Abstract
Background:
Anxiety disorders are prevalent and anxiety symptoms often co-occur with psychiatric disorders. Here, we aimed to identify genomic risk loci associated with anxiety, characterize its genetic architecture, and genetic overlap with psychiatric disorders.
Methods:
We used the largest available GWAS of anxiety (GAD-2 score), schizophrenia, bipolar disorder, major depression, and attention deficit hyperactivity disorder (ADHD). We employed MiXeR and LAVA to characterize the genetic architecture and genetic overlap between the phenotypes. Additionally, conditional and conjunctional false discovery rate analyses were performed to boost the identification of genomic loci associated with anxiety and those shared with psychiatric disorders. Gene annotation and gene set analyses were carried out using OpenTargets and FUMA, respectively.
Results:
Anxiety was polygenic with 8.4k estimated genetic risk variants and overlapped extensively with psychiatric disorders (4.1–7.8k variants). Both MiXeR and LAVA revealed predominantly positive genetic correlations between anxiety and psychiatric disorders. We identified 154 anxiety loci (139 novel) by conditioning on the psychiatric disorders. We identified loci shared between anxiety and major depression (n = 66), bipolar disorder (n = 19), schizophrenia (n = 51), and ADHD (n = 37). Genes annotated to anxiety loci exhibit enrichment for a broader range of biological pathways and differential tissue expression in more diverse tissues than those annotated to the shared loci.
Conclusions:
Anxiety is a highly polygenic phenotype with extensive genetic overlap with psychiatric disorders. These genetic overlaps enabled the identification of novel loci for anxiety and shared loci with psychiatric disorders. The shared genetic architecture may underlie the comorbidity of anxiety, and the identified genetic loci implicate molecular pathways that could become potential drug targets.
Keywords: anxiety, genetic overlap, psychiatric disorder, genetic loci
Introduction
Anxiety is a human emotion and anxiety disorders encompass several diagnostic categories of mental disorders characterized by the core features of excessive fear and anxiousness, or avoidance behaviors (1). Anxiety disorders including generalized anxiety disorder, panic disorder, agoraphobia, social anxiety disorder, and specific phobias are among the leading causes of global disease burden (2). Because of the extensive phenotypic overlap and comorbidity among various anxiety disorders (3), there is a growing recognition that anxiety disorders can be better understood and measured along a continuum rather than as discrete categories (4, 5). Anxiety disorders are frequently comorbid with other psychiatric disorders (3). For example, nearly two-thirds of individuals with anxiety disorders also have concurrent depressive disorders and vice versa (6). Anxiety disorders are also common in individuals with bipolar disorder (BIP) (7, 8), schizophrenia (SCZ) (9), and attention deficit hyperactivity disorder (ADHD) (10, 11). Further, symptoms of anxiety (ANX), not meeting the criteria for anxiety disorder, frequently co-occur with other psychiatric disorders (12, 13). Anxiety disorders and ANX co-occurring with psychiatric disorders have been linked with greater symptom burden, poorer course and outcome, and lower quality of life (14–18). The clinical relevance of co-occurring ANX is highlighted by the inclusion in the diagnostic criteria of ‘with anxious distress’ as a specifier of psychiatric disorders such as major depression (MD), and BIP (1).
The etiology of anxiety disorders is not clearly understood; however, both genetic and environmental factors are involved (1, 3, 4, 19, 20). Genetic susceptibility plays a major role in anxiety disorders (20), with heritability estimates from twin studies ranging between 30 to 50% (21). Genome-wide association studies (GWAS) have identified several genetic loci for anxiety disorders (20, 22, 23) and ANX (24). Furthermore, recent evidence from genetic correlation analyses supports the notion that there is shared genetic liability between psychiatric disorders, and anxiety disorders and ANX (22, 24). However, genetic correlations do not provide a comprehensive overview of the shared genetic architecture between two phenotypes (25, 26). The identification of more genetic loci for anxiety disorders and a better understanding of the shared genetic landscape with other psychiatric disorders can unveil more information about the biological pathways underlying anxiety disorders (27, 28). This is crucial for the development of more effective treatments against anxiety disorders and comorbid conditions. Advances in statistical genetics have improved genetic discoveries for complex diseases (29, 30). The utility of these novel analytical methods in better characterizing the genetic architecture of anxiety disorders and the genetic overlap with psychiatric disorders may add to our understanding of the underlying biological mechanisms.
The standard approach to evaluate shared heritability in complex disorders is to estimate genetic correlations with linkage disequilibrium score regression (LDSC) (31). However, genetic correlation, typically a genome-wide summary measure (30, 32), may conceal shared genetic architecture involving a mixture of concordant and discordant effect directions and does not capture the specific overlapping loci and relevant genes (25, 26, 33). These characteristics can be captured by the application of the bivariate causal mixture model (MiXeR) (25) and the local analysis of covariant association (LAVA) (33). Genetic overlap can be exploited to boost the discovery of specific genetic risk loci for a phenotype (30) and loci shared with other phenotypes by applying conditional false discovery rate (condFDR) and conjunctional FDR (conjFDR) analyses respectively (29, 34).
Given the high comorbidity between ANX and psychiatric disorders, we aimed to quantify and characterize their genetic overlap. To this end, we applied univariate MiXeR to comprehensively characterize the polygenic architecture of ANX measured using a dimensional scale. We also employed bivariate MiXeR and LAVA to assess the genetic overlap with MD, SCZ, BIP, and ADHD. Additionally, we applied condFDR/conjFDR methods to identify genetic loci associated with ANX, and loci shared between ANX and the four psychiatric disorders. Finally, we tested whether polygenic liability to the psychiatric disorders predicted anxiety disorder status in an independent sample.
Methods and materials
Genome-wide association studies (GWAS) data
We used the largest GWAS available for anxiety symptoms (ANX), measured as a dimensional trait using the Generalized Anxiety Disorder 2-item scale (GAD-2), from the Million Veterans Program (MVP) (n = 175,163) (24) The ANX GWAS summary statistics were accessed through the Database of Genotype and Phenotype (dbGaP; phs001672). As the lifetime anxiety disorder GWAS meta-analysis (20) was not powerful enough as indicated by poor model fit in univariate MiXeR (25), the dataset was used for replication of identified ANX loci. We obtained GWAS summary statistics for BIP, ADHD, and SCZ from the Psychiatric Genomics Consortium (PGC) (35–37), and MD from a meta-analysis of the PGC and 23andMe, Inc. (38). All the GWAS comprised only populations of European ancestry (Table 1 and Supplementary Methods in Supplement 1). All GWAS of these datasets have been approved by their respective ethical approval committees.
