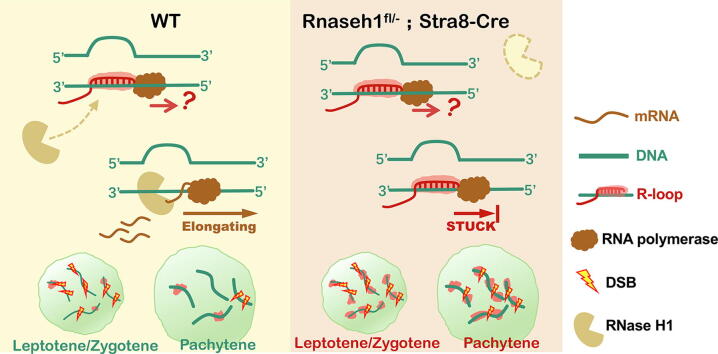
Graphical abstract
Keywords: R-loop, Meiosis, Spermatogenesis, Genome stability, Homologous recombination, DNA repair
Highlights
-
•
Mapped the genomic distribution and dynamic changes of R-loop during the meiotic process.
-
•
The high frequency of R-loop in meiotic cells are usually accompanied by high transcription activity.
-
•
RNase H1 deletion before meiosis entry caused R-loop accumulation with abnormal DSB repairing.
-
•
R-loop accumulated in leptotene/zygotene stage influenced downstream transcription in pachytene stage.
Abstract
Introduction
The R-loop is a naturally formed three-strand nucleic acid structure that recently has been reported to participate in multiple biological processes and helped answer some previously unexplained scientific questions. Meiosis process involves multiple chromatin-related events such as DNA double-stranded breaks (DSB) formation, repairing and transcriptional dynamics.
Objectives
Explore the regulatory roles and physiological functions of R-loops in the mammalian meiosis process.
Methods
In our study, using genome-wide S9.6 CUT & Tag seq, we first mapped the genomic distribution and dynamic changes of R-loop during the meiotic process in mice, from spermatogonia to secondary spermatocytes. And we further explore the role of R-loop in physiological conditions by constructing conditional knockout mice of Rnaseh1, which deleted the R-loop endonuclease before meiosis entry.
Results
R-loop predominantly distributes at promoter-related regions and varies across different meiotic stages. By joint analysis with the corresponding transcriptome, we found that the R-loop was closely related to transcription during the meiotic process. The high frequency of promoter-related R-loop in meiotic cells is usually accompanied by high transcription activity, and we further verified this in the leptotene/zygotene to the pachytene transition process. Moreover, the lack of RNase H1 caused sterility in male mice with R-loop accumulation and abnormal DSB repair in spermatocytes. Further analysis showed that abnormal R-loop accumulation in the leptotene/zygotene stages influenced transcriptional regulation in the pachytene stage.
Conclusion
The mutual regulation of the R-loop and transcription plays an essential role in spermatogenesis. And R-loop is also important for the normal repair process of DSB during meiosis.
Introduction
Meiosis is a complicated process of sexual reproduction. The unique programmed formation and resection of DNA double-strand breaks (DSBs) and subsequent homologous recombination are the most important events of meiosis, which determine accurate chromosome segregation and the generation of new parental alleles [1]. In mammalian meiosis, the resected 3′ single-strand overhangs at DSBs are covered with single-strand DNA (ssDNA) binding proteins RPA and strand-exchange proteins DMC1/RAD51 to search for homologous chromosomes. DSBs are repaired following homologous chromosome synapsis, during which homologous recombination occurs. The DSB repair process requires many essential factors to function cooperatively and is monitored by a strict surveillance mechanism called the meiotic recombination checkpoint [2].
During transcription, the nascent RNA hybridized with the complementary DNA strand to generate an RNA–DNA hybrid duplex and a displaced non-template single DNA strand, this three-strand structure is called an “R-loop” [3]. The R-loop plays multiple physiological roles in cells, for example, by influencing transcription [4], triggering DNA repair response [5] and threatening genome integrity [5], [6], [7], [8], causing replication stress [9], [10], [11] and even changing chromatin landscapes [12]. Recently, Liu et al. reported that RNA-DNA hybrids (R-loop) also contribute to the DSB repair process in U2OS cells [13]. RNA polymerase III catalyzes transcription at DSBs, forming a transient RNA-DNA hybrid to protect the 3’overhang from degradation before RPA binding [13]. However, the potential involvement of R-loops in DSB repair during mammalian germ cell meiosis has not been investigated.
R-loops participate in multiple biological processes and is essential for maintaining the stability of the genome, thus, the formation and resolution of R-loop are proposed to be highly regulated. Several enzymes contribute to the removal of R-loops. The helicases, SETX and Aquarius [14], [15], [16], are considered to have RNA/DNA helicase activity [17]. Additionally, topoisomerase I is reported to prevent the pairing of the nascent RNA with DNA by resolving the negative torsional stress [18], [19]. Moreover, it is considered that RNA processing and exporting factors also contribute to preventing R-loop formation by binding nascent mRNA in time [20], [21]. The most important step is performed by endoribonuclease H, including RNase H1 and RNase H2, which digests the paired RNA strand in R-loops [22], [23], [24]. It has been reported that knockout of Rnaseh1 in mice causes embryonic lethality [25].
The R-loop also participates in the meiotic DSB repair process. In Saccharomyces cerevisiae, the THO complex is responsible for transporting nascent RNA to the cytoplasm. Deletion of Hprl subunit in THO complex caused increased DSB formation and new SPO11-independent DSB, which can be partially rescued by RNase H1 overexpression. In Caenorhabditis elegans, THO mutants cause genome instability and trigger the pachytene checkpoint [26]. In mice, Setx-/- mice showed accumulation of R-loops and persistence of RNA polymerase II with abnormal meiotic sex chromosome inactivation [27]. However, several key questions regarding the role of R-loop in meiosis remain unanswered. In the complicated meiotic process, the formation, removal, and biological importance of the R-loop at each developmental stage are unclear. The interplay among meiotic R-loops, transcription, and DSB formation/repair still needs to be determined. The physiological function of RNase H1, the major R-loop endoribonuclease, in meiosis has not been investigated.
To explore the detailed role of R-loops in meiosis, we used S9.6 CUT & Tag-seq to map the distribution of R-loops in the genome at each stage of meiosis, revealing the distribution of R-loops in the genome under normal physiological conditions. We compared the results of RNA-seq and S9.6 CUT & Tag-seq to analyze the potential relationship between the distribution of R-loops and transcriptional activity. Ultimately, we constructed a germ cell-specific Rnaseh1 knockout mouse strain and analyzed the physiological role of R-loops in meiosis.
Materials and methods
Histology and immunostaining
The testes and cauda epididymis were fixed in Bouin’s solution for histological analysis and 4% paraformaldehyde for immunostaining. After dehydration, samples were embedded in paraffin and sectioned at 5 μm thickness per slice. For histological analysis, slides were deparaffinized, rehydrated, and co-stained with hematoxylin and eosin. For immunofluorescence, sections were boiled in 10 mM sodium citrate buffer (pH 6.0) for 15 min and cooled to room temperature, after washing with ddH2O, blocked samples with 5% BSA for 30 min and then incubated with primary antibodies overnight at 4 °C. After washing, sections were incubated with secondary antibodies for 30 min. mounted with glycerin after washing and then took photos with fluorescence microscopy. The antibodies used are listed in Supplementary Table 2.
Spermatocyte nuclei spreading and staining
Meiotic chromosome spreads were prepared as previously described [28].
