Abstract
This study evaluated a PCR assay for detection of the streptococcal pyrogenic exotoxin B (speB) gene from tissue biopsy specimens of patients with necrotizing fasciitis. speB was detected in specimens from all 10 patients with necrotizing fasciitis due to group A streptococcus. The assay was negative for all 11 patients without culture or serologic evidence of streptococcal infection. These results suggest that the detection of speB by PCR may be useful for confirming group A streptococcal infection when cultures are negative or not available.
Invasive group A streptococcal infections, including streptococcal toxic shock syndrome and necrotizing fasciitis, are associated with considerable morbidity and mortality despite the use of appropriate antibiotics (2, 3, 7, 9). The microbial diagnosis of streptococcal necrotizing fasciitis is generally based on the isolation of group A streptococcus from either soft tissue or blood cultures. However, only about 50% of patients with necrotizing fasciitis are found to be bacteremic (9), and tissue cultures may be negative because empiric antimicrobial therapy is started before specimens for culture are obtained by surgical intervention.
The streptococcal pyrogenic exotoxin B gene is a chromosomally encoded structural gene (speB) associated with pyrogenicity, T-lymphocyte mitogenicity, and the ability to increase susceptibility to endotoxic shock in individuals infected with group A streptococcus (5, 10). Because of its conserved and stable characteristics and its presence in almost all group A streptococci (6, 8, 10, 14), the speB gene was chosen as the target for PCR. In this study, we examined the use of PCR for detection of speB directly from tissue samples of patients being investigated for possible group A streptococcal necrotizing fasciitis and compared PCR detection with conventional culture methods.
Necrotizing fasciitis was determined to be present if there was evidence of soft tissue edema and necrosis at surgery and if histopathology demonstrated necrosis of the superficial fascia with fascial edema and a polymorphonuclear infiltrate (9). Tissue samples and blood cultures were obtained from 10 patients with confirmed group A streptococcal necrotizing fasciitis, from 4 patients with necrotizing fasciitis not due to group A streptococcus, and from 7 patients undergoing elective surgery with resection of fascia or soft tissue but without clinical or surgical evidence of necrotizing fasciitis (Table 1). Soft tissue samples (tissue biopsy specimens and/or swabs) were cultured on 5% Columbia sheep blood agar (PML Microbiologicals, Mississauga, Ontario, Canada), chocolate agar, and fastidious anaerobe broth (Quelab, Montreal, Quebec, Canada) and incubated at 35°C in an atmosphere of 5% CO2 for 3 days. Blood cultures were processed by the BacT/Alert blood culture system (Organon Teknika, Durham, N.C.). Group A streptococci were identified by standard laboratory methods (12). A tissue sample (approximately 100 mg) was obtained from each patient at surgery and was stored at −70°C in a dry, sterile container until used for PCR testing. Acute- and convalescent-phase patient sera, when available, were tested for patients with necrotizing fasciitis and negative cultures, by a streptococcal anti-DNase B antibody assay (Behring Diagnostics, Westwood, Mass.).
TABLE 1.
speB PCR detection results from tissue specimens from 21 patients
| Patient | Classification | Conventional culture result | speB PCR result |
|---|---|---|---|
| 1 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 2 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 3 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 4 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 5 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 6 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 7 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 8 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 9 | Necrotizing fasciitis | Group A streptococcus | Positive |
| 10 | Necrotizing fasciitis | Negative (serology +)a | Positive |
| 11 | Necrotizing fasciitis | Group B streptococcus | Negative |
| 12 | Necrotizing fasciitis | Staphylococcus aureus | Negative |
| 13 | Necrotizing fasciitis | Streptococcus anginosis, Escherichia coli, Proteus mirabilis, Bacteroides fragilis | Negative |
| 14 | Necrotizing fasciitis | No growth | Negative |
| 15 | Control tissueb | Microaerophilic streptococci, Coagulase-negative staphylococci, diphtheroids | Negative |
| 16 | Control tissue | Mixed coliforms | Negative |
| 17 | Control tissue | Coagulase-negative staphylococci, E. coli | Negative |
| 18 | Control tissue | No growth | Negative |
| 19 | Control tissue | No growth | Negative |
| 20 | Control tissue | No growth | Negative |
| 21 | Control tissue | No growth | Negative |
DNase B titer (Behring Diagnostics).
Control tissue samples included ischemic injury with gangrene (one patient), pyoderma gangrenosum (one patient), cellulitis (one patient), and soft tissue with no pathological diagnosis (four patients).
