Graphical abstract
Keywords: Malus, miR408a, BBP, Anthocyanin, Lignin, Copper homeostasis, ROS homeostasis
Highlights
-
•
A crosstalk between copper homeostasis and ROS homeostasis mediated by the miR408a-BBP-LAC3/CSD1 module was revealed under light induction and copper stress in Malus plants.
-
•
The copper homeostasis fluctuation triggers ROS homeostasis variation and participates in modulating anthocyanin accumulationin under excessive copper and copper defficiency in Malus plants.
Abstract
Introduction
The expression of miR408 is affected by copper (Cu) conditions and positively regulates anthocyanin biosynthesis in Arabidopsis. However, the underlying mechanisms by which miR408 regulates anthocyanin biosynthesis mediated by Cu homeostasis and reactive oxygen species (ROS) homeostasis remain unclear in Malus plants.
Objectives
Our study aims to elucidate how miR408a and its target, basic blue protein (BBP) regulate Cu homeostasis and ROS homeostasis, and anthocyanin biosynthesis in Malus plants.
Methods
The roles of miR408a and its target BBP in regulating anthocyanin biosynthesis, Cu homeostasis, and ROS homeostasis were mainly identified in Malus plants.
Results
We found that the BBP protein interacted with the copper-binding proteins LAC3 (laccase) and CSD1 (Cu/Zn SOD superoxide dismutase), indicating a potential crosstalk between Cu homeostasis and ROS homeostasis might be mediated by miR408 to regulate the anthocyanin accumulation. Further studies showed that overexpressing miR408a or suppressing BBP transiently significantly increased the expression of genes related to Cu binding and Cu transport, leading to anthocyanin accumulation under light induction in apple fruit and Malus plantlets. Consistently, opposite results were obtained when repressing miR408a or overexpressing BBP. Moreover, light induction significantly increased the expression of miR408a, CSD1, and LAC3, but significantly reduced the BBP expression, resulting in increased Cu content and anthocyanin accumulation. Furthermore, excessive Cu significantly increased the anthocyanin accumulation, accompanied by reduced expression of miR408a and Cu transport genes, and upregulated expression of Cu binding proteins including BBP, LAC3, and CSD1 to maintain the Cu homeostasis and ROS homeostasis in Malus plantlets.
Conclusion
Our findings provide new insights into the mechanism by which the miR408a-BBP-LAC3/CSD1 module perceives light and Cu signals regulating Cu and ROS homeostasis, ultimately affecting anthocyanin biosynthesis in Malus plants.
Introduction
Anthocyanins are important flavonoid pigments that endow plants with abundant colours [1]. They also play roles by functioning as important antioxidants in defence to various biotic and abiotic stresses [2]. The anthocyanin-rich bilberry (Vaccinium myrtillus L.) extract has protective effects against copper(II) chloride toxicity [3]. Extensive studies have shown that anthocyanin biosynthesis in plants is affected by various environmental factors, such as light, sucrose, and low temperatures, and by micronutrient phosphorus deficiency [4]. In barley, transcriptome analysis revealed that the anthocyanin biosynthesis pathway is involved in Cu tolerance [5]. However, how Cu levels or Cu stress affect anthocyanin biosynthesis remains to be elucidated.
MiRNAs are endogenous single-stranded RNAs in eukaryotes and viruses; mature miRNAs are usually 20–24 nt in length, processed from precursors approximately 64–303 nt in length and highly conserved during evolution [6], [7]. Based on sequence complementarity, miRNAs recognize their mRNA targets and suppress their expression by triggering their degradation and/or translational repression [8], [9], [10]. Increasing evidence supports that miRNAs play critical roles in multiple biological processes, including signal transduction, secondary metabolism, and protection against biotic and abiotic stresses [11], [12]. Several recent studies have shown that miRNAs are involved in anthocyanin biosynthesis, especially under the stress of multiple adverse events. In Arabidopsis, miR828 negatively regulates anthocyanin biosynthesis [13], [14]. In tomatoes, repressing miR858 induces anthocyanin accumulation by modulating SlMYB48-like transcripts [15]. In apple, our recent study revealed that miR7125 positively regulates anthocyanin biosynthesis during light induction [16].
Stress-responsive miRNAs usually exert their effects on target genes related to abiotic stress responses in plants [17]. For example, miR828 promotes lignin biosynthesis and H2O2 accumulation in injured sweet potatoes [18]. In maize, the abundance of miR408 expression was shown to be increased upon hormone depletion during somatic embryogenesis [19], and overexpression of miR319 was shown to enhance tolerance to salt, drought, and cold [20], [21]. Recently, MdWRKY100 expression activated by the miR156a-SPL13 module was demonstrated to regulate salt stress tolerance in apple [22]. In addition, plant miRNAs are also involved in responses to nutrient stresses, such as nitrogen, phosphate, sulfate, iron, and Cu deficiencies [23], [24], [25], [26]. For example, miR399d participated in regulating anthocyanin accumulation under conditions of phosphorus deficiency in Malus crabapple [4]. In maize, miR528 regulates lignin biosynthesis under nitrogen-rich conditions, thus increasing lodging resistance [27].
Cu is one of the seven trace elements necessary for plants growth and an essential cofactor for various Cu-containing enzymes and proteins [28]. Plastocyanin (PC) contains the most abundant Cu in chloroplasts [29], [30]. In Arabidopsis, the Cu content has a direct impact on the activity of Cu/ZnSOD1 [31]. Therefore, the abundance of these Cu-containing proteins and Cu transportation is important for the maintenance of Cu homeostasis in plants [32]. Moreover, Cu has a stabilizing effect on chlorophyll and other pigments to prevent their destruction, especially in adverse environments. Cu deficiency seriously threaten crop yields and quality, and also damage the landscapes of ornamental plants. At present, chemical fertilizers contain almost no Cu, resulting in the aggravation of barren land. Thus, more attention should be given to Cu shortages in agricultural production worldwide. On the other hand, Cu largely exists in contaminated soils, which are toxic to plants at excessive levels. Thus, maintaining Cu homeostasis is crucial for plant to regulate growth and development, and to adapt to the fluctuations of external Cu content. However, the molecular mechanisms implicated in the regulation of Cu homeostasis and the underlying role of miRNAs in Cu stress conditions remain to be elucidated.
