Abstract
In adulthood, sleep-wake rhythms are one of the most prominent behaviors under circadian control. However, during early life, sleep is spread across the 24-hour day. The mechanism through which sleep rhythms emerge, and consequent advantage conferred to a juvenile animal, is unknown. In the second-instar Drosophila larvae (L2), like in human infants, sleep is not under circadian control. We identify the precise developmental time point when the clock begins to regulate sleep in Drosophila, leading to emergence of sleep rhythms in early third-instars (L3). At this stage, a cellular connection forms between DN1a clock neurons and arousal-promoting Dh44 neurons, bringing arousal under clock control to drive emergence of circadian sleep. Last, we demonstrate that L3 but not L2 larvae exhibit long-term memory (LTM) of aversive cues and that this LTM depends upon deep sleep generated once sleep rhythms begin. We propose that the developmental emergence of circadian sleep enables more complex cognitive processes, including the onset of enduring memories.
Formation of a connection between clock and arousal cells controls emergence of sleep rhythms, enabling more persistent memories.
INTRODUCTION
A major unanswered question in chronobiology is how the maturation of circadian clocks is tied to the developmental emergence of behavioral rhythms. Across species, from flies to mammals, central molecular clocks begin cycling during very early developmental periods; however, many behavioral rhythms, such as sleep-wake patterns, do not emerge until later, suggesting that maturation events downstream of the core clock are required to generate rhythmic sleep output (1–10). For example, in mammals, central pacemaker cells of the suprachiasmatic nucleus exhibit synchronous molecular rhythms during gestation (6, 11–15) but sleep rhythms are not present in the early postnatal period. Juvenile sleep in rodents is highly fragmented, with rapid transitions between sleep and wake; sleep/wake bouts eventually become more consolidated as development proceeds (16–18). Likewise, in humans, circadian sleep patterns are increasingly apparent between the ages of 3 and 12 months (19–21). Mechanisms controlling the emergence of sleep rhythms remain largely unknown.
Fundamental insights into the clock neuron network and molecular mechanisms maintaining behavioral rhythms, including daily sleep-wake patterns, come from studies in adult Drosophila melanogaster (22–25). Adult flies, even immediately after eclosion, exhibit mature behavioral and molecular rhythms (7). Therefore, earlier developmental periods must be examined to understand how and when clock control of sleep is initiated. Few studies have examined behavioral rhythms in larvae (10, 26, 27), and sleep during larval stages was only recently characterized (5, 28). Neurons in the larval clock network are functional and encode circadian information as early as the first instar larval stage: Light exposure only during the first instar stage is sufficient to entrain adult locomotor rhythms (7). However, despite the presence of a functioning circadian clock network (7, 27, 29), sleep in second-instar (L2) larvae [48 hours after egg laying (AEL)] is not under clock control; sleep patterns at this stage are unaffected in molecular clock mutants or with other manipulations to disrupt clock function (5). Thus, like mammals, sleep-wake rhythms likely emerge later in development.
Here, we identify the precise developmental time point when the circadian clock begins to regulate sleep in Drosophila, leading to the emergence of sleep rhythms at the early third-instar (L3) stage. We characterize the developmentally timed formation of a circuit motif between larval DN1a clock neurons and arousal-promoting Dh44 neurons, using CCHamide-1 (CCHa1) signaling, which drives sleep rhythms. Last, we find that the circadian control of sleep generates deeper sleep states and with them the onset of more persistent memories.
RESULTS
Rhythmic sleep emerges in early third-instar larvae
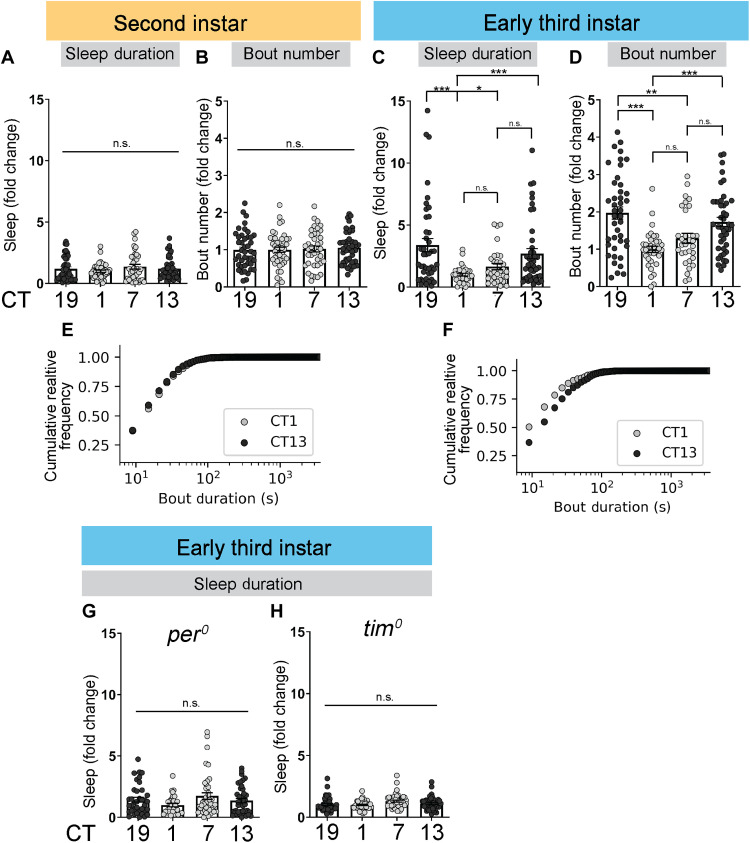
To determine when sleep becomes influenced by the circadian system, we examined sleep under constant conditions at four times across the day [circadian time 1 (CT1), CT7, CT13, and CT19] in developmentally age-matched L2 and early L3 (72 hours AEL) larvae. As expected, there were no differences in sleep across the day in L2 (Fig. 1, A and B; plotted as fold change to account for differences across experimental replicates). However, we observed the emergence of a diurnal sleep pattern in L3, specifically an increase in sleep duration with more sleep bouts during subjective dark times (CT13 and CT19) compared to subjective light times (CT1 and CT7) (Fig. 1, C and D). Analysis of raw sleep values revealed L2 sleep on average ~10 min/hour at all circadian time points, while L3 mean sleep approached nearly 20 min/hour during the subjective night; the increase in sleep at these time points was not accompanied by an overall reduction in activity (fig. S1, A to H). To determine whether sleep bout duration was different at the two CTs, we pooled the total sleep bouts for L2 and L3 larvae at CT1 and CT13 and plotted the cumulative probability distribution. While L2 larvae showed similar bout length distributions at CT1 and CT13 (Fig. 1E), early L3 larvae exhibited a higher proportion of longer sleep bouts at CT13 compared to CT1 (P > 0.5, L2; P < 0.0001, L3; Kolmogorov-Smirnov test; Fig. 1F and figs. S1, C and G, and S2A). Therefore, CT affects both sleep bout number and duration in L3 larvae.
Fig. 1. Rhythmic sleep emerges in early third-instar (L3).
(A to D) Sleep duration and sleep bout number across the day under constant dark conditions in second-instar (L2) (A and B) and L3 (C and D). (E and F) Cumulative relative frequency of sleep bout duration at circadian time 1 (CT1) and CT13 for L2 (E) and L3 (F). (G and H) Sleep duration in L3 clock mutants. The subjective day is shown in light gray circles and the subjective night in black. Sleep metrics represent fold change (normalized to the average value at CT1 for a given panel). (A to D, G, and H) n = 31 to 44 larvae. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests (A to D, G, and H); Kolmogorov-Smirnov test (E and F). For this and all other figures unless otherwise specified, data are presented as means ± SEM; n.s., not significant; *P < 0.05, **P < 0.01, and ***P < 0.001.
To assess whether the daily pattern of sleep in early L3 is dependent on the canonical circadian biological clock, we examined sleep in null mutants for the core clock genes per and tim (30, 31). In both per0 and tim0 mutants, we observed no differences across the day in sleep (Fig. 1, G and H, and figs. S1, I to N, and S2, B to E); moreover, sleep differences across the day were absent in L3 larvae raised in constant light (LL), which disrupts the molecular clock (fig. S2, F and G). Last, cosinor regression confirmed that the temporal pattern of L3 sleep was consistent with daily cycling (P < 0.001, sleep midpoint = CT16.2; fig. S2H). Together, our data define the developmental time point in Drosophila when sleep comes under clock control.
Although L2 sleep is clock-independent, we previously described developmental changes to sleep duration across this larval stage (5). To determine whether L3 sleep likewise undergoes ontogenetic changes, we held CT constant at either a subjective morning (CT5) or subjective evening (CT21) time point and examined sleep duration in newly molted L3 larvae (72 hours AEL; +0 hours), larvae aged 4 hours after molting (76 hours AEL; +4 hours), and larvae aged 8 hours after molting (80 hours AEL; +8 hours). We did not detect sleep differences in larvae from CT5 +0 hours to CT5 +4 hours, but did find a reduction in sleep by CT5 +8-hour sleep (fig. S2I); a similar pattern was evident at CT21 (fig. S2J), suggesting that changes to sleep with development occur during L3, perhaps in anticipation of pupation. Under natural conditions, larvae continue to develop as CT progresses. Is circadian control of sleep relevant in the face of these developmental changes? We allowed for the simultaneous influence of age and CT by focusing on L3 sleep patterns at day-night transitions. L3 exhibited decreased sleep in the night-to-day transition from CT21 to CT1 and increased sleep in the day-to-night transition from CT9 to CT13 (fig. S2, K to M). Thus, sleep continues to cycle in relation to CT even as larvae develop.
