Abstract
Mutation of the MAPK7 gene was related to human scoliosis. Mapk7 regulated the development of limb bones and skulls in mice. However, the role of MAPK7 in vertebral development is still unclear. In this study, we constructed Col2a1-cre; Mapk7f/f transgenic mouse model to delete Mapk7 in cartilage, which displayed kyphosis and osteopenia. Mechanistically, Mapk7 loss decreased MEF2C expression and thus activated PTEN to oppose PI3K/AKT signaling in vertebral growth plate chondrocytes, which impaired chondrocyte hypertrophy and attenuated vertebral ossification. In vivo, systemic pharmacological activation of AKT rescued impaired chondrocyte hypertrophy and alleviated mouse vertebral defects caused by Mapk7 deficiency. Our study firstly clarified the mechanism by which MAPK7 was involved in vertebral development, which might contribute to understanding the pathology of spinal deformity and provide a basis for the treatment of developmental disorders of the spine.
Keywords: Chondrocyte hypertrophy, Growth plate, Kyphosis, MAPK7, MEF2C, Osteopenia, PI3K/AKT, PTEN
Introduction
The vertebra derives from the sclerotome, an embryonic tissue differentiated from somite.1 The sclerotome is composed of pluripotent and mesenchymal stem cells, which organize to form the vertebral primordium and become the vertebra through endochondral bone formation.2 In this process, mesenchymal stem cells gather and differentiate into primary chondrocytes to form the vertebral cartilage template.3 Chondrocytes in the center of the cartilage template proliferate, undergo hypertrophic differentiation, and then form the vertebral growth plate, which includes the resting zone, proliferating zone, and hypertrophic zone.4 Subsequently, blood vessels invade the hypertrophic zone and promote the formation of primary spongiosa to extend the vertebrae. Concurrently, the perichondrium surrounding the primary spongiosa becomes periosteum and undergoes intramembranous ossification to form the bone collar to enlarge the vertebrae.5 The hypertrophic chondrocytes not only secrete a calcified extracellular matrix to form a scaffold for endochondral ossification5 but also transdifferentiate into osteoblasts and are responsible for trabecular bone formation.6, 7, 8 The vertebral growth rate is primarily determined by hypertrophic chondrocytes.9 Therefore, dysfunction of chondrocyte hypertrophy might lead to vertebral malformation.10
Mitogen-activated protein kinase 7 (MAPK7), also known as extracellular signal-regulated kinase 5 (ERK5), encoded by the MAPK7 gene,11 is essential in skeletal development and maintaining bone homeostasis.12 Our previous studies found that MAPK7 mutation was related to scoliosis in humans, and Mapk7 null zebrafish displayed scoliosis and kyphosis deformities.13,14 A recent study demonstrated that the ablation of Mapk7 in mesenchymal cells disturbed endochondral bone formation and attenuated the development of limb bones.15 Our study further clarified that Mapk7 loss in chondrocytes impaired the adaptation of chondrocytes to hypoxia and thus caused defects of limb bone development.16 Moreover, Mapk7 deletion in LepR + BMSC promoted osteoblast differentiation in adult mice.17 However, the role of MAPK7 in vertebral development is still unknown.
In this study, we generated Col2a1-Cre; Mapk7f/f mouse model to delete Mapk7 in cartilage and found severe vertebral defects, including kyphosis and osteopenia. Subsequent analyses revealed that Mapk7 deletion impaired chondrocyte hypertrophy of the vertebral growth plate via inhibiting MEF2C/PTEN/AKT signaling. Our study provides an experimental basis for the treatment of developmental disorders of the spine.
Materials and methods
Animals and in vivo treatment
Mapk7f/f and Col2a1-Cre mouse strains were described in our previous study.16 Mapk7f/f mice were crossed with Col2a1-Cre mice to generate Col2a1-cre; Mapk7f/f (CKO) and Mapk7f/f (WT) mice. The P1 CKO newborn mice were injected with SC79 (0.04 mg/g; Selleck, S7863) intraperitoneally every day for 8 weeks, and the control littermates were treated with equivalent volumes of the vehicle. All mice were raised in an SPF environment. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Sun Yat-sen University (Approval No. SYSU-IACUC-2022-001663).
Primary chondrocyte culture and hypertrophic differentiation
Primary chondrocytes were isolated from the vertebral growth plate of P7 WT and CKO newborn pups. After removing the ligament and disc, we dissected the growth plate tissue from the vertebrae under a stereo microscope. The growth plate tissue was digested with 0.1% collagenase type II (Sigma, V900892-100 MG) overnight at 37 °C. Primary chondrocytes were obtained and seeded at a density of 2.5 × 104 cells/cm2 in a culture dish with DMEM/F-12 (1:1) containing 10% fetal bovine serum (Gibco, 10099-141) and 1% penicillin-streptomycin (Gibco, 15140122). To induce chondrocyte hypertrophy, we supplemented the above culture medium with 1% ITS (Sigma, I3146) and 50 μg/mL ascorbic acid (Sigma, A5960-100G).18 The cultures were maintained in a 37 °C and 5% CO2 incubator.
Whole-mount skeletal staining
We collected the axial skeletons of CKO and WT newborn mice at P0 and P7 for Alcian blue and Alizarin red staining as described previously.19 Briefly, after removing the skin, eyes, internal organs, adipose tissue, and limbs, we fixed the specimens successively in 95% ethanol and acetone overnight at room temperature. Subsequently, the samples were incubated in 0.03% Alcian blue solution (Solarbio, China) overnight at room temperature. After being destained with 70% alcohol, specimens were submerged in 95% alcohol overnight at room temperature and then in 1% KOH solution. When the tissue appeared transparent, we incubated the samples with Alizarin red solution (Solarbio, China) for 12 h at room temperature. Finally, we transferred the specimens to a 50% glycerol: 50% (1%) KOH solution until they were clear and then maintained them in 100% glycerol.
X-ray and micro-CT analysis
Eight-week-old CKO and WT mice were euthanized, and radiographs of skeletons were performed using an X-ray system (Siemens, German) at 50 kV and 300 μA for 10 s. The kyphosis index (KI) was measured from the lateral radiographs as described previously.20 Briefly, KI was defined as the rate of AB and CD. The length of AB was from the posterior edge of the last cervical vertebra to the posterior edge of the sixth lumbar vertebra. The length of CD was the vertical distance from the point of greatest curvature to line AB. Subsequently, we removed the soft tissues, harvested the spine, and fixed them in 1% paraformaldehyde for 36 h. The spine was scanned with Micro-CT (SkyScan1276, Bruker) at a 10-μm resolution with a voltage of 70 kV and a current of 200 μA. The bone parameters of bone volume per tissue volume (BV/TV), trabeculae number (Tb.N.), trabecular thickness (Tb.Th.), and trabecular separation (Tb.Sp.) were used to quantify the trabecular region below the vertebral growth plate.
Histological analysis
For hematoxylin-eosin (H&E) staining, spine tissues of P7 CKO and WT newborn mice were fixed in 4% paraformaldehyde (PFA) for 36 h and decalcified in EDTA for three weeks. The specimens were embedded in paraffin, and 5-μm tissue sections were prepared. Sections were deparaffinized in xylene and rehydrated in graded ethanol. We stained the sections with hematoxylin for 10 min and rinsed them with 1% acid alcohol solution for 2 s. Then, the sections were washed with current water for 20 min and stained with eosin for 5 min. For von Kossa staining, 6-μm frozen slices of spine tissues from P0 mice were incubated with 1% silver nitrate solution and irradiated under a 60 w UV lamp for 1 h. Slices were then washed in distilled water three times and incubated with 5% sodium hyposulfite for 5 min. 0.1% nuclear neutral red staining solution was used to counterstain the nuclei for 5 min. For TRAP staining, 6-μm frozen sections of spine tissues from P7 mice were stained using the Acid Phosphatase, Leukocyte (TRAP) Kit (Sigma, NO387A) according to the manufacturer's instructions. All of the above sections were dehydrated, cleared, and mounted.
