all relevant databases and algorithms were obtained. Various filtering options were used to determine the relationship of the anotation-treated variants with the phenotype. In particular, detailed examination of the variants with a ClinVar pathogenic record was performed. Variants that have not previously been reported in the literature or the Leiden Open Variation Database and the Human Gene Mutation Database have been identified as a novel variant. The variants obtained in the study were evaluated considering the reading quality >Q30 and the confidence score >50. The identified variants were labeled in accordance with the recommendation standards of the American College of Medical Genetics and Genomics.[25] Variants were classified into 5 categories as pathogenic, likely pathogenic, variant of unknown significance (VUS), likely benign and benign. The nomenclature of the mutations found on RB1 gene in the study was done according to the Human Genome Variation Society rules.[26]
2.6. Evaluation of the clinicopathological features
The demographic and clinical data, family history, symptoms, laterality, ocular findings, treatment modalities of the patients with retinoblastoma were evaluated. The clinical diagnoses were confirmed by ophthalmologists and pediatric oncology specialists. The data of the diagnosis, age, gender, stage, sign/symptoms at diagnosis (leukocoria, strabismus and glaucoma status) and treatment history of the patients were obtained from the clinical files. The International Classification of Retinoblastoma was used for staging of intraocular retinoblastoma.[27] All patients had intraocular retinoblastoma. The patient family history of cancer and exposure to risk factors were obtained during the genetic counseling sessions.
2.7. Statistical analysis
All the clinical and genetic data (age, gender, laterality, eye color, stages, leukocoria, strabismus, glaucoma, treatments, genetic inheritance, family history, and ethnicity) were analyzed using the IBM Statistical Package for the Social Sciences Statistics v.20 (SPSS Inc., Chicago, IL) program. All clinical and genetic data were compared with the results of the gene mutation analysis using the Mann–Whitney U test, a non-parametric test for both the patient and their relatives. In addition, linkage analyses were performed between family members in terms of the detected variations. The P value of P < .05 was considered statistically significant.
3. Results
3.1. Clinical and genetic data of the patients
RB1 gene mutation analysis was performed in a total of 219 individuals, consisting of 136 patients with retinoblastoma and 83 healthy family members of RB patients with RB1 mutations. The median age at diagnosis of the 136 patients was 11 (range, 1–80 m) months. At the time of diagnosis, 70 patients (51.5%) were infants (younger than 12 months of age) while 66 (48.5%) were ≥12 months-old.
Retinoblastoma patients diagnosed before the age of 12 months had a significantly higher rate of positive RB1 gene mutation [63.2% (36/70)] compared with the patients who were diagnosed to have the disease at the age of 12 months or older [36.8% (21/66)] (p: 0.021).
The gender distribution of the patients showed that 50.7% (69/136) were male and 49.3% (67/136) were female. RB1 mutation was detected in 42% (29/69) of the boys and in 41.8% (28/67) of girls (p: 0.843).
Of the 136 patients, 84 (61.8%) were diagnosed to have unilateral, 47 (34.5%) were diagnosed have bilateral, 3 (2.2%) were diagnosed to have trilateral disease, and 2 (1.5%) were diagnosed with unilateral retinoma. Mutations were observed in 23 (27.4%) of unilateral RB patients however were detected in 30 (63.8%) of bilateral RB patients. RB1 gene mutation has been detected in all of (100%) trilateral retinoblastoma patients. RB1 gene mutation was detected in 1 patient with retinoma, while no RB1 gene mutation was detected in the other patient. Conversion to bilateral/trilateral disease in patients diagnosed with unilateral disease was not detected.
A total of 136 from 121 different families were examined in the study. Although RB1 gene mutation was detected in 57 individuals in 42 families, no RB1 gene mutation was found in 79 of these families. The data of the families with mutations are given in Table 3. The family members of retinoblastoma patients were screened for the RB1 mutation which was found in the index case of the family. RB1 gene mutation was detected in 19 of the family members. Of these 19 people, 11 were diagnosed with retinoblastoma in childhood. The remaining 8 individuals with RB1 gene mutation were referred to detailed eye examination. Four out of 8 were detected to have already retinoma. No tumor was found in other 4 people.
Table 3.
The pathogenic RB1 gene mutations detected in patients.
| Number of the family | Family ID | Patients number | Family pedigree/degree | Mutations on RB1 gene | ACMG classification status | The types of mutations | dbSNP/ rs codes | Novel mutation status |
|---|---|---|---|---|---|---|---|---|
| 1 | 105 | 650 | Proband | HET, EX:17, c.1567_1568delTT p.(Leu523Lysfs*4) | Pathogenic | Frameshift | - | Novel |
| 105 | 651a | Mother | HET, EX:17, c.1567_1568delTT p.(Leu523Lysfs*4) | Pathogenic | Frameshift | - | Novel | |
| 2 | 125 | 850 | Proband | HET, EX:12, c.1150delC p.(Gln384Asnfs*2) | Pathogenic | Frameshift | - | Novel |
| 125 | 853 | Father | HET, EX:12, c.1150delC p.(Gln384Asnfs*2) | Pathogenic | Frameshift | - | Novel | |
| 3 | 138 | 980 | Proband | HET, EX:3, c.371_372delTA p.(Ile124Argfs*6) | Pathogenic | Frameshift | - | - |
| 4 | 147 | 1070 | Proband | HET, EX:18, c.1784delC p.(Pro595Leufs*16) | Pathogenic | Frameshift | - | Novel |
| 5 | 159 | 1190 | Proband | HET, EX:20, c.2018_2038del p.(His673_Ile680delinsLeu) | Pathogenic | Frameshift | - | Novel |
| 6 | 178 | 1380 | Proband | HET, EX:15, c.525delA p.(Gln176Asnfs*10) | Pathogenic | Frameshift | Novel | |