Dh44 neurons in central brain control arousal in second-instar larvae
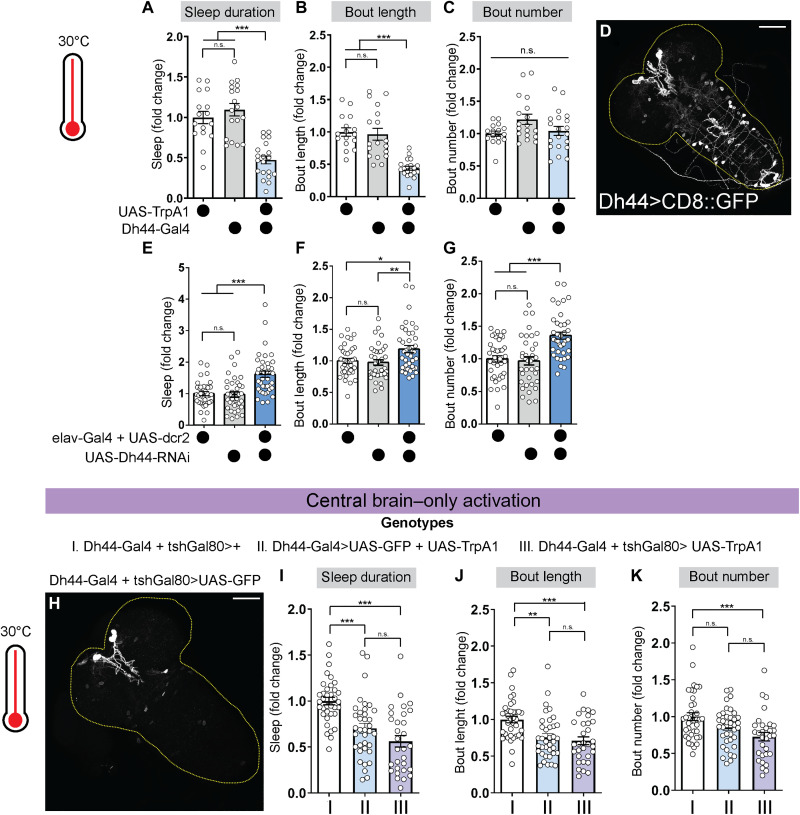
Neuropeptidergic signaling plays a central role in sleep and circadian regulation in adult Drosophila (32, 33). To investigate potential signaling molecules that regulate the emergence of sleep-wake rhythms, we conducted a neural activation screen of peptidergic neurons using the heat-sensitive cation channel, TrpA1 (34). We found that activation of neurons that express the neuropeptide Dh44 caused a substantial reduction in sleep duration in L2 larvae (Fig. 2A and figs. S3A and S4A). Activation of Dh44 neurons resulted in a loss of sleep primarily due to a reduction in sleep bout duration (Fig. 2, B and C, and figs. S3, B and C, and S4, B and C). The decrease in sleep was not due to an increase in feeding rate, as there was no difference in the number of mouth hook contractions normalized to time awake (fig. S4D). Dh44-Gal4 showed restricted expression in the L2 larval nervous system, labeling only a few cells in the brain and ventral nerve cord (VNC) (Fig. 2D). To determine whether these neurons bidirectionally modulate larval sleep, we inhibited Dh44 neurons by expressing the inwardly rectifying potassium channel Kir2.1 (35). Kir2.1 expression in Dh44 neurons increased total sleep duration by increasing bout duration and bout number (fig. S4, E to G). In addition, pan-neuronal knockdown of Dh44 increased sleep duration, bout length, and bout number (Fig. 2, E to G, and figs. S3, D to F, and S4, H and I). Together, our data indicate that Dh44 neurons modulate arousal in L2 larvae and that Dh44 is required for maintaining normal arousal at this stage.
Fig. 2. Dh44 neurons in the brain control arousal in second-instar (L2).
(A to C) Sleep duration, bout length, and bout number in L2 expressing UAS-TrpA1 with Dh44-Gal4 and genetic controls. (D) L2 brain and ventral nerve cord showing green fluorescent protein (GFP) expression in Dh44 neurons (Dh44-Gal4 > UAS-CD8::GFP). (E to G) Sleep duration, bout length, and bout number in L2 expressing UAS-Dh44-RNAi with elav-Gal4 + UAS-dcr2 and genetic controls. (H) L2 brain and ventral nerve cord showing GFP expression in larvae expressing Dh44-Gal4 in the presence of tshGal80 and UAS-GFP. (I to K) Sleep duration, bout length, and bout number in L2 expressing Dh44-Gal4 in the presence of tshGal80 and UAS-TrpA1 with genetic controls. Experiments were performed at ZT1–6. Sleep metrics represent fold change (normalized to the average value of control) in this and subsequent figures. (A to C) n = 16 to 20 larvae; (E to G) n = 37 to 40 larvae; (I to K) n = 30 to 36 larvae. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests. Scale bars, 50 μm.
We next asked which subpopulation (brain or VNC) of Dh44 neurons is sufficient to induce waking. We used intersectional genetic approaches to restrict Dh44-Gal4 expression to either the central brain or VNC. Activation of Dh44 neurons in the brain decreased sleep duration, bout length, and bout number to a similar extent as activation of all Dh44 cells (Fig. 2, H to K, and figs. S3, G to I, and S5, A to C). In contrast, activation of Dh44 neurons in the VNC did not affect sleep (fig. S5, D to J). Thus, Dh44 neurons in the L2 brain promote arousal.
Dh44 neurons connect with DN1a clock neurons in early L3
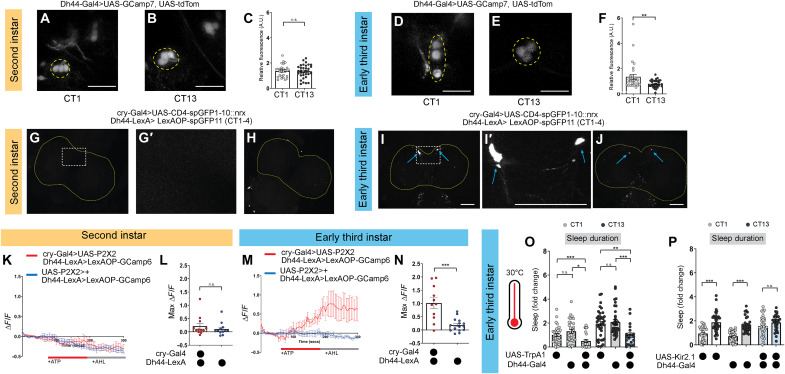
Dh44 neurons are part of the pars intercerebralis, which in the adult is a circadian output center that conveys timing information from the central clock to downstream effectors (24, 36); additionally, in adulthood, Dh44 neural activity cycles across the day in a clock-dependent fashion (37, 38). We wondered whether Dh44 neurons undergo a functional change during development, taking on a circadian role in addition to promoting arousal. To determine whether changes in Dh44 neural activity across the day correlate with the emergence of sleep rhythms in L3, we performed ex vivo calcium imaging of larval brain Dh44 neurons expressing UAS-GCaMP7f. While we observed no calcium activity differences between CT1 and CT13 in L2 (Fig. 3, A to C), by early L3, Dh44 activity was lower at CT13 compared to CT1 (Fig. 3, D to F). Using two-photon imaging of UAS-GCaMP7f–expressing Dh44 neurons in the brain of live L3 larvae, we found that this daily activity pattern also occurs in vivo (fig. S6, A to D). These results are consistent with increased sleep during the night at L3 (less Dh44 arousal activity) and suggest that Dh44 neurons begin to receive clock input at this stage.
Fig. 3. Dh44 neurons anatomically and functionally connect with DN1as in early third-instar (L3).
(A and B) Dh44-Gal4 > UAS-GCaMP7f expression in second-instar (L2) brains at circadian time 1 (CT1) and CT13. (C) GCaMP intensity in Dh44 neurons in L2 at CT1 and CT13. Yellow dotted lines = Dh44 cell bodies. (D and E) Dh44-Gal4 > UAS-GCaMP7f expression in L3 brains at CT1 and CT13. (F) GCaMP intensity in Dh44 neurons in L3 at CT1 and CT13. (G to J) Neurexin-based green fluorescent protein (GFP) reconstitution (GRASP, GFP reconstitution across synaptic partners) between DN1as (cry-Gal4) and Dh44 neurons (Dh44-LexA) in L2 (G and H) and L3 (I and J) brains. Yellow dotted lines = brain; blue arrows = Dh44 cell bodies. Higher magnification of the region of interest in (G) and (I) is shown in (G′) and (I′). (K and M) GCaMP6 signal in Dh44 neurons with the activation of DN1a neurons in L2 (K) and L3 (M). The red bar indicates adenosine 5′-triphosphate (ATP) application and the gray bar indicates artificial hemolymph (AHL) application. (L and N) Maximum GCaMP change (ΔF/F) for individual cells in L2 (L) and L3 (N). (O) Sleep duration in L3 expressing Dh44-Gal4 > UAS-TrpA1 and genetic controls at CT1 and CT13. (P) Sleep duration in L3 expressing Dh44-Gal4 > UAS-Kir2.1 and genetic controls at CT1 and CT13. (C) n = 32 to 37 cells, 10 to 11 brains; (F) n = 30 cells, 10 brains; (G to J) n = 8 to 10 brains; (K to N) n = 9 to 15 cells, 8 brains; (O) n = 20 to 36 larvae; (P) n = 27 to 33 larvae. Unpaired two-tailed Student’s t test (C, F, M, and N); one-way analyses of variance (ANOVAs) followed by Tukey’s multiple comparison tests (O and P). Scale bars, 25 μm (for A, B, D, and E) and, 50 μm (for G to J). A.U. arbitrary units.