MiR408 is a positive regulator of photosynthesis and photomorphogenesis, which is from one of the most evolutionally conserved miRNA families in plants and has been annotated in more than 20 plant species [33], [34]. In Arabidopsis, miR408 was shown to be crucial to the HY5-SPL7 module to coordinate responses to light and Cu, indicating its central role in the response to developmental signals and Cu status in plants, and overexpression of miR408 was shown to increase anthocyanin accumulation in Arabidopsis seedlings [35], [36]. It is worth noting that the target genes of miR408 encode Cu-binding proteins belonging to the phytocyanin family [37], [38], [39], [40]. For example, the LAC13 gene encoding a Cu-containing LAC was identified as a target of miR408 [41]. In rice, miR408 was shown to positively regulate photosynthesis and grain yield by targeting OsUCL8, a phytocyanin protein [42]. Interestingly, overexpressing miR408 exhibited increased abundances of Cu in chloroplasts, increased abundances of PC, higher rates of vegetative growth, and photosynthetic gene induction in transgenic Arabidopsis, tobacco, and rice plants [43]. Moreover, miR408 is also involved in various abiotic stress responses, such as tolerance to salt, cold, oxidative stress, drought, and osmotic stresses, as well as defence responses upon wounding [44], [45], [46], [47]. These studies led to our hypothesis that miR408 bridges light signals to intracellular Cu homeostasis to regulate anthocyanin biosynthesis in response to the fluctuations of external Cu content. Therefore, it is extremely important and essential to clarify the regulatory pathway of Cu homeostasis driven by target genes of miR408, especially under Cu stress conditions.
ROS, acting as important signalling molecules, regulate various biological and stress response processes. The seedling photobleaching phenotype during the dark-to-light transition is largely caused by ROS [48]. ROS also regulate root meristem activity mediated by Abscisic acid (ABA) in mitochondria [49]. Previous studies revealed that light and ROS signalling were bridged by PIF1, PIF3, HY5, and HYH to regulate cell death and the photooxidative response [50]. In radish sprouts, H2O2, NO, and UVR8 participate in UV-B-induced anthocyanin biosynthesis [51], illustrating that ROS are involved in anthocyanin biosynthesis. In Arabidopsis, miR398b regulates ROS homeostasis by inhibiting CSD1 and CSD2 [52]. Substantial amounts of ROS accumulate in plants under Cu stress [53], but the relationship between Cu homeostasis and ROS homeostasis remains unclear. In addition, illustrating how plants regulate Cu homeostasis in response to Cu stress is of great significance. Therefore, we hypothesized that the crosstalk between Cu homeostasis and ROS homeostasis might be mediated by miR408 to regulate the anthocyanin accumulation.
While miR408 has thus far been identified in several plant species, its biological mechanisms of regulating the crosstalk between Cu homeostasis and ROS homeostasis are not clear. In this study, we revealed that miR408a and its target gene, BBP, help to regulate anthocyanin biosynthesis in Malus plants. The expression of miR408a and BBP was induced by Cu stress. Further study revealed that BBP interacts with CSD1 and laccase 3 (LAC3) to regulate Cu homeostasis and ROS homeostasis. Taken together, the results of this study reveal that the miR408a-BBP-LAC3/CSD1 module regulates anthocyanin biosynthesis mediated by the crosstalk between Cu homeostasis and ROS homeostasis in Malus plants. Our findings shed light on the crosstalk between Cu and ROS homeostasis mediated by the miR408a-BBP-LAC3/CSD1 module affects anthocyanin biosynthesis in Malus plants in response to light signals and Cu stress.
Materials and methods
Plant materials and growth conditions
‘Royalty’ leaves were collected at five different developmental stages. The Malus crabapple cultivar ‘Royalty’ and Malus ‘Golden delicious’ plantlets were cultured on propagation medium (pH 5.8, 4.47 g L−1 Murashige and Skoog medium, 30 g L−1 sucrose, 0.4 mg L−1 6-BA, 0.05 mg L−1 NAA, and 6 g L−1 agar) for 30 days at 23 °C under 60–70 % relative humidity and a 16 h light/8h dark photoperiod until treatment. Nicotiana benthamiana plants were grown in vermiculite and nutritive soil (1:1) at 23 °C under 50–60 % relative humidity and a 16 h light/8h dark photoperiod.
The bagged ‘Stolav’ apple fruits at 160 days after blooming were subjected to light treatments after bag removal (12,000 lx, 20 °C, 16 h light/8h dark photoperiod). Apple fruits were sampled at 0, 3, 5, 8, and 12 d after light treatment, and the peel was frozen in liquid nitrogen and preserved under −80 °C.
To achieve different Cu levels, three-week-old uniform Malus plantlets were transferred onto a standard MS medium under the following Cu2SO4 conditions: no Cu (-Cu), normal copper (0.1 µM Cu, Control), 1 µM Cu, 10 µM Cu, and 100 µM Cu. Leaf and stem samples were collected at 1, 3, 5, and 10 days, followed by total RNA isolation, reverse transcription, and RT-qPCR analysis. All samples were frozen in liquid nitrogen and preserved at −80℃ freezer until use.
Total RNA extraction and quantitative real-time PCR (qRT–PCR) analysis
Total RNA was isolated from leaves and stems using an EASYspin Plus Complex Plant RNA Kit (Aidlab: RN38) and reverse-transcribed using Moloney murine leukaemia virus reverse transcriptase (M−MLV RT; TaKaRa, Otsu, Japan) and oligo (dT) primers. A Nano spectrophotometer (IMPLEN, CA, USA) and an RNA Nano 6000 Assay Kit with the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) were used to assess the RNA integrity and purity, respectively. qRT-PCR was performed with SYBR Premix TaqTMII (RR820A; TaKaRa, Otsu, Japan) by a CFX96 Real-Time PCR system (Bio-Rad, Hercules, CA, USA). The 18S ribosomal RNA was used as the internal control (GenBank accession number DQ341382). Relative expression levels were determined using the 2−△△Ct method [54].
