Abstract
A national survey of hepatitis C virus (HCV) infections among dialysis patients in The Netherlands was performed. The study involved 2,653 patients (2,108 hemodialysis patients and 545 chronic ambulatory peritoneal dialysis [CAPD] patients) from 39 of the 49 dialysis centers in the country. Patient sera were analyzed by both serological and molecular methods. Screening by a third-generation enzyme immunoassay (EIA) yielded 79 reactive sera. The presence of anti-HCV antibodies was confirmed in 70 patients by a line immunoassay. All seropositive samples were tested by reverse transcriptase PCR, and 57 samples were found to contain HCV RNA. Of the nine EIA-positive and line immunoassay-negative or indeterminate samples, four were HCV RNA positive. All seronegative samples were screened for the presence of HCV RNA in pools of five sera. Of 2,576 antibody-negative samples, 6 contained HCV RNA. All antibody-positive and RNA-positive samples were also tested by a second serological assay. The prevalence of HCV infections among Dutch dialysis patients as determined by serology or the presence of HCV RNA was 3% (80 of 2,653), i.e., 3.5% (73 of 2,108) in patients treated on hemodialysis and 1.3% (7 of 545) in patients on CAPD. Of these 80 HCV-infected dialysis patients, 67 (84%) were HCV RNA positive. Serological screening alone would have diagnosed only 70 infected patients. Therefore, antibody screening combined with detection of HCV RNA should be considered as the “gold standard” for diagnosing HCV infection in dialysis patients. The prevalence of HCV-infected patients in Dutch dialysis centers ranged from 0 to 8%, suggesting the existence of local risk factors for acquiring HCV infection. Genotyping analysis by reverse hybridization line probe assay revealed the presence of genotypes 1a (23%), 1b (46%), 2 (3%), 2a (13%), 2b (1%), 3a (7%), and 4a (4%). In four (6%) samples multiple genotypes were detected. The genotype distribution of HCV isolates among Dutch dialysis patients was similar to the distribution among nondialysis patients from the Benelux, except for subtype 1a, which was significantly more prevalent among dialysis patients. In only one center, a high prevalence of an uncommon genotype was suggestive of infection from a common source.
Hepatitis C virus (HCV) is the major cause of posttransfusion hepatitis (16). Among blood donors the prevalence of HCV infection varies from less than 1% in western Europe and the United States to approximately 1% in Japan and more than 5% in selected blood donor populations in some African and Asian countries (2, 7, 9, 23, 25). In The Netherlands 0.03 to 0.1% of the healthy donor population has antibodies to HCV (23, 29).
In addition to recipients of blood products, other groups that are frequently exposed to blood, such as hemophiliacs, intravenous drug users, and hemodialysis patients, are at risk (16, 29).
Studies performed in a selected group of dialysis centers showed that the prevalence of HCV infections among hemodialysis patients in various countries is much higher than that among healthy blood donors, ranging from 2 to 6% in northwestern Europe to more than 20% in Japan and over 60% in Saudi Arabia (9, 11, 13, 29). However, these figures may not be representative for a whole country due to selection bias (14).
In the past multiple blood transfusions seemed to be an important risk factor for hemodialysis patients in the acquisition of HCV infection (26). However, it is unlikely that blood transfusions are the only source for recently acquired infections, since screening of blood donors for anti-HCV antibodies has been shown to be highly effective in preventing transmission of HCV (1). A considerable number of HCV-infected hemodialysis patients did not receive blood at all (26).
Hemodialysis can be a risk for transmission of HCV. The length of the period during which patients have been dialyzed appears to be a risk factor for HCV infection independent of blood transfusion (12, 23). Moreover, molecular epidemiological studies have revealed convincing evidence for transmission of HCV between dialysis patients in the same center (3, 21). The frequent sharing of facilities over a prolonged period may result in an accumulated risk (3, 13). Whatever the precise transmission route may be, standard infection control practices reduce the risk of transmission of HCV in dialysis units (13).
Several studies have indicated that serological assays alone are not sufficient for the diagnosis of HCV infection in dialysis patients and that detection of HCV RNA is required to identify all infected patients (5, 13). Partial immunosuppression in dialysis patients, resulting in a poor antibody response, may play a role in this observation (10). Epidemiological studies of dialysis patients which rely on serological screening could therefore underestimate the prevalence of HCV infections considerably (5, 15, 24).