Western blotting
Collected 5 × 104 spermatogenic cells per sample, lysed in loading buffer with β-mercaptoethanol and then boiled for 5 min at 95 °C. SDS-PAGE running and immunoblotting were performed according to the standard protocol of Mini-PROTEAN Tetra Cell System (Bio-Rad, CA, USA). The antibodies are shown in Table S2.
Isolation of spermatogenic cells with FACS
Spermatogenic cells were isolated from 6-week-old male mice according to a previously published protocol [29].
RNA-seq
Spermatocytes were collected (1,500 cells per sample) with FACS as described above. The cDNA synthesis was performed according to the Smart-seq2 method as described previously [30]. 500 pg cDNA were taken for Sequencing libraries construction with TruePrep DNA Library Prep Kit V2 for Illumina (TD503; Vazyme, Nanjing, China). And then sequence with barcoded libraries on the Illumina HiSeq X™ Ten platform.
RNA-seq data analysis
Paired-end 150 bp reads of RNA-seq were aligned to the mm9 reference genome (UCSC) using the STAR alignment tool (version 2.7.3a) with default parameters [31]. Uniquely mapped reads were employed to quantify gene expression using RSEM software (version 1.3.1) [32]. Fragments per kilobase per million mapped reads (FPKM) were calculated to estimate the gene expression levels, normalized for gene length and sequencing depth. The Euclidean distance based on the FPKM values was used to evaluate the similarities across biological replicates. Differential gene expression analysis was carried out using the DESeq2 R package, adjusted P-value < 0.05, and absolute fold change > 2 were used as statistical significances to identify differentially expressed genes (DEGs). The results of the analysis are listed in Supplementary Table 3.
R-loop CUT & Tag library construction
R-loop CUT & Tag were performed as previously described, with some modifications [33]. In brief, 5 × 104 spermatogonia were sorted and washed once in 0.5 mL wash buffer [20 mM Hepes (pH 7.5); 150 mM NaCl; 0.5 mM spermidine; and 1 × protease inhibitors] and were incubated with 10 μL pre-washed concanavalin A–coated magnetic beads in a 1.5 mL microtube at 25 °C for 10 min. Cells were resuspended in 80 μL ice-cold antibody incubation buffer [20 mM HEPES (pH 7.5); 150 mM NaCl; 0.5 mM spermidine; 1 × protease inhibitors; 0.025% digitonin; 0.01% NP-40; and 2 mM EDTA] with 1 μg S9.6 antibody per sample (MABE1095; MilliporeSigma), and then rotated end-over-end at 4 °C overnight. The cells were resuspended in 100 μL of dig-wash buffer [20 mM Hepes (pH 7.5); 150 mM NaCl; 0.5 mM spermidine; 1 × protease inhibitors; 0.025% digitonin) with rabbit anti-mouse IgG (1:100 dilution, Ab6709; ThermoFisher) and rotated end-over-end at 25 °C for 1 h. After washing, home-made pA-Tn5 adapter complexes in dig-300 buffer (20 mM Hepes [pH 7.5]; 300 mM NaCl; 0.5 mM spermidine; 1 × protease inhibitors; 0.01% digitonin] were added to the samples at a final concentration of 1:100 and then rotated end-over-end at 25 °C for 1 h. After washing, the cells were resuspended in 300 μL fragmentation buffer [dig-300 buffer with 10 mM MgCl2] and incubated at 37 °C for 1 h. To stop fragmentation and solubilize DNA fragments, 10 μL of 0.5 M EDTA, 3 μL of 10% SDS and 2.5 μL of 20 mg/mL Proteinase K were added to each sample and incubated at 50 °C for 1 h. After extraction with phenol–chloroform and ethanol precipitation, the libraries were amplified by PCR. The barcoded libraries were pooled and sequenced on the Illumina HiSeq X™ Ten platform to generate 150 bp paired-end reads.
R-loop data analysis
Raw reads of S9.6 CUT & Tag data were trimmed to filter low-quality bases using TrimGalore (version 0.6.4; https://github.com/FelixKrueger/TrimGalore). Trimmed reads were aligned to the mm9 reference genome (UCSC) using Bowtie 2 aligner [34] with parameters “--local -I 10 -X 700.” Unmapped or multi-mapped reads and reads mapped to the mitochondrial DNA (chrM) were discarded. For unique alignments, duplicate reads were marked by “Mark Duplicates” function of Picard software https://broadinstitute.github.io/picard/). Uniquely mapped reads were used to carry out peak calling using MACS2 with a no-local lambda mode [35]. We used input samples as controls for peak calling, and candidate R-loop peaks in WT spermatogonia and spermatocytes were defined by the fold change of IP/input > 2 and adjusted P-value < 0.05. The read coverage of candidate R-loop peaks was normalized by reads per kilobase per million mapped reads (RPKM) using deepTools [36]. A putative “R-loop site” was defined as the peak center with RPKM maxima.
For comparison of R-loop signals across different stages and samples, we performed spike-in normalization through mapping S9.6 reads to the E. coli genome by Bowtie2. CUT&Tag reads were normalized by toal aligned reads (RPKM) and spike-in scaling factors calculated by E. coli% reads. The track files of normalized reads were generated with bamCoverage function of deepTools for visulization.
For comparison of R-loop signals across different samples, we generated read count normalized genome-wide pileup tracks using ‘SPRM’ function of MACS2, then output bigwig files to visualization.
The genomic distribution of R-loop peaks was annotated by “ChIP seeker” R package. Each non-intergenic R-loop peak was assigned to the nearest gene. Dynamic R-loop genes were defined as genes with alterations in signal intensity between leptotene/zygotene and pachytene/diplotene. The results of the analysis are listed in Supplementary Table 4.
Statistical analysis
Student’s t-test and Mann-Whitney-Wilcoxon test were used to compare continuous variables between the two groups (*P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant). Analysis of variance (ANOVA) was used to estimate variance among multiple groups. Fisher’s exact test was used to compare the categorical variables. The P-values in multiple tests were adjusted to the false discovery rate (FDR) using the Benjamini-Hochberg method.