Total miRNAs were reverse transcribed using TransScript miRNA First-strand cDNA Synthesis SuperMix (AT351; TransGen Biotech) according to the manufacturer’s instructions. The transcript levels of mature miR408a were analyzed using miRNA-specific primers and a universal primer (AQ202-01; TransGen Biotech). The U6 gene was used as a internal control. All experiments were performed independently three times.
Construct preparation and Agrobacterium-mediated transient transformation
The miR408a (123 bp, LOC103344813) precursor sequences and MdBBP (372 bp, LOC103402332) coding sequences were cloned from ‘Royalty’ leaves (Table S4). Afterwards, the amplicon was cloned into a pEasy-T1 subcloning vector (TransGen Biotech, Beijing, China) and then transferred into the plant expression vector pCAMBIA1300 with an eGFP label. To reduce miR408a activity and the transcript level of MdBBP, the miR408a precursor and gene-specific fragment of the MdBBP coding region were separately inserted into the pTRV2 vector.
All the constructs were transformed into Agrobacterium tumefaciens (strain GV3101) and grown in a Luria-Bertani medium containing 50 mg L−1 kanamycin and 50 mg L−1 rifampicin. The agrobacterium strains containing the construct were harvested at 5000 rpm for 20 min and resuspended in infiltration buffer (10 mM 2-(N-morpholino) ethanesulfonic acid (MES), 200 µM acetosyringone (As), and 10 mM MgCl2) to a final OD600 of 0.8–1.0. Agrobacterium containing pTRV1 and pTRV2, pTRV2-miR408, and pTRV2-BBP were mixed at a 1:1 volumetric ratio and placed at room temperature for 3 h [55]. For transient transformation of the Malus plantlets, whole explants were subjected to osmotic infection in above agrobacterium suspention under conditions of a vacuum of −0.08 MPa for 90 s. Let the air out after 5 min and repeat again. The phenotypes of infected explants were observed approximately 7 to 10 days later.
RNA ligase mediated rapid amplification of cDNA ends (RLM-RACE)
Modified RLM-RACE was performed to verify the cleavage site of the BBP transcripts using a First Choice RLM-RACE Kit (Invitrogen) according to the manufacturer’s instructions. RLM-RACE was performed as described previously [16].
Determination of lignin content
Lignin quantification was performed using the method described by Bruce & West [56]. All measurements were performed with three biological replicates.
High-performance liquid chromatography (HPLC)
Frozen samples to be measured were ground to a fine powder in liquid nitrogen, quickly weighed (0.2–1 g), and transferred into a 5 mL centrifuge tube, after which 2 mL of the anthocyanin extraction solution (formic acid, methanol and water at a ratio of 1:80:19) was added. All the samples were evaluated with three biological replicates. Anthocyanin contents was detected by HPLC at OD 520 nm using previously published methods [4], [57].
Determination of the Cu content
The fresh samples were dried for 24 h at 70℃ and then ground into powder. Two micrograms of the plant samples were carbonized using a muffle furnace for 5–10 min. After carbonization, the powder was subjected to podzolization for 4–5 h at 560 °C in a resistance furnace. Then, 1 mL of 6 M HCl was added and burned to near dry in a muffle furnace, and 0.1 M HCl was added to a final volume of 50 mL. The Cu content in the plant tissues was analyzed by inductively coupled plasma spectrometry (ICP) and calculated as follows: Cu content = PPM (μg/mL) × V (constant volume, mL)/m (sample weight, g).
Microscopy
For histological analysis, stems 1 cm away from the top bud of ‘Golden Delicious’ plantlets were collected after 10 days of administration of the different copper treatments and fixed in FAA fixative (50 % ethanol: formaldehyde: glacial acetic acid = 18:1:1) at 4 ℃ for more than 24 h. The stem tissues were dehydrated with an ethanol gradient (70 %, 85 %, 95 %, and 100 % ethanol, 60 min per step), dewaxed with xylene, and then embedded in paraffin. The samples were cut into 8-μm-thick horizontally continuous slices with a YD-2508A rotary microtome and dewaxed in dimethylbenzene. The prepared paraffin-embedded sections were observed using a Leica DCF500 microscope.
Yeast one-hybrid assays
A yeast one-hybrid system was used to assay transcriptional activation by the MdBBP protein. The full-length CDS region of MdBBP was cloned and ligated into a pGADT7 vector. The promoter fragments of MdCHS, MdDFR, MdUFGT, MdF3′H, and MdFLS were cloned and ligated into pAbAi. Yeast one-hybrid interactions were detected by selecting yeast that was resistant to Aureobasidin A (AbA).
Yeast two-hybrid assay
Yeast two-hybrid assays were performed using the DUAL membrane system. The MdBBP coding region was cloned into the pBT3-SUC and pBT3-STE vectors. The MdCSD1 and MdLAC3 coding sequences were cloned into the PR3-N vector (expressing DNA-binding domains). The primers used to create these constructs are listed in supporting information (Table S1). All constructs and empty vector plasmids were transferred into the yeast (Saccharomyces cerevisiae) strain AH109 (Clontech, USA). The pTSU2-APP and pNubG-Fe65 vectors were cotransferred into yeast cells as a positive control. Yeast grown on plates containing selective medium lacking Leu and Trp (-L/-T) were activated on plates containing medium lacking Leu, Trp, His and adenine (-L/-T/-H/-A) + X-α-gal (5-bromo-4-chloro-3-indolyl-α-D-galactoside) solid medium plates to detect protein interactions (Clontech, USA).
