Abstract
The incidence of salmonellosis has been increasing in Hong Kong since 1989. The most common Salmonella enterica serotype isolated in 1994 was S. enteritidis. The antimicrobial susceptibilities and molecular epidemiology of 275 S. enteritidis strains isolated in this locality between 1986 and 1996 were studied. Over 99% of the isolates were susceptible to 17 of the 19 antimicrobial agents tested. One isolate harbored an autotransferring plasmid that confers resistance to tetracycline and trimethoprim-sulfamethoxazole. Another isolate harbored a mobilizable plasmid that confers resistance to ampicillin and cephalothin. This isolate was found to produce a β-lactamase with a pI of 5.2. A total of 264 isolates (96%) were found to harbor one to five plasmids, and the majority (254) harbored a 60-kb plasmid. Of these isolates, 94% contained identical 60-kb plasmids. Based on plasmid profiles, plasmid and chromosomal fingerprints, ribotypes, and randomly amplified polymorphic DNA (RAPD) patterns, 170 (62%) isolates were allocated to group 1b. About 90% of isolates had identical or similar DNA fingerprints, ribotypes, and RAPD patterns, suggesting that a predominant clone of S. enteritidis was circulating in Hong Kong during the period being studied.
Despite improved sanitation, salmonellosis remains a major public health concern in many countries. The incidence of salmonella infections has been on the increase over the past 10 years. The organism that is most often responsible for the illness is Salmonella enterica serotype Enteritidis (referred to herein as S. enteritidis) (34–36, 39).
There has been a gradual but significant increase in group D salmonella infections, particularly S. enteritidis, in Hong Kong since 1989 (50). S. enteritidis has become the most-common group D Salmonella isolate and the third-most-common Salmonella serotype from extraintestinal sources in this city.
S. enteritidis is known to cause gastroenteritis and other acute infections. There is, however, little information on its antimicrobial susceptibilities and epidemiology, which would help prevent the spread of the infections and provide data about the best choices for treatment. Our aim in the present study was to investigate the antimicrobial susceptibilities and characteristics of resistant strains and the molecular epidemiology of this organism.
MATERIALS AND METHODS
Bacterial strains.
Two hundred fifty-two nonduplicate isolates of S. enteritidis were obtained from stools, blood, pus, or body fluids of patients admitted to the Prince of Wales Hospital in Hong Kong, China. Twenty-three more isolates were obtained from the stools of patients attending outpatient clinics. All specimens were collected between 1986 and 1996. Ninety-two percent of the strains were isolated from patients within 3 days of their admission to the hospital. The remaining strains were isolated from patients who were hospitalized for more than 3 days and could therefore have been acquired at the hospital. Four such strains were isolated from children under 5 years old. The locations where the infections originated were not known, but there was no apparent clustering of cases.
Identification.
All strains were identified by the API20E system (bioMérieux, Lyon, France) and a slide agglutination test with Salmonella-specific antisera 9-O and g,m-H (Wellcome Diagnostics, Dartford, United Kingdom).
Antimicrobial susceptibilities.
Susceptibilities to 19 antimicrobial agents (ampicillin, cephalothin, cefuroxime, ceftriaxone, cefotaxime, ceftazidime, gentamicin, tobramycin, netilmicin, amikacin, streptomycin, kanamycin, nalidixic acid, ciprofloxacin, ofloxacin, rifampin, trimethoprim-sulfamethoxazole, tetracycline, and chloramphenicol) were tested by determining the MICs by the agar dilution method (Mueller-Hinton agar; Oxoid, Basingstoke, United Kingdom) according to recommendations put forth by the National Committee for Clinical Laboratory Standards (27).
Plasmid profile analysis.
Crude plasmids were extracted as described by Kado and Liu (12). Plasmids were separated on 0.7% agarose gels in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA [pH 8.3]) at 100 V for 4 to 5 h (GNA200 horizontal electrophoresis system; Pharmacia LKB, Uppsala, Sweden) and visualized by staining with 0.5 mg of ethidium bromide per ml.
Plasmid fingerprinting.
Plasmids of the same size were purified by the method of Kado and Liu (12) and digested with EcoRI and EcoRV (Gibco BRL, Gaithersburg, Md.). Individual plasmids in strains that harbored more than one plasmid were purified from agarose gels after electrophoretic separation (Sephaglas BandPrep kit; Pharmacia Biotech Biotechnology, Uppsala, Sweden).
Transferability of resistance plasmids.
Six isolates that were resistant to more than one antibiotic but not to nalidixic acid and rifampin or highly resistant to antibiotics were selected. The isolates were tested for the ability to transfer or mobilize antibiotic resistance (2) with rifampin-resistant Escherichia coli Jp995 and transfer factors X and Δ (a gift from B. Rowe, Colindale, United Kingdom). Donors were counterselected by 64 mg of rifampin per liter in MacConkey agar (Oxoid).
Characterization of β-lactamases.
β-Lactamases were extracted from ampicillin-resistant isolates by sonication, subjected to isoelectric focusing on pH 3.5 to 9.5 LKB Ampholine PAGplates (Pharmacia LKB) on an Ultrophor unit (Pharmacia LKB), and detected by 1 mg of nitrocefin (Oxoid) per liter.
Total DNA fingerprinting.