The present study describes a nationwide survey among dialysis patients in The Netherlands by serological as well as molecular methods to screen for HCV infection. The study had three aims: (i) to assess the prevalence of HCV infection among dialysis patients in the different centers in The Netherlands, (ii) to compare serological and molecular methods for detection of HCV infection, and (iii) to study the genotype distribution of HCV isolates.
MATERIALS AND METHODS
Patients.
Of the 49 dialysis centers in The Netherlands, 39 participated in the study. A total of 2,653 patients, 2,108 on hemodialysis and 545 on chronic ambulatory peritoneal dialysis (CAPD), were treated in these centers (range, 22 to 165 patients per center; mean, 68.0; standard deviation, 29.4).
The study protocol was approved by the medical ethical committees of the participating centers, and all patients gave their informed consent. Serum samples were collected between September 1995 and July 1996. Serum was prepared within 2 h after blood sampling, stored at −20°C, and transported on dry ice. All samples were divided into 0.5-ml aliquots in a separate location to prevent contamination and unnecessary thawing and freezing.
Serology.
All serum samples were tested by the INNO-test HCV Ab III enzyme immunoassay (EIA) (Innogenetics, Antwerp, Belgium) for the presence of antibodies to HCV. Positive samples were examined by the INNO-LIA HCV Ab III (Innogenetics) confirmation assay. All seropositive or RNA-positive samples were also tested by the Ortho HCV 3.0 EIA (Ortho Diagnostic Systems, Neckargemund, Germany). The tests were performed according to the instructions of the manufacturers.
Molecular screening.
All seropositive samples were tested individually for the presence of HCV RNA. To permit the molecular analysis of the large number of seronegative samples, a pooling strategy was developed, similar to the method described by Corcoran et al. (6). This involved the pooling of three to five seronegative serum samples and the analysis of the mixture for the presence of HCV RNA. Twenty-five microliters of each of the five samples were mixed together, and the entire 125-μl pool was used for the assay. For all samples or pools, 125 μl was tested.
In order to monitor the efficacy of the HCV RNA test, an internal control RNA was used in all assays. A PCR fragment, comprising nucleotides −341 to +410 of the HCV RNA genome, obtained from a genotype 1b isolate, was cloned into the pGEM-T plasmid (Promega, Leiden, The Netherlands). A 51-bp insert was introduced into the SphI site at position −63 in the 5′ untranslated region (UTR). RNA transcripts were synthesized from purified recombinant plasmid with the Riboprobe kit and T7 RNA polymerase (Promega) and serially diluted in diethyl pyrocarbonate-treated water containing 1 mg of poly(A)/ml as a carrier. Tenfold dilutions were tested by reverse transcriptase (RT)-PCR, and the detection limit was reproducibly established at 10−10 dilution. A 10−9 dilution (2.5 μl) was used in each assay as an internal control.
HCV RNA was isolated from 125 μl of individual or pooled serum by mixing with 500 μl of lysis buffer (5 M thiocyanate, 0.125 M Tris-HCl [pH 7.4] 0.3 M sodium acetate), freshly supplemented with 100 μg of poly(A)/ml and 1.25% (vol/vol) 2-mercaptoethanol, and 5 μl of 10−9 internal control/ml. After vigorous mixing and incubation at 65°C for 10 min, the samples were cooled on ice and 625 μl of cold isopropanol was added to each sample. The mixtures were centrifuged at 14,000 × g for 20 min at 4°C. Pellets were washed once with 500 μl of cold 80% ethanol. Each pellet was dissolved in 30 μl of RNase-free water. Ten microliters of this solution was used immediately for cDNA synthesis by adding the antisense cDNA primer (20 pmol) and deoxynucleoside triphosphates (1 mM final concentration) followed by denaturation for 2 min at 80°C and cooling on ice. Buffer (final concentrations, 50 mM Tris-HCl [pH 8.0], 3 mM MgCl2, 75 mM KCl, 0.01 M dithiothreitol), 200 U of Moloney murine leukemia virus RT (Gibco-BRL, Breda, The Netherlands), 30 U of RNasin (Promega), and water (RNase free) were added to a final volume of 25 μl. After incubation at 37°C for 60 min, the RT was inactivated at 95°C for 10 min and 75 μl of PCR mixture containing the sense primer, 0.25 U of Taq DNA polymerase (SuperTaq; SphaeroQ, Leiden, The Netherlands), and the appropriate buffer (final concentrations, 10 mM Tris-HCl [pH 9.0], 50 mM KCl, and 2.5 mM MgCl2) were added. The PCR program consisted of a preincubation at 95°C for 1 min followed by 40 cycles of 1 min at 95°C, 1 min at 52°C, and 1 min at 74°C. Nested PCR was performed by the transfer of 1 μl of the first-round PCR product into a new PCR reaction mixture containing nested primers. The PCR products were examined on 2% agarose gels.
