Abstract
Background:
Diabetes mellitus (DM) is a recognized risk factor for dementia. Because DM is a potentially modifiable condition, greater understanding of the mechanisms linking DM to the clinical expression of Alzheimer’s disease dementia may provide insights into much needed dementia therapeutics.
Objective:
In this feasibility study, we investigated DM as a dementia risk factor by examining genome-wide distributions of the epigenetic DNA modification 5-hydroxymethylcytosine (5hmC).
Methods:
We obtained clinical samples from the Rush Memory and Aging Project and used the highly sensitive 5hmC-Seal technique to perform genome-wide profiling of 5hmC in circulating cell-free DNA (cfDNA) from antemortem serum samples and in genomic DNA from postmortem prefrontal cortex brain tissue from 80 individuals across four groups: Alzheimer’s disease neuropathologically defined (AD), DM clinically defined, AD with DM, and individuals with neither disease (controls).
Results:
Distinct 5hmC signatures and biological pathways were enriched in persons with both AD and DM versus AD alone, DM alone, or controls, including genes inhibited by EGFR signaling in oligodendroglia and those activated by constitutive RHOA. We also demonstrate the potential diagnostic value of 5hmC profiling in circulating cfDNA. Specifically, an 11-gene weighted model distinguished AD from non-AD/non-DM controls (AUC=91.8%; 95% CI, 82.9–100.0%), while a 4-gene model distinguished DM-associated AD from AD alone (AUC=87.9%; 95% CI, 77.5–98.3%).
Conclusions:
We demonstrate in this small sample the feasibility of detecting and characterizing 5hmC in DM-associated AD and of using 5hmC information contained in circulating cfDNA to detect AD in high-risk individuals, such as those with diabetes.
Keywords: Diabetes mellitus, Alzheimer’s Disease, dementia, epigenetics, 5-hydroxymethylcytosine, 5hmC, cell-free DNA, cfDNA
INTRODUCTION
The underlying pathobiological mechanisms of Alzheimer’s dementia need further elucidation to improve its diagnosis, treatment, and prevention strategies. One substantial risk factor for dementia, of which Alzheimer’s is the most common type, is diabetes mellitus (DM). DM is associated with morbidity in the brain [1,2], and early features of pre-diabetes, such as hyperinsulinemia, are associated with a decline in memory and increased risk of Alzheimer’s dementia [3]. We and others have shown that DM relates to lower cognition, faster cognitive decline, a doubling of dementia risk, and increased neuropathology of dementia at autopsy [4–6]. While it is known that both genetic and environmental factors play a role in complex diseases like Alzheimer’s dementia, the underlying molecular basis linking DM to the brain pathology of dementia remains mostly unclear.
Epigenetic modifications to DNA, including 5-methylcytosine (5mC) and its oxidative derivative 5-hydroxymethylcytosine (5hmC), are increasingly well studied in health and disease. DNA methylation states are essential for neurodevelopment and cognition and are also modified during aging of the central nervous system [7–9]. Previous studies have implicated aberrant distributions of 5mC and 5hmC modifications in DM-associated pathogenic conditions, including neurodegeneration and Alzheimer’s dementia [10–13]. Groups, including our own, have also demonstrated that genomic profiling of 5hmC in convenient clinical biospecimens, e.g., circulating cell-free DNA (cfDNA), might serve as minimally invasive diagnostic and prognostic tools for DM-associated complications, including nephropathies [14], retinopathies [15], and other vascular complications [16].
To better understand at the molecular level how DM functions as a contributing factor for Alzheimer’s dementia pathologically defined (AD), we performed genome-wide 5hmC profiling in older individuals with and without AD and with and without DM who came to the autopsy to characterize their 5hmC epigenetic signatures using target tissue (dorsolateral prefrontal cortex brain) and blood (serum) biospecimens. We examined differentially modified genomic features (e.g., gene bodies) between conditions and the biological pathways that are associated with AD with or without DM. We further demonstrated the biomarker potential of 5hmC profiles in serum-derived circulating cfDNA as a minimally invasive diagnostic tool for DM-associated AD.
METHODS
Study participants and biospecimens
We obtained biological samples from a subset of deceased and autopsied persons from the Rush Memory and Aging Project (MAP), an epidemiologic longitudinal, clinical-pathologic study, enrolling mostly non-Latino White participants (~90% of the cohort) since 1997 [17]. All participants agreed to annual testing, including a detailed clinical evaluation and blood draw and organ donation at time of death. The study was approved by an Institutional Review Board of the Rush University Medical Center. All participants signed an informed consent, Anatomic Gift Act, and a repository consent allowing their resources to be shared. At the time of analyses, from 2180 participants enrolled, 834 White non-Latinos died and came to autopsy. Among these, 337 participants had the required neuropathologic data for this study, were aged between 70 and 95 years of age at death, had complete demographic data, and had valid data on cognition and Hb A1C, postmortem interval information, as well as a postmortem brain specimen available. Among the 337, 284 had antemortem serum specimens collected within 6 years if death and did not have a rare neurological condition with the potential to influence our study results (e.g., brain tumor). Participants were matched using a Mahalanobis distance-based algorithm, such that 4 groups of 20 selected participants with a similar distribution of age at death (within 5 years) and sex (women/men) each were obtained. The 4 groups were: (1) AD and no DM, referenced in this article as “AD-only,” (2) DM and no AD, referenced in this article as “DM-only,” (3) both AD and DM, referenced in this article as “AD+DM,” and (4) no AD and no DM, the control group, referenced in this article as “controls.”
All MAP participants underwent a baseline and annual clinical evaluation, including a detailed medical history, review of medications, cognitive testing, physical examination, and other data collection, along with a blood draw. DM was identified at baseline and annually thereafter based on self-reported medical history (four questions) and visual inspection of all medication receptacles, as previously published in this and other Rush cohorts [18]. DM is designated as “present” if identified at any evaluation. Blood is collected by a trained and certified phlebotomist. Blood samples are processed using standardized protocols for serum and other components, including using appropriate tubes for collection (e.g., tiger-top serum separator tubes for serum) and single 15 minute centrifugation of samples after 30 minutes (and within 2 hours) of collection [19]. Serum samples are transported to the Rush Alzheimer’s Disease Center (RADC) laboratory, divided into barcoded 0.5ml volume aliquots, and stored in −80°C monitored freezers until use.
At time of death, a rapid autopsy was performed, and the brain was removed using a standardized, rapid autopsy protocol (average postmortem interval [PMI] = 9.6 hours). A systematic neuropathologic evaluation was conducted. After gross inspection and photo documentation, slabs from one cerebral hemisphere were frozen at −80°C, and the other hemisphere fixed in paraformaldehyde prior to dissection for diagnostic, including for AD, and other studies. The presence of AD was identified (present if high or intermediate likelihood AD; not present if low likelihood or no AD) blinded to all clinical data and as previously described [20], using a modified version of the NIA-Reagan neuropathologic criteria based on both neurofibrillary tangles and neuritic plaques [21].
Preparation of samples
For this study, we used a single serum aliquot collected closest to the time of death for the assessment of 5hmC in cfDNA. We also used frozen dorsolateral prefrontal cortex (DLPFC) in this work, because of its role in higher cognition and its vulnerability to neuropathology in AD and related dementias [22]. Serum and postmortem brain specimens were shipped to the University of Chicago for testing. The processing of specimens and sequencing data (see below) yielded a total of 74 serum and 75 brain samples, 69 of each from the same individual, with high-quality data for analysis (Figure 1).
Figure 1. Study design.
