Abstract
R-spondins are secretory proteins localized in the endoplasmic reticulum and Golgi bodies and are processed through the secretory pathway. Among the R-spondin family, RSPO2 has emanated as a novel regulator of Wnt signaling, which has now been acknowledged in numerous in vitro and in vivo studies. Cancer is an abnormal growth of cells that proliferates and spreads uncontrollably due to the accumulation of genetic and epigenetic factors that constitutively activate Wnt signaling in various types of cancer. Colorectal cancer (CRC) begins when cells in the colon and rectum follow an indefinite pattern of division due to aberrant Wnt activation as one of the key hallmarks. Decades-long progress in research on R-spondins has demonstrated their oncogenic function in distinct cancer types, particularly CRC. As a critical regulator of the Wnt pathway, it modulates several phenotypes of cells, such as cell proliferation, invasion, migration, and cancer stem cell properties. Recently, RSPO mutations, gene rearrangements, fusions, copy number alterations, and altered gene expression have also been identified in a variety of cancers, including CRC. In this review, we addressed the recent updates regarding the recurrently altered R-spondins with special emphasis on the RSPO2 gene and its involvement in potentiating Wnt signaling in CRC. In addition to the compelling physiological and biological roles in cellular fate and regulation, we propose that RSPO2 would be valuable as a potential biomarker for prognostic, diagnostic, and therapeutic use in CRC.
Keywords: CRC, Gene fusion, R-Spondins, RSPO2, Therapeutics, Wnt signaling
Introduction
Cancer is generally a heterogenous disease that causes abnormal and uncontrollable cell growth that can invade and spread to other body organs or parts. In 2020, approximately 9,958,133 deaths were attributed to cancer, making it the second most prominent cause of fatalities worldwide. According to WHO reports, the most prevalent cancers in men are lung, colorectal, prostate, liver, and stomach cancers, while, in women, the most common cancers are breast, colorectal, cervix, thyroid, and lung cancers. Cancer mainly develops when normal cells transform into tumor cells via a multi-step process, typically starting with a precancerous lesion and ultimately leading to a malignant tumor. Identifying and understanding such changes in the cancer genome have become crucial for developing targeted therapeutic approaches.1
Colorectal cancer (CRC) is a heterogeneous and acute health problem worldwide. There were 1,931,590 (10%) new cases in 2020,2 making it the third-leading cancer and the second-leading cause of cancer death in the Western world.3 In India, CRC ranks third in men, while it is the fifth most diagnosed cancer in females.2 Several risk factors have been associated with CRC progression, which can be broadly categorized into genetic, epigenetic, lifestyle-related, and environmental factors. Prior research has uncovered some active signal transduction pathways, such as Wnt/β-catenin, receptor tyrosine kinase (RTK), and transforming growth factor-β (TGF-β) in CRC progression.4 In addition, alterations such as mutations, chromosomal structural variants, microsatellite instability, gene fusions, altered gene expression, and epigenetic inactivation in cancer hallmark genes have been implicated in CRC development.5 In the case of CRC, earlier studies have identified alterations in genes such as KRAS, NRAS, c-Myc, APC, TP53, CTNNB1, PIK3CA, and mismatch repair (MMR) genes with significant frequency in the number of CRC cases.6,7 In recent years, researchers have also identified several new genes that serve as a potential therapeutic target (e.g., LGR4-6, RNF43, ZNRF3, RSPO2, ANO5, SLITRK1, NRXN1, and ANK2) and are linked to the CRC development.8,9 Three prominent molecular pathways commonly implicated in CRC are the chromosomal instability pathway (CIN), the microsatellite instability pathway (MSI), and the CpG island methylator phenotype pathway (CIMP). In the case of the CIN pathway, chromosomal instability mainly occurs due to abnormal segregation of chromosomes along with aneuploidy, telomere dysfunction, and irregularity in the DNA damage repair system, which affects genes like APC, KRAS, PI3K, and TP53.10 CIMP pathway involves hypermethylation of CpG islands in promoter regions resulting in transcriptional silencing of regulatory genes such as APC, MLH1, MCC, and many others, leading to their function inactivation.11 MSI pathway primarily involves aberration of DNA mismatch repair system (MMR) either due to hypermethylation in promoter CpG region of MLH1 gene or inactivation of other MMR genes. Irregularity in the MMR system results in the aggregation of unstable microsatellites, thereby progressing CRC development.12
Molecular mechanisms of most genes have been identified; however, it largely remains obscure for a few candidate genes. Moreover, early-stage diagnosis, absence of specific reliable biomarkers, improved survival rate, and better clinical management of CRC still are major challenges that have not improved as desired. It is, therefore, imperative to gain a better understanding of the mechanism that promotes CRC development to find new strategies and therapeutic targets to combat it. This review highlights the following points: (i) Description of R-spondins (RSPOs) with emphasis on the RSPO2 gene; (ii) Effect of aberrant RSPO2 expression in different cancer types; (iii) A significant role of the RSPO2 gene in the regulation and potentiation of the Wnt signaling; (iv) Mechanism of RSPO2 mediated signaling pathways; (v) Establishing a promising role of RSPO2 fusion genes in the initiation and progression of CRC as well as various other cancers; (vi) Application of RSPO2 as a potential biomarker in CRC and the development of a progressive approach to use RSPO2 fusions as a novel therapeutic target.
Historical background, structural form, and general functions of R-spondins
The RSPO family comprises four proteins, i.e., R-spondin 1-4, which are important secreted regulators of the Wnt/β-catenin and Wnt/PCP signaling pathway and govern various functions and are involved in vital cellular processes such as stem cell renewal, morphogenesis and, tumorigenesis. The first member of the RSPO family was characterized as hRSPO3, formerly known as human protein with thrombospondin type I repeat (hPWTSR) from a study of the cDNA library of the human fetal brain in 2002. Later, other RSPO genes were subsequently identified in other species.13, 14, 15 RSPO3 (ENSG00000146374) has three transcript variants regulating various biological processes.15,16 Subsequently, RSPO1 was identified in 2006 when a homozygous mutation in RSPO1 was shown to be involved in sex reversal and hermaphroditism. RSPO1 (ENSG00000169218) possesses 4 transcript variants and has been demonstrated to exhibit a pivotal role in skin differentiation, mammary gland development, ovarian and reproductive system development in addition to skin malignancy (squamous cell carcinoma),17 via the Wnt signaling pathway by acting as a ligand for receptor LGR4/5/6.18,19 The genetic defect of anonychia has led to the discovery of RSPO4 (ENSG00000101282),20,21 which has two transcript variants and was shown to be implicated in stabilizing protein present in the Wnt signaling pathway. Further, the study suggests that its expression was associated with advanced stages of nail development in mammals.22,23 RSPO2 (ENSG00000147655), which is considered a key member of the RSPO family (formerly known as roof plate-specific spondin-2), is a secreted protein that regulates ligand-dependent β-catenin signaling. RSPO2 (alternative names: CRISTIN2, TETAMS2, and HRspo2) is located at q23.1, and consists of 5 coding exons.24 However, Nam et al in 2006 identified 6 exons and 9 transcript variants, five of which are RSPO2 protein-coding transcripts.25 All four R-spondin proteins tend to interact with both receptors i.e. LGR4/5/6 and RNF43/ZNRF3 which results in cells becoming sensitized to the canonical Wnt/β-catenin signaling pathway.14,26 However, there have been concerns as to whether all four R-spondins equally bind to RNF43/ZNRF3 receptor and exhibit Wnt stimulating activities or not. This question was solved in an experiment that found that RSPO2 and RSPO3 are more potent compared to RSPO1 and RSPO4 and have a greater affinity to interact with RNF43/ZNRF3.27,28
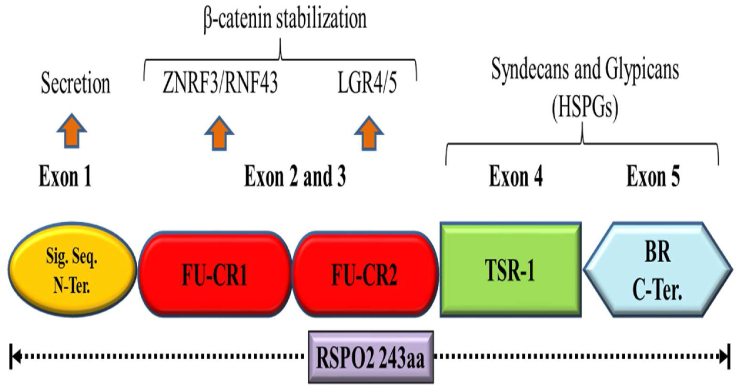
Members of RSPOs share 40%–60% structural homology between domain organizations, and each member consists of five exons in mammals, including humans and mice.15,29,30 Structurally, it consists of an N-terminal signal sequence, two adjacent furin-type cysteine-like domains (FU-CR1 and FU-CR2), conserved thrombospondin type 1 repeat (TSR-1), and followed by a variable C-terminal domain consisting of a positively charged basic amino acid (BR).24,31, 32, 33 Figure 1 illustrates the schematic representation of four R-spondin protein domains and their respective functions and interacting receptors. The four RSPO members with domains (FU-CR1 and FU-CR2) and TSR-1 share significant sequence similarities in mammals, suggesting that both domains serve almost similar functions. The two FU-CR1 and FU-CR2 domains of RSPOs lie close to the N-terminus (90–134 aa in RSPO2) and are solely responsible for Wnt signaling activation. Its deletion leads to the inactivation of downstream Wnt signaling.15,25,34 Wnt amplifying activity of R-spondin can be better understood through interactions between R-spondin-specific domains and various membrane receptors such as LGR4-6, LRP5/6, ZNRF3/RNF43, Frizzled, and heparan sulfate proteoglycans (HSPGs).25 Some pioneering work was carried out earlier, revealing ZNRF3/RNF43 directly interacts with R-spondin proteins via two conserved amino acids, arginine and glutamine present within the FU-CR1 domain.35 The FU-CR2 domain of R-spondin adjacent to FU-CR1 contains two highly conserved phenylalanine that supports direct binding with the high-affinity receptors LGR4-6.36, 37, 38 Interestingly, the mutation caused in FU-CR1 (R66A or Q71A) was shown to interrupt the interaction between RSPO1 and ZNRF3/RNF43, while a mutation affecting FU-CR2 (F106A or F110A) disrupted the contact between RSPO1 and LGR4, suggesting that RSPO binding to both receptors has emerged as a critical regulatory mechanism in activation of Wnt signaling. These experiments have illustrated two things: (i) R-spondin binds to two distinct receptors through different furin-like domains, and (ii) both domains work independently without affecting the interaction with other RSPO receptors.39,40 Moreover, overwhelming evidence also suggests that the substitution of tyrosine and arginine for cysteine residues at 78 and 113 positions within the FU-CR domain could abruptly disrupt the transport of RSPO2 as a secreted protein on the cell surface and extracellular matrix, suggesting that the FU-CR domain is also responsible for the proper secretion of RSPO2.41
Figure 1.
A visual representation of the structural domain organization corresponding to RSPO2. Other RSPO families RSPO1, RSPO3, and RSPO4 consisting of 263, 273, and 234 amino acids respectively share similar gene architecture with RSPO2 (not mapped).
In an experiment, it was shown that the TSR-1 and BR domains of RSPO2 and RSPO3 consist of 61aa (144–204), which supports the interaction with the cell surface heparan sulfate proteoglycans (HSPGs), such as syndecan 4/glypican 3, which alternatively activates the LGR4-6 independent Wnt signaling via associating with ZNRF3/RNF43.42,43 To our knowledge, there are no investigations on the involvement of HSPGs in the potentiation of canonical Wnt signaling. Earlier reports reveal that the TSR-1 domain also assists in RSPO2 binding to BMPR1A and serves as a BMP receptor antagonist.44 In a recent report, an inhibitor peptide (RW dendrimer) against the TSR-1 domain has been used for therapeutic purposes in AML. Furthermore, it was demonstrated that RW dendrimer reduces RSPO2-mediated BMP signaling in human THP-1 AML cells, induces differentiation, and consequently reduces cell growth independent of Wnt signaling.45
Mature human RSPO2 has three described isoforms resulting from alternative splicing and four computationally mapped isoforms and four variants that have not yet been well reported in the pieces of literature and are represented in Table 1. Isoforms two and three contain alternate sequences, which are subsections of natural isoforms. Functionally, Kazanskaya and co-workers have done substantial work and showed that injecting Wnt or β-catenin into the Xenopus embryo induces the expression of RSPO2/3. Moreover, decreased activity of Wnt was observed in the RSPO2 knockdown cell line, indicating that RSPO2 responded to the Wnt/β-catenin in a positive feedback manner.15,46,47 Further, recombinant hRSPO2 remarkably induces the proliferation of epithelial cells in mouse intestines by activating the Wnt/β-catenin signaling cascade.24,48 The R-spondins share some unique and conserved domain structures; these domains support the binding of the RSPO protein with different interactive receptors, thereby elucidating some critical biological functions as given in Table 2.49
Table 1.
