Summary
Tandem mass tags data-dependent acquisition (TMT-DDA) as well as data-independent acquisition-based label-free quantification (LFQ-DIA) have become the leading workflows to achieve deep proteome and phosphoproteome profiles. We present a modular pipeline for TMT-DDA and LFQ-DIA that integrates steps to perform scalable phosphoproteome profiling, including protein lysate extraction, clean-up, digestion, phosphopeptide enrichment, and TMT-labeling. We also detail peptide and/or phosphopeptide fractionation and pre-mass spectrometry desalting and provide researchers guidance on choosing the best workflow based on sample number and input.
For complete details on the use and execution of this protocol, please refer to Koenig et al.1 and Martínez-Val et al.2
Subject areas: Protein Biochemistry, Proteomics, Mass Spectrometry
Graphical abstract
Highlights
-
•
Streamlined, modular, and semi-automated pipeline using TMT-DDA or LFQ-DIA
-
•
Guidance in choosing the best workflow based on sample number and input
-
•
Sensitive phosphoproteome profiling from limited biological material
-
•
Scalable method for high-throughput analysis of larger sample cohorts
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Tandem mass tags data-dependent acquisition (TMT-DDA) as well as data-independent acquisition-based label-free quantification (LFQ-DIA) have become the leading workflows to achieve deep proteome and phosphoproteome profiles. We present a modular pipeline for TMT-DDA and LFQ-DIA that integrates steps to perform scalable phosphoproteome profiling, including protein lysate extraction, clean-up, digestion, phosphopeptide enrichment, and TMT-labeling. We also detail peptide and/or phosphopeptide fractionation and pre-mass spectrometry desalting and provide researchers guidance on choosing the best workflow based on sample number and input.
Before you begin
Cultivation of cell lines
Timing: 5 days
-
1.
Culture cells in the appropriate media supplemented with 10% (v/v) of fetal bovine serum (FBS Gibco, Cat#10270106), 100 μg/mL penicillin and streptomycin (Invitrogen, Cat#15140-122). Here, HeLa (ATCC CCL-2) were cultured in DMEM (Fisher Scientific, Cat#31966047).
-
2.
Keep cell lines in a humidified incubator at 37°C, with 5% CO2 and check regularly for mycoplasma using the EZ PCR Mycoplasma detection Kit (Biological Industries, Cat#20-700-20).
Cultivation of single spheroids
Timing: 5 days
-
3.
Seed 20,000 HCT116 cells (ATCC CCL-247) on ultra-low attachment 96-well plates (Corning Costar, Merck #CLS3474).
-
4.
Culture in 90% of DMEM, supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Gibco, Cat#10270106) and 100 μg /mL penicillin and streptomycin (Invitrogen, Cat#15140-122).
-
5.
Culture spheroids for at least 96 h at 37°C, in a humidified incubator with 5% CO2.
-
6.
Refresh cell medium at least every 48 h, by aspirating half the old medium (making sure not to alter the spheroid) and adding the same amount of fresh medium.
Note: Cell cultivation medium may be stored at 4°C for several weeks.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Cell lines | ||
| Immortalized cancer cell line (HeLa or HCT116) | ATCC CCL-2 or ATCC CCL-247 | |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s modified Eagle’s medium (DMEM) | Fisher Scientific | Cat#31966047 |
| Penicillin/streptomycin | Life Technologies | Cat#15140-122 |
| Fetal bovine serum | Life Technologies | Cat#10270106 |
| Lysyl endopeptidase | Fujifilm Wako | Cat#129-02541 |
| Sequencing grade modified trypsin | Promega | Cat#V511X |
| MagReSyn Hydroxyl (20 mg/mL in 20% EtOH) | ReSyn Biosciences | Cat#MR-HYX010 |
| MagReSyn Ti-IMAC HP (20 mg/mL in 20% EtOH) | ReSyn Biosciences | Cat#MR-THP010 |
| MagReSyn Zr-IMAC HP (20 mg/mL in 20% EtOH) | ReSyn Biosciences | Cat#MR-ZHP010 |
| Triethylammonium bicarbonate (TEAB) | Sigma-Aldrich | Cat#90360 |
| HEPES pH 8.0 | Sigma-Aldrich | Cat#H3375 |
| Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) | Sigma-Aldrich | Cat#C4706 |
| Ammonium bicarbonate (AmmBic) | Sigma-Aldrich | Cat#T7408 |
| Trifluoroacetic acid (TFA) | Sigma-Aldrich | Cat#302031 |
| Formic acid (FA) – for LC-MS | Sigma-Aldrich | Cat#00940 |
| Sodium dodecyl sulfate (SDS) | Fisher Scientific | Cat#10607443 |
| 2-Chloroacetamide (CAA) | Sigma-Aldrich | Cat#C0267 |
| Glycolic acid (GA) | Sigma-Aldrich | Cat# 420581 |
| Acetonitrile (MeCN) – for LC-MS | Sigma-Aldrich | Cat#1.00029 |
| Tris-HCl | Sigma-Aldrich | Cat#PHG0002 |
| Ammonia water (NH4OH 25% aqueous solution) | Sigma-Aldrich | Cat#1054321011 |
| Ethanol (EtOH) > 99.9% | Sigma-Aldrich | Cat#51976 |
| 1-isopropanol or 2-propanol analysis | Sigma-Aldrich | Cat# 1070222511 |
| TMTpro 16-plex Label Reagent | Thermo Fisher Scientific | Cat#A44520 |
| Hydroxylamine hydrochloride (50.0%) | Thermo Fisher Scientific | Cat#90115 |
| Critical commercial assays | ||
| Pierce Microplate BCA Protein Assay Kit - Reducing Agent Compatible | Thermo Fisher Scientific | Cat#23252 |
| EZ PCR Mycoplasma Detection Kit | Biological Industries | Cat#20-700-20 |
| Software and algorithms | ||
| Spectronaut | https://biognosys.com/software/spectronaut/ | |
| MaxQuant | https://www.maxquant.org/ | |
| Perseus | https://maxquant.net/perseus/ | |
| Peptide Collapse plugin | https://github.com/AlexHgO/Perseus_Plugin_Peptide_Collapse | |
| UniProt database human | UniProt | https://www.uniprot.org/proteomes/UP000005640 Proteome ID: UP000005640 |
| Other | ||
| Ultra-low attachment 96-well plates | Corning Costar | Cat#CLS3474 |
| Cell scraper | TPP | Cat#99003 |
| Micro centrifuge (1.5–2 mL tubes) | Various suppliers | N/A |
| Magnetic separator | Thermo Fisher Scientific | Cat#12321D |
| Incubator for cells | Various suppliers | N/A |
| Biological safety cabinet for cell culture | Various suppliers | N/A |
| Chemical safety cabinet | Various suppliers | N/A |
| Eppendorf ThermoMixer F1.5 or equivalent | Eppendorf | Cat#5384000012 |
| SpeedVac concentrator | Thermo Fisher Scientific | Cat#SPD111V-115 |
| Tip Horn for Model 120 Sonic Dismembrator (8-tip probe sonicator) or equivalent dry sonicator | Fisherbrand | Cat#FB4599 |
| NanoDrop 2000 spectrophotometer | Thermo Fisher Scientific | Cat#ND-2000 |
| Computer station with minimum requirements of used Spectronaut version | Various suppliers | N/A |
| KingFisher Flex (Apex, Duo Prime) system | Thermo Fisher Scientific | Cat#A31508 (5400920, 5400110) |
| KingFisher Flex Microtiter deep-well 96 plate | Thermo Fisher Scientific | Cat#95040460 |
| Sep-Pak tC18 96-well plate, 40 mg sorbent per well | Waters | Cat#186003966 |
| Extraction plate manifold for 96-well plates | Waters | Cat#186001831 |
| Multifuge X3 | Thermo Fisher Scientific | Cat#75004500 |
| Carriers for microplates | Thermo Fisher Scientific | Cat#75003617 |
| MultiScreenHTS HV filter plate, 0.45 μm | Merck | Cat#MSHVN4550 |
| KingFisher Flex 96 Heating Block (compatible with deep-well microtiter plate) | Thermo Fisher Scientific | Cat#24075430 |
| KingFisher Flex Microtiter standard 200 μL 96 plate | Thermo Fisher Scientific | Cat#97002540 |
| KingFisher Flex 96 Heating Block (compatible with 200 μL microtiter plate) | Thermo Fisher Scientific | Cat#24075420 |
| C18 stage tips: P200 LoBind pipette tips | Merck; 3M | Cat#CLS4154-4X960EA |
| C18 stage tips: Empore C18 extraction disk | Fisher Scientific | Cat#13110018 |
| UltiMate 3000 high-performance liquid chromatography system | Thermo Fisher Scientific | N/A |
| Acquity CSH C18 1.7 μm × 1 mm × 150 mm column | Waters | Cat#186006935 |
| Evosep One HPLC system | Evosep | N/A |
| Evosep endurance column (15 cm × 150 μm, 1.9 μm) | Evosep | EV1106 |
| Evosep performance column (8 cm × 150 μm, 1.5 μm) | ||
| Evosep performance column (15 cm × 75 μm, 1.9 μm) | Evosep | |
| Evosep | EV1109 | |
| EV1112 | ||
| Aurora Elite column (15 cm × 75 μm, 1.7 μm) | IonOpticks | Cat#AUR3-15075C18-TS |
| Sonation Column Oven PRSO-V2 | ||
| Butterfly heater | Sonation lab solutions | |
| MSWil | PRSO-V2 | |
| Cat#PST-ES-BPH-20 | ||
| Evosep stainless steel emitter 30 μm i.d. | Evosep | EV1086 |
| Evosep fused silica emitter 10 μm i.d. | Evosep | EV1111 |
| Thermo Scientific Exploris 480 mass spectrometer or any other mass spectrometer with high precision MS1 spectrum recording | Thermo Fisher Scientific | |
Materials and equipment
-
•
Stop and go extraction (Stage) tip assembly.
C18 Stage tips: P200 low bind pipette tips (Merck, Cat#CLS4154-4X960EA) with three Empore Disk C18 (CDS, Cat#13110018) cores assembled according to Rappsilber et al.3
-
•
Equipment setup HPLC for offline peptide fractionation.
UltiMate 3000 high-performance liquid chromatography (HPLC) system (Thermo Fisher Scientific) with Chromeleon software. Acquity CSH C18 1.7 μm × 1 mm × 150 mm column (Waters, Cat#186006935).
-
•
Equipment setup mass spectrometer.