Alizarin red staining
Primary chondrocytes were seeded at a density of 1 × 105 cells per well in a 24-well plate overnight and transfected with plasmids using Lipofectamine 3000 (Invitrogen, L3000-015). The cultures were maintained in DMEM/F-12 (1:1) containing 10% FBS, 1% ITS, and 50 μg/mL of ascorbic acid for 21 days. After fixation with 4% PFA for 5 min, the cells were stained with Alizarin red solution for 30 min. An ImmunoSpot Analyser (CTL S6 Ultra, American) was used to visualize the mineralized area.
Immunofluorescence assay
Spine tissues of P7 CKO and WT newborn mice were collected, and 6-μm frozen sections were prepared. The sections were permeabilized with 0.1% Triton X-100 for 5 min and blocked in 5% BSA for 30 min at room temperature. Then, sections were incubated with primary antibodies against mouse MAPK7 (Cell Signaling Technology, 3372s; 1:200), COL10A1 (Abcam, ab49945; 1:500), RUNX2 (Abcam, ab192256; 1:400), MMP13 (Abcam, ab39012; 1:400), PTEN (Cell Signaling Technology, 9559s; 1:400), p-AKT (Cell Signaling Technology, 9018s; 1:200), MEF2C (Cell Signaling Technology, 5030s; 1:200), HIF1α (Cell Signaling Technology, 36169s; 1:200), and Cre recombinase (Abcam, ab216262; 1:500) overnight at 4 °C. The following incubation with the secondary antibody Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen, S11227; 1:1000) was performed at room temperature for 1 h. Nuclei were labeled with DAPI (Sigma, SLCJ8103) for 5 min. The immunofluorescence staining images were captured with fluorescence microscopy (Olympus, BX63, Japan) and quantified using Image J software.
Proliferation and apoptosis analysis
For ethynyl deoxyuridine (EdU) assay, P7 CKO and WT newborn mice were injected intraperitoneally with EdU (ST067, Beyotime; 20 mg/g) and euthanized 4 h later. As described previously, the spine tissues were fixed, and 6-μm frozen sections were prepared. EdU was detected using the BeyoClick™ Cell Proliferation Kit with Alexa Fluor 555 (C0075S, Beyotime) following the manufacturer's instructions. For the TUNEL assay, we fixed spine tissues of P7 CKO and WT mice and then prepared 6-μm frozen slices. Following the manufacturer's manual, we detected apoptotic chondrocytes in the vertebra using the TdT-mediated dUTP Nick-End Labelling kit (C1088, Beyotime). Briefly, slices were incubated with 20 μg/mL proteinase K for 30 min at 37 °C and then transferred to TUNEL detective solution for 1 h at 37 °C, followed by counterstaining and mounting with DAPI (Sigma, SLCJ8103). Fluorescence microscopy (Olympus, BX63, Japan) was used to capture the immunofluorescence staining images.
RNA-seq analysis
We isolated the vertebral growth plate cartilage from P7 CKO newborn mice (n = 3) and WT littermates (n = 3) under a stereo microscope. Total RNA was extracted from the growth plate using TRIzol regent (Invitrogen, Carlsbad, CA). We constructed libraries using TruSeq Stranded mRNA LTSample Prep Kit (Illumina, USA) and sequenced them on the Illumina sequencing platform according to the manufacturer's instructions. Genes with a fold change of 2 and a p-value less than 0.05 were considered significantly differentially expressed. GO enrichment and KEGG pathway enrichment of differentially expressed genes were analyzed using the online tool of DAVID Bioinformatics Resources 2021.21
Real-time quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from the vertebral growth plate or cultured cells using TRIzol regent (Invitrogen, Carlsbad, CA). cDNA was synthesized using UnionScript First-strand cDNA Synthesis Mix for qPCR (with dsDNase) (SR511, Genesand, China) according to the manufacturer's instructions. Subsequently, RT-qPCR was performed on CFX 96 Real-Time PCR System (Bio-Rad) using GS AntiQ qPCR SYBR Green Master Mix (SQ412, Genesand, China). The specific primers used for RT-qPCR were shown in Table S1. Gapdh was used as the reference gene to normalize the transcript levels, and relative gene expression was quantified by the 2−ΔΔCt method.
Western blot analysis
We extracted proteins from the vertebral growth plate tissues or cultured cells using RIPA buffer (Epizyme, PC101) supplemented with protease and phosphatase inhibitors. A BCA protein assay kit (Epizyme, ZJ101) was used to quantify the total proteins. Specific proteins were separated by 10% SDS-PAGE and then transferred to PVDF membranes (Millipore). We used 5% skim milk to block the membranes for 1 h at room temperature and then incubated the membranes with primary antibodies overnight at 4 °C and secondary antibodies for 1 h at room temperature. We used an ECL kit (Epizyme, SQ202) for chemiluminescence detection on a ChemiDoc multiplex imaging system (Bio-Rad). Primary antibodies used for Western blotting were rabbit anti-MAPK7 (Cell Signaling Technology, 3372s; 1:1000), anti-COL10A1 (Abcam, ab58632; 1:1000), anti-RUNX2 (Abcam, ab192256: 1:1000), anti-MMP13 (Abcam, ab19013; 1:1000), anti-Ki67 (Abcam, ab16667; 1:1000), anti-Bax (Abcam, ab182733; 1:1000), anti-caspase 3 (Abcam, ab184787; 1:1000), anti-PTEN (Cell Signaling Technology, 9559s; 1:1000), anti-AKT (Cell Signaling Technology, 75692s; 1:1000), anti-p-AKT (Cell Signaling Technology, 9018s; 1:1000), anti-PI3K (Cell Signaling Technology, 4257t; 1:1000), anti-p-PI3K (Cell Signaling Technology, 4228t; 1:1000), anti-MEF2C (Cell Signaling Technology, 5030s, 1:1000), anti-phospho-MEF2C (Abcam, ab78888; 1:1000, anti-HIF1α (Cell Signaling Technology, 36169s; 1:1000), Anti-Osteopontin (Abcam, ab214050; 1:1000), Anti-Osteocalcin (Abcam, ab93876; 1:1000), and anti-GAPDH (Cell Signaling Technology, 5174s; 1:2000).
Plasmid and transfection
SgRNA-Mapk7 and sgRNA-PTEN were constructed (Tsingke Biotechnology, China) and cloned into pSpCas9(BB)-2A-GFP (PX458) plasmid as described previously.22 The sgRNA sequences were listed in Table S2. The empty vector plasmid was used as the negative control. The plasmid was transfected into primary chondrocytes using Lipofectamine 3000 (Invitrogen, L3000015) reagent according to the manufacturer's protocol. Briefly, primary chondrocytes were plated at a density of 1 × 105 cells/cm2 in a 24-well (or 6-well) culture plate overnight. Lipofectamine 3000 Reagent (1 μL) was diluted with Opti-MEM™ medium (25 μL) (Invitrogen, 31985088). We diluted DNA plasmid (0.5 μg) with Opti-MEM™ medium (25 μL) and P3000 Reagent (1 μL). The diluted plasmid and Lipofectamine 3000 Reagent above were mixed and incubated for 15 min at room temperature. Plasmid-Lipofectamine 3000 complex was added to chondrocytes and maintained for 6 h. Subsequently, we cultured the cells in a medium containing ITS and ascorbic acid, as described previously, to induce hypertrophic differentiation for 21 days and then performed alizarin red staining, Western blotting, and RT-qPCR analysis.
Luciferase reporter assay
293T cells were seeded at a density of 2 × 105 cells/well in a 24-well plate for 24 h and transfected with a luciferase reporter plasmid containing PTEN promoter fragments and renilla control plasmid along with MEF2C expressing plasmid or negative control plasmid (NC), as indicated, using Lipofectamine 3000 (Invitrogen, L3000-015) according to the manufacturer's protocol. After 48-h transfection, cells were lysed, and luciferase and renilla activities were measured using a Dual-Luciferase Reporter Gene Assay Kit (Beyotime, China) on ELISA (Infinite F500, Tecan).
