Abstract
Randomly amplified polymorphic DNA (RAPD) analysis of 33 Paracoccidioides brasiliensis strains from Argentina, Brazil, Colombia, Peru, and Venezuela produced reproducible amplification products which were sufficiently polymorphic to allow differentiation of the strains. Types generated with five primers (OPG 03, OPG 05, OPG 14, OPG 16, and OPG 18) resulted in a high discriminatory index (0.956). The discriminatory index was slightly reduced (0.940) when only two primers (OPG 3 and OPG 14) were used. A dendrogram based on these results showed a high degree of similarity among the strains, and genetic differences were expressed in clusters related to geographical regions but not to pathological features of the disease. With a few exceptions, strains were sorted into five groups by geographical origin as follows: group I, Venezuelan strains; group II, Brazilian strains; group III, Peruvian strains; group IV, Colombian strains; and group V, Argentinian strains. The group containing the most disparate strains was group V (discriminatory index, 0.633); the discriminatory index for the other four groups was 0.824. The use of primer OPG 18 by itself was sufficient to discriminate species specificity, and the use of primer OPG 14 by itself was sufficient to discriminate among the geographical locations of the strains in the sample. This method may be helpful for epidemiological studies of P. brasiliensis.
Molecular techniques are powerful tools for the genomic analysis of many pathogens (4). They have been used to classify strains in order to get more information on host-parasite relationships. In fungi such as Candida albicans (11), Blastomyces dermatitidis (6), and Histoplasma capsulatum (9), differences in restriction fragment length polymorphism (RFLP) of DNA have led to the classification of clinical and soil isolates according to geographical distribution and virulence levels.
Another molecular technique is randomly amplified polymorphic DNA (RAPD) analysis, which is based on the sensitive PCR technique. RAPD analysis has also become very useful for microbial strain identification, providing information comparable to that from RFLP analysis but with the advantage of requiring simpler procedures because of the use of arbitrary primers. This technique has been successfully used to discriminate among isolates of fungi such as Aspergillus fumigatus (2), H. capsulatum (20), and Paracoccidioides brasiliensis (17). For the latter species, it is particularly important to classify strains because this fungus is the causative agent of paracoccidioidomycosis, a disease which affects people in rural areas of Latin America, mainly Brazil, Colombia, and Venezuela, where it constitutes one of the most prevalent systemic mycoses. The disease has several pathologies which have been classified into categories according to type of lesion and patient characteristics (5). This diversity suggests that strain variability plays a role in host-parasite relationships (5). Soares et al. (17) used RAPD analysis to classify seven isolates of P. brasiliensis (five from Brazil and two from Ecuador). They were able to classify the strains into two groups which shared only 35% genomic identity, although both groups were unrelated to the geographical origins of the strains. Soares et al. did not attempt to determine if any relationship existed between genetic pattern and type of pathology.
With this in mind, we initiated a program of RAPD analysis of 33 P. brasiliensis strains from diverse geographical origins and pathologies to study their possible grouping according to these factors.
MATERIALS AND METHODS
Strains.
Strains, their geographical origins, and the lesions from which the fungus was isolated are listed in Table 1.
TABLE 1.