| 7 | 183 | 1550 | Proband | HET, EX:19, c.1939_1940delCT p.(Leu647Phefs*5) | Pathogenic | Frameshift | - | - |
| 8 | 185 | 1570 | Proband | HET, EX:24, c.2508_2509dupTG p.(Glu837Valfs*13) | Pathogenic | Frameshift | - | Novel |
| 9 | 220 | 1920 | Proband | HET, EX:2, c.189delA p.(Lys63Asnfs*2) | Pathogenic | Frameshift | - | Novel |
| 10 | 123 | 830 | Proband | HET, EX:6, c.585G > A p.(Trp195*) | Pathogenic | Nonsense | - | - |
| 11 | 129 | 890 | Proband | HET, EX:18 c.1735C > T p.(Arg579*) | Pathogenic | Nonsense | rs121913305 | - |
| 129 | 892b | Father (Healthy) | HET, EX:18 c.1735C > T p.(Arg579*) | Pathogenic | Nonsense | rs121913305 | - | |
| 129 | 893 | Sibling | HET, EX:18 c.1735C > T p.(Arg579*) | Pathogenic | Nonsense | rs121913305 | - | |
| 12 | 141 | 1010 | Proband | HET, EX:19, c.1954A > T p.(Lys652*) | Pathogenic | Nonsense | - | - |
| 141 | 1011 | Mother | HET, EX:19, c.1954A > T p.(Lys652*) | Pathogenic | Nonsense | - | - | |
| 13 | 148 | 1080 | Proband | HET, EX:2, c.225G > A p.(Trp75*) | Pathogenic | Nonsense | - | - |
| 14 | 154 | 1140 | Proband | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - |
| 154 | 1141a | Mother | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - | |
| 154 | 1143a | Maternal-Grandmother | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - | |
| 154 | 1145 | Maternal-Uncle | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - | |
| 154 | 1147 | Maternal-Cousin (Child of R1145) | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - | |
| 154 | 1149 | Maternal-Cousin (Child of R1145) | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - | |
| 15 | 160 | 1200 | Proband | HET, EX:18, c.1735C > T p.(Arg579*) | Pathogenic | Nonsense | rs121913305 | - |
| 16 | 165 | 1250 | Proband | HET, EX:24, c.2513C > A p.(Ser838*) | Pathogenic | Nonsense | rs1131690908 | - |
| 17 | 177 | 1370 | Proband | HET, EX:14, c.1363C > T p.(Arg455*) | Pathogenic | Nonsense | rs121913302 | - |
| 18 | 204 | 1760 | Proband | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - |
| 19 | 213 | 1850 | Proband | HET, EX:10, c.958C > T p.(Arg320*) | Pathogenic | Nonsense | rs121913300 | - |
| 20 | 214 | 1860 | Proband | HET, EX:4, c.409G > T p.(Glu137*) | Pathogenic | Nonsense | rs121913296 | - |
| 21 | 136 | 960 | Proband | HET, EX:12 c.1215 + 1G > A p.? | Pathogenic | Splice error | rs587776783 | - |
| 22 | 151 | 1110 | Proband | HET, c.265-2A > T p.? | Pathogenic | Splice error | - | - |
| 151 | 1111 | Sibling | HET, c.265-2A > T p.? | Pathogenic | Splice error | - | - | |
| 151 | 1112a | Mother | HET, c.265-2A > T p.? | Pathogenic | Splice error | - | - | |
| 23 | 153 | 1130 | Proband | HET, c.607 + 1G > T p.? | Pathogenic | Splice error | rs587776789 | - |
| 24 | 195 | 1670 | Proband | HET, c.2520 + 3_2520 + 6delGAGT p.? | Pathogenic | Splice error | rs1131690558 | - |
| 25 | 206 | 1780 | Proband | HET, c.1960 + 1G > C p.? | Pathogenic | Splice error | - | Novel |
| 26 | 208 | 1800 | Proband | HET, c.1960 + 1delG p.? | Pathogenic | Splice error | - | - |
| 27 | 216 | 1880 | Proband | HET, c.1390-14A > G p.? | Pathogenic | Splice error | rs9535023 | - |
| 28 | 217 | 1890 | Proband | HET, c.1960 + 1G > A p.? | Pathogenic | Splice error | - | - |
| 29 | 221 | 1930 | Proband | HET, c.607 + 1G > T p.? | Pathogenic | Splice error | rs587776789 | - |
| 221 | 1931b | Father (Healthy) | HET, c.607 + 1G > T p.? | Pathogenic | Splice error | rs587776789 | - | |
| 221 | 1933b | Sibling (Healthy) | HET, c.607 + 1G > T p.? | Pathogenic | Splice error | rs587776789 | - | |
| 30 | 181 | 1500 | Proband | HET, EX:19, c.1960G > C p.(Val654Leu) (Last base of Exon 19) | Pathogenic | Missense | rs483352690 | - |
| 31 | 104 | 640 | Proband | HET, EX:13, c.1332 G > A p.(Gln444=), Last base of Exon 13) | Pathogenic | Synonymous substitution | - | - |
| 104 | 643 | Sibling | HET, EX:13, c.1332 G > A p.(Gln444=), Last base of Exon 13) | Pathogenic | Synonymous substitution | - | - | |
| 32 | 187 | 1590 | Proband | Upstream, HET, c.-198G > A p.? | Pathogenic | Upstream substitution | rs387906521 | - |
| 187 | 1591 | Sibling | Upstream, HET, c.-198G > A p.? | Pathogenic | Upstream substitution | rs387906521 | - | |
| 33 | 109 | 690 | Proband | HET, EX:1-27 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 109 | 693 | Sibling | HET, EX:1-27 DELETION | Pathogenic | Large deletion or duplication | - | - | |
| 34 | 119 | 790 | Proband | HET, EX:21-23 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 35 | 128 | 880 | Proband | HET, EX:7-11 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 36 | 134 | 940 | Proband | HET, EX:1-27 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 134 | 942b | Father (Healthy) | HET, EX:18-27 DELETION | Pathogenic | Large deletion or duplication | - | - | |
| 134 | 943 | Sibling | HET, EX:18-27 DELETION | Pathogenic | Large deletion or duplication | - | - | |
| 37 | 140 | 1000 | Proband | HET, EX:1-17 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 38 | 172 | 1320 | Proband | HET, EX:13 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 39 | 173 | 1330 | Proband | HET, EX:1-27 DELETION + Partial deletion of DLEU1 and PCHD8 gene | Pathogenic | Large deletion or duplication | - | - |
| 40 | 193 | 1650 | Proband | HET, EX:3-27 DELETION | Pathogenic | Large deletion or duplication | - | - |
| 41 | 200 | 1720 | Proband | HET, EX:4-17 DUPLICATION | Pathogenic | Large deletion or duplication | - | - |
| 42 | 201 | 1730 | Proband | HET, EX:1-27 DELETION | Pathogenic | Large deletion or duplication | - | - |
ACMG = American College of Medical Genetics and Genomics, HET = heterozygous, RB1 = retinoblastoma gene.
Retinoma.
Healthy/unaffected.
The evaluation of the eye (iris) color of the patients showed a significantly higher mutation rate in patients with light (green or blue) eye colors than the patients with dark (black or brown) eye colors [71.4% (15/21) vs 36.5% (42/115)], respectively; p: 0.003.