Statistical analysis
All experiments were repeated at least three times with independent samples. The statistical analyses were performed using GraphPad Prism 9.0 software. Two-tailed paired Student's t-test determined statistical significance between two groups and one-way ANOVA with Tukey's multiple comparisons test between multiple groups. A P-value < 0.05 was considered statistically significant. All data were presented as the mean ± standard deviation (SD).
Results
Mapk7 deletion in chondrocytes caused kyphosis and osteopenia in mice
To investigate the role of MAPK7 in vertebral development, we constructed Col2a1-Cre; Mapk7f/f mice (conditional knockout, CKO) by crossing Mapk7f/f mice (wild type, WT) with Col2a1-Cre mice. The expression of Cre recombinase was detected in the growth plate and nucleus pulposus of P7 CKO pups and Mapk7 was efficiently deleted in these tissues, which was confirmed by immunofluorescence staining and Western blotting (Fig. S1A–C). The physical size of P0 CKO mice with no obvious abnormality in appearance was similar to their age-matched control WT littermates (Fig. 1A, C). However, we observed that P7 CKO mice exhibited kyphosis and decreased body length relative to the control WT pups (Fig. 1B, C). Whole-mount skeletal staining of P0 newborn mice showed that the vertebral ossification was delayed in CKO pups with some ossification region of caudal vertebrae missing and a decline of vertebral height compared with the control littermates (Fig. 1D, F). In addition, obvious kyphosis deformity with a reduction of vertebral height, an increase of vertebral width, and malformation of the vertebral arch, was exhibited in P7 CKO newborn mice (Fig. 1E). X-ray images showed that the 8-week-old CKO mice displayed more curved in the thoracic region with a reduction in the kyphosis index (KI) compared with the control WT mice (Fig. 1G, H). Furthermore, Micro-CT was used to analyze the thoracic vertebrae of 8-week-old CKO mice and their control WT mice. The three-dimensional reconstruction of Micro-CT images showed a wedge-shaped deformity of the twelfth and thirteenth thoracic vertebra of CKO mice (Fig. 1I). Moreover, sagittal, transverse, and coronal Micro-CT images showed that the vertebrae of CKO mice were shorter and wider than those of WT mice (Fig. 1J). Further analysis showed that the 8-week-old CKO mice displayed osteopenia, as evidenced by a decrease in bone volume per tissue volume (BV/TV), trabecular number (Tb.N.), and trabecular thickness (Tb.Th.) and an increase in trabecular separation (Tb.Sp.) compared with the control WT mice (Fig. 1K). Collectively, these results demonstrated that MAPK7 was required for normal vertebral development.
Figure 1.
Mapk7 deletion in chondrocytes caused vertebral defects in mice. (A, B) Gross appearance of WT and CKO newborn pups at P0 (A) and P7 (B). Scale bar = 1 cm. (C) Quantification of the body length of WT and CKO mice at indicated ages. (D, E) Alizarin red and Alcian blue staining of the axial skeleton of P0 (D) and P7 (E) WT and CKO newborn mice. The red arrow indicates the lack of Alizarin red staining in some caudal vertebrae of CKO newborns. Scale bar = 1 mm. (F) Statistical analysis of the vertebral height of P0 WT and CKO newborns based on Alizarin red and Alcian blue staining. (G) X-ray images of 8-week-old WT and CKO mice. AB: distance from the posterior edge of the last cervical vertebra to the posterior edge of the sixth lumbar vertebra. CD: vertical distance from the point of greatest curvature to line AB. Scale bar = 1 cm. (H) Quantification of kyphosis of 8-week-old mice according to X-ray images. Kyphosis index (KI) = AB/CD. (I) Three-dimensional reconstruction of micro-computed tomography (Micro-CT) images of the spine of 8-week-old WT and CKO mice. The white arrow indicates the 12th and 13th thoracic vertebrae. Scale bar = 1 mm. (J) Sagittal, transverse, and coronal Micro-CT images of the vertebrae. Scale bar = 1 mm. (K) Quantitative parameters of Micro-CT, including bone volume per tissue volume (BV/TV), trabecular number (Tb.N.), trabecular separation (Tb.Sp.), and trabecular thickness (Tb.Th.). Data were presented as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. ns, no significance.
Mapk7 deficiency impaired chondrocyte proliferation and hypertrophy
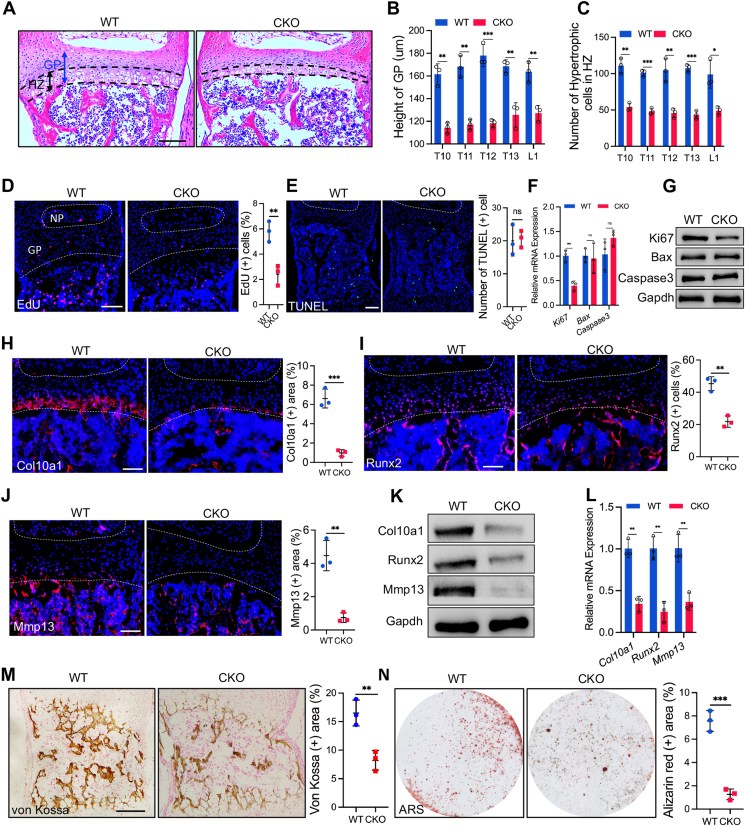
As vertebral development and ossification are regulated by the vertebral growth plate,23 we then performed a histological analysis of the vertebral body of CKO mice and WT littermates to determine whether the vertebral defects in CKO mice were related to dysplasia of the vertebral growth plate. H&E staining showed that the height of the vertebral growth plate of P7 CKO mice was shorter than that of the control mice (Fig. 2A, B), especially the hypertrophic zone (HZ), in which the number of hypertrophic cells was less than that of WT littermates (Fig. 2C). In addition, the hypertrophic chondrocytes were not observed in CKO mice aged 4 weeks compared with the WT control mice (Fig. S2). The narrower hypertrophic zone might result from decreased proliferation, increased apoptosis, or reduced hypertrophic differentiation of chondrocytes. Hence, we first performed EdU and TUNEL assays to analyze the status of chondrocyte proliferation and apoptosis in the vertebral growth plate. We observed a significant reduction in the percentage of EdU-positive cells in the vertebral growth plate of P7 CKO pups (Fig. 2D). Moreover, we isolated the vertebral growth plate from P7 CKO and WT newborns and found that the expression of Ki67, a marker of cell proliferation, was significantly decreased in CKO mice (Fig. 2F, G), which evidenced the depression of chondrocyte proliferation. However, the number of TUNEL-positive cells in the vertebra of CKO mice was similar to that of control WT mice (Fig. 2E). The expression of Bax and caspase 3, two apoptosis-related proteins, in the vertebral growth plate of P7 CKO pups was comparable to that of WT littermates (Fig. 2F, G), indicating that the apoptosis of chondrocytes was not affected by Mapk7 deficiency. Subsequently, we examined the status of chondrocyte hypertrophy in the vertebral growth plate. The expression of Col10a1, Runx2, and Mmp13, which are markers of hypertrophic, early-hypertrophic, and late-hypertrophic chondrocytes, respectively, was significantly decreased in the growth plate of CKO mice relative to their control WT littermates, as evidenced by immunofluorescence staining (Fig. 2H–J), Western blotting (Fig. 2K), and RT-qPCR analysis (Fig. 2L). To investigate whether bone formation in CKO mice was impaired, we performed von Kossa staining of vertebrae. The results showed that mineralization of the vertebral cartilage matrix was decreased in P0 CKO newborn mice (Fig. 2M). In addition, the expression of osteocalcin (OCN), a marker of osteoblasts, was reduced in the vertebrae of CKO mice (Fig. S3). The H&E staining showed that the number of vertebral trabecular bones was significantly reduced in CKO mice aged 8 weeks. To further examine the hypertrophic differentiation potential of chondrocytes, we isolated primary chondrocytes from CKO newborn pups and WT littermates and cultured them as monolayers in a hypertrophic differentiation medium for 21 days. The Alizarin red-positive staining areas of chondrocytes from CKO mice were less than those of WT mice, which suggested reduced maturation of chondrocytes in CKO mice (Fig. 2N). These results revealed that Mapk7 loss impaired the proliferation and hypertrophy of chondrocytes in the vertebral growth plate and delayed bone formation.
