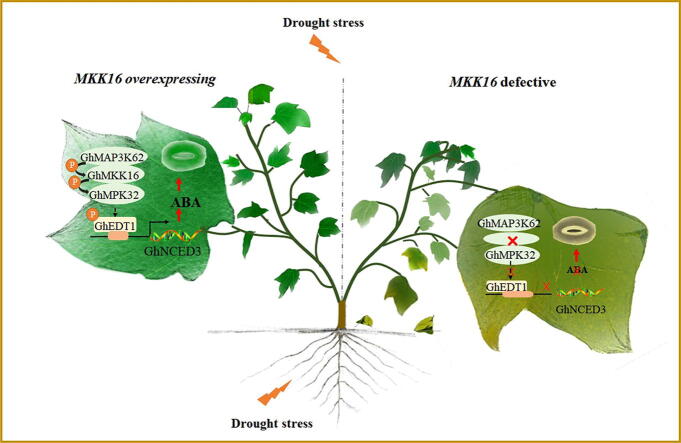
Graphical abstract
Keywords: ABA, Cotton, Drought, GhEDT1, MAPK cascade pathway, Stomatal movement
Highlights
-
•
Cotton GhMKK16 is functionally identified to promotes ABA accumulation, and enhances drought tolerance via regulating stomatal closure under drought stress.
-
•
GhMKK16 mediated stomatal closure and drought tolerance depend on GhMPK32 or GhEDT1.
-
•
GhMKK16 interacts with GhMAP3K62 and GhMPK32 to form a MAPK cascade pathway GhMAP3K62-GhMKK16-GhMPK32.
-
•
GhEDT1 was identified as a downstream transcription factor target of MAPK cascade pathway GhMAP3K62-GhMKK16-GhMPK32.GhEDT1 was found to bind to the promoter of GhNCED3 and to activate its expression, resulting in ABA accumulation.
-
•
It was proposed that the MAPK cascade GhMAP3K62-GhMKK16-GhMPK32 pathway functions in the cotton drought response through ABA-dependent stomatal movement.
Abstract
Introduction
Drought is the principal abiotic stress that severely impacts cotton (Gossypium hirsutum) growth and productivity. Upon sensing drought, plants activate stress-related signal transduction pathways, including ABA signal and mitogen-activated protein kinase (MAPK) cascade. However, as the key components with the fewest members in the MAPK cascade, the function and regulation of GhMKKs need to be elucidated. In addition, the relationship between MAPK module and the ABA core signaling pathway remains incompletely understood.
Objective
Here we aim to elucidate the molecular mechanism of cotton response to drought, with a focus on mitogen-activated protein kinase (MAPK) cascades activating ABA signaling.
Methods
Biochemical, molecular and genetic analysis were used to study the GhMAP3K62-GhMKK16-GhMPK32-GhEDT1 pathway genes.
Results
A nucleus- and membrane-localized MAPK cascade pathway GhMAP3K62-GhMKK16-GhMPK32, which targets and phosphorylates the nuclear-localized transcription factor GhEDT1, to activate downstream GhNCED3 to mediate ABA-induced stomatal closure and drought response was characterized in cotton. Overexpression of GhMKK16 promotes ABA accumulation, and enhances drought tolerance via regulating stomatal closure under drought stress. Conversely, RNAi-mediated knockdown of GhMKK16 expression inhibits ABA accumulation, and reduces drought tolerance. Virus-induced gene silencing (VIGS)-mediated knockdown of either GhMAP3K62, GhMPK32 or GhEDT1 expression represses ABA accumulation and reduces drought tolerance through inhibiting stomatal closure. Expression knockdown of GhMPK32 or GhEDT1 in GhMKK16-overexpressing cotton reinstates ABA content and stomatal opening-dependent drought sensitivity to wild type levels. GhEDT1 could bind to the HD boxes in the promoter of GhNCED3 to activate its expression, resulting in ABA accumulation. We propose that the MAPK cascade GhMAP3K62-GhMKK16-GhMPK32 pathway functions on drought response through ABA-dependent stomatal movement in cotton.
Introduction
Plants are constantly exposed to diverse environmental surroundings during their lifecycles. These environmental factors including biotic and abiotic stresses, such as pest and pathogen infection, drought, salinity, heat and cold, that can adversely affect plant development and productivity [1], [2]. Drought causes a wide range of detrimental effects such as osmotic and oxidative stress, and is a major abiotic stress limiting plant growth and distribution. To survive, plants have evolved multiple mechanisms to cope with drought conditions, such as changes in morphology and physiology [3], [4], [5]. Upon sensing drought, plants activate stress-related signal transduction pathways to effect changes in stress-responsive gene expression patterns that are required for plant adaptation to stress [6], [7]. The ABA and mitogen-activated protein kinase (MAPK) cascade pathways play important roles in stress responses in plants [8].
MAPK cascade pathways are highly conserved in eukaryotes, involved in cell–cell communication and transducing extracellular signals to the nucleus [9]. A typical MAPK cascade module is composed of three kinase components, a MAPK kinase kinase (MAP3K), a MAPK kinase (MKK) and a MAPK (MPK). In general, MAP3Ks are activated by extracellular stimuli through membrane-bound sensors/receptors, then activate downstream MKKs by phosphorylation of their serine/threonine residues, and in turn activate MPKs by MKKs on threonine and tyrosine residues in conserved -TXY- motifs. Finally, the activated MPKs phosphorylate downstream targets such as transcription factors, protein kinases or cytoskeletal proteins [1], [10]. A number of MAPK components and cascades participate in variety of stress response in plants [11]. The first completely described MAPK cascade pathway is the MEKK1-MKK4/5-MPK3/6 cascade in Arabidopsis. This pathway is activated by the flagellin receptor FLS2 and positively regulates innate immunity responses by targeting downstream WRKY22/29 [12]. In contrast the MEKK1-MKK1/2-MPK4/6 negatively regulates innate immunity responses in Arabidopsis [13], and is also involved in responses to oxidative stress [14] and to cold and salt stress [15]. The YDA-MKK4/5-MPK3/6 signal cascade plays a crucial roles in regulating stoma morphology and patterning in Arabidopsis [16], [17]. In rice, OsMAP3K10-OsMKK4-OsMPK6 regulates yield by controlling grain size and weight [18].
MAPK cascades also regulate guard cell signaling, seed germination and plant growth, downstream of ABA signaling. The activities of AtMPK1 and AtMPK2 increase after ABA treatment in Arabidopsis [19], [20], and AtMKK1‐AtMPK6 mediates ROS homeostasis and stress responses via ABA-induced catalase (CAT1) expression. MPK9 and MPK12, which are preferentially expressed in guard cells, mediate ABA‐induced guard cell closure [21]. The MAPK cascade MAP3K17/18-MKK3-MPK1/2/7/14 is activated by the ABA core signaling module to regulate stress responses [22]. ABA-mediated stomatal closure is an active response of plants under drought stress. ABA affects the content of osmotic substances in guard cells, and then regulates stomatal movements by regulating osmotic potential [23]. In addition, changes in plant hormones, CO2 and pH in guard cells, induced by drought stress, can also regulate stomatal opening and closing [24], [25]. A MAPK cascade pathway is involved in regulating stomatal movement and stomatal development, and is an important downstream component of ABA signaling [26], [27]. The role of these kinases, and particularly the relationship between MAPK modules and the ABA core signaling pathway, is of significant interest but remains incompletely understood.
Cotton (Gossypium hirsutum) is a globally important fiber and oil crop. More than half of global cotton is grown in regions with limited irrigation, which significantly limits productivity [28]. Previous studies have shown that MAPK cascade genes are involved in the drought stress response in cotton [29], [30]. Overexpression of the cotton gene GhMPK4 in Arabidopsis decreases tolerance to drought and salt stress [29]. In cotton the GhMAP3K15-GhMKK4-GhMPK6 cascade phosphorylates a WRKY transcription factor, GhWRKY59, which directly regulates GhDREB2 expression, leading to improved drought tolerance in an ABA-independent manner [28]. As the key components with the fewest members in the MAPK cascade, the function and regulation of GhMKKs needs to be elucidated.
In the present study, we show that GhMKK16 regulates stomatal movement through the GhMAP3K62-GhMKK16-GhMPK32 pathway, which in turn targets the transcription factor GhEDT1. GhEDT1 was found to positively regulate drought stress responses and promote ABA accumulation by binding the promoter of, and activating the expression of, GhNCED3, which encode a rate-limiting ABA biosynthesis enzyme. Thus, we identified an ABA-dependent MAPK cascade, GhMAP3K62-GhMKK16-GhMPK32-GhEDT1, regulates drought response by regulating stomatal movement in cotton.
Materials and Methods
Plant materials and drought treatments
Seeds of Upland cotton (Gossypium hirsutum) cultivar YZ1 were used for genetic transformation and VIGS assays. Overexpression lines OE7, OE8 and RNAi lines R8, R25 of GhMKK16 transgenic plants were identified for subsequent assays, and YZ1 was used as control. Cotton plants were grown in controlled environment rooms at 25℃ with 16 h light/8h dark photo-period, and cultured in nutrient soil and Hoagland solution. The trefoil stage plants were used to perform drought treatment with natural drought for 14 days or treated with 15 % PEG 6000 (w/v) in Hoagland solution. The water loss rate was analyzed as described previously [28]. Relative electrical conductivity (REC) assay was carried out as described previously [30]. Three biological replicates were performed in each assay.