Thermo Scientific Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific) connected to an Evosep One (Evosep Biosystems) with pre-programmed gradients 30SPD, 60SPD, 40SPD (WHISPER) and 20SPD (WHISPER). The 30SPD run with an Evosep endurance column (15 cm × 150 μm, 1.9 μm, EV1106) and the 60SPD on an Evosep performance column (8 cm × 150 μm, 1.5 μm, EV1109), both packed with ReproSil-Pur C18 beads by Dr. Maisch and connected to a stainless-steel emitter (30 μm i.d., Evosep, EV1086) without temperature control. The WHISPER gradients run with an IonOpticks Aurora column (15 cm × 75 μm, 1.7 μm, AUR3-15075C18) or an Evosep performance column (15 cm × 75 μm, 1.9 μm, EV-1112) connected to a fused silica emitter (10 μm i.d., Evosep, EV1087). The Evosep columns are mounted on an EASY-Spray source (Thermo Fisher Scientific). 20 and 40SPD are used with a Butterfly heater (MSWil, Cat#PST-ES-BPH-20) and the temperature is set at 40°C. The Aurora IonOpticks column is mounted on a nanoflex source (Thermo Fisher Scientific) and heated with a Sonation Oven PRSO-V2 (Cat#PROSO-V2) at 50°C.
-
•
Cell cultivation medium.
| Reagent | Final concentration | Amount (555 mL) |
|---|---|---|
| Dulbecco’s modified Eagle medium (DMEM) | N/A | 500 mL |
| Penicillin /streptomycin | 1% (v/v) | 5 mL |
| Fetal bovine serum | 10% (v/v) | 50 mL |
Note: Ideal cell medium should be selected based on cell line used for the experiment. Here DMEM is recommended for both HeLa and HCT116 cells.
-
•
SDS lysis buffer.
| Reagent | Final concentration | Amount (10 mL) |
|---|---|---|
| SDS (20% diluted in MilliQ water) | 5% (w/v) | 2,5 mL |
| Tris/HCL pH 8.5 (1 M) | 100 mM | 1 mL |
| Tris (2-carboxyethyl) phosphine (TCEP, 0.5 M diluted in MilliQ water)) | 5 mM | 100 μL |
| Chloroacetamide (CAA, 550 mM diluted in 25 mM AmmBic)) | 10 mM | 180 μL |
| MilliQ water | N/A | 6.72 mL |
CRITICAL: SDS is corrosive and flammable, use dust mask type N95 (US), eyeshield, faceshield and gloves.
-
•
PAC bind buffer.
| Reagent | Final concentration | Amount (10 mL) |
|---|---|---|
| MeCN | 100% (v/v) | 10 mL |
| Up to 700 μL is needed for each sample | ||
CRITICAL: MeCN is highly flammable.
-
•
PAC wash buffer 1.
| Reagent | Final concentration | Amount (150 mL) |
|---|---|---|
| MeCN | 95% (v/v) | 142.5 mL |
| MilliQ water | 7.5 mL | |
| ∼3 mL of 95% MeCN is needed for each sample | ||
CRITICAL: MeCN is highly flammable.
-
•
PAC wash buffer 2.
| Reagent | Final concentration | Amount (100 mL) |
|---|---|---|
| EtOH | 70% (v/v) | 70 mL |
| MilliQ water | 30 mL | |
| ∼2 mL of 70% EtOH is needed for each sample | ||
CRITICAL: EtOH is highly flammable.
-
•
PAC digestion buffer.
| Reagent | Final concentration | Amount (15 mL) |
|---|---|---|
| AmmBic (1 M, pH 8.5) | 50 mM | 750 μL |
| MilliQ water | 14.250 mL | |
| Lys-C (0.1 μg/μL) | 1 μg per 100 μg protein (1:100 ratio). For > 150 μg protein use 1:500 ratio | 10 μL per 100 μg of protein |
| Trypsin (0.1 μg/μL) | 2 μg per 100 μg protein (1:50 ratio). For > 150 μg protein use 1:250 ratio | 20 μL per 100 μg of protein |
| Prepare freshly before each use. ∼ 300 μL PAC digestion buffer is needed for each sample | ||
-
•
Hydroxylamine Hydrochloride.
| Reagent | Final concentration | Amount (1 mL) |
|---|---|---|
| Hydroxylamine Hydrochloride (50.0%) | 1% (v/v) aqueous solution | 20 μL |
| MilliQ water | 980 μL | |
| Prepare freshly before each use. | ||
CRITICAL: Hydroxylamine Hydrochloride is corrosive. Use dust mask type N95 (US), eyeshield, faceshield and gloves.
-
•
Desalting SPE buffer A.
| Reagent | Final concentration | Amount (3 mL) |
|---|---|---|
| 10% TFA | 0.1% (v/v) aqueous solution | 30 μL |
| MilliQ water | 2970 μL | |
| Prepare freshly before each use. ∼ 3 mL desalting buffer A will be required per sample. | ||
CRITICAL: TFA is highly corrosive. Use eyeshield, faceshield, gloves and work under a fume hood. To minimize plastic and polymer contamination, use glass pipettes when working with >50% TFA.
-
•
Desalting SPE buffer B.
| Reagent | Final concentration | Amount (3 mL) |
|---|---|---|
| MeCN | 100% (v/v) | 3000 μL |
| ∼ 2 mL desalting buffer B will be required per sample. | ||
CRITICAL: MeCN is highly flammable.
-
•
Desalting SPE buffer C.
| Reagent | Final concentration | Amount (3 mL) |
|---|---|---|
| MeCN | 40% (v/v) | 1200 μL |
| MilliQ water | 1800 μL | |
| Prepare freshly before each use. ∼ 0.15 mL desalting buffer C will be required per sample. | ||
CRITICAL: MeCN is highly flammable.
-
•
Desalting SPE buffer D.
| Reagent | Final concentration | Amount (3 mL) |
|---|---|---|
| MeCN | 60% (v/v) | 1800 μL |
| MilliQ water | 1200 μL | |
| Prepare freshly before each use. ∼ 0.15 mL desalting buffer D will be required per sample. | ||
CRITICAL: MeCN is highly flammable.
-
•
Microflow buffer A.
| Reagent | Final concentration | Amount (50 mL) |
|---|---|---|
| AmmBic (1 M diluted in MilliQ water) | 5 mM | 250 μL |
| MilliQ water | 49.75 mL | |
| Prepare freshly before each use. | ||
CRITICAL: Filter the buffer using 0.2 μm filter prior use to avoid HPLC lines or column blockages.
-
•
Microflow buffer B.
| Reagent | Final concentration | Amount (50 mL) |
|---|---|---|
| MeCN | 100% (v/v) | 50 mL |
| ∼10 mL 100% MeCN is needed for each run | ||
CRITICAL: MeCN is highly flammable.
-
•
Microflow sample resuspension buffer.
| Reagent | Final concentration | Amount (10 mL) |
|---|---|---|
| AmmBic (1 M diluted in MilliQ water) | 25 mM | 250 μL |
| MilliQ water | 9750 μL | |
| Prepare freshly before each use. | ||
-
•
Phosphopeptide Bind buffer (Ti-IMAC).
| Reagent | Final concentration | Amount (50 mL) |
|---|---|---|
| Glycolic acid (GA) | 1 M | 3.8 g |
| MeCN | 80% (v/v) | 40 mL |
| TFA | 5% (v/v) aqueous solution | 2.5 mL |
| MilliQ water | 7.5 mL | |
| Prepare freshly before each use. ∼1.2 mL of Phosphopeptide Bind buffer (Ti-IMAC) is needed for each sample | ||
CRITICAL: GA is corrosive, use eyeshields, faceshields, gloves, respirator cartridge type N100 (US), type P1 (EN143) respirator filter, type P3 (EN 143) respirator cartridges. MeCN is highly flammable and TFA is corrosive.
-
•
Phosphopeptide Bind buffer (Zr-IMAC).
| Reagent | Final concentration | Amount (50 mL) |
|---|---|---|
| Glycolic acid (GA) | 0.1 M | 0.38 g |
| MeCN | 80% (v/v) | 40 mL |
| TFA | 5% (v/v) aqueous solution | 2.5 mL |
| MilliQ water | 7.5 mL | |
| Prepare freshly before each use. ∼1.2 mL of Phosphopeptide Bind buffer (Zr-IMAC) is needed for each sample |
||
CRITICAL: GA is corrosive, use eyeshields, faceshields, gloves, respirator cartridge type N100 (US), type P1 (EN143) respirator filter, type P3 (EN 143) respirator cartridges. MeCN is highly flammable and TFA is corrosive.
-
•
Phosphopeptide Wash buffer 1.
| Reagent | Final concentration | Amount (20 mL) |
|---|---|---|
| MeCN | 80% (v/v) | 16 mL |
| TFA | 1% (v/v) aqueous solution | 200 μL |
| MilliQ water | 3.8 mL | |
| Prepare freshly before each use. ∼0.5 mL of Phosphopeptide Wash buffer 1 is needed for each sample |
||
CRITICAL: TFA is highly corrosive. Use eyeshield, faceshield, gloves and work under a fume hood. MeCN is highly flammable.
-
•
Phosphopeptide Wash buffer 2.
| Reagent | Final concentration | Amount (20 mL) |
|---|---|---|
| MeCN | 10% (v/v) | 2 mL |
| TFA | 0.2% (v/v) aqueous solution | 40 μL |
| MilliQ water | 17.96 mL | |
| Prepare freshly before each use. ∼0.5 mL of Phosphopeptide Wash buffer 2 is needed for each sample | ||
CRITICAL: TFA is highly corrosive. Use eyeshield, faceshield, gloves and work under a fume hood. MeCN is highly flammable.
-
•
Phosphopeptide Elution buffer.
| Reagent | Final concentration | Amount (10 mL) |
|---|---|---|
| NH4OH 25% aqueous solution | 1% (v/v) | 400 μL |
| MilliQ water | 9.60 mL | |
| Prepare freshly before each use. ∼0.2 mL of Phosphopeptide Elution buffer is needed for each sample | ||
CRITICAL: NH4OH is corrosive and toxic. Use eyeshield, faceshield, gloves and work under a fume hood. MeCN is highly flammable.
-
•
Stage tip equilibration buffer 2 (STEB2).
| Reagent | Final concentration | Amount (20 mL) |
|---|---|---|
| NH4OH 25% aqueous solution | 20 mM | 30 μL |
| 100% MeCN | 20 mL | |
| Prepare freshly before each use. |
-
•
Stage tip equilibration buffer 3 (STEB3).
| Reagent | Final concentration | Amount (40 mL) |
|---|---|---|
| NH4OH 25% aqueous solution | 20 mM | 60 μL |
| MilliQ water | 40 mL | |
| Prepare freshly before each use. |
-
•
Evotip buffer A.
| Reagent | Final concentration | Amount (100 mL) |
|---|---|---|
| LC-MS grade H2O | 99.9% (v/v) | 99.9 mL |
| LC-MS grade FA | 0.1% (v/v) | 100 μL |
-
•
Evotip buffer B.
| Reagent | Final concentration | Amount (100 mL) |
|---|---|---|
| LC-MS grade MeCN | 99.9% (v/v) | 99.9 mL |
| LC-MS grade FA | 0.1% (v/v) | 100 μL |
Note: LC-MS grade H2O or MeCN supplemented with 0.1% FA may be purchased premixed
CRITICAL: MeCN is highly flammable. If premixed LC-MS buffers are not available, use formic acid from ampules and pipette fitted with disposable glass micropipettes (e.g., BRAND disposable BLAUBRAND micropipettes, intraMark).