Bimolecular fluorescence complementation (BiFC) assays
The coding sequences of MdBBP, MdCSD1, and MdLAC3 were cloned and inserted into the plasmids pEarleyGate201-35S/YC and pEarleyGate-35S/YN, respectively. All constructs were introduced into Agrobacterium strain GV3101. The transformed agrobacterium lines were cultured in a Luria-Bertani medium with appropriate antibiotic selection. The agrobacterium thallus were harvested by centrifugation at 5,000 rpm for 10 min, resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 200 µM As, pH 5.7) to a final OD600 of 1.0, and placed in the dark at room temperature for 3 h before infiltration. The agrobacterium suspension was used to infect the leaves of 1-month-old tobacco (Nicotiana benthamiana) plants. After 2–3 days of transformation, the expression of yellow fluorescent protein (YFP) was observed and photographed by laser confocal microscopy (FV1000, Olympus, Tokyo, Japan).
Subcellular localization
The coding sequences of MdBBP and MdCSD1 without the stop codon were amplified and subcloned into a pHBT-eGFP-NOS vector to create a 35S::BBP-eGFP fusion construct and 35S::CSD1-eGFP construct, respectively. The p35S::BBP-eGFP plasmid was introduced into Arabidopsis protoplasts and epidermal onion cells by the particle bombardment method, respectively. The p35S::CSD-eGFP plasmid was introduced into Arabidopsis protoplasts. The p35S::GFP vector served as the control. After 12 h of darkness, eGFP fluorescence was observed with a laser-scanning confocal microscopy (FV1000, Olympus, Tokyo, Japan).
Protein expression and purification
The protein expression and purification of MdMYB16 (HM122617.1) were performed as described previously [16].
Biolayer interferometry (BLI)
Real-time binding assays between dsDNA and protein using BLI were performed using previously published methods [16].
Bioinformatics analysis and phylogenetic analyses
Bioinformatics analysis of MdBBP was performed with the Swiss model [58], [59], MEME [60], TMPred [61], TMHMM2.0, ExPASY-ProtParam and Protscale tool [62], SignalP, and NetPhos 3.1 tools [63]. The amino acid sequences of MdBBP in the National Center for Biotechnology Information database were used for BLAST analysis. The phylogenetic tree was constructed using the neighbour-joining method with MEGA version 6.0 software [64], [65]. The evolutionary distances were computed using the Poisson correction method [66].
Statistical analysis
The diagrams were generated by Adobe Photoshop CS6 and Origin 8.5. The data were plotted as the means ± standard errors (SEs) of three independent biological replicates. Statistical analysis and significance analysis of the data were calculated by SPSS software, version 2018. ANOVA with subsequent Duncan’s test and Student’s test was performed to identify significant differences, and the differences were defined as significant at P < 0.05.
Results
miR408a targets a basic blue protein, MdBBP
To identify the differentially expressed miRNAs that regulate anthocyanin accumulation and leaf colour variations in Malus plants, mature ‘Royalty’ (red) and mature ‘Flame’ (green) leaves were collected and subjected to miRNA sequencing. We found that the abundance of miR408a was significantly upregulated by 3.6-fold in ‘Royalty’ leaves compared with that in ‘Flame’ leaves.
Furthermore, target gene prediction was performed using the psRNATarget website (https://plantgrn.noble.org/psRNATarget/). Among the target genes of miR408a, we selected three predicted target genes whose sequences suggested good complementarity with the miR408a sequence, including MdBBP (LOC103402332), thiamine-repressible mitochondrial transport protein THI74-like (LOC103435824), and endoglucanase 5 (LOC103403402). To confirm the target relationships between miR408a and the above-predicted targets, the cleavage positions mediated by miR408a were analyzed by RLM-RACE. MdBBP was identified as a cleavable target of miR408a, and the cleavage site was located between the tenth and eleventh bases of the complementary sequence.
To investigate the biological function of MdBBP, we cloned the core coding region (CDS) of MdBBP from Malus crabapple ‘Royalty’ and performed bioinformatics analysis. The MdBBP protein contains a transmembrane helix and conserved domains of plantacyanin (Accession: cd11013; Region interval: 30–123) and the cupredoxin superfamily. Plantacyanin is a subclass of the phytocyanin family of blue copper proteins, a ubiquitous family of cupredoxins belonging to the plant type I copper proteins. The position and pattern of 4 conserved residues above the triangle that compose this conserved feature were H(67)-C(107)-H(112)-M(117), denoting the type 1 Cu binding site. Three-dimensional structure prediction of MdBBP showed that it was a monomer, and protein hydrophilicity analysis showed that it is hydrophobin. In addition, it contains two possible transmembrane helices, and one transmembrane helix of MdBBP contains a signalling peptide (1–28 aa) at the N-terminus of MdBBP. Based on these results, we speculated that MdBBP is involved in regulating Cu homeostasis as a nutritional Cu sensor in response to growth and development signals. Moreover, we found that the promoters of miR408a and MdBBP contain many cis-acting elements associated with ABA, light, and MeJA responsiveness, implying that miR408a and MdBBP might play important roles in responses to numerous environmental signals. Given that miR408a promotes anthocyanin accumulation in response to Cu signalling in Arabidopsis, we concentrated on its function and investigated the role of MdBBP in regulating anthocyanin accumulation and Cu homeostasis in Malus plants.