Results
Most R-loops predominantly distribute at promoter regions during spermatogenesis
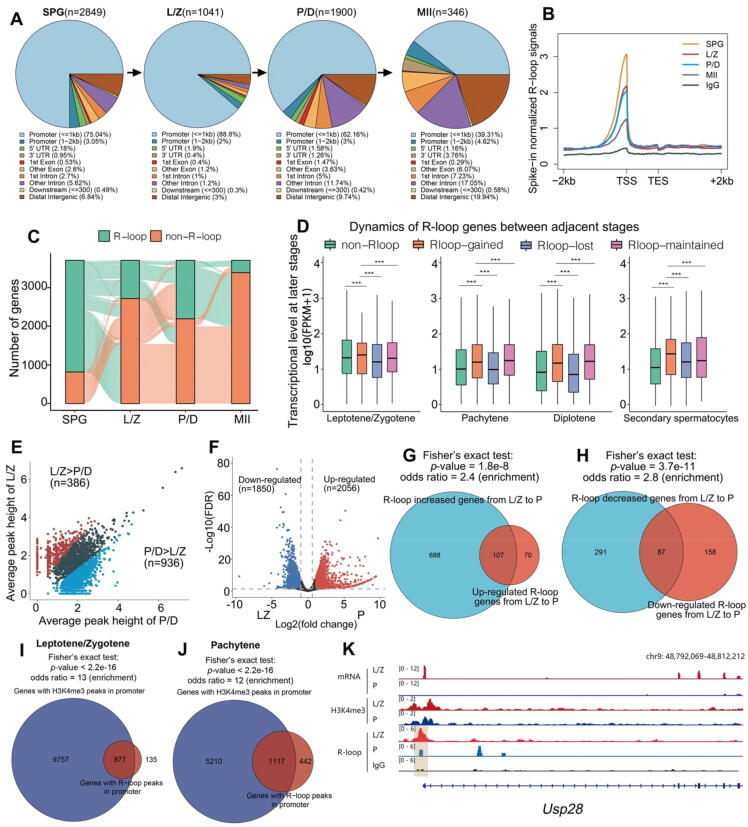
To explore the role of the R-loop in the meiotic process, We first use S9.6 CUT & Tag-seq to detect the distribution and dynamic changes of R-loop in the genome during spermatogenesis. We selected four main stages of spermatogenesis to perform genome-wide R-loop mapping: spermatogonia (SPG), leptotene/zygotene (L/Z), pachytene/diplotene (P/D), and secondary spermatocytes (MII). The purity of the isolated meiotic prophase I cells was verified by SYCP3-γH2AX staining (Fig. S1A). A total of 2,849 R-loop peaks were detected in SPG, more than 1,041 peaks detected in L/Z and 1,900 peaks of P/D, and only 346 peaks were detected in the MII stage. Genomic distribution analysis of the four stages showed that the genomic distributional composition of the R-loop varied across different stages, but the vast majority of R-loop signals were localized at promoter regions, especially in L/Z spermatocytes (Fig. 1A). Further comparison of R-loop signals across the whole transcripts showed that the TSS-related region showed a much stronger signal than other areas. Among the four stages, SPG had the highest signal intensity, and L/Z had slightly higher R-loop signals than P/D, and MII had the lowest signal intensity (Fig. 1B). Furthermore, we utilized heatmaps to show the patterns of R-loop read density across WT spermatogonia/spermatocytes (Fig. S1B). The high frequency of R-loop appearance at the promoter and promoter-related regions implied that the R-loop might be related to the transcriptional process during meiosis.
Fig. 1.
Genomic distribution of R-loop peaks during spermatogenesis. A: Pie charts showing the genomic distribution variation of R-loop peaks during spermatogenesis. SPG, spermatogonia; L/Z, leptotene/zygotene; P/D, pachytene/diplotene; MII, secondary spermatocytes. B: Genomic metaplots showing relative signal intensity comparison of R-loops across the 2 kb window around gene bodies in spermatogonia and spermatocytes. R-loop signals of different stages were normalized based on library sizes and spike-in scaling factors. C: Alluvial plots showing the global dynamics of genes with R-loop and genes without R-loops during spermatocyte development. Each line represents an R-loop gene, defined as a gene-detected R-loop peak in at least one analyzed stage. D: Box plots showing the expression level comparison of different gene clusters grouped by R-loop variation between adjacent stages in meiotic progress. E: Scatter plots showing the comparison of signal intensities of R-loop peaks between leptotene/zygotene and pachytene/diplotene spermatocytes. F: Volcano plots showing differentially expressed genes between leptotene/zygotene and pachytene spermatocytes. Genes upregulated (FC > 2; FDR < 0.05) and downregulated (FC < 1/2; FDR < 0.05) in pachytene spermatocytes are colored red and blue, respectively. G: Venn diagrams showing the overlap of R-loop accumulated genes and transcriptionally upregulated R-loop genes in leptotene/zygotene to pachytene stage. H: Venn diagrams showing the overlap of R-loop decreased genes and transcriptionally downregulated R-loop genes in leptotene/zygotene to pachytene stage. I: Venn diagrams showing the overlap of H3K4me3 peaks in genes promoter regions and R-loop peaks in leptotene/zygotene spermatocytes (Fisher’s exact test; ***P < 0.001). J: Venn diagrams showing the overlap of H3K4me3 peaks in genes promoter regions and R-loop peaks in pachytene spermatocytes (Fisher’s exact test; ***P < 0.001). K: Example snapshot of the relative distribution of the R-loop, H3K4me3, and downstream transcriptional level. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The establishment and clearance of the R-loop on each gene are also dynamic at different stages of the meiotic process (Fig. 1C). According to the dynamic changes of the R-loop signal in adjacent stages, we divided R-loop genes into four groups: non-R-loop, R-loop-gained, R-loop-lost and R-loop maintained. Transcriptional analysis (combined with previous RNA-seq data GSE133636) showed that the R-loop-gained genes were always accompanied with higher transcription than other genes, while the R-loop-lost genes mostly appeared at lower transcriptional levels (Fig. 1D).
Clustering analysis of all replicates revealed that SPG showed more correlations with LZ, but PD showed more correlations with MII (Fig. S1C). It is surprising that L/Z and P/D showed similar intensities but were clustered into different groups (Fig. S1C). We used the L/Z to P/D transition as an example to explore the important role of the R-loop in meiotic developmental progress. Firstly, we analyzed the R-loop dynamics during L/Z to P/D differentiation. Across all detected R-loop peaks between L/Z and P/D, 16.8% of R-loop peaks (total 386 peaks) were higher in L/Z than P/D, but 40.9% of peaks (total 936 peaks) were lower in L/Z (Fig. 1E; S1D). Dynamic R-loop genes were defined as genes that detected differential R-loop peaks between L/Z and P/D. There were also dynamic transcriptional changes during L/Z to P/D transition. A total of 1,850 genes were downregulated, and 2,056 genes were upregulated by 2-fold change screening (Fig. 1F). Above differentially expressed genes (DEGs) detected R-loop peaks in the promoter region were assigned to R-loop DEGs. Further intersection analysis of the dynamic R-loop genes and R-loop DEGs during L/Z to P/D transition showed a significant overlapping ratio (Fig. 1G-H). Briefly, 60% of up-regulated R-loop DEGs overlapped with increased R-loop genes (P-value = 1.8e-8), 35% of down-regulated R-loop DEGs were shared with decreased R-loop genes (P-value = 3.7e-11). Interestingly, we also observed 87% of L/Z R-loop peaks, 72% P/D R-loop peaks were shared with the H3K4me3 peaks in genes promoter regions respectively, which partially supports the close relationship between the R-loop and transcription (Fig. 1I-J). As shown in the genome browser of Usp28, lower levels of R-loop from L/Z to P stage at promoter regions are accompanied by decreasing H3K4me3 modification, following downstream lower transcriptional levels in P (Fig. 1K). Taken together, the R-loop is predominantly distributed at promoter regions and is closely related to the transcriptional process in meiosis.