Table 1:
The GWAS summary statistics data
| Phenotype | Number of cases | Number of controls | Source of data |
|---|---|---|---|
|
| |||
| ANX | 175,163 | - | MVP (24) |
| MD | 246,363 | 561,190 | PGC-MD + 23andMe (38) |
| BIP | 41,917 | 371,549 | PGC-BIP (37) |
| SCZ | 53,386 | 77,258 | PGC-SCZ (36) |
| ADHD | 38,691 | 186,843 | PGC-ADHD (35) |
|
| |||
| Anxiety disordera | 31,977 | 82,114 | ANGST + UKBB + iPSYCH (20) |
GWAS: Genome-wide association study, ANX: Anxiety symptoms, MD: Major depression, BIP: Bipolar disorder, SCZ: Schizophrenia, ADHD: Attention deficit hyperactivity disorder, MVP: Million Veterans Program, PGC: Psychiatric Genomics Consortium, UKBB: UK Biobank, ANGST: Anxiety NeuroGenetics Study, iPSYCH: Initiative for Integrative Psychiatric Research
Replication sample
Independent sample
We obtained genotype data for a total of 95,792 mothers and fathers from the Norwegian Mother, Father, and Child Cohort Study (MoBa) (39). The details of sample collection, genotyping, and quality control are provided elsewhere (40). We obtained ICD-10 psychiatric diagnoses from the Norwegian Patient Registry until 2018. Our cases of anxiety disorder (n = 3,330) included agoraphobia (F40.0, n = 493), social phobia (F40.1, n = 874), panic disorder (F41.0, n = 913), and generalized anxiety disorder (F41.1, n = 1,050). MoBa cohort participants had provided written informed consent at enrollment. We excluded participants who withdrew their consent from the analyses. MoBa study was approved by the Regional Committees for Medical and Health Research Ethics (2016/1226).
Statistical analysis
Assessing Genetic overlap
We performed a series of casual mixture model (MiXeR) analyses (25) to investigate the genetic architecture of ANX and the genetic overlap of ANX with MD, BIP, SCZ, and ADHD. First, we conducted univariate MiXeR analyses to estimate the number of trait-influencing variants explaining 90% of SNP-based heritability after controlling for linkage disequilibrium (LD). These were followed by bivariate MiXeR analyses to estimate the number of SNPs shared between pairs of phenotypes irrespective of effect direction. We also determined the estimated proportion of shared SNPs between two phenotypes out of the total number of SNPs estimated to influence both phenotypes (Dice coefficients) and the fraction of SNPs with concordant effects in the shared component (25) (Supplementary Methods in Supplement 1).
We employed LAVA to estimate local genetic correlations between ANX, and MD, BIP, SCZ, and ADHD (33). LAVA estimates local genetic correlations across 2,495 semi-independent genetic regions of approximately 1Mb and identifies shared genetic regions with their effect directions (33). It takes sample overlap into account by using the genetic covariance intercept from LDSC (31). LAVA estimates the heritability of each of the genetic regions for each of the phenotypes followed by estimation of local genetic covariance between the pairs of phenotypes. We corrected the p-values for local genetic correlations for multiple testing using the Bonferroni method.
Conditional and conjunctional false discovery rates
We generated quantile-quantile (Q-Q) plots where the p-values of single nucleotide polymorphisms (SNPs) in ANX were plotted conditional on three different cut-offs of p-values in the secondary phenotypes (i.e., one of MD, BIP, SCZ, and ADHD). Q-Q plots with successive leftward and upward deviation compared to the null were considered to exhibit cross-trait enrichment (29). We performed condFDR analyses to identify loci associated with ANX. Next, conjFDR analyses identified loci shared between ANX and each of the secondary phenotypes respectively (29, 34). In condFDR analysis, the SNP p-values in the ANX GWAS summary statistics were re-ranked based on their p-values in the GWAS summary statistics of the secondary phenotype. CondFDR leverages the SNPs’ association with the secondary phenotype to boost the power to identify novel SNPs associated with the primary phenotype (i.e., anxiety) (29). This boost in power is contingent on the extent of genetic overlap between the two phenotypes (29, 41).
We then performed inverse condFDR analyses whereby MD, BIP, SCZ, and ADHD were primary phenotypes, and ANX was the secondary phenotype and used both pairs of condFDR results for conjFDR analyses. The conjFDR value for a SNP is defined by taking the maximum of the condFDR and inverse condFDR values for a given pair of phenotypes (29). A threshold of 5% was used as statistically significant for both condFDR and conjFDR p-values. We excluded SNPs within the extended major histocompatibility complex (MHC) region and chromosome 8p23.1 inversion (genome build 19 positions of chr6:25119106 – 33854733 and chr8:7200000 – 12500000, respectively) from the condFDR model fit procedure, but not from the discovery analyses (42). We randomly pruned SNPs in 500 iterations to minimize inflation in fold enrichment. All the p-values were corrected for inflation using a genomic inflation control procedure as described previously (34) (Supplementary Methods in Supplement 1). The cond/conjFDR code is available online (https://github.com/precimed/pleiofdr).
Definition of genomic loci
We designated independent genomic loci according to the functional mapping and gene annotation (FUMA) protocol (43). We specified candidate SNPs as any SNP with condFDR or conjFDR < 0.05, and candidate SNPs with LD r2 < 0.6 with each other as independent significant SNPs. Lead SNPs were defined as independent SNPs with LD r2 < 0.1. The candidate SNPs in LD r2 ≥ 0.6 with a lead SNP delineated the boundaries of a genomic locus. We defined all candidate SNPs positioned within the boundaries of a genomic locus to correspond to a single independent genomic locus. We obtained LD information from the 1000 Genomes Project European reference panel (44). In conjFDR, we interpreted the effect directions by comparing the Z-scores of lead SNPs for each locus in the GWAS summary statistics corresponding to the phenotype. We defined novel risk loci as genomic loci not identified in the GWAS catalog for ANX and anxiety disorder (accessed in October 2022) and in ANX or anxiety disorders GWAS (20, 23, 24).
Consistency of genetic effects in an independent sample
We performed a left-sided binomial test of lead SNPs for concordant effect directions in the discovery (GWAS used for condFDR) and replication datasets (independent samples). For replication, we used independent GWAS summary statistics of lifetime anxiety disorders (20). If the statistics for a lead SNP were not available in the replication GWAS dataset, we used a candidate SNP with the next lowest FDR value.
