Abstract
We describe a case of “Flexispira rappini” bacteremia from a 9-year-old girl who presented with a 5-day history of fever, productive cough, and malaise. A chest X-ray result was compatible with right middle lobe pneumonia. Blood culture grew a gram-negative spiral fusiform bacterium 2 days after the inoculation. Biochemical tests showed the organism to be catalase negative, oxidase positive, sodium hippurate hydrolysis negative, and urea hydrolysis negative. 16S rRNA gene sequencing identified this organism as “F. rappini,” showing a six-base substitution from the type strain. This is the first report of “F. rappini” bacteremia in a human, suggesting that this organism has the potential of causing invasive infection, but its role in pneumonia is uncertain and could be unrelated to the bacteremia.
“Flexispira rappini” is described as a microaerophilic, urease-producing, fusiform, gram-negative bacterium with bipolar sheathed flagella (1). 16S rRNA gene sequence analysis has shown that this organism is closely related to and shares morphological features with some of the newly discovered Helicobacter spp., including H. bilis (4), H. canis (19), and H. acinonyx (3).
“F. rappini” has been isolated from aborted sheep fetuses (2, 8), from the intestinal mucosa of laboratory mice (18), and from the stool samples of two humans with mild chronic diarrhea (17). The organism has also been recovered from the stool of an asymptomatic family member of one of these patients and from his 5-month-old healthy puppy (17). The association of this organism with human gastrointestinal disease is yet to be defined. We describe the first case of “F. rappini” bacteremia suggestive of pneumonia in a child and the use of 16S rRNA gene sequencing to identify the organism.
CASE REPORT
A 9-year-old girl was admitted to hospital with a 5-day history of fever, cough productive of white sputum, decreased oral intake, and general malaise. She had not complained of any diarrhea or abdominal pain. She had a past history of an appendectomy 3.5 years earlier and recurrent chest infections over the last 2 years, which were treated with multiple courses of antibiotics, but which did not require hospitalization. On the day prior to admission, she visited her local doctor, who had made a clinical diagnosis of pneumonia and had suggested transfer to the hospital, but had not started her on any antibiotic treatment.
On examination, she had a temperature of 39.6°C, and had focal chest signs of decreased air entry, inspiratory crepitations, and bronchial breathing in the right middle zone. The abdominal examination was normal. Laboratory investigations revealed hemoglobin of 141 g/liter, leukocyte count of 5.0 (neutrophils, 2.7; lymphocytes, 1.5) × 109/liter, and a platelet count of 248 × 109/liter. C-reactive protein was 47 mg/liter. A blood sample was taken in the Emergency Department, and one pediatric blood culture bottle was inoculated. A chest X ray was performed, and this showed peribronchial thickening and patchy consolidation in the right middle zone, compatible with a diagnosis of pneumonia. No sputum specimen was sent for culture.
The patient was empirically treated with intravenous penicillin at a dose of 1.2 g every 6 h. Within 48 h of starting the antibiotics, her condition had markedly improved, with resolution of her fevers and return of her appetite. The intravenous penicillin was ceased after 72 h, and she was discharged home with a 7-day course of oral erythromycin at 250 mg every 8 h.
The patient was reviewed in the Outpatient Department 3 months later. She had remained well postdischarge, and a repeat X ray was clear. Of interest, a 10-week-old puppy had joined the patient’s household about 6 weeks before she became unwell. The puppy was healthy. There were also two cats in the house; both had lived there for many years and were well.
MATERIALS AND METHODS
The strain was recovered from the inoculated pediatric bottle of the BacT/Alert Microbial Detection System (Organon Technika, Turnhout, Belgium) after 2 days of incubation. The contents of the bottle were subcultured onto a 6% sheep blood agar plate and incubated in a microaerobic atmosphere, and the culture was examined daily for growth.
Biochemical tests for Campylobacter sp., Gram smear morphology, and phase-contrast microscopy were performed by standard methods.
Antibiotic susceptibility testing.
The isolate was tested for susceptibility to ampicillin, amoxicillin-clavulanate, metronidazole, cephalothin, nalidixic acid, imipenem, doxycycline, chloramphenicol, ciprofloxacin, erythromycin, and gentamicin by a disk diffusion test method. A total of 0.1 ml of a suspension of 105 organisms in brain heart infusion broth was evenly spread over the surface of a 6% sheep blood agar plate (Columbia agar base), and the susceptibility test disk (Oxoid, Basingstoke, England) was placed on top of the lawn culture. The plates were incubated under hydrogen-enhanced microaerobic conditions at 37°C. After 48 h, the plates were read and interpreted according to the recommendations of the National Committee for Clinical Laboratory Standards guidelines for gram-negative bacteria (13).
