Abstract
Direct sequencing of the 16S rRNA gene (16S rDNA) of Mycobacterium celatum isolates showed ambiguities, suggesting heterogeneity. Cloned 16S rDNA yielded two copies of the gene, which differed by insertion of a thymine at position 214 and by additional mismatches. Restriction fragment length polymorphism analysis confirmed the presence of two copies of 16S rDNA within the bacterial chromosome.
Mycobacterium celatum has been isolated from samples of immunocompetent patients with pulmonary disease (5) and from patients infected with human immunodeficiency virus in whom it had caused disseminated disease (20). While its resistance to antituberculosis treatment requires rapid and accurate diagnosis, identification of M. celatum by conventional techniques often takes several weeks because of the slow growth of this organism (11). Furthermore, rapid identification techniques, such as commercially available DNA probes, may yield false-positive results for Mycobacterium tuberculosis when they are applied to M. celatum (4, 11). Direct sequencing of the bacterial 16S rRNA gene (16S rDNA) has proven to be a stable and specific marker for mycobacterial identification (15, 16, 24); while the number of copies of 16S rRNA genes may vary among bacterial species, their sequences are assumed to be identical, with only minor differences (14, 21). Fast-growing members of the genus Mycobacterium generally have two identical copies of the 16S rRNA gene (9), while slow growers are thought to have only one (13) and, therefore, yield unambiguous sequence patterns. However, two different 16S rRNA genes were recently detected in a slowly growing mycobacterium belonging to the Mycobacterium terrae complex (19). Here, we report the observation of 16S rDNA heterogeneity in three isolates of the clinically important M. celatum.
Mycobacterial isolates.
Three clinical laboratories each provided one clinical isolate of M. celatum (1732, T322, and MI1581). The isolates were grown on Loewenstein-Jensen medium and examined for growth rate, microscopic colony morphology, and pigmentation. Biochemical tests of all three clinical isolates yielded identical results, which were consistent with those published by Butler et al. (5).
Amplification and sequencing of genomic DNA.
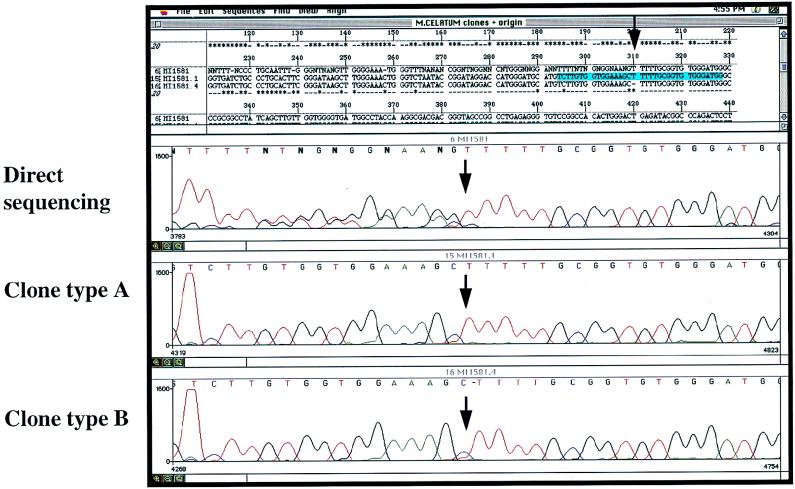
A single colony of each isolate was harvested and chromosomal DNA was extracted as previously described (22). One microgram of bacterial DNA was subjected to PCR with each of two primer pairs, i.e., biotinylated M285 in combination with M264 (15) and primer R247 (GTAGTCCACGCCGTAAACGG) in combination with M261 (15), thus yielding the complete 16S rDNA in two fragments. Nonradioactive sequencing of all three isolates was performed with forward primer M285 and reverse primer M259 (15) by using a commercially available cycle sequencing kit (PRISM Ready Reaction Dye Deoxy Terminator cycle sequencing kit; Applied Biosystems GmbH, Weiterstadt, Germany). Electrophoresis of sequenced DNA and data collection were performed with an automated sequencer (ABI 373A; Applied Biosystems). The results revealed identical patterns, showing heterogeneity consecutive to position 214 within helix 10 (first line of Fig. 1); in position 214, all isolates showed a partial insertion of an additional T (TTTTT instead of TTTT), thus leading to a duplication of the peaks seen on the electropherograms, consecutive to the point of insertion (Fig. 1). Clear peak patterns were obtained for the stretch between the sequencing primer and the insertion. These results were confirmed by manual sequencing (data not shown).