Cells grown overnight in Mueller-Hinton broth were pelleted and washed with 1 ml of TE-1 buffer (50 mM Tris-HCl, 20 mM EDTA [pH 8]). The washed cell pellet was resuspended in a solution containing 540 μl of TE-2 buffer (50 mM Tris-HCl, 10 mM EDTA [pH 8]), 12 μl of a 10-mg/ml RNase solution, 30 μl of 10% sodium dodecyl sulfate (SDS), 12 μl of a 5-mg/ml lysozyme solution, and 6 μl of a 20-mg/ml proteinase K solution and incubated overnight at 56°C. The solution was extracted once with an equal volume of buffered phenol, twice with an equal volume of buffered phenol-chloroform-isoamyl alcohol (25:24:1), and once with an equal volume of chloroform-isoamyl alcohol (24:1). The DNA was precipitated with 16 μl of 5 M NaCl and 2 volumes of ice-cold absolute alcohol. After the mixture was left overnight at −70°C, the DNA pellet was washed with ice-cold 70% alcohol, air dried, and resuspended in 50 μl of distilled water. Ten microliters of the DNA extract (about 4 μg) was digested with 10 U of EcoRV or MluI (Gibco BRL), and the digested fragments were separated on a 0.4% agarose gel for 15 h at 80 V and 9 h at 100 V, respectively (GNA200 horizontal electrophoresis system; Pharmacia LKB).
Ribotyping.
Five microliters (about 2 μg) of total DNA was digested with 5 U of PvuII (Gibco BRL). The digested fragments were separated on 0.8% agarose gels at 100 V for 4 1/2 h and transferred onto nylon membranes (Hybond-N+; Amersham, Buckinghamshire, United Kingdom) in a vacuum blotting system (LKB2016 VacuGene; Pharmacia LKB) according to the manufacturer’s instructions.
A cDNA probe was prepared by reverse transcription of 16S plus 23S rRNA (4 μg/μl; Boehringer, Mannheim, Germany). The probe was labelled with the digoxigenin DNA labelling kit (Boehringer), and avian myeloblastosis virus reverse transcriptase (20 U/ml; Finnzymes, Espoo, Finland) as described by Popovic et al. (32). The labelled probe was purified by passage through a Bio-spin 30 column (Bio-Rad Laboratories Inc., Richmond, Calif.), boiled for 10 min, cooled on ice for 5 min, and then used immediately for hybridization or stored at −20°C until use.
The nylon membrane was prehybridized in 80 ml of hybridization solution (32) at 68°C for 2 h and then hybridized overnight at 68°C in 10 ml of hybridization solution containing the cDNA probe (20 μl). The hybridized membrane was washed twice with washing solution A (2× SSC [1× SSC is 0.15 M NaCl, 0.015 M sodium citrate], 0.1% SDS) at room temperature for 5 min and twice with washing solution B (0.1% SSC, 0.1% SDS) at 68°C for 15 min. Hybridized fragments were detected by the digoxigenin nucleic acid detection kit (Boehringer).
Random-amplified polymorphic DNA (RAPD) analysis.
The 15-mer oligonucleotide (TGA GCA TAG ACC TCA) (28) was used as primer. Amplification reactions were carried out in a 25-μl solution containing PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl), 200 μM (each) dATP, dCTP, dGTP, and dTTP, 1.0 μM primer, 1 U of Taq polymerase (Gibco BRL), and 2.5 mM MgCl2. Approximately 40 pg of DNA was added to the mixture, which was then overlaid with 20 μl of liquid wax (Chill-out 14; MJ Research Inc., Watertown, Mass.). Amplification was carried out in a thermal controller (PTC-100; MJ Research) at 94°C for 7 min, followed by 45 cycles (each) at 94°C for 1 min, 40°C for 1 min, and 72°C for 2 min and a final extension of 72°C for 5 min. Amplified products were electrophoresed on 0.8% gels at 100 V for 2 h (GNA100 horizontal electrophoresis system; Pharmacia LKB).
Determination of relatedness of types.
The percentages of similarity between different chromosomal fingerprints, ribotypes, and RAPD types were calculated by the unweighted pair group method with arithmetic averages (38).
RESULTS
A total of 316 strains of S. enteritidis were isolated from human stool and extraintestinal specimens gathered from the Prince of Wales Hospital between 1986 and 1995. These represented 11% of all gastroenteric salmonellae isolated (2,909). Most of the isolates were from stools; only 16% were from blood, pus, or body fluids. Twenty-eight percent of the isolates were from patients younger than 5 years, and 37% were from patients aged between 20 and 39. The male-to-female ratio was 1:1.13. Most of the isolates (79%) were from samples gathered during the hotter months of the year (May to November).
All S. enteritidis isolates tested (275) were susceptible to the expanded-spectrum cephalosporins (ceftriaxone, cefotaxime, and ceftazidime; MIC range, 0.03 to 0.5 mg/liter), the 4-quinolones (ciprofloxacin and ofloxacin; MIC range, 0.0075 to 1 mg/liter), and chloramphenicol (MIC range, 1 to 8 mg/liter); all isolates were resistant to 1 mg of rifampin per liter. One isolate was resistant to ampicillin (MIC = 512 mg/liter) and cephalothin (MIC = 32 mg/liter), and two isolates were resistant to cefuroxime (MIC = 16 mg/liter). One isolate was resistant to the aminoglycoside tobramycin, and two isolates were resistant to streptomycin, but the MICs were low, 8 to 16 mg/liter. One isolate was resistant to 4 mg of tetracycline per liter, two isolates were resistant to 8 mg of nalidixic acid per liter, and two isolates were resistant to 32 mg of trimethoprim-sulfamethoxazole per liter.