The evaluation of the clinical stages of the patients showed that 44 (32.4%) of the 136 patients with retinoblastoma were diagnosed with retinoblastoma at an early stage (Group A, Group B, Group C), while 92 (67.6%) were diagnosed with advanced stage (Group D or Group E). In bilateral tumors, if any of the eye was in advanced stage, they were recorded as advanced stage patients. 40 (43.5%) out of 57 patients with RB1 mutation were in the advanced stage; 17 (38.6%) were diagnosed at an early stage. The evaluation of the clinical stage and RB1 mutation status showed no statistically significant relationship between the early-stage and advanced-stage patients in terms of RB1 mutation (p: 0.778). The evaluation of the bilateral patients in accordance with the stages showed that 32 out of 47 (68.1%) bilateral patients were diagnosed to have the advanced stage. Leukocoria was detected in 67 out of 92 (72.8%) advanced stage patients (p: 0.03).
29 out of 91 (66.9%) patients who presented to the clinic with the symptoms of leukocoria were detected to have RB1 mutation (p: 0.001). 85 (62.5%) patients presented to the clinic with strabismus and 49 (57.6%) of these patients had esodeviation and 36 (42.4%) had exodeviation. RB1 mutation was found positive in 37 (43.5%) strabismus patients. Glaucoma was observed in 16 (11.8%) at the time of diagnosis, while mutations were found in 7 (43.7%) of these patients.
The evaluation of the treatment options of the patients showed that 49 (36%) received intra-arterial chemotherapy (IAC), 69 (50.7%) received systemic chemotherapy (CT) for chemoreduction, 16 (11.8%) received regional radiotherapy (RT) and 39 (28.7%) received local ophthalmic treatment (LOT) as cryotherapy, thermotherapy, laser therapy and 54 (39.7%) underwent surgery for enucleation of the eye. No statistically significant relationship was found between RB1 gene mutation and the treatment choices of the patients (P ≥ .05). The treatment modalities were recorded so that in the follow-up of the patients, there might be a correlation of exposure to radiation (in patients receiving RT or the ones exposed to medical radiation as in IAC procedures) and RB1 gene mutation as in secondary malignancies. All the demographic, clinical and genetic data of the patients are shown in Table 1.
Table 1.
The gene mutations detected in patients. All the clinical data and RB1 mutation distributions of the patients.
| Clinicopathological features | RB1 mutations | Total n(%) | Significance | |
|---|---|---|---|---|
| No n(%) | Yes n(%) | |||
| Age (mo) median 11 mo range (1–80) | ||||
| <12 mo | 34 (48.6%) | 36 (51.4%) | 70 (100%) | p: 0.021* |
| ≥12 mo | 45 (68.2%) | 21 (31.8%) | 66 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Gender | ||||
| Female | 39 (58.2%) | 28 (41.8%) | 67 (100%) | p: 0.843 |
| Male | 40 (58%) | 29 (42%) | 69 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Laterality | ||||
| Unilateral | 61 (72.6%) | 23 (27.4%) | 84 (100%) | p: 0.000* |
| Bilateral | 17 (36.2%) | 30 (63.8%) | 47 (100%) | |
| Trilateral | 0 (0.0%) | 3 (100%) | 3 (100%) | |
| Unilateral Retinoma | 1 (50%) | 1 (50%) | 2 (100%) | |
| Healthy family members | 79 (95.2) | 4 (4.8%) | 83 (100%) | |
| Total | 158 (72.1%) | 61 (27.9%) | 219 (100%) | |
| Eye laterality for unilateral RB | ||||
| Left | 30 (68.2%) | 14 (31.8%) | 44 (100%) | p: 0.475 |
| Right | 32 (76.2%) | 10 (23.8%) | 42 (100%) | |
| Total | 62 (72.1%) | 24 (27.9%) | 86 (100%) | |
| Eye color | ||||
| Black-brown | 73 (63.5%) | 42 (36.5%) | 115 (100%) | p: 0.003* |
| green-blue | 6 (28.6%) | 15 (71.4%) | 21 (100%) | |
| total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| The Stage | ||||
| Group A,B,C | 27 (61.4%) | 17 (38.6%) | 44 (100%) | p: 0.778 |
| Group D,E | 52 (56.5%) | 40 (43.5%) | 92 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Leukocoria | ||||
| No | 17 (37.8%) | 28 (62.2%) | 45 (100%) | p: 0.001 |
| Yes | 62 (68.1%) | 29 (31.9%) | 91 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Strabismus | ||||
| Esodeviation | 32 (65.3%) | 17 (34.7%) | 49 (100%) | p: 0.182 |
| Exodeviation | 16 (44.4%) | 20 (55.6%) | 36 (100%) | |
| No | 31 (60.8%) | 20 (39.2%) | 51 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Glaucoma | ||||
| No | 70 (58.3%) | 50 (41.7%) | 120 (100%) | p: 0.925 |
| Yes | 9 (56.2%) | 7 (43.8%) | 16 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Treatment | ||||
| IAC | ||||
| Yes | 30 (61.2%) | 19 (38.8%) | 49 (100%) | p: 0.747 |
| No | 49 (56.3%) | 38 (43.7%) | 87 (100%) | |
| CT | ||||
| Yes | 38 (55.1%) | 31 (44.9%) | 69 (100%) | p: 0.587 |
| No | 41 (61.2%) | 26 (38.8%) | 67 (100%) | |
| RT | ||||
| Yes | 7 (43.8%) | 9 (56.2%) | 16 (100%) | p: 0.243 |
| No | 72 (60%) | 48 (40%) | 120 (100%) | |
| Surgery | ||||
| Yes | 33 (61.1%) | 21 (38.9%) | 54 (100%) | p: 0.474 |
| No | 46 (56.1%) | 36 (43.9%) | 82 (100%) | |
| LOTs | ||||
| Yes | 18 (46.2%) | 21 (53.8%) | 39 (100%) | p: 0.095 |
| No | 61 (62.9%) | 36 (37.1%) | 97 (100%) | |
CT = chemotherapy, IAC = intraarterial chemotherapy, LOT = local opthalmic treatment (cryotherapy, thermotherapy, lasertherapy), RB1 = retinoblastoma gene, RT = radiotherapy, surgery = enucleation of eye.
P < .05 is significant.
RB1 mutation screening revealed mutations in 57 (41.9%) of 136 patients. Of these mutations, 45 (78.9%) were in the form of small insertions and deletions and small genetic rearrangements and 12 (21.1%) were in the form of large genetic rearrangements (LGRs). Distribution of RB1 mutation by mutation types is shown in Table 2.
Table 2.