Expression profile analysis and Southern blotting
For stress-related expression analysis, cotton plants (YZ1) at the trefoil stage were treated with 15 % PEG 6000, 100 μM ABA, 200 mM NaCl, 8.8 mM H2O2 and 100 μM MeJA for 1 h, 4 h, 8 h, 12 h. Meanwhile, cotton plants (YZ1) at the trefoil stage were treated with drought stress for 2d, 4d and 6d for drought-induced expression pattern analysis. In addition, six Upland cotton accessions (drought-resistant genotypes: ZY61, ZY63 and ZY434; drought-sensitive genotypes: ZY207, ZY321 and ZY440) were used to determine the expression of GhMKK16 during drought treatment [31]. Leaves were collected at different timepoints and frozen in liquid nitrogen and stored for RNA isolation. Total RNA was extracted using the RNAprep Pure Plant Kit (Cat. #DP441, TIANGEN), then 3 μg of total RNA was reverse transcribed to cDNA using the SuperScript III reverse transcriptase (Cat. No. 18080–093, Invitrogen). qRT-PCR analysis was performed to determine gene expression levels, as described previously [32]. Southern blotting was performed as described previously [33]. The primers are listed in Table S1.
Protein sequences and structure analysis
Homologous protein sequences of GhMKK16 in different species (Gossypium hirsutum, Theobroma cacao, Vitis vinifera, Arabidopsis thaliana, Oryza sativa and Zea mays) were downloaded from NCBI by using the BLAST program. Multiple protein sequence alignments were conducted using ClustalX (version 1.81) and DNAMAN (version 6.0.3.99) software. Protein structure analysis was performed using the online program PROSITE (https://prosite.expasy.org/).
Gene cloning, vectors construction and genetic transformation
The coding sequence (CDS) and RNAi fragments of GhMKK16 (Gh_D11G0703) were cloned and inserted into the vector pK2GW7 and pHellsgate4 respectively using Gateway cloning technology, to generate the overexpressing vector GhMKK16-pK2GW7 and RNAi vector GhMKK16-pHellsgate4. The recombinant overexpression and RNAi vectors were introduced into Agrobacterium tumefaciens EHA105 strain and hypocotyls of YZ1 seedings were transformed. The cotton genetic transformation method was described previously [34].
VIGS assays
VIGS assays were performed as reported previously [35]. Gene fragments (300–500 bp) of GhMAP3K62 (Gh_A10G0115), GhMPK32 (Gh_D05G1825) and GhEDT1 (Gh_A12G2462) from CDS regions were constructed to the vector TRV2. The primers are listed in Table S1. The vector constructs were introduced into Agrobacterium tumefaciens strain GV3101. The recombinant vector TRV:GhMAP3K62 was injected into the cotyledons of wildtype cotton plants, YZ1 (WT/ TRV:GhMAP3K62). The other recombinant vectors TRV:GhMPK32 and TRV:GhEDT1 were infiltrated into fully expanded cotyledons of wildtype cotton plants (WT/TRV:GhMPK32, WT/TRV:GhEDT1) and GhMKK16 overexpression plants, OE7 (OE7/TRV:GhMPK32, OE7/TRV:GhEDT1), respectively. Plants were grown in controlled environment rooms at 25℃ with a 16 h light/8h dark photoperiod. VIGS efficiency was determined two weeks after infiltration, the leaves were collected and frozen in liquid nitrogen for target gene expression analysis. The successfully silenced plants were used for the following drought stress treatments.
Measuring the stomatal aperture and ABA content
10-day-drought stress-treated cotton plants were used to measure stomatal aperture. Stomata in the lower epidermis of newly expanded cotton leaves were imaged using light microscopy (Zeiss Axio Scope A1, Oberkochen, Germany) and scanning electron microscopy (JSM-6390/LV SEM, Jeol, Tokyo, Japan). Stomatal aperture was measured using Digimizer (v4.2.60) software, and at least fifty stomata from each of three different leaves with the same leaf position were measured for each line.
ABA measurements were performed as described [36]. 200 mg of leaf samples were dropped into 400 μL of extraction buffer (10 ng/mL 2H6-ABA in 80 % [v/v] methanol) overnight at 4℃, then centrifuged at 12000 rpm at 4℃ for 20 min. Then the liquid supernatant was moved to new tube, and the precipitate was re-extracted with 100 μL above buffer. The two-step supernatants were combined and quantified using HPLC-MS/MS. 2H6-ABA (Olchemim ltd) was used as an internal standard and ABA was used as external standard.
Subcellular localization
Subcellular localization of proteins was analyzed both in tobacco epidermis and cotton protoplasts. The full-length CDSs of GhMKK16, GhMPK32 and GhEDT1 were each constructed into the vector pMDC43. The primers are listed in Table S1. The recombinant vectors pMDC43-GhMKK16, pMDC43-GhMPK32 and pMDC43-GhEDT1 were electroporated into Agrobacterium tumefaciens GV3101. The GFP-fusion proteins were transiently expressed in tobacco epidermis and cotton protoplast as described previously [32]. The localization of the proteins was observed using a Leica TCS SP2 confocal spectral microscope (Leica, Heidelberg, Germany). CBL1:RFP was used as a plasma membrane marker [37], HY5:RFP was used as a nucleus marker [38].
Y2H assays and in vitro pull-down assays
The Matchmaker Gold Yeast Two-Hybrid system (Cat. No. 630489) was used in (Yeast-two-hybrid) Y2H assays. The full-length CDSs of GhMAP3K62 and GhMPK32 were each constructed into yeast vector pGBKT7 (TaKaRa), and transformed into yeast strain Y2H. The full-length CDSs of GhMKK16 and GhEDT1 were cloned into the vector pGADT7, and introduced into yeast strain Y187. Interactions between different proteins were identified as growth on SD medium, SD -Leu-Trp (SD-2) and SD-Leu-Trp-His-Ade (SD-4) (with X-α-Gal), respectively. The primers are listed in Table S1.
For in vitro pull-down assays, the full-length CDS sequences of GhMPK32 and GhEDT1 were cloned into the vectors PET-28-a (Novagen) and pGEX-4 T-1 (Pharmacia), respectively. The constructs His-GhMPK32 and GST-GhEDT1 were transformed into Escherichia coli BL21 (DE3). Empty GST and recombinant GST-GhEDT1 proteins were used to pull-down the His-GhMPK32. The pull-down proteins were purified with MagneGSTTM Protein Purification System (Promega V8603) and MagneHisTM Protein Purification System (Promega V8550). The pull-down assay was performed as described previously [39]. The primers are listed in Table S1.
BiFC, LCI and Dual‑Luciferase reporter assays
For Bimolecular Fluorescence Complementation (BiFC) assays, the CDSs of GhMAP3K62, GhMKK16, GhMPK32 and GhEDT1 were respectively constructed to the vector pDONR221, and pBiFCt-2in1-CC and pBiFCt-2in1-CN vectors were constructed by Gateway cloning [40]. For the Luciferase Complementation Imaging (LCI) assays, the CDSs of GhMAP3K62, GhMPK32, GhMKK16 and GhEDT1 were respectively constructed into the vectors JW771 and JW772 [41]. The recombinant vectors were transformed into Agrobacterium tumefaciens GV3101. YFP fluorescence in BiFC assays was observed using a Leica TCS SP2 confocal spectral Microsystems laser-scanning microscope. LUC luminescence in LCI assays was observed by CCD camera (Lumazome PyLoN 2048B) [42].
Transient dual-luciferase reporter assays were used to demonstrate the transcriptional activation ability of GhNCED3 by GhEDT1 either in tobacco leaf or cotton protoplasts. 1132 bp GhNCED3 (Gh_D13G1614) promoter was cloned into the vector pGreenII 0800-LUC, and the full-length CDS of GhEDT1 was cloned into vector pGreenII 62-SK, with empty vector pGreenII 62-SK used as negative control. The recombinant vectors were transformed into Agrobacterium tumefaciens strain GV3101. For tobacco leaf, the constructs were mixed in equal volumes and injected into N. benthamiana leaf. After, and the LUC luminescence was detected after 60 to 72 h as described above.
For cotton protoplasts, the effector and reporter vectors (6 μg) were co-transformed into protoplasts using polyethylene glycol 4000, then the transfected protoplast cells were cultured in the dark at room temperature for 16 h. Then the activities of luciferase were detected using the Dual-Luciferase Reporter Assay kits (Promega E1910) and recorded using a Multimode Plate Reader (Perkin-Elmer). All primers used are listed in Table S1.
Y1H assay
Yeast-one-hybrid (Y1H) screening was conducted using the Matchmaker Gold One-Hybrid Library Screening System (Clontech, 630499, PT4102). The 1132 bp promoter of GhNCED3 was cloned into the vector pAbAi, to generate the pAbAi- GhNCED3pro, which then was linearized and recombined into the genome of the yeast. Meanwhile, the CDS of GhEDT1 were cloned into the vector pGADT7. The primers listed in Table S1. pGADT7-GhEDT1 and empty pGADT7 were transformed respectively into yeast strain pAbAi-GhNCED3pro. The transformants were screened on SD/−Leu/AbA* medium.