Step-by-step method details
Sample generation: Cell harvest with single-step lysis, reduction and alkylation
Timing: 0.25 days
CRITICAL: The cells should be grown under sterile conditions in a biological safety cabinet. Ideally, prepare 5 or more replicates per biological condition.
Note: the procedure and lysis buffer are applicable to protein extraction from most mammalian cells by varying the sonication time and intensity to best suit the sample source
-
1.
Preheat SDS lysis buffer to 95°C.
-
2.Add the boiling SDS lysis buffer to the samples:
-
a.If working with adherent cell lines:
-
i.Remove medium from cells and wash them twice with phosphate salt buffered solution (PBS) (Life Technologies, Cat#14190-094).
-
ii.Add boiling SDS lysis buffer directly to the plate and rapidly scrape the cells.
-
iii.Transfer the lysate to an Eppendorf tube. For a P15 cell plate at ∼ 80% confluency, use 300 μL of SDS lysis buffer.
-
i.
-
b.If working with suspension cell lines:
-
i.Transfer the cells to a falcon tube (or a tube appropriate to the cell volume).
-
ii.Pellet the cells by centrifugation at 400 × g for 3 min.
-
iii.Wash the cell pellet with PBS twice and pellet the cells again by centrifugation (400 × g, 3 min).
-
iv.Remove the PBS and add boiling SDS buffer to the pellet. For a T-175, corresponding to about 20 million cells, use 300 μL of SDS lysis buffer.
-
i.
-
c.If working with spheroids:
-
i.Aspirating half the medium and add 150 μL PBS.
-
ii.Repeat this process 4 times to remove traces of media.
-
iii.Transfer the spheroid to an Eppendorf tube and add boiling SDS buffer. For a single spheroid, use 200 μL of SDS lysis buffer.
-
i.
-
a.
CRITICAL: The PAC digestion is concentration dependent. It is recommended to lyse the cells in the lowest possible volume to keep the lysate concentrated and ensure an efficient digestion. The volume of lysis buffer given in this protocol should be scaled with the size of the dish/flask used.
-
3.
Incubate the lysates for 10 min at 95°C with mixing at 850 rpm in ThermoMixer (Eppendorf, Cat#538400001).
Note: During boiling pressure can form inside the tube and lids can randomly open. To avoid this use “safe-lock” Eppendorf. When collecting the samples, carefully release the pressure by opening the lids.
-
4.
Reduce viscosity by sonicating using a micro-tip probe (1 min, 1 s ON, 1 s OFF, 50% amplitude). If samples are in a 96-well format, an 8-tip probe can be used. Problem 1.
-
5.
To clarify the lysate, centrifuge for 5 min as 10,000 g and transfer the supernatant to a clean tube.
Pause point: it is possible to store the cell/ spheroid extracts at −80°C.
Protein Aggregation capture (PAC): On-bead protein precipitation, clean-up and digestion automated on KingFisher Flex (Apex, Duo, Prime) systems
Timing: 1 day
-
6.
Quantify cell-extracted protein concentration using Pierce microplate BCA Protein Assay Kit - Reducing Agent Compatible (Thermo Fisher Scientific, #23227) according to manufacturer’s protocol: (https://www.thermofisher.com/order/catalog/product/23227).
CRITICAL: If using another quantification kit or method make sure it is compatible with reducing agents such as TCEP and detergents such as SDS.
-
7.Prepare PAC buffers as per materials and equipment section:
-
a.PAC Bind buffer: 100% MeCN.
-
b.PAC Wash buffer 1: 95% MeCN.
-
c.PAC Wash buffer 2: 70% EtOH.
-
d.PAC Digest buffer: 50 mM AmmBic supplemented with Lys-C and Trypsin (1:500 and 1:250 respectively for 12 h digest of lysate inputs > 50 μg of protein at 37°C; 1:100 and 1:50 respectively for 6 h digest of lysate inputs < 50 μg of protein) at 37°C.
-
a.
Note: depending on downstream steps and input material the ideal digest buffer may change e.g., for downstream TMT labeling without peptide quantification use 100 mM HEPES buffer for PAC digest whilst for LFQ from low input material (i.e. single spheroid) where PAC is directly coupled to phosphopeptide enrichment it is recommended to utilize 50 mM TEAB as digest buffer instead of AmmBic.
CRITICAL: Add Lys-C and Trypsin fresh just before starting the KingFisher program.
-
8.
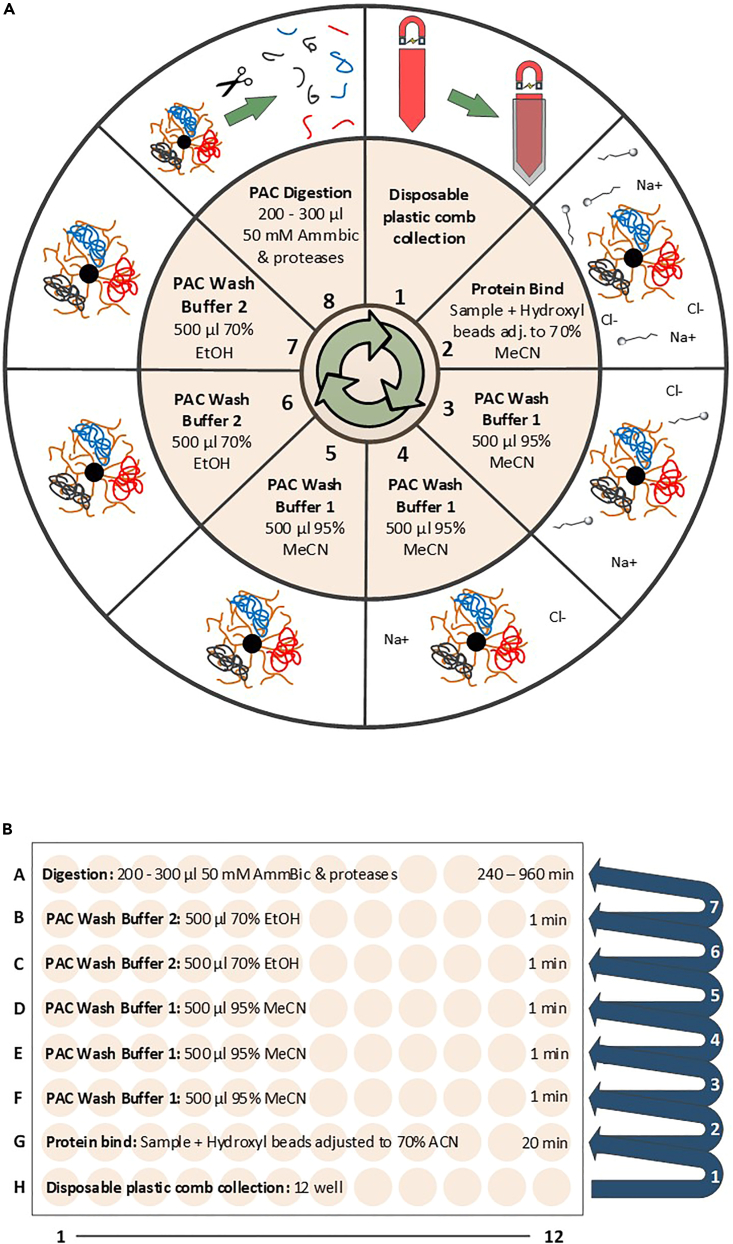
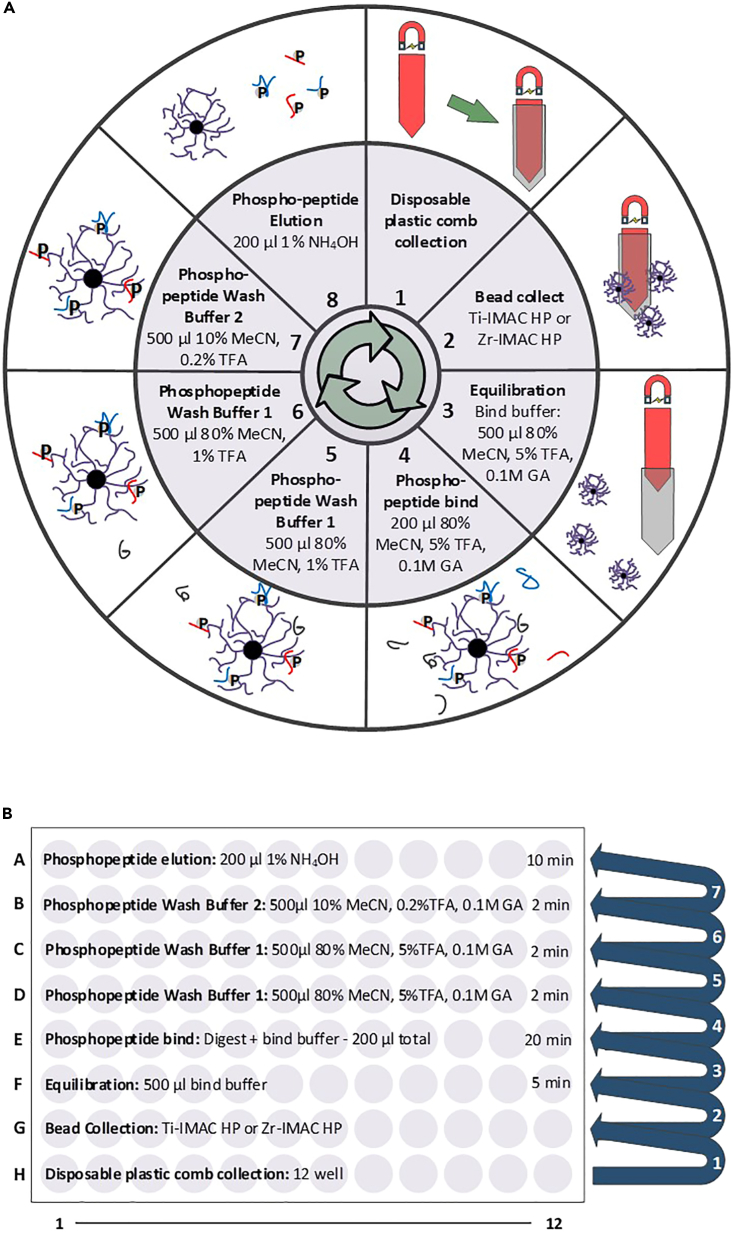
Prepare the KingFisher plates with beads, samples and solutions as per Table 1/ Figure 1A (KingFisher Flex or Apex models) or Table 2/ Figure 1B (KingFisher Duo or Presto models).