β-Lactamase produced by the isolate resistant to ampicillin (MIC > 512 mg/liter) and cephalothin (MIC = 32 mg/liter) had a pI of 5.2. The two isolates that were susceptible to 16 mg of cefuroxime per liter did not produce detectable amounts of β-lactamases.
Six isolates that were resistant to more than one antibiotic were tested for the transferability of their resistance (Table 1). These isolates were from samples collected from inpatients. An isolate which was resistant to tetracycline, trimethoprim-sulfamethoxazole, and rifampin transferred its resistance to an E. coli recipient at frequencies of 1.3 × 10−6 at 28°C and 2.5 × 10−6 at 37°C. This isolate harbored a 105-kb plasmid and a 60-kb plasmid. The tetracycline and trimethoprim-sulfamethoxazole resistance was located on the 105-kb plasmid, since only this plasmid was found in transconjugants resistant to tetracycline and trimethoprim-sulfamethoxazole. Another isolate that was resistant to ampicillin, cephalothin, and rifampin was found to have its ampicillin and cephalothin resistance mobilized by the Δ factor at a frequency of 2.6 × 10−5 at 37°C. As only a single 5.2-kb plasmid (other than the Δ transfer factor) was present in ampicillin- and cephalothin-resistant transconjugants, the ampicillin and cephalothin resistance was attributed to this plasmid. Other isolates which contained one, two, or three plasmids did not transfer their resistances or could not have their resistances mobilized by transfer factors, as resistant transconjugants could not be obtained.
TABLE 1.
Characteristics of multiple-drug-resistant Salmonella enteritidisa
| No. | Yr | Resistance patternb | Resistance plasmid
|
Other plasmids (kb) | pI of βls | ||
|---|---|---|---|---|---|---|---|
| TF at 28°C | TF at 37°C | kb | |||||
| 1 | 1990 | A (>512), CTH (32), RIF (8) | 0 | 2.6 × 10−5c | 5.2 | 60 | 5.2 |
| 2 | 1991 | I (128), SXT (>128), RIF (8) | 1.3 × 10−6 | 2.5 × 10−6 | 105 | 60 | NA |
| 3 | 1991 | SXT (64), RIF (8) | 0 | 0 | ND | 98, 60 | NA |
| 4 | 1991 | CXM (16), RIF (4) | 0 | 0 | ND | 78 | ND |
| 5 | 1992 | CXM (16), S (16), RIF (8) | 0 | 0 | ND | 78 | ND |
| 6 | 1992 | S (16), RIF (4) | 0 | 0 | ND | 78, 3.6, 3.1 | NA |
Abbreviations: Yr, year isolated; TF, transfer frequency; βls, β-lactamase; NA, not applicable; ND, not detected; A, ampicillin; CTH, cephalothin; CXM, cefuroxime; RIF, rifampicin; SXT, trimethoprim-sulfamethoxazole.
Drugs resistance to which was transferred or mobilized are underlined; MICs are shown in parentheses.
Frequency of mobilization.
The number of isolates harboring up to five plasmids with sizes ranging from 1.7 to 105 kb was 264. These isolates were allocated to 21 plasmid profiles (PP1 to PP21). The majority of isolates (226) harbored only one plasmid, and of these, 97% (219) harbored a 60-kb plasmid. Eight percent of isolates harbored two plasmids, and 3% harbored four or five plasmids. Only two isolates harbored three plasmids. The next-most-common large plasmid was 78 kb in size (harbored by eight isolates). Plasmids of other sizes were less common.
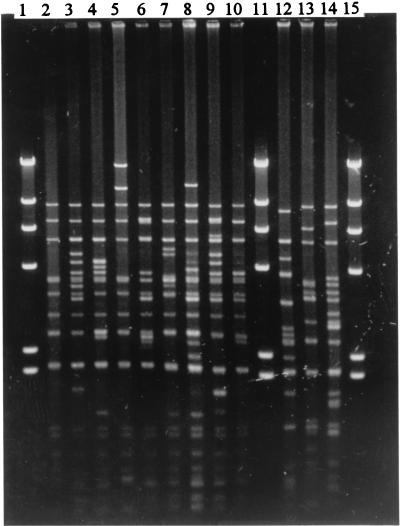
The 60- and 78-kb plasmids were digested with EcoRI and EcoRV, and each restriction pattern was identified by a number after the letters PI (EcoRI digestion) or PV (EcoRV digestion). Nine restriction patterns were seen in the 60-kb plasmid EcoRI digest (PI1 to PI9) and EcoRV digest (PV1 to PV9) (Fig. 1). The PI and PV patterns of the same plasmid were similar; plasmids of one PI pattern frequently had the same PV pattern. The 60-kb plasmids in 240 strains (94% of the 254 60-kb-plasmid-harboring strains) which were isolated from 1987 to 1996 had identical PI1 and PV1 patterns. The plasmids in the remaining 14 strains exhibited the remaining eight patterns. In contrast, the 78-kb plasmids produced three quite different PI (PI10 to PI12) and PV (PV10 to PV12) restriction patterns. Four isolates were found to have restriction pattern PI10-PV10, two isolates were found to have restriction pattern PI11-PV11, and two other isolates had pattern PI12-PV12.