P. brasiliensis strains used in this study
| Strain (ATCCa designation) | Original designation | Year of isolation | Region and country of isolation | Localization or type of lesions in patients |
|---|---|---|---|---|
| Pb73 (ATCC 32071) | C81b | ca. 1970 | Antioquia, Colombia | Unknown |
| Pb9 (ATCC 36324) | 7193c | ca. 1960 | Caracas, Venezuela | Lymph nodes and mucocutaneous lesionsd |
| Pb9RIe | Pb9 | 1997 | Caracas, Venezuela | |
| Pb9RIIf | Pb9 | 1997 | Caracas, Venezuela | |
| Pb300 | T1g,h | 1971 | Miranda, Venezuela | |
| Pb301 | T2g,h | 1972 | Miranda, Venezuela | |
| Pb302 | 6688g | 1979 | Barinas, Venezuela | Skin, lung |
| Pb303 | 7861g | 1980 | Caracas, Venezuela | Skin |
| Pb304 | 5598g | 1988 | Barinas, Venezuela | Skin, mucosa |
| Pb305 | 6182g | 1988 | Caracas, Venezuela | Mouth, lung |
| Pb306 | 7775g | 1991 | Trujillo, Venezuela | Mouth, lung |
| Pb307 | 7987g | 1991 | Guaira, Venezuela | Mouth, lung, skin |
| Pb308 | 9806g | 1994 | Miranda, Venezuela | Mouth, lung |
| Pb309 | 0172g | 1994 | Miranda, Venezuela | Mouth, lung |
| Pb312 | 31189b | 1994 | Antioquia, Colombia | Mouth, lung |
| Pb316 | 32243b | 1994 | Antioquia, Colombia | Lung |
| Pb317 | 29362b | 1993 | Antioquia, Colombia | Mouth |
| Pb320 | Pb696i | 1992 | Entre Rios, Argentina | Bone (6-year-old child) |
| Pb321 | 118975i | 1996 | Formosa, Argentina | Lung |
| Pb322 | Pb-TUi | Unknown | Talavera, Argentina | Uterine neck |
| Pb324 | Pb-Romeroi | 1992 | Argentina | Larynx |
| Pb325 | Pb-Ayalai | 1988 | Argentina | Unknown |
| Pb326 | Pb18j | ca. 1929 | Sao Paulo, Brazil | Unknownk |
| Pb327 | BATj | ca. 1990 | Riberao Preto, Brazil | Unknown |
| Pb328 | FPj | ca. 1990 | Goiania, Brazil | Unknown |
| Pb329 | SSj | ca. 1990 | Goiania, Brazil | Unknown |
| Pb331 | 311RMj | ca. 1990 | Sao Paulo, Brazil | Unknown |
| Pb336 | 622265j | ca. 1990 | Argentina | Bone marrow (child) |
| Pb338 | Bt01j | ca. 1990 | Botucatu, Brazil | Chronic |
| Pb341 | 38RMj | ca. 1990 | Sao Paulo, Brazil | Chronic |
| Pb342 | 259RMj | ca. 1990 | Belo Horizonte, Brazil | Chronic |
| Pb346 | 38550j | ca. 1990 | Niteroi, Brazil | Chronic |
| Pb332 | 4294j | 1996 | Lima, Peru | Unknown |
| Pb333 | 15959j | 1996 | Lima, Peru | Subcutaneous abscess |
| Pb334 | 18749j | 1996 | Lima, Peru | Ganglion |
ATCC, American Type Culture Collection.
Provided by Angela Restrepo, Medellín, Colombia.
Provided by L. Pollak, Instituto Nacional Antituberculoso, Caracas, Venezuela.
See reference 3 for more details.
Obtained from passage of strain Pb9 in mice. See reference 15 for more details.
Obtained from the second passage of strain Pb9RI in mice. See reference 15 for more details.
Provided by M. B. Albornoz, Instituto de Biomedicina, Caracas, Venezuela.
The strain was isolated from soil. See reference 1 for more details.
Provided by Ricardo Negroni and Cristina Iovannitti, Universidad de Buenos Aires, Buenos Aires, Argentina.
Provided by Zoilo Pires de Camargo, Escola Paulista de Medicina, Sao Paulo, Brazil.
See reference 8 for more details.
Growth conditions.
The P. brasiliensis strains were maintained in our laboratory on PYG (peptone, 5 g; yeast extract, 5 g; glucose, 15 g; per liter of distilled water [pH 7.0]) agar slants. They were grown in PYG liquid medium (100 ml of medium in 500-ml Erlenmeyer flasks) after inoculation with 10 ml of a seed culture. The cultures were incubated for 3 days at 37°C with continuous shaking on a gyratory shaker operated at 120 turns min−1 to obtain the yeast phase (18).
DNA preparation.