To test whether larval clock neurons anatomically connect to Dh44 neurons in early L3, we assessed synaptic connections between different populations of clock neurons and Dh44 neurons using neurexin-based green fluorescent protein (GFP) reconstitution across synaptic partners (GRASP) (39–41). At both the L2 and L3 stages, the functional larval clock network is composed of four small ventrolateral neurons (s-LNvs), two anterior dorsal neuron group 1 (DN1as) cells, and two dorsal neuron group 2 (DN2s) cells per brain hemisphere (42–44). Using cry-Gal4 (larval s-LNv/DN1a–specific driver) with Dh44-LexA to express independent GRASP components, we observed reconstituted GFP signal around the cell body and dendrites of Dh44 neurons in early L3 but not L2 (Fig. 3, G to J). We did not observe GRASP signal with pdf-Gal4 (larval s-LNv–specific driver) or per-Gal4 (larval DN2-specific driver) (fig. S6, E and F), suggesting the DN1a-Dh44 connection is key. The Dh44 central brain expression pattern itself does not change between L2 and L3 (fig. S7A). To determine whether DN1as and Dh44 neurons are also functionally connected in early L3, we expressed adenosine 5′-triphosphate (ATP)–gated P2X2 receptors (45) in DN1a neurons and GCaMP6 in Dh44 neurons. When DN1as were activated with ATP application to dissected brains, we observed an increase in calcium in Dh44 neurons in L3, but not in L2 (Fig. 3, K to N). While we cannot rule out the possibility that stronger DN1a activation might elicit some response in Dh44 neurons in L2 as well, our data demonstrate a marked change from L2 to L3. Dh44 neurons, therefore, appear to form anatomical and functional connections with DN1a clock neurons in early L3 larvae, without evidence of such connections before this time. Given the connectivity changes upstream of Dh44 neurons between L2 and L3, we next asked whether the functional consequences of Dh44 neuronal activation likewise change. We found that, as in L2, activation of Dh44 neurons in L3 promotes arousal, and Dh44 inhibition disrupts daytime wakefulness (Fig. 3, O and P, and fig. S7, B to H), supporting the hypothesis that upstream input from the clock is the major developmental switch. Thus, Dh44 neurons become connected to the central clock in early L3, bringing arousal under clock control to drive sleep rhythms.
CCHa1 signaling between DN1as and Dh44 neurons is necessary for sleep rhythms
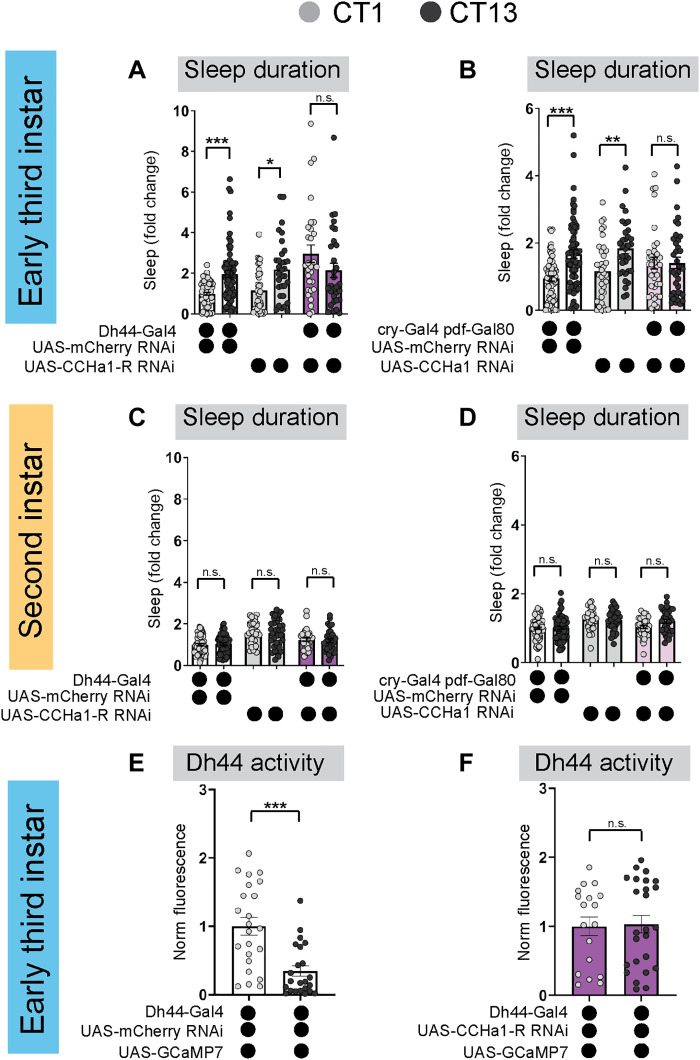
We next sought to determine the molecular signals used in the DN1a-Dh44 circuit to drive L3 sleep rhythms. We conducted an RNA interference (RNAi)–based candidate screen of receptors for five neuropeptides shown to cycle in adult DN1a neurons (46). Sleep duration at CT1 and CT13 was assessed with receptor knockdown in Dh44 neurons, and we found that knockdown of the CCHamide-1 receptor (CCHa1-R) resulted in a loss of rhythmic changes in sleep duration and bout number (Fig. 4A and figs. S8, A and B, and S9, A to C). To determine whether CCHamide-1 release from DN1a neurons is required, we knocked down CCHa1 in DN1as and observed a loss of rhythmic changes in sleep at CT1 and CT13 (Fig. 4B and figs. S8, C and D, and S9, D to F). Manipulations of CCHamide-1 signaling in L2 larvae did not affect sleep duration or bout number at CT1 or CT13 (Fig. 4, C and D, and figs. S8, E to H, and S9, G to L), providing further evidence that this circuit is not yet functional at this stage. To determine whether the diurnal changes in Dh44 neural activity observed in L3 (see Fig. 3, D to F) require CCHa1 signaling, we performed ex vivo calcium imaging of L3 Dh44 neurons expressing UAS-GCaMP7f and UAS-CCHa1-R RNAi. We found that the knockdown of CCHa1-R abolished the diurnal changes in Dh44 activity (Fig. 4, E and F). Together, these findings demonstrate that a developmentally timed circuit connecting DN1a and Dh44 neurons uses CCHa1 neuropeptidergic signaling to drive circadian sleep.
Fig. 4. CCHa1 signaling between DN1a and Dh44 neurons is necessary for sleep rhythms.
(A and B) Sleep duration in third-instar (L3) expressing UAS-CCHa1-R-RNAi with Dh44-Gal4 (A) and UAS-CCHa1-RNAi with cry-Gal4 pdf-Gal80 (DN1as) (B) and genetic controls at circadian time 1 (CT1) and CT13. (C and D) Sleep duration in second-instar (L2) expressing UAS-CCHa1-R-RNAi with Dh44-Gal4 (C) and UAS-CCHa1-RNAi with cry-Gal4 pdf-Gal80 (DN1as) (D) and genetic controls at CT1 and CT13. (E and F) GCaMP intensity (normalized fluorescence) in Dh44 neurons in L3 expressing UAS-CCHa1-R-RNAi with Dh44-Gal4 and genetic controls at CT1 and CT13. Sleep metrics represent fold change (normalized to the average value of control). (A to D) n = 64 to 68 mCherry larvae, 29 to 37 other genotypes; (E and F) n = 18 to 25 cells, 10 brains. Two-way analyses of variance (ANOVAs) followed by Sidak’s multiple comparisons tests (A to D); unpaired two-tailed Student’s t test (E and F). n.s., not significant.