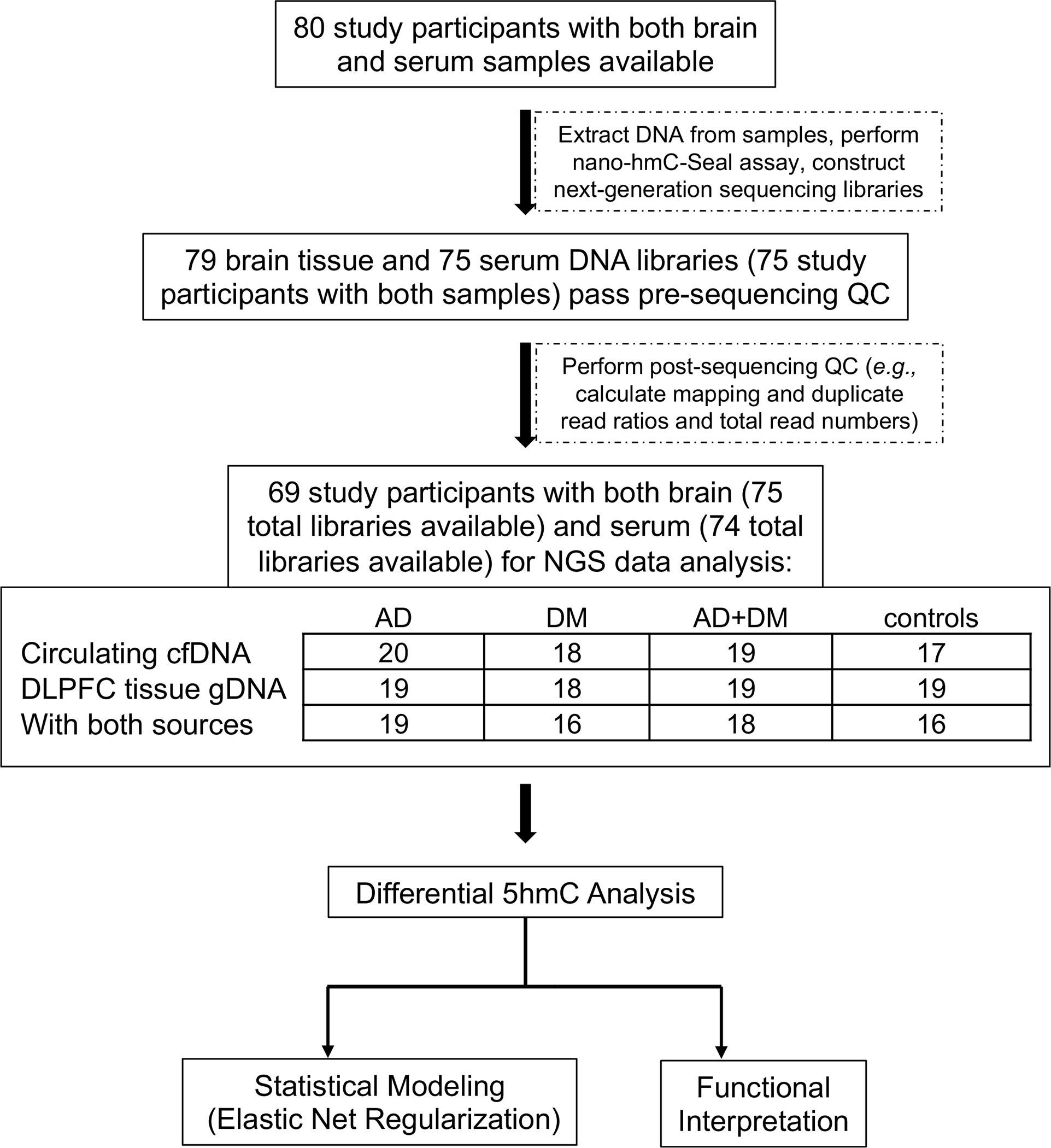
A cohort of age- and sex-matched circulating cfDNA and DLPFC brain tissue gDNA samples from the Memory and Aging Project (Rush University Medical Center) was used to identify genomic distributions of 5hmC in four different disease/control groups (AD-only, DM-only, AD+DM, and controls), to perform differential 5hmC-gene analysis to determine 5hmC-associated biological features in the different groups, and to explore the diagnostic potential of 5hmC in circulating cfDNA for AD using a machine learning approach. AD: Alzheimer’s Disease; DM: Diabetes Mellitus; control: non-AD/non-DM individuals; QC: quality control; DLPFC: dorsolateral pre-frontal cortex; cfDNA: cell-free DNA; gDNA: genomic DNA.
Isolation of DNA and characterization of serum cfDNA
All blood samples and brain samples were processed in a random order. Serum cfDNA was isolated using QIAamp Circulating Nucleic Acid Kit (Qiagen) from 0.5 mL of frozen serum per study subject. Genomic DNA (gDNA) was isolated from ~5 mg of frozen DLPFC using the Monarch Genomic DNA Purification Kit (New England Biolabs). Resulting DNA concentration was measured using the Qubit High Sensitivity dsDNA Assay (Invitrogen).
We addressed the appropriateness of serum-derived cfDNA for use in the 5hmC-Seal assay in four separate ways. First, in a preliminary experiment, we directly compared 5hmC genomic profiles across person-matched plasma-derived cfDNA, serum-derived cfDNA, and white blood cell-derived genomic DNA from a total of 6 different persons (Supplemental Methods). We determined the concordance of genome-wide 5hmC profiling across the different DNA samples and different individuals and found that 5hmC distributions from cfDNA derived from plasma and serum were highly correlated with each other, while WBC gDNA samples formed a clearly separable correlative. This initial examination suggested that serum-derived cfDNA could perform comparably to plasma cfDNA in the 5hmC-Seal assay.
Secondly, we subjected all serum-derived cfDNA samples to analysis on a Fragment Analyzer (Agilent, CA, USA) if there was enough remaining cfDNA after successful NGS library construction (71 of 74 samples). Of these 71 samples, 9 (12.7%) showed signs of appreciable gDNA contamination (i.e., significant amounts of DNA above 1000 bp in length). Though potentially problematic, the specific workflow of the 5hmC-Seal capture technique greatly minimizes the contribution of large-sized DNA fragments to NGS sequencing reads (see below). Ignoring the 9 samples with obvious gDNA contamination, our average recovery of cfDNA per serum sample was 29.2 ± 11.6 ng (median 26.0 ng), which is consistent with an increased amount of circulating cfDNA in older versus younger persons [23].
Next, in the 5hmC-Seal assay, we stress that we do not perform a cfDNA fragmentation step, as the characteristic sizes of cfDNA fragments are already within the recommended length of Illumina sequencing protocols [~100 to ~600 base pairs (bp)]). DNA fragments greater than ~1000 bp in length should be, a) ligated inefficiently to dual index barcode adapters, as used here, and if ligated, b) PCR-amplified inefficiently compared to cfDNA-sized barcoded fragments (~300 - ~650 bp). Thus, larger-sized DNA fragments should not give rise to analyzable sequencing reads in the cfDNA 5hmC-Seal protocol.
Lastly, in our analysis, we ascertained that serum-derived cfDNA 5hmC signals had very similar mapping and enrichment, etc. characteristics to those expected for plasma-derived cfDNA (e.g., Figure 2A–C). Taken together, we find the serum-derived cfDNA used here to be an appropriate analyte for the 5hmC-Seal assay and downstream 5hmC genome-wide characterization.
Figure 2. Genome-wide distributions of 5hmC in circulating cfDNA and brain tissue.
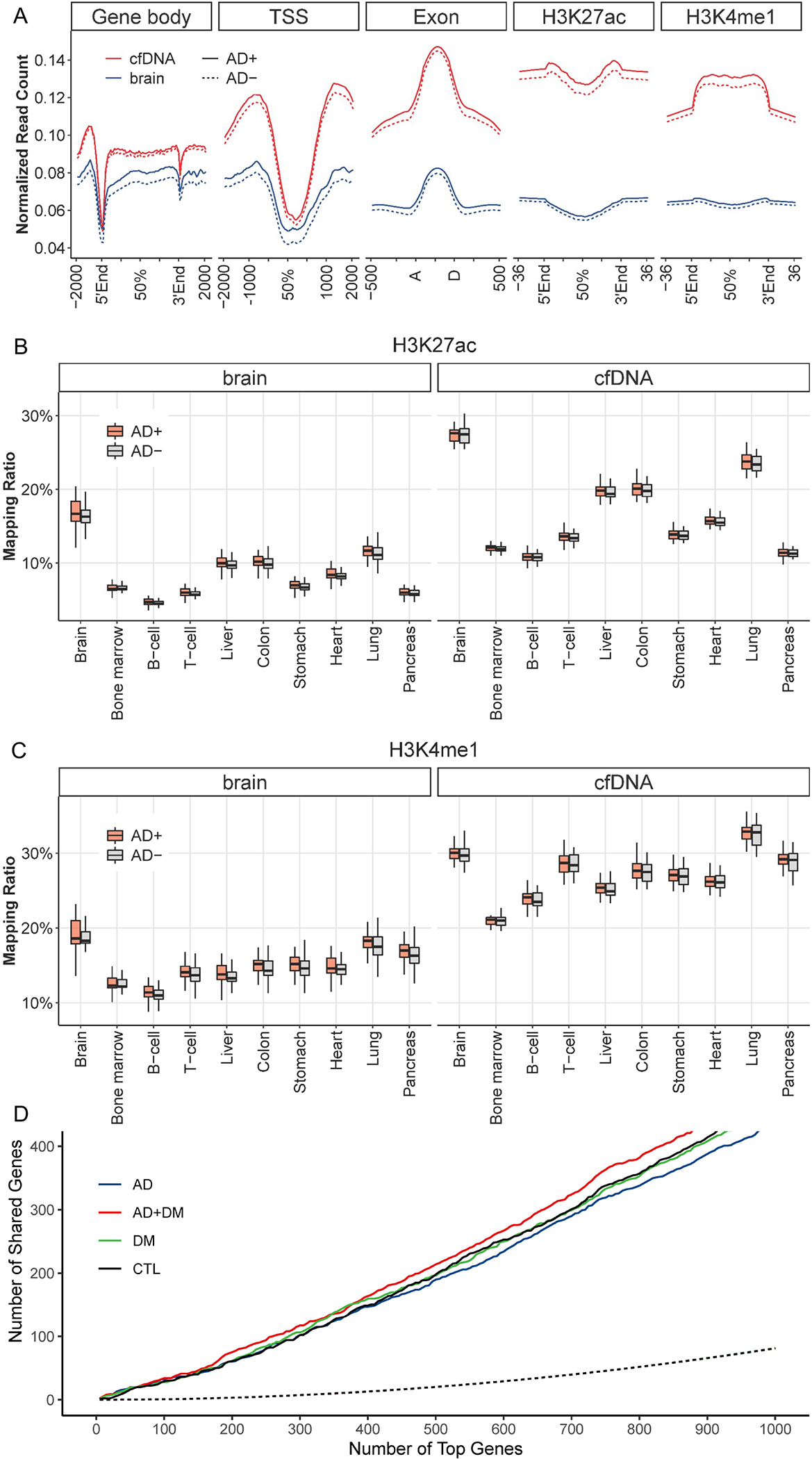
Genome-wide 5hmC profiles are summarized for various genomic feature types, including gene bodies, promoter regions, exons, and enhancers. A. Gene bodies exhibit enrichment of 5hmC-Seal reads compared to flanking regions regardless of AD status. TSS: transcription start site; TES: transcription end site; AD+: AD-only and AD+DM samples; AD-: DM-only and non-AD/non-DM controls. B. and C. Co-localization of 5hmC-Seal reads with histone modifications identified in specific tissues from the Epigenomics Roadmap Project are shown for two enhancer markers: B. H3K27ac; and C. H3K4me1. AD+: AD-only and AD+DM samples; AD-: DM-only and non-AD/non-DM controls. D. 5hmC-Seal reads demonstrate high correlation between person-matched brain tissue and circulating cfDNA samples. Blue, green, red, and black lines indicate shared top genes between cfDNA and brain tissue in AD-only, DM-only, AD+DM, and control samples, respectively. Dotted line indicates the number of simulated shared genes. AD: Alzheimer’s Disease; DM: Diabetes Mellitus; CTL: non-AD/non-DM individuals.