FIG. 1.
EcoRV restriction patterns of Salmonella enteritidis plasmids. Lanes 2 to 10, PV1 to PV9 of the 60-kb plasmid; lanes 12 to 14, PV10 to PV12 of the 78-kb plasmid; lanes 1, 11, and 15, HindIII digest of λ DNA (Gibco BRL) as molecular size marker.
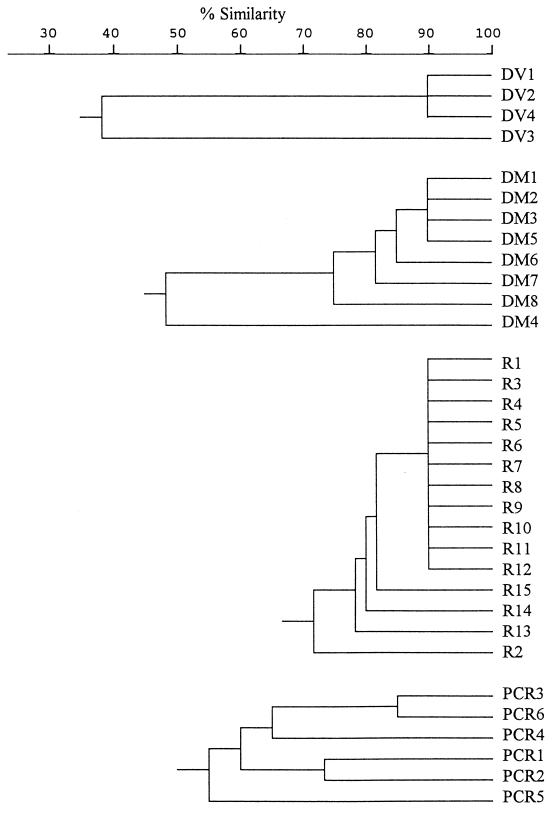
Seven EcoRV fragments ranging from 12.8 to 17.8 kb and six MluI fragments ranging from 20.7 to 51.4 kb were obtained by digesting total DNA with restriction endonucleases EcoRV and MluI. Each restriction pattern was assigned a number following the letters DV (EcoRV digestion) or DM (MluI digestion). All isolates were categorized as one of four DV (DV1 to DV4) types or one of eight DM (DM1 to DM8) types. The relatedness of the chromosomal fingerprints is shown in a dendrogram (Fig. 2). Type DV1, which differed from DV2 by a single band, was seen in 93% of isolates. Ninety-two percent of the isolates were of type DM1. Types DM2 and DM3 differed from DM1 by one fragment and were seen in 3 and 2% of isolates, respectively. The patterns of the remaining types (DM4 to DM8) were less alike; between one and four isolates were one of these types. Most of the isolates (87%) were found to have the pattern DV1-DM1. Only 5, 3, and 2% of isolates were found to have patterns DV2-DM1, DV1-DM2, and DV1-DM3, respectively. Five other patterns contained one to four isolates each.
FIG. 2.
Dendrogram with chromosomal fingerprints obtained after EcoRV or MluI digestion (DV or DM, respectively), ribotypes (R), and RAPD (PCR).
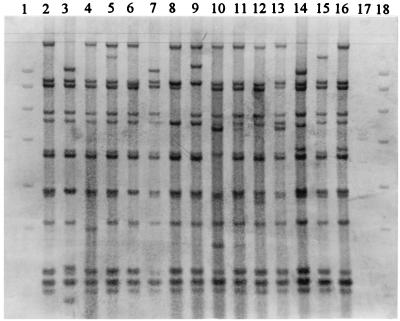
The isolates were allocated to 15 ribotypes (R1 to R15) by PvuII restriction. The patterns were similar, differing only in one to three fragments (Fig. 3). The number of fragments which contained rRNA genes was 13 to 15, and their sizes ranged from 1.8 to 14.0 kb. R1 was the most-frequent type (91% of isolates). Only one to four isolates were found in each of the remaining ribotypes.
FIG. 3.
PvuII ribotypes of Salmonella enteritidis. Lanes 2 to 16, R1 to R15; digoxigenin-labelled DNA molecular size markers III (21, 5.2, 5.0, 4.3, 3.5, 2.0, 1.9, and 1.6 kb) and VII (8.0, 7.1, 6.0, 4.8, 3.5, 2.7, 1.9, 1.8, and 1.5 kb) (both from Boehringer) are used in lanes 1 and 17 and lane 18, respectively.
Six different RAPD types (PCR1 to PCR6), each with 2 to 11 fragments ranging from 0.5 to 4.0 kb, were observed among the 275 isolates. Ninety-five percent of isolates had pattern PCR1; one to five isolates were found in each of the remaining RAPD types.