Chromosomal DNA extraction, PCR, 16S rRNA gene sequencing, and rRNA sequence alignment.
The extraction and purification of chromosomal DNA from the plate culture were performed as previously described (21). DNA amplification of small subunit rRNA genes was performed by using the consensus terminal primers as previously described (20). The full-length product (∼1,500 nucleotides [nt]) was sequenced directly by using terminal primers and internal primers specific for 16S rRNA genes (9) in an automatic DNA sequencer (model 373A; Applied Biosystems, Inc., Foster City, Calif.) by using a dye-labelled deoxy termination method (Taq Dye-Deoxy Terminator Cycle Sequencing Kit; Applied Biosystems, Inc.).
Nucleotide sequence accession number.
The rRNA gene sequence of the isolate obtained in this study has been lodged with the GenBank database under accession no. AF034135.
RESULTS
Two days after inoculation of a pediatric bottle of the BacT/Alert Blood Culture System, the bottle flagged positive. Gram staining of the blood culture revealed no organisms, and the blood culture was subcultured onto two 6% horse blood agar plates, a MacConkey agar plate, and a chocolate agar plate. One of the inoculated horse blood agar plates was incubated under anaerobic conditions, while the remaining three plates were incubated under 5% CO2 at 35°C for 2 days. However, no growth was detected after 2 days of incubation.
The blood culture bottle was reloaded into the BacT/Alert Blood Culture System for a further 24 h, and it again flagged positive. A wet preparation of the blood culture revealed organisms with darting motility. The Gram smear showed a long, thin, but slightly curved gram-negative rod. Because a Campylobacter sp. was suspected, a set of plates similar to the ones described above were inoculated and incubated at 42°C under microaerobic conditions (Gas Generating Kit, Campylobacter System BR 056A; Oxoid). Growth was detected after 2 days of incubation.
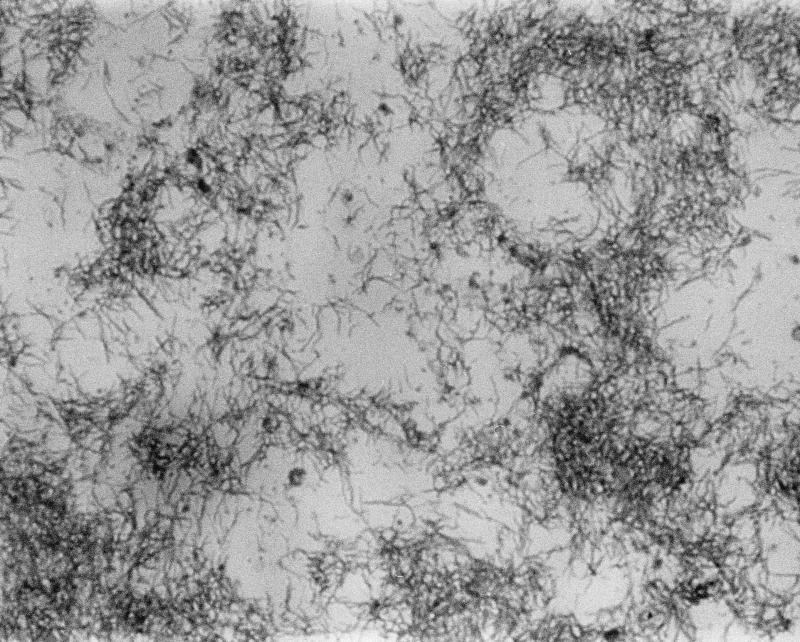
The organism took 2 days to grow into a greyish, wavy, glistening colony 1 to 2 mm in diameter with an entire edge. Growth was detected at 37 and 42°C under microaerobic conditions and in the presence of enhanced H2, but not at 25°C. Gram staining of the colony showed a gram-negative, slightly curved fusiform bacterium (Fig. 1). Biochemical tests showed the organism was oxidase positive, but it had negative reactions to catalase, nitrate reduction, urea hydrolysis, and sodium hippurate hydrolysis. It was sensitive to nalidixic acid (30-μg disc), but resistant to cephalothin (30-μg disc). The organism was fastidious and did not survive upon subsequent subculture. Further tests or repeated testing of some of the biochemical reactions were not possible. Samples from the first and second subcultures, which had been stored at −70°C, were retrieved and were found to be no longer viable.
FIG. 1.
Light microscopy photomicrograph of a Gram smear of a culture of the clinical isolate of “F. rappini” showing characteristic slightly curved fusiforms. Magnification, ×1,000.