Polygenic risk scores
In MoBa, we restricted the polygenic risk score (PRS) analyses to individuals of European ancestry selected based on genotype principal components (PCs) as described elsewhere (45). We used a kinship coefficient greater than 0.05 to exclude one of the related pairs of study participants while prioritizing individuals with anxiety disorders. When two related individuals had an anxiety disorder diagnosis, one of them was selected. We used PRSice (46) to calculate PRSs at different p-value thresholds (i.e., 5e-8, 1e-7, 1e-6, 1e-5, 1e-4, 1e-3, 1e-2, 5e-2, 1e-1, 5e-1, 1) using the GWAS summary statistics for ANX, MD, BIP, ADHD, and SCZ (Table 1). Subsequently, we extracted the first PC for each PRS across all p-value thresholds (47). Next, we used logistic regression to estimate PRS association with anxiety disorder using models that included PRS of the disorder, and age, sex, and the first 10 genotype PCs as covariates. The combined model included the PRSs for all the psychiatric disorders and the covariates.
Functional annotations and gene set analyses
We performed functional gene mapping for lead SNPs from cond/conjFDR using the OpenTarget platform (https://genetics.opentargets.org/) (48). For each lead SNP, we selected the gene with the highest overall score. We used genes mapped to the lead SNPs using the OpenTarget platform for gene set analyses conducted using the GENE2FUNC analyses in FUMA. We also obtained Combined Annotation Dependent Depletion (CADD) scores which show how deleterious the SNP is on protein function (49), and RegulomeDB scores which predicted the regulatory function of the SNP from FUMA (50). We performed gene set analyses for genes mapped to lead SNPs identified condFDR and conjFDR analyses. We obtained the expression of genes identified in 54 different human tissues using Genotype-Tissue Expression (GTEx) (51).
Results
Polygenicity and genetic overlap
The MiXeR analyses showed that ANX is a polygenic trait with 8.4k ± 0.8k (mean ± SD) trait-influencing variants contributing to 90% of its heritability. We estimated the SNP heritability of ANX to be 5.6 ± 0.4% (Table 2). The other psychiatric disorders were also polygenic with the following estimated numbers of trait-influencing variants: MD 13.9k ± 0.4k, BIP 8.6k ± 0.2k, SCZ 9.6k ± 0.2k, and ADHD 7.7k ± 0.4k (Supplement 2: Tables S1 – S5), as reported previously (26, 35).
Table 2:
SNP heritability and genetic correlation parameters between anxiety and psychiatric disorders from LD score regression analyses
| Phenotypes | SNP heritability (SE) | Genetic correlation (rg, SE) | p-value (rg) |
|---|---|---|---|
|
| |||
| ANX | 0.056 (0.004) | 1.00 | - |
| MD | 0.065 (0.002) | 0.63 (0.03) | 8.97e-94 |
| BIP | 0.197 (0.007) | 0.23 (0.04) | 4.53e-11 |
| SCZ | 0.384 (0.013) | 0.31 (0.03) | 7.61e-28 |
| ADHD | 0.168 (0.008) | 0.47 (0.04) | 1.03e-38 |
SNP: Single nucleotide polymorphism, LD: Linkage disequilibrium, SE: Standard error, rg: Genetic correlation, ANX: Anxiety symptoms, MD: Major depression, BIP: Bipolar disorder, SCZ: Schizophrenia, ADHD: Attention deficit hyperactivity disorder.
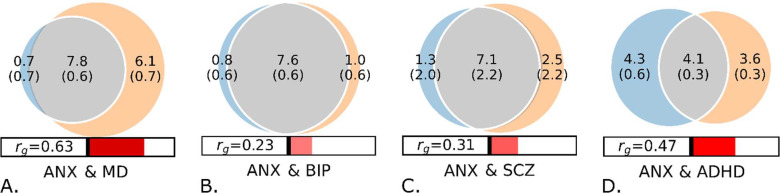
In the bivariate MiXeR analyses, ANX exhibited a huge genetic overlap with psychiatric disorders as demonstrated by the estimated number of shared variants with MD (7.8k ± 0.6k), BIP (7.6k ± 0.6k), SCZ (7.1k ± 0.2k), and ADHD (4.1k ± 0.3k). The Dice coefficients also indicated a substantial overlap with MD (69.6%), BIP (89.6%), SCZ (79.6%), and a lesser extent with ADHD (51.3%). The estimates for genetic overlap between ANX and MD may be unreliable due to suboptimal model fit based on the Akaike Information Criterion. The majority of trait-influencing variants shared between ANX and each of the psychiatric disorders had concordant effect directions, with 92% for ADHD, 86% for MD, 60% for BIP, and 68% for SCZ (Figure 1, Supplement 2: Tables S6 – S9).
Figure 1.
A – D: Bivariate MiXeR – Genome-wide genetic overlap between anxiety symptoms (ANX), and major depression (MD)*, bipolar disorder (BIP), schizophrenia (SCZ), and attention deficit hyperactivity disorder (ADHD). rg: genetic correlation. The numbers indicate estimates of trait-influencing variants in thousands. * Unreliable model fit, interpret with caution.
Local genetic correlation estimates from LAVA showed mixed directions with the majority of regions with nominal p < 0.05 having positive genetic correlations between ANX and all four psychiatric disorders (Supplement 1: Figure S1). A few positively correlated regions remained significant after Bonferroni correction of the p-values with five positively correlated loci between ANX and MD (Figure 2). Genome-wide genetic correlations using LDSC also showed a significant positive genetic correlation (rg) between ANX and all four psychiatric disorders.
Figure 2.
A – D: LAVA – Volcano plots of local genetic correlation coefficients (rho) with −log10 p-values for each locus. Dark red dots represent significantly correlated loci after Bonferroni correction. ANX: Anxiety symptoms, ADHD: Attention deficit hyperactivity disorder, BIP: Bipolar disorder, MD: Major depression, SCZ: Schizophrenia.
Cross-trait polygenic enrichment
We examined the Q-Q plots for cross-trait polygenic enrichment between ANX and psychiatric disorders. The Q-Q plots SNP p-values for ANX exhibited upward and leftward deviation when conditioned on progressively smaller p-value strata from each of MD, BIP, SCZ, and ADHD (Supplement 1: Figure S2). The pattern of Q-Q plots was consistent with the presence of polygenic enrichment between ANX and each of the psychiatric disorders.
Identification of genetic loci for anxiety
We leveraged the cross-trait enrichment and genetic overlap between ANX and psychiatric disorders to identify genomic risk loci for ANX. The condFDR analyses identified 154 unique loci associated with ANX (condFDR < 0.05), and 139 of these loci (90%) were novel (Supplement 3: Tables S10 – S13). The lead SNPs of the identified ANX-loci showed a significant en masse concordance of effect directions in the independent GWAS of lifetime anxiety disorders (Table 3).