For PCR aimed at the 5′ UTR, antisense primer HCV19 (GTGCACGGTCTACGAGACCT; positions −1 to −20) and sense primer HCV35 (TTGGCGGCCGCACTCCACCATRRATCACTCCCC; positions −319 to −297) (underlined sequences are not HCV specific) were used in the first round. Primers NCR3 (GGGGCGGCCGCCACCATRRATCACTCCCCTGTGAGG; positions −315 to −289) and NCR4 (CACTCTCGAGCACCCTATCAGGCAGTACC; positions −66 to −47) were used in the nested PCR reaction. All positive HCV RNA results were confirmed on fresh aliquots. The HCV RNA method was evaluated by testing the proficiency panel as described by Zaaijer et al. (33).
If the internal control RNA was not amplified, the sample was spiked with a 10-fold-higher amount of internal control RNA. If the internal control RNA remained undetectable, the sample was considered to be inhibitory for RT-PCR and was excluded from further analysis.
Genotyping analysis.
For genotyping analysis, the reverse hybridization line probe assay (INNO-LiPA HCV; Innogenetics) was used. This method allows discrimination between the major types and subtypes of HCV based on sequence heterogeneity within the 5′ UTR (27). The efficacy of this method for genotyping HCV isolates in western Europe has been reported earlier (30).
Statistical analysis.
The significance of differences was analyzed by the chi-square test and Fisher’s exact test.
RESULTS
Serum samples were collected from a total of 2,653 dialysis patients from 39 dialysis centers distributed evenly over The Netherlands. These patients represent 68% of all Dutch dialysis patients registered at the beginning of this study. Ten centers did not participate for logistic reasons. The collected sera were subjected to serological screening and confirmation as well as molecular analysis to determine the presence of HCV. The results are summarized in Table 1. A total of 79 sera reacted positively in the INNO-test EIA. Seventy of 79 (89%) EIA-positive results could be confirmed by line immunoassay (LIA). Thus, 70 (2.6% of 2,653) confirmed seropositive dialysis patients were identified. In 57 (81.4%) of the confirmed seropositive samples, HCV RNA was detected. For nine patients, EIA was positive but LIA was either negative (four of nine) or indeterminate (five of nine). HCV RNA was detected in four of these nine EIA-positive patients whose results were not confirmed by LIA. In two samples LIA was negative, and in two LIA was indeterminate.
TABLE 1.
Detection of HCV infection among 2,653 Dutch dialysis patients
| Test results
|
No. of patients
|
||
|---|---|---|---|
| EIA | LIA | Total | PCR positive |
| Positive | Positive | 70 | 57 |
| Positive | Indeterminate | 5 | 2 |
| Positive | Negative | 4 | 2 |
| Negative | 2,574 | 6 | |
| Total no. | 2,653 | 67 | |
All 2,574 anti-HCV antibody-negative sera were tested by RT-PCR divided among 533 pools. The pooling strategy was evaluated by testing serum samples from one of the dialysis centers. The sera were tested in pools and individually, and all HCV-infected patients were identified by both methods. Among the 533 pools of seronegative sera, 6 yielded positive signals, and testing of the individual sera resulted in 6 HCV RNA-positive, seronegative samples. In 10 seronegative pools the internal control could not be amplified, indicating the presence of inhibitory substances. Based on HCV RNA detection alone, a total of 67 HCV-infected dialysis patients were identified. The presence of HCV RNA was confirmed in all cases by independent retesting of a fresh aliquot from each serum.