Based on the pattern of plasmid profiles, plasmid and chromosomal fingerprints, ribotypes, and RAPD types, all isolates could be divided into 28 groups (1 to 28; Table 2). Isolates of the same DNA fingerprints, ribotypes, and RAPD types belonged to the same group. Each group was further subdivided according to plasmid profiles and plasmid fingerprints. These subgroups were assigned alphabetical suffixes to the main group to indicate relatedness. There were 20 subgroups within group 1 (1a to 1t), 4 in group 2 (2a to 2d), and 3 in group 3 (3a to 3c).
TABLE 2.
Pattern combination of plasmid profiles, plasmid and chromosomal fingerprints, ribotypes, and RAPD typesa
| Pattern | PP | PI/PV | DV | DM | R | PCR | No. of strains isolated in indicated yr
|
No. of strains from OP | Total (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1986 | 1987 | 1988 | 1989 | 1990 | 1991 | 1992 | 1993 | 1994 | 1995 | |||||||||
| 1a | Nil | DV1 | DM1 | R1 | PCR1 | 2 | 3 | 2 | 7 (3) | |||||||||
| 1b | PP1 | PI1/PV1 | DV1 | DM1 | R1 | PCR1 | 1 | 1 | 7 | 13 | 7 | 23 | 31 | 27 | 44 | 16 | 170 (62) | |
| 1c | PP1 | PI2/PI2 | DV1 | DM1 | R1 | PCR1 | 2 | 3 | 5 (2) | |||||||||
| 1d–1t | Various | Various | DV1 | DM1 | R1 | PCR1 | 1 | 1 | 1 | 5 | 1 | 6 | 7 | 11 | 1 | 34 (12) | ||
| 2a–2d | Various | Various | DV1 | DM2 | R1 | PCR1 | 2 | 2 | 2 | 2 | 8 (3) | |||||||
| 3a–3c | Various | Various | DV2 | DM1 | R1 | PCR1 | 2 | 5 | 4 | 2 | 13 (5) | |||||||
| 4 | PP1 | PI1/PV1 | DV1 | DM3 | R1 | PCR1 | 1 | 1 | 2 | 1 | 5 (2) | |||||||
| 5–28 | Various | Various | DV1–DV4 | DM1–DM8 | R1–R15 | PCR1–PCR6 | 1 | 1 | 10 | 3 | 5 | 1 | 2 | 6 | 4 | 33 (12) | ||
| Total | 1 | 3 | 4 | 10 | 27 | 17 | 30 | 40 | 48 | 72 | 23 | 275 (100) | ||||||
Abbreviations: PP, plasmid profile; PI/PV, plasmid fingerprint; DV, chromosomal fingerprint by EcoRV restriction; DM, chromosomal fingerprint by MluI restriction; R, ribotype; PCR, RAPD pattern; OP, outpatients.
Most of the isolates belonged to group 1b (62%) and were isolated from specimens obtained from inpatients and outpatients between 1987 and 1996. Isolates of groups 2, 3, and 4 (9%) differed only in plasmid and DNA fingerprints, while those of groups 5 to 28 had more varied molecular patterns.
DISCUSSION
Most of our S. enteritidis isolates were susceptible to the antibiotics tested. All were susceptible to the expanded-spectrum cephalosporins, the fluoroquinolones, and chloramphenicol, and more than 99% were susceptible to ampicillin, cephalothin, cefuroxime, aminoglycosides, nalidixic acid, tetracycline, and trimethoprim-sulfamethoxazole. The proportion of resistant strains appears to have remained unchanged over the study period. Our findings confirm those of earlier studies indicating that most strains were susceptible to a wide range of antimicrobial agents (26, 35, 37, 47).
Ampicillin resistance in S. enteritidis was usually due to TEM-type β-lactamases encoded by genes on a 34-, 60-, or 100-MDa plasmid (10, 43). Our only ampicillin-resistant isolate, which was also resistant to cephalothin, produced a β-lactamase of pI 5.2. The resistance was encoded on a 5.2-kb plasmid. Five β-lactamases with identical pIs (5.2) have been reported. These are HMS-1, TEM-30, TEM-31, TEM-35, and TEM-36 (22, 44, 52). Further studies on substrate profile, inhibition profile, and molecular mass may reveal which β-lactamase was produced by this isolate.
Plasmid-mediated resistance to tetracycline and trimethoprim-sulfamethoxazole or ampicillin and cephalothin was expressed in two isolates. Antibiotic resistance in other isolates was usually low and probably encoded by genes on the chromosome.
Previous studies showed that most S. enteritidis strains contained plasmids of up to 140 MDa in size (26, 37, 39) and that more than 70% of S. enteritidis strains harbored a serotype-specific plasmid of 35 to 40 MDa (11). Our findings, indicating that 96% of isolates harbored plasmids, were similar. Twenty-one plasmid profiles were identified. Profiles showing the 60-kb (36-MDa) plasmid were seen in 80% of isolates. The EcoRI or EcoRV restriction patterns of the 60-kb plasmid in 94% of isolates were identical, and those of the remaining isolates were similar. The only other large and commonly seen plasmid (78 kb) was found in eight isolates. This plasmid exhibited three different fingerprints.