Figure 2.
Mapk7 loss impaired chondrocyte proliferation and hypertrophy. (A) Hematoxylin–Eosin (H&E) staining of the vertebral growth plate of P7 WT and CKO newborn mice. GP, growth plate. HZ, hypertrophic zone. Scale bar = 100 μm. (B, C) Quantification of the height of growth plate (B) and the number of hypertrophic cells in the hypertrophic zone (C) based on H&E staining of the vertebral growth plate of P7 WT and CKO newborns. (D) EdU assay of the vertebral growth plate of P7 WT and CKO newborn mice and quantification of EdU-positive cells (right panel). NP, nucleus pulposus. GP, growth plate. Scale bar = 100 μm. (E) TUNEL assay of the vertebra of P7 WT and CKO newborns and quantification of TUNEL-positive cells (right panel). Scale bar = 100 μm. (F, G) mRNA (F) and protein (G) levels of Ki67, Bax, and caspase 3 in the vertebral growth plate of P7 WT and CKO mice. (H, I, J) Immunofluorescence staining and quantification (right panel) of Col10a1 (H), Runx2 (I), and Mmp13 (J) in the vertebral growth plate of P7 WT and CKO mice. (K, L) Protein (K) and mRNA (L) levels of Col10a1, Runx2, and Mmp13 in the vertebral growth plate of P7 WT and CKO mice. (M) Von Kossa staining of the vertebra of P0 WT and CKO newborns and quantification (right panel) of von Kossa staining. Scale bar = 100 μm. (N) Alizarin red staining (ARS) of primary chondrocytes cultured as monolayer in a hypertrophic differentiation medium for 21 days. The right panel was the quantification of ARS. Data were shown as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. ns, no significance.
Mapk7 loss inhibited chondrocyte hypertrophy via regulating PTEN/PI3K/AKT signaling
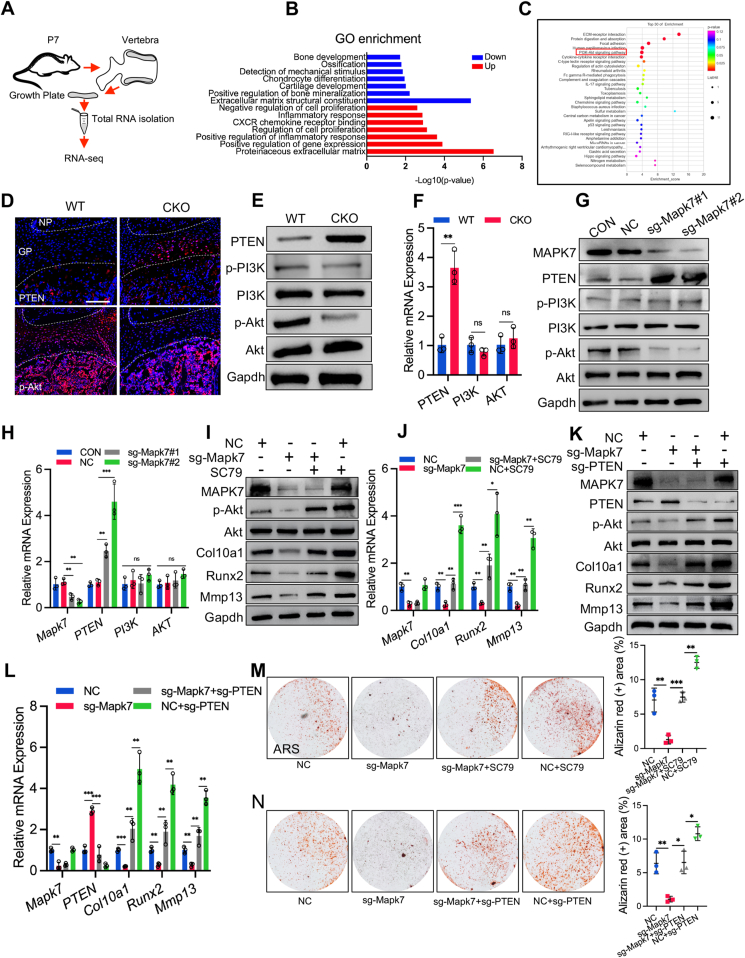
To examine the molecular mechanism of vertebral growth plate defects induced by Mapk7 deletion in chondrocytes, we isolated the vertebral growth plate cartilage from P7 CKO newborn pups and WT littermates and then performed RNA-sequencing (RNA-seq) analysis (Fig. 3A). GO enrichment analysis showed that the down-regulated genes in CKO mice were mainly related to cartilage development and ossification (Fig. 3B). KEGG pathway analysis revealed significant enrichment of PI3K/AKT signaling (Fig. 3C). It was reported that PI3K/AKT signaling plays an essential role in maintaining chondrocyte hypertrophy during endochondral bone formation.24 Hence, we hypothesized that the impaired chondrocyte hypertrophy might result from the down-regulation of PI3K/AKT signaling in CKO mice. To elucidate this hypothesis, we first detected the expression of PI3K, phospho-PI3K (p-PI3K), AKT, and phospho-AKT (p-AKT) in the vertebral growth plate of P7 CKO and WT mice. The expression of PI3K, p-PI3K, and AKT showed no significant difference between WT and CKO mice, but the protein level of p-AKT significantly declined in CKO mice (Fig. 3E, F). Previous studies found that PTEN, a tumor suppressor, opposed PI3K/AKT activation25 and regulated cartilage development.26 We found that PTEN expression was significantly increased in the growth plate of CKO mice, which was confirmed by immunofluorescence staining (Fig. 3D), Western blotting (Fig. 3E), and RT-qPCR (Fig. 3F). Our in vitro study further identified that knockdown of MAPK7 by Cas9/sgRNA-Mapk7 (sg-Mapk7) in primary chondrocytes led to up-regulation of PTEN and down-regulation of p-AKT (Fig. 3G, H). These findings suggested that MAPK7 regulated PTEN/AKT signaling in chondrocytes.
Figure 3.