Total genomic DNA was extracted as described by Relman et al. (11). Briefly, 100 mg of tissue was digested in 5 ml of proteinase K (Boehringer Mannheim, Laval, Quebec, Canada) solution consisting of 500 mM Tris-HCl (pH 9.0), 20 mM EDTA (pH 9.0), 10 mM NaCl, 1% sodium dodecyl sulfate, and 1 mg of proteinase K per ml at 60°C for 48 to 72 h. RNase A (4 μg/ml; Sigma Chemicals, St. Louis, Mo.) was added, and the solution was incubated at 25°C for 2 h. DNA was extracted by standard phenol-chloroform extraction and was precipitated and resuspended in 50 μl of sterile, distilled, deionized water according to the methods of Sambrook et al (13). PCR was performed in a 25-μl volume, with 1× PCR buffer (Perkin-Elmer Cetus, Norwalk, Conn.) containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 200 μM (each) deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Gibco-BRL, Burlington, Ontario, Canada), 0.2 μM (each) primer (Gibco-BRL), and approximately 25 ng of DNA. Thermocycling conditions in a GeneAmp 9600 thermocycler (Perkin-Elmer Cetus) were as follows: 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for each cycle. The primer sequences used for speB were as previously published (6, 14) (SPEB-1, 5′ GTC AAC ATG CAG CTA CAG GA 3′; SPEB-2, 5′ AAT ACC AAC ATC AGC CAT CA 3′), giving a PCR product of 257 bp. PCR amplicons were visualized on a 2% agarose gel after being stained with ethidium bromide and photographed under UV illumination. After PCR amplification, the 257-bp fragment was cleaned (Geneclean II; Bio 101, Vista, Calif.) and digested with HinfI (New England BioLabs, Mississauga, Ontario, Canada), yielding restriction fragment lengths of 204 and 53 bp, respectively. PCR products were visualized on a 4% agarose gel (Metaphor agarose; FMC BioProducts, Rockland, Maine) after being stained with ethidium bromide and photographed under UV illumination.
Of 14 patients with necrotizing fasciitis, 9 had group A streptococcus isolated from soft tissue (8 patients) and/or blood cultures (5 patients). PCR for speB was positive for tissue samples obtained from all of these patients (Table 1). One patient (patient 10) with necrotizing fasciitis had negative tissue and blood cultures, possibly due to prior treatment with broad-spectrum antibiotics (ampicillin, clindamycin, and gentamicin). However, a convalescent-phase serum sample was positive for anti-DNase B antibody, as was the PCR assay for speB. Three patients had necrotizing fasciitis due to microorganisms other than group A streptococcus. The PCR assay was negative for all of these patients, including one whose tissue and blood cultures grew group B streptococcus (patient 11). The other patient with necrotizing fasciitis and negative cultures (patient 14) had negative anti-DNase B serology, and the tissue sample was also negative by PCR for speB. All seven patients undergoing elective surgical resection of soft tissue without evidence of necrotizing fasciitis had negative PCR assays for speB.
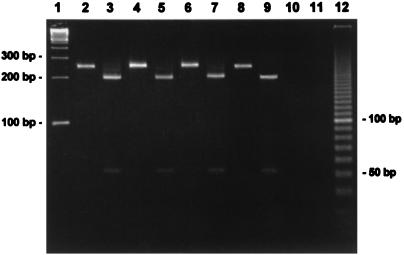
All speB-positive samples were subjected to restriction endonuclease digestion with HinfI to ensure specificity of the speB PCR and to confirm and validate amplicon integrity. The 257-bp speB fragment has a single HinfI internal restriction site, and the restriction fragment length polymorphism patterns obtained were the same as those predicted from the published nucleotide sequence of speB (14). Figure 1 shows an agarose gel with representative PCR amplification fragments and restriction fragment length polymorphisms of speB-positive tissue samples.
FIG. 1.
Agarose gel showing representative PCR amplification fragments and restriction fragment length polymorphisms of speB-positive tissue samples. Lane 1, 100-bp ladder (Gibco-BRL); lanes 2, 4, and 6, speB-positive samples; lanes 3, 5, and 7, amplicons from lanes 2, 4, and 6 digested with HinfI; lanes 8 and 9, Streptococcus pyogenes ATCC 19615 (positive control); lane 10, Streptococcus pneumoniae ATCC 49619 (negative control); lane 11, reagent control; lane 12, 10-bp ladder (Gibco-BRL).