With the combined strategy of serology and HCV RNA detection, a total of 80 HCV-positive dialysis patients were identified, 73 of 2,108 (3.5%) hemodialysis patients and 7 of 545 (1.3%) CAPD patients (odds ratio, 2.76; 95% confidence interval, 1.26 to 6.02). Serum samples from 72 of the INNO-test-positive patients and from the 6 seronegative, RNA-positive patients were also tested by the Ortho EIA. The results are shown in Table 2. Of the six RNA-positive, INNO-test-negative samples, one was positive by the Ortho EIA. Of the 72 INNO-test-positive sera, 64 were also positive by the Ortho EIA and 8 were negative. Of these eight INNO-test-positive, Ortho EIA-negative samples, four contained HCV RNA. Among 62 HCV RNA-positive samples that were tested by both EIAs, INNO-test and Ortho EIA detected 56 and 52, respectively.
TABLE 2.
Comparison of INNO-test HCV Ab III EIA, Ortho HCV 3.0 EIA, and RT-PCR results
| INNO-test | Ortho
|
||||
|---|---|---|---|---|---|
| Positive
|
Negative
|
Total | |||
| PCR positive | PCR negative | PCR positive | PCR negative | ||
| Positive | 52 | 12 | 4 | 4 | 72 |
| Negative | 1 | 0 | 5 | 0 | 6 |
| Total | 53 | 12 | 9 | 4 | 78 |
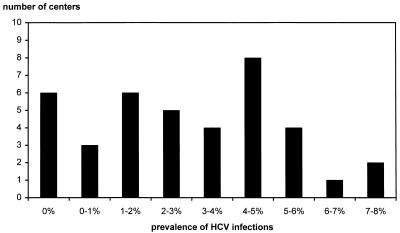
The distribution of HCV-infected patients among the dialysis centers is shown in Fig. 1. The prevalence of HCV infection was 7 to 8% in three centers. In six dialysis units no HCV-positive patients were found. There was no significant relationship between the population size (total number of treated patients) and the prevalence of HCV-infected patients in the dialysis centers (r = 0.53).
FIG. 1.
Distribution of HCV-positive sera among 39 dialysis centers in The Netherlands.
For HCV RNA-positive samples genotyping by the reverse hybridization line probe assay was performed. The results are summarized in Table 3. Genotype 1b is the most prevalent genotype (46%), but genotypes 1a (24%), 2a (13%), 2b (1%), 3a (7%), and 4a (4%) were also found. One HCV RNA-positive sample could not be genotyped. Multiple genotypes were detected in four patients (1a plus 1b, twice; 2 plus 4a, once; and 1a plus 2, once).
TABLE 3.
Distribution of HCV genotypes among Dutch dialysis patients
The majority of the centers with HCV-infected patients showed no obvious cluster of identical genotypes. In one center (50 patients) genotype 1a was observed in three patients. In two other centers, with 100 and 165 patients, respectively, genotype 1b was observed in four patients. In one center (128 patients) four patients had genotype 2a, and preliminary results of phylogenetic analysis of these four isolates suggested infection from a single source (data not shown).
DISCUSSION
The prevalence of HCV infections among dialysis patients is generally much higher than that among healthy blood donors (9, 23). Studies in selected dialysis centers from different countries all over the world revealed that prevalences range from 2 to 3% to 60% (11, 23, 29). To a certain extent this may reflect the different prevalences of HCV-infected individuals among the general population in these countries. However, the dialysis process itself and the level of hygienic standards may influence the risk of HCV infection (23, 26). This may explain differences found between dialysis centers in one country (14). In order to assess the prevalence of HCV infection among dialysis patients in The Netherlands, we conducted a nationwide epidemiological survey. Serum samples were obtained from 2,653 patients, 68% of all Dutch dialysis patients, who were treated in 39 dialysis centers.
Diagnosis of HCV infections is usually based on detection of specific HCV antibodies by EIA followed by a confirmation assay such as the LIA (17). This approach is convenient for large-scale screening. Using an antibody-screening assay that combines antigens from the core and the NS3 region as well as from the NS5 region along with the confirmation assay, we found 70 of 2,653 (2.6%) dialysis patients were seropositive. There was a good correlation between the INNO-test and the Ortho serological tests. The Ortho EIA detected one of six HCV RNA-positive samples that were negative by the INNO-test. On the other hand, the INNO-test detected eight samples that were negative by the Ortho test, and four of these contained HCV RNA. The INNO-test detected more HCV RNA-positive sera than the Ortho test.