Onset of sleep rhythmicity facilitates long-term memory
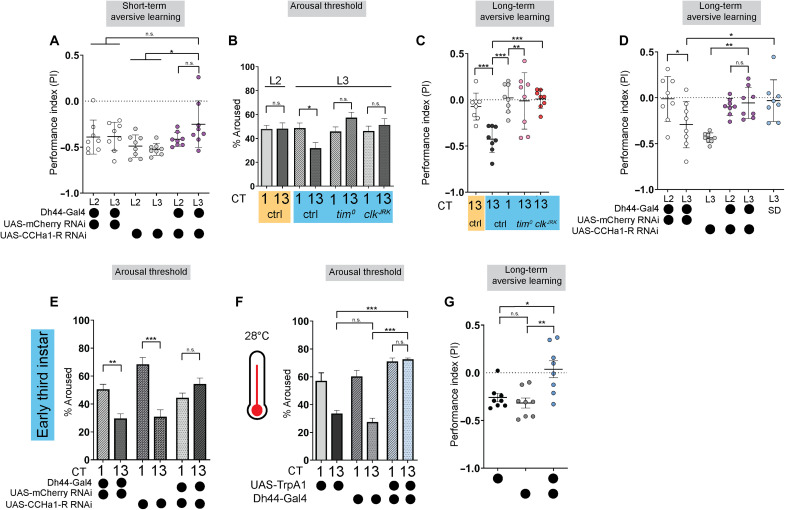
What advantages are conferred to an animal with the onset of circadian sleep patterns? Sleep is a critical regulator of memory formation and consolidation during development (47, 48). We first asked if sleep rhythms are necessary for short-term memory (STM). We used a two-odor reciprocal olfactory conditioning paradigm and tested aversive memory performance in both L2 and L3 immediately after training (49–53). Naïve preferences for odor and aversive stimuli were largely stable between L2 and L3 for each genetic manipulation (fig. S10, A to C). We found that both L2 and L3 control animals showed STM with similar performance indices (Fig. 5A), indicating that the presence of sleep rhythms is not a prerequisite. Knockdown of CCHa1-R in Dh44 neurons had no effect on STM in early L3 (Fig. 5A), providing further evidence that circadian control of sleep is not required.
Fig. 5. Sleep rhythms facilitate long-term memory.
(A) Short-term aversive memory performance in second-instar (L2) and third-instar (L3) expressing Dh44-Gal4 > CCHa1-R-RNAi and genetic controls. (B) Arousal threshold in L2 controls (yellow shaded), L3 controls, and L3 tim0 and clkJRK mutants (blue shaded) at circadian time 1 (CT1) and CT13. (C) Long-term aversive memory performance in L2 and L3 controls and L3 tim0 and clkJRK mutants. (D) Long-term aversive memory performance in L2 and L3 genetic controls (Dh44-Gal4 > mCherry RNAi); L3 CCHa1-R-RNAi/+; L2 and L3 expressing Dh44-Gal4 > CCHa1-R-RNAi; and in sleep-deprived (SD) L3 genetic controls (Dh44-Gal4 > mCherry RNAi). (E) Arousal threshold in L3 genetic controls and L3 expressing Dh44-Gal4 > CCHa1-R-RNAi at CT1 and CT13. (F) Arousal threshold in L3 genetic controls and L3 expressing Dh44-Gal4 > UAS-TrpA1 at CT1 and CT13 at 28°C. (G) Long-term aversive memory performance in L3 expressing Dh44-Gal4 > UAS-TrpA1 and genetic controls at 28°C. (A, C, D, and G) n = 8 performance indices (PIs) (240 larvae) per genotype; (B and E) n = 200 to 430 sleep episodes, 36 larvae per genotype; (F) n = 120 to 200 sleep episodes, 18 larvae per genotype. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests (A, C, F, and G); one-way ANOVAs followed by Sidak’s multiple comparisons tests (B and D); two-way ANOVAs followed by Sidak’s multiple comparison tests (E). n.s., not significant.
Deep sleep stages are important for long-term memory (LTM) consolidation in adulthood across species (54–56). While STM was similar in L2 and L3, we wondered if developmental sleep differences might be relevant for more enduring memories. We therefore asked if clock control of sleep modulates specific sleep features, such as sleep depth, that are beneficial to memory. To assess sleep depth, we examined the arousal threshold in sleeping L2 and L3 animals at CT1 or CT13. While L2 showed similar levels of responsiveness to a low-intensity light stimulus irrespective of the time of day, sleeping L3 were less responsive during the subjective night (Fig. 5B), consistent with deeper sleep during this time. L3 did not show a time-of-day difference in the percentage of larvae that spontaneously awaken without a stimulus (fig. S10D). Providing further evidence of increased sleep depth in L3 during the night, L3 exhibited a higher probability of transitioning from a wake state to sleep at CT13 compared to CT1 and decreased likelihood of exiting a sleep state; L2 showed no differences in these measures across the day (fig. S10, E and F) (57). Moreover, we observed that deeper sleep in L3 at CT13 was absent in clock mutants (Fig. 5B). These data demonstrate that in addition to an increase in sleep duration at night, L3 also sleep more deeply in a clock-dependent manner.
To test whether circadian induction of deeper sleep in L3 provides advantages to memory function, we assessed a protein synthesis-dependent aversive LTM based on three odor-quinine training cycles (fig. S11A) (49, 58). We found that L2 did not show LTM, but early L3 exhibited a strong persistent memory of the aversive cue with training and testing at CT12 to CT15 (Fig. 5C). We did not observe LTM in early L3 at CT1 to CT4 (Fig. 5C), suggesting that deeper sleep stages are necessary for LTM in this paradigm. We then tested if LTM in early L3 is clock-dependent by examining memory performance in tim0 and clkJRK mutants (59) and found that both mutations rendered L3 unable to show LTM (Fig. 5C). Critically, L3 and L2 tim0 and clkJRK mutants still showed STM (fig. S11, B and F), arguing against a more generalized inability to exhibit memories in clock mutants; moreover, we detected no differences in baseline preference to odors or quinine with regard to larval stage or clock mutants (fig. S11, C to E and G to I). We next examined whether the knockdown of CCHa1-R in Dh44 neurons, which specifically disrupts sleep rhythms in L3 (see Fig. 4A), alters LTM. First, as expected, given that CCHa1-R knockdown has no effect on sleep in L2, we found L2 RNAi control larvae did not show that LTM and CCHa1-R knockdown had no further effect. By contrast in L3, the knockdown of CCHa1-R in Dh44 neurons blocked LTM (Fig. 5D). Consistent with deeper sleep being necessary for LTM, L3 Dh44 > CCHa1-R RNAi larvae failed to exhibit the increased arousal threshold normally observed during the night (Fig. 5E). We tested if sleep itself is necessary for LTM in early L3 by using a high-intensity light stimulus to sleep-deprived larvae during the 1.5 hours following training (fig. S11J) and found that sleep deprivation between training and testing disrupted LTM (Fig. 5D). Moreover, mild activation of Dh44 neurons in L3 was associated with impaired deep sleep without a major reduction in sleep duration, also leading to loss of LTM (Fig. 5, F and G, and fig. S12, A to G). Our data indicate that sleep rhythms emerge in early L3 larvae to promote deeper sleep at night, which facilitates LTM.
DISCUSSION
The core molecular clock begins cycling during nascent developmental stages (6–10), but numerous behavioral rhythms, including sleep-wake cycles, are not apparent until far later. Here, we discover in Drosophila that precisely timed completion of a circuit motif allows information to flow from the central clock network to behavioral output cells, generating sleep rhythms. Our findings suggest that unchecked arousal is a primordial driver of state changes between sleep and wake during early life until clock input brings arousal under circadian control. A key molecular signal in this bridge, CCHa1, is known to be involved in circadian intercellular coordination as well as gut-to-brain signaling (60, 61), and our results demonstrate an additional clock network-extrinsic role for CCHa1/CCHa1-R in behavioral rhythms. The mammalian homolog of CCHa1-R, a gastrin-releasing peptide receptor, is also implicated in intrinsic clock network coordination (62). Coupled with the fact that—as in flies—sleep-clock loci do not exhibit mature circuit connectivity in young rodents (16), both the circuit and molecular findings described here might be evolutionarily conserved.
Our functional data support a model in which CCHa1 release from DN1as is excitatory to Dh44 neurons, promoting daytime arousal in L3; disrupted CCHa1 signaling between these cells is associated with loss of deep sleep at night. We note that with regard to raw sleep values, the major change from L2 to L3 in iso31 flies that coincides with the development of this arousal circuit is actually an increase in night sleep, not an increase in daytime arousal (fig. S1, A and E). This precise pattern appears to be background-dependent, as control flies from other experiments do show an increase in daytime wakefulness with maturation from L2 to L3 (fig. S9, A, D, G, and J). How might the emergence of a new arousal cue lead to more sleep at night without apparent less sleep during the day in some cases? We speculate that the quality of daytime experience might be enriched, which is known to affect subsequent sleep (63, 64); additionally, the baseline set point of sleep and Dh44 neural activity might change from L2 to L3, so that a direct comparison of raw values across development obscures a relative reduction in daytime sleep in L3. Last, beyond a connection with the clock, larval Dh44 cells also receive extensive sensory inputs (65) that likely further modulate the arousal state in a complex manner in conjunction with circadian regulation. Regardless, our findings underscore a role specifically for sleep rhythms in LTM: Inappropriate day sleep dissipates sleep drive, limiting cognitive processes dependent upon developmentally emergent sleep features at night. The pattern of sleep depth, rather than duration, appears to be most critical, as we describe manipulations that disrupt sleep rhythms through an increase in day sleep, a decrease in night sleep, or a mixture of the two, all of which ultimately impair night sleep depth and LTM.