RNase H1 deletion in pre-meiotic germ cells causes spermatogenic defects
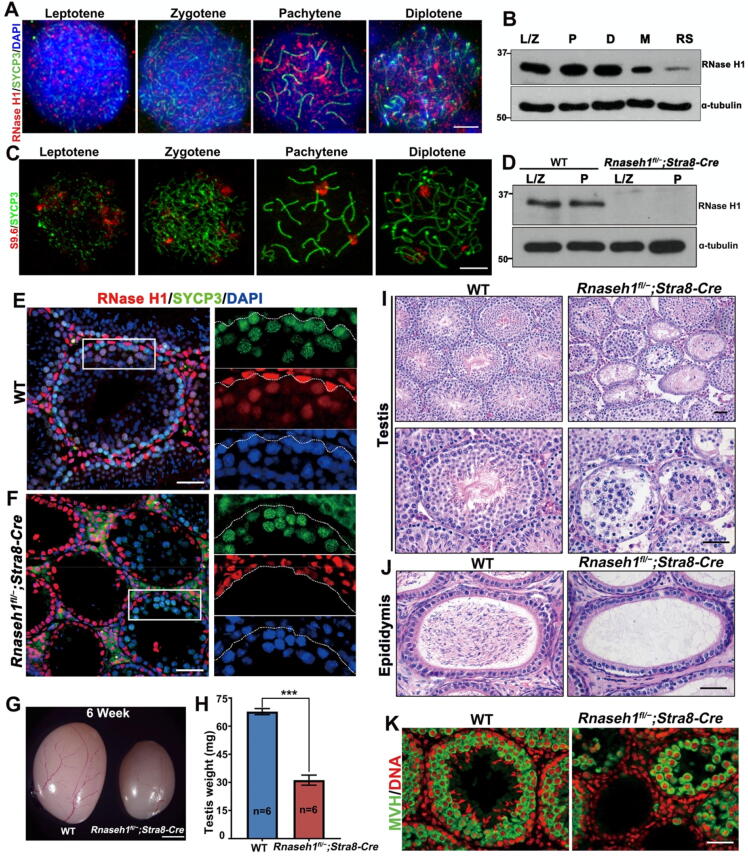
The endoribonuclease, RNase H1 plays a crucial role in the removal of R-loops. Thus, we selected RNase H1 as a starting point to explore the biological function of R-loops during meiotic progression. During the first wave of spermatogenesis, spermatocytes and spermatids of various developmental stages gradually appear in the seminiferous tubular lumen. We selected mouse testes at 10, 12, 16, 28, and 42 days to specify different developmental stage cells. Immunofluorescence staining of RNase H1 and phosphorylated histone H2A.X (γH2AX) in wild type (WT) mouse testes at different stages showed that RNase H1 protein is mainly detected in the nucleus of Sertoli cells and spermatogonia, and is highly expressed in meiotic prophase spermatocytes, but is eliminated in the round and elongated spermatids (Fig. S2A). We also verified the RNase H1 expression pattern by immunohistochemistry in 6-week-old testes slices (Fig. S2B). In the published single-cell RNA-seq dataset [29], the transcriptional level of Rnaseh1 gradually increased from the preleptotene stage and remained at a low level in diplotene and later stages (Fig. S2C). We also determined the relative localization of RNase H1 on chromatin by immunofluorescent staining of RNase H1 and synaptonemal complex protein 3 (SYCP3) on spermatocyte chromatin spread, RNase H1 localized diffusely over the entire chromatin (Fig. 2A). Moreover, western blotting results further confirmed the expression of RNase H1 in specific subset spermatocytes isolated by flow cytometry (Fig. 2B). In contrast, we found that the R-loop localizes in clusters on chromatin in spermatocytes by S9.6 immunofluorescent staining on spermatocyte chromatin spreads (Fig. 2C). High expression levels in meiotic prophase I and chromatin association imply that RNase H1 may be involved in meiosis.
Fig. 2.
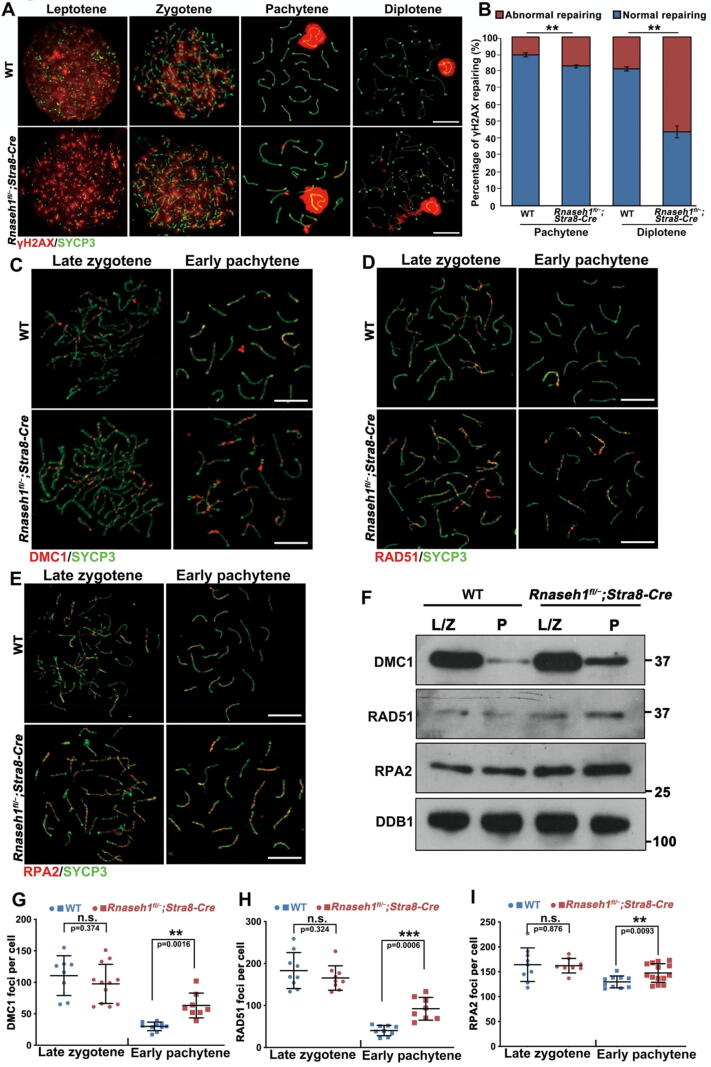
RNase H1 deletion in pre-meiotic germ cells causes spermatogenic defects. A: RNase H1 immunofluorescence staining on spermatocyte spreads from WT mice. DNA was counterstained with DAPI. Scale bar: 10 µm. B: Western blot of ribonuclease H1 (RNase H1) in spermatogenic cells isolated from WT mice. L/Z, leptotene/zygotene; P, pachytene; D, diplotene; M, meiotic metaphase; RS, round spermatids. C: S9.6 immunofluorescence staining on WT mice spermatocyte spreads. Scale bar: 10 µm. D: Western blot of RNase H1 in germ cells from WT and Rnaseh1fl/−;Stra8-Cre mice. E-F: Left tri-color panel, Immunofluorescent co-staining of SYCP3 and RNase H1 in testes of 6-week-old WT (A) and Rnaseh1fl/−;Stra8-Cre (B) mice. DNA was counterstained with DAPI. Scale bar: 50 µm. The right three panels show enlarged images of each channel. Meiotic cells of the lumen are divided out from spermatogonia and Sertoli cells with a white dotted line. G: Gross morphology of testes from Rnaseh1fl/−;Stra8-Cre and WT littermate mice. Scale bar: 1 mm. H: Weights of testis from 6-week-old Rnaseh1fl/−;Stra8-Cre and WT littermate mice. Error bars, s.e.m. ***P < 0.001 (two-tailed Student’s t-test). I-J: H&E staining results showing testis (F) and epididymis (G) histology of 6-week-old WT and Rnaseh1fl/−;Stra8-Cre mice. Scale bar: 50 µm. K: MVH immunofluorescence staining in the testes of 6-week-old WT and Rnaseh1fl/−;Stra8-Cre mice. DNA was counterstained with DAPI. Scale bar: 50 µm.