DNA preparation was performed by using a modification of the method of Raeder and Broda (13). Briefly, P. brasiliensis cultures were filtered (Whatman no. 1 filter paper) and thoroughly washed with a sterile solution of 20 mM EDTA. Cells in the filter paper were wrapped with aluminum foil and dipped in liquid N2 for 5 min and then lyophilized. Afterwards, cells were ground to a fine powder by mechanical maceration. Ground material (50 mg) was homogenized in 0.5 ml of extracting buffer (0.2 M Tris-HCl [pH 8.5], 0.25 M NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate). Extraction was carried out with phenol-chloroform (7:3 vol/vol; 500 μl), and the extracts were mixed carefully and centrifuged at 4°C at 14,000 × g for 1 h. The aqueous phase was removed, and RNA was discarded by treatment with 10-mg/ml RNase for 45 min at 37°C. An equal volume of chloroform was added for extraction, and the suspension was centrifuged again at 14,000 × g for 10 min. DNA was precipitated from the aqueous phase by addition of an equal volume of cold isopropanol. After a short centrifugation (5 to 10 s), the pellet was washed with 70% ethanol and dried and then resuspended in 10 mM Tris-HCl–1 mM EDTA (30 to 80 ml).
RAPD analyses.
Five primers designated OPG 03, OPG 05, OPG 14, OPG 16, and OPG 18 (Operon Biotechnology) were used (Table 2). RAPD analysis was carried out essentially as described by Williams et al. (19) with minor modifications. Every RAPD reaction mixture contained 10 ng of genomic DNA; 0.24 μM primer; 100 μM (each) dATP, dCTP, dGTP, and dTTP; and 0.75 U of Taq DNA polymerase (Gibco BRL) in the PCR buffer (final volume, 25 μl). After the solutions were mixed, the tubes containing the mixtures were placed in a PTC-100 programmable thermal controller (MJ Research, Inc.) for 2 min at 94°C, followed by 40 cycles of 94°C for 1 min, 30°C for 2 min, and 72°C for 2 min and a final extension period of 72°C for 7 min. Randomly amplified products were analyzed by electrophoresis on a 1.2% agarose gel in Tris-borate-EDTA buffer (0.5 M Tris, 0.5 M boric acid, 10 mM EDTA [pH 8.0]) and visualized by ethidium bromide staining. The molecular size standards used were those derived from bacteriophage λ DNA digested with either HindIII or PstI (Sigma).
TABLE 2.
DI of primers used with the P. brasiliensis strains
| OPG primer | Nucleotide sequence (5′ to 3′) | % G+C | DI |
|---|---|---|---|
| 03 | GAGCCCTCCA | 70 | 0.912 |
| 05 | CTGAGACGGA | 60 | 0.853 |
| 14 | GGATGAGACC | 60 | 0.907 |
| 16 | AGCGTCCTCC | 70 | 0.750 |
| 18 | GGCTCATGTG | 60 | 0.729 |
Analysis of data from RAPD.
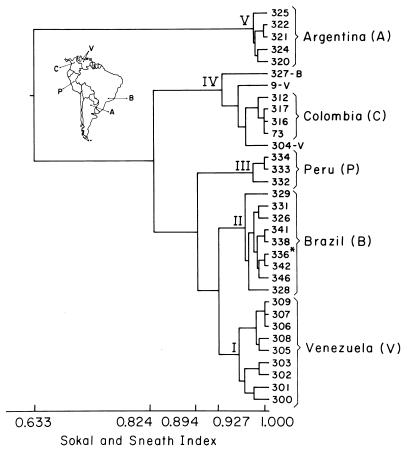
The discriminatory index (DI) from primers (Table 2) was worked out according to the procedure detailed by Hunter and Gaston (7). The dendrogram was determined by calculating Sokal and Sneath indices (SSI) for the strains (16) (Fig. 2).
FIG. 2.
Dendrogram of genetic relationships among the P. brasiliensis strains used in this study. Capital letters in parentheses refer to geographical origins of the strains (inset). ∗, strain 336 was isolated from an Argentinian girl living in the vicinity of Brazil; sample was obtained from Z. P. Camargo, Sao Paulo, Brazil.