Table 3:
En masse test of concordance of effect directions between genetic variants identified for ANX (condFDR) and corresponding variants in independent GWAS of anxiety disorders
| CondFDR Phenotype pairs | ANX-loci (n) | SNPs (n) in the replication dataset | SNPs (n) with concordant effects | p-value* |
|---|---|---|---|---|
|
| ||||
| ANX | MD | 83 | 79 | 69 | 2.77e-12 |
| ANX | BIP | 41 | 38 | 34 | 3.02e-07 |
| ANX | SCZ | 70 | 66 | 54 | 8.47e-08 |
| ANX | ADHD | 59 | 59 | 47 | 2.56e-06 |
GWAS: Genome-wide association study, SNP: Single nucleotide polymorphism, FDR: False discovery rate, ANX: Anxiety symptoms, MD: Major depression, BIP: Bipolar disorder, SCZ: Schizophrenia, ADHD: Attention deficit hyperactivity disorder,
one-sided binomial test
Genomic loci shared between anxiety and psychiatric disorders
We identified 113 genomic loci shared between ANX and psychiatric disorders. Notably, ANX and MD had 66 jointly associated loci (conjFDR < 0.05) of which 62 had a concordant effect direction (94%), and 33 were novel loci for MD. There were 51 loci shared between ANX and SCZ with 46 loci having concordant effect direction (90%). Fourteen of the 19 shared loci between ANX and BIP (74%), and 35 of the 37 loci (95%) shared between ANX and ADHD had concordant effect directions (Supplement 3: Tables S14 – S17). Many of the joint loci were shared across different pairs of traits in conjFDR analysis (Supplement 1: Figure S3).
Polygenic risk scores
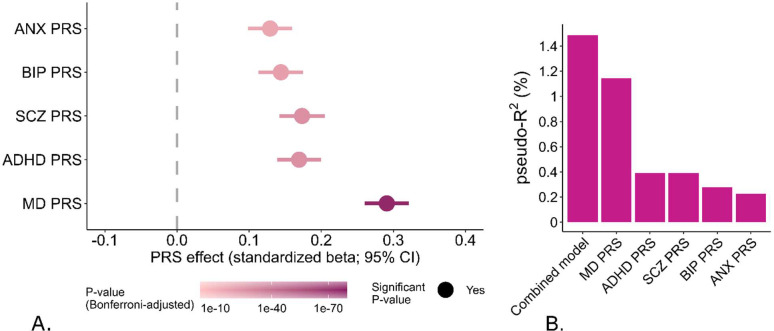
The PRS for each of the psychiatric disorders and ANX positively associated with anxiety disorders Bonferroni corrected p < 1.0e-12, MD PRS had a larger effect estimate than all other PRSs. Nagelkerke’s R2 showed that MD PRS had a stronger association with anxiety disorders (1.28%) than ANX PRS (0.29%) (Figure 3A–B).
Figure 3.
A – B: Logistic regression – The association between polygenic risk scores (PRS) of various psychiatric disorders and anxiety disorders in MoBa parents. A – models for the PRS of anxiety symptoms (ANX), attention deficit hyperactivity disorder (ADHD), bipolar disorder (BIP), major depression (MD), and covariates. B – Nagelkerke’s R2 shows the difference in the percentage of prediction of anxiety traits by each PRS over a base model that includes age, sex, and genotype principal components.
Functional annotations and gene set analyses
Most of the lead SNPs in the ANX-loci were either intronic or intergenic while five of the lead SNPs were exonic: rs61740399 (YTHDF3), rs1792 (FARP1), rs473267 (DOPEY1; PGM3), rs11507683 (EXD3), and rs9988 (NUP85) (Supplement 3: Tables S10 – S13). Similarly, most lead SNPs in shared loci were intronic or intergenic except a SNP shared between ANX and MDD - rs4434138 (STAB1), and another shared between ANX and SCZ - rs473267 (DOPEY1; PGM3) were also exonic. A few SNPs shared between ANX, and MDD (rs4434138, rs17149653, rs5030889), ANX and SCZ (rs473267, rs7858, rs17288409), and ANX and ADHD (rs56403421) had a CADD score >12.37 (Supplement 3: Tables S14 – S17).
Gene set analyses of genes annotated to loci identified for ANX with condFDR revealed enrichment of biological processes relevant to neurodevelopment and synaptic biology including neuron development, synaptic signaling, synapse organization, and regulation of trans-synaptic signaling. Other enriched biological processes included fear response, and glutamatergic and GABAergic synaptic transmissions (Supplement 4: Table S18). Genes annotated to loci shared between ANX and psychiatric disorders were enriched for biological processes such as neuron development, synaptic signaling, and regulation of neuron differentiation (Supplement 1: Figure S4, Supplement 4: Table S19). The top enriched cellular components for genes annotated to loci identified for ANX as well as those shared with psychiatric disorders converge to synapse, synaptic membrane, and synapse part (Supplement 4: Tables S20 and S21).
Enrichment analysis of the genes annotated to ANX loci showed that they are differentially expressed in various tissues of the nervous system, cardiovascular system, gastrointestinal system, and genitourinary system (Figure 4). In contrast, enrichment analysis of the genes annotated to the loci shared between ANX and the psychiatric disorders showed differential tissue expression in parts of the brain and minor salivary glands (Supplement 1: Figure S5).
Figure 4.
FUMA functional annotation – Tissue enrichment for differential gene expression (DEG) in 54 GTEx tissue types of genes mapped to genomic loci associated with anxiety symptoms (condFDR < 0.05; Supplement 3)
Discussion
Here, we showed that ANX is a highly polygenic phenotype with over eight thousand trait-influencing genetic variants, with extensive genetic overlap with other psychiatric disorders beyond their positive genetic correlations. We also demonstrated that the polygenic liabilities for ANX, MD, SCZ, and ADHD predicted a lifetime clinical diagnosis of anxiety disorders in an independent population-based sample. Overlapping trait-influencing variants were the largest for ANX and MD, BIP, and SCZ (70 – 90%), and a relatively smaller overlap between ANX and ADHD (51%). The proportion of variants with concordant effects within the shared component was the highest for ADHD (92%) and lowest for BIP (59%). Local genetic correlations revealed a mixture of effect directions with predominantly positively correlated loci. We identified 139 novel genetic loci associated with ANX, and unique 113 genetic loci shared between ANX and psychiatric disorders and a similar pattern of effect directions, with a predominance of concordant effects. Downstream analyses revealed genes annotated to ANX loci exhibited enrichment for a wider range of biological pathways and differential tissue expression in more diverse tissues than those annotated to the shared loci.