To determine the role of Rnaseh1 during mammalian meiotic prophase, we used a conditional knockout approach in which a Rnaseh1 floxed line was crossed with Stra8-Cre knock-in mice expressing CRE from A1 spermatogonia onward [37] (Fig. S2D). Western blot results also verified that RNase H1 was undetectable in spermatocytes isolated by flow cytometry from Rnaseh1fl/−;Stra8-Cre mice (Fig. 2D). We verified the knockout efficiency by immunofluorescent co-staining of RNase H1 and SYCP3 on testicular sections of 6-week-old WT and conditional knockout mice (Fig. 2E, F). As shown in the enlarged image, no RNase H1 signal was detected in SYCP3 marked spermatocytes (Fig. 2F).
The Rnaseh1fl/−;Stra8-Cre males were able to copulate normally but were completely sterile. Testes of 6-week-old Rnaseh1fl/−;Stra8-Cre mice were significantly smaller than those of the control littermates (Fig. 2G, H). Histological examinations of 6-week-old Rnaseh1 knockout mice testes showed that nearly a quarter of the seminiferous tubules were hollow, while most tubules contained spermatocytes arrested at the pachytene stage and apoptotic cells (Fig. 2I). Consistent with this, no sperm was found in the epididymis of adult Rnaseh1fl/−;Stra8-Cre mice (Fig. 2J). Immunofluorescence staining of mouse VASA homolog (MVH) in the testes of 6-week-old WT and Rnaseh1fl/−;Stra8-Cre mice confirmed that fewer germ cell layers and smaller lumen formed in knockout mice testes than WT testes (Fig. 2K). Moreover, co-staining with GATA4 and c-Kit showed no significant change in Sertoli cells and spermatogonia development after Rnaseh1-deletion (Fig. S2E). These results suggest that RNase H1 plays an essential role in spermatogenesis and male fertility.
RNase H1 deletion causes pachytene arrest with accumulated R-loops and abnormal DSB repairing
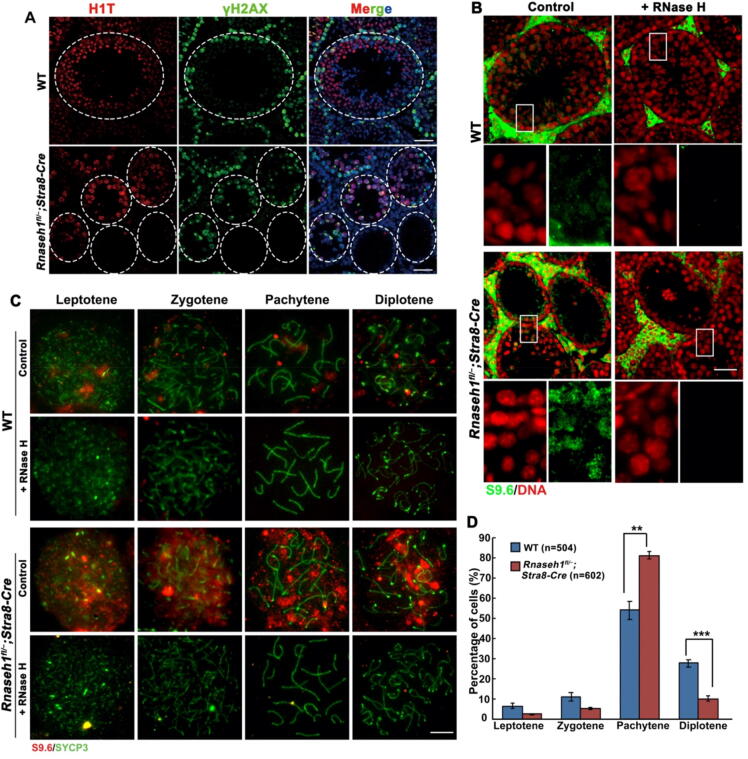
To further specify the stage at which the spermatogenetic cells of Rnaseh1fl/−;Stra8-Cre mice were arrested, we performed immunofluorescent co-staining of testis-specific histone H1 (H1T), which marks spermatocytes from mid-pachytene and beyond, and phosphorylated H2AX (γH2AX), which marks unrepaired DNA lesions and sex body in pachynema/diplonema. The lumen of Rnaseh1fl/−;Stra8-Cre mice testicular sections were full of H1T-positive cells with sex body marked γH2AX signal, implying that the knockout spermatogenic process is arrested at the pachytene/diplotene stage (Fig. 3A). Immunofluorescent staining of S9.6 on both testicular sections (Fig. 3B) and spermatocyte chromatin spread (Fig. 3C) showed that these arrested cells had significantly increased R-loops compared to WT spermatocytes. And exogenous RNase H digestion abolished the R-loop signal in both kinds of spermatocytes (Fig. 3B, C). Further analysis of the arrested spermatocytes based on γH2AX and SYCP3 co-staining showed that most of the arrested cells were pachytene spermatocytes (Fig. 3D), and some pachytene and diplotene spermatocytes with unrepaired DSBs on autosomes (Fig. 4A, B), while DSBs were completely repaired on autosomes in WT spermatocytes. We further analyzed the detailed DSB repair process by immunofluorescent staining (Fig. 4C-E) and western blot (Fig. 4F) of single-strand binding proteins-RAD51, DMC1, and RPA2. Foci counting on fluorescent images (Fig. 4G-I) and the western blot (Fig. 4F) showed that more foci persisted in Rnaseh1-null pachytene spermatocytes than in WT spermatocytes. Collectively, these results indicate that Rnaseh1 deletion caused the arrest of spermatogenesis at the defective pachytene stage with increasing R-loops and an abnormal DSB repair process.
Fig. 3.
RNase H1 deletion in pre-meiotic germ cells causes pachytene arrest with increased R-loops. A: Immunofluorescent co-staining of H1T and γH2AX in the testes of 6-week-old WT and Rnaseh1fl/−;Stra8-Cre mice. DNA was counterstained with DAPI. Scale bars: 50 µm. B: S9.6 immunofluorescent staining in the testes of 6-week-old WT and Rnaseh1fl/−;Stra8-Cre mice. DNA was counterstained with DAPI. Scale bars: 50 µm. The below panels are enlarged images. C: S9.6 immunofluorescence staining on spermatocyte spreads made from WT and Rnaseh1fl/−;Stra8-Cre mice. Scale bars: 10 µm. D: Spermatocyte stage proportion in WT and Rnaseh1fl/−;Stra8-Cre mice based on SYCP3 and γH2AX immunofluorescence staining of spermatocyte spreads. The number of analyzed spermatocytes is indicated (n). Error bars, s.e.m.. n.s., non-significant. **P < 0.01. ***P < 0.001 (two-tailed Student’s t-test). The experiment was repeated at least 3 times.
Fig. 4.
Rnaseh1 deletion impairs the DSB repair process in spermatogenic cells. A: Immunofluorescence staining of SYCP3 and γH2AX on WT and Rnaseh1fl/−;Stra8-Cre spermatocyte spreads. Scale bars: 10 µm. B: The proportion of autosomes DSB repaired and unrepaired ratio in WT and Rnaseh1fl/−;Stra8-Cre spermatocytes. Error bars, s.e.m. **P < 0.01 (two-tailed Student’s t-test). The experiment was repeated at least 3 times. C-E: Immunofluorescence staining of DMC1 (A), RAD51 (B), and RPA2 (C) on spermatocyte spreads made from WT and Rnaseh1fl/−;Stra8-Cre mice. Scale bars: 10 µm. F: Western blot of DSB repairing-related proteins in spermatogenic cells isolated from adult WT and Rnaseh1fl/−;Stra8-Cre mice. L/Z, leptotene/zygotene; P, pachytene. G-I: Signal quantification of DMC1 (E), RAD51 (F), and RPA2 (G) foci number per cell at different meiotic stages. Error bars, s.e.m. ***P < 0.001 (two-tailed Student’s t-test). n.s., non-significant. Above experiments were repeated at least 3 times.