5hmC-Seal assay and high-throughput sequencing
For individual serum samples in which less than 10 ng DNA was extracted (6 of 80 samples), we used all available recovered DNA in the 5hmC-Seal assay. For the remaining samples, 10 ng cfDNA and 100 ng gDNA were used in the 5hmC-Seal assay, following our established protocols [24,25]. In particular, circulating cfDNA is already fragmented (<200 bp) in peripheral serum and can be barcoded without further fragmentation. To obtain sequencing-amenable ~200 bp fragments of gDNA from brain samples, we performed enzymatic fragmentation using the KAPA HyperPlus Kit (Roche). Then, after ligation of unique dual index barcoded sequencing adapters, DNA fragments proceeded through the 5hmC-Seal protocol to construct sequencing libraries. Briefly, T4 bacteriophage β-GT was used to couple an engineered azide-conjugated glucose moiety to the hydroxyl group of 5hmC in input DNA. The azide group was chemically coupled to biotin with a biotin-PEG-linked alkyne using a “Click chemistry” cycloaddition reaction. Next, streptavidin beads were used for affinity pull-down of biotin-labeled, 5hmC-containing DNA fragments. Captured DNA fragments were PCR-amplified (15 total cycles for cfDNA and 11 total cycles for genomic DNA) and then purified using AMPure Beads (Beckman Coulter). The 5hmC-Seal libraries were subject to a Fragment Analyzer (Agilent) and qPCR (KAPA Universal Library Quantification Kit, Roche), followed by paired-end sequencing (PE50) on the Illumina NovaSeq 6000 platform at the University of Chicago Genomics Core Facility (Chicago, Illinois). On average, we obtained ~40 million uniquely mapped reads per library. Additional sequencing metrics are provided in the Supplemental Methods.
Bioinformatic processing
We followed the bioinformatic pipelines of our previous publications [24,26]. Briefly, adapter sequences were removed from raw sequencing reads using Trimmomatic [27]. Low-quality bases at the 5’ (phred score <5) and 3’ (5 bp-sliding window phred score <15) were trimmed to a minimum length of 30 bp. Sequencing reads were aligned to the human genome reference (hg19) using Bowtie2 with end-to-end alignment mode [28]. Read pairs were concordantly aligned with fragment length ≤ 500 bp and an average ≤ 1 ambiguous base and up to four mismatched bases per 100 bp length. Alignments with Mapping Quality Score ≥ 10 were counted for overlap with GENCODE [29] gene bodies using featureCounts [30] without strand information. 5hmC profiles were summarized for enhancer markers H3K4me1 and H3K27ac derived from various tissues from the Roadmap Epigenomics Project [31]. In total, two obvious cfDNA sample outliers were removed based on principal components analysis (PCA). Raw fragment counts were normalized using DESeq2 [32], which performs an internal normalization that corrects for library size. Batch effect was corrected with the ComBat tool [33].
Comparing 5hmC profiles between circulating cfDNA and brain-derived gDNA
For biospecimens derived from persons with AD, we evaluated the correlation of 5hmC profiles between the two different DNA sources: serum-derived circulating cfDNA and brain-derived gDNA. All genes were ranked by average 5hmC modification levels in cfDNA and brain tissue-derived gDNA separately, followed by Pearson’s correlation test using a null distribution generated by random sampling (N=10,000). We also evaluated the relationships between person-matched cfDNA and brain tissue samples under different conditions (e.g., AD-only, DM-only, AD+DM, and controls). For each condition, the most variable genes (e.g., top 100, 500) in cfDNA were evaluated for overlap with the most variable genes in brain tissue. The observed number of overlapped genes between the two sources was compared with a null distribution generated by random sampling using the hypergeometric test.
Differential analysis, statistical modeling, and pathway exploration
We performed differential analysis systematically in cfDNA and brain tissue-derived gDNA samples for gene bodies between (1) AD-only vs. controls, (2) AD-only vs. AD+DM, (3) DM-only vs. AD+DM, using logistic regression models, adjusting for age and sex. In order to adjust for multiple comparisons, a null distribution of the p-value was generated for each comparison by permutation (e.g., randomly sampling the sample label 10,000 times) following the max T procedure [34]. An adjusted p-value < 0.05 was considered significant. The adjusted p-value <0.05 and fold-change >20% cutoffs were used to identify candidates for feature selection and modeling for each comparison. Feature selection was performed using elastic net regularization to fit a multivariable logistic regression model using the glmnet library (v2.0–16) in R [35]. The selected features were used to build a final logistic model for each comparison. We calculated a weighted ad-score for each sample under each model: ad-score=∑βk×Gk, where βk is the logistic model coefficient and Gk is the normalized 5hmC level for the kth model gene, following our previous publications [24,26]. Given our small sample size, we used the leave-one-out cross-validation (LOOCV) procedure to evaluate the performance of selected features in all samples to generate the AUCs in different comparisons (e.g., AD vs. CTL, AD vs. AD+DM, and AD+DM vs. DM) to determine if the selected features were significantly affected by any potential outlier sample. The 95% confidence intervals (CI) were computed using pROC [36]. We used Metascape [37] to explore potential functions of the differential features (i.e., genes) in the different comparisons, requiring a minimum of 5% input genes for each term (or 4 total genes, if 5% was less than 4), and p-value < 0.01.
RESULTS
Table 1 provides data for four demographic variables of the study participants. All participants were non-Latino, White. The average age at blood draw across assayed subjects was 86.2 ± 3.6 years, average age at death was 88 ± 3.5 years, 60.5% were male, and average education level was 15.2 ± 2.8 years (equivalent to some college).
Table 1.
Summary of the study participants and biospecimens.
| Variable/Summary | Age, blood draw - yrs (SD) | Age, death - yrs (SD) | Sex - male (%) | Education - yrs (SD) |
|---|---|---|---|---|
| AD | 86.2 (4.3) | 88.4 (4.0) | 12 (60.0) | 14.9 (2.4) |
| DM | 86.1 (3.7) | 88.2 (3.9) | 12 (60.0) | 14.6 (2.5) |
| AD+DM | 86.3 (2.6) | 88.4 (2.5) | 12 (60.0) | 14.9 (3.1) |
| Controls | 86.3 (3.8) | 88.5 (3.6) | 12 (60.0) | 16.5 (2.8) |
| TOTAL | 86.2 (3.6) | 88.4 (3.5) | 48 (60.0) | 15.2 (2.8) |
Abbreviations: SD, standard deviation; AD, Alzheimer’s Disease (based on neuropathologic criteria as described in the text); DM, Diabetes Mellitus; Controls, individuals with neither AD nor DM.
Genomic distribution of 5hmC modifications by functional genomic feature
Using the 5hmC-Seal technique followed by next-generation sequencing, we profiled the genome-wide 5hmC in circulating cfDNA and DLPFC-derived gDNA in 79 individuals comprising four groups: 19 individuals with neither AD nor DM (“controls”), 20 individuals with both AD and DM (“AD+DM”), 20 individuals with AD and no DM (“AD-only”), and 20 individuals with DM and no AD (“DM-only”). From these persons, we generated a total of 74 5hmC-enriched DNA libraries from cfDNA samples and 75 5hmC-enriched DNA libraries from tissue gDNA samples of sufficient quality for analysis (Figure 1).
We plotted normalized read counts by physical location within genic features (e.g., gene bodies and transcription start sites) and sites of known histone modifications that mark active enhancers (i.e., H3K27ac and H3K4me1) in brain tissue and cfDNA samples separately (Figure 2A) to illustrate the relative distribution of 5hmC across different genic features. We found that 5hmC-containing reads were enriched in gene bodies relative to flanking regions and were also enriched for the genomic locations of known H3K27ac and H3Kme1 enhancer marks (Figure 2A), consistent with previous observations of 5hmC distributions in various biospecimens [38].