MdBBP interacts with both MdLAC3 and MdCSD1
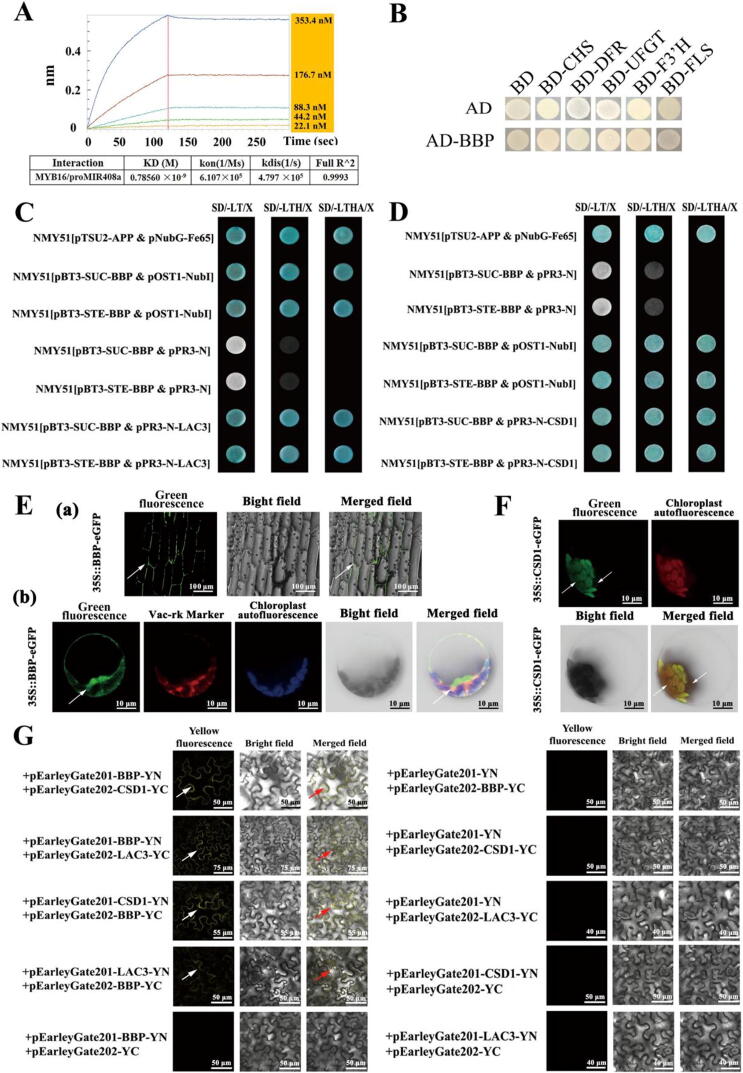
To determine whether MdMYB16, a repressor of anthocyanin biosynthesis, interacts with the miR408a promoter, we determined the kinetic values and binding affinities of the MdMYB16 protein for miR408a promoter elements using biolayer interferometry (BLI). As shown in Fig. 1A, MdMYB16 is bound to the element GGTAGGTA (+1761) within the miR408a promoter. In addition, the equilibrium dissociation constant (KD) for MYB16 and this element of the miR408a promoter was 0.7856 nM. These results indicated that MdMYB16 interacts with the miR408a promoter. Accordingly, we speculated that MdMYB16 is an upstream regulator of miR408a. To investigate the mechanism by which MdBBP regulates anthocyanin biosynthesis, a yeast one-hybrid assay was carried out to verify whether MdBBP could directly interact with the promoters of anthocyanin biosynthesis genes which encoding chalcone isomerase (CHS), dihydroflavonol reductase (DFR), UDP-glucosyltransferase (UFGT), flavonoid 3′ hydroxylase (F3′H), and flavonol synthetase (FLS), respectively. However, no positive interactions between them were observed (Fig. 1B).
Fig. 1.
BBP interacts with both LAC3 and CSD1. (A) Dynamic curves showing the affinity of MYB16 for fragments of the miR408a promoter. (B) BBP interacted with the promoter of anthocyanin genes in yeast one-hybrid assays. (C) BBP interacted with LAC3 in yeast two-hybrid assays. (D) BBP interacted with CSD1 in yeast two-hybrid assays. (E) Subcellular localization of BBP. (F) Subcellular localization of CSD1. (G) BBP interacted with LAC3 and CSD1 in BiFC assays.
To elucidate the mechanism by which the miR408-MdBBP module regulates anthocyanin biosynthesis, we predicted the potential interaction proteins of BBP using STRING online software (https://string-db.org/cgi/input.pl). According to the predicted scores, CSD1 and LAC3 were selected as potential target proteins. Laccase is the last enzyme of the lignin biosynthesis pathway and is important for the process by which lignin monomers are polymerized into macromolecular heteropolymers. CSD1, a key enzyme responsible for reducing ROS in plants subjected to oxidative stress, plays an important role in regulating the redox potential in cells. Subsequently, we cloned these genes, and the subcellular localization of the protein was predicted. The prediction results showed that BBP was localized in the cell membrane using Plant-mPLoc software (https://www.csbio.sjtu.edu.cn/bioinf/plant-multi/#) [67], vocuole, cytoplasm, and outside the cell (https://cello.life.nctu.edu.tw/; https://wolfpsort.hgc.jp/). CSD1 might be localized not only in the cell, such as chloroplasts (https://www.csbio.sjtu.edu.cn/bioinf/plant-multi/#), nucleus, cytoplasm, but also outside the cell (https://cello.life.nctu.edu.tw/; https://wolfpsort.hgc.jp/). LAC3 might be localized in the cell membrane and outside the cell (https://www.csbio.sjtu.edu.cn/bioinf/plant-multi/#; https://cello.life.nctu.edu.tw/;). As shown in Fig. 1E (a) and 1(b), BBP was localized in cell membrane and tonoplast. CSD1 was localized in chloroplast (Fig. 1F).
To explore whether BBP interacts with LAC3 and CSD1, we used yeast two-hybrid (Y2H) assays and found that BBP interacted with both LAC3 and CSD1. As shown in Fig. 1C, BBP-AD and LAC3-BD were cotransformed into Y2H yeast strains that grew successfully on -LT, -LTH, and -LTHA selective media in the presence of X-α-gal. Conversely, yeast cotransformed with BBP-AD and BD grew on -LT selective media but not -LTHA selective media. As shown in Fig. 1D, BBP-AD and CSD1-BD were cotransformed into Y2H yeast strains that grew successfully on -LT, -LTH, and -LTHA selective media containing the substrate X-α-gal.
To further confirm the interaction between BBP and LAC3, as well as that between BBP and CSD1, we used a BiFC system in which YFP fluorescence was reconstituted. As shown in Fig. 1G, 35S: BBP-CYFP and 35S: LAC3-NYFP were cotransformed into tobacco leaves, and clear YFP fluorescence was observed, indicating coexpression of BBP-CYFP and LAC3-NYFP. Moreover, when 35S: BBP-NYFP was combined with 35S: LAC3-CYFP, clear YFP fluorescence was also captured, indicating that BBP was capable of interacting with LAC3. Conversely, when 35S: NYFP was combined with 35S: BBP-CYFP and 35S: BBP-NYFP was combined with 35S: CYFP, no fluorescence was observed.