Distribution of pathogenic RB1 mutation according to mutation types.
| Type of mutations | Number of RB1 mutation n(%) |
|---|---|
| Pathogenic indels and small genetic rearrangements | 45 (78.9%) |
| Splice error | 11 (19.3%) |
| Frameshift | 11 (19.3%) |
| Nonsense | 18 (31.6%) |
| Missense | 1 (1.8%) |
| Synonymous substitution | 2 (3.5%) |
| Upstream substitution/Promoter Mutation | 2 (3.5%) |
| Pathogenic large genetic rearrangements | 12 (21.1%) |
| Total | 57 |
RB1 = retinoblastoma gene.
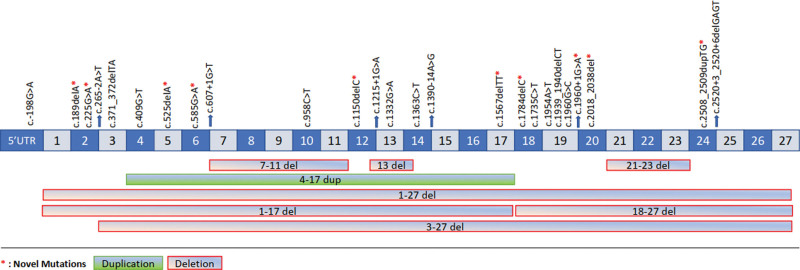
The frameshift mutations were found in 11 patients (19.3%), nonsense in 18 patients (31.6%), 11 patients with splice error (19.3%), and in 1 patient with missense (1.8%), synonymous substitution in 2 patients (3.5%), upstream substitution/promoter in 2 patients (3.5%), 12 patients with large rearrangement (21.1%) mutations. Distribution of mutation types according to diagnosis are shown in Figure 2. The detailed gene mutations detected in all patients are shown in Table 3. Also, Figure 3 demonstrates the distribution of RB1 mutations detected in the gene sequence over the exon and intronic regions.
Figure 2.
The distribution of the mutation types in accordance with the diagnosis.
Figure 3.
The distribution of mutations detected in the RB1 gene sequence on the exon and intronic regions. RB1 = retinoblastoma gene.
No RB1 gene mutation was detected in 79 (58.1%) patients and these patients were considered non-hereditary for RB1 gene mutations (Table 4).
Table 4.
The genetic inheritance of the RB1 mutation in the cohort.
| Genetic inheritance | RB1 mutations | Total n(%) | Significance | |
|---|---|---|---|---|
| No n(%) | Yes n(%) | |||
| De novo/heritable** | ||||
| 0 (0%) | 44 (100%) | 44 (32.4%) | p: 0.000* | |
| Hereditary/familial*** | ||||
| 0 (0%) | 13 (100%) | 13 (9.5%) | ||
| Non-hereditary for RB1 gene mutations**** | ||||
| 79 (100%) | 0 (0%) | 79 (58.1%) | ||
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
RB1 = retinoblastoma gene.
P < .05 is significant.
De novo: no family history of retinoblastoma, but the patient was a carrier of RB1 gene mutation and was the first index case.
A family history of retinoblastoma and the patient was a carrier of the RB1 gene mutation.
*No family history of retinoblastoma, and the patient was not carry RB1 gene mutation.
The examination of the family trees of the patients showed that retinoblastoma history was present in the families of 31 patients (22.8%). The investigation of patients with mutations in accordance with the other cancer histories in the family showed that 96 patients (70.6%) had other types of cancers such as brain, breast, lung, thyroid, prostate cancer, and leukemia in their family. In addition, 93 (68.4%) out of 136 patients were detected to have individuals working in risky jobs in their family. The RB1 mutation frequency of these families was 39.8% (Table 5).
Table 5.
Distribution of mutations according to family history and occupational groups.
| Family history | RB1 mutations | Total n(%) | Significance | |
|---|---|---|---|---|
| No n(%) | Yes n(%) | |||
| RB family | ||||
| Yes | 5 (16.1%) | 26 (83.9%) | 31 (100%) | p: 0.000* |
| No | 74 (70.5%) | 31 (29.5%) | 105 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| Other tumors in family | ||||
| Yes | 63 (65.6%) | 33 (34.4%) | 96 (100%) | p: 0.006* |
| No | 16 (40%) | 24 (60%) | 40 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
| RAOG family | ||||
| Yes | 56 (60.2%) | 37 (39.8%) | 93 (100%) | p: 0.461 |
| No | 23 (53.5%) | 20 (46.5%) | 43 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
OT = other tumors (brain tumor, breast cancer, lung cancer, thyroid cancer, prostate cancer), RB = retinoblastoma, RAOG = risk assessment of occupational groups.
P < .05 is significant.
The ethnic origins of the 136 patients showed that 113 were Turkish-origin (%83.1), 9 Caucasus-origin (%6.6), 8 Arabian-origin (5.9%), 4 Balkan-origin (2.9%), 2 were Iranian-origin (%1.5) (Table 6). The evaluation of the RB1 gene mutation status in accordance with their ethnic origin revealed a statistical difference (p: 0.02). The number of sub-ethnic groups was quite inadequate, therefore, this statistical significance should be confirmed by larger studies in larger population groups.
Table 6.
The distributions of mutations’ percentages in accordance with the ethnicity in our study.
| Ethnicity | RB1 mutations | Total n(%) | Significance | |
|---|---|---|---|---|
| No n(%) | Yes n(%) | |||
| Turkish-origin | 71 (62.8%) | 42 (37.2%) | 113 (100%) | p: 0.02* |
| Caucasus-origin | 4 (44.4%) | 5 (55.6%) | 9 (100%) | |
| Arabian-origin | 1 (12.5%) | 7 (87.5%) | 8 (100%) | |
| Balkan-origin | 2 (50%) | 2 (50%) | 4 (100%) | |
| Iranian-origin | 1 (50%) | 1 (50%) | 2 (100%) | |
| Total | 79 (58.1%) | 57 (41.9%) | 136 (100%) | |
RB1 = retinoblastoma gene.