Electrophoretic mobility shift assays (EMSA)
For EMSA, the CDS of GhEDT1 was constructed into the vector pGEX-4 T-1, and the GST-tagged protein was induced and expressed in E. coli, and then affinity purified by chromatographic column. GST protein was used as a negative control. Two probes (P1 and P2, including the HD binding sites) of the GhNCED3 promoter, and two mutant probes (mutation of the HD binding site in P1 and P2) were synthesized, and ampified using biotin-labelled primers (Table S1). Nonlabelled probes were used as competitors. Binding and competition reactions were carried out using the EMSA/Gel-Shift Kit (Beyotime, Shanghai, China). Signals were captured by X-ray film.
Phosphorylation assay of MAPK cascade proteins
For protein kinase assays, the CDS of GhMAP3K62, GhMKK16, GhMPK32 and GhEDT1 were respectively cloned into the vector pET-28-a. The CDS of GhMKK16 was cloned into the vector pGEX-4 T-1. The 6 × His tagged proteins were purified from Escherichia coli BL21 (DE3) using a His purification column (Ni-NTA Agarose, QIAGEN) and GST-tagged proteins purified using a Glutathione S-transferase Column (Pierce Glutathione Agarose, Thermo). For Phos-tag assays, 2 μg of each protein was incubated in a final volume of 30 mL of kinase buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 1 mM ATP), and then incubated in a water bath for 45 min at 30 °C. The phosphoproteins were separated in 12 % (w/v) Mn2+-Phos-tag-SDS-PAGE gels (50 μM Phos-tag-acrylamide, 100 μM MnCl2). Immunoblotting and subsequent analyses were performed according to standard techniques. Different from Phos-tag assays, ATP was replaced by 2 μCi of [γ-32P]ATP [PerkinElmer; 3000 Ci/ mmol 10 mCi/ml] for isotope phosphorylation assays in vitro. The reactions were stopped by adding 5 × SDS loading buffer and heating at 100 °C for 5 min in a metal bath. And the phosphorylation of recombinant proteins was analyzed by autoradiography. Phos-tag SDS-PAGE (Phosbind Acrylamide, APExBIO) was used to detect the MAPK cascade and [γ-32P] ATP was used to detect phosphorylation of GhEDT1 by GhMPK32.
Results
Identification and expression analysis of GhMKK16
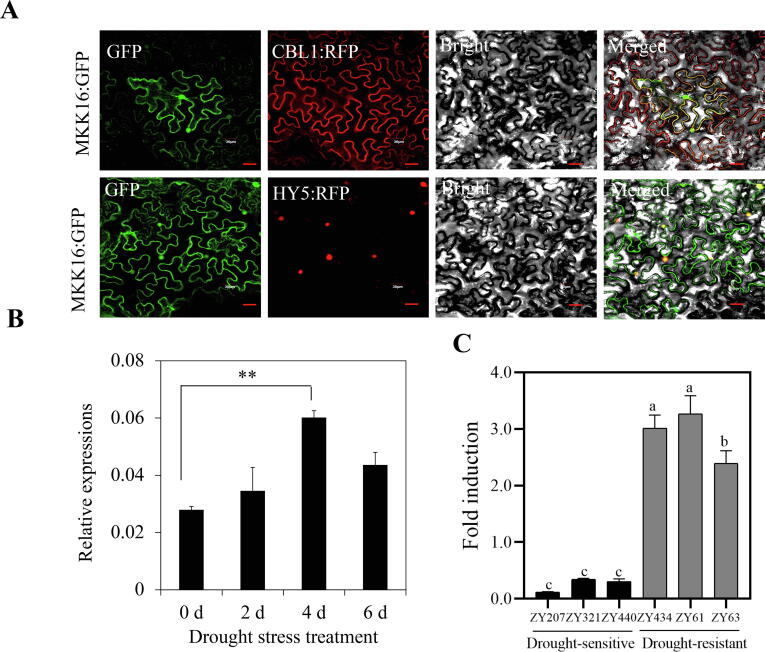
As core components of the MAPK cascade pathway, GhMKK proteins participate in several plant stress responses. In a previous study, twenty GhMKK genes were identified in cotton. The phylogenetic analysis showed that GhMKK16 (Gh_D11G0703) was closely related to Arabidopsis AtMKK3, a group Ⅱ MKK gene [30]. The full-length CDS of GhMKK16 is 1557 nucleotides and encodes a protein of 518 amino acids, containing a protein kinase domain and an NTF2 domain (Fig. S1A). Multiple sequence alignments show that the GhMKK16 protein contains 11 conserved subdomains, four conserved S/T-X3-5-S/T motifs and one active site D(I/L/V)K motif (Fig. S1B). Subcellular localization in N. benthamiana leaf epidermis and cotton protoplasts suggested that GhMKK16 showed a nucleus and plasma membrane localization (Fig. 1A; Fig. S1C).
Fig. 1.
Subcellular localization of GhMKK16 and expression profile of GhMKK16 treated by drought stress. (A) The subcellular localization of GhMKK16:GFP protein. GFP fluorescence was observed after transiently expressing GhMKK16:GFP protein in tobacco epidermal cells. CBL1 and HY5 were used as plasma membrane and nucleus markers respectively. Scale bar = 30 μm. (B) Expression of GhMKK16 in drought-treated cotton. (C) Expression of GhMKK16 in different cotton accessions after drought stress treatment. Expression of GhMKK16 in (B) and (C) were determined by qRT-PCR. GhUB7 was used as the internal reference. Values in (B, C) were means ± SE (n = 3), significant differences analysis were using Student’ s t-test, **P-value < 0.01. And different lower-case letters a–c above columns represent significant differences among columns (LSD multiple comparisons, P-value < 0.05).
GhMKK16 was significantly induced by drought stress in Upland cotton (YZ1) (Fig. 1B). We analysed the expression of GhMKK16 in six accessions (ZY207, ZY321 and ZY440; ZY434, ZY61 and ZY63) with different drought tolerances after drought stress treatment. The results show that the fold change in GhMKK16 expression was higher in drought-resistant genotypes than in drought-sensitive genotypes after drought stress treatment (Fig. 1C). qRT-PCR analysis confirmed that GhMKK16 was induced by treatment with 15 % PEG 6000, 100 mM H2O2 and phytohormones (ABA and MeJA) (Fig. S2). These findings indicate that GhMKK16 might be involved in the cotton response to drought stress and might participate in phytohormone-mediated signaling.
GhMKK16 positively regulates drought tolerance in cotton at seedling stage
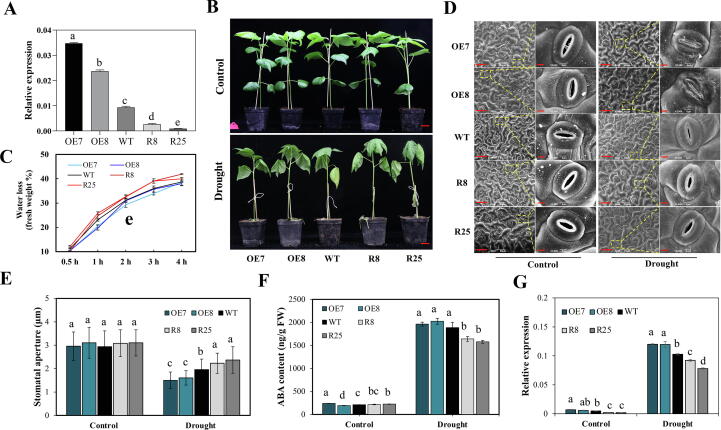
To elucidate the function of GhMKK16 in cotton under drought conditions, we generated a series of transgenic lines, i.e. two overexpression lines (OE7 and OE8) and two RNAi lines (R8 and R25). Each was demonstrated to have a single transgene insertion (OE8 was a single copy line isolated from T1 generation with multiple copies inserted) by Southern blotting (Fig. S3A), and the expression levels of GhMKK16 increased to 3.7- fold and 2.5-fold in OE7 and OE8 compared to WT plants, while decreased to 28.3 % (R8) and 9.7 % (R25) of the level in WT plants (Fig. 2A). Then these lines were used for subsequent assays.
Fig. 3.