When 35S: BBP-CYFP and 35S: CSD1-NYFP were cotransformed, and clear YFP fluorescence was obtained, indicating coexpression of BBP-CYFP and CSD1-NYFP. Moreover, when 35S: BBP-NYFP was combined with 35S: CSD1-CYFP, clear YFP fluorescence was also observed. In summary, BBP is capable of interacting with LAC3 and CSD1 in vivo, and the interaction might occurs on the cell membrane and tonoplast. Thus, we hypothesized that BBP is critical for the regulation of Cu homeostasis and that it transmits Cu deficiency or Cu poisoning signals from the cell membrane to the cytoplasm or nucleus. We next focused on LAC3 and CSD1 as potential downstream effectors of the miR408-BBP module in response to Cu conditions.
MiR408a positively regulates anthocyanin biosynthesis in Malus plants
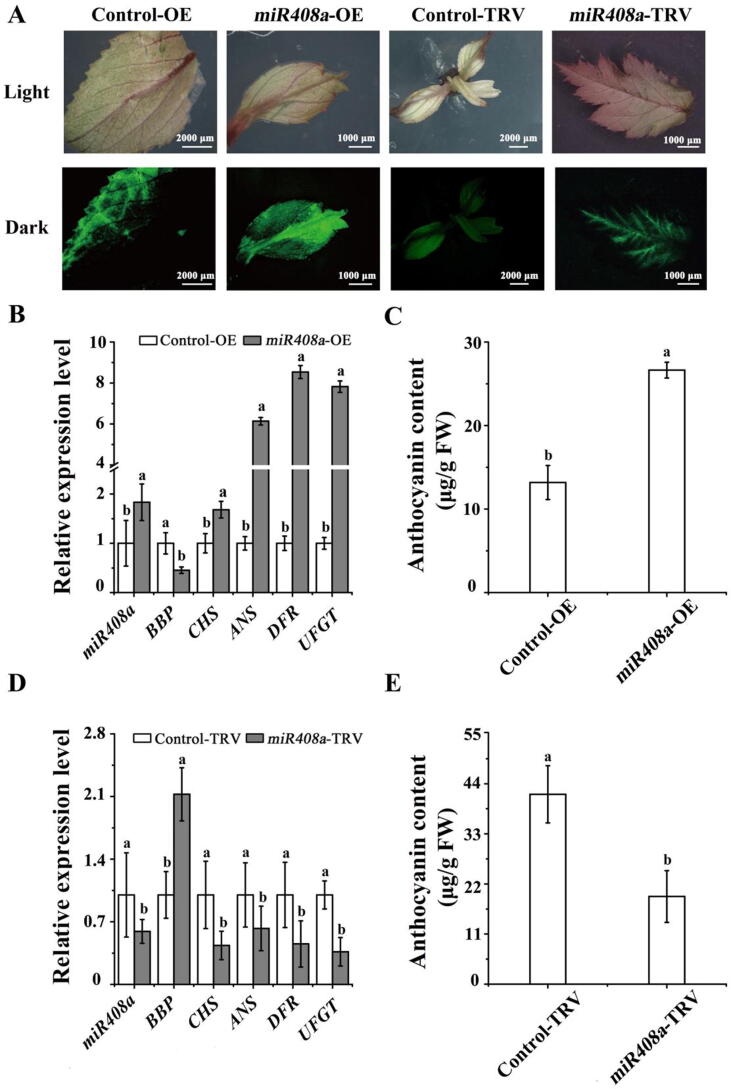
To investigate whether the modulation of miR408a affects anthocyanin accumulation under normal Cu conditions, we constructed vectors overexpressing miR408a (OE-miR408a) and silencing miR408a (TRV-miR408a). As shown in Fig. 2A, the expression level of mature miR408a increased significantly in leaves when miR408a was instantaneously overexpressed. Consequently, the levels of BBP transcripts decreased significantly, which further verified the target relationship between miR408a and BBP (Fig. 2B). Transient overexpression of miR408a resulted in red leaves and higher anthocyanin contents compared to those in the controls (Fig. 2C). Consistently, the transcription levels of genes encoding CHS, anthocyanin reductase (ANS), DFR, and UFGT in the anthocyanin pathway were substantially increased (Fig. 2B). However, silencing miR408a yielded seedlings that were poorly coloured and had lower anthocyanin contents (Fig. 2E). The expression levels of miR408a and BBP were downregulated and upregulated significantly, indicating effective silencing of miR408a (Fig. 2D). These results were accompanied by significant reductions in the expression levels of genes related to the anthocyanin pathway, including CHS, ANS, DFR, and UFGT (Fig. 2D).
Fig. 2.
Characterization of the transient transformation of OE-miR408a and TRV-miR408a in ‘Royalty’. (A) Phenotypes of leaves infected with different vectors. (B) qRT–PCR analysis results showing the gene expression levels infected with OE-miR408a. (C) Anthocyanin contents in leaves infected with OE-miR408a. (D) qRT–PCR analysis showing relative gene expression following infection with TRV-miR408a. (E) Anthocyanin contents in leaves infected with TRV-miR408a. Error bars denote the SD of three replicate measurements. Different lowercase letters above the bars indicate significant differences (P < 0.05) calculated using one-way ANOVA.