FIG. 1.
Graphic plot of the direct sequencing (lane 1) of PCR-amplified 16S rDNA and cloned 16S rDNA (clones 1 and 4) of the clinical isolate MI1581, obtained by automated sequencing. The stretch of consecutive T’s indicates the start of ambiguities in the pattern obtained by direct sequencing with reverse primer M259.
RFLP.
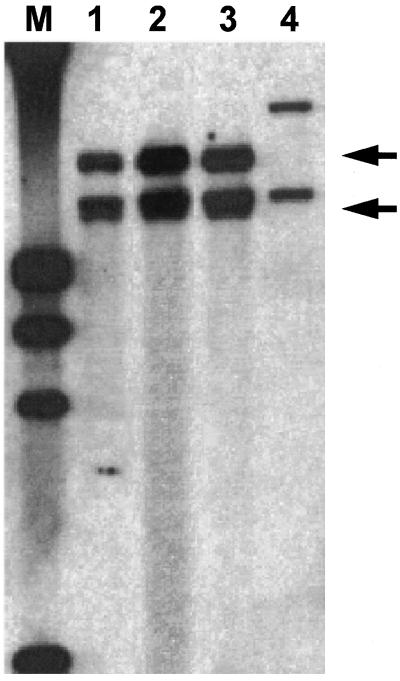
Single colonies of each isolate were grown for as many as 3 weeks in 5 ml of Middlebrook 7H9 broth supplemented with 0.05% Tween 80. The cultures were centrifuged, resuspended in 0.85% NaCl, and inactivated by heat (30 min at 80°C). Thereafter, bacterial DNA was extracted with cetytrimethylammonium bromide as described elsewhere (26). After digestion with 10 U of PvuII (Boehringer Mannheim GmbH, Mannheim, Germany), fragments were electrophoresed (1% agarose gel), transferred by Southern blotting onto a nylon membrane, and hybridized to a PCR-generated, genus-specific, digoxigenin-labelled probe (positions 9 to 341) (23). In the absence of a known PvuII restriction site within the 16S rDNA of M. celatum, restriction fragment length polymorphism (RFLP) patterns of all three isolates yielded two bands of equal sizes (Fig. 2).
FIG. 2.
RFLP analysis. Southern blot of genomic DNAs of all three clinical isolates of M. celatum, i.e., 1732 (lane 1), T322 (lane 2), and MI1581 (lane 3), which were digested with PvuII and hybridized with a 16S rDNA-specific probe. The rapidly growing Mycobacterium hassiacum (lane 4) is shown as a reference. Digoxigenin-labelled DNA molecular weight marker III (Boehringer Mannheim GmbH) was applied (lane MW).
Cloning of 16S rDNA amplification products.
The complete 16S rDNA from M. celatum was amplified from single colonies of isolate MI1581 with primers M285 and M261. Amplicons were directly cloned into a pCR 2.1 vector (Invitrogen Corp., San Diego, Calif.) and transformed into competent cells (TOP10F′; Invitrogen Corp.). Plasmid DNA extracted from 20 transformed colonies yielded seven interpretable sequences of both hypervariable regions (>500 bp). In contrast to direct sequencing, two unambiguous but slightly different types of sequences were obtained; types A and B showed four and five consecutive thymine residues, respectively, at positions 210 to 214 (Fig. 1 and 3). Both types could be further differentiated within positions 78 to 85. When the sequences of both types of clones were compared with published sequences for M. celatum, type A was identical to M. celatum type 3 (MC3RNA16S [GenBank accession no. Z46664]) and type B resembled M. celatum type 1 (MCRGDSA [GenBank accession no. L08169]), differing only at positions 77 to 79 (additional CCT) and 110 (A/G) (Fig. 3).
FIG. 3.
Alignment of a portion of 16S rDNA sequences (M. celatum types 1 (L08169) and 3 (Z46664) with clones derived from MI1581).