Figure 2B–C illustrates the co-localization of gDNA- and cfDNA-derived 5hmC sequencing reads with enhancer markers from the Epigenomics Roadmap Project [31], providing insight into the distribution patterns of 5hmC in these samples and the potential tissue-of-origin features of circulating cfDNA. The mapping ratio indicates the proportion of 5hmC-Seal reads mapped to the histone modification marks derived from a particular tissue. Analysis of the co-localization of 5hmC and enhancer marks with regard to tissue type confirmed that 5hmC profiles from brain tissue gDNA were enriched in brain-derived histone modification marks compared to other tissue types and white blood cells. 5hmC profiles from cfDNA also produced strong signals of co-localization with brain-derived histone modifications. This finding suggests that cfDNA from brain tissue can be detected within the circulation system [39], and the detection of additional tissues reflects the heterogeneous anatomical origins of cfDNA. Of note, for the genes with the highest variability in 5hmC modification levels, the number of genes overlapped between them was significantly higher than the null expectation from random sampling (hypergeometric test, p<0.0001 for the top 1,000 modified genes in cfDNA) (Figure 2D), suggesting a correlation between person-matched brain tissue and cfDNA samples (Supplemental Figure S1).
Differentially modified gene bodies associated with AD and DM
We identified 222 gene bodies differentially modified for 5hmC between persons with AD-only and controls from circulating cfDNA (Figure 3A). Hierarchical clustering of the most differentially modified loci was generally able to separate individuals with AD-only versus controls. Similarly, we identified 199 gene bodies differentially 5hmC-modified between AD-only and AD+DM individuals (Figure 3B) and 201 gene bodies modified between DM-only and AD+DM individuals (Figure 3C). These features were generally able to separate individuals with either single disease (AD or DM) from those with both diseases. Supplemental Table S1 provides the gene bodies identified as differentially 5hmC-modified between conditions.
Figure 3. Differential analysis and hierarchical clustering using circulating cfDNA suggests strong correlations between 5hmC distribution and disease states.
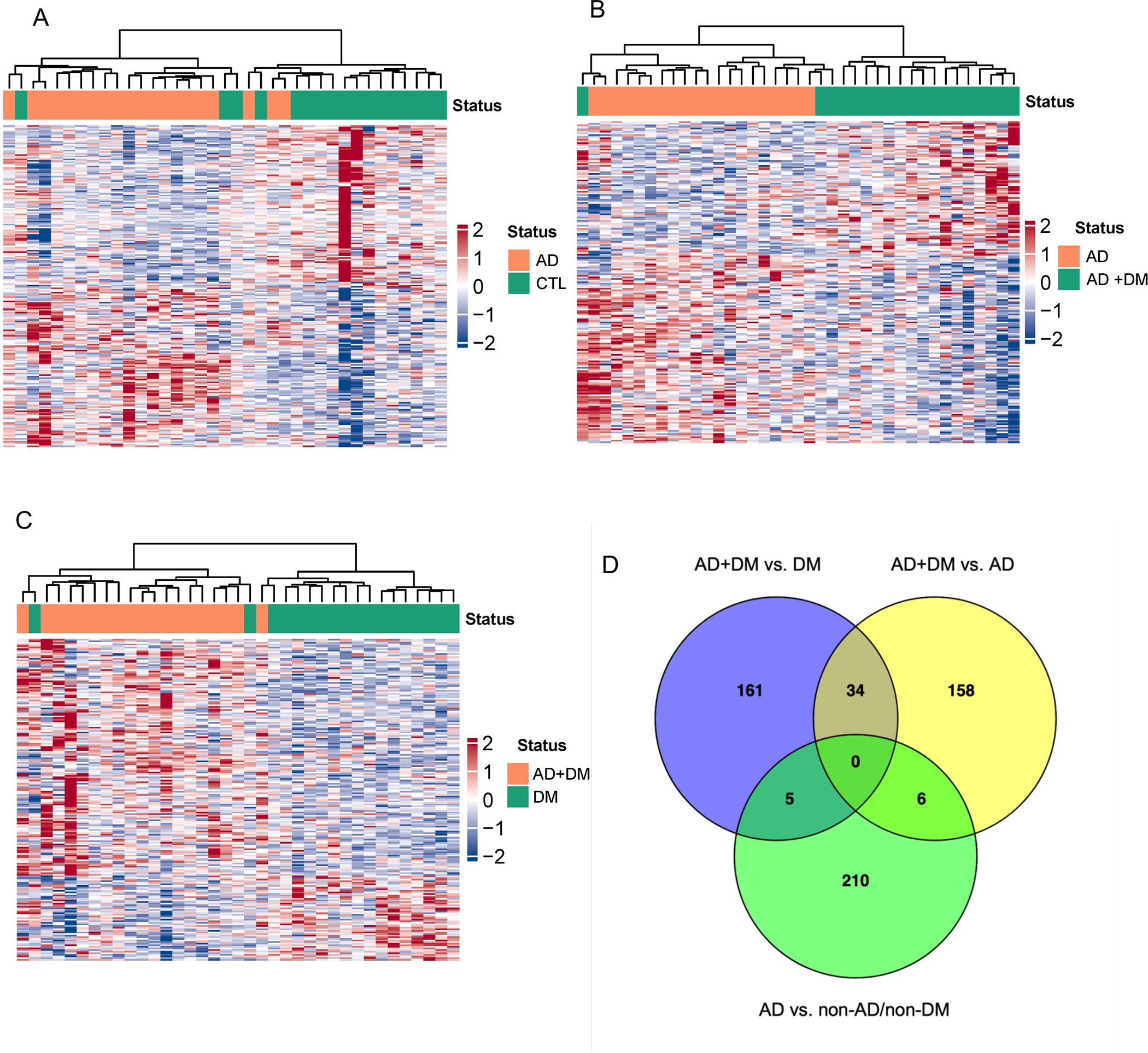
Differential analysis using multivariable logistic regression detects gene bodies differentially 5hmC-modified in cfDNA for hierarchical clustering: A. AD-only vs. controls (CTL); B. AD-only vs. AD+DM; and C. DM-only vs. AD+DM. D. Venn diagram of the differential genes identified in three comparisons: AD+DM vs. DM-only; AD+DM vs. AD-only; AD-only vs. non-AD/non-DM controls. Abbreviations as above and in main text.
Exploring the commonalities in 5hmC distribution between AD and DM may help to uncover common biology that renders DM an important risk factor for AD. We identified 34 differentially 5hmC-modified genes in circulating cfDNA in persons with AD+DM versus both AD-only and DM-only (Figure 3D). Notably, all 34 genes displayed consistent up- or down-enrichment levels across both AD+DM comparisons versus single disease (23 genes with higher 5hmC levels and 13 with lower levels in AD+DM vs. both single diseases), suggesting that they behave similarly in AD+DM versus underlying conditions. Interestingly, we identified only 5 differential genes in common between ‘AD-only vs. controls’ (which should reveal genes aberrantly 5hmC-modified in AD) and ‘DM-only vs. AD+DM’ (which should reveal genes aberrantly 5hmC modified in AD in a DM background). The paucity of shared AD-associated 5hmC-modified genes in these different backgrounds (non-DM versus DM) potentially hints at a substantial effect of DM upon the genes subject to aberrant 5hmC regulation in AD.
We also used data from DLPCF brain tissue to identify 52 differentially 5hmC-modified genes between AD-only and controls, 54 genes between AD-only and AD+DM, and 300 genes between DM-only and AD+DM (Supplemental Figure S2A–C). Though the top-ranking differential genes appeared to be different between cfDNA and brain tissue, our simulation analysis suggested that the most differential genes identified in cfDNA significantly overlapped with those identified in brain tissue compared to a null distribution by random sampling (Supplemental Figure S3). Only a small number of differential genes from each comparison were found to overlap with any other comparison, again hinting at an important influence of DM on AD and brain biology (Supplemental Figure S2D). Supplemental Table S1 lists the gene bodies differentially 5hmC-modified between conditions.
Functional relevance of differentially modified gene bodies associated with AD
To further explore how 5hmC epigenetic modifications may illuminate biological features underlying AD and DM, we examined the differentially 5hmC-modified genes identified from circulating cfDNA in each of three comparisons, AD-only vs. controls, DM-only vs. AD+DM, and AD-only vs. AD+DM, in gene ontology/pathway analysis (Figure 4A; Supplemental Table S2). We eliminated redundant, overly broad, and cancer-specific terms and then ascertained the overlap of remaining annotations (Figure 4B). We uncovered distinct biological pathways for all three comparisons, as well as some common terms. Gene annotation terms unique to AD vs. controls included ‘genes associated with high-confidence PAX3-FOXO1 enhancer sites’ and ‘aberrant gene expression in senescent cells with forced cell cycle dysregulation.’ Similarly, for AD-only vs. AD+DM, ‘positive correlation with GABAergic interneuron density in prefrontal cortex’ and ‘polymeric cytoskeletal fiber’ were uniquely significant, while ‘hematopoietic/lymphoid organ development’ and ‘upregulated in PHF8 (a histone demethylase) knockdown’ were unique to ‘DM-only vs. AD+DM.’
Figure 4. Gene annotation analyses of differential genes using circulating cfDNA suggests distinct biological pathways affected in persons with AD in different backgrounds.