4. Discussion
RB1 loss has been reported to cause non-proliferative retinoma and subsequently triggers the progression of retinoblastoma with increased genomic instability.[28] Up to 45% of retinoblastomas have been reported to develop due to a mutation of the RB1 gene and is inherited to next generations, and these cases were shown to mostly have bilateral retinoblastoma.[29] In the present study, the general RB1 gene mutation rate was 41.9%. Our data have concordance with the literature.[30] Researchers reported in the literature that patients with bilateral retinoblastoma account for about 20% to 40% of all patients with retinoblastoma.[31,32] We found this rate as 34.5% which is consistent with the literature data. The percentage value of RB1 mutations in the germline of the patients with bilateral/trilateral retinoblastoma was found as 66% in our cohort somewhat smaller than the rate of 90% in the literature.[33] The reason behind this low rate in our study population was probably due to gene pool differences of the population. However, the rate of germline RB1 mutation in our population was so similar with retinoblastoma patients in the Vietnamese population. In the particular population, the detection rate of germline mutations in bilateral and unilateral cases with mutations were 81.8% and 30.0%, respectively.[34] Germline RB1 mutations were detected in 60% (31/52) of patients in the population of Thailand.[35] The detection rates in the bilateral and unilateral cases were 100.0% (7/7) and 25.0% (3/12), respectively in Malaysian population.[36] Germline mutation rate was found in 7 out of 36 sporadic unilateral RB patients (20%) in Tunisian population.[37] Germline mutations were identified in 8 out of 9 patients with bilateral/ trilateral retinoblastoma (89%) and in 5 out of 25 sporadic unilateral patients (20%) in the population of Argentina.[38] However, it is noteworthy that the germline RB1 gene mutation ratio is different in various populations. The germline RB1 gene mutation rate in the Turkish population is 100%, 63.8%, and 27.4% in trilateral, bilateral, and unilateral cases, respectively. In the Turkish Population, the overall mutation rates are within the range seen in different populations. Since a large proportion of the patients in our cohort were sequenced with Sanger method, we suggested that the mutation might be overlooked, and 6 patients with bilateral retinoblastoma without RB1 gene mutation were sequenced with multigene panel using NGS. As a result of the second screening with Sanger method of the patients with no RB1 gene mutation showed that no RB1 gene mutation was found in sequencing with NGS in these patients. Therefore, Sanger Sequencing and NGS were provided similar results in germline DNA examinations. In addition, in patients sequenced with Sanger Sequencing, approximately ±150–200 nucleotides were examined in exon-intron junctions by entering up to introns. This ratio was ±10 nucleotides in NGS. Also, almost all of the 5’-UTR region is examined in the Sanger study, while only a small part of this region is examined with NGS. Although there is no difference between Sanger and NGS in germline examinations, there is variability in tumor tissue examinations. Mutations in different genes were observed in 6 patients diagnosed with bilateral RB, except for the RB1 gene mutation. These genes are FGFR4, NQO1, ACADS CX3CR1, GBE1, KRT85, and TYR, which are suggested to possibly have an effect on the pathogenesis of the disease in these patients with no germline RB1 gene mutations, or that differences in RB1 gene methylation and expression status may be effective, as we are investigating in patients without RB1 gene mutation.[7–9] The presence of a RB1 gene mutation in all 3 patients with trilateral RB underlines the role of the RB1 gene in trilateral disease.
Bilateral patients are known to have commonly been diagnosed at an earlier age compared with the age in unilateral patients.[31,32] In our study, 51.5% of our patients were diagnosed before the age of 12 months. The mutation rate in patients diagnosed before the age of 12 months was found as 51.4% significantly higher than the older patients. In a study where 4351 patients from 153 countries mostly consisting of low middle-income countries (LMIC) were evaluated; the median age at diagnosis was reported as 30.5 months (interquartile range, 18.3–45.9), which may be due to late diagnosis of the patients.[39] In our study, the median age at diagnosis was 11 (range, 1–80 m) months. This shows that our patients were diagnosed at an earlier age.
In the literature, leukocoria reported by families at the time of first diagnosis in patients with retinoblastoma is approximately 66.7%. In our study, this rate was 66.9%, similar to the literature.[39] While strabismus status reported at the time of diagnosis was 14.1% in the literature,[39] we found the rate as 62.5% in our study group. These rates are also proof of how the clinical findings of retinoblastoma play a decisive role at the time of initial diagnosis. In addition, although the stage at diagnosis of the tumor was insignificant for correlation with RB1 gene mutation; some presenting symptoms such as leukocoria which is mostly related to advanced stage was found to correlate with RB1 gene mutation. This finding needs further investigation to be assessed in larger series.
One another important finding in our study was that 4.8% of healthy individuals in families with retinoblastoma and who have never been diagnosed with retinoblastoma were mutation carriers as a result of genetic screening. After 8 mutation carriers (R651, R892, R1141, R1143, R1112, R1931, R1933, R942) were referred for detailed fundoscopic examination as recommended for genetic counseling, retinomas were detected in 4 of these individuals (R651, R1141, R1143, R1112). This suggests that retinoblastoma may actually have developed in childhood and that the stalled progression of tumor over time in these healthy looking individuals (R892, R1931, R1933, R942). It is unknown why these mutations cause an aggressively progressive tumor in their offsprings and have a stalled progression profile. Non-progressive retinal lesions observed in patients with known RB1 gene mutations in the literature are called “spontaneous regression/stalled progression of retinoblastoma.” This term suggests a malignant growth shrink perhaps in response to certain host defense mechanisms.[40] More detailed genetic and clinical studies are needed to clarify this issue.
Genetic inheritance in retinoblastoma can occur either hereditary or de novo, as in familial RB.[41] In familial RB, it shows autosomal dominant inheritance with one of the affected parents. When de novo occurs, the phenotype is observed only in the child, not in the parents.[42,43] The de novo mutation rate has been reported as approximately 75% in the literature.[42] It is noteworthy that in 32.4% of patients, the mutation occurred de novo when other family members consisting of parents, siblings were evaluated for a known mutation in order to understand the genetic inheritance of the disease within the scope of our study.
Although no mutation was found in the parents of proband in 3 families, detection of a mutation in the sibling was important in our study. This can be explained by germline mosaicism in parents who carry no mutations as germline.[44] According to germline mosaicism, the individual is not affected if only the germ cells mutate, while mosaicism can pass to the offspring and affect them.[45] In clinical practice, mosaic embryos can be identified using both invasive (amniocentesis and chorionic villus sampling) and noninvasive prenatal genetic diagnostic methods up to the first 8 to 10 weeks.[46] Prenatal genetic diagnosis should be recommended to families with germline mosaicism.
Due to cultural differences, until recently cancer was considered a “death toll” so parents did not want their children nor relatives to know about the cancer diagnosis in LMICs. In FN154 family; the proband (R1140) was found to have a bilateral RB, the mother (R1141) was found to have a retinoma. The detailed searching of the family history convinced with the importance that will help to prevent new diseases in the family, the uncle (R1145) was reported with unilateral RB, who in fact was found to have the disease in our follow up. He was married and had a baby aged 15 days old. The baby (R1147) was immediately examined and was found to have bilateral retinoblastoma. The grandmother (R1143) who had an eye enucleated due to a trauma was convinced to question for the reason of enucleation and after forty-five years she had learned that the procedure was done due to unilateral RB. This case emphasizes the importance of genetic counseling and educating of the population that cancer is not fatal, and early diagnosis is possible or can be totally prevented in some cases. Late diagnosis of many cases could have been prevented if the families explained the fact to their children, other family members and healthcare professionals.