Advances in technology have made direct sequencing of 16S rDNA a powerful tool for the identification of bacteria in medical microbiology. 16S rDNA sequence analysis has become a standard in bacterial identification and in systematic bacterial taxonomy (1, 16). In mycobacteriology, this approach has been particularly valuable in clinical diagnosis, since slow growth hampers diagnosis by conventional tests (2, 10). Furthermore, 16S rDNA sequencing has contributed to the description of many new species of this family, among them M. celatum (3, 5, 15).
16S rDNA analysis for mycobacterial identification depends on the assumption that the sequences obtained from reference strains represent functional rRNA molecules typical of their taxa (12, 27). Although the level of intraspecific sequence variability is assumed to be low, some species are represented in international databases by an unexpectedly high number of deviating sequences (e.g., 11 different 16S rDNA sequences for Mycobacterium paratuberculosis). Some authors have traced these deviations back to errors in laboratory procedures, e.g., to sequencing techniques (7) or to subspecies variations (14, 21). However, recent findings indicate the presence of heterogeneity between different copies of 16S rDNA in Escherichia coli (6) and in Thermospora bispora (25), as well as in a slowly growing mycobacterium resembling M. terrae (19).
In M. celatum, we suspected heterogeneity when identical sequence patterns were obtained in different laboratories from clinical strains from different patients; all patterns were characterized by the same ambiguities starting at position 214, regardless of the strand sequenced. However, the phenotypical features of all strains were consistent with M. celatum type 1. Cloning of 16S rDNA finally resolved the ambiguities, yielding two types of unambiguous sequences. Interestingly, the two types were almost identical to the reference sequences for M. celatum type 1 (5) and M. celatum type 3 (3), respectively. We take it for granted that analysis of single colonies from pure isolates ruled out contamination. Thus, our results indicate the presence of two different 16S rRNA genes within the genome of M. celatum.
Little is known about heterogeneity in bacterial 16S rDNA. When present, distinct types of 16S rDNA seemed to be equally expressed (19, 25), suggesting their functionality. With regard to the origin of divergent 16S rDNAs, lateral gene transfer has been discussed (18) but has not yet revealed unsuspected relationships to other species (19, 25). However, in the present study both sequences obtained by cloning were identical with either one of the GenBank reference sequences for subtypes 1 and 3 of M. celatum, which may not be distinguished by conventional techniques (3, 5).
With regard to unambiguous identification of mycobacteria, sequence diversity in 16S rRNA genes may raise a problem. In agreement with Clayton et al. (7), we would recommend that to define a new bacterial or mycobacterial taxon, one should investigate multiple isolates. Sequence analysis of cloned genes may complement the characterization of ambiguous positions, and possible heterogeneity should be indicated in the published reference sequence by using ambiguity codes such as IUPAC (Nomenclature Committee of Biochemistry) (8). Thus, the quality of sequence data in public databases such as GenBank, EMBL, or the Ribosomal Database Project (17) will improve and lead to higher accuracy in the identification of clinically important bacteria, including mycobacteria, via 16S rDNA sequence analysis.
Acknowledgments
We thank Mohamed Rifai and Norbert Lehn for their active support and gratefully acknowledge the excellent technical assistance of Birgit Haber, Isabelle Jan, and E. Schmidt.
The work of S. Emler was supported by the Fondation Lancardis du Centre Valaisan de Pneumologie.