RESULTS
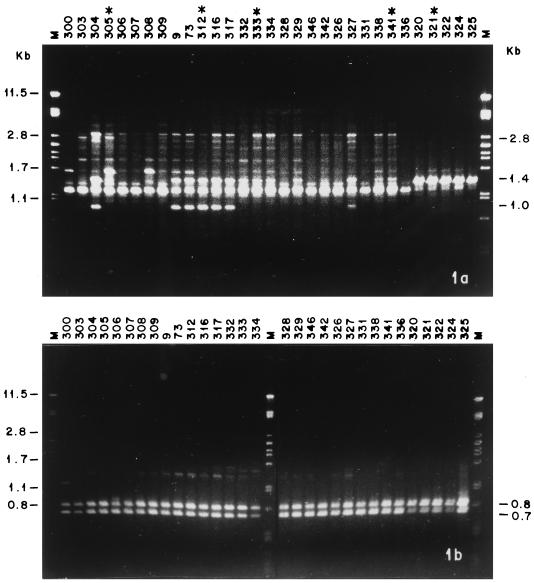
A total of 82 reproducible amplification products were sufficiently polymorphic to allow the differentiation of the strains under study. Depending on the primer, 2 to 10 bands were separated, ranging in size from 0.5 to 3.9 kb. Of these bands 11 patterns generated by Primer OPG 14 separated isolates into patterns according to geographical origin (Fig. 1a). The following strains were representative of the strains grouped by geographical origin: Pb305, Venezuela; Pb312, Colombia; Pb321, Argentina; Pb333, Peru; and Pb341, Brazil. A high DI (0.956) was obtained when all the primers were used (Table 2), although the best results were obtained with primers OPG 03, OPG 05, OPG 14, and OPG 16, which generated 39 different patterns. The use of these four primers enabled discrimination among all 33 of the isolates under study. The use of one (OPG 14), two (OPG 03 and OPG 14), or three primers (OPG 03, OPG 05, and OPG 14) led to a discriminatory index of 0.907, 0.940, or 0.954, respectively. The use of primer OPG 18 (Fig. 1b) generated two bands (molecular sizes, 0.72 and 0.83 kbp) which were common to all samples tested.
FIG. 1.
Comparison of RAPD profiles obtained with genomic DNAs from P. brasiliensis strains with primers OPG 14 (a) and OPG 18 (b). Numbers at left indicate the sizes of the standards in lanes M); numbers at right indicate the sizes of some bands in the samples. ∗, representative strain of each group. 305, group I (Venezuela); 312, group IV (Colombia); 333, group III (Peru); 341, group II (Brazil); 321, group V (Argentina).
With the use of the SSI (16), a dendrogram was built which showed a high degree of similarity among the strains (Fig. 2). Genetic differences were expressed in clusters which were related to geographical regions but not to pathologies. To set them, an SSI of 0.633 was used. This resulted in the identification of five groups as follows (see inset in Fig. 2 for geographical locations): I, Venezuelan isolates; II, Brazilian strains and strain 336, isolated from an Argentinian child living close to the border with Brazil (C. Iovannitti, personal communication); III, Peruvian isolates; IV, strains Pb9 and Pb304, from Caracas and Barinas, Venezuela (north central and southwestern Venezuela, respectively), isolate Pb327 from Brazil, and all strains from Colombia (Antioquia Department); and (V) all Argentinian isolates. Group V was the most disparate group (SSI, 0.633); the SSI for groups I to IV was 0.824.
DISCUSSION
Little is known about the extent of genetic variations within P. brasiliensis. Soares et al. (17) performed RAPD analyses of five strains from Brazil and two strains from Ecuador. With the help of five primers, they distinguished two groups which shared 35% genomic identity. One of the groups could be divided into two subgroups which shared 81% genetic similarity. They could not correlate the data for the two groups with other data such as growth or origin of isolates.