Mapk7 deficiency inhibited chondrocyte hypertrophy by regulating PTEN/PI3K/AKT signaling. (A) Schematic of the procedure for isolating vertebral growth plate cartilage for RNA-seq analysis. (B) GO enrichment analysis of the differentially expressed genes for biological processes. (C) KEGG enrichment analysis of differentially expressed genes for signaling pathways. (D) Immunofluorescence staining for localization of PTEN and p-AKT in the vertebral growth plate of P7 WT and CKO newborn mice. Scale bar = 100 μm. (E) Western blot analyses of PTEN, PI3K, p-PI3K, AKT, and p-AKT in the vertebral growth plate of P7 WT and CKO mice. (F) mRNA expression of PTEN, PI3K, and AKT in the vertebral growth plate of P7 WT and CKO newborn mice. (G, H) Protein levels of MAPK7, PTEN, PI3K, p-PI3K, AKT, and p-AKT (G) and mRNA levels of MAPK7, PTEN, PI3K, and AKT (H) in primary chondrocytes transfected with a plasmid expressing Cas9/sgRNA-Mapk7 (sg-Mapk7) or negative control plasmid (NC). (I, J) Primary chondrocytes with Mapk7 deficiency were treated with SC79 (4 μg/mL) and induced to hypertrophic differentiation for 21 days, followed by Western blotting (I) for expression of MAPK7, p-AKT, AKT, Col10a1, Runx2, and Mmp13 and RT-qPCR analyses for indicated mRNA levels (J). (K, L) Primary chondrocytes with MAPK7 knockdown were transfected with a plasmid expressing Cas9/sgRNA-PTEN (sg-PTEN), followed by Western blotting for expression of indicated proteins (K) and QT-PCR analyses for indicated mRNA levels (L). (M, N) Alizarin red staining (ARS) of primary chondrocytes with indicated treatments for 21 days and quantification of ARS (right panel). Data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. ns, no significance.
To further determine whether MAPK7 regulated chondrocyte hypertrophy via PTEN/AKT signaling, we transfected primary chondrocytes with a plasmid encoding Cas9/sgRNA-Mapk7 (sg-Mapk7) to knockdown MAPK7 and then stimulated these cells to hypertrophic differentiation for 21 days. The decreased expression of p-AKT, Col10a1, Runx2, Mmp13, OPN, and OCN caused by Mapk7 deficiency was significantly elevated by SC79 (4 μg/mL), an activator of AKT (Fig. 3I, J; Fig. S4A, B). In addition, the knockdown of PTEN by Cas9/sgRNA-PTEN (sg-PTEN) also considerably rescued the down-regulation of p-AKT, Col10a1, Runx2, and MMP13 in Mapk7-deficient chondrocytes (Fig. 3K, L). Moreover, the impaired maturation of chondrocytes caused by Mapk7 deficiency was reversed by AKT activation (Fig. 3M) or PTEN knockdown (Fig. 3N), as evidenced by increasing calcified nodules in alizarin red staining. Accordingly, MAPK7 regulated chondrocyte hypertrophy via PTEN/AKT signaling.
Activation of AKT alleviated vertebral defects in Mapk7-deficient mice
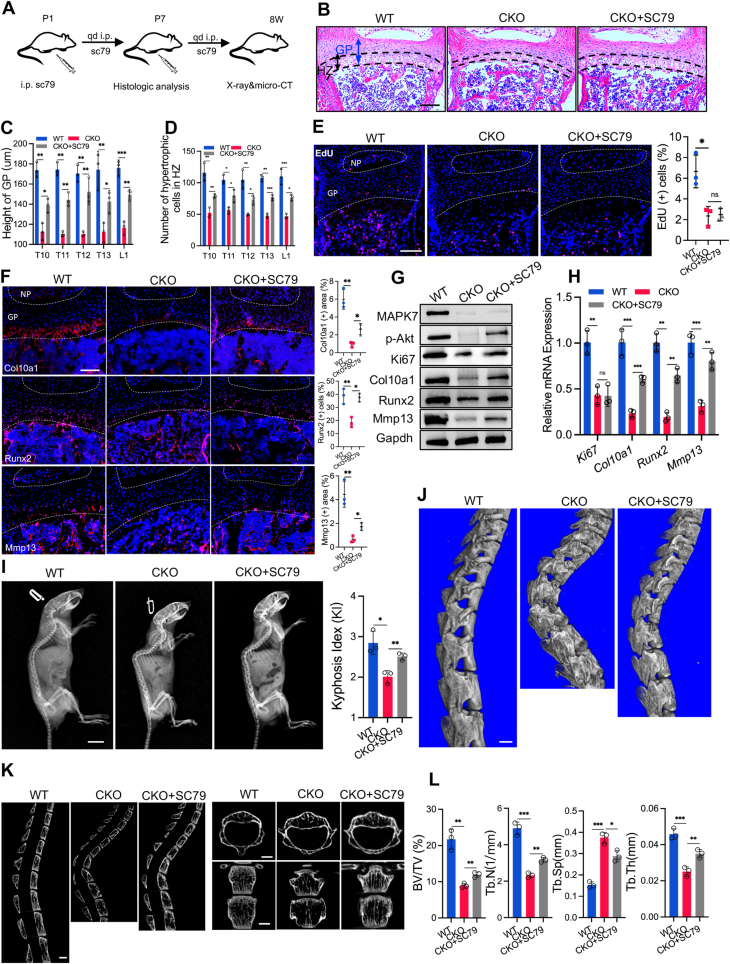
To investigate whether activation of AKT was able to attenuate the vertebral defects caused by Mapk7 deletion in chondrocytes, we intraperitoneally injected P1 CKO newborn mice with SC79 (40 mg/kg) every day to enhance the activity of AKT (Fig. 4A). The control littermates were treated with the same dose of DMSO, a solvent of SC79. H&E staining showed that the height of the vertebral growth plate and the number of hypertrophic cells in P7 CKO mice were significantly increased after 6 times SC79 treatments compared with the control CKO littermates (Fig. 4B–D). However, the reduced chondrocyte proliferation in CKO mice was not rescued by SC79 (Fig. 4E), which was also confirmed by Western blotting (Fig. 4G) and RT-qPCR analysis (Fig. 4H). Consistent with the previous in vitro study results, the decreased expression of Col10a1, Runx2, and Mmp13 in CKO mice was elevated by AKT activator, as certified by immunofluorescence staining (Fig. 4F), Western blotting (Fig. 4G), and RT-qPCR analysis (Fig. 4H). In addition, X-ray images showed that activation of AKT partially alleviated the kyphosis deformity of 8-week-old CKO mice, as evidenced by improvement of kyphosis index (KI) by SC79 (Fig. 4I). Furthermore, Micro-CT analysis of the thoracic vertebra revealed that the wedge-shaped deformity of the T12 and T13 vertebra of 8-week-old CKO mice was attenuated by SC79 (Fig. 4J, K). Further analysis showed that the vertebral bone loss in CKO mice was partially rescued by AKT activator (Fig. 4K), as CKO mice treated with SC79 displayed an increase in bone volume bone per tissue volume (BV/TV), trabeculae number (Tb.N.), and trabecular thickness (Tb.Th.) and a decrease in trabecular separation (Tb.Sp.) (Fig. 4L). These results suggested that activation of AKT partially alleviated the impaired chondrocyte hypertrophy and vertebral defects resulting from Mapk7 deletion in chondrocytes.
Figure 4.
Activation of AKT alleviated vertebral defects caused by Mapk7 deficiency. (A) Schematic of the experimental procedure of CKO newborn mice with SC79 treatment. i.p., intraperitoneal injection. qd, every day. (B) H&E staining of the vertebra of P7 WT newborns and CKO littermates with or without SC79 treatment. Scale bar = 100 μm. (C, D) Quantification of the height of the vertebral growth plate and the number of hypertrophic cells in the hypertrophic zone (D) based on H&E staining. (E) EdU staining of the vertebral growth plate of P7 WT newborn pups and CKO littermates with or without SC79 treatment and quantification of EdU staining (right panel). Scale bar = 100 μm. (F) Immunofluorescence (IF) staining of Col10a1, Runx2, and Mmp13 in the vertebral growth plate and quantification of IF staining (right panels). Scale bar = 100 μm. (G, H) Vertebral growth plate tissues were isolated from P7 WT newborns and CKO littermates with or without SC79 treatment and analyzed for protein levels of MAPK7, p-AKT, Ki67, Col10a1, Runx2, and Mmp13 (G) and indicated mRNA levels (H). (I) X-ray images of 8-week-old mice and quantification of kyphosis (right panel). Scale bar = 1 cm. (J) Three-dimensional reconstruction of Micro-CT images of the spine of 8-week-old mice. Scale bar = 1 mm. (K) Sagittal, transverse, and coronal Micro-CT images of the vertebrae. Scale bar = 1 mm. (L) Quantification of bone parameters of Micro-CT scanning, including BV/TV, Tb.N., Tb.Sp., and Tb.Th. Data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. ns, no significance.