Because of the relative instability of extrachromosomal elements in a bacterial cell, total DNA was used to elucidate the epidemiology of S. enteritidis. DNA fingerprinting, ribotyping, and RAPD analysis showed that more than 90% of isolates had the same patterns, regardless of where and when they were isolated. When the patterns obtained by all the methods used in this study were combined, 62% of the isolates were found to be identical and to belong to group 1b. Isolates in other subgroups of group 1 and those in groups 2 to 4 were probably variants of isolates of group 1b. They differed from those of group 1b only in plasmid content or DNA fingerprints (DV1-DM2, DV2-DM1, or DV1-DM3). DNA fingerprints DV1 and DV2 and DM1, DM2, and DM3 were similar. The isolates in these groups (groups 1 to 4) were from specimens collected from inpatients and outpatients between 1987 and 1996. It is therefore possible that a predominant clone and a few related clones of S. enteritidis were circulating in Hong Kong during this period. Since a strain isolated in 1986 belonged to a very different genotype, the upsurge of the incidence of S. enteritidis in 1989 may have been due to the spread of strains isolated in 1987.
Studies from other parts of the world support the hypothesis that there is a clonal relationship between different S. enteritidis phage types (18). In the United Kingdom and other parts of Europe, S. enteritidis phage type 4 was responsible for most food-borne diseases in humans (3–5, 7, 13, 30, 31, 41, 46). Although there was an increase in the incidence of infections caused by S. enteritidis phage type 24 in England and Wales, this was thought to be part of the epidemic spread of phage type 4, because phage type 24 was derived from phage type 4 by acquisition of a drug resistance plasmid (10). In the United States, Germany, Canada, and Switzerland, S. enteritidis phage type 8 was responsible for most of the sporadic human and animal cases (13, 25, 39, 45) or outbreaks (15, 42). Strains circulating in Malaysia or Italy were also clonally related (16, 40).
Although phage typing has been used primarily for the epidemiological study of S. enteritidis (13, 25, 39, 45), it is of little use in places where a small number of phage types predominate. Environmental conditions can affect the sensitivity of bacteria to infection by phages. S. enteritidis phage type 4 has been shown to convert to phage type 7 by losing lipopolysaccharide (6), to phage type 9a by phage receptor mutation (33), and to phage type 24 by acquiring a drug resistance plasmid (10). Therefore, we did not phage type our strains.
Plasmid analysis and ribotyping have been used by others to study the epidemiology of bacterial strains (17, 21, 42, 43). Methods such as pulsed-field gel electrophoresis are the most discriminatory for studying the epidemiology of S. enteritidis (29). Unfortunately, these methods require expensive equipment and may not be affordable in smaller laboratories. In this study, we used traditional horizontal electrophoretic equipment, low-concentration agarose gels, and unique gel-running conditions to separate the DNA fragments. To further increase the discriminatory power of our method, specially chosen restriction endonucleases were used to produce fragments that were well separated on agarose gels. Good separation of bands >12 and >20 kb was achieved with EcoRV- and MluI-digested fragments, respectively. The MluI-digested fragments produced nine different patterns, while the EcoRV-digested fragments produced only three. Therefore, MluI is a better enzyme to use for discriminating S. enteritidis isolates than EcoRV.
RAPD is a recently developed PCR-based technique for detecting genomic diversity among different organisms (1, 14, 20, 23, 51) and has been useful for studying the epidemiology of S. enteritidis (9, 19). However, different primers have to be tested first, and conditions of PCR must be optimized for each organism studied (8). Suitable primers should have a G+C content greater than 40% (49). The discriminatory power of this method can be increased by using two primers (24, 48).
The present study showed that a single method cannot be relied upon for discriminating between S. enteritidis strains. Isolates with the same DNA fingerprints and PCR types (groups 9 to 21) can belong to different ribotypes. Those with the same DNA fingerprints and ribotyping patterns (groups 25 to 28) can exhibit different PCR patterns. It is therefore necessary to combine the results of different molecular typing methods for reporting the epidemiology of S. enteritidis.
To summarize, most S. enteritidis isolates were susceptible to all the antibiotics tested except rifampin. Ninety-six percent harbored plasmids, and 91% of these had an identical 60-kb plasmid. As 88% of isolates had identical or similar DNA fingerprints, ribotypes, and RAPD patterns, there was probably a predominant clone of S. enteritidis circulating in Hong Kong.
ACKNOWLEDGMENTS
We thank K. L. E. Ling for reviewing the manuscript.