We hypothesize that in immature stages of life, metabolic demand is high and nutritional storage capabilities are low, necessitating rapid alterations between sleep and feeding. At this developmental time, clock control of sleep and resultant consolidated sleep periods would be disadvantageous and risk malnourishment. With growth, as prolonged periods without feeding become more sustainable, circadian sleep emerges through a newly formed clock-to-arousal neuronal circuit. In flies, this rhythmic sleep is deeper at night, unlocking more sophisticated brain functions. Maturation of sleep patterns in human infancy likewise occurs coincidentally with enhanced cognitive capabilities (66–68). Our data suggest a direct link between these events, with the onset of daily sleep rhythms and deeper sleep states enabling persistent memories.
METHODS
Fly stocks
The following lines have been maintained as laboratory stocks or were obtained from A. Sehgal: iso31, clockJrk (59), per0 (31), tim0 (30), Dh44VT-Gal4 (VT039046) (69), cry-Gal4 pdf-Gal80 (70), UAS-TrpA1 (34), UAS-Kir2.1 (35), elav-Gal4 UAS-dcr2, UAS-mCherry RNAi, VDRC KK control (60100), tsh-Gal80 UAS-TrpA1, tsh-Gal80 UAS-mCD8::GFP, UAS-GCaMP7f UAS-tdTom (71), LexAOP-sp-GFP11; UAS-CD4-spGFP1–10::nrxn (39), pdf-Gal4 (72), LexAOP-GCaMP6 UAS-P2X2 (45). Dh44-LexA (80703), per-Gal4 (7127), cry-Gal4 (24774), CCHa1-R RNAi (51168), CNMaR RNAi (57859), Gyc76D RNAi (57315), pdfr RNAi (38347), Dh44-R1 RNAi (28780), and CCHa1 RNAi (57562) were from the Bloomington Drosophila Stock Center (BDSC). Dh44 KK RNAi (108473) was from the Vienna Drosophila Resource Center (VDRC). tub>Gal80>; tsh-LexA LexAOP-Flp; UAS-TrpA1 and tub>Gal80>; tsh-LexA LexAOP-Flp; UAS-GFP were gifts from W. Grueber.
Larval rearing and sleep assays
Adult flies were maintained on a standard molasses-based diet (8.0% molasses, 0.55% agar, 0.2% Tegosept, 0.5% propionic acid) at 25°C on a 12:12 light:dark (LD) cycle. For experiments performed in the subjective evening (CT12 to CT21), adult flies were maintained on a 12:12 reverse dark:light (DL) cycle. To collect synchronized L2 and L3 larvae, adult flies were placed in an embryo collection cage (Genesee Scientific, catalog no. 59-100) and eggs were laid on a petri dish containing 3% agar, 2% sucrose, and 2.5% apple juice with yeast paste on top (for L2) or a molasses-based diet with yeast paste on top (for L3). Animals developed on this media for 2 days (for L2) or 3 days (for L3). To examine sleep in either L2 or L3 larvae at constant conditions, petri dishes were moved to constant darkness (DD) after the first day of entrainment.
Sleep assays for L2 larvae were performed using the LarvaLodge and image acquisition parameters as described previously (5). Briefly, molting L2 larvae were placed into individual wells of the LarvaLodge containing 120 μl of 3% agar and 2% sucrose media covered with a thin layer of yeast paste. Sleep assays in molting L3 larvae were performed using a modified LarvaLodge, with 20 rounded wells of 20-mm diameter and 1.5-mm depth. Molting L3 larvae were placed into individual wells of the modified LarvaLodge containing 95 μl of 3% agar and 2% sucrose media covered with a thin layer of yeast paste. The LarvaLodge was covered with a transparent acrylic sheet and placed into a DigiTherm (Tritech Research) incubator at 25°C for imaging. Experiments were performed in the dark. For thermogenetic experiments, adult flies were maintained at 22°C. Larvae were then placed into the LarvaLodge (as described above) which was moved into a DigiTherm (Tritech Research) incubator at 30°C for imaging.
For analysis of L3 sleep across development while holding CT constant, adult flies were maintained in two separate incubators on a 12:12 LD cycle set 4 hours apart. Newly molted L3 larvae were selected 1 hour before the desired CT (for +0 time points) and placed into individual wells of the LarvaLodge for sleep analysis. For the +4 and +8 time points, newly molted L3 larvae were selected 4 or 8 hours before the desired CT, and then allowed to develop on a petri dish containing a standard molasses-based diet with yeast paste in DD for either 4 hours (+4) or 8 hours (+8). Larvae were then placed into individual wells of the LarvaLodge and experiments were performed as described above.
For analysis of L3 sleep as both CT and developmental age proceed across the day, adult flies were maintained in a single incubator on a 12:12 LD cycle. Newly molted L3 larvae were selected and placed into individual wells of the LarvaLodge for sleep analysis (for the first time point) or allowed to develop on a petri dish containing a standard molasses-based diet with yeast paste in DD for either 4 hours or 8 hours (for the later time points). Larvae were then placed into individual wells of the LarvaLodge and experiments were performed as described above.
For analysis of sleep in animals raised in LL, larvae on petri dishes in egg lay chambers were entrained to a 12:12 LD or a reverse 12:12 DL cycle for 24 hours. Then, the petri dishes were moved to LL conditions for 48 hours. Molting L3 larvae were moved to the dark for adaptation for 3 hours before placing larvae into individual wells of the adapted LarvaLodge. Sleep assays were then conducted in DD.
LarvaLodge image acquisition and processing
Images were acquired every 6 s with an Imaging Source DMK 23GP031 camera (2592 × 1944 pixels, The Imaging Source, USA) equipped with a Fujinon lens (HF12.55A-1, 1:1.4/12.5 mm; Fujifilm Corp., Japan) with a Hoya 49mm R72 Infrared Filter. We used IC Capture (The Imaging Source) to acquire time-lapse images. All experiments were carried out in the dark using infrared light-emitting diode (LED) strips (Ledlightsworld LTD, 850-nm wavelength) positioned below the LarvaLodge.
Images were analyzed using custom-written MATLAB software [see (28) and (5)]. Temporally adjacent images were subtracted to generate maps of pixel value intensity change. A binary threshold was set such that individual pixel intensity changes that fell below 40 grayscale units within each well were set equal to zero (“no change”) to eliminate noise. For L3, the threshold was set to 45 to account for a larger body size. Pixel changes greater than or equal to the threshold value were set equal to one (“change”). The activity was then calculated by taking the sum of all pixels changed between images. Sleep was defined as an activity value of zero between frames. For L2 sleep experiments done across the day, total sleep was summed over 6 hours beginning 2 hours after the molt to L2. For sleep experiments performed at certain CTs, total sleep in the second hour after the molt to the second (or third) instar was summed. For all experiments, sleep metrics were normalized to the average value for the control for a given biological replicate.
For experiments examining raw sleep or development and circadian interactions (figs. S1 and S2), activity values were normalized to account for developmental differences in body size. We adjusted the binary thresholding values to define sleep episodes as less than a 2% change in activity.
Sleep bout distribution analysis
Sleep bout distribution analysis was performed as described previously (73). Briefly, we calculated the duration of every sleep bout for the second hour of the experiment for L2 and L3 larvae at CT1 and CT13. All sleep bouts for a given condition were pooled together. The pooled data were then used to generate cumulative relative frequency plots. The cumulative relative frequency for any given sleep episode duration (in seconds) was calculated as the total number of bouts equal to and shorter than that sleep episode duration divided by the total number of all bouts. The resulting cumulative relative frequency was plotted on the y axis and the x axis represented the continuous time interval with a bin size of 6 s. For comparing sleep bout distributions, Kolmogorov-Smirnov tests were used.
Circadian analysis
To assess whether sleep in L3 larvae followed a circadian pattern, normalized sleep duration at CT1, -7, -13, and -19 was fit with a standard cosinor regression model (74) in R (v4.0.5).
Normalized sleep was fit to the following equation
An F test evaluated the null hypothesis A = B = 0 (P = 0.00014). Sleep acrophore (midpoint) is given by tan−1 (B/A) = CT 16.23. Log-transforming normalized sleep data, for variance stabilization, before regression analysis resulted in a similarly significant result.
Calculation of p(wake) and p(doze)
p(wake) and p(doze) were calculated as described previously (57). p(doze) is defined as the number of wake (active) to sleep (quiescent) transitions divided by the number of frames awake (active). p(wake) is defined as the number of sleep (quiescent) to wake (active) transitions divided by the number of frames asleep (quiescent).
Feeding behavior analysis
Newly molted L2 larvae were placed in individual wells of the LarvaLodge containing 120 μl of 3% agar and 2% sucrose media covered with a thin layer of yeast paste. Larvae were then imaged continuously with a Sony HDR-CX405 HD Handycam camera (B&H Photo, catalog no. SOHDRCX405) for 10 min. The number of mouth hook contractions (feeding) was counted manually over the imaging period and divided by the time awake.