Antibody testing may prove useful to measure present or past infections, irrespective of the actual infectivity of the patients (32). However, patients who have cleared the virus may gradually lose their antibodies, and consequently, antibody screening will not detect all past infections (26, 28). Detection of HCV RNA permits direct detection of the presence of the virus and also permits detection of infectivity during the seronegative window, immediately after infection (13, 19, 32). Detection of HCV RNA may be more reliable than serology in detecting ongoing HCV infections in dialysis patients, who may not mount an adequate antibody response (5, 10, 13). However, detection of HCV RNA by PCR is still laborious, requires specific expertise and facilities, and is usually only used to confirm positive serology (33).
In the present study RT-PCR was used to screen for the presence of HCV RNA in all 2,653 serum samples by using a pooling strategy. HCV RNA was detected in 67 (2.5%) samples. Of these, 57 were confirmed as seropositive. In six samples HCV RNA was detected by RT-PCR in the absence of antibodies. These cases may be considered either as patients with recent infections, where the sample was obtained during the seronegative-window phase, or as patients with impaired immune responses (4, 13). In one of these six cases, antibodies were detected by the Ortho test but not by the INNO-test. In four of the six cases, HCV RNA could also be detected by RT-PCR, aimed at the hypervariable part of the E2 region (data not shown), indicating that these patients were truly HCV infected.
In conclusion, by using both serology and PCR, 80 HCV-positive patients (3% of a total of 2,653 patients) could be identified. The combination of serological and molecular methods resulted in the most accurate estimation of the number of HCV infections among Dutch dialysis patients. Using only antibody assays in this population, 10 of 80 (12.5%) HCV-positive patients would have been missed.
The confirmation blot assay (LIA) could not confirm the result of the screening EIA in nine cases, yielding an indeterminate result five times and a negative result four times. Since four of the unconfirmed serological results were HCV RNA positive, this shows the need for molecular diagnostic methods, but also the limitations of this confirmation assay. The confirmation assay alone adds little to solve the problem of inconclusive results of antibody assays, especially for immunocompromised patients (18).
This is the first study in which large-scale pooled screening was employed, and we have shown that in this way it is feasible to use PCR with a large patient population. Pooling of sera has reduced the cost considerably and allowed us to identify six HCV RNA-positive but seronegative patients. We designed a system with pools of three to five sera spiked with an internal control to detect inhibition of amplification. The use of an internal RNA control not only permits control of sensitivity in every test but also reveals the presence of inhibitory factors.
The seroprevalence of 2.6% found in the present study is higher than the 2% observed for Dutch dialysis patients studied in a period just before universal screening of blood donors was introduced in 1991 (29). However, these data cannot be compared directly, and the observed difference may be explained by the use of more sensitive antibody assays in the present study (20). Since the number of transfusions to dialysis patients has been decreased by the use of erythropoietin, and routine screening of blood donors for HCV infection has been used in The Netherlands since 1991, the risk of HCV transmission to dialysis patients through blood transfusion has decreased significantly. Consequently, nosocomial transmission of HCV in the dialysis center may remain the most important risk factor for HCV infection in the future (22, 23).
For follow-up analysis all dialysis patients will be sampled again in 1998 and tested by serological and molecular assays. This will permit an estimation of the incidence of HCV infections in Dutch dialysis centers.
The prevalence of HCV infection among hemodialysis patients was 3.5%, compared to 1.3% in CAPD patients. The statistically significant difference between these patient groups indicates the increased risk of nosocomial transmission of HCV for hemodialysis patients compared to that for CAPD patients.
We have found six centers (15%), with an average of 60 dialysis patients, without any HCV-infected patients. Three centers, with an average of 93 patients, had a prevalence of HCV-positive patients of more than 6% (range, 7 to 8%). The nonrandom distribution of HCV-infected individuals among the centers indicates that local factors may play a role in the epidemiology of HCV. This is in accordance with the finding that the size of a dialysis center (i.e., the total number of patients treated) was not related to the prevalence of HCV infections.