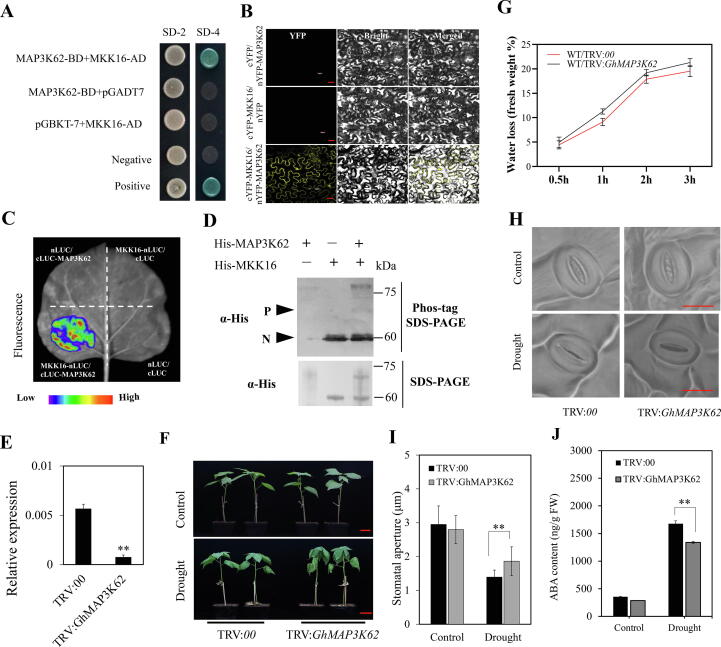
GhMAP3K62 interacts with GhMKK16 and involved in improving drought tolerance in cotton. (A) Y2H assay for GhMAP3K62-GhMKK16 interaction. SD-2 (-Trp/-Leu), SD-4 (-Trp/-Leu/-His/-Ade). (B) BiFC assay between GhMAP3K62-nYFP and GhMKK16-cYFP in tobacco epidermal cells. Bars = 30 μm. (C) LCI analysis of GhMKK16-nLUC and cLUC-GhMAP3K62 in tobacco leaves. (D) The phosphorylation of His-GhMKK16 by His-GhMAP3K62, monitored by western blot on Phos-tag gel. α-His: anti-His antibody. The phosphorylated form (P) and nonphosphorylated form (N) are shown. SDS-PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis. (E) Relative expression levels of GhMAP3K62 in silenced or control plants. (F) Phenotypes of GhMAP3K62-silenced plants under drought stress treatments. Plants at trefoil stage in soil were exposed to drought stress for two weeks. Bars = 3 cm. (G) Water loss in TRV:GhMAP3K62 and TRV:00 plants at room temperature. (H) Light microscopy images of stomata from the abaxial epidermis of cotton leaves in TRV:GhMAP3K62 and TRV:00 plants under drought conditions. Bars = 20 μm. (I) Stomatal aperture in TRV: GhMAP3K62 and TRV:00 plants grown under normal watering or drought stress. (J) ABA content in TRV: GhMAP3K62 and TRV:00 plants grown under normal watering or drought stress. Values in figures (E, G, I) represent means ± SE (n = 3). Significant differences analysis was done by Student’s t-test, ** P-value < 0.01.
Fig. 2.
GhMKK16 positively regulates drought tolerance in cotton. (A) Relative expression levels of GhMKK16 in GhMKK16 transgenic plants. OE7 and OE8 are overexpression lines, R8 and R25 are RNAi lines. (B) Phenotypes of transgenic and WT plants under drought stress treatments. Plants at trefoil stage in soil were exposed to drought stress for two weeks. Bars = 3 cm. (C) Water loss of transgenic and WT plants at room temperature. (D) Scanning electron microscopy images of stomata from the abaxial epidermis of cotton leaves in transgenic and WT plants under normal or drought conditions. Bars in 450X was 50 μm, Bars in 3000X was 5 μm. (E) Relative expression levels of GhABF1 in GhMKK16 transgenic and WT plants under control and drought stress conditions. (F) Relative expression levels of GhABF2 in GhMKK16 transgenic and WT plants under control and drought stress conditions. (G) Relative expression levels of GhNCED3 in GhMKK16 transgenic and WT plants under control and drought stress conditions. Plants in (E, F and G) were incubated with normal or drought stress condition, respectively. Values in (A, C, D, E, F and G) represent means ± SE (n = 3). Different lower-case letters a–e above columns represent significant differences among columns (LSD multiple comparisons, P-value < 0.05).
To investigate drought stress tolerance of these transgenic cotton lines, trefoil stage seedings were exposed to drought stress by withholding water. The results showed that OE7 and OE8 plants exhibit enhanced drought tolerance compared to WT, while R8 and R25 wilt prematurely, after 14 days withdrawal of water (Fig. 2B). We further analyzed the water loss in WT and transgenic plants during dehydration by comparing the water content of detached leaves. More water loss was evident in R8 and R25 than in WT plants, and plants overexpressing GhMKK16 showed decreased water loss in leaves (Fig. 2C). Meanwhile, hydroponic culture with 15 % PEG 6000 was used to simulate drought stress. Results show that R8 and R25 plants were more severely affected, while OE7 and OE8 plants grew better than WT plants in PEG solution (Fig. S3B). The relative electrical conductivity in cotton leaves coincided with the phenotypes of transgenic and WT plants treated by drought stress and PEG simulated drought stress (Fig. S3C). These results suggest that GhMKK16 positively regulates drought stress tolerance in cotton.
GhMKK16 regulates stomatal movement and ABA homoeostasis in cotton
To explore the cellular basis for the accelerated water loss in GhMKK16 RNAi lines and decreased water loss in overexpression lines, stomatal aperture on the abaxial epidermis of cotton leaves was measured by light and scanning electron microscopy (Fig. 2D; Fig. S4A). No significant difference in stomatal aperture was observed between transgenic and WT plants under normal water condition. While, a reduced stomatal aperture was observed in OE7 and OE8 lines and an increased stomatal aperture were observed in R8 and R25 lines compared to WT plants after drought stress treatment (Fig. 2E). These data suggest that the expression level of GhMKK16 determines stomatal aperture in cotton, which results in the observed differences in plant drought tolerance.
Fig. 4.
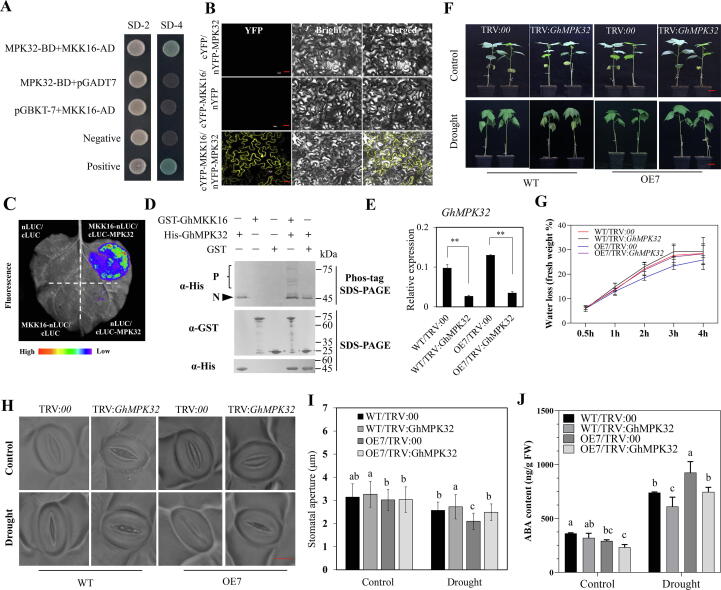
GhMKK16 role in drought response depends on GhMPK32. (A) Y2H assays showing the interaction relationships between GhMKK16 and GhMPK32. SD-2 (-Trp/-Leu), SD-4 (-Trp/-Leu/-His/-Ade). (B) BiFC assay between GhMKK16-cYFP and GhMPK32-nYFP in tobacco epidermal cells. Bars = 30 μm. (C) LCI analysis of GhMKK16-nLUC and cLUC-GhMPK32 in tobacco leaves. (D) The phosphorylation of His-GhMPK32 by GST-GhMKK16, monitored by western blot on Phos-tag gel. α-His: anti-His antibody; α-GST: anti-GST antibody. The phosphorylated form (P) and non-phosphorylated form (N) are shown. SDS-PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis. (E) Relative expression of GhMPK32 in GhMPK32-silenced plants in WT and OE7 backgrounds. (F) Phenotypes of GhMPK32-silenced plants in WT and OE7 backgrounds under drought stress. Plants at trefoil stage in soil were exposed to drought stress for two weeks. Bars = 3 cm. (G) Water loss in TRV:GhMPK32 and TRV:00 plants at room temperature. (H) Light microscopy images of stomata from the abaxial epidermis of cotton leaves in GhMPK32-silenced plants in WT and OE7 backgrounds plants under drought and control conditions. Bars = 10 μm. (I) Stomatal aperture in TRV: GhMPK32 and TRV:00 plants grown under normal watering or drought stress. (J) ABA content in TRV: GhMPK32 and TRV:00 plants under WT and OE7 backgrounds, respectively. Plants were grown under standard watering drought stress. Values in figures (E, G, I, J) represent means ± SE (n = 3). Significant difference analysis was done by Student’s t-tests (**P-value < 0.01) in E. And different lower-case letters a–c above columns represent significant differences among columns (LSD multiple comparisons, P-value < 0.05) in I, J.
The phytohormone ABA plays crucial role in the regulation of stomatal movement. Therefore, we determined the ABA concentrations in cotton leaves from GhMKK16 transgenic lines and WT plants. The results showed that all transgenic and WT plants maintained relatively low levels of ABA under standard watering conditions, but following drought stress for 10 days, increased ABA concentrations were detected in all plants. Significantly, higher ABA concentrations were detected in OE7 and OE8 plants, and lower ABA levels in R8 and R25 plants, compared with WT plants (Fig. 2F). Additionally, qRT-PCR results showed that the expression of ABA biosynthesis and signaling pathway genes GhNCED3, GhABF1 and GhABF2 was highly induced in OE7 and OE8 plants compared with WT plants, whereas the expression of these genes was at lower levels in R8 and R25 lines compared with WT (Fig. 2G, Fig. S4B). These results suggest that GhMKK16 regulates stomatal aperture in response to drought stress through effects on ABA biosynthesis.