Susceptibility testing of the organism with a disk diffusion test showed the organism to be sensitive to most antibiotics, including ciprofloxacin, gentamicin, erythromycin, doxycycline, amoxicillin-clavulanate, metronidazole, imipenem, and chloramphenicol. Because the organism was not viable upon subculture, we were unable to determine the MICs of various antibiotics for the organism.
The 16S rRNA gene sequence of our isolate was determined (GenBank accession no. AF034135) and was compared to others in the sequence databases. The closest match was to the 16S rRNA gene of “F. rappini” NADC 1937 (GenBank accession no. M88138), which showed only six base substitutions. These occurred at positions 18 (A to C), 427 (A to T), 450 (T to A), 1099 (T to C), 1293 (A to G), and 1325 (A to C). However, the sequences of the four “F. rappini” strains in the databases were incomplete, and there were a number of gaps and undetermined nucleotides (N’s). Three of these are only partial sequences of less than 1,000 nt. The most complete sequence (of strain NADC 1937) is 1,421 nt long, but it contains 25 uncertain bases (gaps and N’s). The sequence determined for our isolate is unambiguous and is 1,488 nt long.
There was some difficulty in assigning the closest relative of our isolate, because the 16S rRNA gene sequence of “F. rappini” is a member of a closely related group, mostly classified as Helicobacter spp., and because the quality of the 16S rRNA gene sequences in the databases is variable. Other similar sequences include those of H. bilis ATCC 51631 (11 base differences over 1430 nt), Helicobacter cinaedi ATCC 35683 (17 base differences over 1,439 nt), and H. canis NCTC 12739 (24 base differences over 1,408 nt).
DISCUSSION
In this report, we describe the first case of “F. rappini”-associated bacteremia suggestive of pneumonia in a child. The presence of clinical indications and radiological evidence of pneumonia when the organism was isolated from the bloodstream may suggest this organism as a cause of this condition, but we are uncertain. The organism may cause transient bacteremia. Antibiotic therapy resulted in resolution of symptoms and clinical improvement. However, the response to penicillin is not compatible with the organism’s in vitro resistance to cephalothin. The clinical isolate was sensitive to erythromycin in vitro, but the response to antibiotics may be coincidental.
Previous reports indicate that this organism causes mild gastroenteritis, but our patient had no gastrointestinal symptoms. The origin of “F. rappini” infection in our patient is not clear. In view of the fact that a previous report demonstrated “F. rappini” in the feces of an asymptomatic puppy of an infected patient, we postulate that the most likely source of infection was from contact with the household’s recently acquired puppy. Unfortunately, because no attempt was made to culture this organism from any of the domestic pets in the patient’s household, we cannot be certain whether the puppy or other animals, such as cats, may also harbor this organism.
It is known that Helicobacter spp. have a wide range of niches and can be found in mammals, including rodents (4, 5, 11), cheetahs (3), hamsters (7), ferrets (6), cats (10), dogs (19), and humans (12, 14). “F. rappini” has been isolated from aborted ovine fetuses (2, 8), laboratory mice (18), dogs, and humans (1, 17). Other reservoirs for this infection have yet to be found.
Limited biochemical tests performed for this organism showed that it has similar morphological and biochemical reactions to the previously reported isolates, with one exception. Our isolate tested negative for urea hydrolysis, compared to all other isolates, which produced rapid positive reactions. We are uncertain whether our isolate represents a variant of the reported strains or whether our test of urea hydrolysis yielded a false-negative reaction because the isolate was already dead. The original laboratory which referred the isolate did not perform the urea hydrolysis reaction test. They reported similar results in the other tests. Unfortunately, the original BacT/Alert bottle was discarded after growth of a plate culture.
“F. rappini” is a fastidious organism. One of the three isolates described by Archer et al. (1) was subsequently lost during biochemical tests. Our isolate lasted two subcultures, despite the fact that the primary plate showed such abundant growth and that great care and conscious efforts were made not to leave it in the air longer than 2 h during testing. In spite of this, several attempts to revive it failed. Similar problems occurred with the type strain of “F. rappini” obtained from the American Type Culture Collection, even though two repeated isolates were sent when requested.
Like most campylobacters and helicobacters, “F. rappini” is biochemically inert in standard laboratory tests, and there are only a limited range of phenotypic tests available for identification and differentiation. These groups of organisms usually require more advanced techniques for definitive identification. Because we have encountered great difficulties in growing the type strain to be used for testing, we were unable to use a number of recognized methods, such as DNA-DNA hybridization, whole-cell protein banding profiles, or gas-liquid chromatography of fatty acids, to characterize this organism.