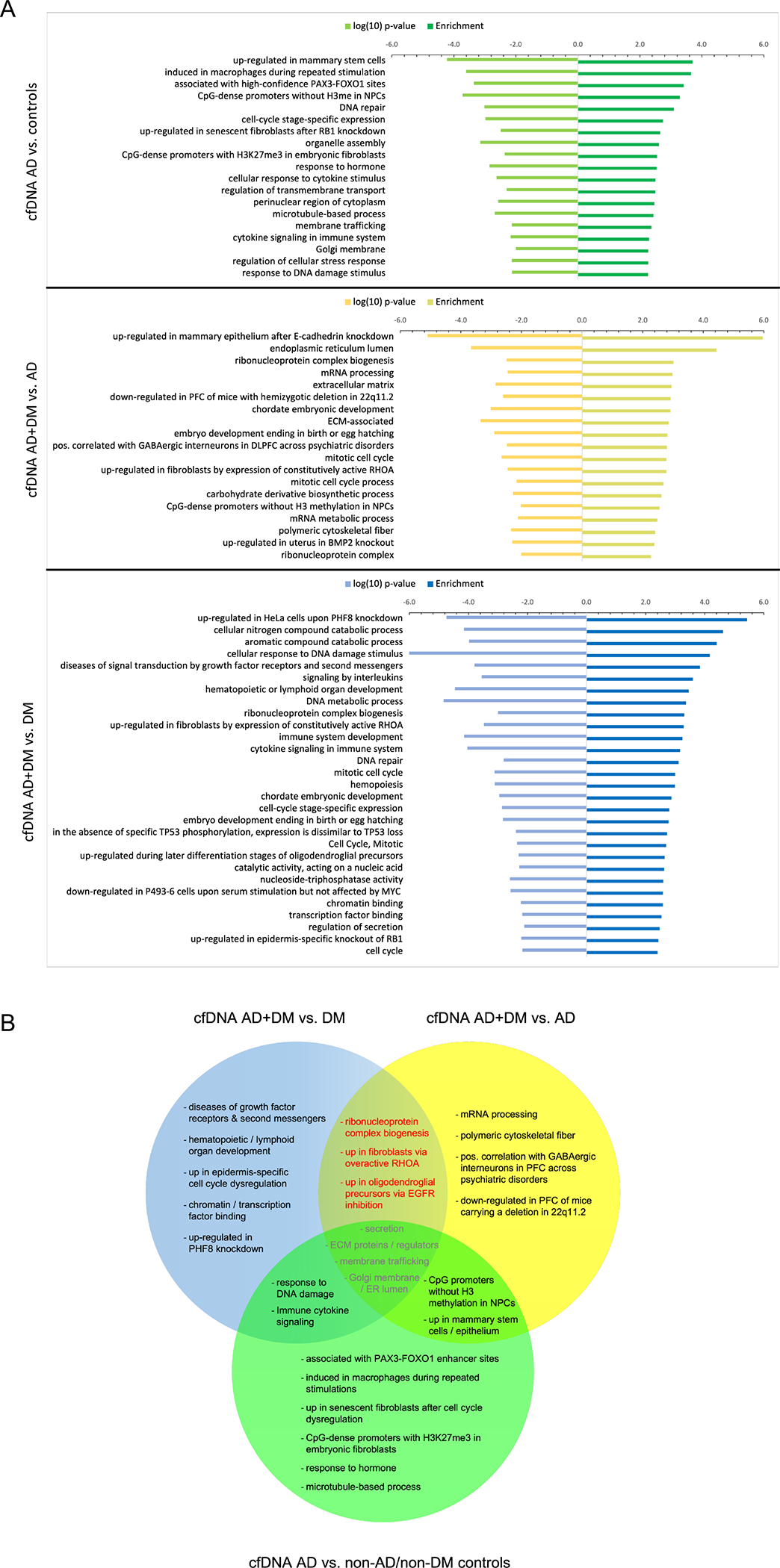
A. B. Comparison of gene annotation terms associated with differential genes reveals similarities and differences in associated biological pathways for the different disease and control backgrounds. Dashed box highlights annotation terms in common to the two comparisons of persons with AD+DM versus each single disease (≥ 5% of input genes and p-value <0.01). PFC, pre-frontal cortex; ECM, extracellular matrix; ER, endoplasmic reticulum; NPCs, neural precursor cells; H3me, methylation of histone H3. AD: Other abbreviations as above and in main text.
Further, several annotation terms, ‘ribonucleoprotein complex biogenesis,’ ‘up in fibroblasts transformed by constitutively active RHOA,’ and ‘up in oligodendroglial precursors during EGFR inhibition,’ were exclusive to the overlap between ‘AD-only vs. AD+DM’ and ‘DM-only vs. AD+DM.’ As significant pathways common to persons with AD+DM, but absent in either single disease, should reveal biological functions specifically affected in persons with both AD and DM, alteration of these particular processes might be particularly important for the specific pathology of DM-associated AD.
We also performed gene annotation analyses for differentially modified genes in DLPFC brain tissue (Supplemental Figure S4A). No exact terms overlapped between any of the three comparisons (Supplemental Figure S4B). However, notably, several annotations found in ‘AD+DM vs. DM-only’ and ‘AD+DM vs. AD-only’ are involved directly or indirectly with immune system function, including ‘neutrophil degranulation, ‘up in activated macrophages,’ ‘enriched in pseudopodia,’ ‘lytic vacuole/lysosome,’ and ‘cilium assembly.’
Machine learning to explore the diagnostic potential of 5hmC in cfDNA
To explore the possibility of using 5hmC epigenomic profiles derived from circulating cfDNA as a tool for AD screening and diagnosis, we used elastic net regularization under multivariable logistic regression models to create a final panel of 11 genes differentially modified between persons with AD-only and controls (Figure 5A), 4 genes between persons with AD-only and AD+DM (Figure 5B), and 4 genes between persons with DM-only and AD+DM (Figure 5C; Supplemental Table S1). Specifically, the LOOCV procedure using these final panels produced AUCs of 91.8% for distinguishing persons with AD-only from controls (95% CI 82.9–100.0%; Figure 5A), 87.9% for distinguishing persons with AD-only from those with both AD and DM (95% CI, 77.5–98.3%; Figure 5B), and 82.7% for distinguishing persons with DM-only from those with both AD and DM (95% CI, 67.6–97.9%; Figure 5C). Notably, several gene components in the final model have been implicated in AD or identified as potential therapeutic targets in previous studies, such as CXCL9 [40], ERLIN2 [41], POLDIP2 [42] between AD-only and controls; SENP8 [43] between AD-only and AD+DM; and BCR [44] between DM-only and AD+DM.
Figure 5. Machine-learning suggests diagnostic potential of 5hmC in cfDNA for AD.
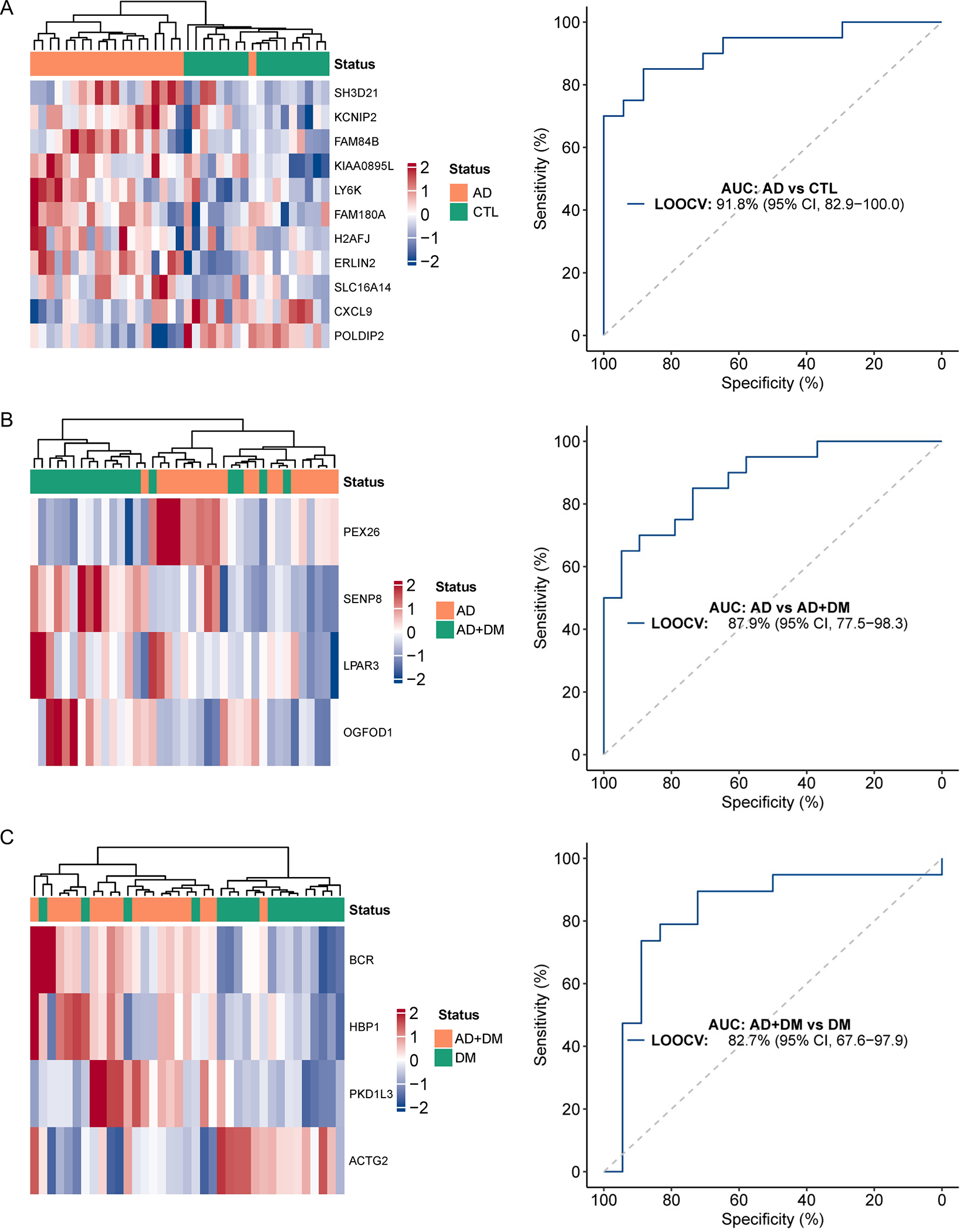
Statistical modeling using an elastic net regularization and multivariable logistic regression approach identifies: A. a weighted model comprised of 11 gene bodies that distinguishes AD from controls (CTL); B. a 4-gene weighted model that distinguishes AD from AD+DM; and C. a 4-gene weighted model that distinguishes DM from AD+DM. The AUC (area under the curve) and 95% CI (confidence interval) indicate performance of selected features evaluated by the LOOCV (leave-one-out cross-validation) procedure. Abbreviations as above and in main text.