Note: MagReSyn Hydroxyl microparticles do not have to be pre-equilibrated prior to use and can be added directly with storage solution (20% EtOH) to the KingFisher plate. When calculating the volume of MeCN (bind solvent) to be added in order to initiate protein capture (final of 70% MeCN required) the MagReSyn Hydroxyl bead volume can be ignored, and calculated based only on sample volume (see Table 3).
Table 1.
Plates with their contents used for the automated PAC procedure on KingFisher Flex or Apex models
| KingFisher 96 deep-well plate order | |||
|---|---|---|---|
| Plate number and name | μL/ well | Buffer | Time per step (min) |
| 1: Tip plate | |||
| 2: MagReSyn Hydroxyl, Sample lysate, Bind solvent | Varies based on sample volume | 70% MeCN final | 20 |
| 3: Wash | 500 | Wash buffer 1 | 2.5 |
| 4: Wash | 500 | Wash buffer 1 | 2.5 |
| 5: Wash | 500 | Wash buffer 1 | 2.5 |
| 6: Wash | 500 | Wash buffer 2 | 2.5 |
| 7: Wash | 500 | Wash buffer 2 | 2.5 |
| 8: Digest | 200–300 | Digest buffer | 120–960 |
Figure 1.
Overview of automated PAC-based protein capture, clean-up and digestion
(A and B) (A) KingFisher Flex /Apex layout (processing of up to 96 samples in parallel) or (B) KingFisher Duo /Presto layout (processing of up to 12 samples in parallel).
Table 2.
Plates with their contents used for the automated PAC procedure on KingFisher Duo and Presto models
| KingFisher 96 deep-well plate order | |||
|---|---|---|---|
| Plate row number and name | μL/ well | Buffer | Time per step (min) |
| H: Tip plate | |||
| G: MagReSyn Hydroxyl, sample lysate, bind solvent | Varies based on sample volume | 70% MeCN final | 20 |
| F: Wash | 500 | Wash buffer 1 | 1 |
| E: Wash | 500 | Wash buffer 1 | 1 |
| D: Wash | 500 | Wash buffer 1 | 1 |
| C: Wash | 500 | Wash buffer 2 | 1 |
| B: Wash | 500 | Wash buffer 2 | 1 |
| A: Digest | 200–300 | Digest buffer | 240–960 |
Table 3.
PAC bead amount and Bind solvent volume based on protein lysate amount and volume
| Lysate amount (μg) | Lysate volume (μL) | Bead amount (μg) | Bead volume (μL) | Bind solvent volume (μL) (100% MeCN) |
|---|---|---|---|---|
| 5 | 40 | 200 | 10 | 92 |
| 10 | 40 | 200 | 10 | 92 |
| 20 | 40 | 200 | 10 | 92 |
| 50 | 100 | 200 | 10 | 230 |
| 100 | 200 | 200 | 10 | 460 |
| 200 | 300 | 400 | 20 | 700 |
| 1000 | 300 | 2000 | 100 | 700 |
Note: for low protein amounts (<5 μg) it is recommended to reduce the digestion volume to 200 μL and digest a maximum of 6 h.
Note: The recommended protein to bead ratio is 1:2 (for protein amounts ≥ 100 μg). For protein amounts < 100 μg use a constant Hydroxyl bead volume of 10 μL (see Table 3).
CRITICAL: When dispensing the beads make sure storage container is frequently mixed to minimize beads from settling and thus ensure equal amount of beads is distributed in each well. For large scale experiments beads can be dispensed using multi-channel pipette. Frequent mixing using up/down pipetting is recommended in this case in order to keep beads in suspension.
Note: MagReSyn Hydroxyl is formulated at 20 mg/mL in 20% EtOH
CRITICAL: For KingFisher deep well plates total volume (lysate and bind solvent) should not exceed 1 mL.
-
9.
Run the KingFisher method and once complete add formic acid to a final of 2% FA, or TFA to a final of 1%, to each sample in order to quench Lys-C / Trypsin activity and stop the digestion by reducing the pH to below 4. Problem 2.
Note: Formic acid is recommended for low input lysate and thus digest amounts (≤ 50 μg) where direct phosphopeptide enrichment is performed without desalting. TFA is recommended in cases where desalting will be performed prior to phosphopeptide enrichment (lysate/ digest amount ≥ 50 μg).
Note: For manual sample processing using PAC follow the same steps as described above using 2 mL microcentrifuge tubes together with a suitable magnetic separator such as DynaMag-2 Magnet (Thermo Fisher Scientific, Cat#12321D). During the wash steps keep the beads on the magnetic separator. As a fail-safe, to prevent SDS contamination that may coat the side walls of the microcentrifuge tubes, transfer the protein-bead aggregates to new 2 mL microcentrifuge tubes after the second wash and before continuing with washes three to five.
CRITICAL: Sample can be clarified by centrifugation (10 min, 10,000 g) in particular if white precipitates are observed. These are likely due to residual nucleic acid or lipid contamination.
-
10.
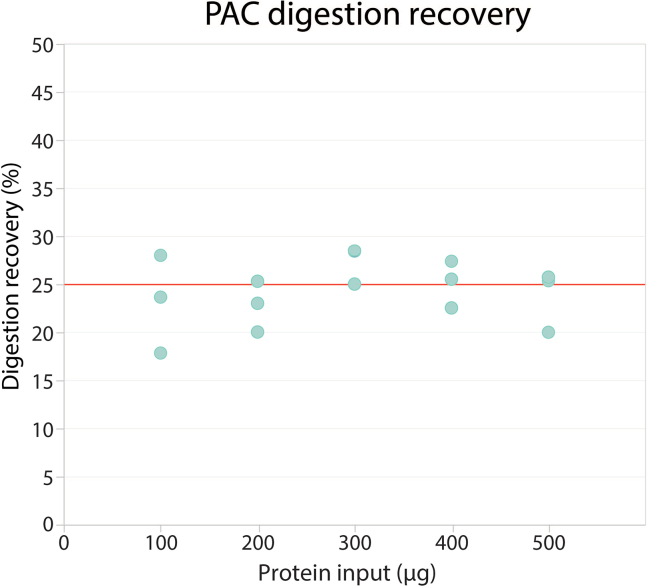
For label-free proteome analysis, transfer the equivalent of 750 ng of peptide digest directly to Evotips (Step 57). No quantification is performed on the peptide digest, instead we consider the recovery from the PAC digestion to be 25% (for 100 μg of protein input, we would recover 25 μg of peptides) (Figure 2) and load accordingly.
-
11.Keep the remaining peptides and proceed to:
-
a.If input digest > 50 μg and/or TMT-based scheme will be used, continue to step 12 for peptide desalting.Note: It is possible to proceed directly to TMT labeling, without desalting. If so, it is important that the lysis and digest buffers are amine free i.e. utilize TEAB or HEPES instead of Tris-HCl or AmmBic, and the digest is not acidified in step 9. Please note that quantification of peptide amount is not accurate without first desalting. This can affect the ability to do equal TMT channel loading, which will in-turn increase the technical variability of the TMT workflow.
-
b.If input digest amount is ≤ 50 μg and the DIA label-free scheme will be used, concentrate the peptide in a SpeedVac at 45°C (1–4 h) until volume is < 20 μL. Then, proceed directly to phosphopeptide enrichment (step 37). Problem 3.Note: Extended drying times can result in phosphopeptide losses. In laboratories where freeze-drying equipment is available this could be utilized instead of vacuum drying.
 CRITICAL: if proceeding to phosphopeptide enrichment it is recommended to avoid complete peptide drying of your peptide sample, but rather only reduce the sample volume to ∼ 20 μL, followed by dilution using phosphopeptide enrichment bind buffer (final 200 μL).
CRITICAL: if proceeding to phosphopeptide enrichment it is recommended to avoid complete peptide drying of your peptide sample, but rather only reduce the sample volume to ∼ 20 μL, followed by dilution using phosphopeptide enrichment bind buffer (final 200 μL). Pause point: it is possible to store acidified peptides at −20°C.
Pause point: it is possible to store acidified peptides at −20°C.
-
a.
Figure 2.
PAC with desalting digestion recovery for input amounts ranging from 100 μg to 500 μg HeLa lysate
The protein concentration was measured using BCA and the peptide concentration was estimated after peptide desalting using absorbance at 280 nm.
Desalting
Timing: 2 h
Note: desalting is necessary before TMT-labeling if the digest buffer is not amine free and if TMT channel load needs to be adjusted by performing peptide quantification. It is further recommended for non-labeled peptides where digest amounts is ≥ 50 μg. Below this amount, one can proceed directly to phosphopeptide enrichment (step 37).
-
12.Prepare the following solutions for desalting as per materials and equipment section:
-
a.SPE buffer A: 0.1% TFA (stable up to 4 weeks at 4°C).
-
b.SPE buffer B: 100% MeCN.
-
c.SPE buffer C: 40% MeCN.
-
d.SPE buffer D: 60% MeCN.
-
a.
-
13.
Desalt peptides using Sep-Pak tC18 96-well plate (Waters, Cat#186003966).
-
14.
Condition the plate by adding 1 mL of SPE Buffer B (100% MeCN).
-
15.
Wash out the MeCN by adding 1 mL of SPE buffer A.
-
16.
Repeat step 15 for a total of 3 times.
-
17.
Load the pre-acidified samples, maximum volume 1 mL and run through Sep-Pak tC18 bed.
-
18.
Wash by adding 1 mL of SPE buffer A (0.1% TFA).
-
19.
Repeat step 18 for a total of 3 washes.
Pause point: Peptides can be stored on the Sep-Pak at +4°C.
-
20.
Place an empty sample collection plate (Thermo Fisher Scientific KingFisher, Cat#95040450) below the Sep-Pak tC18 plate. Elute peptides by adding 150 μL SPE buffer C (40% MeCN), followed by 150 μL of SPE Buffer D (60% MeCN).
CRITICAL: acetonitrile concentration > 60% can result in elution of contaminants and interference with downstream steps such as phosphopeptide enrichment.
Note: to run each buffer through the packed Sep-Pak tC18 bed place in a 96-well extraction plate manifold adaptor (Waters, Cat#186001831) and apply vacuum as per manufacturer instructions. Alternatively, a swing-out rotor centrifuge (Multifuge X3, Thermo Scientific, Cat#75004500) with microplate adaptor (Thermo Scientific, Cat#75003795) can be used. In this case, spin the plates for 1 min at 500 g.