Summary of alternatively spliced isoforms, computationally derived isoforms, and natural variants of human RSPO2.
| Alternatively spliced | |||||
|---|---|---|---|---|---|
| isoform name & tissue expressed | Size (amino acid) | UniProt ID | Position | Comments | Reference |
|
243 | Q6UXX9-1 | – | – | 50 |
|
176 | Q6UXX9-2 | 1–67 VSP_018321 | Missing in isoform 2 | 51 |
|
179 | Q6UXX9-3 | 32–95 VSP_018322 & 143 VSP_018323 | ASYVS NRCAR → G missing in isoform 3 | 51 |
| Computationally derived isoforms | |||||
| E5RH25_HUMAN | 121 | E5RH25 | – | – | 52 |
| E5RGU9_HUMAN | 76 | E5RGU9 | – | – | 52 |
| A0A590UJ52_HUMAN | 38 | A0A590UJ52 | – | – | 52 |
| RSPO2_HUMAN | 243 | Q6UXX9 | – | – | 52,53 |
| Variants | |||||
| Natural variant VAR_081036 | 1 | VAR_081036 | 69 | Change in sequence R → C in HHRRD results in decreased ability to amplify WNT3A signaling. | 54 |
| Natural variant VAR_081037 | 174 | VAR_081037 | 70–243 | Amino acid sequences 70–243 are missing in TETAMS2 and are characterized by complete loss of amplification of WNT3A signaling. | 54 |
| Natural variant VAR_081038 | 107 | VAR_081038 | 137–243 | The following amino acids are missing in TETAMS2, resulting in these variants | 54 |
| Natural variant VAR_026247 | 1 | VAR_026247 | 186 | Variations in sequences are observed at 186 aa L → P | 50,51 |
aa: amino acid; HHRRD: humerofemoral hypoplasia with radiotibial ray deficiency; TETAMS2: tetraamelia syndrome 2.
Table 2.
R-spondins and their important interactive receptors.
| Serial number | R-spondin | Receptor | Function | Reference |
|---|---|---|---|---|
| 1 | RSPO | LGR4/5/6 | Potentiates Wnt signaling pathway | 38 |
| 2 | RSPO | LRP5/6 | LRP5/6 phosphorylation and activation lead to the stabilization of β-catenin | 55,56 |
| 3 | RSPO | Frizzled | Activates canonical and non-canonical Wnt signaling through phosphorylation of the disheveled gene | 25 |
| 4 | RSPO2/3 | ZNFR3/RNF43 | Auto-ubiquitination and membrane clearance of ZNRF3/RNF43 and Wnt signaling pathway potentiation | 35 |
| 5 | RSPO2/3 | Syndecan 4 | Induces non-canonical Wnt/PCP signaling | 57 |
| 6 | RSPO | Glypicans | RSPO-enhanced Wnt signaling. | 57,58 |
| 7 | RSPO 1 | Kremen | Disrupts DKK1/Kremen-mediated internalization and enhances Wnt signaling | 59 |
Functional roles of RSPO2 in morphogenesis and early developmental processes
RSPO2 is a key regulator of the Wnt signaling pathway and plays a pivotal role in various development processes and morphogenesis. In a study, researchers have established the role of RSPO2 in regulating the expression of genes like Msx1 and Msx2 in mandibular branchial ARCH1 (BA1) and thereby affecting the craniofacial development process and morphogenesis.60 It was also reported that RSPO2 plays a pivotal role in myogenesis, as any mutation in RSPO2 genes gradually decreased myf5 expression in the limbs.61
Evidence from previous research suggests that up-regulated RSPO2 plays a crucial role in osteoblast differentiation by activating the β-catenin protein. In addition, in vitro and in vivo experiments in mice confirmed that RSPO2 can also act as an important modulator for osteoblastogenesis and bone mineralization.62,63 The molecular genetics approach revealed that defects in the RSPO2 gene (homozygous truncating mutation) lead to congenital abnormalities, e.g., tetraamelia syndrome 2 (TETAMS2) — an autosomal recessive disorder characterized by the absence of limbs and rudimentary appendages along with other deformities and humerofemoral hypoplasia with radio tibial ray deficiency (HHRRD) exhibiting severe limb malformation.54 Additionally, human RSPO2 was shown to be linked with Dupuytren's disease which was confirmed by a genome-wide association study.64
Despite the wealth of literature mentioned above, we attempted to investigate more information based on curated databases, experimental evidence, text mining, co-localization, and gene neighbourhood, in which some of the putative interactors of RSPO2 have been proposed. Many of the roles of RSPO2 in mouse tissues are conserved in humans. Loss of RSPO2 function in mice and humans induces defects in limb outgrowth attributable to a loss of signaling from the limb apical ectodermal ridge.42,54,65, 66, 67, 68 RSPO2 loss in the lung induces aplasia in both species due to the inability to maintain lung epithelial progenitors.36,42,54,67 In the ovary, RSPO2 is expressed in developing oocytes.69,70 RSPO2 heterozygous mice exhibit polycystic ovaries65 and age-dependent sterility.42,68 In humans, polycystic ovaries have been associated with mutations in the transcription factor NOBOX recently shown to regulate RSPO2 gene expression.70,71 Gene targeting approach and genetic studies revealed that rspo2 was also implicated in negative regulation of the odontogenesis,67 osteoblast differentiation,62,72 keratinocyte proliferation,73 trachea cartilage morphogenesis,66,67 and cochlear development.48 These results suggest the idea that there is a lot of functional conservation of RSPO2 between the two species.
Driving potential of RSPO2 in tumorigenesis of various other cancers
Most cancers experience deregulation in the Wnt/β-catenin signaling, in which R-spondins are one of the significant oncogenic drivers in tumor development. Interestingly, the extensive literature survey shows that any deviation from the normal molecular activities of Wnt signaling could increase the risk of tumor formation and metastasis.74 Deviation from the normal function of RSPO2 due to gene rearrangements, fusions, amplification, and hypomethylation can lead to overexpression of RSPO2, consequently leading to an up-regulation of the Wnt response.8 Therefore, to probe the activity of RSPO2, a number of investigators have explored the impact of RSPO2 on the pathophysiology of different types of cancers which are being discussed as follows.
Breast cancer
Coussy and co-workers 2017 have elucidated the expression of RSPO2 in breast cancer (BC) patients as well as in cell lines using real-time PCR (qRT-PCR). They also investigated the involvement of RSPO2 in regulating Wnt signaling and cell proliferation activity using standard techniques. It was also found that a fusion transcript with RSPO2 was overexpressed in patients, particularly in metastatic BC, triple-negative BC, and cell lines such as HBCc-15, MDA-MB-468, MDA-MB-231, and HEK-293. Overexpression of RSPO2 was also associated with poor survival rates in BC patients. Furthermore, extensive mechanism-based studies confirm its potential role in activating the Wnt/β-catenin signaling pathway.75 Conboy and colleagues reported that RSPO2 and RSPO3 act as oncogenes in a subset of colon and breast cancer. In silico analysis in a subset of 1048 breast tumor samples and 111 controls extracted from the TCGA dataset predicted a deficient RSPO2 mRNA expression in most of the control as well as tumor tissues. Further, it has also been concluded that the knockdown of RSPO2 results in decreased Wnt signaling and proliferation of the human breast cancer cell BT-549.76
Liver cancer
A good overview of earlier work in this area was provided by Yin et al in 2017, in which a high level of RSPO2 was observed in the liver cancer cell lines Huh7, HepG2, and Hep3B when compared to normal cell line QSG7701. RSPO2 gene overexpression in QGY7703 cells showed increased proliferation and migration. Although this gain in function was well versed with RSPO2, VEGF, c-Myc, CTNNB1, and STAT3 expression, which indicated activation of Wnt signaling.77 Up-regulation of RSPO2 in liver cancer cell lines was contrary to what was observed in colon and breast cancer cell lines.75,76 Conceptually similar work has been carried out by Conboy et al in 2019, highlighting in the literature that hepatomegaly and liver cancer were progressively induced by the overexpression of RSPO2 and loss of the TP53 protein through activation of the Wnt signaling pathway. Additionally, it has been concluded that oncogenic activation of RSPO2 leads to the initiation and progression of hepatocellular carcinoma through the Hippo/Yap mediated pathway, suggesting that RSPO2 may be the new and potential target in liver cancer therapy.78 There are a number of research studies that have focussed extensively on considering the Hippo/Yap pathway involved in liver regeneration and hepatocyte differentiation.79 In the case of liver cancer, RSPO2-mediated deregulation of Hippo/Yap signaling leads to proliferation and inhibition of apoptosis, resulting in tumor progression. The correlation of elevated RSPO2 expression with Wnt and Hippo/Yap signaling in numerous other aggressive tumors was investigated and documented. Experimental evidence indicates that RSPO2 is regulated by the overactivation of micro-RNA493 (miR-493) leading to an inhibition of cell proliferation, migration, invasion, and motility in HCC cell lines, while reversal of this restores the tumorigenic activity in miR-493 treated HCC cell lines, suggesting that RSPO2 is involved in hepatocellular carcinoma progression and can be used as a potential target in the treatment of HCC.80 However, conflicting results have been reported in the literature regarding the oncogenic action of RSPO2 in HCC. The study by Zheng et al in 2020 addressed the previous reports and demonstrated that RSPO2 expression is down-regulated in HCC specimens. In addition, they also establish that knockdown of the RSPO2 genes induces tumorigenic properties, while the overexpression of RSPO2 has been correlated with reduced proliferation and invasion in HCC cell lines by modulating the MAPK signaling pathway.81
Gastric cancer
Similar to the pathogenesis of HCC, a close relationship between up-regulated RSPO2 in gastric cancer (GC) has been demonstrated, representing that higher RSPO2 levels increase cell proliferation, migration, and transition from epithelial to mesenchymal in GC specimens and cancer cell lines (AGS and BGC-823).82
Pancreatic cancer
Nowadays, a property, such as cancer stemness, has been prominently exploited to understand how it involves tumor seeding by escaping the natural defense system and therapeutic regimens. Researchers have explored the cancer stemness property and shown its impact on the development and progression of pancreatic ductal adenocarcinoma driven by the high Wnt activity. In this regard, higher expression of RSPO2 in a subpopulation of pancreatic cancer cells tends to induce high Wnt activity, which is likely associated with drug resistance, metastasis, and poor survival rates, suggesting cancer stemness has a tumor-promoting property that could be therapeutically targeted by disrupting its stemness inducing factors and pathways.83
Lung cancer
The exact function of RSPOs has not been well defined in lung carcinoma, which accounts for one of the top cancers worldwide. Although, an integrated bioinformatics approach suggested that patients with lung cancer had lower levels of RSPO1, RSPO2, and RSPO3 than normal. In addition, the Kaplan–Meier graph for overall survival analysis indicated that up-regulated RSPO1, RSPO2, and RSPO3 prolonged the survival rate of patients with lung cancer.84 The expression level of RSPO2 at transcript and protein levels was also measured in lung adenocarcinomas cell lines such as A549, NCI-H1792, and NCI-H226, suggesting its expression was down-regulated in lung adenocarcinoma and repressed the tumor-promoting activity.85
Screening and further characterization have shown that increased or decreased expression level of RSPO2 has been associated with the development and progression of other types of cancer such as tongue squamous cell carcinoma,86 pituitary adenoma,87 ovary cancer,88 and nasopharyngeal carcinoma89 following the different involved molecular mechanisms and pathways.