DISCUSSION
Here, we demonstrate that 5hmC epigenetic modifications to DNA can be detected and characterized in DM-associated AD from both brain tissue and circulating cfDNA. We uncover genes and biological pathways that may be important specifically for the pathology of DM-associated AD, including ribonucleoprotein complex biogenesis, constitutive activity of RHOA GTPase, and action of oligodendrocyte precursor cells in response to EGFR inhibition. We further demonstrate the feasibility of using 5hmC information contained in circulating cfDNA as a tool for the detection of AD in high-risk individuals, such as those with diabetes. We identify panels containing modest numbers of differentially 5hmC-modified genes that demonstrate considerable ability to distinguish persons with either single disease (AD-only or DM-only) from those with both AD and DM. Of note, the 5hmC discriminating models were found to contain genes relevant to AD and/or DM disease states. Finally, while most previously published studies employing the 5hmC-Seal technique have utilized plasma-derived cfDNA as input material, in the current study, we show the feasibility of 5hmC profiling in cfDNA derived from serum samples. This technical development provides a key advance for widely applying 5hmC-Seal, offering more flexibility for future research.
Previous work has implicated epigenetic contributors, including 5hmC, in DM and diabetic complications, as well as in Alzheimer’s dementia [45]. 5hmC is important for dynamically regulating chromatin states and gene expression and is closely associated with open chromatin, reflecting positive gene expression [46,47]. Thus, the specific locations of 5hmC modifications in the genome can provide information about the identity of loci likely transcriptionally activated, offering insights into the aberrantly regulated genes driving disease. Here, we found that the majority of differentially 5hmC-modified genes were not shared between persons with AD in different biological backgrounds, suggesting that AD pathogenesis might be influenced by different biological mechanisms depending on underlying condition. Indeed, between ‘AD vs. controls’ and ‘AD+DM vs. DM-only,’ the latter which should provide a sensitized background for investigating DM-associated AD, only 5 of ~200 genes were shared between the two comparisons.
Though largely of hematopoietic origin, the remaining cfDNA is derived from a number of characteristic and heterogeneous anatomical sources in healthy individuals. Given that DM is a system-wide disorder, and that brain-derived DNA can be detected within circulating cfDNA [39], it is not entirely surprising that we observed a larger differential 5hmC signal between conditions using cfDNA, which interrogates multiple tissues, rather than brain tissue gDNA. Further, the solid tissue responsible for contributing the largest amount (~10%) of cfDNA in healthy individuals is the vascular endothelium [23]. Vasculature is very often compromised in persons with DM, and it is also increasingly understood as significant in the pathogenesis of AD. For these reasons, we might expect cfDNA to be a rich source of differential 5hmC modifications between persons with AD, with DM, and healthy individuals versus brain tissue.
Evaluating the differential 5hmC genes and associated pathways exclusive to persons with AD+DM, and not present in persons with either single disease, might allow elucidation of the biological functions important specifically for DM-associated Alzheimer’s dementia. Here, we uncovered 34 differentially 5hmC-modified genes from cfDNA and an additional 8 from DLPFC brain tissue that are common to persons with AD+DM. Importantly, all of the genes identified from cfDNA exhibit the same up- or down- direction of enrichment in both comparisons (i.e., AD+DM vs. AD and AD+DM vs. DM), increasing our confidence that these genes operate similarly across disease comparisons and allowing cohesive pathway analysis. Using the 34 genes, we identified three unanticipated gene annotation terms. Of these, the roles of oligodendrocytes and white matter EGFR signaling [48,49] and also ribonucleoprotein complexes [50–52], such as the spliceosome and telomerase complex, are under-studied areas in AD, yet increasingly appreciated as important for its pathology. The small GTPase RhoA has been reasonably well-studied in AD [53,54] and in DM [55,56] independently, but no studies to our knowledge have explored how shared aberrant RhoA-mediated signaling could impact DM-associated cognitive dysfunction. We also observed that annotations related to the processing and secretion of molecules membrane-bound and/or external to the cell were in common to all three comparisons (AD+DM vs. AD, AD+DM vs. DM, and AD-only vs. controls), perhaps highlighting the key roles that secretion and its requisite protein processing play in both diseases [57–59].
Our ability to separate persons based on disease condition using differential 5hmC gene analysis was diminished in DLPFC gDNA relative to circulating cfDNA. This may reflect the modest number of DM-instigated changes to brain tissue versus other tissues of the body, small sample size, or other factors. Using brain tissue 5hmC, we uncovered a small number of differential genes that overlap between the different disease and control groups. Consequently, we uncovered gene annotation terms unique to each disease/control comparison, including “up in radial glia relative to neurons” for ‘AD-only vs. controls,’ “metabolism of lipids” for ‘DM-only vs. AD+DM,’ and “transcriptional regulation by TP53” for ‘AD-only vs. AD+DM.’ Interestingly, we also uncovered significant gene annotation terms regarding primary endothelial and hematopoietic stem progenitor cells among the brain tissue comparisons. The manifestation of these terms may reflect the presence of vasculature within our DLPFC brain tissue samples, thereby allowing evaluation of differentially 5hmC-modified genes in a known physiological contributor to Alzheimer’s dementia, the brain vasculature.
Alzheimer’s dementia remains an underdiagnosed condition, and it has been estimated that average time to diagnosis is delayed 2–3 years after symptom onset. Current screening and diagnostic procedures, such as PET scans and lumbar punctures, are time-consuming, invasive, expensive, and not widely available. Circulating cfDNA might be used to create diagnostic and/or prognostic disease assays that use simple blood draws for detecting molecular changes associated with AD, even in the absence of clinical symptoms. Assayable from cfDNA, 5hmC is a sensitive and powerful analyte to probe for disease monitoring, as it is abundant in the genome, chemically stable, and closely associated with disease states. Other studies have shown that AD-associated levels of 5hmC can be detected at preclinical stages, indicating that 5hmC should be a viable analyte by which to monitor Alzheimer’s dementia onset and progression [60,61]. Moreover, as epigenetic changes like alterations in 5hmC distribution are influenced by environment, they may be particularly appropriate for assessing the onset, progression, and/or treatment response of multifactorial and lifestyle- and environmentally-impacted diseases like Alzheimer’s dementia and DM [62–64]. Our findings here lay the groundwork for further research on utilizing 5hmC in circulating cfDNA as a diagnostic biomarker or disease monitoring tool, with the ultimate goal of identifying individuals at risk for, or with early-stage, DM-associated Alzheimer’s dementia for timely intervention and improved clinical outcomes. Future work will seek to replicate and expand these findings in larger and more diverse samples.
Limitations of the Study
There are several limitations to the current study. First, sample size was relatively small. Larger studies in this and other cohorts are underway to confirm the 5hmC diagnostic models for AD and the genomic 5hmC signatures identified here as associated with AD. Secondly, the current study included mostly individuals of European ancestry, and future studies of more diverse populations will allow us to evaluate population differences and health disparities. Thirdly, although our cross-sectional analyses allowed us to identify 5hmC signatures that correlate with AD with or without DM, for the ultimate goal of developing a tool for early Alzheimer’s dementia detection and intervention, a longitudinal study framework will best help us evaluate the predictive ability of our models by examining the time to Alzheimer’s dementia onset. Fourthly, this study did not take into account several potential confounders and other important variables that may play a role in the relation of 5hmC signatures to metabolism and brain function. We are planning follow-up research in a larger sample that will consider socio-demographic factors such as age, sex, and race, as well as common co-occurring medical conditions in DM (e.g., hypertension, obesity), especially as we and others have shown their relation to brain impairment and pathology [65–67]. Fifthly, there might be limitations in comparing results from our study with those of other studies using different designs and/or methods, for instance with cfDNA data generated from plasma or from samples that were “double-spun” (e.g., once at low speed and once at high speed). Finally, future systems biological studies that integrate, for example, transcriptomics, genome-wide association data, and detailed phenotype and lifestyle data, will help elucidate the interactions of epigenetic contributors to Alzheimer’s dementia with gene expression, genetic background, and environmental influences.