We found that ANX is a highly polygenic phenotype, and this is important as common genetic variants contribute to a large portion of the heritability of anxiety disorders (20). We also found that common genetic variants in ANX exhibit low discoverability and hence require very large sample size studies for genome-wide discoveries. This may partly explain the missing heritability – the huge difference between the SNP heritability we found or reported in other GWASs (20, 24), and heritability reported in twins (21). Our comprehensive characterization of the genetic overlap between ANX, and MD, BIP, SCZ, and ADHD disorders using methods agnostic to effect directions such as bivariate MiXeR expand our understanding beyond genetic correlations (24). Genetic correlations alone show strong overlap of ANX with internalizing disorders (24) whereas our results from MiXeR revealed extensive overlap with externalizing disorders as well. Further, the higher genetic correlation between ANX and ADHD despite a smaller genetic overlap compared to that of ANX and BIP is probably accounted for by the similar effect directions for the majority of shared genetic variants with ADHD (25).
By leveraging genetic overlap with psychiatric disorders, we obtained an over 30-fold boost in the identification of genetic loci for ANX. The identification of a larger number of loci and the annotated genes revealed a wide range of biological pathways potentially involved in the pathophysiology of ANX (43). Genes annotated to ANX loci are enriched for pathways of neurodevelopment and synaptic signaling. In particular, enrichment for ‘regulation of glutamatergic synaptic transmission’ and ‘GABAergic synaptic transmission’ support the role of the specific neurotransmitters in the pathophysiology of ANX (52, 53). Conceivably, the large number of shared genetic loci identified between ANX, and psychiatric disorders suggest shared biological pathways related to neurodevelopment. These highlight the role of neurodevelopmental factors in the pathophysiology of anxiety (54). We speculate that such shared biological pathways may underlie the higher prevalence of anxiety among individuals with psychiatric disorders than in the general population.
Notably, the genes annotated to ANX loci showed differential tissue expression in much broader range of tissues including the brain, gastrointestinal, cardiovascular, and endocrine tissues while those of the shared loci were enriched mainly in brain tissue (Figure 4 and Figure S5). While these may be due to comorbidity between anxiety disorders and medical conditions (55, 56), we argue that ANX has stronger somatic component involving various organ systems than other psychiatric disorders. Also, the genetic risk for ANX may influence risk through a more diverse set of tissues than other psychiatric disorders as demonstrated in animal models (57).
Five of the novel loci identified for ANX rs11507683 [EXD3], rs61740399 [YTHDF3], rs1792 [FARP1], rs9988 [NUP85], rs473267 [PGM3; DOPEY1] are in the exonic region of the respective genes. In particular, FARP1 is a protein-coding gene that has been reported to play a role in the regulation of excitatory synapse development and dendritic extensions (58, 59). Several of the identified genetic loci are associated with high deleteriousness as indicated by CADD scores (Supplement 3: Tables S10 – S13). Notably, four of these loci have previously been linked to neurodevelopmental processes rs7858 [NFIB] and rs78642714 [SYT3] (60, 61), and psychiatric disorders such as SCZ rs61687445 (DRD2) and rs11062166 (CACNA1C) (62, 63).
We found five shared loci, rs12023347 [RP4–598G3.1], rs7585730 [AC068490.2], rs55789728 [MAD1L1], rs7155889 [TRAF3], and rs375241 [ASIC2 /ACCN1] common to all four conjFDR analyses. Notably, the locus annotated to ASIC2 is novel for all five psychiatric phenotypes (Supplement 3: Tables S14 – S17). ASIC2 is a coding gene for a sodium ion channel protein located on the cell membranes and has biased expression in parts of the brain such as the hippocampus, amygdala, striatum, cortex, and cerebellum (64). In preclinical studies, ASIC2 was shown to be involved in neurotransmission and plays a role in behavioral changes of drug abuse (65, 66). An intronic variant of ASIC2 was associated with response to treatment with lithium in patients with BIP (67). Furthermore, genetic studies in humans of a closely related gene ASIC1a have previously demonstrated a link between variants, panic disorder (68), and amygdala volume (69). Although ASIC2 is less well studied or lacked adequate power (70) compared to ASIC1a, there is evidence suggesting that ASIC2 works in concert and regulates neuroplasticity in the brain (71, 72). ASICs are considered potential drug targets in various somatic and psychiatric conditions (64, 72, 73).
The tool used to measure ANX i.e., GAD-2 as a dimensional score in the primary GWAS has the advantage of capturing several of the anxiety disorders as well as subsyndromal ANX (74) both of which have important clinical implications. The recognition of concomitant symptoms of anxiety is highlighted by the addition of a diagnostic specifier - anxious distress - in DSM-5 (1, 75, 76). Anxious distress can have a negative impact on the severity and clinical outcome of primary psychiatric disorders (75, 77). Our findings on the shared genetic architecture of ANX and MD, BIP, SCZ, and ADHD provide insight into the underlying molecular pathways and may assist in improving the treatment of not only comorbid anxiety disorders but also for optimizing treatment in co-occurring ANX. The demonstration that polygenic liability for psychiatric disorders predicting anxiety disorders lends further evidence for a shared genetic risk. Our results suggest the genetic risk for anxiety may involve pathophysiology in neurodevelopment and neurotransmission, and the former might be common to anxiety and other psychiatric disorders.
We acknowledge that the primary phenotype ANX GWAS (24) was a dimensional trait defined based on self-reported GAD-2 and referred to symptoms experienced in the preceding two weeks rather than a lifetime clinical diagnosis of specific anxiety disorders. The vast majority of the study participants were males which may limit the identification of female-specific genetic loci of ANX. Nevertheless, we replicated the genetic loci identified for ANX in an independent GWAS of a predominantly female study population (20). Since individuals with a history of diagnosis of depression were not excluded from the ANX GWAS, the genetic overlap between ANX and MD could partly be due to comorbidity. We applied our analyses to GWAS data from populations of European ancestry and therefore, generalizations cannot be made to other ancestries. As multi-ancestry GWAS data become available, our methods can be applied to improve genetic discoveries for anxiety.
In conclusion, our investigation of the genetic architecture of symptoms of anxiety revealed a high polygenicity and low discoverability which partly explains the missing heritability of anxiety. There was also a large genetic overlap between anxiety and psychiatric disorders, which enabled the identification of 139 novel anxiety loci and 113 shared loci. The shared genetic architecture may underlie the high burden of anxiety symptoms in individuals with other psychiatric disorders. The genes annotated to anxiety loci implicated a broader range of biological pathways as well as differential tissue expression in more diverse tissues than the shared loci. Our findings advance our understanding of the pathophysiology of anxiety that occurs alone or concomitantly with other psychiatric disorders and may help in the identification of potential drug targets. Further research is needed to investigate the genetic underpinnings of specific types of anxiety disorders.