Mapk7 deficiency up-regulated PTEN expression by inhibiting MEF2C activity in chondrocytes
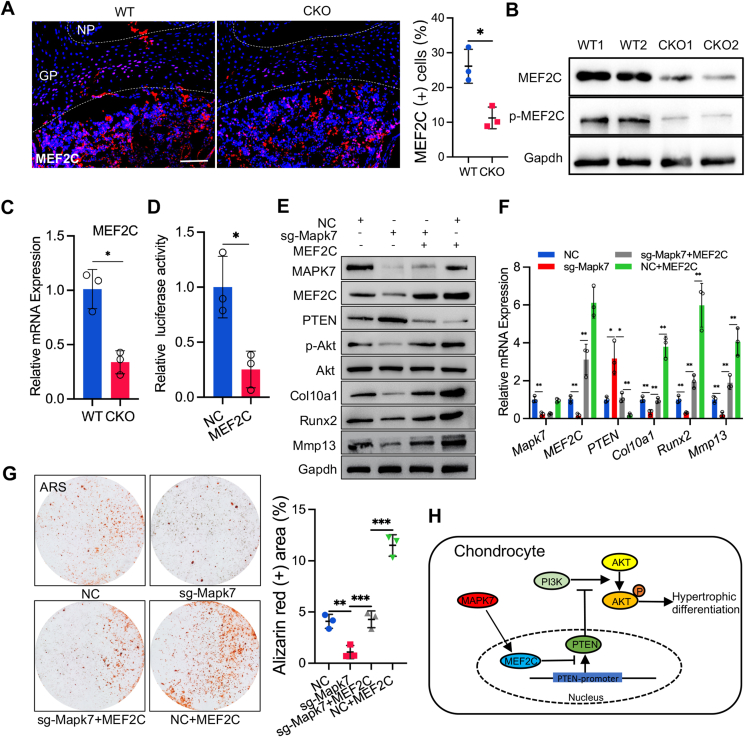
Next, we investigated the mechanism by which MAPK7 regulated PTEN expression. MAPK7 was reported to transactivate MEF2C,27 a transcription factor essential for endochondral bone formation.28 MEF2C mutant mice displayed defects in chondrocyte hypertrophy and ossification,29 which was similar to the phenotype of CKO mice. In addition, PTEN expression was reported to be inhibited by c-JUN,25 a transcription factor positively regulated by MEF2C.12 Hence, we deduced that MEF2C might be involved in the up-regulation of PTEN caused by Mapk7 deletion in chondrocytes. To elucidate this hypothesis, we first detected the expression of MEF2C in the vertebral growth plate of P7 CKO newborn mice and WT littermates. Immunofluorescence staining showed that MEF2C expression significantly decreased in the vertebral growth plate of CKO mice relative to the control WT littermates (Fig. 5A). Western blotting (Fig. 5B) and RT-qPCR analysis (Fig. 5C) further confirmed that MEF2C phosphorylation and its transcriptional activity was inhibited in CKO mice. Subsequently, to determine whether MEF2C regulated PTEN activity, we transfected 293T cells with PTEN promotor-driven luciferase reporter plasmid and MEF2C-expressing plasmid. The luciferase activity was significantly down-regulated by MEF2C overexpression, indicating that MEF2C inhibited the activity of PTEN (Fig. 5D). To explore whether MEF2C overexpression could alleviate the impaired chondrocyte hypertrophy caused by Mapk7 deficiency, we transfected primary chondrocytes with a plasmid encoding Cas9/sgRNA-Mapk7 (sg-Mapk7) or negative control plasmid (NC) with and without MEF2C-expressing plasmid and then induced these cells to hypertrophic differentiation for 21 days. We found that the increased expression of PTEN and decreased expression of p-AKT, Col10a1, Runx2, and Mmp13 caused by Mapk7 loss were significantly reversed by MEF2C overexpression (Fig. 5E, F). Moreover, alizarin red staining showed that the reduced maturation of Mapk7-deficient chondrocytes was rescued by MEF2C overexpression (Fig. 5G). These findings demonstrated that Mapk7 deficiency up-regulated PTEN expression by inhibiting MEF2C activity in chondrocytes, which down-regulated PI3K/AKT signaling and thus impaired chondrocyte hypertrophy (Fig. 5H).
Figure 5.
Mapk7 loss up-regulated PTEN expression via inhibiting MEF2C activity in chondrocytes. (A) Immunofluorescence (IF) staining of MEF2C in the vertebral growth plate of P7 WT and CKO newborn mice and quantification of IF staining (right panel). Scale bar = 100 μm. (B, C) Vertebral growth plate tissues were isolated from P7 WT and CKO pups and analyzed for protein levels of MEF2C and p-MEF2C (B) and mRNA levels of MEF2C (C). (D) Dual-Luciferase reporter analysis of the effect of MEF2C on PTEN promotor activity in 293T cell line. (E, F) Primary chondrocytes with Mapk7 deletion were transfected with a plasmid expressing MEF2C and induced to hypertrophic differentiation for 21 days, followed by Western blotting (E) for expression of MAPK7, MEF2C, p-AKT, AKT, Col10a1, Runx2, and Mmp13 and RT-qPCR analyses for indicated mRNA levels (F). (G) Alizarin red staining (ARS) of primary chondrocytes with indicated treatments for 21 days and quantification of ARS (right panel). (H) Schematic role of MAPK7 in this study. In the vertebral growth plate, MAPK7 transactivated MEF2C to inhibit the activity of PTEN, which up-regulated PI3K/AKT signaling and promoted chondrocyte hypertrophy. Data were shown as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Discussion
In this study, we first showed that MAPK7 is required for vertebral development in mice. Ablation of Mapk7 in chondrocytes resulted in remarkable vertebral developmental dysplasia, including kyphosis and osteopenia. More importantly, Mapk7 deficiency reduced chondrocyte hypertrophy of the vertebral growth plate, which impaired endochondral bone formation and thus caused vertebral defects.
The kyphotic spine deformity in CKO mice was similar to Scheuermann's disease in humans. Scheuermann's disease, also known as juvenile kyphosis, is the most common cause of kyphosis in adolescence,30 which is not apparent at birth but develops gradually in adolescence. There are many common characteristics between patients with Scheuermann's disease and CKO mice, such as wedge-shaped deformity of vertebrae,30 abnormity of vertebral growth plate,31 and juvenile osteoporosis.32 However, the etiopathogenesis of Scheuermann's disease has not been elucidated. It was reported that genetic factors33 and mechanical stress34 contributed to the pathophysiology of this disease. It will be interesting to determine whether MAPK7 is involved in the pathological process of Scheuermann's disease, which will be investigated in our future study. This kyphotic spine deformity mouse might provide an animal model to study the pathogenesis and treatment of some human kyphotic disorders.