16S rRNA gene sequencing was used in this study to identify the organism. This method allows identification of the organism without the prior knowledge of the gene sequence and without the need for a culture or the DNA of a type strain. The organism was lost during testing, but sufficient DNA was extracted prior to the loss of a culture to enable us to continue with further analysis. The presence of more than 5,000 16S rRNA gene sequences of bacteria, including reference strains, in the databases that are readily available and accessible allows genus and species identification of bacterial pathogens. As demonstrated in this case and by many previous studies, comparative 16S rRNA sequence analysis is a useful and powerful tool for identification, classification, and phylogenetic study of bacteria, especially for those that are difficult to grow or that are uncultivable (15, 16).
REFERENCES
- 1.Archer J R, Romero S, Ritchie A E, Hamacher M E, Steiner B M, Bryner J H, Schell R F. Characterization of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:101–105. doi: 10.1128/jcm.26.1.101-105.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryner J H, Ritchie A E, Pollet L, Kirkbride C A, Collins J E. Experimental infection and abortion of pregnant guinea pigs with a unique spirillum-like bacterium isolated from aborted ovine fetuses. Am J Vet Res. 1987;48:91–95. [PubMed] [Google Scholar]
- 3.Eaton K A, Dewhirst F E, Radin M J, Fox J G, Paster B J, Krakowka S, Morgan D R. Helicobacter acinonyx sp. nov., isolated from cheetahs with gastritis. Int J Syst Bacteriol. 1993;43:99–106. doi: 10.1099/00207713-43-1-99. [DOI] [PubMed] [Google Scholar]
- 4.Fox J G, Yan L L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Jr, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrappings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox J G, Edrise B M, Cabot E B, Beaucagr V C, Murphy J C, Prostak K S. Campylobacter-like organisms isolated from gastric mucosa in ferrets. Am J Vet Res. 1986;47:236–239. [PubMed] [Google Scholar]
- 7.Gebhart C J, Fennell C I, Murtagh M P, Stamm W E. Campylobacter cinaedi is a normal intestinal flora in hamsters. J Clin Microbiol. 1989;27:1692–1694. doi: 10.1128/jcm.27.7.1692-1694.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirkbride C A, Gates C E, Collins J E, Ritchie M S. Ovine abortion associated with an anaerobic bacterium. J Am Vet Med Assoc. 1985;186:789–791. [PubMed] [Google Scholar]
- 9.Lane D J. 16/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 10.Lee A, Hazell S L, O’Rourke J L, Kouprach S. Isolation of a spiral shaped bacterium from the cat stomach. Infect Immunol. 1988;56:2843–2850. doi: 10.1128/iai.56.11.2843-2850.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee A, Phillips M W, O’Rourke J L, Paster B J, Dewhirst F E, Fraser G J, Fox J G, Sly L I, Romaniuk P J, Trust T J, Kourprach S. Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int J Syst Bacteriol. 1992;42:27–36. doi: 10.1099/00207713-42-1-27. [DOI] [PubMed] [Google Scholar]
- 12.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for antimicrobial susceptibility testing for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Quinn T C, Goodell S E, Fennell C, Wang S-P, Schuffler M D, Holmes K K, Stamm W E. Infection with Campylobacter jejuni and campylobacter-like organisms in homosexual men. Ann Intern Med. 1984;101:187–192. doi: 10.7326/0003-4819-101-2-187. [DOI] [PubMed] [Google Scholar]
- 15.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkin L S. The agent of bacillary angiomatosis—an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 16.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 17.Romero S, Archer J R, Hamacher M E, Bologna S M, Schell R F. Case report of an unclassified microaerophilic bacterium associated with gastroenteritis. J Clin Microbiol. 1988;26:142–143. doi: 10.1128/jcm.26.1.142-143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauer D B, Ghori N, Falkow S. Isolation of “F. rappini” from laboratory mice. J Clin Microbiol. 1993;31:2709–2714. doi: 10.1128/jcm.31.10.2709-2714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanley J, Linton D, Buenens A P, Dewhirst F E, Owen R J, Porter A, On S L W, Costas M. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J Gen Microbiol. 1993;139:2495–2504. doi: 10.1099/00221287-139-10-2495. [DOI] [PubMed] [Google Scholar]
- 20.Tee W, Dyall-Smith M, Woods W, Eisen D. Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J Clin Microbiol. 1996;34:1760–1764. doi: 10.1128/jcm.34.7.1760-1764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 2.4.1–2.4.5. [Google Scholar]