Regulatory effect of RSPO on Wnt signaling
It is a well-known fact that Wnt signaling plays an integral role in normal biological processes during embryonic development and tissue homeostasis; moreover, this pathway also regulates cell survival, migration, polarity, differentiation, and proliferation. Various Wnt (Wnt1/3/5a/5b/6/10, etc.) ligands bind to transmembrane protein frizzled (fzd1/2/4/5/6, etc.) and activate two types of important pathways, canonical (β-catenin dependent) and non-canonical (β-catenin independent). The canonical Wnt/β-catenin signaling pathway is mainly involved in regulating various cellular processes like differentiation, proliferation, and morphogenesis which play a vital role in cell progression and functioning. Further, several epigenetic alterations and genetic mutations in canonical pathways contribute directly to carcinogenesis.90
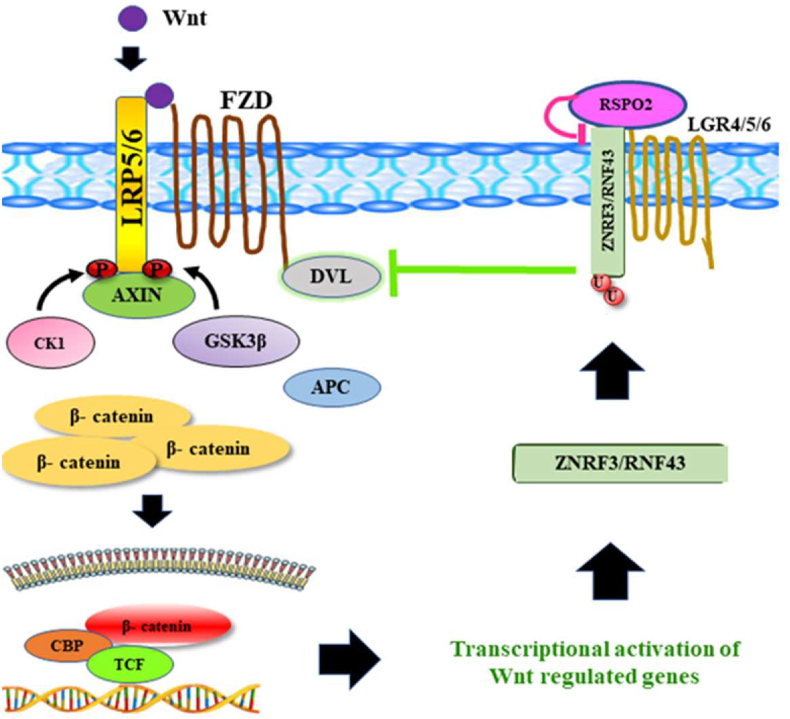
Several lines of studies confirmed that R-spondins play a stimulating role in the canonical Wnt/β-catenin-signaling, as they exhibit direct interactions with engagement receptor LGR 4/5/6 and simultaneously bind to the extracellular domain of ZNRF3/RNF43.27 Formation of ZNRF3/RNF43, RSPO and LGR 4/5/6 complex results in auto-ubiquitination and degradation of efficacy receptor ZNRF3/RNF43 through a negative feedback loop35,91 which leads to an enhanced Wnt signaling through the accumulation of Frizzled receptors on the cell surface and thereby resulting in increased β-catenin accumulation.35,74,92 Conversely, regulation of Frizzled turnover can also be explained by a molecular mechanism by which ZNRF3 and RNF43 interact with Dishevelled (DVL) and significantly reduces cell surface expression of Frizzled receptors and LRPs, as well as promote ubiquitination and degradation in the absence of R-spondin.35,93 In the case of cancer initiation and progression, cells require sustained Wnt signaling, which is only possible when the mutation occurs in ZNRF3/RNF43 or RSPO2/3 overexpression/translocation occurs. This signaling process is graphically depicted in Figure 2.
Figure 2.
This diagram represents a regular Wnt pathway occurring in normal cells. Activation of Wnt signaling is initiated by the binding of Wnt ligand to the Frizzled-LRP5/6 co-receptor complex, resulting in phosphorylation of Dishevelled (DVL) and LRP5/6, which in turn causes inhibition of GSK3β and further inactivation of the destruction complex AXIN, GSK3β, APC, and CK1. Collectively, Wnt ligand stimulation relocalizes the destruction complex to the plasma membrane where their inhibition remains controversial. Inactivating the destruction complex leads to the cytoplasmic stabilization of β-catenin, and thus, stabilized β-catenin moves to the nucleus and replaces the repressive transcription complex with the active transcription factors like TCF and CBP to induce the expression of Wnt-responsive target genes. However, Wnt signaling is regulated in a negative feedback manner through RSPO signaling. Expression of the ZNRF3/RNF43 degrades Frizzled receptors assisted by DVL protein through ubiquitination. As a part of feedback regulation, the antagonistic effect of ZNRF3/RNF43 on Wnt signaling is counteracted by binding of RSPO ligand to LGR4/5-ZNRF3/RNF43 to form multiple ligand-receptor complexes, inducing degradation and ubiquitination of ZNRF3/RNF43, which inhibits the degradation of Frizzled and achieves a high level of β-catenin. Hence, the level of β-catenin is regulated with the help of this cyclic loop.
The crystal structure and mutation analysis predicted a clear observation regarding interactions between R-spondin and its complex, which helps understand the molecular insights into Wnt/β-catenin pathway regulation. Furthermore, the crystal structure of the complex is essential to correlate with their binding affinities for Wnt regulatory elements, which has been discussed extensively by many authors.26,46,47 However, it was also noted that R-Spondins are not directly involved in inducing the activity of the Wnt pathway, but they tend to up-regulate activity by enhancing the concentration of Frizzled/LRP receptors present at the plasma membrane. In a different observation by Park and co-workers, a significant negative association was observed between RSPO2 and LGRs in potentiating Wnt receptors without affecting the ZNRF3 activity.92 Further research suggested that instead of interacting with its obligate receptors (LGRs), RSPOs (RSPO2 and RSPO3) follow an alternate mechanism of action in activating the canonical Wnt/β-catenin signaling pathway. However, still, the mechanism is obscure about how RSPO2 and RSPO3 potentiate the Wnt signaling in LGR4-6 lacking cells. Probably, this may be explained by two possible mechanisms: (i) they work by engaging the heparan sulfate proteoglycans (HSPGs) as alternate co-receptors for RSPOs; (ii) RSPOs follow a distinct mechanism that differs in its use of either HSPGs or LGRs for Wnt potentiation.58,94
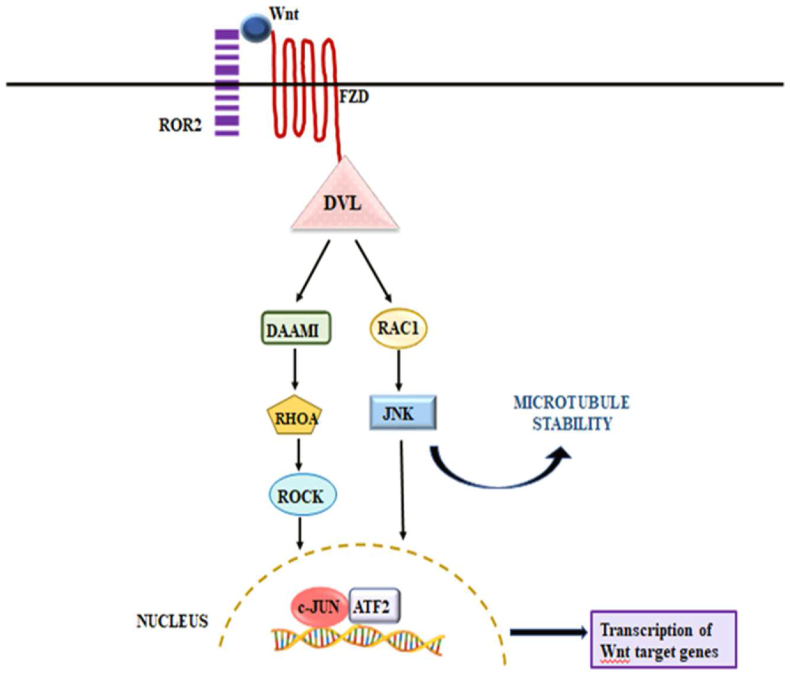
Indeed, the strength of Wnt signaling is controlled by various mechanisms directing the proper signaling module in different pathophysiological conditions. Despite the growing biological importance of R-spondin-induced canonical Wnt/β-catenin signaling, its significance in potentiating Wnt/PCP pathway should not be disregarded as well. It is worth mentioning that, in the non-canonical Wnt/PCP signaling, Wnt utilizes different coreceptors ROR1/2, RYK, or PTK7 depending on the cellular system to stimulate signal transduction mechanism that is independent of β-catenin and plays a vital role in regulating development processes, enhancing tissue movement and facilitating cancer cells motility during metastasis (Fig. 3).95
Figure 3.
Wnt/PCP signaling. The Wnt ligand binds to Frizzled and ROR2 receptor complex which activates its downstream effector DVL and RAC1, resulting in the phosphorylation of JNK. Simultaneously, the activated DVL also initiates the de-inhibition of DAAM1 cytoplasmic protein which stimulates RHO thereby promoting the induction of ROCK. Phosphorylated JNK contributes to microtubule stability and along with ROCK activates the c-JUN and ATF2 genes, resulting in the transcription of Wnt-targeted genes. ATF2, activating transcription factor 2; DAAM1, Disheveled associated activator of morphogenesis 1; DVL, Dishevelled; FZD, Frizzled; JNK, c-Jun N-terminal kinase; RAC1, Rac family small GTPase 1; Rho, Rhodopsin; ROCK, Rho-associated coiled-coil containing protein kinase; ROR2, receptor tyrosine kinase-like orphan receptor 2.
At this point, it is of interest to notice that R-spondin proteins also activate the Wnt/PCP signaling through two well-recognized proteins, Frizzled (FZD1/2/4/5) and Dishevelled (DVL) that share a critical role in both the pathways.96 Interestingly, DVL, associated with FZD and ZNRF3/RNF43, serves as an adaptor protein and knockout of DVL markedly reduces the ubiquitination and degradation of FZD receptors, indicating that DVL negatively regulates Wnt signaling.93,97 Since the discovery of the RSPO/ZNRF3/RNF43 module and FZD turnover has emerged as an essential regulatory mechanism of Wnt/β-catenin signaling, but the critical role of ZNRF3/RNF43 is not clearly understood in Wnt/PCP signaling. It is clear from previous research undertaken that the function of ZNRF3/RNF43 is necessary to activate Wnt/PCP signaling which was further confirmed in genetic studies of zebrafish embryos. The in vitro study identified that knockout and overexpression of ZNRF3 deregulate the Wnt/PCP signaling.35 Another challenging aspect of Wnt/PCP activation was mediated through RSPO3 and syndecan 4 in the Xenopus embryo. The RSPO3 promotes the Wnt/PCP signaling cascade by interacting with syndecan 4 dependent endocytosis of Wnt5a-FZD7 complex and transduces via downstream target protein DVL and JNK which are required for gastrulation and head cartilage morphogenesis.43,98,99 Collectively, mounting literature suggests that activation of both the pathways Wnt/β-catenin and Wnt/PCP are significantly mediated by RSPO/ZNRF3/RNF43 and HSPGs. Notably, aberrant/constitutive activation of Wnt/PCP signaling is also implicated in various cancers, primarily acting as initiating event in the development and progression of CRC, although not explained in this review.100, 101, 102 In the last few years, substantial advancements have been made in identifying RSPO members, particularly RSPO2, as a new biomarker and potential therapeutic target for cancers including CRC.58,103, 104, 105
RSPO2: induce stemness for colorectal cancer stem cells
Cancer stem cells (CSCs) are pluripotent subpopulations of tumor origin that can renew themselves and can initiate tumorigenesis. It is noteworthy that at the time of tumor formation the origins of CSCs come either from the differentiated cells or from adult tissue stem cells. The evidence to date suggests that several regulatory molecules and signaling pathways such as cytokines, miRNAs, Hedgehog, Notch, and Wnt/β-catenin could control the properties of CSCs. Several subsequent studies stated that the small intestine and colon,106 stomach,107 and hair follicle108 have Wnt-dependent resident stem cells. Work in this arena is fairly advanced, and the signatures of colon cancer stem cells (CCSCs) have been identified, which are the major molecular drivers for colon cancer. Self-renewal and long-term maintenance of CCSCs required stem cell markers like LGR5 that have been shown to display validated markers of stem cell niche and have the propensity to behave like stem cells of the colon and intestinal crypts.108,109 The expression of LGR5 has been extensively investigated to find its importance for the proliferation of CCSCs under the influence of RSPO2.30 Furthermore, cancer stemness has been thoroughly reviewed in the differentiation and progression of CRC. The complex of RSPO2–ZNRF3–RNF43 regulates intestinal stem cell activity, most likely through the involvement of the Wnt/β-catenin pathway, which enables the proper differentiation and development of different organs.108, 109, 110 However, the loss of control over the normal function of this complex is critical for the self-renewal activity of stem cells and organ development and often leads to uncontrolled activation of the Wnt pathway, resulting in the progression of CRC.111, 112, 113
Concerning this specific concept, Zhang et al in 2016 demonstrated that a high level of RSPO2 mediates the enrichment of the LGR5-positive cells and markedly increases their expression, which was assessed in the HCT116 human colon cancer cell line. Unsurprisingly, there was a considerably reduced expression of LGR5 post siRNA silencing of RSPO2. Literature and results show that invasion and migration are crucial to CCSCs. One of the key findings of this study was that RSPO2 was proposed to be a potent growth factor for CCSCs and it was implicated in increased invasiveness and migration of HCT116 colon cancer cells via the epithelial to mesenchymal transition (EMT) pathway.110 Similar work was also carried out by Klauzinska and co-workers in which it was demonstrated that RSPO2 adversely affects the proliferation and invasiveness of mammary epithelial cells.114 In addition to the LGR5, ASCL2, LRIG1, NANOG, OCT4, ALDH1, and TERT are also crucial stem cell marker genes that require signals in the form of RSPO3 to respond to high levels of Wnt signaling for the stem cell activity.115 This was consistent with the fact that RSPO expression maintains the intestinal stem cell crypts, and particularly RSPO1109 and RSPO2110,116 promote the proliferation and migration of LGR5-positive intestinal stem cells, suggesting that RSPO1 and RSPO2 have strong mitogenic activity on LGR5-positive cells. It is also demonstrated that intestinal crypts bearing proliferation and migration of LGR5-positive cells due to gene fusion events of EMC2 and RSPO2, which play a critical role in tumor development.116 However, this is a matter of ongoing discussion. Future improvements are expected to result in an improved understanding of RSPOs, particularly RSPO2 in CSCs and cellular reprogramming. We suggest that more studies should be concerned with understanding the regulation of CSCs in tumorigenicity and bringing the CSCs to the clinical platform which would help identify the best possible therapeutic options for targeting CSCs in the clinical setting.