CKO newborn mice displayed delayed vertebral ossification and these vertebral defects were associated with a narrower zone of hypertrophic chondrocytes in the vertebral growth plate. It was likely that the impaired chondrocyte hypertrophy delayed the formation of ossification and thus compromised vertebral development. Previous studies have demonstrated that impaired chondrocyte hypertrophy might result in vertebral deformities.29,35 In the current study, AKT activator treatment significantly ameliorated the impaired chondrocyte hypertrophy and spinal deformity of the CKO mice, strongly indicating that the impaired chondrocyte hypertrophy contributed to kyphosis. Similar impaired chondrocyte hypertrophy was also observed in mice with PI3K/AKT signaling inactivation, which displayed defects in endochondral bone formation.24,36 Conversely, loss of PTEN in chondrocytes up-regulated PI3K/AKT signaling and caused more extended hypertrophic zones of the growth plate and increased trabeculae.26,37 This was consistent with our findings that activation of PI3K/AKT signaling rescued the impaired chondrocyte hypertrophy in CKO mice. These studies suggested that the impaired chondrocyte hypertrophy in CKO mice might be attributed to the inactivation of PI3K/AKT signaling. It seemed that the impaired chondrocyte hypertrophy caused by the down-regulation of PI3K/AKT signaling was mediated by Runx2, a transcription factor regulating chondrocyte hypertrophy and osteoblast differentiation.38 Runx2 mutant mice exhibited a slowdown of chondrocyte hypertrophy and an absence of endochondral ossification,39 which was similar to the CKO mice. Moreover, PI3K/AKT signaling promoted terminal chondrocyte differentiation by enhancing the DNA-binding ability of Runx2.40 In our study, Mapk7 deficiency decreased the expression of Runx2, which was up-regulated by activation of PI3K/AKT signaling. Our findings, combined with previous studies, suggested that the down-regulation of PI3K/AKT signaling might cooperate with Runx2 to inhibit the hypertrophic differentiation of chondrocytes in mice with Mapk7 deletion.
It was reported that PTEN/AKT signaling regulates the expression of HIF1α,26 a transcription factor mediating hypoxia to maintain the adaptation of chondrocytes to hypoxic conditions. Our previous study found that Mapk7 deletion in chondrocytes down-regulated HIF1α and thus decreased chondrocyte proliferation and hypertrophy in the growth plate of limb bones.16 In the current study, chondrocyte proliferation in the vertebral growth plate was also reduced. However, the expression of HIF1α in the vertebral growth plate showed no significant difference between WT and CKO mice (Fig. S5A, B). In addition, the reduced proliferation of chondrocytes in CKO mice was not rescued by the AKT activator. These results indicated that the change in PTEN/AKT signaling might not contribute to the decreased proliferation of chondrocytes and might not regulate the expression of HIF1α in the vertebral growth plate of CKO mice. Furthermore, a delay in endochondral ossification was not observed in the long bones of Col2a1-cre; Mapk7f/f mice.16 These results suggested that MAPK7 played different roles in the development of long bones and vertebrae, which might be attributed to the fact that limb bones were derived from somatic mesoderm and vertebrae from somites, and both were regulated by different signaling pathways.2,41 Moreover, MEF2C was also a transcription factor that positively regulated cell proliferation12 except for the negative regulation of PTEN expression in our study. This might explain why activation of AKT by SC79 increased chondrocyte hypertrophy in CKO mice but did not rescue the decreased proliferation of chondrocytes. Further investigation is needed to determine whether other signaling pathways might be involved in down-regulating chondrocyte proliferation in CKO mice.
Nkx3.1-Cre; Mapk7f/f mice also displayed similar spinal deformity and osteopenia.42 However, the loss of bone mass in Nkx3.1-Cre; Mapk7f/f adult mice was associated with increased bone resorption due to the deletion of Mapk7 in osteoclast lineage precursors, which promoted the formation of osteoclasts. This was different from our CKO mice, in which the bone loss was mainly caused by impaired endochondral bone formation with no change in bone resorption (Fig. S6A, B). A recent study found that LepR-Cre; Mapk7f/f mice displayed an increase in bone formation,17 which was opposite to the phenotypes of decreased bone mass in CKO mice. These inconsistent phenotypes in Mapk7-deficient mice might be attributed to the fact that osteoblasts mainly originated from chondrocytes of the growth plate in adolescence and then from LepR+ BMSCs after adolescence.43 In our study, terminal hypertrophic differentiation was significantly attenuated in CKO mice, thus causing the decreased formation of osteoblasts from chondrocytes. Conversely, the loss of Mapk7 in LepR+ BMSCs increased osteoblast formation in adult mice and thus promoted bone formation.17 These studies suggested that MAPK7 plays different roles in bone formation at different ages. Nevertheless, the function of MAPK7 in the transdifferentiation of chondrocytes into osteoblasts remains to be determined in our future study.
Interestingly, there were many other phenotypes displayed in CKO mice that might not be explained by impaired chondrocyte hypertrophy. First, the width of the vertebral body and long bone16 of CKO mice was wider than that of WT mice, which was similar to the phenotype of long bones of prx1-cre; Mapk7f/f mice.15 Recently, cell lineage tracing studies revealed that Col2-expressing skeletal stem cells in the growth plate contributed to chondrocytes during endochondral bone development.44,45 Skeletal stem cells were similar to mesenchymal cells,46 in which Mapk7 deletion increased chondrogenic differentiation.15,47 Hence, it is possible that Mapk7 might also be deleted in Col2-expressing skeletal stem cells and thus promotes the formation of chondrocytes to enlarge the vertebrae and long bones. Second, kyphosis deformity was not observed in P0 CKO mice but gradually appeared in one week. Likely, mechanical stress might also be involved in this process. During bone development, mechanical stimulation affects bone growth by regulating the growth plate function.48,49 Furthermore, it was reported that MAPK7 could be up-regulated in chondrocytes by mechanical stress to maintain the homeostasis of cartilage. Considering that the mechanical stress in newborn vertebrae might be different from that in fetal mice, it could be that Mapk7 loss in chondrocytes might decrease the adaptability of chondrocytes to mechanical stimulation, which might affect the proliferation or hypertrophic differentiation of chondrocytes. Further studies are needed to elucidate these hypotheses.
However, the possibility that the decreased proliferation of chondrocytes in the vertebral growth plate might also contribute to impaired chondrocyte hypertrophy could not be ruled out, which requires further investigation.
In conclusion, the findings of this study demonstrated that MAPK7 was essential for vertebral development. Mapk7 deficiency in chondrocytes reduced chondrocyte hypertrophy of the vertebral growth plate by down-regulating MEF2C/PTEN/AKT signaling and thus impaired endochondral bone formation. Our findings might contribute to understanding the mechanism by which MAPK7 regulates spinal deformities and osteopenia.
Author contributions
C.W. and H.L. performed experiments, analyzed data, and wrote the manuscript. D.Z., X.Y., Z.L., Y.C., S.Z., D.S., B.Z., C.L., and L.T. performed experiments. C.X. and P.S. designed experiments and acquired the funds.
Conflict of interests
The authors declare no competing interests.
Funding
This work was supported by the National Natural Science Foundation of China (No. 92068105, 82172376, 82072385).
Data availability
The RNA-seq data from this publication have been uploaded to the Gene Expression Omnibus (GEO) database and assigned the GEO accession number GSE218095.
Acknowledgements
We would like to express our gratitude to all the members of the Su lab and the Xu lab for the stimulating discussions. We also thank Prof. Xiaochun Bai from Southern Medical University for providing us with Col2a1-Cre mice.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2023.02.012.
Contributor Information
Caixia Xu, Email: supq@mail.sysu.edu.cn.