-
21.
Pool the two eluates and dry using a SpeedVac concentrator at 45°C (1–4 h).
CRITICAL: if proceeding to phosphopeptide enrichment it is recommended to avoid complete peptide drying but rather reduce sample volume to ∼ 20 μL followed by dilution using phosphopeptide enrichment bind buffer (200 μL).
TMT labeling
Timing: 2 h
Note: if performing LFQ data acquisition, proceed directly to phosphopeptide enrichment (step 37)
CRITICAL: it is important to randomize the samples in order to minimize labeling bias. Refer to Brenes et al.4 for indications of proper experiment allocation in a TMT setup.
-
22.
Quantify the peptides using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Cat#ND-2000) at 280 nm to ensure equal peptide loading per TMT channel.
-
23.
Re-suspend TMT reagent stock (5 mg, Thermo Scientific, Cat#A44520) in 200 μL 100% MeCN.
CRITICAL: Aliquots of TMT reagents were stored dry at −80°C. Prior to labeling the peptides, the TMT reagents were resuspended in MeCN and stored on ice until use.
CRITICAL: Unlike in the manufacturer's protocol, HPLC grade acetonitrile and not anhydrous acetonitrile is used for the resuspension of the TMT tags and the dilution of the samples. The use of non-anhydrous acetonitrile hasn’t affected the performance of the labeling in this protocol.
-
24.
Add HEPES (1 M, pH 8.0) to each sample to a final concentration of 100 mM and check that the pH is > 8. Add MeCN to a final concentration of 50%.
-
25.
Add 1 μL TMT reagent per 10 μg of sample, vortex and quickly centrifuge the mixture to gather the solution at the bottom of the tube. Incubate for 1 h at 24°C. Problem 4.
-
26.
Quench the labeling reaction by adding 1% (v/v) hydroxylamine (Sigma Aldrich, Cat#V001331) in 1:1 ratio to TMT reagent, vortex and incubate for 15 min at 24°C.
-
27.
Acidify the samples with 10% TFA (to final 1% TFA), check that the pH is < 2, and then pool the samples.
-
28.Evaporate the MeCN and concentrate the pooled peptides using a SpeedVac concentrator at 45°C (1–4 h).
-
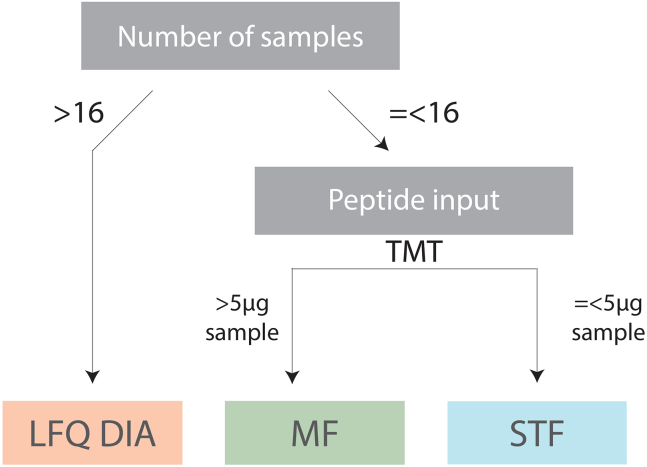
a.When > 5 μg peptide per TMT channel was used, proceed to high pH Microflow-based fractionation (MF) of TMT-labeled peptides (step 29).
-
b.When ≤ 5 μg peptide per TMT channel was used, proceed to phosphopeptide enrichment (step 37) followed by high pH Stage tip-based fractionation (STF) of TMT-labeled phosphopeptides (step 43).
-
a.
CRITICAL: if proceeding to phosphopeptide enrichment it is recommended to avoid complete peptide drying but rather reduce sample volume to <20 μL followed by dilution using phosphopeptide enrichment bind buffer (final of 200 μL).
CRITICAL: if labeling low peptide inputs per TMT channel it is recommended to perform the reaction in 0.5 mL Eppendorf (Eppendorf, Cat#0030121503) with a sample volume as low as possible.5
High pH microflow-based fractionation (MF) of TMT-labeled peptides
Timing: 4 h
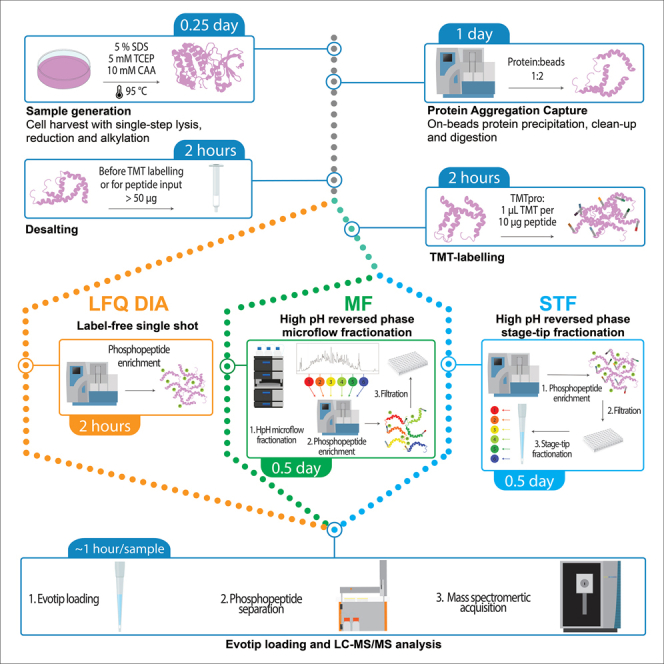
Note: this is a recommended step when > 5 μg peptide is labeled per TMT channel (Figure 3). If peptide amount is ≤ 5 μg peptide per TMT channel proceed to phosphopeptide enrichment (step 37) followed by stage tip-based phosphopeptide fractionation (step 43) prior LC-MS analysis.
Figure 3.
Decision tree for helping in the selection of the ideal sample preparation approach, i.e., no fractionation with LFQ DIA or TMT-labeling followed by microflow fractionation (MF) or stage-tip fractionation (STF)
-
29.Prepare the following solutions for microflow fractionation as per materials and equipment section:
-
a.Microflow buffer A: 5 mM AmmBic.
 CRITICAL: pass the 5 mM AmmBic buffer through 0.45 μm filter before use.
CRITICAL: pass the 5 mM AmmBic buffer through 0.45 μm filter before use. -
b.Microflow buffer B: 100% MeCN.
-
a.
-
30.
Resuspend samples from step 28 a) in 20 μL of Microflow sample resuspension buffer (25 mM AmmBic) and check that pH is at approximately 8 using pH paper.
CRITICAL: if pH has not reached 8, add another 1 μL of 1 M AmmBic. It is important to keep final volume ≤30 μL in order to avoid exceeding the maximum injection volume of the sample loop.
-
31.
Load 16–200 μg of TMT-labeled peptides on a reversed-phase Acquity CSH C18 1.7 μm × 1 mm × 150 mm column (Waters, Cat#186006935) coupled to UltiMate 3000 high-performance liquid chromatography (HPLC) system (Thermo Fisher Scientific) with Chromeleon software.
-
32.
Apply a flow rate of 30 μL/min, with column oven temperature set to 40°C, and gradient formed by microflow buffer A (5 mM AmmBic) and microflow buffer B (100% MeCN).
-
33.
Elute peptides via multi-step gradient as follows: 0–10 min 6.5% B–15% B, 10–59.5 min 15% B–30% B, 59.5–67 min 30% B–65% B, 67–70 min 65% B–80% B, 70–77 min 80% B, 78–87 min 6.5% B.
-
34.
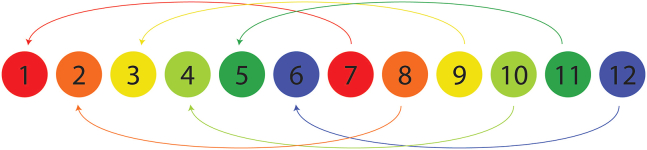
Collect twelve fractions with concatenation (from min 1 to 72, 1 min of collection period per well).
-
35.
Add 40 μL of 10% formic acid.
Manually concatenate into six fractions by pooling every 6th fraction together (1 and 7, 2 and 8, 3 and 9, 4 and 10, 5 and 11, 6 and 12) as per Figure 4.
-
36.
Concentrate the desalted peptides using a SpeedVac concentrator at 45°C (1–4 h) and proceed to phosphopeptide enrichment (step 37).
CRITICAL: it is recommended to avoid complete peptide drying but rather reduce sample volume to ∼20 μL followed by dilution using phosphopeptide enrichment bind buffer (final of 200 μL).
CRITICAL: It is recommended to reduce the volume of the fractions after acidification, so that the combined volume of the two fractions to concatenate do not exceed the total volume of the well.
Figure 4.
Schematic representation of the fraction concatenation after microflow fractionation (MF)
Phosphopeptide enrichment automated on KingFisher Flex (Apex, Duo, Prime) systems
Timing: 2 h
-
37.Prepare all solutions freshly before starting the enrichment as per materials and equipment section:
-
a.Phosphopeptide Bind buffer (Ti-IMAC HP): 1 M glycolic acid in 80% MeCN, 5% TFA.
-
b.Phosphopeptide Bind buffer (Zr-IMAC HP): 0.1 M glycolic acid in 80% MeCN, 5% TFA.
-
c.Phosphopeptide Wash buffer 1: aqueous solution of 80% MeCN and 1% TFA.
-
d.Phosphopeptide Wash buffer 2: aqueous solution of 10% MeCN and 0.2% TFA.
-
e.Phosphopeptide Elution buffer: 1% NH4OH in ddH2O.
-
a.
CRITICAL: the elution buffer must be prepared fresh and preferably stored in glass container. Conical polypropylene tubes can leach polymer when NH4OH is stored, in particular at higher concentrations.
-
38.
Dilute peptide (or TMT-labeled peptides) from steps 11b, 21, 36 to a final of 200 μL using phosphopeptide bind buffer: 0.1 M (or 1 M) glycolic acid in 80% MeCN, 5% TFA and vortex briefly to re-suspend.
CRITICAL: complete resuspension of TMT-labeled peptides is assisted by first adding 5% TFA followed by 80% MeCN, 1 M (or 0.1 M in the case of Zr-IMAC enrichment) glycolic acid.
-
39.Prepare the KingFisher plates with beads, samples and solutions as per Table 4/ Figure 5A (KingFisher Flex or Apex) or Table 6/ Figure 5B (KingFisher Duo or Presto). Problem 5.
Table 6.