This study was supported in part by a research grant from the Faculty of Medicine, The Chinese University of Hong Kong.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson E S, Threlfall E J. The characterization of plasmids in the enterobacteria. J Hyg. 1974;71:619–631. doi: 10.1017/s0022172400023718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. Salmonella in eggs. PHLS Microbiol Digest. 1989;6:1–9. [Google Scholar]
- 4.Bautsch W. NotI macrorestriction analysis suggests a clonal relationship of Salmonella enterica ser. enteritidis phage lysotype 4 strains. Infection. 1993;21:328–330. doi: 10.1007/BF01712457. [DOI] [PubMed] [Google Scholar]
- 5.Chart H, Threlfall E J, Rowe B. Virulence of Salmonella enteritidis phage type 4 is related to possession of a 38 Mda plasmid. FEMS Microbiol Lett. 1989;58:299–304. doi: 10.1016/0378-1097(89)90057-8. [DOI] [PubMed] [Google Scholar]
- 6.Chart H, Rowe B, Threlfall E J, Ward L R. Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves loss of lipopolysaccharide with concomitant loss of virulence. FEMS Microbiol Lett. 1989;66:37–40. doi: 10.1016/0378-1097(89)90073-6. [DOI] [PubMed] [Google Scholar]
- 7.Cowden J M, Chisholm D, O’Mahony M, Lynch D, Mawer S L, Spain G E, Ward L, Rowe B. Two outbreaks of Salmonella enteritidis phage type 4 infection associated with the consumption of fresh shell-egg products. Epidemiol Infect. 1989;103:47–52. doi: 10.1017/s095026880003034x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellsworth D L, Rittenhouse K D, Honeycutt R L. Artifactual variation in randomly amplified polymorphic DNA banding patterns. BioTechniques. 1993;14:214–217. [PubMed] [Google Scholar]
- 9.Fadl A A, Nguyen A V, Khan M I. Analysis of Salmonella enteritidis isolates by arbitrary primed PCR. J Clin Microbiol. 1995;33:987–989. doi: 10.1128/jcm.33.4.987-989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost J A, Ward L R, Rowe B. Acquisition of a drug-resistance plasmid converts Salmonella enteritidis phage 4 to phage type 24. Epidemiol Infect. 1989;103:243–248. doi: 10.1017/s0950268800030594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmuth R, Stephan R, Bunge C, Hoog B, Steinbeck A, Bulling E. Epidemiology of virulence-associated plasmids and other membrane protein patterns within seven common Salmonella serovars. Infect Immun. 1985;48:175–182. doi: 10.1128/iai.48.1.175-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katouli M, Seuffer R H, Wollin R, Kuhn I, Moliby R. Variations in biochemical phenotypes and phage types of Salmonella enteritidis in Germany 1989-92. Epidemiol Infect. 1993;111:199–207. doi: 10.1017/s0950268800056909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauppinen J, Mantyjarvi R, Katila M. Random amplified polymorphic DNA genotyping of Mycobacterium malmoense. J Clin Microbiol. 1994;32:1827–1829. doi: 10.1128/jcm.32.7.1827-1829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khakhria R, Duck D, Lior H. Distribution of Salmonella enteritidis phage types in Canada. Epidemiol Infect. 1991;106:25–32. doi: 10.1017/s0950268800056417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagatolla C, Dolzani L, Tonin E, Lavenia A, Michele M D, Tommasini T, Monti-Bragadin C. PCR ribotyping for characterizing Salmonella isolates of different serotypes. J Clin Microbiol. 1996;34:2440–2443. doi: 10.1128/jcm.34.10.2440-2443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landeras E, Gonzales-Hevia M A, Alzugaray R, Mendoza M C. Epidemiological differentiation of pathogenic strains of Salmonella enteritidis by ribotyping. J Clin Microbiol. 1996;34:2294–2296. doi: 10.1128/jcm.34.9.2294-2296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebisch B, Schwarz S. Molecular typing of Salmonella enterica subsp. enterica serovar Enteritidis isolates. J Med Microbiol. 1996;44:52–59. doi: 10.1099/00222615-44-1-52. [DOI] [PubMed] [Google Scholar]
- 19.Lin A W, Usera M A, Barrett T J, Goldsby R A. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J Clin Microbiol. 1996;34:870–876. doi: 10.1128/jcm.34.4.870-876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makino S I, Okada Y, Maruyama T, Kaneko S, Sasakawa C. PCR-based random amplified polymorphic DNA fingerprinting of Yersinia pseudotuberculosis and its practical applications. J Clin Microbiol. 1994;32:65–69. doi: 10.1128/jcm.32.1.65-69.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinetti G, Altwegg M. rRNA gene restriction patterns and plasmid analysis as a tool for typing Salmonella enteritidis. Res Microbiol. 1990;141:1151–1162. doi: 10.1016/0923-2508(90)90088-8. [DOI] [PubMed] [Google Scholar]
- 22.Matthew M, Hedges R W, Smith J T. Types of β-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979;138:657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazurier S I, Wernars K. Typing of Listeria strains by random amplification of polymorphic DNA. Res Microbiol. 1992;143:499–505. doi: 10.1016/0923-2508(92)90096-7. [DOI] [PubMed] [Google Scholar]
- 24.Micheli M R, Bova R, Calissano P, D’Ambrosis E. Randomly amplified polymorphic DNA fingerprinting using combinations of oligonucleotide primers. BioTechniques. 1993;15:388–390. [PubMed] [Google Scholar]