REFERENCES
- 1.Amann R, Ludwig W, Schleifer K-H. Identification of uncultured bacteria: a challenging task for molecular taxonomists. ASM News. 1994;60:360–365. [Google Scholar]
- 2.Böttger E C, Teske A, Kirschner P, Bost S, Chang H R, Beer V, Hirschel B. Disseminated “Mycobacterium genavense” infection in patients with AIDS. Lancet. 1992;340:76–80. doi: 10.1016/0140-6736(92)90397-l. [DOI] [PubMed] [Google Scholar]
- 3.Bull T J, Shanson D C, Archard L C, Yates M D, Hamid M E, Minnikin D E. A new group (type 3) of Mycobacterium celatum isolated from AIDS patients in the London area. Int J Syst Bacteriol. 1995;45:861–862. doi: 10.1099/00207713-45-4-861. [DOI] [PubMed] [Google Scholar]
- 4.Butler W R, Connor S P O, Yakrus M A, Gross W. Cross-reactivity of genetic probe for detection of Mycobacterium tuberculosis with newly described species Mycobacterium celatum. J Clin Microbiol. 1994;32:536–538. doi: 10.1128/jcm.32.2.536-538.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler W R, Connor S P O, Yakrus M A, Smithwick R W, Plikaytis B B, Moss C W, Floyd M M, Woodley C L, Kilburn J O, Vadney F. Mycobacterium celatum sp. nov. Int J Syst Bacteriol. 1993;43:539–548. doi: 10.1099/00207713-43-3-539. [DOI] [PubMed] [Google Scholar]
- 6.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 7.Clayton R, Granger Sutton A, Hinkle P S, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 8.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domenech P, Menendez M C, Garcia M J. Restriction fragment length polymorphisms of 16S rRNA genes in the differentiation of fast-growing mycobacterial species. FEMS Microbiol Lett. 1994;116:19–24. doi: 10.1111/j.1574-6968.1994.tb06669.x. [DOI] [PubMed] [Google Scholar]
- 10.Emler S, Böttger E C, Broers B, Cassis I, Perrin L, Hirschel B. Growth-deficient mycobacteria in patients with AIDS: diagnosis by analysis of DNA amplified from blood or tissue. Clin Infect Dis. 1995;20:772–775. doi: 10.1093/clinids/20.4.772. [DOI] [PubMed] [Google Scholar]
- 11.Emler S, Praplan P, Rohner P, Auckenthaler R, Hirschel B. Disseminated infection with Mycobacterium celatum. Schweiz Med Wochenschr. 1996;126:1062–1065. [PubMed] [Google Scholar]
- 12.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Balch W E, Tanner R S, Magrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Stahl D A, Luehrsen K R, Chen K N, Woese C R. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 13.Ji Y E, Colston M J, Cox R A. The ribosomal RNA (rrn) operons of fast-growing mycobacteria: primary and secondary structures and their relation to rrn operons of pathogenic slow-growers. Microbiology. 1994;140:2829–2840. doi: 10.1099/00221287-140-10-2829. [DOI] [PubMed] [Google Scholar]
- 14.Kirschner P, Kiekenbeck M, Meissner D, Wolters J, Böttger E. Genetic heterogeneity within Mycobacterium fortuitum complex species: genotypic criteria for identification. J Clin Microbiol. 1992;30:2772–2775. doi: 10.1128/jcm.30.11.2772-2775.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschner P, Meier A, Böttger E C. Genotypic identification and detection of mycobacteria: facing novel and uncultured pathogens. In: Persing D H, Tenover F, White T J, Smith T F, editors. Diagnostic molecular microbiology. Washington, D.C: American Society of Microbiology; 1993. pp. 173–190. [Google Scholar]
- 16.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F C, Böttger E. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T, Marsh T L, Woese C. The ribosomal database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mylvaganam S, Dennis P P. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ninet B, Monod M, Emler S, Pawlowski J, Metral C, Rohner P, Auckenthaler R, Hirschel B. Two different 16S rRNA genes in a mycobacterial strain. J Clin Microbiol. 1996;34:2531–2536. doi: 10.1128/jcm.34.10.2531-2536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piersimoni C, Tortoli E, De Sio G. Disseminated infection due to Mycobacterium celatum in patient with AIDS. Lancet. 1994;344:332. doi: 10.1016/s0140-6736(94)91369-2. [DOI] [PubMed] [Google Scholar]
- 21.Portaels F, Fonteyne P-A, de Beenhouwer H, de Rijk P, Guédenon A, Hayman J, Meyers W M. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J Clin Microbiol. 1996;34:962–965. doi: 10.1128/jcm.34.4.962-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reischl U, Pulz M, Ehret W, Wolf H. Pcr-based detection of mycobacteria in sputum samples using a simple and reliable DNA extraction protocol. BioTechniques. 1994;17:844–845. [PubMed] [Google Scholar]
- 23.Reischl U, Ruger R, Kessler C. Nonradioactive labeling and high-sensitive detection of pcr products. Mol Biotechnol. 1994;1:229–240. doi: 10.1007/BF02921691. [DOI] [PubMed] [Google Scholar]
- 24.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zwang Z S, Ramanan N. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J Bacteriol. 1997;179:3270–3276. doi: 10.1128/jb.179.10.3270-3276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman I G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Interscience Publishing Associates and John Wiley & Sons; 1992. pp. 2.10–2.12. [Google Scholar]
- 27.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]