R-loops accumulate in Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes
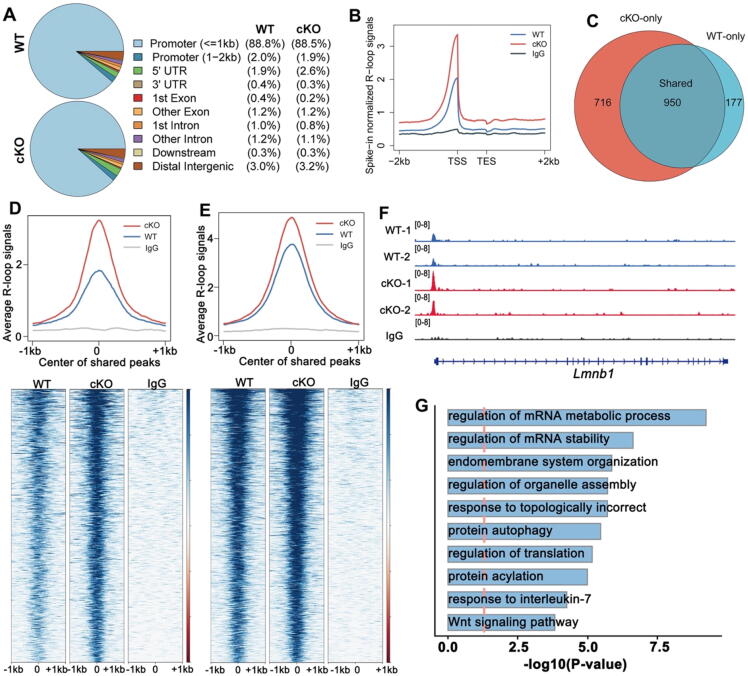
To explore the dynamic changes in R-loop signals in Rnaseh1-deleted spermatocytes, we performed S9.6 CUT & Tag-seq of leptotene/zygotene spermatocytes in WT and Rnaseh1fl/−;Stra8-Cre (cKO) mice respectively. The biological replicates showed a high correlation and repeatability (Fig. S3A–C). The genomic distribution of the R-loop was similar to that of the WT in cKO spermatocytes (Fig. 5A), but the signal intensity at the promoter regions was higher than the WT (Fig. 5B). However, cKO spermatocytes showed significantly increased R-loop signals compared to WT spermatocytes (Fig. S3D). Overlap analysis of WT and cKO R-loop peaks showed that 950 peaks were shared between WT and cKO spermatocytes, but there were 716 cKO-only R-loop peaks and 177 wt-only peaks (Fig. 5C). In summary, 819 genes possessed a higher R-loop in cKO spermatocytes (Fig. S3E). Further analysis of the cKO-only peaks and the shared peaks showed that cKO R-loop signals were higher than those in WT L/Z spermatocytes in both peak regions (Fig. 5D-E), demonstrating the significant accumulation of R-loops in cKO spermatocytes. An example gene is displayed on a snapshot (Fig. 5F). Gene ontology analysis of these genes was significantly enriched in terms related to the regulation of mRNA metabolic processes and mRNA stability (Fig. 5G), which supports our conclusion that the R-loop is closely related to transcription in the meiotic process.
Fig. 5.
R-loops accumulate in Rnaseh1fl/−;Stra8-Cre spermatocytes. A: Pie charts showing the genomic distribution of R-loops in leptotene/zygotene spermatocytes of WT and Rnaseh1fl/−;Stra8-Cre mice.B: Genomic metaplots showing relative signal intensity comparison of R-loops from the transcription start sites (TSSs) to transcription end sites (TESs) in WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes. C: Venn diagram showing the overlap of R-loop peaks in WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes (Fisher’s exact test; ***P < 0.001). D: Average R-loop read density and heatmap of WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes in the 1 kb window around the center of cKO-only peaks (D) and shared R-loop peaks. Loci are ordered by the descending R-loop intensities in cKO samples. E: Average R-loop read density and heatmap of WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes in the 1 kb window around the center of shared R-loop peaks. Loci are ordered by the descending R-loop intensities in cKO samples. F: Snapshots of accumulated R-loops on example genes. G: Gene ontology analysis of R-loop accumulated genes in Rnaseh1-null spermatocytes.
Rnaseh1 deletion resulted in transcriptional dysregulation in meiotic prophase I spermatocytes
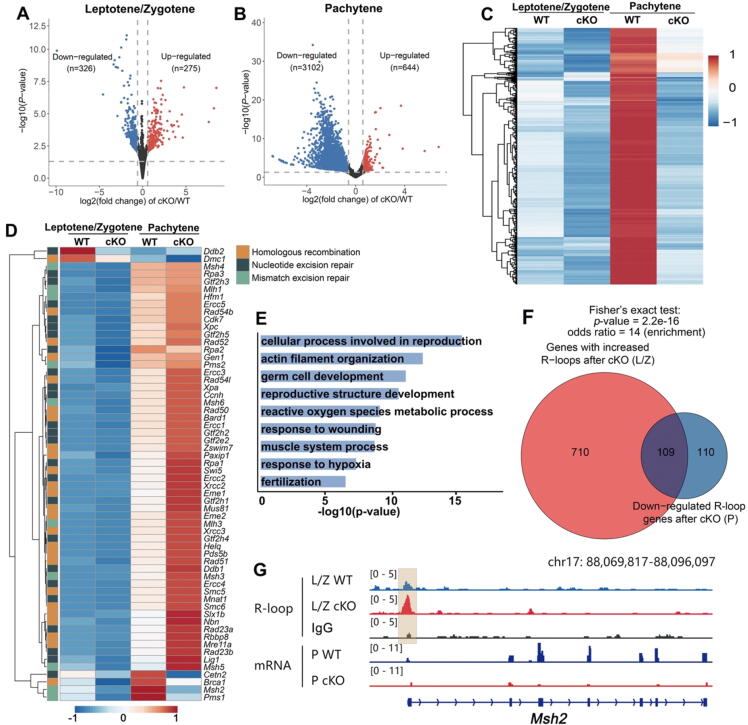
To further explore the influence of increased R-loops on transcription, we performed RNA–seq analysis on WT and Rnaseh1-null spermatocytes at the leptotene/zygotene and pachytene stages. Gene expression levels were assessed as fragments per kilobase of transcript per million mapped reads (FPKM). All samples were analyzed in triplicate, and the replicates showed high correlations (Fig. S3F). As shown in the snapshot, there are no signals detected in the exon2 and exon3 of Rnaseh1, verifying the efficient Rnaseh1 knockout (Fig. S3G). The number of down-regulated genes was higher than that of upregulated genes in Rnaseh1-null spermatocytes at both stages (Fig. 6A-B), especially in the pachytene stage, and a total of 3,102 genes were down-regulated (Fig. 6B). Further analysis of the down-regulated genes in pachytene revealed that all of these 3,102 down-regulated genes displayed a similar pattern; they were upregulated from L/Z to P in WT spermatocytes, implying that these genes are physiologically essential genes during L/Z to P development (Fig. 6C). On the other hand, most genes related to DSB repair and the recombination processes were upregulated in Rnaseh1-null spermatocytes at the pachytene stage (Fig. 6D), presumably due to the compensation effect of unrepaired DSBs. Gene ontology analysis showed that downregulated genes in the pachytene stage were significantly enriched in terms related to germ cell development and reproduction (Fig. 6E).