Supplementary Material
Acknowledgments
We are grateful to all the families who participated in MoBa. MoBa is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We would like to thank the research participants and employees of 23andMe, the Psychiatric Genomic Consortium (Schizophrenia, Bipolar Disorder, and Depression Working Groups), the UK Biobank, the Million Veteran Program, and MoBa for making this work possible. This work was performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT) (tsd-drift@usit.uio.no).
Funding
We gratefully acknowledge support from the American National Institutes of Health (NS057198, EB00790), the Research Council of Norway (RCN) (296030, 324252, 324499, 300309, 273291, 223273), the South-East Norway Regional Health Authority (2022–073), and KG Jebsen Stiftelsen (SKGJ-MED-021). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreements No 847776, 964874, and 801133 under the Marie Skłodowska-Curie grant.
List of acronyms
- ADHD
Attention Deficit Hyperactivity Disorder
- ANX
Anxiety symptoms
- BIP
Bipolar Disorder
- CADD
Combined Annotation Dependent Depletion
- CondFDR
Conditional FDR
- ConjFDR
Conjunctional FDR
- dbGAP
Database of Genotype and Phenotype
- FDR
False Discovery Rate
- FUMA
Functional Mapping and Annotations
- GO
Gene Ontology
- GAD-2
Generalized Anxiety Disorder 2-item scale
- GTEx
Genotype-Tissue Expression
- GWAS
Genome-Wide Association Study
- LD
Linkage Disequilibrium
- LDSC
Linkage Disequilibrium Score Regression
- MiXeR
bivariate causal mixture model
- MD
Major Depression
- MHC
major histocompatibility complex
- MVP
Million Veterans Program
- MoBa
Mor og Barn (Norwegian Mothers and Children Cohort
- PGC
Psychiatric Genomics Consortium
- Q-Q
Quantile-Quantile
- SCZ
Schizophrenia
- SNP
Single Nucleotide Polymorphism
Footnotes
Competing interests
Ole A. Andreassen is a consultant for cortechs.ai and has received speaker’s honoraria from Lundbeck, Janssen, and Sunovion. Srdjan Djurovic has received speaker’s honoraria from Lundbeck. Anders M. Dale is a founder of and holds equity in CorTechs Labs and serves on its scientific advisory board. He is also a member of the Scientific Advisory Board of Human Longevity, Inc., and receives research funding from General Electric Healthcare and Medtronic, Inc. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict-of-interest policies. The other authors have no conflicts of interest to declare.
Ethics approval and consent to participate
The MoBa cohort has initially been approved by the Norwegian Data Protection Agency and The Regional Committees for Medical and Health Research Ethics (REK) in Norway and is currently regulated by the Norwegian Health Registry Act. Previously published datasets have been approved by their respective ethical approval committees. This research was conducted according to the Helsinki Declaration.
Availability of data and materials
GWAS of anxiety can be accessed at https://www.ncbi.nlm.nih.gov/gap/, dbGaP Study Accession phs001672, All PGC data are available at https://www.med.unc.edu/pgc/download-results/, Full GWAS summary statistics for the 23andMe DEP dataset will be available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Interested investigators should email dataset-request@23andme.com and reference this paper for more information. Access to the MoBa data can be obtained by applying to the Norwegian Institute of Public Health (NIPH). Restrictions apply regarding the availability of the MoBa data, and therefore, it is not publicly available. Access can be given after approval provided that the applications are consistent with the consent provided by participants. Detailed information on the application can be found on the NIPH website at https://www.fhi.no/en/studies/moba/. The cond/conjFDR code is available online at https://github.com/precimed/pleiofdr.
References
- 1.APA (2013): American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders. Fifth ed. Arlington, VA: American Psychiatric Association. [Google Scholar]
- 2.Collaborators GBDMD (2022): Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. 9:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penninx BW, Pine DS, Holmes EA, Reif A (2021): Anxiety disorders. Lancet. 397:914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koen N, Stein D.J. (2020): Chapter 86: Core dimensions of anxiety disorders. In: Geddes JR, Andreasen N.C., Goodwin G.M., editor. New Oxford Textbook of Psychiatry, Third Edition ed. New York, USA: Oxford University Press, pp 897–904. [Google Scholar]
- 5.Meier SM, Deckert J (2019): Genetics of Anxiety Disorders. Curr Psychiatry Rep. 21:16. [DOI] [PubMed] [Google Scholar]
- 6.Lamers F, van Oppen P, Comijs HC, Smit JH, Spinhoven P, van Balkom AJ, et al. (2011): Comorbidity patterns of anxiety and depressive disorders in a large cohort study: the Netherlands Study of Depression and Anxiety (NESDA). J Clin Psychiatry. 72:341–348. [DOI] [PubMed] [Google Scholar]
- 7.Yapici Eser H, Kacar AS, Kilciksiz CM, Yalcinay-Inan M, Ongur D (2018): Prevalence and Associated Features of Anxiety Disorder Comorbidity in Bipolar Disorder: A Meta-Analysis and Meta-Regression Study. Front Psychiatry. 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yapici Eser H, Taskiran AS, Ertinmaz B, Mutluer T, Kilic O, Ozcan Morey A, et al. (2020): Anxiety disorders comorbidity in pediatric bipolar disorder: a meta-analysis and meta-regression study. Acta Psychiatr Scand. 141:327–339. [DOI] [PubMed] [Google Scholar]
- 9.Achim AM, Maziade M, Raymond E, Olivier D, Merette C, Roy MA (2011): How prevalent are anxiety disorders in schizophrenia? A meta-analysis and critical review on a significant association. Schizophr Bull. 37:811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller-Thomson E, Carrique L, MacNeil A (2022): Generalized anxiety disorder among adults with attention deficit hyperactivity disorder. J Affect Disord. 299:707–714. [DOI] [PubMed] [Google Scholar]
- 11.Melegari MG, Bruni O, Sacco R, Barni D, Sette S, Donfrancesco R (2018): Comorbidity of Attention Deficit Hyperactivity Disorder and Generalized Anxiety Disorder in children and adolescents. Psychiatry Res. 270:780–785. [DOI] [PubMed] [Google Scholar]
- 12.Temmingh H, Stein DJ (2015): Anxiety in Patients with Schizophrenia: Epidemiology and Management. CNS Drugs. 29:819–832. [DOI] [PubMed] [Google Scholar]
- 13.Kauer-Sant’Anna M, Kapczinski F, Vieta E (2009): Epidemiology and management of anxiety in patients with bipolar disorder. CNS Drugs. 23:953–964. [DOI] [PubMed] [Google Scholar]
- 14.Spoorthy MS, Chakrabarti S, Grover S (2019): Comorbidity of bipolar and anxiety disorders: An overview of trends in research. World J Psychiatry. 9:7–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wetherell JL, Palmer BW, Thorp SR, Patterson TL, Golshan S, Jeste DV (2003): Anxiety symptoms and quality of life in middle-aged and older outpatients with schizophrenia and schizoaffective disorder. J Clin Psychiatry. 64:1476–1482. [DOI] [PubMed] [Google Scholar]