Fusion products promote aberrant RSPO2 expression
Gene fusions are a strong source of driver mutations in different human malignancies, often due to chromosomal rearrangements (duplication, translocation, inversion, and deletion) that result in fusion between genes. In some cases, gene fusions serve as the genetic events that provide insight into the disease development and prognosis. Approximately fifty thousand cases are reported due to chromosomal aberrations, particularly translocations, which are often strongly associated with distinct tumor entities.117 Gene fusions are a potent oncogenic factor clinically relevant in many types of malignancies, exerting their oncogenic action either through overexpression of a gene or the creation of a hybrid of two genes.118 Importantly, gene fusions are identified as actionable targets in multiple tumor types and are ideally suited for diagnostic purposes. There has been extensive interest in this matter recently, and more groups are working to explore the spectrum of gene fusion products in distinct types of cancer including CRC. However, the biological impact of genomic aberration in solid tumors is not frequently observed and accounts for less than 1%,117 whereas an increasing number of morbidities is associated with hematological malignancy and childhood sarcomas in which fusions are being recognized as an important diagnostic biomarker and promising determinants of cancer.
Accumulating evidence suggests that genomic rearrangement plays an essential role in the pathogenesis of cancer. In the case of CRC, extensive literature and experimental findings indicate that RSPO2 gene fusion leads to an increase in the expression of genes and possibly modulates the Wnt signaling to promote cell proliferation, differentiation, and survival. Notably, it is reported that the entire RSPO-fusion product in CRCs is independent of the APC mutations, suggesting RSPO alone can potentiate the signaling to induce tumorigenesis.119 A well-recognized fusion between EIF3E–RSPO2 has been reported in CRC and NSCLC (information regarding NSCLC is given near the end of this section), except for the exon number. The pioneering work by Seshagiri and co-workers stated that recurrent fusion products of RSPO2 (EIF3E–RSPO2) and other members of R-spondin are the most preferred Wnt regulators and are considered to be potent genetic drivers of CRC.120 A detailed study showed that recurrent fusion between exon 1 of EIF3E and exon 2 of RSPO2, exon 1 of EIF3E, and exon 3 of RSPO2, were highly recognized in tumor samples. In the case of the RSPO2 fusion, they suggested that EIF3E (e1) and RSPO2 (e2) were expected to produce a functional RSPO2 product, while the fusion transcript of EIF3E (e1) and RSPO2 (e3) fusion transcript are not expected to have functional RSPO2 protein, which was further confirmed by sequencing. Their findings also indicated that RSPO fusions were mutually exclusive with APC mutations, as RSPO fusion-positive tumors showed up-regulation of target genes of the Wnt pathway, similar to tumors that carry APC mutations. These results pave the way for new likely therapeutic strategies to use R-spondin fusions as striking targets for antibody-based therapy in CRC patients, and for the use of RSPO2 fusions to target various downstream components of the Wnt signaling pathway.120 To review the demographic occurrence of RSPO fusion, Shinmura et al 2014 conducted a comparative study on CRC and NSCLC patients in the Japanese population. Using qRT-PCR and sequencing techniques, they examined RSPO fusions in various primary CRCs, while in the case of primary lung carcinomas the expression of RSPO fusions was not detected. Moreover, this study also suggests that RSPO mRNA expression is down-regulated in most CRCs but up-regulated in CRCs containing the RSPO fusion transcripts. However, in the evaluation of 75 CRC samples, only 3 showed an RSPO fusion, with EIF3E–RSPO2 found in 2 CRC samples, while only 1 sample expressed the other RSPO fusion transcript. In addition, a novel fusion was detected in their study, suggesting a fusion of RSPO2 through a 351 bp insertion unrelated to EIF3E or RSPO2. Additionally, with the help of immunohistochemical (IHC) and mutational analysis, it was suggested that the RSPO fusion had a CDX2 cell lineage expressed in CRC and was positive for mismatch repair protein (MMR) expression, which had the wild-type APC allele. They concluded that the RSPO fusion occurred recurrently in a patient with CRC in the Japanese population and was caused by activation of the Wnt signaling pathway.121
Later, a functional study was conducted in animal models (in mice) to observe the biological consequences of RSPO2 fusion. CRISPR/Cas9 genome editing tool was used for RSPO2 gene rearrangement in the mouse intestine, which was demonstrated by endogenous expression of fusion transcript EIF3E–RSPO2. It provided direct evidence of hyperplasia and tumor progression in the intestine.122 Work in this arena was heavily influenced by Hashimoto et al who found in 2019 that the RSPO2 fusion is linked with the traditional serrated adenoma (TSA), a condition characterized by the premalignant potential that gives rise to CRC. Their study agrees well with the existing studies on EIF3E–RSPO2, which was detected in three out of six lesions with RSPO2 overexpression in patients with TSA. In addition, qRT-PCR identified a novel PIEZO1–RSPO2 fusion in one TSA patient. One of the most significant findings of this study concerns all four spectra of the RSPO2 fusion in TSA, which also harbor KRAS mutation.123 However, their earlier outcome for RSPO2 characterization detected only one new fusion, i.e., NRIP1–RSPO2 in which 129 TSAs were involved.124 Nanostring assay of RSPO2 fusion transcript and expression level from 207 patient-derived xenografts (PDX) of colon, pancreatic, lungs, and liver cancer-derived samples were tested and found mixed expression level of RSPO2. The nanostring assay identified a highly elevated oncogenic EMC2–RSPO2 fusion gene with exon1 of EMC2 and exon2 of RSPO2, which was responsible for tumor growth and metastasis. In addition, other fusion events are also identified in colon tumor samples that contained a copy high fusion gene, of which the PVT1 gene exon1–RSPO2 gene exon2 produced a functional full-length protein with increased RSPO2 expression, while the fusion event between PVT1 exon1–RSPO2 exon3 due to the premature translation termination of RSPO2 exon3 results in a non-functional protein.116 Recent theoretical work by Conboy and colleagues supported the above finding and reported that in a minor CRC cohort, 2 of 434 RSPO2 mRNA level was four-fold higher compared to normal samples. After careful consideration, it was deduced that an exon imbalance leads to the predicted high level of RSPO2 mRNA in the samples, which was required for the overexpressed fusion transcript of FLJ31306–RSPO2 and CASC19–RSPO2. Much of the earlier work centers on studying the determination of higher RSPO2 expression due to gene rearrangement, copy number variations, and epigenetic changes. However, in a major cohort of 434 CRCs and 41 normal samples, RNA-seq. data show that 422/434 (97.2%) CRCs were free of gene rearrangement and had a four-fold decrease in RSPO2 expression.76 Contrary, the initial work published by researchers reported that the small population of CRC patients express low levels of RSPO2 mRNA that could be due to promoter hypermethylation.15,120,121 A summary of the detailed analysis of the fusion transcript in CRC in addition to the other cancer is given in Table 3. Thus, the outcome of various experiments leads to the conclusion that most CRC patients with RSPO2 fusions had elevated expression compared to normal, indicating RSPO2 behaves like an oncogene and is an alternative Wnt modulator. Prospective studies are needed to explore altered RSPO2 fusion as a predictive biomarker in pre-clinical and clinical models and can be an ideal druggable target against CRC. Significantly, it has been proposed that Wnt pathway inhibitors are an ideal choice to provide an effective treatment for RSPO2-dependent CRC.76
Table 3.
Fusion transcripts and their biological alterations of different types of cancers.
| Serial number | Cancer | Source | Fusion genes | Biological alterations | Chromosomal location | Reference |
|---|---|---|---|---|---|---|
| 1 | Gastric Cancer | Patient-derived xenograft samples | EMC2(e1)–RSPO2(e2) | Fusion at 450 bp in RSPO2-e2) | 8q23.1; 8q23.1 | 116 |
| HNF4G(e1)–RSPO2(e2) | Fusion at 447 bp in HNF4G and 490 bp in RSPO2) | 8q21.13; 8q23.1 | 116 | |||
| Colorectal cancer | PVT1(e1)–RSPO2(e2) | Fusion occurs between PVT1(e1) at 202 bp and RSPO2 (e2) at 490 bp resulting functional protein | 8q24.21; 8q23.1 | 116 | ||
| PVT1(e1)–RSPO2(e3) | Fusion occurs at 753 bp between both the gene, producing non-functional protein due to premature termination | 8q24.21; 8q23.1 | 116 | |||
| 2 | Lung Cancer | Human tissue | EIF3E(e1)–RSPO2(e1) | Deletion | 8q23.1; 8q23.1 | 132 |
| 3 | Liver Cancer | Human tissue | RSPO2(e1)–rearranged HCA | 46.4 kb microdeletion on chromosome 8q23.1 | – | 133 |
| 4 | Colorectal Cancer | Human tissue | EIF3E(e1)–RSPO2(e2) | Deletion (at 158 kb) | 8q23.1; 8q23.1 | 120 |
| EIF3E(e1)–RSPO2(e3) | Deletion (at 113 kb) | 8q23.1; 8q23.1 | ||||
| 5 | Colorectal Cancer | Human tissue | EIF3E(e1)–RSPO2(e1) | 351-bp insertion | 8q23.1; 8q23.1 | 121 |
| 6 | Colorectal Cancer | Patient-derived xenograft mice | PVT1(e1)–RSPO2(e2) | Fusion at 202 bp in PVT1 and 490 bp in RSPO2 | 8q24.21; 8q23.1 | 116 |
| PVT1(e1)–RSPO2(e3) | Fusion at 202 bp in PVT1 and 753 bp in RSPO2 | 8q24.21; 8q23.1 | ||||
| 7 | Colorectal cancer | Human tissue | CASC19–RSPO2 | Loss of border element CCCTC-binding factor | 8q24.21; 8q23.1 | 76 |
| FLJ31306–RSPO2 | Loss of border element CCCTC-binding factor | 14q23.1; 8q23.1 | ||||
| 8 | Traditional serrated adenoma | Human tissue | NRIP1(e2/3)–RSPO2(e2) | Fusion result lack of NRIP1 coding region with no loss of RSPO2 region | 21q21.1; 8q23.1 | 124 |
| 9 | Traditional serrated adenoma | Human tissue | EIF3E–RSPO2 | Recurrent genetic alterations | 8q23.1; 8q23.1 | 123 |
| PIEZO1(e1)–RSPO2(e2) | Recurrent genetic alterations | 16q24.3; 8q23.1 | ||||
| PIEZO1(e1)–RSPO2(e3) | Fusion generates in-frame chimeric protein | 16q24.3; 8q23.1 |
CASC19: cancer susceptibility 19; e: Exon; bp: base pair; EMC2: ER membrane protein complex subunit 2; EIF3E: eukaryotic translation initiation factor 3 subunit E; HNF4G: hepatocyte nuclear factor 4 gamma; kb: kilobase; PIEZO1: piezo type mechanosensitive ion channel component 1; PTPRK: protein tyrosine phosphatase receptor type K; PVT1: Pvt1 oncogene.