To further analyze the relatedness of HCV isolates in dialysis units, all HCV RNA-positive samples were genotyped. The prevalences of the different genotypes among dialysis patients were compared to genotyping data from 315 nondialysis patients in the Benelux (Belgium, Netherlands, and Luxembourg) that were obtained earlier (Table 3) (31). The genotype distributions appear to be similar, except for the prevalences of subtype 1a (P = 0.005) and patients with multiple genotypes (P = 0.03), which were more prevalent among dialysis patients. Genotype 5a was not found in our study group.
HCV isolates belonging to the same genotype found within one center can be studied for molecular relatedness by sequence analysis. We have found four centers with more than two patients with the same genotype. In one center four patients were infected with genotype 2a strains. Since the prevalence of this genotype in The Netherlands is relatively low, this could suggest infection from a common source (8). Preliminary results from sequence analysis further supported this finding (data not shown).
Data presented in this study indicate that the prevalence of HCV infections in Dutch dialysis centers is relatively low compared with those shown by data from other countries. The genotype distribution is comparable to that for nondialysis patients. However, the differences among centers indicate that local factors could play a role in causing HCV infection. Since new HCV infections still occur in dialysis patients and routes of transmission are unknown, screening by both serological and molecular methods at regular intervals is necessary to identify infected patients and to study HCV transmission among dialysis patients. Pooling sera for HCV RNA detection as described in our study may facilitate this screening regimen.
ACKNOWLEDGMENTS
This study was supported by the Nier Stichting Nederland, project C94.1416.
We thank all participating dialysis centers for their valuable contributions to this study and M. Hoekstra for assisting in the statistical analysis.
REFERENCES
- 1.Aach R D, Stevens C E, Hollinger F B, Mosley J W, Peterson D A, Taylor P E, Johnson R G, Barbosa L H, Nemo G J. Hepatitis C virus infection in post-transfusion hepatitis. An analysis with first- and second-generation assays. N Engl J Med. 1991;325:1325–1329. doi: 10.1056/NEJM199111073251901. [DOI] [PubMed] [Google Scholar]
- 2.al-Faleh F Z, Ramia S, Arif M, Ayoola E A, al-Rashed R S, al-Jeffry M, Hossain A, el-Hazmi M. Profile of hepatitis C virus and the possible modes of transmission of the virus in the Gizan area of Saudi Arabia: a community-based study. Ann Trop Med Parasitol. 1995;89:431–437. doi: 10.1080/00034983.1995.11812972. [DOI] [PubMed] [Google Scholar]
- 3.Allander T, Gruber A, Naghavi M, Beyene A, Soderstrom T, Bjorkholm M, Grillner L, Persson M A. Frequent patient-to-patient transmission of hepatitis C virus in a haematology ward. Lancet. 1995;345:603–607. doi: 10.1016/s0140-6736(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 4.Bjoro K, Froland S S, Yun Z, Samdal H H, Haaland T. Hepatitis C infection in patients with primary hypogammaglobulinemia after treatment with contaminated immune globulin. N Engl J Med. 1994;331:1607–1611. doi: 10.1056/NEJM199412153312402. [DOI] [PubMed] [Google Scholar]
- 5.Bukh J, Wantzin P, Krogsgaard K, Knudsen F, Purcell R H, Miller R H. High prevalence of hepatitis C virus (HCV) RNA in dialysis patients: failure of commercially available antibody tests to identify a significant number of patients with HCV infection. Copenhagen Dialysis HCV Study Group. J Infect Dis. 1993;168:1343–1348. doi: 10.1093/infdis/168.6.1343. [DOI] [PubMed] [Google Scholar]