Our results led to separation of the strains into five groups arranged according to geographical zones. The strains in groups I to III showed the most similarity (SSI, 0.894), followed by the strains in groups I to IV (SSI, 0.824). The strains in group V were the most disparate (SSI, 0.633). The Brazilian strains shared differential bands with the Argentinian and the Colombian-Venezuelan strains and had a higher similarity to the latter group (group IV). At the same time, the Brazilian showed specific bands that were not shared with any of the other strains under study. Our two reference strains (Pb9 and Pb73, from Venezuela and Colombia, respectively) have been kept in our laboratory for more than 30 years. They clustered together in group IV, with the remaining Colombian strains, and were very similar in their RAPD patterns, suggesting a Colombian origin or, for strain Pb9, an origin in Venezuela close to the border with Colombia (3). There were no changes in the RAPD patterns for strain Pb9 when DNA was used which had been prepared from a sample recovered from lesions which developed after 4 weeks in mice inoculated intraperitoneally with strain Pb9 (strains Pb9RI and Pb9RII) (strains were prepared according to the procedure described by San-Blas et al. [15]). The other reference strain (Pb73, of Colombian origin) has remained clustered to the other Colombian strains despite its long maintenance in the laboratory. This stability has also been reported for H. capsulatum strains (10). Some bands present in Pb73 were not observed in Pb9 (e.g., a 3.42-kb band [OPG 14] and a 1.00-kb band [OPG 16]) and others were present in Pb9 but not in Pb73 (e.g., a 3.33-kb band [OPG 14]) (primers used are in brackets).
Our data allow the differentiation of strains according to the geographical zone of their isolation, with a few exceptions, namely, strains Pb9, -304, -327, and -336. This technique could be of use when studying the epidemiology of paracoccidioidomycosis, as a single primer (OPG 14) generated a DI high enough (0.907) to be used as the sole primer to place strains according to geographical origin. As with P. brasiliensis, H. capsulatum samples obtained in New York, N.Y., from Puerto Rican AIDS patients could also be differentiated according to their geographical origin, except that for the latter case RFLP patterns of the fungal genomes were used (9). On the other hand, primer OPG 18 generated two bands which were common to all P. brasiliensis samples, suggesting their possible use as genetic markers. This is currently under study.
So far, it has not been possible to correlate virulence of strains or pathology of disease with any P. brasiliensis genetic pattern generated by RAPD analysis, RFLP, or another typing method. This was also the case in this work since strain Pb9, which has a low level of virulence, was indistinguishable in its RAPD patterns from those of the more virulent strains Pb9RI and Pb9RII, which were derived from Pb9 after passage through mice, following a protocol described elsewhere (15). Instead, this kind of correlation has been achieved for A. fumigatus (12), which has been separated into virulent and avirulent strains by means of RAPD analysis. With this fungus the use of one primer (OPQ 06) generated a reproducible amplification product that enabled distinction between two groups, according to the presence or absence of a 0.95-kb fragment that correlated with the nature of the infection (noninvasive or invasive). Also, Cryptococcus neoformans varieties and serotypes were differentiated by means of RAPD analysis (14). And in the typing of H. capsulatum by RFLP analysis with a nuclear gene, it was possible to separate avirulent and virulent isolates from North America into different classes (9).
Multiple factors are likely to contribute to the high variability in the genome of P. brasiliensis and other fungi. For example, the diversity observed in these clinical isolates suggests that they are resident in many soil types or microclimates. Hence, genetic drift and unique selection pressures impacting on the organism in various environmental niches may influence their genetic structure in populations, which may be reflected in the geographical typing of the P. brasiliensis strains under study.
Therefore, this method opens up new approaches for epidemiological studies of P. brasiliensis.
ACKNOWLEDGMENTS
We thank the suppliers of the strains identified in Table 1 and Belisario Moreno for preparing strains Pb9RI and Pb9RII.