Similarly, there were several other cancers where genomic rearrangements were identified as consequences of the origin of cancer. Karkera et al worked on identifying fusion transcripts in lung cancer patients in their study. They evaluated 324 samples from non-small cell lung cancer patients, including 197 squamous and 127 adenocarcinoma subtypes. Using the TaqMan qRT-PCR-based approach, they identified three fusion transcripts including EIF3E (e1)–RSPO2 (e1), PTPRK (e1)–RSPO3 (e2), and PTPRK (e7)–RSPO3 (e2) which were exclusively present in squamous subtype of NSCLC. EIF3E(e1)–RSPO2(e1) was identified in 1% of the NSCLC samples and resulted in the production of the functional RSPO2 protein, while an in-frame fusion of PTPRK(e1)–RSPO3(e2) was observed in 2% of the samples which led to the replacement of the RSPO3 secretion signal sequence with that of PTPRK, while the entire coding sequence of RSPO3 was retained. Their findings helped in establishing the role of RSPO fusions in down-regulating the Wnt/β-catenin signaling in lung cancer patients and the development of its therapeutic potential in NSCLC.132 Furthermore, various other fusion products like ETVG–RUNX1, BCR–ABL1 in acute lymphocytic leukemia,125,126 FGFR3–TACC3 in bladder cancer,127,128 TMPRSS2–ERG, TMPRSS2–ETV1, TMPRSS2–ETV4 in prostate cancer,129,130 and ETV6–NTRK3 in secretory breast carcinoma131 have also been successfully acknowledged.
RSPO2: therapeutic potential and clinical translation
Therapeutic approach targeting Wnt signaling in colorectal cancer
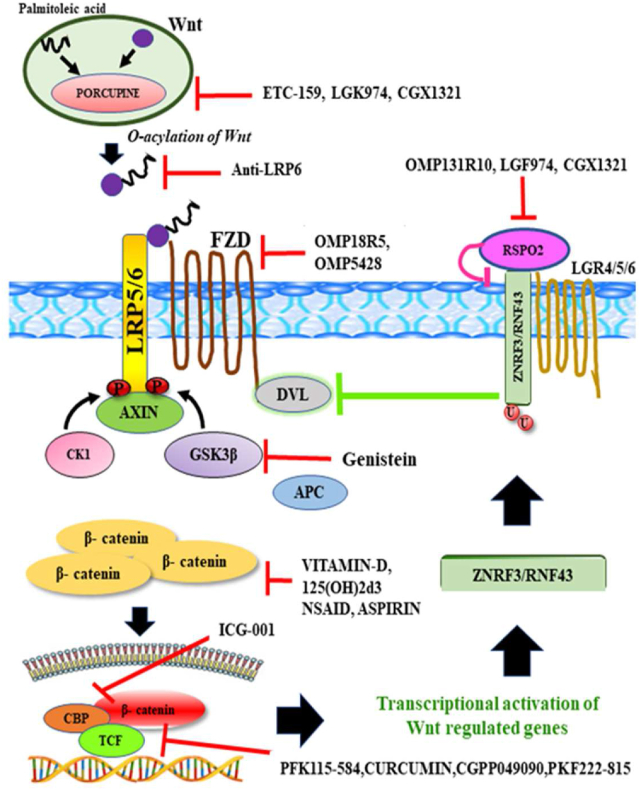
Aberrant changes or mutations in Wnt pathway-related genes may result in the accumulation of β-catenin which may lead to tumorigenesis.134 Hence, a therapeutic approach directly targeting Wnt-dependent signaling components or RSPO2-induced signaling may be a feasible option for CRC treatment, as shown in Figure 4.
Figure 4.
Illustration of the Wnt/β-catenin and RSPO signaling with therapeutic intervention which comprises various inhibitors acting on different components in the case of colorectal cancer.
The only prominent and restorative treatment for CRC is surgery, but for cases where the disease has reached an advanced stage, a combination of various therapies like chemotherapy, biotherapy, and radiotherapy can be used as an effective treatment approach. An amalgamation of chemotherapy such as FOLFIRI, XELOX/CAPOX, FOLFOX, and FOLFOXIRI with various therapeutic antibodies like cetuximab or bevacizumab has been shown to increase the survival rate in CRC patients. In addition, therapeutic approaches for CRC patients involving the Wnt/β-catenin signaling pathway mainly include natural product-based compounds, small molecules, and drugs.135
Natural product-based compounds that may inhibit Wnt signaling mainly include vitamin D, 125(OH)2D3, curcumin, genistein, and resveratrol. Vitamin D, 125(OH)2D3 results in the binding of β-catenin with vitamin D receptor; this binding leads to up-regulation of E-cadherin expression, which causes a decrease in β-catenin activity.136 Small molecule inhibitors like ETC-159, a PORCN-targeted Wnt antagonist, inhibit PORCN activity, thereby blocking Wnt/β-catenin signaling.137 Another popular PORCN inhibitor is LGK974 which deregulates the secretion of Wnt3A and inhibits the growth of tumor samples.69,138 Therapeutic regimens using anti-LRP6 antibodies have been found to play a major role in regulating the Wnt signaling pathway and are proposed to have a therapeutic effect on several cancers.139 In addition to the above-discussed inhibitors various other groups of inhibitory compounds targeting the Wnt signaling pathway have been briefly discussed in Table 4.
Table 4.
The inhibitors targeting the Wnt pathway and their targeting components in colorectal cancer.
| Serial number | Category | Drug | Molecular target | Drug development stage | Reference |
|---|---|---|---|---|---|
| 1 | Natural compound | Vitamin D, 125(OH)2d3 | β-catenin | Clinical | 136 |
| Curcumin | Tcf/β-catenin | Phase I | 140 | ||
| Genistein | GSK3β | Phase I | 141 | ||
| 2 | Existing drug | Aspirin | β-catenin | Phase III | 142 |
| NSAID | β-catenin | Clinical | 143 | ||
| Salinomycin | LRP5/6 | Pre-clinical | 144 | ||
| 3 | Antibody | OMP18R5 | Frizzled | Discovery | 145 |
| OMP5428 | Frizzled | Discovery | 145 | ||
| OMP131R10 | RSPO-fusion | Phase I/II | 146 | ||
| OMP131R10 | Intracellular regulatory component | Clinical | 146 | ||
| Anti-LRP6 | Wnt | Discovery | 147 | ||
| 4 | Small molecules | ETC-159 | Porcupine | Discovery | 137 |
| LGK974 | Porcupine | Phase I | 69 | ||
| RSPO-fusion | Clinical | 122 | |||
| PKF115-584 | Tcf/β-catenin | Discovery | 148 | ||
| CPG049090 | Tcf/β-catenin | Discovery | 148 | ||
| PKF222-815 | Tcf/β-catenin | Discovery | 148 | ||
| ICG-001 | CBP/β-catenin | Discovery | 149 | ||
| CGX1321 | Porcupine | Discovery | 116 | ||
| RSPO-fusion | Discovery | 116 | |||
| Foxy-5 | Frizzled | Phase II | 150 | ||
| JW55 | TNKS1/2 | Pre-clinical | 151 | ||
| IWR-1 | TNKS1/2 | Pre-clinical | 152 | ||
| 2,4-Diamino-quinazolines | β-catenin/Tcf-4 | Pre-clinical | 153 |
GSK3β: glycogen synthase kinase 3 beta; NSAID: nonsteroidal anti-inflammatory drugs; Tcf: transcription factor 7-like 2; TNKS1/2: tankyrase 1/2.
Targeting RSPO2 fusion products: an alternative approach to maximize the therapeutic effects
Fusion products are widely used as prognostic and diagnostic markers for assessing various cancers and are also involved in monitoring responses to various other molecular therapies. With the discovery of rare fusion gene products in cancer patients, it becomes easier to develop a personalized form of medicine in which treatments are adapted to the molecular characteristics of each patient. For such molecular therapies, fusion gene products are an ideal target as they are highly specific to cancerous cells.154 A comparative analysis of other cancers along with CRC highlights the therapeutic effect of each drug or therapy on different cancer types. While describing the role of the RSPO2 fusion gene in liver cancer,133 it was highlighted that RSPO2 fusion-positive hepatocellular adenoma displayed activation of WNT signaling due to increased levels of RSPO2 protein, nuclear accumulation of β-catenin, and transcriptional activation of β-catenin target genes. Further, it was postulated that the appropriate reason for β-catenin signaling activation in hepatocellular adenoma could be due to recurrent RSPO2 gene rearrangement and this rearrangement may lead to oncogenic activation of the Wnt signaling in HCA cases. They proposed that a prominent Wnt secretory inhibitor like a porcupine inhibitor could be used as a potent therapeutic strategy for targeting RSPO fusion transcripts in liver cancers and their cumulative diagnostic analysis might help in screening hepatocellular carcinoma patients who may get an advantage from the clinical approach. Notably, one such PORCN inhibitor is CGX1321 which acts remarkably against the Wnt pathway.116 Authors in their study identified EMC2 exon1–RSPO2 exon 2 gene fusions in GA0007 gastric cancer sample, HNF4G exon3–RSPO2 exon2 fusion in GA3055 gastric cancer sample, and PVT1 exon1–RSPO2 exon2 fusion and PVT1 exon1–RSPO2 exon3 fusion in CR3056 CRC sample. They also examined the gene function of the RSPO2–EMC2 fusion gene and confirmed its role in tumor progression, oncogenesis, and metastasis, along with suggesting that excessive RSPO2 promotes LGR5+ cell proliferation and migration. On administering a daily dose of CGX1321 to the PDX tumor-bearing mice, they established that mice models obtained from GA007, CR3056, and GA3055 tumors showed a response towards the inhibitor treatment, while gastric cancer sample GA108 did not show the expected result. Hence, their results suggest that the administration of CGX1321 inhibitors can effectively target tumors exhibiting RSPO2 fusions in gastric cancer and CRC patients. Inhibitors that directly deregulate the functioning of Wnt signaling are often recommended for treating various types of cancer. A well-known Wnt/β-catenin pathway inhibitor WNT974 has been found to deregulate Wnt signaling in ovarian cancer patients as it causes a decrease in phosphorylation of LRP6 and AXIN2 expression. RSPO2 fusions were not quite evident in the case of ovarian cancer; hence no proper correlation between WNT974 and RSPO2 fusion inhibition was established.155
It has been observed that RSPO2 and fusion transcripts of RSPO2 are found in most CRC cases, up-regulating the Wnt pathway's activity.76 Several studies have identified various fusion-based therapeutic approaches targeting CRC cases. In a study, the role of inhibitor LGK974 was successfully established in treating RSPO-fusion cancers by recreating chromosomal rearrangement in mice with the help of a genome-editing tool CRISPR/Cas9. EIF3E–RSPO2 fusion transcript (formed due to an intra-chromosomal deletion between intron 1 of EIF3E and intron 1 of RSPO2). They engineered RSPO fusions comprising RSPO2 in vivo in mouse intestines and observed that these fusions initiated tumor progression. On subjecting this endogenous RSPO2 rearrangement (R26-rtTA/EIF3E-RSP02) with LGK974, a significant reduction in intestinal lesions was observed and it did not alter the normal morphology and functioning of normal intestinal cells. Thus, their results suggest that RSPO fusion, especially EIF3E–RSPO2 derived tumors are susceptible to LGK974 and PORCN inhibitors may serve as a potent anti-tumor drug for treating the RSPO fusion-based subset of CRCs.122 Growth in various human tumor types can be easily inhibited by blocking Wnt signaling with the help of a specific antibody. The mAb-based drugs and porcupine PORCN inhibitors have proved to be quite effective in regulating the Wnt signaling pathway. Various human-based monoclonal antibody drugs such as humanized monoclonal antibody (mAb) drugs, such as anti-FZD1/2/5/7/8 mAb (vantictumab/OMP-18R5), anti-FZD5 mAb (IgG-2919), anti-FZD10 antibody-drug conjugate (ADC) (OTSA101-DTPA-90Y), anti-LGR5 ADC (mAb-mc-vc-PAB-MMAE), and anti-RSPO3 mAb (rosmantuzumab/OMP-131R10) have also been developed, which interacts with Wnt signaling regulators and mainly target the interaction of Wnt with its co-receptors.156 With the analysis of different therapeutic approaches used in colorectal and other cancer types, we can conclude that inhibitors targeting the Wnt pathway are equally effective with cancers containing RSPO2-fusion transcripts as they directly or indirectly hinder its interaction with Wnt regulators. The antibody-based approach and use of various chemically synthesized drugs may serve great therapeutic significance as they inhibit anti-tumor activities of cancerous cells containing R-spondin fusions.