Patients with hereditary retinoblastoma are at the risk of developing secondary malignancies such as osteosarcoma, soft tissue sarcomas or melanomas due to the mutation found in the second copy of the RB1 gene.[47] The incidence of secondary primary tumors have been reported to have increased over than 50% in individuals with retinoblastoma undergoing external beam radiation therapy.[48] Therefore, radiation exposure including X-rays, computed tomography (CT) scans, and external beam radiotherapy should be avoided as possible to minimize the lifetime risk of secondary cancers in patients who are known carry the RB1 gene mutations. The Global Retinoblastoma Study Group evaluated the treatment modalities of 4351 patients in their study in 2020. They reported that 71.1% of the patients received external beam radiotherapy, 27.3% received plaque brachytherapy, and 46.1% received intra-ophthalmic arterial chemotherapy. In addition, 71.1% of the patients were stated to have received CT scans for diagnostic procedures.[39] Radiation exposure occurs due to the fluoroscopy used in external beam radiotherapy, plaque brachytherapy and intra-ophthalmic arterial chemotherapy. The study of the Global Retinoblastoma Study Group reported that radiotherapy has currently been widely used in some countries, in which other modalities may be preferred. Therefore, evaluation of the risks of secondary tumors is recommended when making treatment choices, particularly in RB1 mutation carrier patients. 16 patients in our cohort received radiotherapy. The time period for radiotherapy-induced radiation to form secondary cancer is approximately 10 to 20 years. Therefore, all germline mutated cases are under follow-up for secondary cancers.
The literature search showed a significant relationship between the risky occupational groups (welding, metal industry, military service, machinists, jewelers, etc) in the family with germline RB1 gene mutation.[49] In our study, we have noted that ancestors of the 64.9% of the patients had risky occupational groups in their families, however no significant correlation with the RB1 mutation rate was noted.
Detection of a relationship between the eye color of the RB patients and RB1 mutations was interesting. It is noteworthy that 15.4% of the RB patients who underwent mutation screening had green-blue eyes, and 71.4% of these patients had RB1 gene mutations. Estimated eye color distribution rates in the world population are 70% for black-brown eyes; it is around 10% for green-blue eyes. Other colors (hazel, amber, gray, red/violet or heterochromia) are seen in the remaining 20% of the population. Since the dominant eye color is brown in the Turkish population, the ratio of black-brown eyes is over 90%. People with brown eyes have a lower incidence of eye cancer, macular degeneration and diabetic retinopathy. Ophthalmologists are not exactly sure about the cause however suggest that melanin pigment might provide higher protection for brown eyes.[50] Our study results showed that both the green-blue eyes ratios of patients with RB were remarkable compared to the standards in the Turkish population, and the mutation distributions in these patients were quite higher. The relationship between eye color, melanin pigment and RB1 gene mutation distribution in patients with retinoblastoma needs further assessment in larger groups.
The evaluation of the types of RB1 gene mutations stated that small genetic re-arrangements are observed in 78.9% of the cases. Although the large genetic rearrangement rate was reported as 9% in the literature,[30] we found as 21.1% in our study. This shows that screening for RB1 gene mutations only in the regions at the exon and exon-intron boundaries is not adequate to investigate the genetic background of the disease. Therefore, CNV or MLPA analysis should be performed in patients with retinoblastoma to assess the status of the loss of heterozygosity status.
The evaluation of the mutation types of the patients showed that the mutations that caused the formation of stop codons such as nonsense, frameshift were most frequently observed. It is also noteworthy that mutations that cause exon deletion or addition such as splice error and large rearrangement mutations are often observed in these patients. These rates are similar to the numbers in the literature.[30] Furthermore, researchers in the literature also indicated that splice errors cause incomplete penetrance. c.607 + 1G > T p.? detected in the study of Klutz et al and causing incomplete penetration.[51,52] Mutation was also seen in a family (FN221) in our study. In addition, we found 8 novel mutations, which have not yet been identified in the international databases.
The results of our study in a total of 136 patients diagnosed with retinoblastoma, provide data on all exon, exon-intron junction regions and large deletions and duplications screening of the RB1 gene.
RB1 gene mutation screening is a time-consuming and expensive procedure. In Türkiye, all treatment and most diagnostic modalities are reimbursed by the Government. However, most genetic and molecular tests are not fully reimbursed for the patients and are not reimbursed at all for the parents. However, considering the socioeconomic status of the patients’ families, it is not possible for pay out of their pocket. We strived to create funds through research grants and nongovernmental voluntary organizations to perform RB1 gene mutation screenings in our institution. It is crucial to choose the individuals to be tested correctly and to identify the people at risk in family for using the funds appropriately. In this regard, we provide genetic counseling to both patients and their families and refer them to genetic testing in our genetic counseling outpatient clinic. In addition, in case of genetic testing and detection of mutations both in the patient and their families, healthy embryo selection with the preimplantation genetic diagnosis method will help to prevent the disease. Thus, the prevention of some genetically transmitted cases of retinoblastoma in the population may be enabled and unnecessary health spendings may also be prevented. This study involves one of the largest cohorts of genetic testing in LMIC. The results of this study may play a role in advising the reimbursement on these tests in the related health care systems in LMIC.
In conclusion, we found the rate of RB1 gene mutation as 41.9% in children with retinoblastoma in Türkiye. The mutation was significantly higher in patients with bilateral/trilateral retinoblastoma in comparison to the levels in patients with unilateral retinoblastoma; in infants in comparison to older children; in patients with green/blue eye color in comparison to black/brown eye color. In this cohort, 8 novel mutations of the RB1 gene were found using the Leiden Open Variation Database and the Human Gene Mutation Database. The ophthalmologic examination of the family members of all retinoblastoma patients and the molecular genetic analysis of the family members of all bilateral, and unilateral patients who were found to have mutations is important in genetic counseling and possible assisted in vitro fertilization (IVF) in order to prevent retinoblastoma in the offspings when parents plan for another child. However, our previous study and the results of the present study showed that examining the RB1 gene alone is not sufficient when performing germline screening in patients with retinoblastoma and their families. For this reason, ideal approach will be to examine using multigene panels and NGS as a method. In addition, such studies will enable the determination of gene pathways that cause retinoblastoma disease by accumulating and meta-analyzing the data of many families screened with NGS in the future. This will enable the discovery of additional genes associated with hereditary retinoblastoma in families with retinoblastoma in which no RB1 gene mutation is detected.