GhMAP3K62 regulates drought tolerance by phosphorylating GhMKK16 in cotton
To elucidate the regulatory mechanism of GhMKK16 in the drought stress response in cotton, yeast two-hybrid (Y2H) assays were performed to screen for potential interacting proteins. A GhMAP3K protein GhMAPK3K62 was identified as interacting with GhMKK16 (Fig. 3A). To verify the interaction between GhMAP3K62 and GhMKK16 protein in vivo, BiFC and LCI assays were performed by transient co-expressing both proteins in tobacco leaf epidermal cells. Strong YFP fluorescence signals were observed in the nucleus and plasma membrane (Fig. 3B), and the strong interaction signals were similarly detected between GhMKK16 and GhMAP3K62 proteins in LCI assays (Fig. 3C). To verify the relationships between GhMAP3K62 and GhMKK16 proteins, we expressed and purified them in prokaryotic cells and then carried out Phos-tag phosphorylation assays in vitro. The results showed that GhMKK16 could be phosphorylated by GhMAP3K62 (Fig. 3D).
To find out the possible role of GhMAP3K62 in cotton response to drought stress, we used VIGS to knock down the expression of GhMAP3K62 and carried out drought tolerance assays. We constructed a recombinant pTRV2 vector, TRV:GhMAP3K62, and the empty recombinant pTRV2 vector (TRV:00) was used as a control. The expression of GhMAP3K62 was detected by qRT-PCR (Fig. 3E). Plants at the trefoil stage were exposed to drought stress for two weeks. The results showed that TRV:GhMAP3K62 plants showed more severe wilting than control plants TRV:00 (Fig. 3F). The relative electrical conductivity of TRV:GhMAP3K62 plants was significantly higher than for TRV:00 plants (Fig. S5A), consistent with the observed phenotype of TRV:GhMAP3K62 plants after drought stress. These results indicate that GhMAP3K62 act as a positive regulator in cotton tolerance to drought.
Fig. 5.
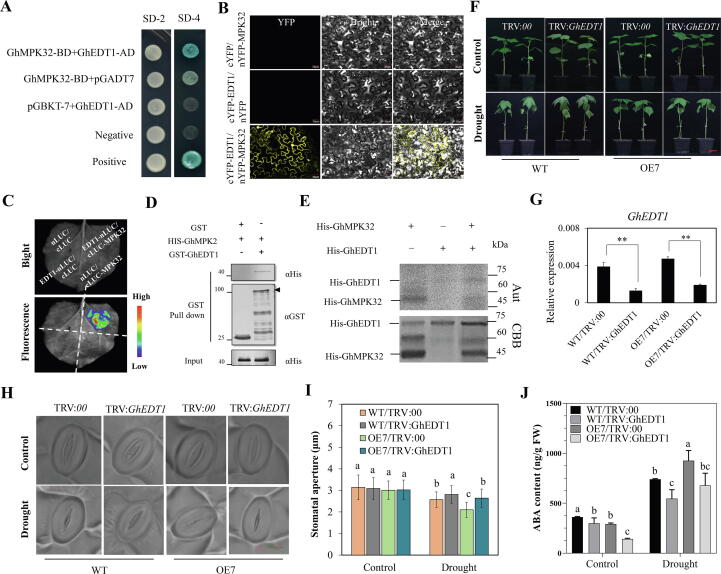
GhEDTI physically interacts with GhMPK32 and positively regulates drought response in cotton. (A) Y2H assays showing the interaction relationships between GhEDT1 and GhMPK32. SD-2 (-Trp/-Leu), SD-4 (-Trp/-Leu/-His/-Ade). (B) BiFC assay between GhEDT1-cYFP and GhMPK32-nYFP in tobacco epidermal cells. Bars = 30 μm. (C) LCI assays of GhEDT1-nLUC with cLUC-GhMPK32 in tobacco leaves. (D) Pull-down assays for GhEDT1 and GhMPK32 proteins. His-GhMPK32 protein was pulled down by GST-GhEDT1 protein, GST was used as a negative control. (E) In vitro autophosphorylation assays to determine the phosphorylation of His-GhEDT1 by His-GhMPK32. PGhEDT1: Phosphorylated protein of GhEDT1; PGhMPK32: Phosphorylated protein of GhMPK32. Aut: Autoradiograph. CBB: Coomassie brilliant blue. (F) Phenotypes of GhEDT1-silenced plants in WT and OE7 backgrounds under drought conditions. Plants at the trefoil stage in soil were exposed to drought stress for two weeks. Bar s = 3 cm. (G) Relative expression of GhEDT1 in GhEDT1-silenced plants in WT and OE7 backgrounds. (H) Light microscopy images of stomata from the abaxial epidermis of cotton leaves in GhEDT1-silenced plants in WT and OE7 backgrounds plants under control and drought stress conditions. Bars = 10 μm. (I) Stomatal aperture in TRV: GhEDT1 and TRV:00 plants grown under normal watering or drought stress. (J) ABA content in TRV: GhMPKEDT1 and TRV:00 plants in WT and OE7 backgrounds, respectively. Plants were grown under standard watering or drought stress. The values in figures (G, I, J) represent means ± SE (n = 3). Significant difference analysis was done by Student’s t-tests (**P-value < 0.01) in G. And different lower-case letters a–c above columns represent significant differences among columns (LSD multiple comparisons, P-value < 0.05) in I, J.
Moreover, silencing of GhMAP3K62 caused increased water loss from droughted cotton leaves (Fig. 3G). We observed the phenotypes of stomata on the abaxial epidermis of leaves of TRV:GhMAP3K62 and TRV:00 plants (Fig. 3H; Fig. S5B). Stomatal aperture in TRV:GhMAP3K62 plants was larger than in TRV:00 plants after drought stress treatment (Fig. 3I), indicating that GhMAP3K62 promotes stomatal closure during drought.
We measured ABA concentrations in the leaves of TRV:GhMAP3K62 and TRV:00 plants. The results showed that silencing GhMAP3K62 reduced the accumulation of ABA under drought stress (Fig. 3J). In according with that, the expression of GhNCED3, GhABF1 and GhABF2 were less induced in TRV:GhMPK32 plants than in controls under drought stress (Fig. S5C), which suggest that GhMAP3K62 positively regulates the expression of ABA related genes under drought stress.
GhMPK32 acts downstream of GhMKK16, and regulates drought tolerance in cotton
From the result of Y2H assays, we found GhMPK32 might be one of the potential downstream targets of GhMKK16 (Fig. 4A). GhMPK32 is a group III member of the GhMPK family, and subcellular localization analysis showed that GhMPK32 localizes to the plasma membrane and nucleus (Fig. S6A-B). Both BiFC assays (Fig. 4B) and LCI assays (Fig. 4C) also indicate that GhMPK32 directly interacts with GhMKK16. GST-GhMKK16 and His-GhMPK32 were used to carry out Phos-tag phosphorylation assays, and the results demonstrated that GhMKK16 specifically phosphorylates GhMPK32 in vitro (Fig. 4D).
Fig. 6.
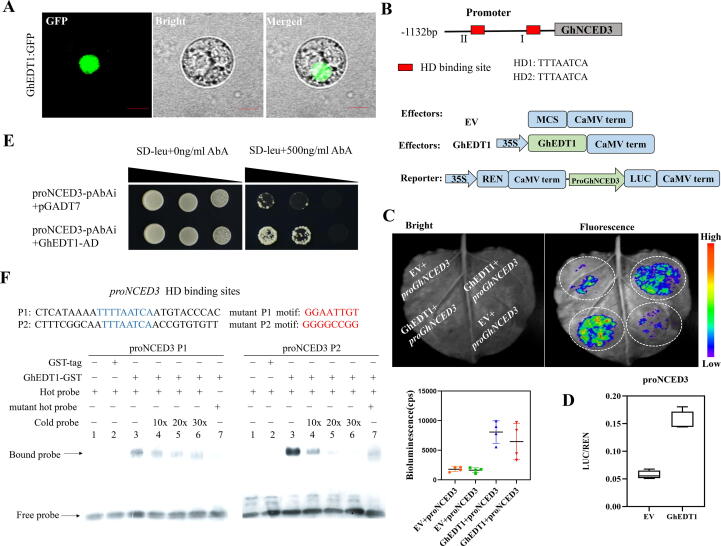
GhEDT1 binds to the promoter of GhNCED3 to activate its expression and promote ABA synthesis. (A) Subcellular localization of GhEDT1 in cotton protoplasts. Bars = 10 μm. (B) Distribution diagram of HD binding sites contained in GhNCED3 promoter region. Schematic diagram of effector and reporter constructs. The empty vector (EV) was used as a control. LUC signals are shown in the image. (C) The intensity of fluorescence in dual-luciferase reporter assays. The bioluminescence reflects the trans-activation ability of GhEDT1. (D) The transient dual-luciferase reporter assays in cotton (YZ1) protoplasts. The ratio of firefly luciferase (LUC) to renilla luciferase (REN) reflects the trans-activation ability of GhEDT1. (E) Yeast-one-hybrid assays revealed that GhEDT1 could bind the GhNCED3 promoter region. The target fragment containing two HD binding sites was cloned into the pAbAi vector, and the full-length coding sequence (CDS) of GhEDT1 was fused to the pGADT7 vector. Interaction was determined on selective medium lacking Leu in the presence of 500 ng ml−1 aureobasidin A (SD/−Leu + AbA500). Different colony dots of the yeast represent different dilution times. (F) EMSA assay of the DNA binding activity of GhEDT1 for the GhNCED3 promoter. P1 and P2 probes were labelled with biotin as hot probes and incubated with recombinant GhEDT1-GST protein. Unlabeled probes as cold probes were added to compete with biotin-labelled probe. Mutation probes as control. GST protein was used as negative control. Values represent means ± SE (n = 3).