RSPO2: a new prognostic biomarker in colorectal cancer
According to the NIH definition, a biological marker (biomarker) is described as a distinctive characteristic that is accurately measured and assessed as an indicator of various processes like normal biological or pathogenic processes, or it could be examined as a pharmacological response to a particular therapeutic action.157 Tumorigenesis mainly depends on factors like epigenetic changes, gene expression changes, and environmental factors.158 Suitable therapeutic approach and prognosis may result in certain changes in genes and protein expression levels which may serve as a potential biomarker in the case of cancers including CRC. With the help of a suitable biomarker, it would become easy to develop a personalized form of therapy for patients considered at high risk and appropriate treatment can be provided to such patients so that disease advancement can be controlled.159 In most CRC cases, chromosomal rearrangements in RSPO protein result in the formation of fusion transcripts, leading to an aberrant increase in Wnt signaling. Such mutations may be a potent biomarker for selecting patients with optimum response against Wnt inhibitors.137 In the case of lung cancer, particularly lung adenocarcinoma, Wu and colleagues in 2019 suggested that high levels of RSPO might help improve the overall survival of lung cancer patients; hence such R-spondins might serve as a potential biomarker and analytical factor for these patients.84 The only demerit of their work was that the results were not validated clinically in lung cancer cell lines. While establishing the role of RSPO in CRC, Hao et al stated that R-spondin translocation in CRC is one of the potent prognostic biomarkers for detecting Wnt-dependent tumors. Identifying such translocations helps obtain a striking target for inhibiting the Wnt pathway using antibodies. These RSPO2/RSPO3 antibodies also serve as an efficient analytical biomarker depicting tumor progression and development.91 Ter Steege and Bakker et al in 2021 in their study highlighted the biomarker potential of RSPOs and their role in predicting therapy responsiveness.49
Either the RSPO ligand is directly targeted to inhibit the Wnt signaling pathway, or PORCN inhibitors (PORCNi) are used. With the application of PORCNi, the active Wnt ligands are not being secreted which causes impairment of RSPO ligand to exhibit any effect on Wnt signaling.49 Hence, the efficiency of PORCNi can be determined by taking the genetic alterations in RSPO as inclusion criteria in the case of CRC; therefore, alterations in RSPOs may serve as a biomarker for PORCNi response in these cases. Further, Chong et al 2018 examined different types of RSPO2 fusions in PDX tumor samples and established their role in tumor development. It was evident with the help of their result that RSPO2 fusions perform oncogenic functions. RSPO2 fusion tumor samples were treated with a Wnt inhibitor which resulted in the inhibition of the growth of such tumor patients.116 Hence, these results suggest that RSPO2 fusion products exhibit prominent biomarker properties which would help in patient selection in CRC and gastric cancers that are Wnt-dependent.
Inhibiting Wnt signaling may induce severe complications
To begin, we introduced a few concepts relevant to Wnt signaling that regulate multiple physiological processes by maintaining the adult stem cell niche in response to R-spondin. Then, we briefly discussed how Wnt signaling is aberrantly activated particularly in CRC under the influence of RSPO2, attracts attention, and offers enormous promise in cancer research with multiple druggable targets for effective therapeutics. Knowingly, therapeutic agents/drugs targeting the Wnt pathway are unlikely to elicit any adverse effects or toxicity. However, certain risks or concerns remain in targeting this highly dependent signaling cascade for maintenance of the stem cell niche, development of an array of organs, tissues, and intestinal homeostasis. Note that, after injury, maintenance of the somatic stem cell niche and regenerative processes are only possible by activating the developmental signal transduction pathway, specifically the Wnt pathway, and the fact that targeting the Wnt cascade might disturb the stem cell homeostasis. Furthermore, embryonic patterning during initial development is also affected by the devastating nature of Wnt inhibitors, as evidenced by the characterization of several genetic and congenital defects.160 In particular, small molecules cause congenital defects by rapidly modulating developmental pathways: dorsomorphin affects embryogenesis, and thalidomide is characterized by disruption of embryonic patterning and limb reduction defects in addition to other developmental abnormalities that can also be predictable, which limits the use of defined inhibitors in patients.161 Mice treated with tankyrase inhibitors develop toxicity and weight loss, in contrast, porcupine inhibitors do not cause adverse effects on the gut in the course of cancer treatment.162,163 Despite the mention of such adverse effects as alopecia, fatigue, bone loss or breakage, muscle cramps, and dysgeusia are also documented as a result of targeting the signal transduction pathway including Wnt. Nevertheless, these unfavorable impacts are generally reversible upon cessation of treatment. Thus, these observations indicate that small molecule inhibitors or drugs targeting important developmental signals utilized by normal tissues and cancer stem cells may pose issues in normal homeostasis.
Conclusions and future perspectives
Wnt signaling plays a vital role in the proper differentiation of various tissues and is also involved in adult homeostasis. Moreover, it maintains the intestinal stem cell niche at the bottom of the crypts of the adult intestine. Notably, abnormal deviation in the Wnt/β-catenin pathway leads to CRC by promoting cell proliferation, survival, and invasion, activated by driver genes such as APC, CTNNB1, RNF43, AXIN1/2, and RSPO2/3. However, little is known about CRC and its molecular etiology, which is considered the most complex malignancy for clinical management. Recently, a breakthrough in targeting the upstream component of Wnt signaling was achieved with the discovery of R-spondin2 in the case of CRC. Thus, in this review, the crucial molecular factor RSPO2 was thoroughly investigated, which will help to decipher the RSPO2 altered expression and pathway deregulation during the phenotypic presentation of CRC and thus offer the possibility of secondary prevention of CRC. Altered RSPO2 expression and rearrangement can be harnessed to develop biomarkers and targeted therapies in Wnt-dependent tumors with much effort, there are very few FDA-approved drugs and inhibitors that can be routinely incorporated into clinical trials that target the Wnt pathway and its upstream components for CRC. Despite several efforts, many challenges and problems are still associated with developing specific and efficient inhibitors targeting Wnt signaling in CRC.
Thus, this review leads to some valuable conclusions, the most important of which are outlined as follows: (i) R-spondins are secretory proteins and can function as Wnt signaling regulators; (ii) Signal transduction through Wnt broadly regulates cell proliferation, migration, differentiation, and polarity during development and homeostasis; (iii) Aberrant Wnt signaling and RSPO2 are implicated in many forms of human cancer including CRC; (iv) RSPO2 and fusion transcripts of RSPO2 tend to be ideal for molecular therapy as they are precise to cancerous cells and have also been explored as prognostic & diagnostic indicators in CRC; (v) Therapeutic approaches (natural products, small molecules, and drugs) directly targeting Wnt-dependent signaling components or RSPO2 signaling offer promising anti-cancer strategies for CRC treatment.
Author contributions
Ankit Srivastava: conceptualization, data analysis, writing - original draft, and literature review; Deeksha Rikhari: literature review and drafting, and writing- original draft; Sameer Srivastava: supervision, conceptualization, and review & editing.
Conflict of interests
All the authors declare they have no conflicts.
Acknowledgements
The authors are thankful to the Director, Motilal Nehru National Institute of Technology, Allahabad, India for providing research facilities. The authors are also thankful to the Ministry of Human Resource and Development, Govt. of India, New Delhi, for providing scholarship during this tenure.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Ferlay J., Lam F., Colombet M., et al. International Agency for Research on Cancer; Lyon, France: 2020. Global Cancer Observatory: Cancer Today. [Google Scholar]
- 2.Ferlay J., Colombet M., Soerjomataram I., et al. Cancer statistics for the year 2020:an overview. Int J Cancer. 2021;149(4):778–789. doi: 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 3.World Cancer Research Fund International. Accessed May 15, 2022. https://www.wcrf.org/.
- 4.Srivastava A., Rai S., Bisht D., et al. Protein Kinase Inhibitors. Elsevier; Amsterdam: 2022. Targeting the altered tyrosine kinases in colorectal cancer: from inhibitors to drugs; pp. 361–391. [Google Scholar]
- 5.Srivastava A., Rai S., Singh M.P., et al. In: Computational Intelligence in Oncology. Raza K, editor. Springer; Singapore: 2022. Computational intelligence-based gene expression analysis in colorectal cancer: a review; pp. 387–410. [Google Scholar]
- 6.Willett C.G., Chang D.T., Czito B.G., et al. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh M.P., Rai S., Suyal S., et al. Genetic and epigenetic markers in colorectal cancer screening: recent advances. Expert Rev Mol Diagn. 2017;17(7):665–685. doi: 10.1080/14737159.2017.1337511. [DOI] [PubMed] [Google Scholar]
- 8.Dong X., Liao W., Zhang L., et al. RSPO2 suppresses colorectal cancer metastasis by counteracting the Wnt5a/Fzd7-driven noncanonical Wnt pathway. Cancer Lett. 2017;402:153–165. doi: 10.1016/j.canlet.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Singh M.P., Rai S., Singh N.K., et al. Transcriptomic landscape of early age onset of colorectal cancer identifies novel genes and pathways in Indian CRC patients. Sci Rep. 2021;11 doi: 10.1038/s41598-021-91154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markowitz S.D., Bertagnolli M.M. Molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Sohaily S., Biankin A., Leong R., et al. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol. 2012;27(9):1423–1431. doi: 10.1111/j.1440-1746.2012.07200.x. [DOI] [PubMed] [Google Scholar]
- 12.Mundade R., Imperiale T.F., Prabhu L., et al. Genetic pathways, prevention, and treatment of sporadic colorectal cancer. Oncoscience. 2014;1(6):400–406. doi: 10.18632/oncoscience.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J.Z., Wang S., Tang R., et al. Cloning and identification of a cDNA that encodes a novel human protein with thrombospondin type I repeat domain, hPWTSR. Mol Biol Rep. 2002;29(3):287–292. doi: 10.1023/a:1020479301379. [DOI] [PubMed] [Google Scholar]
- 14.Kamata T., Katsube K.I., Michikawa M., et al. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim Biophys Acta. 2004;1676(1):51–62. doi: 10.1016/j.bbaexp.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Kazanskaya O., Glinka A., del Barco Barrantes I., et al. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev Cell. 2004;7(4):525–534. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 16.Chang L.S., Kim M., Glinka A., et al. The tumor suppressor PTPRK promotes ZNRF3 internalization and is required for Wnt inhibition in the Spemann organizer. Elife. 2020;9 doi: 10.7554/eLife.51248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parma P., Radi O., Vidal V., et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38(11):1304–1309. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 18.de Lau W., Barker N., Low T.Y., et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 19.Ruffner H., Sprunger J., Charlat O., et al. R-Spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaydon D.C., Ishii Y., O'Toole E.A., et al. The gene encoding R-spondin 4 (RSPO4), a secreted protein implicated in Wnt signaling, is mutated in inherited anonychia. Nat Genet. 2006;38(11):1245–1247. doi: 10.1038/ng1883. [DOI] [PubMed] [Google Scholar]
- 21.Ishii Y., Wajid M., Bazzi H., et al. Mutations in R-spondin 4 (RSPO4) underlie inherited Anonychia. J Invest Dermatol. 2008;128(4):867–870. doi: 10.1038/sj.jid.5701078. [DOI] [PubMed] [Google Scholar]
- 22.Bergmann C., Senderek J., Anhuf D., et al. Mutations in the gene encoding the wnt-signaling component R-spondin 4 (RSPO4) cause autosomal recessive Anonychia. Am J Hum Genet. 2006;79(6):1105–1109. doi: 10.1086/509789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitz C.S., van Steensel M., Frank J., et al. The Wnt signalling ligand RSPO4, causing inherited anonychia, is not mutated in a patient with congenital nail hypoplasia/aplasia with underlying skeletal defects. Br J Dermatol. 2007;157(4):801–802. doi: 10.1111/j.1365-2133.2007.08059.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim K.A., Zhao J., Andarmani S., et al. R-Spondin proteins: a novel link to beta-catenin activation. Cell Cycle. 2006;5(1):23–26. doi: 10.4161/cc.5.1.2305. [DOI] [PubMed] [Google Scholar]