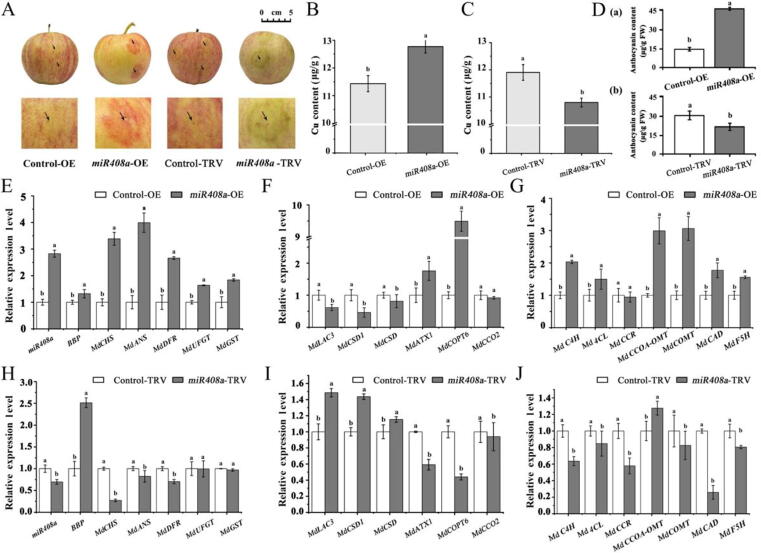
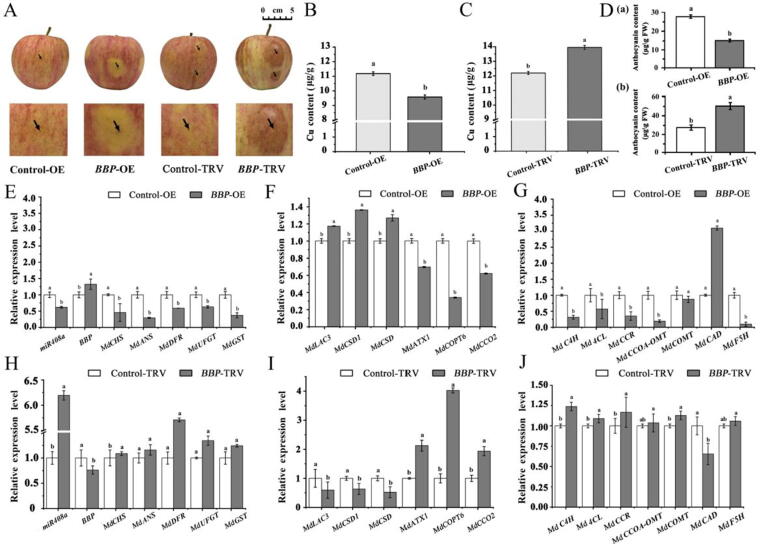
To further verify the roles of miR408 in regulating anthocyanin metabolism, OE-miR408a and TRV-miR408a vectors were individually injected into the apple fruit. As shown in Fig. 3A, apple fruit with miR408a overexpression showed a red appearance surrounding the infection site. Simultaneously, the mature miR408a level and Cu content increased significantly (Fig. 3B). In contrast, miR408a silencing reduced the colouration and Cu content in apple fruit (Fig. 3C). Consistent with the phenotype, the anthocyanin content increased significantly in miR408-OE but decreased significantly in TRV-miR408a (Fig. 3D (a) and (b)). As expected, the expression levels of CHS, ANS, DFR, UFGT, and GST increased significantly in apple fruit infected with miR408a-OE (Fig. 3E). Whereas, the expression levels of CHS, ANS, and DFR decreased significantly (Fig. 3F). Notably, MdBBP, MdLAC3, MdCSD1, and MdCSD expression in miR408a-OE apple fruit decreased significantly, as well as the expression of MdATX1 and MdCOPT6, which are involved in copper transportation, might be influenced by increased Cu content (Fig. 3E and 3G).
Fig. 3.
Characterization of the transient transformation of miR408a-OE and TRV-miR408a in apple fruit. (A) Phenotypic comparison of apple fruit infected with different vectors. (B) Comparison of Cu content measured for miR408a-OE and Control-OE. (C) Comparison of Cu content measured for miR408a-TRV and Control-TRV. (D) Anthocyanin contents in apple fruit peel infected with different vectors. (E and H) Quantitative analysis of miR408a, MdBBP, MdCHS, MdANS, MdDFR, MdUFGT, and MdGST transcript levels. (F and I) Quantitative analysis of MdLAC3, MdCSD1, MdATX1, MdCOPT6, and MdCCO2. (G and J) Expression of lignin pathway-related genes in apple fruit pericarp infected with different vectors. Error bars denote the standard deviations of three replicate measurements. Different lowercase letters denotes significant differences (ANOVA, P < 0.05).
In TRV-miR408a, due to the reduction in miR408a levels, elevated levels of the MdBBP transcript were observed (Fig. 3H). MdLAC3 and MdCSD1 expression in TRV-miR408a also increased significantly, which might be due to the synergistic effect due to the interaction among MdBBP, MdLAC3, and MdCSD1. The expression of MdATX1, MdCOPT6, and MdCCO2 decreased significantly, which is consistent with the decreased Cu content in TRV-miR408a (Fig. 3H).
Moreover, in miR408-OE, except for the cinnamoyl-coenzyme A reductase gene (CCR), the expression of genes encoding Cinnamate 4-hydroxylase (C4H), 4-coumarate: CoA ligase (4CL), caffeoyl-CoA-O-methyltransferase (CCoA-OMT), caffeic acid O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD) in the lignin pathway increased significantly in miR408-OE (Fig. 4I). However, silencing miR408a down-regulated C4H, 4CL, CCR, COMT, CAD, and ferulate 5 hydroxylase (F5H) but not CCoA-OMT. Taken together, these results suggest that miR408a is a positive regulator of anthocyanin accumulation and Cu content in Malus plants.