Conclusions
We performed genome-wide mapping of 5hmC in circulating cfDNA and brain tissue samples, and we identified distinct 5hmC signatures and biological pathways in persons with DM-associated AD compared to persons with only AD or only DM. These pathways may be particularly important for future investigation into the pathogenesis of DM-associated Alzheimer’s dementia. Our findings help lay the foundation for future development of 5hmC-based tests using routine blood draws for Alzheimer’s dementia diagnosis or prognosis, which could be integrated into the management of high-risk individuals, such as those with DM, or utilized as a screening tool for the early detection of Alzheimer’s dementia in the general population. For devastating, currently, poorly treated conditions like AD and associated dementias, early detection of at-risk persons from among those with potentially modifiable diseases, such as DM, offers a powerful opportunity for targeted intervention and improved cognitive and societal outcomes.
Supplementary Material
Acknowledgements
We thank the participants of the Rush MAP, as well as the staff and faculty of the RADC. We thank the University of Chicago Genomics Facility (RRID:SCR_019196) for DNA library quality assessment and high-throughput DNA sequencing. C.H. is an investigator of the Howard Hughes Medical Institute. This article is subject to HHMI’s Open Access to Publications policy. HHMI lab heads have previously granted a nonexclusive CC BY 4.0 license to the public and a sublicensable license to HHMI in their research articles. Pursuant to those licenses, the author-accepted manuscript of this article can be made freely available under a CC BY 4.0 license immediately upon publication.
Funding
This study was supported by grants from the National Institutes of Health: RF1 AG074549, RF1 AG059621, R01 NS084965, and R33 CA269100. MAP is supported by R01 AG17917 and AG052476. The funding bodies had no role in the design of the study and collection, analysis, interpretation of data, or in writing the manuscript.
Footnotes
Competing interests
CH was the scientific founder of Epican Genetech, which obtained a license from the University of Chicago to develop the 5hmC-Seal technique for clinical applications. WZ was an advisor of Epican Genetech, and he received research support from the company.
Ethics approval and consent to participate
We obtained biological samples from a subset of deceased and autopsied persons from the Rush Memory and Aging Project (MAP). The study was approved by an Institutional Review Board of the Rush University Medical Center. All participants signed an informed consent, Anatomic Gift Act, and a repository consent allowing their resources to be shared.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its additional files]. Raw and processed sequencing data supporting the conclusions of this article are available in the NCBI GEO repository, Accession No. GSE203337 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203337. MAP resources can be requested at www.radc.rush.edu.
REFERENCES
- [1].Antal B, McMahon LP, Sultan SF, Lithen A, Wexler DJ, Dickerson B, Ratai E-M, Mujica-Parodi LR (2022) Type 2 diabetes mellitus accelerates brain aging and cognitive decline: Complementary findings from UK Biobank and meta-analyses. Elife 11, e73138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ortiz GG, Huerta M, González-Usigli HA, Torres-Sánchez ED, Delgado-Lara DL, Pacheco-Moisés FP, Mireles-Ramírez MA, Torres-Mendoza BM, Moreno-Cih RI, Velázquez-Brizuela IE (2022) Cognitive disorder and dementia in type 2 diabetes mellitus. World J Diabetes 13, 319–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Blázquez E, Hurtado-Carneiro V, LeBaut-Ayuso Y, Velázquez E, García-García L, Gómez-Oliver F, Ruiz-Albusac JM, Ávila J, Pozo MÁ (2022) Significance of Brain Glucose Hypometabolism, Altered Insulin Signal Transduction, and Insulin Resistance in Several Neurological Diseases. Front Endocrinol (Lausanne) 13, 873301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Arvanitakis Z, Wilson RS, Bienias JL, Evans DA, Bennett DA (2004) Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Arch Neurol 61, 661–666. [DOI] [PubMed] [Google Scholar]
- [5].Simó R, Ciudin A, Simó-Servat O, Hernández C (2017) Cognitive impairment and dementia: a new emerging complication of type 2 diabetes-The diabetologist’s perspective. Acta Diabetol 54, 417–424. [DOI] [PubMed] [Google Scholar]
- [6].Arvanitakis Z, Wang H-Y, Capuano AW, Khan A, Taïb B, Anokye-Danso F, Schneider JA, Bennett DA, Ahima RS, Arnold SE (2020) Brain Insulin Signaling, Alzheimer Disease Pathology, and Cognitive Function. Ann Neurol 88, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wheater ENW, Stoye DQ, Cox SR, Wardlaw JM, Drake AJ, Bastin ME, Boardman JP (2020) DNA methylation and brain structure and function across the life course: A systematic review. Neurosci Biobehav Rev 113, 133–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cui D, Xu X (2018) DNA Methyltransferases, DNA Methylation, and Age-Associated Cognitive Function. Int J Mol Sci 19, E1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harman MF, Martín MG (2020) Epigenetic mechanisms related to cognitive decline during aging. J Neurosci Res 98, 234–246. [DOI] [PubMed] [Google Scholar]
- [10].Zhao J, Zhu Y, Yang J, Li L, Wu H, De Jager PL, Jin P, Bennett DA (2017) A genome-wide profiling of brain DNA hydroxymethylation in Alzheimer’s disease. Alzheimers Dement 13, 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fetahu IS, Ma D, Rabidou K, Argueta C, Smith M, Liu H, Wu F, Shi YG (2019) Epigenetic signatures of methylated DNA cytosine in Alzheimer’s disease. Science Advances 5, eaaw2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Poon CH, Tse LSR, Lim LW (2020) DNA methylation in the pathology of Alzheimer’s disease: from gene to cognition. Ann N Y Acad Sci 1475, 15–33. [DOI] [PubMed] [Google Scholar]
- [13].Pellegrini C, Pirazzini C, Sala C, Sambati L, Yusipov I, Kalyakulina A, Ravaioli F, Kwiatkowska KM, Durso DF, Ivanchenko M, Monti D, Lodi R, Franceschi C, Cortelli P, Garagnani P, Bacalini MG (2021) A Meta-Analysis of Brain DNA Methylation Across Sex, Age, and Alzheimer’s Disease Points for Accelerated Epigenetic Aging in Neurodegeneration. Front Aging Neurosci 13, 639428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang Y, Zeng C, Yang K, Xu S, Zhang Z, Cai Q, He C, Zhang W, Liu S-M (2022) Genome-wide Analysis Reflects Novel 5-Hydroxymethylcytosines Implicated in Diabetic Nephropathy and the Biomarker Potential. Extracell Vesicles Circ Nucl Acids 3, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Han L, Chen C, Lu X, Song Y, Zhang Z, Zeng C, Chiu R, Li L, Xu M, He C, Zhang W, Duan S (2021) Alterations of 5-hydroxymethylcytosines in circulating cell-free DNA reflect retinopathy in type 2 diabetes. Genomics 113, 79–87. [DOI] [PubMed] [Google Scholar]
- [16].Yang Y, Zeng C, Lu X, Song Y, Nie J, Ran R, Zhang Z, He C, Zhang W, Liu S-M (2019) 5-Hydroxymethylcytosines in Circulating Cell-Free DNA Reveal Vascular Complications of Type 2 Diabetes. Clinical Chemistry 65, 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA (2018) Religious Orders Study and Rush Memory and Aging Project. J Alzheimers Dis 64, S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arvanitakis Z, Wilson RS, Li Y, Aggarwal NT, Bennett DA (2006) Diabetes and function in different cognitive systems in older individuals without dementia. Diabetes Care 29, 560–565. [DOI] [PubMed] [Google Scholar]
- [19].Arvanitakis Z, Brey RL, Rand JH, Schneider JA, Capuano AW, Yu L, Leurgans SE, Bennett DA, Levine SR (2015) Relation of Antiphospholipid Antibodies to Postmortem Brain Infarcts in Older Persons. Circulation 131, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC, Wilson RS (2006) Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 66, 1837–1844. [DOI] [PubMed] [Google Scholar]
- [21].(1997) Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer’s Disease. Neurobiol Aging 18, S1–2. [PubMed] [Google Scholar]
- [22].Arnold SE, Hyman BT, Flory J, Damasio AR, Van Hoesen GW (1991) The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer’s disease. Cereb Cortex 1, 103–116. [DOI] [PubMed] [Google Scholar]
- [23].Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, Samet Y, Maoz M, Druid H, Arner P, Fu K-Y, Kiss E, Spalding KL, Landesberg G, Zick A, Grinshpun A, Shapiro AMJ, Grompe M, Wittenberg AD, Glaser B, Shemer R, Kaplan T, Dor Y (2018) Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun 9, 5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li W, Zhang X, Lu X, You L, Song Y, Luo Z, Zhang J, Nie J, Zheng W, Xu D, Wang Y, Dong Y, Yu S, Hong J, Shi J, Hao H, Luo F, Hua L, Wang P, Qian X, Yuan F, Wei L, Cui M, Zhang T, Liao Q, Dai M, Liu Z, Chen G, Meckel K, Adhikari S, Jia G, Bissonnette MB, Zhang X, Zhao Y, Zhang W, He C, Liu J (2017) 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 27, 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han D, Lu X, Shih AH, Nie J, You Q, Xu MM, Melnick AM, Levine RL, He C (2016) A Highly Sensitive and Robust Method for Genome-wide 5hmC Profiling of Rare Cell Populations. Mol Cell 63, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, Shi G, Ge Y, Gao P, Yang Y, Ke A, Xiao L, Dong R, Zhu Y, Yang X, Wang J, Zhu T, Yang D, Huang X, Sui C, Qiu S, Shen F, Sun H, Zhou W, Zhou J, Nie J, Zeng C, Stroup EK, Zhang X, Chiu BC-H, Lau WY, He C, Wang H, Zhang W, Fan J (2019) Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 68, 2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Trimmomatic: a flexible trimmer for Illumina sequence data | Bioinformatics | Oxford Academic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, Baren J van, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ (2012) GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- [31].Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Yaping Liu, Coarfa C, Alan Harris R, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, David Hawkins R, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Scott Hansen R, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M (2015) Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127. [DOI] [PubMed] [Google Scholar]