Aversive olfactory conditioning
We used an established two-odor reciprocal olfactory conditioning paradigm with 10 mM quinine (quinine hydrochloride, EMSCO/Thermo Fisher Scientific, catalog no. 18-613-007) as a negative reinforcement to test STM or LTM performance in L2 and early L3 larvae (49) at CT1 to CT4 or CT12 to CT15. Experiments were conducted on assay plates (100 × 15 mm, Genesee Scientific, catalog no. 32-107) filled with a thin layer of 2.5% agarose containing either pure agarose (EMSCO/Thermo Fisher Scientific, catalog no. 16500-500) or agarose plus reinforcer. As olfactory stimuli, we used 10 μl of amyl acetate (AM; Sigma-Aldrich, catalog no. STBF2370V, diluted 1:50 in paraffin oil, Sigma-Aldrich, catalog no. SZBF140V) and octanol (OCT; Thermo Fisher Scientific, catalog no. SALP564726, undiluted). Odorants were loaded into the caps of 0.6-ml tubes (EMSCO/Thermo Fisher Scientific, catalog no. 05-408-123) and covered with parafilm (EMSCO/Thermo Fisher Scientific, catalog no. 1337412). For naïve preferences of odorants, a single odorant was placed on one side of an agarose plate with no odorant on the other side. A group of 30 larvae were placed in the middle. After 5 min, individuals were counted on the odorant side, the non-odorant side, or in the middle. The naïve preference was calculated by subtracting the number of larvae on the non-odorant side from the number of larvae on the odorant side and then dividing by the total number of larvae. For naïve preference of quinine, a group of 30 larvae was placed in the middle of a half agarose–half quinine plate. After 5 min, individuals were counted on the quinine side, the agarose side, or in the middle. The naïve preference for quinine was calculated by subtracting the number of larvae on the quinine side from the number of larvae on the agarose side and then dividing by the total number of larvae. Larvae were trained by exposing a group of 30 larvae to AM while crawling on an agarose medium plus quinine reinforcer. After 5 min, larvae were transferred to a fresh petri dish containing agarose alone with OCT as an odorant (AM+/OCT). A second group of 30 larvae received reciprocal training (AM/OCT+). Three training cycles were used for all experiments. For LTM, larvae were transferred after training onto agarose plates with a small piece of Kimwipe moistened with tap water and covered in dry active yeast (LabScientific, catalog no. FLY804020F). Larvae were then kept in the dark for 1.5 hours before testing memory performance. Training and retention for thermogenetic experiments were conducted at 28°C (for Fig. 5). For STM, larvae were immediately transferred after training onto test plates (agarose plus reinforcer) on which AM and OCT were presented on opposite sides of the plate. After 5 min, individuals were counted on the AM side, the OCT side, or in the middle. We then calculated a preference index (PREF) for each training group by subtracting the number of larvae on the conditioned stimulus side from the number of larvae on the unconditioned stimulus side. For one set of experiments, we calculated two PREF values: (i) PREFAM+/OCT = (#AM − #OCT)/# TOTAL and (ii) PREFAM/OCT+ = (#OCT − #AM)/# TOTAL. We then took the average of each PREF value to calculate an associative performance index (PI) as a measure of associative learning. PI = (PREFAM+/OCT + PREFAM/OCT+)/2.
Arousal and sleep deprivation
Blue light stimulation was delivered as described in (5) using two high-power LEDs (Luminus Phatlight PT-121, 460-nm peak wavelength, Sunnyvale, CA) secured to an aluminum heat sink. The LEDs were driven at a current of 0.1 A (low intensity) (for arousal experiments) or 1 A (high intensity) (for sleep deprivation experiments). For sleep deprivation, we used a high-intensity stimulus for 30 s every 2 min for 1 hour beginning the second hour after the molt to L3. Undisturbed control animals were placed in a separate incubator during deprivation. For arousal experiments, we used a low-intensity stimulus for 4 s every 2 min for 1 hour beginning the second hour after the molt to the second (or third) instar. We then counted the number of larvae that showed an activity change in response to a stimulus. The percentage of animals that moved in response to the stimulus was recorded for each experiment. For each genotype, at least four biological replicates were performed. We then plotted the average percentage across all replicates. To compare arousal thresholds in L2 larvae to those in larger L3 larvae at different CT times, we normalized the activity values for L3 to their relative body size. We used the standard binary thresholding to define sleep episodes and then subtracted 35 pixels to achieve a similar degree of change in response to the light stimulus. To calculate the percentage of early L3 that spontaneously awaken in the absence of a stimulus, we counted the number of larvae that showed an activity change in the alternating 2 min between light stimuli for 1 hour beginning at the second hour after the mold to L3.
Immunohistochemistry & imaging
Brains were dissected in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min at room temperature. Following 3× 20 min washes in PBS with Triton X-100 (PBST), brains were incubated with primary antibody at 4°C overnight. Following 3× 20 min washes in PBST, brains were incubated with secondary antibody at 4°C overnight. Following 3× 20 min washes in PBST, brains were mounted in VECTASHIELD (Vector Laboratories). Primary antibodies included Rabbit anti-GFP (1:200; A-11122, Thermo Fisher Scientific), Mouse anti-GFP (for GRASP dissections) (1:200; EMSCO/Thermo Fisher Scientific, catalog no. G6539-.2ML), and Rabbit anti-Dh44 [1:500; Johnson et al. (75)]. Secondary antibodies included Alexa Fluor donkey anti-rabbit 488 (1:500; The Jackson Laboratory) and goat anti-mouse 488 (1:500; The Jackson Laboratory). Brains were visualized and imaged with a Leica SP8 confocal microscope.
P2X2 activation and GCaMP imaging
For all live imaging experiments (GCaMP and P2X2), brains were dissected in artificial hemolymph (AHL) buffer consisting of (in millimolar): 108 NaCl, 5 KCl, 2 CaCl2, 8.2 MgCl2, 4 NaHCO3, 1 NaH2PO4-H2O, 5 trehalose, 10 sucrose, 5 Hepes (pH 7.5). Brains were placed on a small glass coverslip (Carolina Cover Glasses, Circles, 12 mm, catalog no. 633029) in a perfusion chamber filled with AHL. Solutions were perfused over the brains using a gravity-fed ValveLink perfusion system.
For baseline GCaMP7f imaging at CT0 and CT12, brains were chosen for imaging based on the UAS-tdTomato signal (except for experiments in Fig. 4, E and F). AHL buffer was perfused over the brains throughout imaging. Twelve-bit images were acquired with a 40× water immersion objective at 256 × 256 pixel resolution. Z Stacks were acquired every 30 s for 10 min. Image processing and measurement of fluorescence intensity were performed in ImageJ. A max intensity Z projection of each time step and smooth thresholding was used for analysis. Regions of interest (ROIs) were manually drawn in ImageJ to encompass individual tdTomato-positive cell bodies and mean fluorescence intensities were measured for each ROI at each time point. GCaMP7f signal was normalized to the tdTomato signal (FGCaMP7f/tdTom) for each ROI. For each cell, the average FGCaMP7f/tdTom over the 10-min imaging period was used as a measure of relative fluorescence (A.U., arbitrary units). All analysis was done blind to experimental conditions.
For P2X2 imaging, dissections were performed at CT12 to CT15, and AHL buffer was perfused over the brains for 1 min of baseline GCaMP6 imaging, then ATP was delivered to the chamber by switching the perfusion flow from the channel containing AHL to the channel containing 2.5 mM ATP in AHL, pH 7.5. ATP was perfused for 2 min and then AHL was perfused for 2 min. Twelve-bit images were acquired with a 40× water immersion objective at 256 × 256 pixel resolution. Z Stacks were acquired every 5 s for 3 min. Image processing and measurement of fluorescence intensity were performed in ImageJ. A max intensity Z projection of each time step and smooth thresholding was used for analysis. ROIs were manually drawn in ImageJ to encompass individual GCaMP-positive cell bodies and mean fluorescence intensities were measured for each ROI at each time point. For each cell body, fluorescence traces over time were normalized using this equation: ΔF/F = (Fn − F0)/F0, where Fn is the fluorescence intensity recorded at time point n, and F0 is the average fluorescence value during the 1-min baseline recording. Maximum GCaMP change (ΔF/F) for individual cells was calculated using this equation: ΔF/Fmax = (Fmax − F0)/F0, where Fmax is the maximum fluorescence intensity value recorded during ATP application, and F0 is the average fluorescence value during the 1-min baseline recording. All analysis was done blind to experimental conditions.
For in vivo imaging, early L3 larvae were anesthetized with diethyl ether as described previously (76). Briefly, larvae were placed in a plastic Eppendorf tube cup on a glass dish and were exposed to ether vapor for up to 10 min. Anesthetized larvae were immobilized between two glass slides elevated by four layers of scotch tape. Baseline GCaMP7f imaging at CT0 and CT12 was performed using a Bruker 2P Plus two-photon resonant scanning microscope, a 16× Nikon objective (0.8 numerical aperture), and a Coherent Chameleon Ultra II laser (925 nm, 30-mW average power). Images (512 × 512 pixels) were acquired as Z stacks (1-μm step) at a resolution of 1.8 pixels/μm. Each stack slice was generated by averaging 32 frames (acquired at 30 Hz). Multiple stacks were collected over 10 min. Analysis was conducted using ImageJ and MATLAB. For each animal, Z-stack slices with target neurons in focus were used for analysis; circular ROIs were drawn around visible cell bodies to extract mean tdTomato and GCaMP signal values. Images without resolvable tdTomato signal were excluded from the analysis. Data are reported as the ratio of mean GCaMP and tdTomato and value for each cell body.
Cycloheximide treatment
Early L3 larvae were fed 75 μl of 35 mM cycloheximide (Sigma-Aldrich, catalog no. 01810-1G) in 5% sucrose or 75 μl of 5% sucrose alone (ctrl) for 20 hours before the experiment (49). Before the experiment, larvae were transferred to an empty petri dish and washed with tap water before training and testing.