Fig. 6.
RNA-seq analyses of leptotene/zygotene and pachytene spermatocytes derived from WT and Rnaseh1fl/−;Stra8-Cre mice. A-B: Volcano plots showing differentially expressed genes of leptotene/zygotene and pachytene spermatocytes. Genes upregulated (FC > 2; FDR < 0.05) and downregulated (FC < 1/2; FDR < 0.05) in spermatocytes are colored red and blue, respectively. C: Dynamic profiling of the 3,102 downregulated genes in Rnaseh1fl/−;Stra8-Cre pachytene spermatocytes. D: Dynamic profiling of DNA damage repair and homologous recombination-related genes across different stages in WT and Rnaseh1fl/−;Stra8-Cre spermatocytes. E: Gene ontology analysis of differentially expressed genes in Rnaseh1-null pachytene spermatocytes. F: Venn diagram showing the overlap of genes with accumulated R-loops in Rnaseh1-null leptotene/zygotene spermatocytes and down-regulated R-loop genes in pachytene spermatocytes. L/Z, leptotene/zygotene; P, pachytene. G: Example snapshots of the increased R-loops and downregulated transcription in Rnaseh1-null spermatocytes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Further analysis showed 109 genes with accumulated R-loops in leptotene/zygotene but downregulated transcription in pachytene stages (Fig. 6F). Accumulated R-loop at promoter regions was accompanied by decreased downstream transcription in cKO spermatocytes (Fig. 6G). Additionally, visualization of several candidate genes that were previously reported to be critical for the meiotic process and spermatogenesis were presented (Fig. S4). Taken together, RNase H1 is essential for the meiotic process by removing R-loops in the DSB repair process, while safeguarding normal transcriptional processes. Most importantly, major changes during spermatogenesis of WT and Rnaseh1 null mice were deduced (Fig. S5).
Discussion
Currently, there are two popular technologies for R-loop mapping: one is DRIP-Seq and its derivative technology, whose principle is immunoprecipitation with the S9.6 monoclonal antibody followed by high-throughput DNA sequencing [12], [38]; Another one is R-ChIP, this method requires exogenous overexpression of catalytically inactive RNaseH1 but can bind to R-loop, and then through chromatin immunoprecipitation and performs high-throughput sequencing [39]. The most commonly used method relies on the S9.6 antibody. The S9.6 antibody equally binds the DNA.RNA heteropolymer and poly(I).poly(dC), but 100-fold higher levels of poly(A).poly(dT) are necessary for similar binding efficiency, which is a structural characteristic of the R-loop [40]. However, the second method relies on exogenous RNase H1 overexpression and is not applicable under physiological conditions, such as in vivo meiotic cells. In addition, exogenous overexpression of catalytically inactive RNaseH1 will interfere with the normal function of endogenous RNaseH1 and induce more R-loop accumulation. In our study, we optimized the CUT & Tag-seq of the S9.6 antibody for genomic R-loop mapping with just 50,000 cells per sample. For a long time, it is difficult to obtain a sufficient number of samples for deep sequencing is one of the obstacles in the field of germ cell research. The requirement of fewer cells makes it more amenable to other precious samples. We mapped the R-loop distribution at different developmental stages with FACS-isolated spermatogenic cells for the first time. To make a systematic comparison of the role of R-loop in meiotic cells, we also collected samples of spermatids for R-loop mapping but failed at library construction because the R-loop signals were too low to enrich. The chromatin condensation and transcriptional silencing in spermatids may be the reason, which partially reflects that the R-loop is closely related to transcriptional status. Owing to R-loops having an important impact on both physiological and pathological processes, we believe our data sources of R-loop profiling have broad interest to researchers in multiple fields.
There have been many studies on genome-wide R-loops in other species. In Arabidopsis, R-loops are enriched in gene promoters and gene bodies and are highly associated with non-coding RNA and repetitive genomic regions [41]. In Saccharomyces cerevisiae, the R-loops form predominantly over rDNA highly transcribed genes and Ty1 and Ty2 retrotransposons [42], [43]. Several different sequencing technologies have been used to explore the R-loop in various cell types of human-derived cells previously. A recent article reported that, through comparison, higher confidence of R-loop localization was distributed at promoter regions and associated with GC skew [44]. Similarly, in mammalian spermatogenic cells, the R-loops formed mostly at promoter regions during the whole meiotic process and varied in different genes between adjacent stages. The high proportion of promoter region distribution also supports that the R-loop participates in spermatogenesis by interacting with transcription, which is consistent with the conclusion of our Rnaseh1 conditional knockout mice.
There have been some studies on the function of the R-loop in meiosis. RNA/DNA helicase Senataxin deletion arrested spermatogenesis at the pachytene stage with abnormal DSB repair and defects in meiotic sex chromosome inactivation (MSCI), which is consistent with the XY body localization in spermatocytes. Similarly, RNase H1 deletion also caused abnormal DSB repair, which contributes to the presumption that the R-loop may take part in the meiotic DSB repair process. There are also some differences. RNase H1 has a diffusion distribution on the chromosome and RNase H1-null spermatocytes with DSB defects are distributed across the whole genome and not limited to the XY body. The complicated R-loop clearing process involves many proteins with multiple functions. For example, another endonuclease RNase H2A may also play an important role or participate in different biological processes during meiosis.
Collectively, we first mapped the genomic distribution and dynamic changes of R-loop during the meiotic process and revealed the relationship between R-loop and transcription in mammalian meiotic cells. Moreover, Our research also found that R-loops may participate in the meiotic DSB repair process. The selection of DSB hotspot sites is based on H3K4me3 as a signal-targeting guide. In the L/Z stage, R-loop peaks and H3K4me3 peaks have a high overlap ratio, which implies that the R-loop may also play a role in the DSB generation process. Combined with the abnormal DSB repair process in RNase H1-null spermatocytes, it is possible that the DSB sites in knock-out spermatocytes might have changed. Further exploration of the specific relationship with DSB sites requires genome-wide sequencing for joint analysis, such as DMC1-seq or END-seq, which needs further study.
Conclusion
A triple-stranded structure named “R-loop,” formed by the incorporation of RNA into DNA, has been reported to participate in a variety of biological processes and exerts its special structural function. However, this has not been reported in mammalian meiosis. For the first time, we mapped the genomic distribution of R-loops at four continuous meiotic stages (SPG, L/Z, P/D, and MII cells). By comparing the transcriptional variation during the meiotic process, we found that the dynamic R-loops in meiosis were closely related to transcription, which was consistent with the high peak of H3K4me3 distribution. We also verified that in Rnaseh1 conditional knockout mice, abnormal accumulation of R-loop affects the transcription of downstream functional genes and causes transcriptional dysregulation. Additionally, we found that the R-loop plays an important role in the meiotic DSB repair process.
Compliance with Ethics Requirements
All Institutional and National Guidelines for the care and use of animals (fisheries) were followed.