- [34].Westfall PH, Young SS (1993) Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment | Wiley. [Google Scholar]
- [35].Friedman JH, Hastie T, Tibshirani R (2010) Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- [36].Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cui X-L, Nie J, Ku J, Dougherty U, West-Szymanski DC, Collin F, Ellison CK, Sieh L, Ning Y, Deng Z, Zhao CWT, Bergamaschi A, Pekow J, Wei J, Beadell AV, Zhang Z, Sharma G, Talwar R, Arensdorf P, Karpus J, Goel A, Bissonnette M, Zhang W, Levy S, He C (2020) A human tissue map of 5-hydroxymethylcytosines exhibits tissue specificity through gene and enhancer modulation. Nat Commun 11, 6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gaitsch H, Franklin RJM, Reich DS (2022) Cell-free DNA-based liquid biopsies in neurology. Brain awac438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Krauthausen M, Kummer MP, Zimmermann J, Reyes-Irisarri E, Terwel D, Bulic B, Heneka MT, Müller M (2015) CXCR3 promotes plaque formation and behavioral deficits in an Alzheimer’s disease model. J Clin Invest 125, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Teranishi Y, Hur J-Y, Gu GJ, Kihara T, Ishikawa T, Nishimura T, Winblad B, Behbahani H, Kamali-Moghaddam M, Frykman S, Tjernberg LO (2012) Erlin-2 is associated with active γ-secretase in brain and affects amyloid β-peptide production. Biochem Biophys Res Commun 424, 476–481. [DOI] [PubMed] [Google Scholar]
- [42].Kim Y, Park H, Nah J, Moon S, Lee W, Hong S, Kam T-I, Jung Y-K (2015) Essential role of POLDIP2 in Tau aggregation and neurotoxicity via autophagy/proteasome inhibition. Biochemical and Biophysical Research Communications 462, 112–118. [DOI] [PubMed] [Google Scholar]
- [43].Chen Y, Neve RL, Liu H (2012) Neddylation dysfunction in Alzheimer’s disease. J Cell Mol Med 16, 2583–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tanaka Y, Sabharwal L, Ota M, Nakagawa I, Jiang J-J, Arima Y, Ogura H, Okochi M, Ishii M, Kamimura D, Murakami M (2018) Presenilin 1 Regulates NF-κB Activation via Association with Breakpoint Cluster Region and Casein Kinase II. J Immunol 201, 2256–2263. [DOI] [PubMed] [Google Scholar]
- [45].Hu Z, Jiao R, Wang P, Zhu Y, Zhao J, De Jager P, Bennett DA, Jin L, Xiong M (2020) Shared Causal Paths underlying Alzheimer’s dementia and Type 2 Diabetes. Sci Rep 10, 4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Péron CL, Madigou T, Durand E, Froguel P, Staels B, Lefebvre P, Métivier R, Eeckhoute J, Salbert G (2012) Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Research 40, 8255–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR (2012) Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res 22, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rivera AD, Azim K, Macchi V, Porzionato A, Butt AM, De Caro R (2022) Epidermal Growth Factor Pathway in the Age-Related Decline of Oligodendrocyte Regeneration. Front Cell Neurosci 16, 838007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Brun A, Englund E (1986) A white matter disorder in dementia of the Alzheimer type: A pathoanatomical study. Annals of Neurology 19, 253–262. [DOI] [PubMed] [Google Scholar]
- [50].Hsieh Y-C, Guo C, Yalamanchili HK, Abreha M, Al-Ouran R, Li Y, Dammer EB, Lah JJ, Levey AI, Bennett DA, De Jager PL, Seyfried NT, Liu Z, Shulman JM (2019) Tau-Mediated Disruption of the Spliceosome Triggers Cryptic RNA Splicing and Neurodegeneration in Alzheimer’s Disease. Cell Rep 29, 301–316.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Spilsbury A, Miwa S, Attems J, Saretzki G (2015) The role of telomerase protein TERT in Alzheimer’s disease and in tau-related pathology in vitro. J Neurosci 35, 1659–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bai B (2018) U1 snRNP Alteration and Neuronal Cell Cycle Reentry in Alzheimer Disease. Front Aging Neurosci 10, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Socodato R, Portugal CC, Canedo T, Rodrigues A, Almeida TO, Henriques JF, Vaz SH, Magalhães J, Silva CM, Baptista FI, Alves RL, Coelho-Santos V, Silva AP, Paes-de-Carvalho R, Magalhães A, Brakebusch C, Sebastião AM, Summavielle T, Ambrósio AF, Relvas JB (2020) Microglia Dysfunction Caused by the Loss of Rhoa Disrupts Neuronal Physiology and Leads to Neurodegeneration. Cell Rep 31, 107796. [DOI] [PubMed] [Google Scholar]
- [54].Casey CS, Atagi Y, Yamazaki Y, Shinohara M, Tachibana M, Fu Y, Bu G, Kanekiyo T (2015) Apolipoprotein E Inhibits Cerebrovascular Pericyte Mobility through a RhoA Protein-mediated Pathway. J Biol Chem 290, 14208–14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Matoba K, Takeda Y, Nagai Y, Sekiguchi K, Yokota T, Utsunomiya K, Nishimura R (2020) The Physiology, Pathology, and Therapeutic Interventions for ROCK Isoforms in Diabetic Kidney Disease. Front Pharmacol 11, 585633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Flentje A, Kalsi R, Monahan TS (2019) Small GTPases and Their Role in Vascular Disease. Int J Mol Sci 20, E917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Biankin AV, Waddell N, Kassahn KS, Gingras M-C, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch A-M, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Lynn Fink J, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Scott Mead R, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Kakkar N, Zhao F, Qing Wu Y, Wang M, Muzny DM, Fisher WE, Charles Brunicardi F, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao M-S, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LFA, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ling C (2020) Epigenetic regulation of insulin action and secretion - role in the pathogenesis of type 2 diabetes. J Intern Med 288, 158–167. [DOI] [PubMed] [Google Scholar]
- [59].Annadurai N, De Sanctis JB, Hajdúch M, Das V (2021) Tau secretion and propagation: Perspectives for potential preventive interventions in Alzheimer’s disease and other tauopathies. Exp Neurol 343, 113756. [DOI] [PubMed] [Google Scholar]
- [60].Bradley-Whitman MA, Lovell MA (2013) Epigenetic changes in the progression of Alzheimer’s disease. Mech Ageing Dev 134, 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lardenoije R, Roubroeks JAY, Pishva E, Leber M, Wagner H, Iatrou A, Smith AR, Smith RG, Eijssen LMT, Kleineidam L, Kawalia A, Hoffmann P, Luck T, Riedel-Heller S, Jessen F, Maier W, Wagner M, Hurlemann R, Kenis G, Ali M, del Sol A, Mastroeni D, Delvaux E, Coleman PD, Mill J, Rutten BPF, Lunnon K, Ramirez A, van den Hove DLA (2019) Alzheimer’s disease-associated (hydroxy)methylomic changes in the brain and blood. Clin Epigenet 11, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Leenen FAD, Muller CP, Turner JD (2016) DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenetics 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Griñán-Ferré C, Izquierdo V, Otero E, Puigoriol-Illamola D, Corpas R, Sanfeliu C, Ortuño-Sahagún D, Pallàs M (2018) Environmental Enrichment Improves Cognitive Deficits, AD Hallmarks and Epigenetic Alterations Presented in 5xFAD Mouse Model. Front Cell Neurosci 12, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Papale LA, Madrid A, Zhang Q, Chen K, Sak L, Keleş S, Alisch RS (2022) Gene by environment interaction mouse model reveals a functional role for 5-hydroxymethylcytosine in neurodevelopmental disorders. Genome Res 32, 266–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Arvanitakis Z, Bennett DA, Wilson RS, Barnes LL (2010) Diabetes and Cognitive Systems in Older Black and White Persons. Alzheimer Dis Assoc Disord 24, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Arvanitakis Z, Capuano AW, Bennett DA, Barnes LL (2018) Body Mass Index and Decline in Cognitive Function in Older Black and White Persons. The Journals of Gerontology: Series A 73, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Arvanitakis Z, Capuano AW, Lamar M, Shah RC, Barnes LL, Bennett DA, Schneider JA (2018) Late-life blood pressure association with cerebrovascular and Alzheimer disease pathology. Neurology 91, e517–e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its additional files]. Raw and processed sequencing data supporting the conclusions of this article are available in the NCBI GEO repository, Accession No. GSE203337 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203337. MAP resources can be requested at www.radc.rush.edu.