Statistical analysis
All statistical analysis was done in GraphPad (Prism). For comparisons between two conditions, two-tailed unpaired t tests were used. For comparisons between multiple groups, ordinary one-way analyses of variance (ANOVAs) followed by Tukey’s multiple comparison tests were used. For comparisons between different groups in the same analysis, ordinary one-way ANOVAs followed by Sidak’s multiple comparisons tests were used. For comparisons between time and genotype, two-way ANOVAs followed by Sidak’s multiple comparisons tests were used. For comparison of p(wake) and p(doze) values, a two-sided Wilcoxon rank sum test was used. For comparison of the GCaMP signal in CCHa1 experiments, the Mann-Whitney test was used. For comparison of sleep bout distributions, Kolmogorov-Smirnov tests were used. *P < 0.05, **P < 0.01, and ***P < 0.001. Representative confocal images are shown from at least 8 to 10 independent samples examined in each case. For live imaging, ~15 to 20 neurons in 5 to 10 brains were used for each condition.
Acknowledgments
We thank members of the Kayser Laboratory, Raizen Laboratory, A. Sehgal, D. Raizen, and other members of the Penn Chronobiology and Sleep Institute for helpful discussions and input. Figure S6A was created with Biorender.com.
Funding: This work was supported by NIH DP2NS111996, a Burroughs Wellcome Career Award for Medical Scientists, an Alfred P. Sloan Fellowship, and a March of Dimes Foundation Award to M.S.K.; NIH T32HL007713 and a Hartwell Foundation Fellowship to A.R.P.; NIH R15GM128170 to D.J.C.; NIH R01AG068577 to R.C.A.; and grants from the German Research Foundation (project numbers 426722269, 432195391, and 441181781) to A.S.T.
Author contributions: Conceptualization: A.R.P., A.S.T., D.J.C., and M.S.K. Investigation: A.R.P., L.Z., P.D.M., M.S., R.C.A., and B.S. Writing—original draft: A.R.P. and M.S.K. Writing—review and editing: All authors. Project supervision and funding: M.S.K.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
REFERENCES AND NOTES
- 1.S. Coons, C. Guilleminault, Development of sleep-wake patterns and non-rapid eye movement sleep stages during the first six months of life in normal infants. Pediatrics 69, 793–798 (1982). [PubMed] [Google Scholar]
- 2.J. Louis, C. Cannard, H. Bastuji, M. J. Challamel, Sleep ontogenesis revisited: A longitudinal 24-hour home polygraphic study on 15 normal infants during the first two years of life. Sleep 20, 323–333 (1997). [DOI] [PubMed] [Google Scholar]
- 3.M. Mirmiran, Y. G. H. Maas, R. L. Ariagno, Development of fetal and neonatal sleep and circadian rhythms. Sleep Med. Rev. 7, 321–334 (2003). [DOI] [PubMed] [Google Scholar]
- 4.M. S. Blumberg, A. J. Gall, W. D. Todd, The development of sleep-wake rhythms and the search for elemental circuits in the infant brain. Behav. Neurosci. 128, 250–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. Szuperak, M. A. Churgin, A. J. Borja, D. M. Raizen, C. Fang-Yen, M. S. Kayser, A sleep state in Drosophila larvae required for neural stem cell proliferation. Elife 7, e33220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.V. Carmona-Alcocer, J. H. Abel, T. C. Sun, L. R. Petzold, F. J. Doyle 3rd, C. L. Simms, E. D. Herzog, Ontogeny of circadian rhythms and synchrony in the suprachiasmatic nucleus. J. Neurosci. 38, 1326–1334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Sehgal, J. Price, M. W. Young, Ontogeny of a biological clock in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 89, 1423–1427 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. R. Poe, K. D. Mace, M. S. Kayser, Getting into rhythm: Developmental emergence of circadian clocks and behaviors. FEBS J. 289, 6576–6588 (2022). [DOI] [PubMed] [Google Scholar]
- 9.D. Vallone, K. Lahiri, T. Dickmeis, N. S. Foulkes, Start the clock! Circadian rhythms and development. Dev Dyn 236, 142–155 (2007). [DOI] [PubMed] [Google Scholar]
- 10.E. O. Mazzoni, C. Desplan, J. Blau, Circadian pacemaker neurons transmit and modulate visual information to control a rapid behavioral response. Neuron 45, 293–300 (2005). [DOI] [PubMed] [Google Scholar]
- 11.D. Landgraf, C. E. Koch, H. Oster, Embryonic development of circadian clocks in the mammalian suprachiasmatic nuclei. Front Neuroanat 8, 143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. M. Reppert, W. J. Schwartz, Maternal coordination of the fetal biological clock in utero. Science 220, 969–971 (1983). [DOI] [PubMed] [Google Scholar]
- 13.S. M. Reppert, W. J. Schwartz, The suprachiasmatic nuclei of the fetal rat: Characterization of a functional circadian clock using 14C-labeled deoxyglucose. J. Neurosci. 4, 1677–1682 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. Sladek, A. Sumova, Z. Kovacikova, Z. Bendova, K. Laurinova, H. Illnerova, Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U.S.A. 101, 6231–6236 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.H. Shimomura, T. Moriya, M. Sudo, H. Wakamatsu, M. Akiyama, Y. Miyake, S. Shibata, Differential daily expression of Per1 and Per2 mRNA in the suprachiasmatic nucleus of fetal and early postnatal mice. Eur. J. Neurosci. 13, 687–693 (2001). [DOI] [PubMed] [Google Scholar]
- 16.A. J. Gall, W. D. Todd, M. S. Blumberg, Development of SCN connectivity and the circadian control of arousal: A diminishing role for humoral factors? PLOS ONE 7, e45338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. G. Frank, N. F. Ruby, H. C. Heller, P. Franken, Development of circadian sleep regulation in the rat: A longitudinal study under constant conditions. Sleep 40, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. G. Frank, H. C. Heller, Development of diurnal organization of EEG slow-wave activity and slow-wave sleep in the rat. Am. J. Physiol. 273, R472–R478 (1997). [DOI] [PubMed] [Google Scholar]
- 19.H. P. Roffwarg, J. N. Muzio, W. C. Dement, Ontogenetic development of the human sleep-dream cycle. Science 152, 604–619 (1966). [DOI] [PubMed] [Google Scholar]
- 20.K. F. Davis, K. P. Parker, G. L. Montgomery, Sleep in infants and young children. J. Pediatr. Health Care 18, 65–71 (2004). [DOI] [PubMed] [Google Scholar]
- 21.S. H. Sheldon, Sleep in infants and children, in Sleep: A Comprehensive Handbook (John Wiley & Sons, 2006), pp. 507–510. [Google Scholar]
- 22.F. Guo, M. Holla, M. M. Diaz, M. Rosbash, a circadian output circuit controls sleep-wake arousal in Drosophila. Neuron 100, 624–635.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 23.A. Lamaze, P. Kratschmer, K. F. Chen, S. Lowe, J. E. C. Jepson, A wake-promoting circadian output circuit in Drosophila. Curr. Biol. 28, 3098–3105.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 24.D. J. Cavanaugh, J. D. Geratowski, J. R. Wooltorton, J. M. Spaethling, C. E. Hector, X. Zheng, E. C. Johnson, J. H. Eberwine, A. Sehgal, Identification of a circadian output circuit for rest: Activity rhythms in Drosophila. Cell 157, 689–701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. N. King, A. Sehgal, Molecular and circuit mechanisms mediating circadian clock output in the Drosophila brain. Eur. J. Neurosci. 51, 268–281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.E. Z. Asirim, T. H. Humberg, G. L. Maier, S. G. Sprecher, Circadian and genetic modulation of visually-guided navigation in Drosophila Larvae. Sci. Rep. 10, 2752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. C. Keene, E. O. Mazzoni, J. Zhen, M. A. Younger, S. Yamaguchi, J. Blau, C. Desplan, S. G. Sprecher, Distinct visual pathways mediate Drosophila larval light avoidance and circadian clock entrainment. J. Neurosci. 31, 6527–6534 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. A. Churgin, M. Szuperak, K. C. Davis, D. M. Raizen, C. Fang-Yen, M. S. Kayser, Quantitative imaging of sleep behavior in Caenorhabditis elegans and larval Drosophila melanogaster. Nat. Protoc. 14, 1455–1488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. Helfrich-Forster, The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc. Res. Tech. 62, 94–102 (2003). [DOI] [PubMed] [Google Scholar]
- 30.R. J. Konopka, S. Benzer, Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. Sehgal, J. L. Price, B. Man, M. W. Young, Loss of circadian behavioral rhythms andperRNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1606 (1994). [DOI] [PubMed] [Google Scholar]
- 32.O. T. Shafer, A. C. Keene, The regulation of Drosophila sleep. Curr. Biol. 31, R38–R49 (2021). [DOI] [PubMed] [Google Scholar]