VIGS technology was employed to study the function of GhMPK32 by suppressing its expression in both WT and OE7 backgrounds (Fig. 4E). Plants at the trefoil stage were exposed to drought stress for two weeks. WT/TRV:GhMPK32 plants exhibited decreased drought tolerance compared to WT/TRV:00 plants (Fig. 4F). Correspondingly, the relative electrical conductivity of TRV:GhMPK32 plants was significantly higher than in TRV:00 plants (Fig. S7A). Furthermore, knockdown of GhMPK32 expression in OE7 plants led to increased sensitivity to drought stress compared to OE7/TRV:00 plants, which had similar drought-resistance phenotypes to WT plants (Fig. 4F).
Fig. 7.
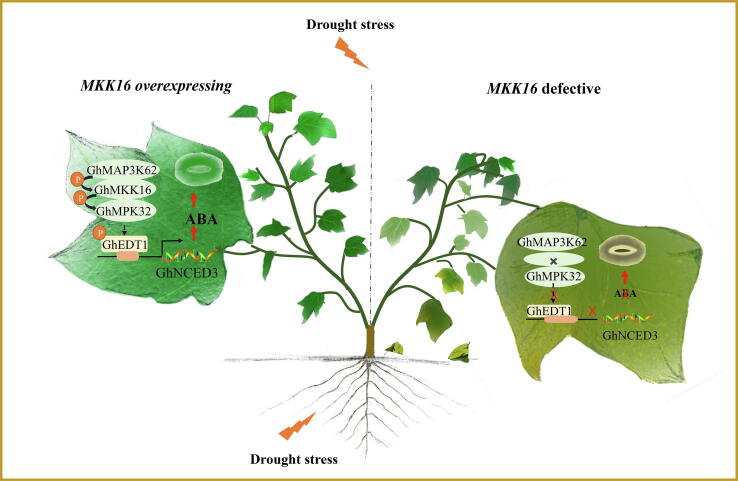
A schematic model of the regulatory network of GhMKK16 response to drought stress in cotton. GhMKK16 positively regulates cotton drought response as a core module in GhMAP3K62-GhMKK16-GhMPK32 MAPK cascade. This MAPK cascade targets and phosphorylates the nuclear-localized transcription factor GhEDT1, to activate downstream GhNCED3 to mediate ABA-induced stomatal closure and drought response in cotton.
It was also found that silencing of GhMPK32 caused an increased leaf water loss compared to wild type (Fig. 4G), which was associated with increased stomatal aperture after drought stress, with similar levels to WT/TRV:00 plants (Fig. 4I). By silencing GhMPK32 in GhMKK16-overexpression line OE7, the water loss rate of isolated leaves was decreased compared with OE7 line (Fig. 4G). Consistent with this, stomatal aperture increased following silencing GhMPK32 compared with the OE7/TRV:00 (Fig. 4I). When the expression of GhMPK32 was inhibited, the overexpression line had a similar drought-resistance level to WT plants in terms of water loss rate and stomatal aperture in isolated leaves (Fig. 4G-4H). These results suggest that GhMPK32 acts downstream of GhMKK16 to positively regulate drought stress tolerance in cotton through regulated stomatal closure.
We next determined ABA concentrations in the leaves of TRV:GhMPK32 and TRV:00 plants. Suppression of GhMPK32 expression in WT and OE7 significantly decreased ABA accumulation in leaves after drought stress, and WT/TRV:00 plants and OE7/TRV:GhMPK32 plants have almost the same ABA concentration (Fig. 4J). qRT-PCR analysis showed that the expression of GhNCED3, GhABF1 and GhABF2 were induced to lower levels in TRV:GhMPK32 plants than in WT under drought stress condition (Fig. S7C). This suggest that GhMPK32 positively regulates the expression of ABA-related genes, such as ABA biosynthesis gene, therefore regulates stomatal closure.
GhEDT1 was phosphorylated by GhMPK32, and positively regulates drought tolerance in cotton
To further explore the mechanism by which GhMPK32 regulates the drought stress response, we performed Y2H assays to identify potential interacting proteins. It was found that GhMPK32 directly interacts with GhEDT1, a transcription factor (Fig. 5A). BiFC results also confirmed that GhEDT1 and GhMPK32 can interact in vivo (Fig. 5B). In support of this, interaction signals were also detected between GhEDT1 and GhMPK32 in LCI assays (Fig. 5C). In in vitro GST pull-down assays, His-GhMPK32 protein was incubated with GST-GhEDT1 and GST, and it was found that GST-GhEDT1 pulls down His-GhPK32, while GST alone did not (Fig. 5D). These results indicate that GhEDT1 interacts with GhMPK32 both in vitro and in vivo.
GhMPK32 has a kinase domain that is important for the interaction betweenGhEDT1 and GhMPK32. Therefore, it is necessary to determine whether GhEDT1 is the phosphorylation target of GhMPK32. To achieve this, we established a protein phosphorylation system in vitro using the [γ-32P]ATP, using purified GhEDT1 (fusion His tag at the N-terminal) as the substrate, and purified GhMPK32 (fusion His tag at the N-terminal) as the kinase. The result showed that GhMPK32 has kinase activity and GhEDT1 has no phosphorylation activity, but GhEDT1 was phosphorylated by GhMPK32 in vitro (Fig. 5E).
To verify the function of GhEDT1 in plants suffering from drought stress, VIGS suppression (verified by qRT-PCR) was performed in WT and OE7 backgrounds (Fig. 5F). Seedlings at the trefoil stage were exposed to drought stress. Results show that silencing GhEDT1 expression led to increased water loss, reducing resistance to drought stress. Overexpression lines had drought-resistant levels of WT plants when the expression of GhEDT1 was suppressed (Fig. 5G). Accordingly, the relative electrical conductivity was determined to assess the degree of leaf damage, and more severe damage occurred in TRV:GhEDT1 plants than in TRV:00 plants (Fig. S8A). Significant differences were also found in stomatal aperture under drought stress between GhEDT1-silenced plants and control plants (Fig. 5I; Fig. S8C). Larger stomatal apertures were observed on GhEDT1-silenced plant leaves compared to control plants, while no significant differences in stomatal aperture were displayed between WT/TRV:00 and OE7/TRV:GhEDT1 plants (Fig. 5H-I). These results suggest that GhEDT1 acts downstream of GhMKK16 forming a MAPK cascade GhMAP3K62-GhMKK16-GhMPK32, which modulates drought tolerance through regulated stomatal movement.
GhEDT1 transactivates GhNCED3 to mediate ABA accumulation in cotton
We measured ABA concentrations in leaves of TRV:GhEDT1 and TRV:00 plants in WT and OE7 backgrounds, respectively. Under standard watering, the accumulated ABA levels in TRV:GhEDT1 plants were lower than that in TRV:00 plants in the backgrounds of both WT and OE7. After 10 days of drought treatment, ABA concentrations increased both in TRV:GhEDT1 and TRV:00 plants, and 36 % higher levels were detected in WT/TRV:00 plants compared to WT/TRV:GhEDT1 plant. Similar results were seen for OE7/TRV:00 and OE7/TRV:GhEDT1 plants. Moreover, under drought stress, the ABA content in OE7/TRV:00 plants was significantly higher than thatin WT/TRV:00 plants (Fig. 5J). qRT-PCR results showed that the expression of GhNCED3, GhABF1 and GhABF2 was induced less in TRV:GhEDT1 compared to TRV:00 plants during drought stress (Fig. S8D).
GhEDT1 encodes a homeodomain leucine zipper (HD-Zip) protein belonging to the homeodomain (HD)-START transcription factor family. The subcellular localization of GhEDT1-GFP in cotton protoplasts and N. benthamiana epidermis showed that it was located in the nucleus (Fig. 6A, Fig. S8E). GhEDT1 regulates gene expression by binding directly to the promoters of genes with HD binding sites. Here, we found two HD binding sites in the promoter of GhNCED3 (Fig. 6B). GhNCED3 is involved in ABA biosynthesis, and to determine whether GhEDT1 could regulate the expression of GhNCED3, the transient dual-luciferase reporter assays, using 1132 bp promoter sequence of GhNCED3, were performed in tobacco leaf epidermis and cotton protoplasts (Fig. 6B). pGreenII 0800-LUC containing the GhNCED3 promoter and pGreenII 62-SK with or without the GhEDT1 coding region were transiently expressed in tobacco epidermal cells, and results showed significantly higher bioluminescence intensity than that the control (Fig. 6C). Transient dual-luciferase reporter assays in cotton plants (YZ1) protoplasts, using firefly luciferase (LUC) and renilla luciferase (REN), showed that the Luc / Ren ratio was three times that of the control group (Fig. 6D). These results show that the expression of GhNCED3 was activated by GhEDT1.