- 25.Morris J G, Jr, Dwyer D M, Hoge C W, Stubbs A D, Tilghman D, Groves C, Israel E, Libonati J. Changing clonal patterns of Salmonella enteritidis in Maryland: evaluation of strains isolated between 1985 and 1990. J Clin Microbiol. 1992;30:1301–1303. doi: 10.1128/jcm.30.5.1301-1303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair U S, Saeed A M, Muriana P M, Kreisle R A, Barrett B, Sinclair C L, Fleissner M L. Plasmid profile and resistance to antimicrobial agents among Salmonella enteritidis isolated from human beings and poultry in the midwestern United States. J Am Vet Med Assoc. 1995;206:1339–1344. [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standards M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 28.Nguyen A V, Khan M I, Lu Z. Amplification of Salmonella chromosomal DNA using the polymerase chain reaction. Avian Dis. 1994;38:119–126. [PubMed] [Google Scholar]
- 29.Olsen J E, Skov M N, Threlfall E J, Brown D J. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J Med Microbiol. 1994;40:15–22. doi: 10.1099/00222615-40-1-15. [DOI] [PubMed] [Google Scholar]
- 30.Paul J, Batchelor B. Salmonella enteritidis phage type 4 and hen’s eggs. Lancet. 1988;ii:1421. doi: 10.1016/s0140-6736(88)90607-1. [DOI] [PubMed] [Google Scholar]
- 31.Pohl P, Lintermans P, Martin M, Couturier M. Epidemiological study of Salmonella enteritidis strains of animal origin in Belgium. Epidemiol Infect. 1991;106:11–16. doi: 10.1017/s0950268800056399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popovic T, Bopp C, Olsvik Ø, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell N G, Threlfall E J, Chart H, Schofield S L, Rowe B. Correlation of change in phage type with pulsed field profile and 16 rrn profile in Salmonella enteritidis phage types 4, 7, and 9a. Epidemiol Infect. 1995;114:403–411. doi: 10.1017/s0950268800052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampling A. Salmonella enteritidis five years on. Lancet. 1993;342:317–318. doi: 10.1016/0140-6736(93)91466-y. [DOI] [PubMed] [Google Scholar]
- 35.Rampling A, Anderson J C, Upson R, Peters E, Ward L R, Rowe B. Salmonella enteritidis phage type 4 infection of broiler chickens: a hazard to public health. Lancet. 1989;ii:436–438. doi: 10.1016/s0140-6736(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigue D C, Tauxe R V, Rowe B. International increase in Salmonella enteritidis: a new pandemic? Epidemiol Infect. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigue D C, Cameron D N, Puhr N D, Brenner F W, St. Louis M E, Wachsmuth I K, Tauxe R V. Comparison of plasmid profiles, phage types, and antimicrobial resistance patterns of Salmonella enteritidis isolates in the United States. J Clin Microbiol. 1992;30:854–857. doi: 10.1128/jcm.30.4.854-857.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sneath P H A. Computer taxonomy. Methods Microbiol. 1972;7A:29–98. [Google Scholar]
- 39.Stubbs A D, Hickman-Brenner F W, Cameron D N, Farmer J J., III Differentiation of Salmonella enteritidis phage type 8 strains: evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns and biotyping. J Clin Microbiol. 1994;32:199–201. doi: 10.1128/jcm.32.1.199-201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thong K L, Ngeow Y F, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timoney J F, Shivaprasad H L, Baker R C, Rowe B. Egg transmission after infection of hens with Salmonella enteritidis phage type 4. Vet Rec. 1989;125:600–601. [PubMed] [Google Scholar]
- 42.Usera M A, Popovic T, Bopp C A, Strockbine N A. Molecular subtyping of Salmonella enteritidis phage type 8 strains from the United States. J Clin Microbiol. 1994;32:194–198. doi: 10.1128/jcm.32.1.194-198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vatopoulos A S, Mainas E, Balis E, Threlfall E J, Kanelopoulou M, Kalapothaki V, Malamou-Lada H, Legakis N J. Molecular epidemiology of ampicillin-resistant clinical isolates of Salmonella enteritidis. J Clin Microbiol. 1994;32:1322–1325. doi: 10.1128/jcm.32.5.1322-1325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vedel G, Beleaouaj A, Gilly L, Labia R, Philippon A, Nevot P, Paul G. Clinical isolates of Escherichia coli producing TRI β-lactamases: novel TEM-enzymes conferring resistance to β-lactamase inhibitors. J Antimicrob Chemother. 1992;30:449–462. doi: 10.1093/jac/30.4.449. [DOI] [PubMed] [Google Scholar]
- 45.Wachsmuth K. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis. 1986;8:682–689. doi: 10.1093/clinids/8.5.682. [DOI] [PubMed] [Google Scholar]
- 46.Ward L R, Rowe B, De Sa J D H. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward L R, Threlfall E J, Rowe B. Multiple drug resistance in salmonellae in England and Wales: a comparison between 1981 and 1988. J Clin Pathol. 1990;43:563–566. doi: 10.1136/jcp.43.7.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong S S Y, Yuen K Y, Yam W C, Lee T Y, Chau P Y. Changing epidemiology of human salmonellosis in Hong Kong, 1982–1993. Epidemiol Infect. 1994;113:425–434. doi: 10.1017/s0950268800068436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woods J P, Kersulyte D, Tolan R W, Jr, Berg C M, Berg D E. Use of arbitrary primed polymerase chain reaction analysis to type disease and carrier strains of Neisseria meningitidis isolated during a university outbreak. J Infect Dis. 1994;169:1384–1389. doi: 10.1093/infdis/169.6.1384. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X Y, Bordon F, Sirot D, Kitzis M-D, Gutmann L. Emergence of clinical isolates of Escherichia coli producing TEM-1 derivatives or an OXA-1 β-lactamase conferring resistance to β-lactamase inhibitors. Antimicrob Agents Chemother. 1994;38:1085–1089. doi: 10.1128/aac.38.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]