- 33.C. Dubowy, A. Sehgal, Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205, 1373–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.F. N. Hamada, M. Rosenzweig, K. Kang, S. R. Pulver, A. Ghezzi, T. J. Jegla, P. A. Garrity, An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R. A. Baines, J. P. Uhler, A. Thompson, S. T. Sweeney, M. Bate, Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21, 1523–1531 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. N. King, A. F. Barber, A. E. Smith, A. P. Dreyer, D. Sitaraman, M. N. Nitabach, D. J. Cavanaugh, A. Sehgal, A peptidergic circuit links the circadian clock to locomotor activity. Curr. Biol. 27, 1915–1927.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. Bai, Y. Lee, C. T. Hsu, J. A. Williams, D. Cavanaugh, X. Zheng, C. Stein, P. Haynes, H. Wang, D. H. Gutmann, A. Sehgal, A conserved circadian function for the neurofibromatosis 1 gene. Cell Rep. 22, 3416–3426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Cavey, B. Collins, C. Bertet, J. Blau, Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci. 19, 587–595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Y. Chen, O. Akin, A. Nern, C. Y. Tsui, M. Y. Pecot, S. L. Zipursky, Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.P. Fan, D. S. Manoli, O. M. Ahmed, Y. Chen, N. Agarwal, S. Kwong, A. G. Cai, J. Neitz, A. Renslo, B. S. Baker, N. M. Shah, Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154, 89–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.E. H. Feinberg, M. K. Vanhoven, A. Bendesky, G. Wang, R. D. Fetter, K. Shen, C. I. Bargmann, GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008). [DOI] [PubMed] [Google Scholar]
- 42.M. Kaneko, C. Helfrich-Forster, J. C. Hall, Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: Newly identified pacemaker candidates and novel features of clock gene product cycling. J. Neurosci. 17, 6745–6760 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Kaneko, J. C. Hall, Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422, 66–94 (2000). [DOI] [PubMed] [Google Scholar]
- 44.T. Liu, G. Mahesh, J. H. Houl, P. E. Hardin, Circadian activators are expressed days before they initiate clock function in late pacemaker neurons from Drosophila. J. Neurosci. 35, 8662–8671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Z. Yao, A. M. Macara, K. R. Lelito, T. Y. Minosyan, O. T. Shafer, Analysis of functional neuronal connectivity in the Drosophila brain. J. Neurophysiol. 108, 684–696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D. Ma, D. Przybylski, K. C. Abruzzi, M. Schlichting, Q. Li, X. Long, M. Rosbash, A transcriptomic taxonomy of Drosophila circadian neurons around the clock. Elife 10, e63056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Konrad, S. Seehagen, S. Schneider, J. S. Herbert, Naps promote flexible memory retrieval in 12-month-old infants. Dev. Psychobiol. 58, 866–874 (2016). [DOI] [PubMed] [Google Scholar]
- 48.S. Seehagen, C. Konrad, J. S. Herbert, S. Schneider, Timely sleep facilitates declarative memory consolidation in infants. Proc. Natl. Acad. Sci. U.S.A. 112, 1625–1629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. Widmann, M. Artinger, L. Biesinger, K. Boepple, C. Peters, J. Schlechter, M. Selcho, A. S. Thum, Genetic dissection of aversive associative olfactory learning and memory in Drosophila larvae. PLOS Genet. 12, e1006378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.B. Gerber, R. F. Stocker, The Drosophila larva as a model for studying chemosensation and chemosensory learning: A review. Chem. Senses 32, 65–89 (2007). [DOI] [PubMed] [Google Scholar]
- 51.B. Gerber, T. Hendel, Outcome expectations drive learned behaviour in larval Drosophila. Proc Biol Sci 273, 2965–2968 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. El-Keredy, M. Schleyer, C. Konig, A. Ekim, B. Gerber, Behavioural analyses of quinine processing in choice, feeding and learning of larval Drosophila. PLoS One 7, e40525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.K. Neuser, J. Husse, P. Stock, B. Gerber, Appetitive olfactory learning in Drosophila larvae: Effects of repetition, reward strength, age, gender, assay type and memory span. Anim. Behav. 69, 891–898 (2005). [Google Scholar]
- 54.L. Marshall, J. Born, The contribution of sleep to hippocampus-dependent memory consolidation. Trends Cogn. Sci. 11, 442–450 (2007). [DOI] [PubMed] [Google Scholar]
- 55.A. P. Vorster, J. Born, Sleep and memory in mammals, birds and invertebrates. Neurosci. Biobehav. Rev. 50, 103–119 (2015). [DOI] [PubMed] [Google Scholar]
- 56.U. Dag, Z. Lei, J. Q. Le, A. Wong, D. Bushey, K. Keleman, Neuronal reactivation during post-learning sleep consolidates long-term memory in Drosophila. Elife 8, e42786 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.T. D. Wiggin, P. R. Goodwin, N. C. Donelson, C. Liu, K. Trinh, S. Sanyal, L. C. Griffith, Covert sleep-related biological processes are revealed by probabilistic analysis in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 117, 10024–10034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. Khurana, M. B. Abu Baker, O. Siddiqi, Odour avoidance learning in the larva of Drosophila melanogaster. J. Biosci. 34, 621–631 (2009). [DOI] [PubMed] [Google Scholar]
- 59.R. Allada, N. E. White, W. V. So, J. C. Hall, M. Rosbash, A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804 (1998). [DOI] [PubMed] [Google Scholar]
- 60.Y. Fujiwara, C. Hermann-Luibl, M. Katsura, M. Sekiguchi, T. Ida, C. Helfrich-Forster, T. Yoshii, The CCHamide1 neuropeptide expressed in the anterior dorsal neuron 1 conveys a circadian signal to the ventral lateral neurons in Drosophila melanogaster. Front. Physiol. 9, 1276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.I. Titos, D. Rogulja, A gut-secreted peptide controls arousability through modulation of dopaminergic neurons in the brain. bioRxiv 10.1101/2020.08.31.275552 (2020). 10.1101/2020.08.31.275552. [DOI]
- 62.I. N. Karatsoreos, R. D. Romeo, B. S. McEwen, R. Silver, Diurnal regulation of the gastrin-releasing peptide receptor in the mouse circadian clock. Eur. J. Neurosci. 23, 1047–1053 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.J. M. Donlea, N. Ramanan, P. J. Shaw, Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.S. R. Lone, S. Potdar, M. Srivastava, V. K. Sharma, Social experience is sufficient to modulate sleep need of Drosophila without increasing wakefulness. PLoS One 11, e0150596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Huckesfeld, P. Schlegel, A. Miroschnikow, A. Schoofs, I. Zinke, A. N. Haubrich, C. M. Schneider-Mizell, J. W. Truman, R. D. Fetter, A. Cardona, M. J. Pankratz, Unveiling the sensory and interneuronal pathways of the neuroendocrine connectome in Drosophila. Elife 10, e65745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Bachevalier, Ontogenetic development of habit and memory formation in primates. Ann. N. Y. Acad. Sci. 608, 457–484 (1990). [DOI] [PubMed] [Google Scholar]
- 67.K. Hartshorn, C. Rovee-Collier, P. Gerhardstein, R. S. Bhatt, T. L. Wondoloski, P. Klein, J. Gilch, N. Wurtzel, M. Campos-de-Carvalho, The ontogeny of long-term memory over the first year-and-a-half of life. Dev. Psychobiol. 32, 69–89 (1998). [PubMed] [Google Scholar]
- 68.H. Hayne, J. Boniface, R. Barr, The development of declarative memory in human infants: Age-related changes in deferred imitation. Behav. Neurosci. 114, 77–83 (2000). [DOI] [PubMed] [Google Scholar]
- 69.A. Jenett, G. M. Rubin, T. T. Ngo, D. Shepherd, C. Murphy, H. Dionne, B. D. Pfeiffer, A. Cavallaro, D. Hall, J. Jeteret al. , A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D. Stoleru, Y. Peng, J. Agosto, M. Rosbash, Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868 (2004). [DOI] [PubMed] [Google Scholar]
- 71.H. Dana, Y. Sun, B. Mohar, B. K. Hulse, A. M. Kerlin, J. P. Hasseman, G. Tsegaye, A. Tsang, A. Wong, R. Patel, J. J. Macklin, Y. Chen, A. Konnerth, V. Jayaraman, L. L. Looger, E. R. Schreiter, K. Svoboda, D. S. Kim, High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019). [DOI] [PubMed] [Google Scholar]
- 72.S. C. Renn, J. H. Park, M. Rosbash, J. C. Hall, P. H. Taghert, A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999). [DOI] [PubMed] [Google Scholar]
- 73.W. Li, Z. Wang, S. Syed, C. Lyu, S. Lincoln, J. O'Neil, A. D. Nguyen, I. Feng, M. W. Young, Chronic social isolation signals starvation and reduces sleep in Drosophila. Nature 597, 239–244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.R. Refinetti, G. C. Lissen, F. Halberg, Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res 38, 275–325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.E. C. Johnson, O. T. Shafer, J. S. Trigg, J. Park, D. A. Schooley, J. A. Dow, P. H. Taghert, A novel diuretic hormone receptor in Drosophila: Evidence for conservation of CGRP signaling. J. Exp. Biol. 208, 1239–1246 (2005). [DOI] [PubMed] [Google Scholar]
- 76.P. Kakanj, S. A. Eming, L. Partridge, M. Leptin, Long-term in vivo imaging of Drosophila larvae. Nat. Protoc. 15, 1158–1187 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12