CRediT authorship contribution statement
Yu Jiang: Formal analysis, Investigation, Writing – original draft, Visualization. Fei Huang: Software, Formal analysis, Visualization. Lu Chen: Methodology, Formal analysis, Investigation, Data curation, Writing – review & editing. Jia-Hui Gu: Methodology, Investigation, Data curation. Yun-Wen Wu: Software, Investigation. Meng-Yan Jia: Investigation. Zhen Lin: Validation. Yong Zhou: Investigation. Yan-Chu Li: Investigation. Chao Yu: Resources. Ming-Han Tong: Resources. Li Shen: Conceptualization, Validation, Resources, Writing – review & editing, Supervision, Funding acquisition. Heng-Yu Fan: Conceptualization, Validation, Resources, Writing – review & editing, Supervision, Funding acquisition, Project administration. Qian-Qian Sha: Conceptualization, Validation, Resources, Data curation, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31930031; 82101731; 31890781), the National Key Research and Development Program of China (2021YFC2700100; 2021YFA1100300), the National Ten-Thousands Talents Program of China, the Natural Science Foundation of Zhejiang Province, China (D22C68649), and Start-up Funding of Guangdong Second Provincial General Hospital [YY2019-001].
We thank Ying-Ying Huang (Core Facilities, School of Medicine, Zhejiang University) for help with cell sorting. We are grateful to our colleagues at the core facility of the Life Sciences Institute for assistance with cell imaging analysis. And we also appreciate Prof. Qing-Hua Shi (The University of Science and Technology of China) for the kind gift of H1T antibody.
Animals
All mice used in this study were maintained with a C57BL/6J genetic background. The Stra8-Cre knock-in mouse line was previously reported (29) and was kindly provided by Prof. Min-Han Tong. The Rnaseh1 floxed mouse strain was constructed by GemPharmaTech Co. (strain number: T014737; Nanjing, China). Primers used for PCR genotyping are listed in Supplementary Table 1. All mice were housed in the pathogen-free facility of the Laboratory Animal Center of Zhejiang University. The experimental procedures were approved by the Zhejiang University Institutional Animal Care and Research Committee (Approval # ZJU20210252 to HYF), and mouse care and use were conducted in accordance with the relevant guidelines and regulations. All applicable institutional and/or national guidelines for the care and use of animals were followed.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.10.016.
Contributor Information
Li Shen, Email: li_shen@zju.edu.cn.
Heng-Yu Fan, Email: hyfan@zju.edu.cn.
Qian-Qian Sha, Email: shaqianqian@zju.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
RNA-seq data were deposited in the NCBI Gene Expression Omnibus database under accession number GSE190430 with token elwxoucgzfgjzsr. ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus database under the accession number GSE193401 with token anmfgkoetvmntip. UCSC Browser URL:
https://genome.ucsc.edu/s/FeiHuang/mm9_Spermatogenesis_Rloop_profiling
References
- 1.Hunter N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol. 2015;7(12). [DOI] [PMC free article] [PubMed]
- 2.Longhese M.P., Bonetti D., Guerini I., Manfrini N., Clerici M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 2009;8(9):1127–1138. doi: 10.1016/j.dnarep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Allison D.F., Wang G.G. R-loops: formation, function, and relevance to cell stress. Cell Stress. 2019;3(2):38–46. doi: 10.15698/cst2019.02.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12(3):711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Cristini A., Ricci G., Britton S., Salimbeni S., Huang S.-Y., Marinello J., et al. Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks. Cell Rep. 2019;28(12):3167–3181.e6. doi: 10.1016/j.celrep.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguilera A., García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Skourti-Stathaki K., Proudfoot N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28(13):1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niehrs C., Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol. 2020;21(3):167–178. doi: 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamperl S., Cimprich K.A. Conflict resolution in the genome: how transcription and replication make it work. Cell. 2016;167(6):1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang K.S., Hall A.N., Merrikh C.N., Ragheb M., Tabakh H., Pollock A.J., et al. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell. 2017;170(4):787–799.e18. doi: 10.1016/j.cell.2017.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170(4):774-86.e19. [DOI] [PMC free article] [PubMed]
- 12.Sanz L., Hartono S., Lim Y., Steyaert S., Rajpurkar A., Ginno P., et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell. 2016;63(1):167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Hua Y.u., Wang J., Li L., Yuan J., Zhang B.o., et al. RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell. 2021;184(5):1314–1329.e10. doi: 10.1016/j.cell.2021.01.048. [DOI] [PubMed] [Google Scholar]
- 14.Sollier J., Stork C.T., García-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56(6):777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skourti-Stathaki K., Proudfoot N.J., Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42(6):794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mischo H.E., Gómez-González B., Grzechnik P., Rondón A.G., Wei W.u., Steinmetz L., et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41(1):21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairman-Williams M.E., Guenther U.P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sordet O., Redon C.E., Guirouilh‐Barbat J., Smith S., Solier S., Douarre C., et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009;10(8):887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuduri S., Crabbé L., Conti C., Tourrière H., Holtgreve-Grez H., Jauch A., et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11(11):1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Niu T., Manley J.L. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13(12):2108–2115. doi: 10.1261/rna.734407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.-C., Guan A., et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35(2):228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. Febs j. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121(7):1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Nowotny M., Cerritelli S.M., Ghirlando R., Gaidamakov S.A., Crouch R.J., Yang W. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. Embo j. 2008;27(7):1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima WF, Murray HM, Damle SS, Hart CE, Hung G, De Hoyos CL, et al. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res. 2016;44(11):5299-312. [DOI] [PMC free article] [PubMed]
- 26.Castellano-Pozo M., García-Muse T., Aguilera A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep. 2012;13(10):923–929. doi: 10.1038/embor.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becherel OJ, Yeo AJ, Stellati A, Heng EY, Luff J, Suraweera AM, et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013;9(4):e1003435. [DOI] [PMC free article] [PubMed]
- 28.Dia F., Strange T., Liang J., Hamilton J., Berkowitz K.M. Preparation of Meiotic Chromosome Spreads from Mouse Spermatocytes. JoVE. 2017;(129):e55378. doi: 10.3791/55378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28(9):879–896. doi: 10.1038/s41422-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 31.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 2013;29(1):15-21. [DOI] [PMC free article] [PubMed]
- 32.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10(1):1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9) doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Z., Hsu P.J., Xing X., Fang J., Lu Z., Zou Q., et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27(10):1216–1230. doi: 10.1038/cr.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahba L., Costantino L., Tan F.J., Zimmer A., Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30(11):1327–1338. doi: 10.1101/gad.280834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L., Chen J.-Y., Zhang X., Gu Y., Xiao R., Shao C., et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell. 2017;68(4):745–757.e5. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89(1):123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 41.Xu W., Xu H., Li K., Fan Y., Liu Y., Yang X., et al. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat Plants. 2017;3(9):704–714. doi: 10.1038/s41477-017-0004-x. [DOI] [PubMed] [Google Scholar]
- 42.Hartono S.R., Malapert A., Legros P., Bernard P., Chédin F., Vanoosthuyse V. The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J Mol Biol. 2018;430(3):272–284. doi: 10.1016/j.jmb.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan Y.A., Aristizabal M.J., Lu P.Y.T., Luo Z., Hamza A., Kobor M.S., et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10(4):e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin R, Zhong X, Zhou Y, Geng H, Hu Q, Huang Z, et al. R-loopBase: a knowledgebase for genome-wide R-loop formation and regulation. Nucleic Acids Res. 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited in the NCBI Gene Expression Omnibus database under accession number GSE190430 with token elwxoucgzfgjzsr. ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus database under the accession number GSE193401 with token anmfgkoetvmntip. UCSC Browser URL:
https://genome.ucsc.edu/s/FeiHuang/mm9_Spermatogenesis_Rloop_profiling