Plate with contents used for the automated phosphopeptide enrichment on KingFisher Duo or Presto modelsKingFisher 96 deep-well plate order
Plate row number and name μL/ well Buffer Time per step (min) H: Tip plate G: MagReSyn Ti-IMAC HP or Zr-IMAC HP beads (1:2 ratio bead to peptide) Varies based on peptide amount 20% EtOH F: Microsphere equilibration 500 Phosphopeptide Bind buffer 5 E: Peptides (or TMT-labeled peptides) 200 Phosphopeptide Bind buffer 20 D: Wash 500 Phosphopeptide Bind buffer 2 C: Wash 500 Phosphopeptide Wash Buffer 1 2 B: Wash 500 Phosphopeptide Wash Buffer 2 2 A: Phosphopeptide elution 200 Phosphopeptide Elution Buffer 10
Note: MagReSyn Ti-IMAC HP and Zr-IMAC HP microparticles do not have to be pre-equilibrated prior use and can be added directly in storage solution (20% EtOH) to the KingFisher plate.
Note: MagReSyn Ti-IMAC HP and Zr-IMAC HP microparticles perform similarly in terms of capacity as well as selectivity but for high sensitivity applications (digest amounts < 100 μg) Zr-IMAC HP is preferred. Zr-IMAC HP also has a longer shelf life (18 months) in comparison to Ti-IMAC HP (6 months).
CRITICAL: When dispensing beads make sure storage container is frequently mixed to minimize beads from settling and thus ensure and equal amount of beads is distributed in each well. For large scale experiments beads can be dispensed using multi-channel pipette. Frequent mixing using up/down pipetting is recommended in this case in order to keep beads in suspension.
CRITICAL: for high sensitivity applications i.e. digest amount < 20 μg it is beneficial to loop the KingFisher method for two sequential enrichments (without changing buffers and beads). This increases the overall recovery of phosphopeptides.
Note: MagReSyn Ti-IMAC HP and Zr-IMAC HP are formulated at 20 mg/mL in 20% EtOH
-
40.
Run the KingFisher method and when complete add 40 μL of 10% TFA to each elution well. Problems 6 and 7.
CRITICAL: Prepare 10% TFA solution freshly in a chemical safety hood before using it. Phosphopeptides are not stable in alkaline conditions. Therefore, the solutions should be acidified immediately after elution.
CRITICAL: If proceeding to Stage tip-based phosphopeptide fractionation (step 43), do not acidify the eluates.
-
41.
Transfer samples to a MultiScreenHTS HV Filter Plate, 0.45 μm (Merck, Cat#MSHVN4550) set-up with a King Fisher standard (200 μL) 96 well collection plate (Thermo Fisher Scientific, Cat# 97002540).
-
42.
Centrifuge for 1 min at 500 x g to collect filtered samples and proceed to Stage-tip based fractionation (if input for phosphopeptide enrichment was TMT-labeled peptide with <5 μg per channel) or to Evotip loading (step 57).
CRITICAL: Eluate filtration is important in order to remove any residual beads as well as other insoluble material that can lead to blockages of Evotip and/or LC system.
Note: For manual phosphopeptide enrichment follow the steps as described above using 2 mL microcentrifuge tubes together with a suitable magnetic separator such as DynaMag-2 Magnet (Thermo Fisher Scientific, Cat# 12321D). A deviation from the KingFisher workflow is during the elution step where for manual processing two sequential elutions of 80 μL using 1% NH4OH are recommended. Each elution is acidified using 10% TFA then pooled together before proceeding to step 41.
Table 4.
Plates with their contents used for the automated phosphopeptide enrichment on KingFisher Flex or Apex models
| KingFisher 96 deep well plate order | |||
|---|---|---|---|
| Plate number and name | μL/ well | Buffer | Time per step (min) |
| 1: Tip plate | |||
| 2: MagReSyn Ti-IMAC HP or Zr-IMAC HP beads (1:2 ratio bead to peptide) | Varies based on peptide amount Table 5 | 20% EtOH | |
| 3: Microsphere equilibration | 500 | Phosphopeptide Bind buffer | 5 |
| 4: Peptides (or TMT-labeled peptides) | 200 | Phosphopeptide Bind buffer | 20 |
| 5: Wash | 500 | Phosphopeptide Bind buffer | 2 |
| 6: Wash | 500 | Phosphopeptide Wash Buffer 1 | 2 |
| 7: Wash | 500 | Phosphopeptide Wash Buffer 2 | 2 |
| 8: Phosphopeptide elution | 200 | Phosphopeptide Elution Buffer | 10 |
Figure 5.
Overview of automated phosphopeptide enrichment
(A and B) (A) KingFisher Flex /Apex layout (processing of up to 96 samples in parallel) or (B) KingFisher Duo Prime /Presto layout (processing of up to 12 samples in parallel).
Table 5.
Recommended peptide to bead ratios for phosphopeptide enrichment
| Digest amount (μg) | Peptide : Bead ratio | Bead amount (μg) | Bead volume (μL) |
|---|---|---|---|
| 2.5 | 1:40 | 100 | 5 |
| 10 | 1:10 | 100 | 5 |
| 20 | 1:5 | 100 | 5 |
| 50 | 1:2 | 100 | 5 |
| 100 | 1:2 | 200 | 10 |
| 200 | 1:2 | 400 | 20 |
| 500 | 1:2 | 1000 | 50 |
| 1000 | 1:2 | 2000 | 100 |
High pH stage tip-based fractionation of TMT-labeled phosphopeptides
Timing: 4 h
Note: this is a recommended step when ≤ 5 μg peptide per TMT channel was used (step 22) followed by phosphopeptide enrichment (step 37) (Figure 3).
-
43.
Prepare stage tips by packing three discs of CDS Empore C18 (Fisher Scientific, Cat#13110018) into low-bind 200 μL tips (Merck, Cat#CLS4154-4X960EA).
-
44.
Place tips into centrifugation adaptors (GL-Sciences, Cat#5010-21514) with a collecting tube below. For collecting fractions, use low bind tubes.
CRITICAL: ensure the C18 discs are packed with equal pressure. Monitor the flow to check if this is the case.
CRITICAL: make sure Stage tips do not dry across the various steps.
-
45.Prepare all solutions required for Stage tip fractionation freshly before starting the enrichment as per materials and equipment section:
-
a.Stage tip equilibration buffer 1 (STEB 1): 100% MeOH.
-
b.Stage tip equilibration buffer 2 (STEB 2): 20 mM NH4OH in 100% MeCN.
-
c.Stage tip equilibration buffer 3 (STEB 3): 20 mM NH4OH in MilliQ water.
-
a.
CRITICAL: ensure pH is > 8 using pH paper.
-
46.Prepare the elution buffers required as described in Table 7:
-
a.Stage tip elution buffer 1: 20 mM NH4OH in 4% MeCN.
-
b.Stage tip elution buffer 2: 20 mM NH4OH in 8% MeCN.
-
c.Stage tip elution buffer 3: 20 mM NH4OH in 12% MeCN.
-
d.Stage tip elution buffer 4: 20 mM NH4OH in 20% MeCN.
-
e.Stage tip elution buffer 5: 20 mM NH4OH in 80% MeCN.
-
a.
-
47.Equilibrate each Stage tip by consecutively loading:
-
a.50 μL of Stage tip equilibration buffer 1 (100% MeOH), centrifuge for 2 min, 1500 × g.
-
b.50 μL of Stage tip equilibration buffer 2 (20 mM NH4OH in 100% MeCN), centrifuge for 2 min, 1500 × g.
-
c.100 μL of Stage Tip equilibration buffer 3 (20 mM NH4OH in ddH2O), centrifuge for 2 min, 1500 × g.
-
d.100 μL of Stage Tip equilibration buffer 3 (20 mM NH4OH in ddH2O), centrifuge for 2 min, 1500 × g.
-
a.
CRITICAL: step 47c can be used as a reference to assess the quality of stage tip packing. With optimal packing, the content of the tip should elute in 2 min.
-
48.
Load TMT-labeled phosphopeptides (step 42) on the pre-equilibrated stage tip, and centrifuge for 3 min, 800 × g.
-
49.
Collect flow through in 2.0 mL Eppendorf tube.
-
50.
Reload the flow through.
-
51.
Repeat steps 49. and 50. for a total of two re-loads.
-
52.
Elute peptides successively using 50 μL Stage tip elution buffers 1–5 and collect each elution separate in 1.5 mL Eppendorf tubes, centrifuge 2 min, 1500 × g.
-
53.
Pool elutions 4 (20 mM NH4OH in 20% MeCN) and 5 (20 mM NH4OH in 80% MeCN).
-
54.
Add formic acid to reach pH ∼ 2 and evaporate the MeCN in a speed vac concentrator at 40°C.
-
55.
Resuspend the samples in 0.1% FA to have equal volumes before Evotip loading. Check the pH again to make sure the phosphopeptides are in an acidic environment.
-
56.
Proceed to Evotip loading (step 57).
Table 7.
Stage tip phosphopeptide fractionation elution buffers (for 5 mL)
| Elution buffer | Volume STEB 3 (mL) | Volume STEB 2 (mL) |
|---|---|---|
| 1 | 4.8 | 0.2 |
| 2 | 4.6 | 0.4 |
| 3 | 4.4 | 0.6 |
| 4 | 4.0 | 1.0 |
| 5 | 1.0 | 4.0 |
Evotip loading
Timing: 10 min
-
57.
Prepare all solutions required for Evotip loading freshly before starting the enrichment as per materials and equipment section.
-
58.
Wash Evotips by pipetting 20 μL of Evotip buffer B (80% MeCN with 0.1% FA) into each tip.
-
59.
Centrifuge Evotips with appropriate counterbalance at 800 × g for 60 s.
-
60.
Fill wells of a 96-well plate with 100 μL of 100% LC-MS grade 1-isopropanol or 2-propanol.
-
61.
Place Evotip adaptor rack on top of the 96-well plate to condition each Evotip by soaking in the solution for a minimum of 10 s. Visually inspect that the tips have changed from bright to pale white.
CRITICAL: Soak Evotips for additional time if color change is not observed. This step may take 30–60 s.
-
62.
Equilibrate the Evotips by pipetting 20 μL of Evotip buffer A (H2O with 0.1% FA) into each tip, while keeping the tips in the adaptor rack. Place the Evotips back into their original tray.
-
63.
Centrifuge Evotips with appropriate counterbalance at 800 × g for 60 s.
-
64.
Transfer the samples into a corresponding Evotip.
-
65.
Centrifuge Evotips with appropriate counterbalance at 500 × g for 120 s.
-
66.
Wash the Evotips by pipetting 20 μL of Evotip buffer A into each tip.
-
67.
Centrifuge Evotips with appropriate counterbalance at 800 × g for 60 s.
-
68.
Preserve the Evotips (keep wet) by pipetting 200 μL of Evotip buffer A into each tip.
-
69.
Briefly centrifuge Evotips with appropriate counterbalance at 800 × g for 10 s.