Moreover, Y1H experiments were conducted to determine whether GhEDT1 could bind to the promoter of GhNCED3 and regulate its expression. The promoter of GhNCED3 contains two possible HD binding sites, which are located in the promoter region at − 1009-bp to − 1001-bp and − 678-bp to − 670-bp (Fig. 6B). After ruling out its self-activation, partial promoter regions of GhNCED3 containing two HD binding sites were selected as bait and introduced into the pAbAi linearization vector. The full-length CDS of GhEDT1 was fused with the vector pGADT7 to activate the expression of AD (protein with activation domain) reporter gene, and pGADT7 empty vector was used as negative control. The results showed that GhEDT1 could bind to HD binding sites in the GhNCED3 promoter (Fig. 6E).
Furthermore, EMSA technology was applied to validate the DNA binding capacity of GhNCED3 promoter by GhEDT1. The recombinant GhEDT1-GST proteins were affinity purified on a Glutathione S-transferase Column. As a result, GST alone could not bind to the GhNCED3 promoter, while GhEDT1-GST was found to directly bind to the biotin-labelled P1 and P2 probes, which was gradually reduced upon the addition of unlabeled competitive probes (Fig. 6F). Moreover, very weak signals were found when biotin-labelled mutant P1 and P2 probes substituting the P1 and P2 probes. These results suggest that GhEDT1 can enhance the transcriptional activity of GhNCED3 in vivo.
Discussion
The MAPK cascade pathway has been regarded as a conserved model in stress alleviated signal transduction pathway [11]. In the current study, GhMKK16, one of the core components in this pathway, is activated rapidly under drought stress, and performs differently in Upland Cotton accessions with different drought stress sensitivities (Fig. 1). We further delineated a three-tiered core signaling module comprised of GhMAP3K62, GhMKK16, and GhMPK32 in cotton, that is involved in the drought stress response through the regulation of stomatal movements. GhMKK16 is a homolog of AtMKK3 in Arabidopsis. Several studies have shown that AtMKK3 regulates drought tolerance through different MAPK cascades. The complete pathway AtMAP3K17/18-AtMKK3-AtMPK1/2/7/14 is involved in timing of senescence, stomatal signaling and drought resistance in Arabidopsis [22], [43], [44]. GhMAP3K62 is the homologue of AtMAP3K18 in Arabidopsis. Similar to our results, knockdown of the expression of AtMAP3K18 significantly decreased drought tolerance in Arabidopsis [43]. GhMPK32 is a homologue of AtMPK2 in Arabidopsis, which is activated by ABA in an AtMKK3-dependent manner [22], [45]. Here, we found that GhMPK32 protein interacts with GhMKK16 protein in cotton to regulate stomatal aperture and water loss (Fig. 4). Our results suggest that the GhMAP3K62-GhMKK16-GhMPK32 cascade pathway is conserved in cotton and Arabidopsis, to regulate drought tolerance in plants.
As a multi-components functional module, MAPK cascade signaling involves in different biological processes via the phosphorylation of different downstream targets, including but not limited to membrane-localized transporters, cytosolic enzymes, and nuclear-localized transcription factors[9], [12], [26], [44], [46]. Although plenty of targets have been explored until now, the relationship between MAPK cascade pathway and ABA homeostasis has been little illustrated. In the current study, RNAi- or VIGS-mediated reduced expression of GhMKK16 or GhMPK32 led to significantly decreased ABA content and GhNCED3, GhABF1 and GhABF2 expression in cotton (Fig. 2, Fig. 3, Fig. 4), indicative of the relationship between MAPK cascade pathway and ABA homeostasis.
As a key plant stress-signaling phytohormone, ABA accumulates under drought stress. Drought stress-responsive genes are regulated by ABA-dependent and ABA-independent pathways [47]. In the early process before endogenous ABA accumulation, the ABA-independent pathway plays a role in perception and transduction of the signal to activate the regulatory system in response to drought stress [48]. DRE transcription factors, such as DREB2, play key roles in ABA-independent gene expression in response to drought stress [48]. In the ABA-dependent pathway, ABA is perceived by the ABA receptor (PYR/PYL/RCAR), and then the ABA–receptor complex binds to and deactivates PP2Cs, thereby releasing self-active SnRK2s, which phosphorylate multiple downstream targets such as AREB/ABFs transcription factors to finally trigger ABA-induced physiological responses[49], [50]. Most studies have shown that MAPK cascade signaling is induced by ABA [22], [43], [44], [45]. Recently, a MAPK-like (MPKL) protein was identified that regulates drought response by suppressing ABA biosynthesis [8]. However, the role of these kinases, and particularly the relationship between MAPK modules and the ABA core signaling pathway, is of significant interest but has remained largely unresolved.
In the current study, the expression of three genes in the GhMAP3K62-GhMKK16-GhMPK32 pathway was induced by ABA. Therefore, we hypothesized that the GhMAP3K62-GhMKK16-GhMPK32 pathway might act downstream of ABA signaling, to participate in the drought stress response of cotton. We showed that ABA homeostasis and related gene expression in cotton leaves could be regulated by affecting the expression of genes in this pathway. Under drought stress, the GhMAP3K62-GhMKK16-GhMPK32 pathway, following activation by the ABA signal, could also activate the expression of GhNCED3 through the downstream target GhEDT1 to promote ABA synthesis, mediating feedback regulation of the ABA signal. Therefore, we propose that the GhMAP3K62-GhMKK16-GhMPK32 pathway is not only regulated by ABA, but also affects ABA synthesis in a feedback way to regulate the response of cotton to drought stress (Fig. 7).
Previous studies have shown that MAPK cascades regulate stomatal closure and density [44], [51], [52]. Stomata are essential for gas and water exchange on plant leaf [53], [54]. Here, we show that GhEDT1 is identified as a downstream transcription factor target of GhMPK32 (Fig. 5). Previous studies have shown that AtEDT1 is a transcriptional activator of CIPK3, NCED3 and ERECTA [55], and enhances drought tolerance in Arabidopsis and other species through improved root architecture and water use efficiency, leading to increased crop yield [56], [57], [58], [59]. In this study, overexpression and defective of GhMKK16 led to extremes of stomatal behavior under drought stress, with wild type as the intermediate type. Silencing of GhMAP3K62, GhMPK32 and GhEDT1, respectively, led to a more open stomatal type compared with the wild type, and decreased ABA content in cotton leaves, which was rescued to the wild type level in overexpression lines. Our data show that the signal cascade is composed of the traditional three-tiered MAPK cascade leading to phosphorylation of the GhEDT1 transcription factor, which directly regulates the expression of GhNCED3 in response to drought stress. This information provides a new understanding of the interaction between the MAPK cascade and ABA signaling in response to drought stress.
Compliance with ethics requirements
This research work does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by funding from the National Key Project of Research and the Development Plan of China (2018YFD1000907) and Fundamental Research Funds for the Central Universities (2662020ZKPY011).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.11.002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Ning J., Li X.H., Hicks L.M., Xiong L.Z. A Raf-Like MAPKKK Gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010;152(2):876–890. doi: 10.1104/pp.109.149856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J.K. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103(35):12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W.-X., Oono Y., Zhu J., He X.-J., Wu J.-M., Iida K., et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20(8):2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., Hara N., et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genet. 2013;45(9):1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- 6.Mehlmer N., Wurzinger B., Stael S., Hofmann-Rodrigues D., Csaszar E., Pfister B., et al. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010;63(3):484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong L.M., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu D., Chang Y., Pei T., Zhang X., Liu L., Li Y., et al. MAPK-like protein 1 positively regulates maize seedling drought sensitivity by suppressing ABA biosynthesis. Plant J. 2020;102(4):747–760. doi: 10.1111/tpj.14660. [DOI] [PubMed] [Google Scholar]