Acknowledgments
We thank to our patients and their relatives for donating blood samples for the research and also to the technicians Turkan Sen Ferhadoglu and Arzu Burnuva for their effort in processing and collecting the samples. We confirm that the persons give permission to be named.
Author contributions
Conceptualization: Demet Akdeniz Odemis, Rejin Kebudi, Sema Buyukkapu Bay, Samuray Tuncer, Hulya Yazici.
Data curation: Demet Akdeniz Odemis, Seda Kilic Erciyas, Gozde Kuru Turkcan, Sema Buyukkapu Bay, Hulya Yazici.
Formal analysis: Demet Akdeniz Odemis, Gozde Kuru Turkcan, Hulya Yazici.
Funding acquisition: Demet Akdeniz Odemis, Rejin Kebudi, Hulya Yazici.
Investigation: Demet Akdeniz Odemis, Rejin Kebudi, Jamila Bayramova, Seda Kilic Erciyas, Gozde Kuru Turkcan, Seref Bugra Tuncer, Betul Celik, Busra Kurt Gultaslar, Samuray Tuncer, Hulya Yazici.
Methodology: Demet Akdeniz Odemis, Jamila Bayramova, Seda Kilic Erciyas, Gozde Kuru Turkcan, Ozge Sukruoglu Erdogan, Betul Celik, Busra Kurt Gultaslar, Hulya Yazici.
Project administration: Demet Akdeniz Odemis, Rejin Kebudi, Hulya Yazici.
Resources: Demet Akdeniz Odemis, Rejin Kebudi, Gozde Kuru Turkcan, Sema Buyukkapu Bay, Samuray Tuncer, Hulya Yazici.
Software: Demet Akdeniz Odemis, Gozde Kuru Turkcan, Ozge Sukruoglu Erdogan, Busra Kurt Gultaslar, Hulya Yazici.
Supervision: Demet Akdeniz Odemis, Rejin Kebudi, Hulya Yazici.
Validation: Demet Akdeniz Odemis, Hulya Yazici.
Visualization: Demet Akdeniz Odemis, Hulya Yazici.
Writing – original draft: Demet Akdeniz Odemis, Rejin Kebudi, Betul Celik, Sema Buyukkapu Bay, Hulya Yazici.
Writing – review & editing: Demet Akdeniz Odemis, Rejin Kebudi, Hulya Yazici.
Abbreviations:
- CNV
- copy number variation
- LMIC
- low middle-income countries
- MLPA
- multiplex ligation probe amplification
- NGS
- next generation sequencing
- RB1 =
- retinoblastoma gene
This work was supported by the Scientific Research Projects Coordination Unit of Istanbul University (Project number: BYP-2016-20546). The study was approved by the Local and Clinical Research Ethics Committee of Istanbul University (Ethical approval no: 2013/252); according to the tenets of the Declaration of Helsinki (JAMA 1997; 277:925-926).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
How to cite this article: Akdeniz Odemis D, Kebudi R, Bayramova J, Kilic Erciyas S, Kuru Turkcan G, Tuncer SB, Sukruoglu Erdogan O, Celik B, Kurt Gultaslar B, Buyukkapu Bay S, Tuncer S, Yazici H. RB1 gene mutations and genetic spectrum in retinoblastoma cases. Medicine 2023;102:36(e35068).
Contributor Information
Rejin Kebudi, Email: rejinkebudi@yahoo.com.
Jamila Bayramova, Email: jamilabayramova6@gmail.com.
Seda Kilic Erciyas, Email: sedaklc@gmail.com.
Gozde Kuru Turkcan, Email: gzzdekuru@gmail.com.
Seref Bugra Tuncer, Email: sbtuncer@yahoo.com.
Ozge Sukruoglu Erdogan, Email: sukruogluozge@gmail.com.
Betul Celik, Email: celikbetul6@gmail.com.
Busra Kurt Gultaslar, Email: bbusrakurt92@gmail.com.
Sema Buyukkapu Bay, Email: semabbay@yahoo.com.tr.
Samuray Tuncer, Email: sbtuncer@yahoo.com.
Hulya Yazici, Email: hulyayazici67@gmail.com.
References
- [1].Demet Akdeniz SBT, Yazici H. Retinoblastoma (RB) gene pathway and cancer. Turkish J Oncol. 2014;29:173–80. [Google Scholar]
- [2].Pandey AN. Retinoblastoma: an overview. Saudi J Ophthalmol. 2014;28:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Leal-Leal C, Flores-Rojo M, Medina-Sansón A, et al. A multicentre report from the Mexican Retinoblastoma Group. Br J Ophthalmol. 2004;88:1074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Canturk S, Qaddoumi I, Khetan V, et al. Survival of retinoblastoma in less-developed countries impact of socioeconomic and health-related indicators. Br J Ophthalmol. 2010;94:1432–6. [DOI] [PubMed] [Google Scholar]
- [5].Young JL, Jr., Ries LG, Silverberg E, et al. Cancer incidence, survival, and mortality for children younger than age 15 years. Cancer. 1986;58(2 Suppl):598–602. [DOI] [PubMed] [Google Scholar]
- [6].Cebeci Z, Tuncer S, Kebudi R. Clinical features and long-term follow-up of patients with retinoblastoma in Turkish children older than 5 years of age. J Ophthalmol. 2020;2020:8148013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Akdeniz D, Tuncer SB, Kebudi R, et al. Investigation of new candidate genes in retinoblastoma using the TruSight One “clinical exome” gene panel. Mol Genet Genomic Med. 2019;7:e785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yazici H, Wu H-C, Tigli H, et al. High levels of global genome methylation in patients with retinoblastoma. Oncol Lett. 2020;20:715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Akdeniz Odemis D, Tuncer SB, Adamnejad Ghafour A, et al. FGFR4 c.1162G > A (p.Gly388Arg) polymorphism analysis in Turkish patients with retinoblastoma. J Oncol. 2020;2020:9401038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–7. [DOI] [PubMed] [Google Scholar]
- [11].Knudson AG, Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309:458–60. [DOI] [PubMed] [Google Scholar]
- [13].McEvoy J, Nagahawatte P, Finkelstein D, et al. RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget. 2014;5:438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kooi IE, Mol BM, Massink MPG, et al. Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes. Sci Rep. 2016;6:25264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lohmann D. Retinoblastoma. in Diseases of DNA Repair, S.I. Ahmad, Editor. 2010. New York: Springer New York, NY. p. 220–7. [Google Scholar]
- [16].de Jong MC, Kors WA, de Graaf P, et al. Trilateral retinoblastoma: a systematic review and meta-analysis. Lancet Oncol. 2014;15:1157–67. [DOI] [PubMed] [Google Scholar]
- [17].Soliman SE, Racher H, Zhang C, et al. Genetics and molecular diagnostics in retinoblastoma--An update. Asia Pac J Ophthalmol (Phila). 2017;6:197–207. [DOI] [PubMed] [Google Scholar]
- [18].Bornfeld N, Biewald E, Bauer S, et al. The interdisciplinary diagnosis and treatment of intraocular tumors. Dtsch Arztebl Int. 2018;115:106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jagadeesan M, Khetan V, Mallipatna A. Genetic perspective of retinoblastoma: From present to future. Indian J Ophthalmol. 2016;64:332–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yun J, Li Y, Xu C-T, et al. Epidemiology and Rb1 gene of retinoblastoma. Int J Ophthalmol. 2011;4:103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bethesda M. Retinoblastoma treatment (PDQ®)–Health Professional Version. 2022; Available at: https://www.cancer.gov/types/retinoblastoma/hp/retinoblastoma-treatment-pdq.