CRITICAL: Evotips loaded with phosphopeptides should not be stored more than one week before analysis. Add enough Evotip buffer A to the bottom of the tray such that the bottoms of the Evotips are submerged and store at 4°C with the lid closed until analysis.
CRITICAL: Following phosphopeptide enrichment (step 42), the sample volume is > 200 μL. This can be directly loaded on Evotips but reduce the centrifuge speed (step 65) to 500 × g for 2 min, instead of the 800 × g for 1 min that is recommended for loading 20 μL of peptides.
LC-MS/MS analysis
CRITICAL: To resolve the reporter ions using TMT, the minimum MS2 resolution required is 30k. We recommend to use 45k unless turbo TMT (11-plex) or turbo TMTpro (16-plex) is available.
CRITICAL: We recommend choosing the Evosep method and the MS method based on the input amount to be analyzed. The MS method parameters described below are for analysis on an Orbitrap Exploris 480 and may differ for other instruments.
LC and gradients
-
70.
Replace Evosep One solvent as required (A: LC-MS grade H2O with 0.1% FA, B: LC-MS grade MeCN with 0.1% FA).
-
71.
Select Evosep method and column based on input amount loaded into the column (Table 8):
Table 8.
Reference table for Evosep gradients and column set-up
| Gradient | Column | Emitter | Input (into column) |
|---|---|---|---|
| 30SPD | Evosep endurance column (15 cm × 150 μm, 1.9 μm beads, EV1106) | Stainless-steel emitter (30 μm i.d., Evosep, EV1086) | >500 ng tryptic peptides or 200 μg of enriched phosphopeptides |
| 60SPD | Evosep performance column (8 cm × 150 μm, 1.5 μm beads, EV1109) | Stainless-steel emitter (30 μm i.d., Evosep, EV1086) | >100 ng tryptic peptides or 100 μg of enriched phosphopeptides |
| 20 SPD | IonOpticks Aurora column (15 cm × 75 μm, 1.7 μm cat#AUR3-15075C18-TS) or Evosep performance column (15 cm × 75 μm, 1.9 μm beads, EV1112) | Fused silica emitter (10 μm i.d., Evosep, EV1087). Only for Evosep column. | <50 ng tryptic peptides or <50 μg of enriched phosphopeptides |
| 40SPD | IonOpticks Aurora column (15 cm × 75 μm, 1.7 μm cat#AUR3-15075C18-TS) or Evosep performance column (15 cm × 75 μm, 1.9 μm beads, EV1112) | Fused silica emitter (10 μm i.d., Evosep, EV1087). Only for Evosep column. | <50 ng tryptic peptides or <50 μg of enriched phosphopeptides |
When using IonOpticks Aurora columns, use a Nanoflex source with a Sonation Column Oven PRSO-V2 (Sonation lab solutions, Cat#PRSO-V2) set at 50°C.
When using Evosep performance columns, use an EasySpray source with an externally controlled Butterfly heater (MSWil, Cat#PST-ES-BPH-20) at 40°C. Problems 8 and 9.
-
72.
Place Evotips in a 96 position Evotip rack in the order that they will be acquired on the mass spectrometer.
CRITICAL: when using Nanoflex source, adjust the emitter position so that it’s not too far (results in reduced and instable spray) or not too close (results in reduced spray) to the orifice.
CRITICAL: Before running Evosep Whisper methods, prepare fresh buffers, run a full solvent exchange, leak test, calibrate the pumps and do not move the Evosep system during the whole maintenance and sample acquisition.
MS parameters
-
73.Setup the mass spectrometer method for the Orbitrap Exploris 480 with the following parameters:
-
a.Positive ion mode.
-
b.Spray voltage at 1.8 kV with a silica emitter, 2.0 kV with a steel emitter.
-
c.Heated capillary temperature at 275°C.
-
d.Funnel RF level at 40.
-
e.Acquire data in profile mode.
-
a.
Note: Data can also be acquired in centroid mode also. For 30K (or higher) MS2 resolution this is preferred.
Data-dependent acquisition (DDA) for TMT
-
74.Edit the following parameters in a DDA method:
-
a.Set full MS resolution at 60,000 at m/z 200 with a maximum injection time of 25 ms.
-
b.Set the MS1 mass range at 350–1400 with an AGC target at 300%.
-
c.Set the HCD fragment spectra resolution at 45,000 with a maximum injection time of 86 ms, using the Top10 method.Note: Using Turbo-TMT with a MS2 resolution of 15,000, we recommend a top12 method with a maximum injection time of 22 ms.
-
d.Set AGC target value at 200% and the intensity threshold at 2e5.
-
e.Set the isolation window at 1.3 m/z and the normalized collision energy at 35%.
 CRITICAL: a higher collision energy of 35% is necessary to achieve efficient fragmentation of TMT reporter ions.
CRITICAL: a higher collision energy of 35% is necessary to achieve efficient fragmentation of TMT reporter ions.
-
a.
Data-independent acquisition (DIA) for LFQ
-
75.Edit the following parameters in a DDA method:
-
a.Set full MS resolution at 120,000 at m/z 200 with an injection time of 45 ms.
-
b.Set the MS2 resolution at 45,000 with an injection time of 86 ms and an AGC target of 1000%.
-
c.Set the HCD collision energy at 27%.
-
d.Set the MS2 mass range at 472–1143.
-
a.
CRITICAL: 17 windows of 39.5 m/z scanning from 472 to 1143 m/z with an overlap of 1 Da are recommended for DIA analysis on the Orbitrap Exploris 480 for phosphoproteomics samples. These settings result in a cycle time of 2 s However, alternative DIA window schemes may be optimized and applied. If the number of points per peak needs to be increased, the cycle time can be reduced to 1 s.
Data processing
LFQ phosphopeptide enriched data processing using directDIA
Note: Here we briefly describe the processing of DIA raw files using spectrum centric, also known as library-free approach using Spectronaut6 directDIA workflow. Other software such as DIA-NN,7 DIA-umpire8 and PECAN9 may be used for library-free DIA data processing.
-
76.
Launch Spectronaut software.
-
77.
Select the “Set-up DirectDIA Analysis from File” option.
-
78.
Select the raw data files to be extracted.
-
79.
Select the FASTA file for the library-free search. You can select more than one FASTA file (e.g., Human FASTA and contaminants).
-
80.
Select the “BGS Phospho PTM Workflow” search parameters.
Note: “BGS Phospho PTM Workflow” contains preset data extraction parameters necessary to process DIA raw data generated from phosphopeptide enriched samples (i.e.: phosphorylation on Ser, Thr and Tyr as variable modification). For non-enriched samples apply the “BGS Factory settings (default)” parameters.
Note: The default “Probability cutoff” setting in the PTM workflow tab is 0.75. For a more confident localization of the phosphorylation site, you can increase this cut-off to 0.99.
-
81.
Optional: Specify the biological conditions in order to set the groups for statistical analysis post data extraction.
-
82.
Optional: Specify a Gene annotation file to add GO terms to the identified proteins post analysis.
-
83.
Optional: Select additional DDA or DIA raw data files generated from phosphopeptide enriched samples.
Note: Preferably the DDA or DIA raw data should have been generated with similar chromatographic gradients, columns and mass spectrometry settings as the data being extracted.
-
84.When the search is done, the report needs to be exported. Two strategies are available:
-
a.Exporting the site report generated from Spectronaut:Click in the Report tab. In the “PTM Site Report” (on the left), select the default pivot report (BSG Factory Report) and click on “Export Report”.
 CRITICAL: The lack of protein grouping in this report will generate a table containing redundancy.
CRITICAL: The lack of protein grouping in this report will generate a table containing redundancy. -
b.Exporting the custom-made table as described in the study by Martinez-Val et al.10 and collapsing it to obtain unique phosphosites using a Perseus plugin described in the study by Bekker-Jensen et al.11 available in GitHub (https://github.com/AlexHgO/Perseus_Plugin_Peptide_Collapse).
-
a.
TMT-labeled phosphopeptide data processing
Note: Here we briefly describe the processing of DDA raw files using MaxQuant.
-
85.
Launch MaxQuant software.
-
86.
Load the raw files to be searched.
-
87.
In the “Group-specific parameters” tab, select “Reporter ion MS2” as a type. Click on the TMT-plex used in the experiment. TMT-16 plex is available in MaxQuant from version 2.1.
-
88.
Add Phospho(STY) as a variable modification.
-
89.
Load the FASTA file in the “Global parameters” tab.
-
90.
Indicate the number of processors and start the search.
-
91.
When the search is done, open the Perseus software and load the Phospho (STY)Sites.txt table. Keep the default importation parameters. Set the reporter intensity columns as “Main”. The number of columns to import correspond to your number of raw file times the number of channels set up in the search times 3 (maximum multiplicity).
-
92.
Click on “Modifications” -> “Expand site table” to have table at the unique phosphosite level.
-
93.
Filter the table to remove reverse hit, contaminants. Only keep class 1 phosphosite by filtering the table based on the localization probability ( > 0.75).
CRITICAL: When searching several raw files, the localization probability added by default represents the maximal localization probability from any raw file.
Expected outcomes
-
•
Table 9 shows the expected depth using both TMT based workflows described in the protocol (STF and MF) using 1, 2.5 and 12.5 μg of peptide per channel. The fractions obtained from 12.5 μg of peptides per sample were acquired using 20SPD while all the other samples were analyzed with the 40SPD gradient. All the samples were separated on an Evosep column (EV1112) and acquired in DDA mode.
-
•
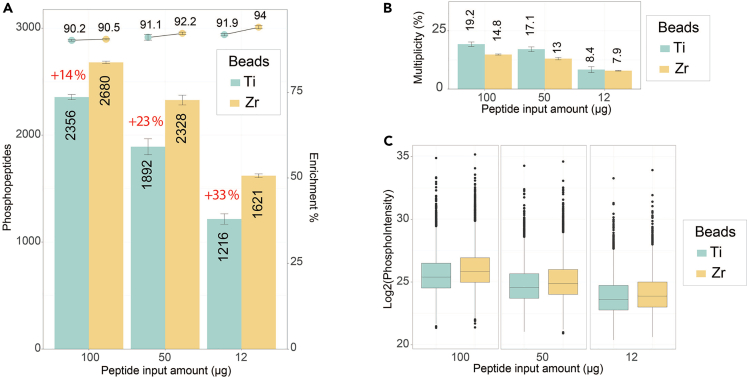
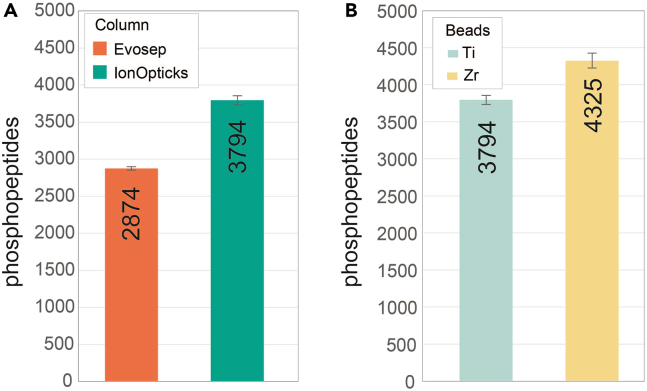
Using MagReSyn Zr-IMAC HP instead of Ti-IMAC HP enables a gain in phosphopeptide identifications especially for input amounts equal of below 100 μg (Figures 6A–6C). This benefit can be further boosted in extremely low input amounts (2.5 μg) when combining DIA and nanoflow chromatography using IonOpticks Aurora columns (Figures 7A and 7B).