A variety of pathogenic bacteria have been isolated from patients with necrotizing fasciitis. Approximately 10% of cases are reported to be caused by group A streptococcus (1), although the incidence of streptococcal necrotizing fasciitis and of other forms of invasive group A streptococcal infections has increased in the past few years (2, 3, 7, 9). However, group A streptococcus may not always be identified, either because tissue samples are not sent for culture or because growth of the organism is inhibited if empiric antimicrobial therapy has been started before surgical samples or blood cultures are obtained. It is important to establish whether the necrotizing fasciitis is due to group A streptococcus for patient management, prophylaxis of close contacts, and epidemiologic investigations. Optimal therapy includes the use of appropriate antimicrobials and may eventually also include immune system modulation (8). Invasive group A streptococcal infections may be associated with a significantly increased risk of secondary infections in household contacts (3, 4). Consequently, chemoprophylaxis for close contacts of patients with group A streptococcal necrotizing fasciitis may be recommended (3, 4). There are also public health and infection control implications that would make a reliable diagnostic test for invasive group A streptococcal infection useful, especially in the setting of a hospital or nursing home outbreak of severe soft tissue infection.
The results of this study indicate that PCR for detection of the speB gene from tissue samples is both sensitive and specific compared to conventional methods for confirming group A streptococcal infection. The PCR assay was positive for all nine patients with culture-confirmed streptococcal necrotizing fasciitis (patients 1 to 9) and was negative for all 11 patients without any evidence of soft tissue infection due to group A streptococcus, including those with necrotizing fasciitis due to other bacteria. speB was detected by PCR for one patient (patient 10) with necrotizing fasciitis whose blood and tissue cultures were negative possibly because broad-spectrum antibiotics had already been started; this patient’s group A streptococcal infection was confirmed serologically. Although the number of patients evaluated is small, these results indicate that PCR detection of the speB gene in tissue samples of patients with necrotizing fasciitis may be a useful adjunct to conventional culture methods for use in select cases.
Acknowledgments
This work was supported, in part, by a grant from the Canadian Bacterial Diseases Network.
We thank L. Cook for excellent secretarial services.
REFERENCES
- 1.Brook I, Frazier E H. Clinical and microbiological features of necrotizing fasciitis. J Clin Microbiol. 1995;33:2382–2387. doi: 10.1128/jcm.33.9.2382-2387.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carapetis J, Robins-Browne R, Martin D, Shelby-James T, Hogg G. Increasing severity of invasive group A streptococcal disease in Australia: clinical and molecular epidemiological features and identification of a new virulent M-nontypeable clone. Clin Infect Dis. 1995;21:1220–1227. doi: 10.1093/clinids/21.5.1220. [DOI] [PubMed] [Google Scholar]
- 3.Davies H D, McGeer A, Schwartz B, Green K, Cann D, Simor A E, Low D E the Ontario Group A Streptococcal Study Group. Invasive group A streptococcal infections in Ontario, Canada. N Engl J Med. 1996;335:547–554. doi: 10.1056/NEJM199608223350803. [DOI] [PubMed] [Google Scholar]
- 4.Gamba M-A, Martinelli M, Schaad H J, Streuli R A, DiPersio J, Matter L, Ris H-B, Marchal F, Kaplan E L, Stevens D L, Malinverni R. Familial transmission of a serious disease-producing group A streptococcus clone: case reports and literature review. Clin Infect Dis. 1997;24:1118–1121. doi: 10.1086/513636. [DOI] [PubMed] [Google Scholar]
- 5.Hauser A R, Schlievert P M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J Bacteriol. 1990;172:4536–4542. doi: 10.1128/jb.172.8.4536-4542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauser A R, Stevens D L, Kaplan E L, Schlievert P M. Molecular analysis of pyrogenic exotoxins from Streptococcus pyogenes isolates associated with toxic shock-like syndrome. J Clin Microbiol. 1991;29:1562–1567. doi: 10.1128/jcm.29.8.1562-1567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoge C W, Schwartz B, Talkington D F, Breiman R F, MacNeill E M, Englender S J. The changing epidemiology of invasive group A streptococcal infections and the emergence of streptococcal toxic shock-like syndrome. A retrospective population-based study. JAMA. 1993;269:384–389. [PubMed] [Google Scholar]
- 8.Kapur V, Majesky M W, Li L-L, Black R A, Musser J M. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul R, McGeer A, Low D E, Green K, Schwartz B, Simor A E Ontario Group A Streptococcal Study. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Am J Med. 1997;103:18–24. doi: 10.1016/s0002-9343(97)00160-5. [DOI] [PubMed] [Google Scholar]
- 10.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. N Engl J Med. 1990;323:1573–1579. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 12.Ruoff K L. Streptococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 299–307. [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. 1989. , p. E.3–E.4 and E.10–E.11. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 14.Tyler S D, Johnson W M, Huang J C, Ashton F E, Wang G, Low D E, Rozee K R. Streptococcal erythrogenic toxin genes: detection by polymerase chain reaction and association with disease in strains isolated in Canada from 1940 to 1991. J Clin Microbiol. 1992;30:3127–3131. doi: 10.1128/jcm.30.12.3127-3131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]