- 6.Corcoran G D, Brink N S, Millar C G, Garson J A, Waite J, Deaville R, Thompson F D, Tedder R S. Hepatitis C virus infection in haemodialysis patients: a clinical and virological study. J Infect. 1994;28:279–285. doi: 10.1016/s0163-4453(94)91793-0. [DOI] [PubMed] [Google Scholar]
- 7.Darwish M A, Raouf T A, Rushdy P, Constantine N T, Rao M R, Edelman R. Risk factors associated with a high seroprevalence of hepatitis C virus infection in Egyptian blood donors. Am J Trop Med Hyg. 1993;49:440–447. doi: 10.4269/ajtmh.1993.49.440. [DOI] [PubMed] [Google Scholar]
- 8.Fabrizi F, Lunghi G, Guarnori I, Raffaele L, Erba G, Pagano A, Locatelli F. Hepatitis C virus genotypes in chronic dialysis patients. Nephrol Dial Transplant. 1996;11:679–683. doi: 10.1093/oxfordjournals.ndt.a027359. [DOI] [PubMed] [Google Scholar]
- 9.Fujiyama S, Kawano S, Sato S, Shimada H, Matsushita K, Ikezaki N, Nakano T, Sato T. Changes in prevalence of anti-HCV antibodies associated with preventive measures among hemodialysis patients and dialysis staff. Hepato-Gastroenterology. 1995;42:162–165. [PubMed] [Google Scholar]
- 10.Goldbloom S E, Reed W P. Host defenses and immunologic alterations associated with chronic hemodialysis. Ann Intern Med. 1980;93:597–613. doi: 10.7326/0003-4819-93-4-597. [DOI] [PubMed] [Google Scholar]
- 11.Huraib S, al-Rashed R, Aldrees A, Aljefry M, Arif M, al-Faleh F A. High prevalence of and risk factors for hepatitis C in haemodialysis patients in Saudi Arabia: a need for new dialysis strategies. Nephrol Dial Transplant. 1995;10:470–474. doi: 10.1093/ndt/10.4.470. [DOI] [PubMed] [Google Scholar]
- 12.Irie Y, Hayashi H, Yokozeki K, Kashima T, Okuda K. Hepatitis C infection unrelated to blood transfusion in hemodialysis patients. J Hepatol. 1994;20:557–559. doi: 10.1016/s0168-8278(05)80506-9. [DOI] [PubMed] [Google Scholar]
- 13.Jadoul M, Cornu C, van Ypersele de Strihou C. Incidence and risk factors for hepatitis C seroconversion in hemodialysis: a prospective study. The UCL Collaborative Group. Kidney Int. 1993;44:1322–1326. doi: 10.1038/ki.1993.385. [DOI] [PubMed] [Google Scholar]
- 14.Keur I, Schneeberger P, Van der Graaf Y, Vos J, van Dijk W, van Doorn L J. Risk factors for HCV-infection in two dialysis units in the Netherlands. Neth J Med. 1997;50:97–101. doi: 10.1016/s0300-2977(96)00092-7. [DOI] [PubMed] [Google Scholar]
- 15.Kuhns M, De Medina M, McNamara A, Jeffers L J, Reddy K R, Silva M, Ortiz Interian C, Jimenez M, Schiff E R, Perez G. Detection of hepatitis C virus RNA in hemodialysis patients. J Am Soc Nephrol. 1994;4:1491–1497. doi: 10.1681/ASN.V471491. [DOI] [PubMed] [Google Scholar]
- 16.Kuo G, Choo Q L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, et al. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 17.Maertens, G., F. Dekeyser, A. Van Geel, F. Sablon, F. Bosman, M. Zrein, and D. Pollet. Confirmation of HCV-antibodies by line immuno assay INNO-LIA HCV Ab III. In J. Y. N. Lau (ed.), Methods in molecular biology: hepatitis C, in press. [DOI] [PubMed]
- 18.Pawlotsky J M, Bastie A, Pellet C, Remire J, Darthuy F, Wolfe L, Sayada C, Duval J, Dhumeaux D. Significance of indeterminate third-generation hepatitis C virus recombinant immunoblot assay. J Clin Microbiol. 1996;34:80–83. doi: 10.1128/jcm.34.1.80-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters T, Mohr L, Scheiffele F, Schlayer H J, Preisler S, Berthold H, Gerok W, Rasenack J. Antibodies and viremia in acute post-transfusion hepatitis C: a prospective study. J Med Virol. 1994;42:420–427. doi: 10.1002/jmv.1890420416. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto N, Enomoto N, Marumo F, Sato C. Prevalence of hepatitis C virus infection among long-term hemodialysis patients: detection of hepatitis C virus RNA in plasma. J Med Virol. 1993;39:11–15. doi: 10.1002/jmv.1890390104. [DOI] [PubMed] [Google Scholar]