- 25.Nam J.S., Turcotte T.J., Smith P.F., et al. Mouse cristin/R-spondin family proteins are novel ligands for the frizzled 8 and LRP6 receptors and activate β-catenin-dependent gene expression. J Biol Chem. 2006;281(19):13247–13257. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- 26.Chen P.H., Chen X., Lin Z., et al. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27(12):1345–1350. doi: 10.1101/gad.219915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zebisch M., Xu Y., Krastev C., et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun. 2013;4:2787. doi: 10.1038/ncomms3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moad H.E., Pioszak A.A. Reconstitution of R-spondin: LGR4:ZNRF3 adult stem cell growth factor signaling complexes with recombinant proteins produced in Escherichia coli. Biochemistry. 2013;52(41):7295–7304. doi: 10.1021/bi401090h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K.A., Kakitani M., Zhao J., et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309(5738):1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 30.Raslan A.A., Yoon J.K. R-spondins: multi-mode WNT signaling regulators in adult stem cells. Int J Biochem Cell Biol. 2019;106:26–34. doi: 10.1016/j.biocel.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Alowolodu O., Johnson G., Alashwal L., et al. Intrinsic disorder in spondins and some of their interacting partners. Intrinsically Disord Proteins. 2016;4(1) doi: 10.1080/21690707.2016.1255295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Y.R., Yoon J.K. The R-spondin family of proteins: emerging regulators of WNT signaling. Int J Biochem Cell Biol. 2012;44(12):2278–2287. doi: 10.1016/j.biocel.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Lau W.B.M., Snel B., Clevers H.C. The R-spondin protein family. Genome Biol. 2012;13(3):242. doi: 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K.A., Wagle M., Tran K., et al. R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol Biol Cell. 2008;19(6):2588–2596. doi: 10.1091/mbc.E08-02-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao H.X., Xie Y., Zhang Y., et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 36.Hein R.F.C., Wu J.H., Holloway E.M., et al. R-SPONDIN2+ mesenchymal cells form the bud tip progenitor niche during human lung development. Dev Cell. 2022;57(13):1598–1614.e8. doi: 10.1016/j.devcel.2022.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glinka A., Dolde C., Kirsch N., et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12(10):1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Zhang W., Mulholland M.W. LGR4 and its role in intestinal protection and energy metabolism. Front Endocrinol. 2015;6:131. doi: 10.3389/fendo.2015.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Y., Zamponi R., Charlat O., et al. Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep. 2013;14(12):1120–1126. doi: 10.1038/embor.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J.G., Huang C., Yang Z., et al. Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G-protein-coupled receptors (LGRs) J Biol Chem. 2015;290(4):2455–2465. doi: 10.1074/jbc.M114.599134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S.J., Yen T.Y., Endo Y., et al. Loss-of-function point mutations and two-furin domain derivatives provide insights about R-spondin2 structure and function. Cell Signal. 2009;21(6):916–925. doi: 10.1016/j.cellsig.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell S.M., Schreiner C.M., Wert S.E., et al. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135(6):1049–1058. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- 43.Ohkawara B., Glinka A., Niehrs C. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev Cell. 2011;20(3):303–314. doi: 10.1016/j.devcel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Lee H., Seidl C., Sun R., et al. R-spondins are BMP receptor antagonists in Xenopus early embryonic development. Nat Commun. 2020;11:5570. doi: 10.1038/s41467-020-19373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H., Sun R., Niehrs C. Uncoupling the BMP receptor antagonist function from the WNT agonist function of R-spondin 2 using the inhibitory peptide dendrimer RWd. J Biol Chem. 2022;298(2) doi: 10.1016/j.jbc.2022.101586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D., Huang B., Zhang S., et al. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 2013;27(12):1339–1344. doi: 10.1101/gad.219360.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng W., De Lau W., Forneris F., et al. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 2013;3(6):1885–1892. doi: 10.1016/j.celrep.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Mulvaney J.F., Yatteau A., Sun W.W., et al. Secreted factor R-Spondin 2 is involved in refinement of patterning of the mammalian cochlea. Dev Dyn. 2013;242(2):179–188. doi: 10.1002/dvdy.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ter Steege E.J., Bakker E.R.M. The role of R-spondin proteins in cancer biology. Oncogene. 2021;40(47):6469–6478. doi: 10.1038/s41388-021-02059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ota T., Suzuki Y., Nishikawa T., et al. Complete sequencing and characterization of 21, 243 full-length human cDNAs. Nat Genet. 2004;36(1):40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 51.Gerhard D.S., Wagner L., Feingold E.A., et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14(10B):2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nusbaum C., Mikkelsen T.S., Zody M.C., et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439(7074):331–335. doi: 10.1038/nature04406. [DOI] [PubMed] [Google Scholar]
- 53.Clark H.F., Gurney A.L., Abaya E., et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13(10):2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szenker-Ravi E., Altunoglu U., Leushacke M., et al. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature. 2018;557(7706):564–569. doi: 10.1038/s41586-018-0118-y. [DOI] [PubMed] [Google Scholar]
- 55.Bian J., Dannappel M., Wan C., et al. Transcriptional regulation of Wnt/β-catenin pathway in colorectal cancer. Cells. 2020;9(9):2125. doi: 10.3390/cells9092125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Q., Yokota C., Semenov M.V., et al. R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and β-catenin signaling. J Biol Chem. 2007;282(21):15903–15911. doi: 10.1074/jbc.M701927200. [DOI] [PubMed] [Google Scholar]
- 57.Li N., Spetz M.R., Ho M. The role of glypicans in cancer progression and therapy. J Histochem Cytochem. 2020;68(12):841–862. doi: 10.1369/0022155420933709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebensohn A.M., Rohatgi R. R-spondins can potentiate WNT signaling without. LGRs. Elife. 2018;7 doi: 10.7554/eLife.33126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Binnerts M.E., Kim K.A., Bright J.M., et al. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc Natl Acad Sci U S A. 2007;104(37):14700–14705. doi: 10.1073/pnas.0702305104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin Y.R., Turcotte T.J., Crocker A.L., et al. The canonical Wnt signaling activator, R-spondin2, regulates craniofacial patterning and morphogenesis within the branchial arch through ectodermal–mesenchymal interaction. Dev Biol. 2011;352(1):1–13. doi: 10.1016/j.ydbio.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han X.H., Jin Y.R., Seto M., et al. A WNT/β-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J Biol Chem. 2011;286(12):10649–10659. doi: 10.1074/jbc.M110.169391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arima M., Hasegawa D., Yoshida S., et al. R-spondin 2 promotes osteoblastic differentiation of immature human periodontal ligament cells through the Wnt/β-catenin signaling pathway. J Periodontal Res. 2019;54(2):143–153. doi: 10.1111/jre.12611. [DOI] [PubMed] [Google Scholar]
- 63.Knight M.N., Karuppaiah K., Lowe M., et al. R-spondin-2 is a Wnt agonist that regulates osteoblast activity and bone mass. Bone Res. 2018;6:24. doi: 10.1038/s41413-018-0026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dolmans G.H., Werker P.M., Hennies H.C., et al. Wnt signaling and Dupuytren's disease. N Engl J Med. 2011;365(4):307–317. doi: 10.1056/NEJMoa1101029. [DOI] [PubMed] [Google Scholar]
- 65.Bell S.M., Schreiner C.M., Hess K.A., et al. Asymmetric limb malformations in a new transgene insertional mutant, footless. Mech Dev. 2003;120(5):597–605. doi: 10.1016/s0925-4773(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 66.Aoki M., Kiyonari H., Nakamura H., et al. R-spondin2 expression in the apical ectodermal ridge is essential for outgrowth and patterning in mouse limb development. Dev Growth Differ. 2008;50(2):85–95. doi: 10.1111/j.1440-169X.2007.00978.x. [DOI] [PubMed] [Google Scholar]
- 67.Yamada W., Nagao K., Horikoshi K., et al. Craniofacial malformation in R-spondin2 knockout mice. Biochem Biophys Res Commun. 2009;381(3):453–458. doi: 10.1016/j.bbrc.2009.02.066. [DOI] [PubMed] [Google Scholar]
- 68.Nam J.S., Turcotte T.J., Yoon J.K. Dynamic expression of R-spondin family genes in mouse development. Gene Expr Patterns. 2007;7(3):306–312. doi: 10.1016/j.modgep.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 69.Liu J., Pan S., Hsieh M.H., et al. Targeting wnt-driven cancer through the inhibition of porcupine by LGK974. Proc Natl Acad Sci U S A. 2013;110(50):20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Cian M.C., Gregoire E.P., Le Rolle M., et al. R-spondin2 signaling is required for oocyte-driven intercellular communication and follicular growth. Cell Death Differ. 2020;27(10):2856–2871. doi: 10.1038/s41418-020-0547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouilly J., Beau I., Barraud S., et al. R-spondin2, a novel target of NOBOX: identification of variants in a cohort of women with primary ovarian insufficiency. J Ovarian Res. 2017;10:51. doi: 10.1186/s13048-017-0345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedman M.S., Oyserman S.M., Hankenson K.D. Wnt11 promotes osteoblast maturation and mineralization through R-spondin 2. J Biol Chem. 2009;284(21):14117–14125. doi: 10.1074/jbc.M808337200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chua A.W.C., Ma D., Gan S.U., et al. The role of R-Spondin2 in keratinocyte proliferation and epidermal thickening in keloid scarring. J Invest Dermatol. 2011;131(3):644–654. doi: 10.1038/jid.2010.371. [DOI] [PubMed] [Google Scholar]
- 74.Carmon K.S., Gong X., Lin Q., et al. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108(28):11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coussy F., Lallemand F., Vacher S., et al. Clinical value of R-spondins in triple-negative and metaplastic breast cancers. Br J Cancer. 2017;116(12):1595–1603. doi: 10.1038/bjc.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conboy C.B., Vélez-Reyes G.L., Rathe S.K., et al. R-spondins 2 and 3 are overexpressed in a subset of human colon and breast cancers. DNA Cell Biol. 2021;40(1):70–79. doi: 10.1089/dna.2020.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin X., Yi H., Wang L., et al. R-spondin 2 promotes proliferation and migration via the Wnt/β-catenin pathway in human hepatocellular carcinoma. Oncol Lett. 2017;14(2):1757–1765. doi: 10.3892/ol.2017.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conboy C.B., Vélez-Reyes G.L., Tschida B.R., et al. R-spondin 2 drives liver tumor development in a yes-associated protein-dependent manner. Hepatol Commun. 2019;3(11):1496–1509. doi: 10.1002/hep4.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fitamant J., Kottakis F., Benhamouche S., et al. YAP inhibition restores hepatocyte differentiation in advanced HCC, leading to tumor regression. Cell Rep. 2015;10(10):1692–1707. doi: 10.1016/j.celrep.2015.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y., Ge K., Lu J., et al. MicroRNA-493 suppresses hepatocellular carcinoma tumorigenesis through down-regulation of anthrax toxin receptor 1 (ANTXR1) and R-spondin 2 (RSPO2) Biomed Pharmacother. 2017;93:334–343. doi: 10.1016/j.biopha.2017.06.047. [DOI] [PubMed] [Google Scholar]
- 81.Zheng C., Zhou F., Shi L.L., et al. R-spondin2 suppresses the progression of hepatocellular carcinoma via MAPK signaling pathway. Mol Cancer Res. 2020;18(10):1491–1499. doi: 10.1158/1541-7786.MCR-19-0599. [DOI] [PubMed] [Google Scholar]
- 82.Zhang H., Han X., Wei B., et al. RSPO2 enhances cell invasion and migration via the WNT/β-catenin pathway in human gastric cancer. J Cell Biochem. 2019;120(4):5813–5824. doi: 10.1002/jcb.27867. [DOI] [PubMed] [Google Scholar]
- 83.Ilmer M., Boiles A.R., Regel I., et al. RSPO2 enhances canonical Wnt signaling to confer stemness-associated traits to susceptible pancreatic cancer cells. Cancer Res. 2015;75(9):1883–1896. doi: 10.1158/0008-5472.CAN-14-1327. [DOI] [PubMed] [Google Scholar]
- 84.Wu L., Zhang W., Qian J., et al. R-spondin family members as novel biomarkers and prognostic factors in lung cancer. Oncol Lett. 2019;18(4):4008–4015. doi: 10.3892/ol.2019.10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Q., Xu Z. miR-196b-5p promotes proliferation, migration and invasion of lung adenocarcinoma cells via targeting RSPO2. Cancer Manag Res. 2020;12:13393–13402. doi: 10.2147/CMAR.S274171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Al-Samadi A., Salo T. Understanding the role of the R-spondin 2-LGR4 system in tongue squamous cell carcinoma progression. EBioMedicine. 2019;44:8–9. doi: 10.1016/j.ebiom.2019.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhong S., Wu B., Li J., et al. T5224, RSPO2 and AZD5363 are novel drugs against functional pituitary adenoma. Aging. 2019;11(20):9043–9059. doi: 10.18632/aging.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tocci J.M., Felcher C.M., García Solá M.E., et al. R-spondin-mediated WNT signaling potentiation in mammary and breast cancer development. IUBMB Life. 2020;72(8):1546–1559. doi: 10.1002/iub.2278. [DOI] [PubMed] [Google Scholar]
- 89.Wang Z., Wang Y., Ma X., et al. RSPO2 silence inhibits tumorigenesis of nasopharyngeal carcinoma by ZNRF3/Hedgehog-Gli1 signal pathway. Life Sci. 2021;282 doi: 10.1016/j.lfs.2021.119817. [DOI] [PubMed] [Google Scholar]