This study was supported by grants from the International Centre for Genetic Engineering and Biotechnology (ICGEB) [grant no. CRP/VEN95-01(h1)] and Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICIT) (Caracas, Venezuela) (grant no. PI-100, PI-96001292, and S1-96000156).
REFERENCES
- 1.Albornoz M B. Paracoccidioidomycosis. Proceedings of the First Pan American Symposium. Scientific publication no. 254. Washington, D.C: Pan American Health Organization; 1972. Isolation of Paracoccidioides brasiliensis from rural soil in Venezuela; pp. 71–75. [Google Scholar]
- 2.Aufauvre-Brown A, Cohen J, Holden D W. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J Clin Microbiol. 1992;30:2991–2993. doi: 10.1128/jcm.30.11.2991-2993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbonell L M, Pollak L. Ultrastructura del Paracoccidioides brasiliensis en cultivos de la fase levaduriforme. Mycopathol Mycol Appl. 1963;19:184–204. doi: 10.1007/BF02051247. [DOI] [PubMed] [Google Scholar]
- 4.Einsenstein B I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J Infect Dis. 1990;161:595–602. doi: 10.1093/infdis/161.4.595. [DOI] [PubMed] [Google Scholar]
- 5.Franco M F. Host-parasite relationship in paracoccidioidomycosis. J Med Vet Mycol. 1987;25:5–18. doi: 10.1080/02681218780000021. [DOI] [PubMed] [Google Scholar]
- 6.Fraser V J, Keath E J, Powderly W G. Two cases of blastomycosis from a common source: use of DNA restriction analysis to identify strains. J Infect Dis. 1991;163:1378–1381. doi: 10.1093/infdis/163.6.1378. [DOI] [PubMed] [Google Scholar]
- 7.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2469. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashino S S, Singer-Vermes L M, Calich V L G, Burger E. Alterations in the pathogenicity of one Paracoccidioides brasiliensis isolate do not correlate with its in vitro growth. Mycopathologia. 1990;111:173–180. doi: 10.1007/BF02282801. [DOI] [PubMed] [Google Scholar]
- 9.Keath E J, Kobayashi G S, Medoff F G. Classification of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersulyte D, Woods J P, Keath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magee B B, D’Souza T M, Magee P T. Strain and species identification by restriction fragment length polymorphisms in the ribosomal DNA repeat of Candida species. J Bacteriol. 1987;169:1639–1643. doi: 10.1128/jb.169.4.1639-1643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondon P, Thélu J, Lebeau B, Ambroise-Thomas P, Grillot R. Virulence of Aspergillus fumigatus strains investigated by random amplified polymorphic DNA analysis. J Med Microbiol. 1995;42:299–303. doi: 10.1099/00222615-42-4-299. [DOI] [PubMed] [Google Scholar]
- 13.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 14.Ruma P, Chen S C A, Sorrell T C, Brownlee A G. Characterization of Cryptococcus neoformans by random DNA amplification. Lett Appl Microbiol. 1996;23:312–316. doi: 10.1111/j.1472-765x.1996.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 15.San-Blas G, San-Blas F, Serrano L E. Host-parasite relationships in the yeastlike form of Paracoccidioides brasiliensis strain IVIC Pb9. Infect Immun. 1977;15:343–346. doi: 10.1128/iai.15.2.343-346.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sneath P H A, Sokal R R. Numerical taxonomy, the principles and practice of numerical classification. San Francisco, Calif: W. H. Freeman and Co.; 1973. pp. 230–234. [Google Scholar]
- 17.Soares C M A, Mollinari Madlum E E W I, da Silva S P, Pereira M, Felipe M S S. Characterization of Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:505–507. doi: 10.1128/jcm.33.2.505-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorais-Landáez F, San-Blas G. Localization of β-glucan synthetase in membranes of Paracoccidioides brasiliensis. J Med Vet Mycol. 1993;31:421–426. [Google Scholar]
- 19.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woods J P, Kersulyte D, Goldman W E, Berg D E. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J Clin Microbiol. 1993;31:463–464. doi: 10.1128/jcm.31.2.463-464.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]