- 21.Sampietro M, Badalamenti S, Salvadori S, Corbetta N, Graziani G, Como G, Fiorelli G, Ponticelli C. High prevalence of a rare hepatitis C virus in patients treated in the same hemodialysis unit: evidence for nosocomial transmission of HCV. Kidney Int. 1995;47:911–917. doi: 10.1038/ki.1995.136. [DOI] [PubMed] [Google Scholar]
- 22.Schlipkoter U, Gladziwa U, Cholmakov K, Weise A, Rasshofer R, Lorbeer B, Luz N, Deinhardt F, Roggendorf M. Prevalence of hepatitis C virus infections in dialysis patients and their contacts using a second generation enzyme-linked immunosorbent assay. Med Microbiol Immunol. 1992;181:173–180. doi: 10.1007/BF00202057. [DOI] [PubMed] [Google Scholar]
- 23.Schneeberger P M, Vos J, van Dijk W C. Prevalence of antibodies to hepatitis C virus in a Dutch group of haemodialysis patients related to risk factors. J Hosp Infect. 1993;25:265–270. doi: 10.1016/0195-6701(93)90112-d. [DOI] [PubMed] [Google Scholar]
- 24.Seelig R, Renz M, Bottner C, Seelig H P. Hepatitis C virus infections in dialysis units: prevalence of HCV-RNA and antibodies to HCV. Ann Med. 1994;26:45–52. doi: 10.3109/07853899409147326. [DOI] [PubMed] [Google Scholar]
- 25.Shakil A O, Conry-Cantilena C, Alter H J, Hayashi P, Kleiner D E, Tedeschi V, Krawczynski K, Conjeevaram H S, Sallie R, Di Bisceglie A M. Volunteer blood donors with antibody to hepatitis C virus: clinical, biochemical, virologic, and histologic features. The Hepatitis C Study Group. Ann Intern Med. 1995;123:330–337. doi: 10.7326/0003-4819-123-5-199509010-00002. [DOI] [PubMed] [Google Scholar]
- 26.Simon N, Courouce A M, Lemarrec N, Trepo C, Ducamp S. A twelve year natural history of hepatitis C virus infection in hemodialyzed patients. Kidney Int. 1994;46:504–511. doi: 10.1038/ki.1994.301. [DOI] [PubMed] [Google Scholar]
- 27.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Hauverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka E, Kiyosawa K, Nakatsuji Y, Inoue Y, Miyamura T, Chiba J, Furuta S. Clinical significance of antibodies to nonstructural and core proteins of hepatitis C virus in posttransfusion hepatitis patients during long-term follow-up. J Med Virol. 1993;39:318–324. doi: 10.1002/jmv.1890390411. [DOI] [PubMed] [Google Scholar]
- 29.van der Poel C L, Reesink H W, Mauser-Bunschoten E P, Kaufmann R H, Leentvaar-Kuypers A, Chamuleau R A, Schaasberg W, Bakker E, Exel-Oehlers P J, Theobalds I, et al. Prevalence of anti-HCV antibodies confirmed by recombinant immunoblot in different population subsets in The Netherlands. Vox Sang. 1991;61:30–36. doi: 10.1111/j.1423-0410.1991.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 30.van Doorn L J, Kleter B, Stuyver L, Maertens G, Brouwer H, Schalm S, Heijtink R, Quint W. Analysis of hepatitis C virus genotypes by a line probe assay and correlation with antibody profiles. J Hepatol. 1994;21:122–129. doi: 10.1016/s0168-8278(94)80148-7. [DOI] [PubMed] [Google Scholar]
- 31.van Doorn L J, Kleter B, Stuyver L, Maertens G, Brouwer J T, Schalm S, Heijtink R A, Quint W. Sequence analysis of hepatitis C virus genotypes 1 to 5 reveals multiple novel subtypes in the Benelux countries. J Gen Virol. 1995;76:1871–1876. doi: 10.1099/0022-1317-76-7-1871. [DOI] [PubMed] [Google Scholar]
- 32.Vrielink H, van der Poel C L, Reesink H W, Zaaijer H L, Scholten E, Kremer L C, Cuypers H T, Lelie P N, van Oers M H. Look-back study of infectivity of anti-HCV ELISA-positive blood components. Lancet. 1995;345:95–96. doi: 10.1016/s0140-6736(95)90063-2. [DOI] [PubMed] [Google Scholar]
- 33.Zaaijer H L, Cuypers H T, Reesink H W, Winkel I N, Gerken G, Lelie P N. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]