Table 9.
Expected number of phosphopeptides quantified using offline fractionation TMT-based approaches
| Input amount (μg digest/sample) | Stage-tip fractionation (STF) | Microflow fractionation (MF) | LFQ DIA |
|---|---|---|---|
| 12.5 | 9856 | 13250 | 7000–9000 |
| 2.5 | 5485 | 4684 | |
| 1 | 4577 | 4342 |
Figure 6.
Expected performance of MagReSyn Ti-IMAC HP and Zr-IMAC HP for TMT-based analysis from bulk HeLa lysate
(A) Comparison between MagReSyn Ti-IMAC-HP and Zr-IMAC-HP beads for phosphopeptide enrichment. The data was acquired in DDA mode with 15k MS2 resolution and, using a 60SPD Evosep gradient for all amounts. The identifications represents the median of 3 replicates per conditions, the error-bars represent the standard deviation.
(B) Barplot showing the phosphopeptide multiplicity as a function of the peptide input amount for both Ti-IMAC-HP and Zr-IMAC-HP. The multiplicity corresponds to the percentage of multiphosphorylated peptides.
(C) Boxplot displaying the measured phosphopeptides intensity as a function of the peptide input amount for both Ti-IMAC-HP and Zr-IMAC-HP beads.
Figure 7.
Expected performance of column and IMAC bead type DIA based phosphopeptide analysis using WHISPER 40SPD Evosep gradient
(A) Comparative bar plot assessing the effect of the column type (Evosep (EV1112) vs. IonOpticks (AUR3-15075C18-TS)) on the number of phosphopeptides identified when using 2.5 μg of peptide input for phosphopeptide enrichment with MagReSyn Ti-IMAC-HP beads with the 40 SPD gradient (n = 3).
(B) Comparative bar plot assessing the effect of the beads type (Ti-IMAC-HP vs. Zr-IMAC-HP) on the number of phosphopeptides (class I) identified when using 2.5 μg of peptide input for phosphopeptide enrichment using an IonOpticks column (n = 3) for phosphopeptide separation. Adapted from Martinez-Val et al.2
Limitations
-
•
The buffers are currently manually aliquoted, but for a fully automated set-up capable of increased throughput further, plates may be filled using a microplate reagent dispenser or a KingFisher Presto could be integrated with liquid handling systems such as Hamilton for a fully automated, hands-off, solution.
-
•
The Ti-IMAC HP beads have a relatively short shelf-life of 6 months and thus for large and in particular longitudinal studies Zr-IMAC HP is recommended for phosphopeptide enrichment due to its extended shelf-life of 18 months.
Troubleshooting
Problem 1
Samples are still viscous after sonication.
Potential solution
The DNA content in the sample is too high. Sonicate multiple times until you can properly pipette the solution.
Alternatively, incubate samples with 1 μL (25 units) of Benzonase (stock solution: 2500 units in 100 μL) per 0.5 million cells and 2 mM MgCl2 for 30 min at 37°C.
Problem 2
White insoluble material detected post PAC on-bead digestion.
Potential solution
The precipitate is likely due to residual nucleic acid and/or lipid contamination and can be removed by centrifuging the samples for 10 min at 10,000 x g and transferring the clarified supernatant to a new Eppendorf tube.
Problem 3
Poor peptide recovery from PAC workflow.
Potential solution
Ensure protein concentration assay has not over-estimated total protein amount per sample post the extraction.
If sample concentration is below 0.1 μg/μL, increase the protein to bead ratio from 1:4 to 1:12.
Ensure that the pH of the lysate is between 7.5 and 8.5.
Problem 4
Poor TMT labeling efficiency.
Potential solution
Utilize buffers without primary amines such as TEAB or HEPES and desalt the samples prior TMT labeling.
Use fresh TMT reagent aliquots, or increase the ratio TMT:peptide labeling if the TMT aliquots might have been stored for too long.
Problem 5
MagReSyn Hydroxyl, Ti-IMAC HP or Zr-IMAC HP bead volumes differ across wells.
Potential solution
When dispensing beads into KingFisher plates ensure the frequent mixing of the storage containers to make sure beads do not settle. When dispensing in multiple wells, one can make use of a multi-channel pipette with beads in storage solution aliquoted in appropriate storage reservoir. In this case, between each dispensing cycle, beads can be mixed by aspirating the bead suspension to minimize settling and ensure an equivalent volume of beads is picked up and subsequently dispensed.
Problem 6
Poor phosphopeptide recovery.
Potential solution
Check the digest recovery by performing peptide quantification after desalting, to ensure the correct peptide amount is loaded for phosphopeptide enrichment. Another option is to analyze the proteome in the MS and estimate if the peptide amount loaded on the tip corresponds to the observed TIC.
Do not dry peptides and phosphopeptides pre or post phosphopeptide enrichment.
Prepare Evotips with phosphopeptides immediately after finishing the phosphopeptides enrichment protocol. Ensure that the pH of the phosphopeptides is ∼2 before loading them into Evotips. Preferably analyze the samples on Evotips immediately and do not store them for more than 7 days prior to MS-analysis.
Prepare fresh elution buffer and store in a glass container. Storage in polypropylene tubes can result in leaching of polymers. This can be verified by complete vacuum drying of the eluted phosphopeptides (or the elution buffer on its own). A translucent pellet on drying is indicative of polymer contamination.
For high sensitivity applications, i.e., where the digest amount is < 20 μg, it is beneficial to run the KingFisher method for two sequential enrichments (without changing buffers and beads). This increases the overall phosphopeptide recovery.
Problem 7
Poor phosphopeptide enrichment.
Potential solution
Check that the pH of the digest (after dilution with phosphopeptide bind buffer) is below 2. Keep a ratio of 20 μL peptides and 200 μL phosphopeptide bind buffer.
Problem 8
Spray not stable or emitter blocked.
Potential solution
Use 20 μm emitters instead of 10 μm. After connecting the column to the emitter, a back pressure increases of > 10 bar on Pump D (pressure channel) indicates a partial blockage and the emitter should be replaced.
Problem 9
Irreproducible peptide retention time with Evosep WHISPER method.
Potential solution
Prepare fresh buffers, perform a full solvent exchange and perform calibration of pumps ABCD. Ensure the Evosep is stable and is not moved before and during acquisition using WHISPER gradients.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stoyan Stoychev (sstoychev@resynbio.com).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
Work at the Novo Nordisk Foundation Center for Protein Research (CPR) is funded in part by a generous donation from the Novo Nordisk Foundation (NNF14CC0001). This work has also been funded as part of the EPIC-XS project under the grant agreement no. 823839 funded by the Horizon 2020 program of the European Union and supported by the European Research Council through ERC-Synergy grant 810057-HighResCells. C.K. is supported by the Marie Skłodowska Curie European Training Network “PUSHH” (grant no. 861389).
Author contributions
A.M.-V. and C.K.: conceptualization, investigation, writing – original draft, writing – review and editing. P.N. and S.S.: writing – original draft, writing – review and editing. J.J. and J.V.O.: writing – review and editing, supervision, funding acquisition.
Declaration of interests
P.N., S.S., and J.J. work for ReSyn Biosciences that supplied some of the reagents for digest preparation and phosphopeptide enrichment utilized in this workflow. S.S. further works for Evosep Biosystems whose LC system is utilized as part of the LC-MS setup.
Contributor Information
Ana Martinez-Val, Email: ana.mdval@cpr.ku.dk.
Stoyan Stoychev, Email: sstoychev@resynbio.com.
Data and code availability
The published articles, Koenig et al.1 and Martínez-Val et al.,2 include all datasets analyzed during this study.
References
- 1.Koenig C., Martinez-Val A., Franciosa G., Olsen J.V. Optimal analytical strategies for sensitive and quantitative phosphoproteomics using TMT-based multiplexing. Proteomics. 2022;22 doi: 10.1002/pmic.202100245. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Val A., Fort K., Koenig C., Van der Hoeven L., Franciosa G., Moehring T., Ishihama Y., Chen Y.J., Makarov A., Xuan Y., Olsen J.V. Hybrid-DIA: intelligent data acquisition integrates targeted and discovery proteomics to analyze phospho-signaling in single spheroids. Nat. Commun. 2023;14:3599. doi: 10.1038/s41467-023-39347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 4.Brenes A., Hukelmann J., Bensaddek D., Lamond A.I. Multibatch TMT Reveals False Positives, Batch Effects and Missing Values. Mol. Cell. Proteomics. 2019;18:1967–1980. doi: 10.1074/mcp.RA119.001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zecha J., Satpathy S., Kanashova T., Avanessian S.C., Kane M.H., Clauser K.R., Mertins P., Carr S.A., Kuster B. TMT Labeling for the Masses: A Robust and Cost-efficient, In-solution Labeling Approach. Mol. Cell. Proteomics. 2019;18:1468–1478. doi: 10.1074/mcp.TIR119.001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruderer R., Bernhardt O.M., Gandhi T., Miladinović S.M., Cheng L.-Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsou C.-C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A.-C., Nesvizhskii A.I. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods. 2015;12:258. doi: 10.1038/nmeth.3255. 64, 7 p following 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting Y.S., Egertson J.D., Bollinger J.G., Searle B.C., Payne S.H., Noble W.S., MacCoss M.J. PECAN: library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat. Methods. 2017;14:903–908. doi: 10.1038/nmeth.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Val A., Bekker-Jensen D.B., Hogrebe A., Olsen J.V. Data Processing and Analysis for DIA-Based Phosphoproteomics Using Spectronaut. Methods Mol. Biol. 2021;2361:95–107. doi: 10.1007/978-1-0716-1641-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Bekker-Jensen D.B., Bernhardt O.M., Hogrebe A., Martinez-Val A., Verbeke L., Gandhi T., Kelstrup C.D., Reiter L., Olsen J.V. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 2020;11:787. doi: 10.1038/s41467-020-14609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published articles, Koenig et al.1 and Martínez-Val et al.,2 include all datasets analyzed during this study.