Peiqiang Su, Email: xucx3@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Scaal M. Early development of the vertebral column. Semin Cell Dev Biol. 2016;49:83–91. doi: 10.1016/j.semcdb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Williams S., Alkhatib B., Serra R. Development of the axial skeleton and intervertebral disc. Curr Top Dev Biol. 2019;133:49–90. doi: 10.1016/bs.ctdb.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackie E.J., Ahmed Y.A., Tatarczuch L., et al. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40(1):46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Long F., Ornitz D.M. Development of the endochondral skeleton. Cold Spring Harbor Perspect Biol. 2013;5(1) doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 6.Yang G., Zhu L., Hou N., et al. Osteogenic fate of hypertrophic chondrocytes. Cell Res. 2014;24(10):1266–1269. doi: 10.1038/cr.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., von der Mark K., Henry S., et al. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10(12) doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Tsang K.Y., Tang H.C., et al. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A. 2014;111(33):12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallett S.A., Ono W., Ono N. Growth plate chondrocytes: skeletal development, growth and beyond. Int J Mol Sci. 2019;20(23):6009. doi: 10.3390/ijms20236009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda S., Bonnamy J.P., Owen M.J., et al. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15(4):467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nithianandarajah-Jones G.N., Wilm B., Goldring C.E.P., et al. ERK5: structure, regulation and function. Cell Signal. 2012;24(11):2187–2196. doi: 10.1016/j.cellsig.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Paudel R., Fusi L., Schmidt M. The MEK5/ERK5 pathway in health and disease. Int J Mol Sci. 2021;22(14):7594. doi: 10.3390/ijms22147594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T., Chen C., Xu C., et al. Mutant MAPK7-induced idiopathic scoliosis is linked to impaired osteogenesis. Cell Physiol Biochem. 2018;48(3):880–890. doi: 10.1159/000491956. [DOI] [PubMed] [Google Scholar]
- 14.Gao W., Chen C., Zhou T., et al. Rare coding variants in MAPK7 predispose to adolescent idiopathic scoliosis. Hum Mutat. 2017;38(11):1500–1510. doi: 10.1002/humu.23296. [DOI] [PubMed] [Google Scholar]
- 15.Iezaki T., Fukasawa K., Horie T., et al. The MAPK Erk5 is necessary for proper skeletogenesis involving a Smurf-Smad-Sox9 molecular axis. Development. 2018;145(14) doi: 10.1242/dev.164004. [DOI] [PubMed] [Google Scholar]
- 16.Yang X., Zhong D., Gao W., et al. Conditional ablation of MAPK7 expression in chondrocytes impairs endochondral bone formation in limbs and adaptation of chondrocytes to hypoxia. Cell Biosci. 2020;10:103. doi: 10.1186/s13578-020-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie T., Fukasawa K., Yamada T., et al. Erk5 in bone marrow mesenchymal stem cells regulates bone homeostasis by preventing osteogenesis in adulthood. Stem Cell. 2022;40(4):411–422. doi: 10.1093/stmcls/sxac011. [DOI] [PubMed] [Google Scholar]
- 18.Hall K.C., Hill D., Otero M., et al. ADAM17 controls endochondral ossification by regulating terminal differentiation of chondrocytes. Mol Cell Biol. 2013;33(16):3077–3090. doi: 10.1128/MCB.00291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigueur D., Lyons K.M. Whole-mount skeletal staining. Methods Mol Biol. 2014;1130:113–121. doi: 10.1007/978-1-62703-989-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laws N., Hoey A. Progression of kyphosis in mdx mice. J Appl Physiol (1985) 2004;97(5):1970–1977. doi: 10.1152/japplphysiol.01357.2003. [DOI] [PubMed] [Google Scholar]
- 21.Huang D.W., Sherman B.T., Tan Q., et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ran F.A., Hsu P.D., Wright J., et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleming A., Kishida M.G., Kimmel C.B., et al. Building the backbone: the development and evolution of vertebral patterning. Development. 2015;142(10):1733–1744. doi: 10.1242/dev.118950. [DOI] [PubMed] [Google Scholar]
- 24.Ikegami D., Akiyama H., Suzuki A., et al. Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development. 2011;138(8):1507–1519. doi: 10.1242/dev.057802. [DOI] [PubMed] [Google Scholar]
- 25.Worby C.A., Dixon J.E. Pten. Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 26.Yang G., Sun Q., Teng Y., et al. PTEN deficiency causes dyschondroplasia in mice by enhanced hypoxia-inducible factor 1α signaling and endoplasmic reticulum stress. Development. 2008;135(21):3587–3597. doi: 10.1242/dev.028118. [DOI] [PubMed] [Google Scholar]
- 27.Kato Y., Kravchenko V.V., Tapping R.I., et al. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16(23):7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dy P., Wang W., Bhattaram P., et al. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell. 2012;22(3):597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold M.A., Kim Y., Czubryt M.P., et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12(3):377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Scheuermann H.W. The classic: kyphosis dorsalis juvenilis. Clin Orthop Relat Res. 1977;(128):5–7. [PubMed] [Google Scholar]
- 31.Lowe T.G., Line B.G. Evidence based medicine: analysis of Scheuermann kyphosis. Spine. 2007;32(19 Suppl):S115–S119. doi: 10.1097/BRS.0b013e3181354501. [DOI] [PubMed] [Google Scholar]
- 32.Bradford D.S., Brown D.M., Moe J.H., et al. Scheuermann's kyphosis: a form of osteoporosis? Clin Orthop Relat Res. 1976;118:10–15. [PubMed] [Google Scholar]
- 33.Damborg F., Engell V., Andersen M., et al. Prevalence, concordance, and heritability of Scheuermann kyphosis based on a study of twins. J Bone Joint Surg Am. 2006;88(10):2133–2136. doi: 10.2106/JBJS.E.01302. [DOI] [PubMed] [Google Scholar]
- 34.Palazzo C., Sailhan F., Revel M. Scheuermann's disease: an update. Jt Bone Spine. 2014;81(3):209–214. doi: 10.1016/j.jbspin.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Ueta C., Iwamoto M., Kanatani N., et al. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J Cell Biol. 2001;153(1):87–100. doi: 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulici V., Hoenselaar K.D., Gillespie J.R., et al. The PI3K pathway regulates endochondral bone growth through control of hypertrophic chondrocyte differentiation. BMC Dev Biol. 2008;8:40. doi: 10.1186/1471-213X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh S.C., Chen N.T., Lo S.H. Conditional loss of PTEN leads to skeletal abnormalities and lipoma formation. Mol Carcinog. 2009;48(6):545–552. doi: 10.1002/mc.20491. [DOI] [PubMed] [Google Scholar]
- 38.Kim J.M., Yang Y.S., Park K.H., et al. A RUNX2 stabilization pathway mediates physiologic and pathologic bone formation. Nat Commun. 2020;11:2289. doi: 10.1038/s41467-020-16038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida C.A., Yamamoto H., Fujita T., et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18(8):952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita T., Azuma Y., Fukuyama R., et al. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol. 2004;166(1):85–95. doi: 10.1083/jcb.200401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salhotra A., Shah H.N., Levi B., et al. Mechanisms of bone development and repair. Nat Rev Mol Cell Biol. 2020;21(11):696–711. doi: 10.1038/s41580-020-00279-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loveridge C.J., van 't Hof R.J., Charlesworth G., et al. Analysis of Nkx3.1:Cre-driven Erk5 deletion reveals a profound spinal deformity which is linked to increased osteoclast activity. Sci Rep. 2017;7 doi: 10.1038/s41598-017-13346-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shu H.S., Liu Y.L., Tang X.T., et al. Tracing the skeletal progenitor transition during postnatal bone formation. Cell Stem Cell. 2021;28(12):2122–2136.e3. doi: 10.1016/j.stem.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Ono N., Ono W., Nagasawa T., et al. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16(12):1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuhashi K., Ono W., Matsushita Y., et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563(7730):254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan C.F., Seo E., Chen J., et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160(1–2):285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bobick B.E., Matsche A.I., Chen F.H., et al. The ERK5 and ERK1/2 signaling pathways play opposing regulatory roles during chondrogenesis of adult human bone marrow-derived multipotent progenitor cells. J Cell Physiol. 2010;224(1):178–186. doi: 10.1002/jcp.22120. [DOI] [PubMed] [Google Scholar]
- 48.Stokes I.A., Mente P.L., Iatridis J.C., et al. Enlargement of growth plate chondrocytes modulated by sustained mechanical loading. J Bone Joint Surg Am. 2002;84(10):1842–1848. doi: 10.2106/00004623-200210000-00016. [DOI] [PubMed] [Google Scholar]
- 49.Lee D., Erickson A., Dudley A.T., et al. Mechanical stimulation of growth plate chondrocytes: previous approaches and future directions. Exp Mech. 2019;59(9):1261–1274. doi: 10.1007/s11340-018-0424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data from this publication have been uploaded to the Gene Expression Omnibus (GEO) database and assigned the GEO accession number GSE218095.