- 16.Buonocore M, Bosia M, Bechi M, Spangaro M, Cavedoni S, Cocchi F, et al. (2017): Targeting anxiety to improve quality of life in patients with schizophrenia. Eur Psychiatry. 45:129–135. [DOI] [PubMed] [Google Scholar]
- 17.Kim SW, Berk L, Kulkarni J, Dodd S, de Castella A, Fitzgerald PB, et al. (2014): Impact of comorbid anxiety disorders and obsessive-compulsive disorder on 24-month clinical outcomes of bipolar I disorder. J Affect Disord. 166:243–248. [DOI] [PubMed] [Google Scholar]
- 18.Titone MK, Freed RD, O’Garro-Moore JK, Gepty A, Ng TH, Stange JP, et al. (2018): The role of lifetime anxiety history in the course of bipolar spectrum disorders. Psychiatry Res. 264:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(2013): American Psychatric Association: Diagnostic and Statistical Manual of Mental Disorders. Fifth ed. Arlington, VA: American Psychiatric Association. [Google Scholar]
- 20.Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, et al. (2020): A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 25:3292–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hettema JM, Neale MC, Kendler KS (2001): A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 158:1568–1578. [DOI] [PubMed] [Google Scholar]
- 22.Ohi K, Otowa T, Shimada M, Sasaki T, Tanii H (2020): Shared genetic etiology between anxiety disorders and psychiatric and related intermediate phenotypes. Psychol Med. 50:692–704. [DOI] [PubMed] [Google Scholar]
- 23.Wendt FR, Pathak GA, Deak JD, De Angelis F, Koller D, Cabrera-Mendoza B, et al. (2022): Using phenotype risk scores to enhance gene discovery for generalized anxiety disorder and posttraumatic stress disorder. Mol Psychiatry. 27:2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, et al. (2020): Reproducible Genetic Risk Loci for Anxiety: Results From approximately 200,000 Participants in the Million Veteran Program. Am J Psychiatry. 177:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frei O, Holland D, Smeland OB, Shadrin AA, Fan CC, Maeland S, et al. (2019): Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun. 10:2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindley G, Frei O, Shadrin AA, Cheng W, O’Connell KS, Icick R, et al. (2022): Charting the Landscape of Genetic Overlap Between Mental Disorders and Related Traits Beyond Genetic Correlation. Am J Psychiatry. 179:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craske MG, Stein MB, Eley TC, Milad MR, Holmes A, Rapee RM, et al. (2017): Anxiety disorders. Nat Rev Dis Primers. 3:17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smoller JW (2020): Anxiety Genetics Goes Genomic. Am J Psychiatry. 177:190–194. [DOI] [PubMed] [Google Scholar]
- 29.Smeland OB, Frei O, Shadrin A, O’Connell K, Fan CC, Bahrami S, et al. (2020): Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 139:85–94. [DOI] [PubMed] [Google Scholar]
- 30.Andreassen OA, Hindley GFL, Frei O, Smeland OB (2023): New insights from the last decade of research in psychiatric genetics: discoveries, challenges and clinical implications. World Psychiatry. 22:4–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. (2015): An atlas of genetic correlations across human diseases and traits. Nat Genet. 47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe K, Stringer S, Frei O, Umicevic Mirkov M, de Leeuw C, Polderman TJC, et al. (2019): A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 51:1339–1348. [DOI] [PubMed] [Google Scholar]
- 33.Werme J, van der Sluis S, Posthuma D, de Leeuw CA (2022): An integrated framework for local genetic correlation analysis. Nat Genet. 54:274–282. [DOI] [PubMed] [Google Scholar]
- 34.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, et al. (2013): Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 92:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demontis D, Walters GB, Athanasiadis G, Walters R, Therrien K, Nielsen TT, et al. (2023): Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet. 55:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trubetskoy V, Pardinas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. (2022): Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 604:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. (2021): Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 53:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. (2019): Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 22:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, et al. (2016): Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol. 45:382–388. [DOI] [PubMed] [Google Scholar]
- 40.Jaholkowski P, Shadrin AA, Jangmo A, Frei E, Tesfaye M, Hindley GFL, et al. (2023): Associations Between Symptoms of Premenstrual Disorders and Polygenic Liability for Major Psychiatric Disorders. JAMA Psychiatry. 80:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahrami S, Hindley G, Winsvold BS, O’Connell KS, Frei O, Shadrin A, et al. (2022): Dissecting the shared genetic basis of migraine and mental disorders using novel statistical tools. Brain. 145:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartzman A, Lin X (2011): The effect of correlation in false discovery rate estimation. Biometrika. 98:199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe K, Taskesen E, van Bochoven A, Posthuma D (2017): Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. (2015): A global reference for human genetic variation. Nature. 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corfield EC, Frei O, Shadrin AA, Rahman Z, Lin A, Athanasiu L, et al. (2022): The Norwegian Mother, Father, and Child cohort study (MoBa) genotyping data resource: MoBaPsychGen pipeline v.1. bioRxiv.2022.2006.2023.496289. [Google Scholar]
- 46.Choi SW, O’Reilly PF (2019): PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coombes BJ, Ploner A, Bergen SE, Biernacka JM (2020): A principal component approach to improve association testing with polygenic risk scores. Genet Epidemiol. 44:676–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ochoa D, Hercules A, Carmona M, Suveges D, Baker J, Malangone C, et al. (2022): The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014): A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. (2012): Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. (2017): Genetic effects on gene expression across human tissues. Nature. 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones SK, McCarthy DM, Vied C, Stanwood GD, Schatschneider C, Bhide PG (2022): Transgenerational transmission of aspartame-induced anxiety and changes in glutamate-GABA signaling and gene expression in the amygdala. Proc Natl Acad Sci U S A. 119:e2213120119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nasir M, Trujillo D, Levine J, Dwyer JB, Rupp ZW, Bloch MH (2020): Glutamate Systems in DSM-5 Anxiety Disorders: Their Role and a Review of Glutamate and GABA Psychopharmacology. Front Psychiatry. 11:548505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer HC, Lee FS (2019): Translating Developmental Neuroscience to Understand Risk for Psychiatric Disorders. Am J Psychiatry. 176:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kariuki-Nyuthe C, Stein DJ (2014): Anxiety and Related Disorders and Physical Illness. In: Sartorius N, Holt RIG, Maj M, editors. Comorbidity of Mental and Physical Disorders: S.Karger AG, pp 0. [Google Scholar]