- 90.Mehdawi L.M., Prasad C.P., Ehrnström R., et al. Non-canonical WNT5A signaling up-regulates the expression of the tumor suppressor 15-PGDH and induces differentiation of colon cancer cells. Mol Oncol. 2016;10(9):1415–1429. doi: 10.1016/j.molonc.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hao H.X., Jiang X., Cong F. Control of Wnt receptor turnover by R-spondin-ZNRF3/RNF43 signaling module and its dysregulation in cancer. Cancers. 2016;8(6):54. doi: 10.3390/cancers8060054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park S., Cui J., Yu W., et al. Differential activities and mechanisms of the four R-spondins in potentiating Wnt/β-catenin signaling. J Biol Chem. 2018;293(25):9759–9769. doi: 10.1074/jbc.RA118.002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koo B.K., Spit M., Jordens I., et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 94.Dubey R., van Kerkhof P., Jordens I., et al. R-spondins engage heparan sulfate proteoglycans to potentiate WNT signaling. Elife. 2020;9 doi: 10.7554/eLife.54469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhan T., Rindtorff N., Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Torres M.A., Yang-Snyder J.A., Purcell S.M., et al. Activities of the Wnt-1 class of secreted signaling factors are antagonized by the Wnt-5A class and by a dominant negative cadherin in early Xenopus development. J Cell Biol. 1996;133(5):1123–1137. doi: 10.1083/jcb.133.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiang X., Charlat O., Zamponi R., et al. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3Ubiquitin ligases. Mol Cell. 2015;58(3):522–533. doi: 10.1016/j.molcel.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 98.Voloshanenko O., Schwartz U., Kranz D., et al. β-catenin-independent regulation of Wnt target genes by RoR2 and ATF2/ATF4 in colon cancer cells. Sci Rep. 2018;8:1–14. doi: 10.1038/s41598-018-20641-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flores-Hernández E., Velázquez D.M., Castañeda-Patlán M.C., et al. Canonical and non-canonical Wnt signaling are simultaneously activated by Wnts in colon cancer cells. Cell Signal. 2020;72 doi: 10.1016/j.cellsig.2020.109636. [DOI] [PubMed] [Google Scholar]
- 100.Bienz M., Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103(2):311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 101.Srivastava A., Sharan S. Sebaceous gland carcinoma of ocular region in India: a brief literature review for disease management. Lat Am J Ophthalmol. 2021;4:4. [Google Scholar]
- 102.Lowther W., Wiley K., Smith G.H., et al. A new common integration site, Int7, for the mouse mammary tumor virus in mouse mammary tumors identifies a gene whose product has furin-like and thrombospondin-like sequences. J Virol. 2005;79(15):10093–10096. doi: 10.1128/JVI.79.15.10093-10096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Theodorou V., Kimm M.A., Boer M., et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39(6):759–769. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 104.Katoh M. Canonical and non-canonical WNT signaling in cancer stem cells and their niches: cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review) Int J Oncol. 2017;51(5):1357–1369. doi: 10.3892/ijo.2017.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Starr T.K., Allaei R., Silverstein K.A.T., et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barker N., van Es J.H., Kuipers J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 107.Barker N., Huch M., Kujala P., et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6(1):25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 108.Jaks V., Barker N., Kasper M., et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40(11):1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 109.Sato T., Vries R.G., Snippert H.J., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 110.Zhang S., Han X., Wei B., et al. RSPO2 enriches LGR5+ spheroid colon cancer stem cells and promotes its metastasis by epithelial-mesenchymal transition. Am J Transl Res. 2016;8(2):354–364. [PMC free article] [PubMed] [Google Scholar]
- 111.Pilati P., Mocellin S., Bertazza L., et al. Prognostic value of putative circulating cancer stem cells in patients undergoing hepatic resection for colorectal liver metastasis. Ann Surg Oncol. 2012;19(2):402–408. doi: 10.1245/s10434-011-2132-2. [DOI] [PubMed] [Google Scholar]
- 112.Valladares-Ayerbes M., Blanco-Calvo M., Reboredo M., et al. Evaluation of the adenocarcinoma-associated gene AGR2 and the intestinal stem cell marker LGR5 as biomarkers in colorectal cancer. Int J Mol Sci. 2012;13(4):4367–4387. doi: 10.3390/ijms13044367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shenoy A.K., Fisher R.C., Butterworth E.A., et al. Transition from colitis to cancer: high Wnt activity sustains the tumor-initiating potential of colon cancer stem cell precursors. Cancer Res. 2012;72(19):5091–5100. doi: 10.1158/0008-5472.CAN-12-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klauzinska M., Baljinnyam B., Raafat A., et al. Rspo2/Int7 regulates invasiveness and tumorigenic properties of mammary epithelial cells. J Cell Physiol. 2012;227(5):1960–1971. doi: 10.1002/jcp.22924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vermeulen L., De Sousa E Melo F., van der Heijden M., et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12(5):468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 116.Li C., Cao J., Zhang N., et al. Identification of RSPO2 fusion mutations and target therapy using a porcupine inhibitor. Sci Rep. 2018;8 doi: 10.1038/s41598-018-32652-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 118.Mertens F., Johansson B., Fioretos T., et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 119.Sansom O.J., Reed K.R., Hayes A.J., et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18(12):1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seshagiri S., Stawiski E.W., Durinck S., et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shinmura K., Kahyo T., Kato H., et al. RSPO fusion transcripts in colorectal cancer in Japanese population. Mol Biol Rep. 2014;41(8):5375–5384. doi: 10.1007/s11033-014-3409-x. [DOI] [PubMed] [Google Scholar]
- 122.Han T., Schatoff E.M., Murphy C., et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun. 2017;8 doi: 10.1038/ncomms15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hashimoto T., Ogawa R., Yoshida H., et al. EIF3E-RSPO2 and PIEZO1-RSPO2 fusions in colorectal traditional serrated adenoma. Histopathology. 2019;75(2):266–273. doi: 10.1111/his.13867. [DOI] [PubMed] [Google Scholar]
- 124.Sekine S., Ogawa R., Hashimoto T., et al. Comprehensive characterization of RSPO fusions in colorectal traditional serrated adenomas. Histopathology. 2017;71(4):601–609. doi: 10.1111/his.13265. [DOI] [PubMed] [Google Scholar]
- 125.Huettner C.S., Zhang P., Van Etten R.A., et al. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat Genet. 2000;24(1):57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- 126.Klein F., Feldhahn N., Herzog S., et al. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene. 2006;25(7):1118–1124. doi: 10.1038/sj.onc.1209133. [DOI] [PubMed] [Google Scholar]
- 127.Williams S.V., Hurst C.D., Knowles M.A. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nassar A.H., Lundgren K., Pomerantz M., et al. Enrichment of FGFR3-TACC3 fusions in patients with bladder cancer who are young, Asian, or have never smoked. JCO Precis Oncol. 2018;2 doi: 10.1200/PO.18.00013. PO.18.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tomlins S.A., Mehra R., Rhodes D.R., et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66(7):3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 130.Winnes M., Lissbrant E., Damber J.E., et al. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17(5):1033–1036. [PubMed] [Google Scholar]
- 131.Tognon C., Knezevich S.R., Huntsman D., et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 132.Karkera J., Martinez G., Bell K., et al. Abstract B03: identification of R-spondin fusions in NSCLC. Clin Cancer Res. 2014;20(2_Supplement):B03. [Google Scholar]
- 133.Longerich T., Endris V., Neumann O., et al. RSPO2 gene rearrangement: a powerful driver of β-catenin activation in liver tumours. Gut. 2019;68(7):1287–1296. doi: 10.1136/gutjnl-2018-317632. [DOI] [PubMed] [Google Scholar]
- 134.MacDonald B.T., Tamai K., He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cheng X., Xu X., Chen D., et al. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–481. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]
- 136.Pálmer H.G., González-Sancho J.M., Espada J., et al. Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. J Cell Biol. 2001;154(2):369–387. doi: 10.1083/jcb.200102028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Madan B., Ke Z., Harmston N., et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35(17):2197–2207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tian D., Shi Y., Chen D., et al. The Wnt inhibitor LGK-974 enhances radiosensitivity of HepG2 cells by modulating Nrf2 signaling. Int J Oncol. 2017;51(2):545–554. doi: 10.3892/ijo.2017.4042. [DOI] [PubMed] [Google Scholar]
- 139.Gong Y., Bourhis E., Chiu C., et al. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Patel K.R., Brown V.A., Jones D.J.L., et al. Clinical pharmacology of resveratrol and its metabolites in colorectal cancer patients. Cancer Res. 2010;70(19):7392–7399. doi: 10.1158/0008-5472.CAN-10-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Patel B.B., Sengupta R., Qazi S., et al. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer. 2008;122(2):267–273. doi: 10.1002/ijc.23097. [DOI] [PubMed] [Google Scholar]
- 142.Cuzick J., Thorat M.A., Bosetti C., et al. Estimates of benefits and harms of prophylactic use of aspirin in the general population. Ann Oncol. 2015;26(1):47–57. doi: 10.1093/annonc/mdu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arber N., Eagle C.J., Spicak J., et al. Celecoxib for the prevention of colorectal adenomatous polyps. N Engl J Med. 2006;355(9):885–895. doi: 10.1056/NEJMoa061652. [DOI] [PubMed] [Google Scholar]
- 144.Klose J., Trefz S., Wagner T., et al. Salinomycin: anti-tumor activity in a pre-clinical colorectal cancer model. PLoS One. 2019;14(2) doi: 10.1371/journal.pone.0211916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gurney A., Axelrod F., Bond C.J., et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109(29):11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ahmad Farooqi A., de la Roche M., Djamgoz M.B.A., et al. Overview of the oncogenic signaling pathways in colorectal cancer: mechanistic insights. Semin Cancer Biol. 2019;58:65–79. doi: 10.1016/j.semcancer.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 147.Ettenberg S.A., Charlat O., Daley M.P., et al. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proc Natl Acad Sci U S A. 2010;107(35):15473–15478. doi: 10.1073/pnas.1007428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lepourcelet M., Chen Y.N.P., France D.S., et al. Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell. 2004;5(1):91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 149.Eguchi M., Nguyen C., Lee S.C., et al. ICG-001, a novel small molecule regulator of TCF/beta-catenin transcription. Med Chem. 2005;1(5):467–472. doi: 10.2174/1573406054864098. [DOI] [PubMed] [Google Scholar]
- 150.Osman J., Bellamkonda K., Liu Q., et al. The WNT5A agonist Foxy5 reduces the number of colonic cancer stem cells in a xenograft mouse model of human colonic cancer. Anticancer Res. 2019;39(4):1719–1728. doi: 10.21873/anticanres.13278. [DOI] [PubMed] [Google Scholar]
- 151.Waaler J., Machon O., Tumova L., et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012;72(11):2822–2832. doi: 10.1158/0008-5472.CAN-11-3336. [DOI] [PubMed] [Google Scholar]
- 152.Lee S.C., Kim O.H., Lee S.K., et al. IWR-1 inhibits epithelial-mesenchymal transition of colorectal cancer cells through suppressing Wnt/β-catenin signaling as well as survivin expression. Oncotarget. 2015;6(29):27146–27159. doi: 10.18632/oncotarget.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen Z., Venkatesan A.M., Dehnhardt C.M., et al. 2, 4-Diamino-quinazolines as inhibitors of β-catenin/Tcf-4 pathway: potential treatment for colorectal cancer. Bioorg Med Chem Lett. 2009;19(17):4980–4983. doi: 10.1016/j.bmcl.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 154.Annala M.J., Parker B.C., Zhang W., et al. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340(2):192–200. doi: 10.1016/j.canlet.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Boone J.D., Arend R.C., Johnston B.E., et al. Targeting the Wnt/β-catenin pathway in primary ovarian cancer with the porcupine inhibitor WNT974. Lab Invest. 2016;96(2):249–259. doi: 10.1038/labinvest.2015.150. [DOI] [PubMed] [Google Scholar]
- 156.Katoh M., Katoh M. Molecular genetics and targeted therapy of WNT-related human diseases (Review) Int J Mol Med. 2017;40(3):587–606. doi: 10.3892/ijmm.2017.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Group B.D.W. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 158.Fearon E.R. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 159.Sarma M.K., Ningthoujam R., Panda M.K., et al. In: Raza K, Dey. N eds. Translational Bioinformatics Applications in Healthcare. CRC Press; Boca Raton, FL: 2021. Translational healthcare system through bioinformatics; pp. 3–21. [Google Scholar]
- 160.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13(7):513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sakata T., Chen J.K. Chemical 'Jekyll and Hyde's: small-molecule inhibitors of developmental signaling pathways. Chem Soc Rev. 2011;40(8):4318–4331. doi: 10.1039/c1cs15019g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lau T., Chan E., Callow M., et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013;73(10):3132–3144. doi: 10.1158/0008-5472.CAN-12-4562. [DOI] [PubMed] [Google Scholar]
- 163.Proffitt K.D., Madan B., Ke Z., et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 2013;73(2):502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]