- 9.Li K., Yang F., Zhang G., Song S., Li Y., Ren D., et al. AIK1, A Mitogen-Activated Protein Kinase, Modulates Abscisic Acid Responses through the MKK5-MPK6 Kinase Cascade. Plant Physiol. 2017;173(2):1391–1408. doi: 10.1104/pp.16.01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez M.C., Petersen M., Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 11.Xu J., Zhang S.Q. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015;20(1):56–64. doi: 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415(6875):977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 13.Kong Q., Qu N.a., Gao M., Zhang Z., Ding X., Yang F., et al. The MEKK1-MKK1/MKK2-MPK4 Kinase Cascade Negatively Regulates Immunity Mediated by a Mitogen-Activated Protein Kinase Kinase Kinase in Arabidopsis. Plant Cell. 2012;24(5):2225–2236. doi: 10.1105/tpc.112.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitzschke A., Djamei A., Bitton F., Hirt H. A Major Role of the MEKK1-MKK1/2-MPK4 Pathway in ROS Signalling. Mol Plant. 2009;2(1):120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., et al. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15(1):141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 16.Meng X.Z., Wang H.C., He Y.X., Liu Y.D., Walker J.C., Torii K.U., et al. A MAPK Cascade Downstream of ERECTA receptor-like protein kinase regulates arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell. 2012;24(12):4948–4960. doi: 10.1105/tpc.112.104695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H.C., Ngwenyama N., Liu Y.D., Walker J.C., Zhang S.Q. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19(1):63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu R., Duan P., Yu H., Zhou Z., Zhang B., Wang R., et al. Control of Grain Size and Weight by the OsMKKK10-OsMKK4-OsMAPK6 Signaling Pathway in Rice. Mol Plant. 2018;11(6):860–873. doi: 10.1016/j.molp.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Umezawa T., Sugiyama N., Takahashi F., Anderson J.C., Ishihama Y., Peck S.C., et al. Genetics and Phosphoproteomics Reveal a protein phosphorylation network in the abscisic acid signaling pathway in arabidopsis thaliana. Sci Signal. 2013;6(270):13. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 20.Xing Y., Jia W.S., Zhang J.H. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54(3):440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- 21.Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci U S A. 2009;106(48):20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danquah A., de Zélicourt A., Boudsocq M., Neubauer J., Frei dit Frey N., Leonhardt N., et al. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015;82(2):232–244. doi: 10.1111/tpj.12808. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y.Q., Assmann S.M. Metabolite Transporter Regulation of ABA Function and Guard Cell Response. Mol Plant. 2014;7(10):1505–1507. doi: 10.1093/mp/ssu093. [DOI] [PubMed] [Google Scholar]
- 24.Chater C.C.C., Oliver J., Casson S., Gray J.E. Putting the brakes on: abscisic acid as a central environmental regulator of stomatal development. New Phytol. 2014;202(2):376–391. doi: 10.1111/nph.12713. [DOI] [PubMed] [Google Scholar]
- 25.Riemann M., Dhakarey R., Hazman M., Miro B., Kohli A., Nick P. Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses. Front Plant Sci. 2015;6:16. doi: 10.3389/fpls.2015.01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danquah A., de Zelicourt A., Colcombet J., Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32(1):40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Hõrak H., Sierla M., Tõldsepp K., Wang C., Wang Y.-S., Nuhkat M., et al. A Dominant Mutation in the HT1 Kinase Uncovers Roles of MAP Kinases and GHR1 in CO2-Induced Stomatal Closure. Plant Cell. 2016;28(10):2493–2509. doi: 10.1105/tpc.16.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F.J., Li M.Y., Wang P., Cox K.L., Duan L.S., Dever J.K., et al. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017;215(4):1462–1475. doi: 10.1111/nph.14680. [DOI] [PubMed] [Google Scholar]
- 29.Wang N.N., Zhao L.L., Lu R., Li Y., L Xb. Cotton mitogen-activated protein kinase 4 (GhMPK4) confers the transgenic Arabidopsis hypersensitivity to salt and osmotic stresses. Plant Cell Tiss Org Cult. 2015;123:619–632. doi: 10.1007/s11240-015-0865-5. [DOI] [Google Scholar]
- 30.Chen L., Sun H., Wang F., Yue D., Shen X., Sun W., et al. Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14-GhMKK11-GhMPK31 pathway is involved in the drought response in cotton. Plant MolBiol. 2020;103(1-2):211–223. doi: 10.1007/s11103-020-00986-0. [DOI] [PubMed] [Google Scholar]
- 31.Li B., Chen L., Sun W., Wu D.i., Wang M., Yu Y.u., et al. Phenomics-based GWAS analysis reveals the genetic architecture for drought resistance in cotton. Plant Biotechnol J. 2020;18(12):2533–2544. doi: 10.1111/pbi.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng J.W., Yang X.Y., Sun W.N., Miao Y.H., He L.R., Zhang X.L. The Calcium Sensor CBL2 and Its Interacting Kinase CIPK6 Are Involved in Plant Sugar Homeostasis via Interacting with Tonoplast Sugar Transporter TST2(1) Plant Physiol. 2020;183(1):236–249. doi: 10.1104/pp.19.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P., Zhang J., Sun L., Ma Y., Xu J., Liang S., et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol J. 2018;16(1):137–150. doi: 10.1111/pbi.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin S., Zhang X., Nie Y., Guo X., Liang S., Zhu H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant. 2006;50(4):519–524. doi: 10.1007/s10535-006-0082-5. [DOI] [Google Scholar]
- 35.Gao W., Long L.u., Zhu L.-F., Xu L.i., Gao W.-H., Sun L.-Q., et al. Proteomic and Virus-induced Gene Silencing (VIGS) Analyses Reveal That Gossypol, Brassinosteroids, and Jasmonic acid Contribute to the Resistance of Cotton to Verticillium dahliae. Mol Cell Proteomics. 2013;12(12):3690–3703. doi: 10.1074/mcp.M113.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H.B., Li X.H., Xiao J.H., Wang S.P. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods. 2012;8:12. doi: 10.1186/1746-4811-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batistic O., Sorek N., Schultke S., Yalovsky S., Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell. 2008;20(5):1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin F., Jiang Y., Li J., Yan T., Fan L., Liang J., et al. B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorphogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation. Plant Cell. 2018;30(9):2006–2019. doi: 10.1105/tpc.18.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., et al. SINAT E3 Ligases Control the Light-Mediated Stability of the Brassinosteroid-Activated Transcription Factor BES1 in Arabidopsis. Dev Cell. 2017;41(1):47–58.e4. doi: 10.1016/j.devcel.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grefen C., Blatt M.R. A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC) Biotechniques. 2012;53(5):311–314. doi: 10.2144/000113941. [DOI] [PubMed] [Google Scholar]
- 41.Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. Negative Regulation of Anthocyanin Biosynthesis in Arabidopsis by a miR156-Targeted SPL Transcription Factor. Plant Cell. 2011;23(4):1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Q., Zhu L., Zhang X., Guan Q., Xiao S., Min L., et al. GhCPK33 Negatively Regulates Defense against Verticillium dahliae by Phosphorylating GhOPR3. Plant Physiol. 2018;178(2):876–889. doi: 10.1104/pp.18.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Cai H., Liu P.u., Wang C., Gao H., Wu C., et al. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem Biophys Res Commun. 2017;484(2):292–297. doi: 10.1016/j.bbrc.2017.01.104. [DOI] [PubMed] [Google Scholar]
- 44.Wang C., Lu W., He X., Wang F., Zhou Y., Guo X., et al. The Cotton Mitogen-Activated Protein Kinase Kinase 3 Functions in Drought Tolerance by Regulating Stomatal Responses and Root Growth. Plant Cell Physiol. 2016;57(8):1629–1642. doi: 10.1093/pcp/pcw090. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L., Xi D., Li S., Gao Z., Zhao S., Shi J., et al. A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant MolBiol. 2011;77(1-2):17–31. doi: 10.1007/s11103-011-9788-7. [DOI] [PubMed] [Google Scholar]
- 46.Li C., Hisamoto N., Nix P., Kanao S., Mizuno T., Bastiani M., et al. The growth factor SVH-1 regulates axon regeneration in C. elegans via the JNK MAPK cascade. Nat Neurosci. 2012;15(4):551–557. doi: 10.1038/nn.3052. [DOI] [PubMed] [Google Scholar]
- 47.Yao T., Zhang J., Xie M., Yuan G.L., Tschaplinski T.J., Muchero W., et al. Transcriptional Regulation of Drought Response in Arabidopsis and Woody Plants. Front Plant Sci. 2021;11:12. doi: 10.3389/fpls.2020.572137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida T., Mogami J., Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17(12):3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park S.-Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai G.H., Wang G.D., Wang L., Liu Y., Pan J.W., Li D.Q. A maize mitogen-activated protein kinase kinase, ZmMKK1, positively regulated the salt and drought tolerance in transgenic Arabidopsis. J Plant Physiol. 2014;171(12):1003–1016. doi: 10.1016/j.jplph.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Zhu Q., Shao Y., Ge S., Zhang M., Zhang T., Hu X., et al. y A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat Plants. 2019;5(4):414–423. doi: 10.1038/s41477-019-0396-x. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y.J., Duursma R.A., Farrior C.E., Medlyn B.E., Feng X. Optimal stomatal drought response shaped by competition for water and hydraulic risk can explain plant trait covariation. New Phytol. 2020;225(3):1206–1217. doi: 10.1111/nph.16207. [DOI] [PubMed] [Google Scholar]
- 54.McKown K.H., Bergmann D.C. Stomatal development in the grasses: lessons from models and crops (and crop models) New Phytol. 2020;227(6):1636–1648. doi: 10.1111/nph.16450. [DOI] [PubMed] [Google Scholar]
- 55.Guo X.-Y., Wang Y., Zhao P.-X., Xu P., Yu G.-H., Zhang L.-Y., et al. AtEDT1/HDG11 regulates stomatal density and water-use efficiency via ERECTA and E2Fa. New Phytol. 2019;223(3):1478–1488. doi: 10.1111/nph.15861. [DOI] [PubMed] [Google Scholar]
- 56.Yu H., Chen X.i., Hong Y.-Y., Wang Y., Xu P., Ke S.-D., et al. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell. 2008;20(4):1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu L., Chen X.i., Wang Z., Wang S., Wang Y., Zhu Q., et al. Arabidopsis Enhanced Drought Tolerance1/HOMEODOMAIN GLABROUS11 Confers Drought Tolerance in Transgenic Rice without Yield Penalty. Plant Physiol. 2013;162(3):1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu L.-H., Wu S.-J., Peng Y.-S., Liu R.-N., Chen X.i., Zhao P., et al. Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol J. 2016;14(1):72–84. doi: 10.1111/pbi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng G.S., Fan C.Y., Di S.K., Wang X.M., Xiang C.B., Pang Y.Z. Over-Expression of Arabidopsis EDT1 Gene Confers Drought Tolerance in Alfalfa (Medicago sativa L.). Front. Plant Sci. 2017;8:14. doi: 10.3389/fpls.2017.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.