- [22].Kutluk MT, Yesilipek A. Pediatric cancer registry Turkey: 2009-2016 (TPOG & TPHD). J Clin Oncol. 2017;35(15_Suppl):e22015–e22015. [Google Scholar]
- [23].Kebudi R, Alkaya DU. Epidemiology and survival of childhood cancer in Turkey. Pediatr Blood Cancer. 2021;68:e28754. [DOI] [PubMed] [Google Scholar]
- [24].Illumina VariantStudio Data Analysis Software. 2016.
- [25].Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].HGVS Variant Description Nomenclature. Available at: https://www.hgvs.org/ [Access date May 1, 2020].
- [27].Berry JL, Murphree AL. Retinoblastoma: staging and grouping. In: Berry J, Kim J, Damato B, Singh A, eds. Clinical Ophthalmic Oncology. Cham: Springer; 2019. [Google Scholar]
- [28].Dimaras H, Khetan V, Halliday W, et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet. 2008;17:1363–72. [DOI] [PubMed] [Google Scholar]
- [29].Xie Y, Xu XL, Wei WB. The RB1 mutation spectrum and genetic management consultation in pediatric patients with retinoblastoma in Beijing, China. Risk Manag Healthc Policy. 2021;14:3453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dommering CJ, Mol BM, Moll AC, et al. RB1 mutation spectrum in a comprehensive nationwide cohort of retinoblastoma patients. J Med Genet. 2014;51:366–74. [DOI] [PubMed] [Google Scholar]
- [31].Draper GJ, Sanders BM, Brownbill PA, et al. Patterns of risk of hereditary retinoblastoma and applications to genetic counselling. Br J Cancer. 1992;66:211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Abramson DH, Frank CM, Susman M, et al. Presenting signs of retinoblastoma. J Pediatr. 1998;132:505–8. [DOI] [PubMed] [Google Scholar]
- [33].Lan X, Xu W, Tang X, et al. Spectrum of RB1 germline mutations and clinical features in unrelated Chinese patients with retinoblastoma. Front Genet. 2020;11:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kiet NC, Khuong Le Thai, Minh DD, et al. Spectrum of mutations in the RB1 gene in Vietnamese patients with retinoblastoma. Mol Vis. 2019;25:215–21. [PMC free article] [PubMed] [Google Scholar]
- [35].Rojanaporn D, Boontawon T, Chareonsirisuthigul T, et al. Spectrum of germline RB1 mutations and clinical manifestations in retinoblastoma patients from Thailand. Mol Vis. 2018;24:778–88. [PMC free article] [PubMed] [Google Scholar]
- [36].Mohd Khalid MK, Yakob Y, Md Yasin R, et al. Spectrum of germ-line RB1 gene mutations in Malaysian patients with retinoblastoma. Mol Vis. 2015;21:1185–90. [PMC free article] [PubMed] [Google Scholar]
- [37].Ayari-Jeridi H, Moran K, Chebbi A, et al. Mutation spectrum of RB1 gene in unilateral retinoblastoma cases from Tunisia and correlations with clinical features. PLoS One. 2015;10:e0116615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Parma D, Ferrer M, Luce L, et al. RB1 gene mutations in Argentine retinoblastoma patients. Implications for genetic counseling. PLoS One. 2017;12:e0189736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Global Retinoblastoma Study G, et al. Global retinoblastoma presentation and analysis by National Income Level. JAMA Oncol. 2020;6:685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gallie BL, Ellsworth RM, Abramson DH, et al. Retinoma: spontaneous regression of retinoblastoma or benign manifestation of the mutation? Br J Cancer. 1982;45:513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kamihara J, Bourdeaut F, Foulkes WD, et al. Retinoblastoma and neuroblastoma predisposition and surveillance. Clin Cancer Res. 2017;23:e98–e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tanwar M, Balaji S, Vanniarajan A, et al. Parental age and retinoblastoma-a retrospective study of demographic data and genetic analysis. Eye (Lond). 2022;36:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mehta M, Sethi S, Pushker N, et al. Retinoblastoma. Singapore Med J. 2012;53:128–35; quiz 136. [PubMed] [Google Scholar]
- [44].Queremel Milani D.A. and Chauhan P.R.. Genetics, Mosaicism, in StatPearls. 2022: Treasure Island (FL). [PubMed] [Google Scholar]
- [45].Miles B. and Tadi P.. Genetics, Somatic Mutation, in StatPearls. 2022: Treasure Island (FL). [PubMed] [Google Scholar]
- [46].Verma RS, Kleyman SM, Conte RA. Chromosomal mosaicisms during prenatal diagnosis. Gynecol Obstet Invest. 1998;45:12–5. [DOI] [PubMed] [Google Scholar]
- [47].Dommering CJ, Marees T, van der Hout AH, et al. RB1 mutations and second primary malignancies after hereditary retinoblastoma. Fam Cancer. 2012;11:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wong FL, Boice JD, Abramson DH, et al. Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA. 1997;278:1262–7. [DOI] [PubMed] [Google Scholar]
- [49].Omidakhsh N, Hansen J, Ritz B, et al. Parental occupation and risk of childhood retinoblastoma in Denmark. J Occup Environ Med. 2021;63:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mukamal R. Why are brown eyes most common? 2017. Available at: https://www.aao.org/eye-health/tips-prevention/why-are-brown-eyes-most-common [Access date Apr 07, 2017].
- [51].Klutz M, Brockmann D, Lohmann DR. A parent-of-origin effect in two families with retinoblastoma is associated with a distinct splice mutation in the RB1 gene. Am J Hum Genet. 2002;71:174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kanber D, Berulava T, Ammerpohl O, et al. The human retinoblastoma gene is imprinted. PLoS Genet. 2009;5:e1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]