- 56.Sareen J, Cox BJ, Clara I, Asmundson GJ (2005): The relationship between anxiety disorders and physical disorders in the U.S. National Comorbidity Survey. Depress Anxiety. 21:193–202. [DOI] [PubMed] [Google Scholar]
- 57.Parent MB, Ferreira-Neto HC, Kruemmel AR, Althammer F, Patel AA, Keo S, et al. (2021): Heart failure impairs mood and memory in male rats and down-regulates the expression of numerous genes important for synaptic plasticity in related brain regions. Behav Brain Res. 414:113452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheadle L, Biederer T (2012): The novel synaptogenic protein Farp1 links postsynaptic cytoskeletal dynamics and transsynaptic organization. J Cell Biol. 199:985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuang B, Su YS, Sockanathan S (2009): FARP1 promotes the dendritic growth of spinal motor neuron subtypes through transmembrane Semaphorin6A and PlexinA4 signaling. Neuron. 61:359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schanze I, Bunt J, Lim JWC, Schanze D, Dean RJ, Alders M, et al. (2018): NFIB Haploinsufficiency Is Associated with Intellectual Disability and Macrocephaly. Am J Hum Genet. 103:752–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dean C, Dunning FM, Liu H, Bomba-Warczak E, Martens H, Bharat V, et al. (2012): Axonal and dendritic synaptotagmin isoforms revealed by a pHluorin-syt functional screen. Mol Biol Cell. 23:1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Schizophrenia Working Group of the Psychiatric Genomics C, et al. (2014): Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 19:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schizophrenia Working Group of the Psychiatric Genomics C (2014): Biological insights from 108 schizophrenia-associated genetic loci. Nature. 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storozhuk M, Cherninskyi A, Maximyuk O, Isaev D, Krishtal O (2021): Acid-Sensing Ion Channels: Focus on Physiological and Some Pathological Roles in the Brain. Curr Neuropharmacol. 19:1570–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuller MJ, Gupta SC, Fan R, Taugher-Hebl RJ, Wang GZ, Andrys NRR, et al. (2023): Investigating role of ASIC2 in synaptic and behavioral responses to drugs of abuse. Front Mol Biosci. 10:1118754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kreple CJ, Lu Y, Taugher RJ, Schwager-Gutman AL, Du J, Stump M, et al. (2014): Acid-sensing ion channels contribute to synaptic transmission and inhibit cocaine-evoked plasticity. Nat Neurosci. 17:1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Squassina A, Manchia M, Borg J, Congiu D, Costa M, Georgitsi M, et al. (2011): Evidence for association of an ACCN1 gene variant with response to lithium treatment in Sardinian patients with bipolar disorder. Pharmacogenomics. 12:1559–1569. [DOI] [PubMed] [Google Scholar]
- 68.Gregersen N, Dahl HA, Buttenschon HN, Nyegaard M, Hedemand A, Als TD, et al. (2012): A genome-wide study of panic disorder suggests the amiloride-sensitive cation channel 1 as a candidate gene. Eur J Hum Genet. 20:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smoller JW, Gallagher PJ, Duncan LE, McGrath LM, Haddad SA, Holmes AJ, et al. (2014): The human ortholog of acid-sensing ion channel gene ASIC1a is associated with panic disorder and amygdala structure and function. Biol Psychiatry. 76:902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hettema JM, An SS, Neale MC, van den Oord EJ, Kendler KS, Chen X (2008): Lack of association between the amiloride-sensitive cation channel 2 (ACCN2) gene and anxiety spectrum disorders. Psychiatr Genet. 18:73–79. [DOI] [PubMed] [Google Scholar]
- 71.Zha XM, Costa V, Harding AM, Reznikov L, Benson CJ, Welsh MJ (2009): ASIC2 subunits target acid-sensing ion channels to the synapse via an association with PSD-95. J Neurosci. 29:8438–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sivils A, Yang F, Wang JQ, Chu XP (2022): Acid-Sensing Ion Channel 2: Function and Modulation. Membranes (Basel). 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heusser SA, Pless SA (2021): Acid-sensing ion channels as potential therapeutic targets. Trends Pharmacol Sci. 42:1035–1050. [DOI] [PubMed] [Google Scholar]
- 74.Kroenke K, Spitzer RL, Williams JB, Monahan PO, Lowe B (2007): Anxiety disorders in primary care: prevalence, impairment, comorbidity, and detection. Ann Intern Med. 146:317–325. [DOI] [PubMed] [Google Scholar]
- 75.Gaspersz R, Lamers F, Kent JM, Beekman AT, Smit JH, van Hemert AM, et al. (2017): Longitudinal Predictive Validity of the DSM-5 Anxious Distress Specifier for Clinical Outcomes in a Large Cohort of Patients With Major Depressive Disorder. J Clin Psychiatry. 78:207–213. [DOI] [PubMed] [Google Scholar]
- 76.Zimmerman M, Kerr S, Balling C, Kiefer R, Dalrymple K (2020): DSM-5 anxious distress specifier in patients with bipolar depression. Ann Clin Psychiatry. 32:157–163. [PubMed] [Google Scholar]
- 77.Gaspersz R, Nawijn L, Lamers F, Penninx B (2018): Patients with anxious depression: overview of prevalence, pathophysiology and impact on course and treatment outcome. Curr Opin Psychiatry. 31:17–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS of anxiety can be accessed at https://www.ncbi.nlm.nih.gov/gap/, dbGaP Study Accession phs001672, All PGC data are available at https://www.med.unc.edu/pgc/download-results/, Full GWAS summary statistics for the 23andMe DEP dataset will be available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Interested investigators should email dataset-request@23andme.com and reference this paper for more information. Access to the MoBa data can be obtained by applying to the Norwegian Institute of Public Health (NIPH). Restrictions apply regarding the availability of the MoBa data, and therefore, it is not publicly available. Access can be given after approval provided that the applications are consistent with the consent provided by participants. Detailed information on the application can be found on the NIPH website at https://www.fhi.no/en/studies/moba/. The cond/conjFDR code is available online at https://github.com/precimed/pleiofdr.