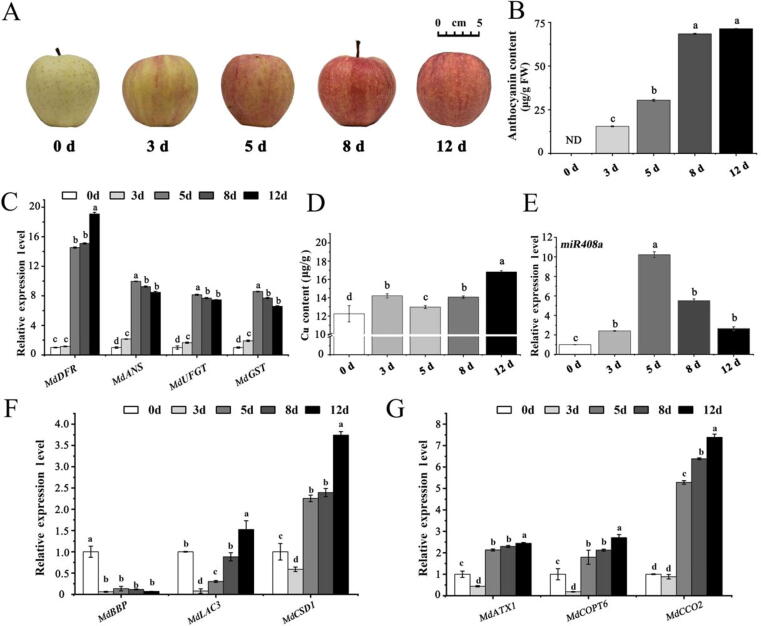
Fig. 4.
Characterization of apple fruit coloration after light treatment. (A) Phenotypes of apple fruit coloration. (B) Total anthocyanin contents in the samples. ND, not detected. (C) qRT–PCR analysis showing the relative expression of genes involved in the anthocyanin biosynthesis pathway. (D) Comparison of Cu content in the samples. (E) Relative fold changes in the abundance of miR408a in apple pericarp upon exposure to light. (F) Transcript levels of MdBBP, MdLAC3, and MdCSD1 in apple pericarp. (G) Transcript levels of MdATX1, MdCOPT6, and MdCCO2 in apple pericarp. Error bars indicate standard deviations (SD). a, b, and c indicate significant differences. Different lowercase letters denote significant differences (ANOVA, P < 0.05).
The miR408-BBP module regulates Cu homeostasis and anthocyanin biosynthesis in response to light signals in apple fruit
As shown in Fig. 4A and Fig. 4B, as the number of days of light increased, the apple colouration was significantly intensified in association with a significant increase in anthocyanin content. Consistently, as shown in Fig. 4C, among the anthocyanin biosynthesis genes, the expression of DFR, ANS, UFGT, and glutathione S-transferase (GST) increased significantly. Interestingly, both the abundance of miR408a and the Cu content were significantly upregulated under light induction (Fig. 4D and 4E). Moreover, the levels of Cu-binding protein gene transcripts, including BBP, LAC3, and CSD1, decreased significantly (Fig. 4F). To elucidate the relationship between Cu homeostasis and anthocyanin accumulation under light induction, the expression profiles of related structural genes involved in Cu transportation were determined using qRT-PCR. As shown in Fig. 4G, ATX1, COPT6, and CCO2 expressions were substantially upregulated 5 days after light treatment. These results are thought-provoking because they imply a positive correlation between Cu content and anthocyanin biosynthesis.
BBP negatively regulates the Cu homeostasis and anthocyanin biosynthesis in Malus plants
To investigate the roles of MdBBP in regulating Cu homeostasis and anthocyanin metabolism, we constructed an MdBBP overexpression vector (BBP-OE) and an MdBBP silencing vector (BBP-TRV). As shown in Fig. 5A, 5B, 5D(a), apple fruit overexpressing BBP showed reduced anthocyanin accumulation around the injection sites, accompanied by a significant decrease in the Cu content. Compared to BBP-OE, the opposite effects in apple fruit were observed in BBP-TRV (Fig. 5A, 5C, 5D (b)). As expected, the expression levels of CHS, DFR, UFGT, and GST decreased significantly in apple fruit infected with BBP-OE (Fig. 5E). However, the expression levels of CHS, ANS, DFR, UFGT, and GST increased significantly in apple fruit infected with BBP-TRV (Fig. 5H). Moreover, BBP expression was upregulated and downregulated significantly in apple fruit infected with BBP-OE or BBP-TRV, respectively (Fig. 5F). These results indicated that BBP prohibits anthocyanin biosynthesis, which might be due to the negative regulation of Cu homeostasis. Interestingly, the expression levels of MdLAC3, MdCSD1, and MdCSD in BBP-OE cells increased significantly (Fig. 5F). The expression of MdATX1, MdCOPT6, and MdCCO2 decreased significantly in BBP-OE, which is consistent with the decreased Cu content in BBP-OE (Fig. 5F). In addition, in BBP-OE, except for CAD, the expression of genes C4H, 4CL, CCR, CCoA-OMT, COMT, and CAD in the lignin pathway decreased significantly (Fig. 5G), indicating that BBP might negatively regulate phenylpropane metabolism by negatively regulating the Cu homeostasis.
Fig. 5.
Characterization of the transient transformation of BBP-OE and BBP-TRV in apple fruit. (A) Phenotypic comparison of apple fruit infected with different vectors. (B) Comparison of Cu content measured for Control-OE and BBP-OE (C) Comparison of Cu content measured for Control-TRV and BBP-TRV. (D) Anthocyanin contents in apple fruit peel infected with different vectors. (E and H) Quantitative analysis of miR408a, MdBBP, MdCHS, MdANS, MdDFR, MdUFGT, and MdGST transcript levels. (F and I) Quantitative analysis of MdLAC3, MdCSD1, MdATX1, MdCOPT6, and MdCCO2. (G and J) Expression of lignin pathway-related genes in apple fruit pericarp infected with different vectors. Error bars denote the standard deviations of three replicate measurements. Different lowercase letters denote significant differences (ANOVA, P < 0.05).
In BBP-TRV, MdLAC3, MdCSD1, and MdCSD expression decreased significantly (Fig. 5I). Furthermore, the expression of MdATX1, MdCOPT6, and MdCCO2 were substantially upregulated, which is consistent with the increased Cu content. Apart from CAD, the expression of C4H, 4CL, CCR, CCoA-OMT, and COMT in the lignin pathway increased significantly (Fig. 5J). Taken together, these results indicate that BBP prohibits anthocyanin biosynthesis by negatively regulating the Cu homeostasis.
The miR408a-BBP-LAC3/CSD1 module is involved in anthocyanin and lignin metabolism during the leaf colour transformation in Malus plants
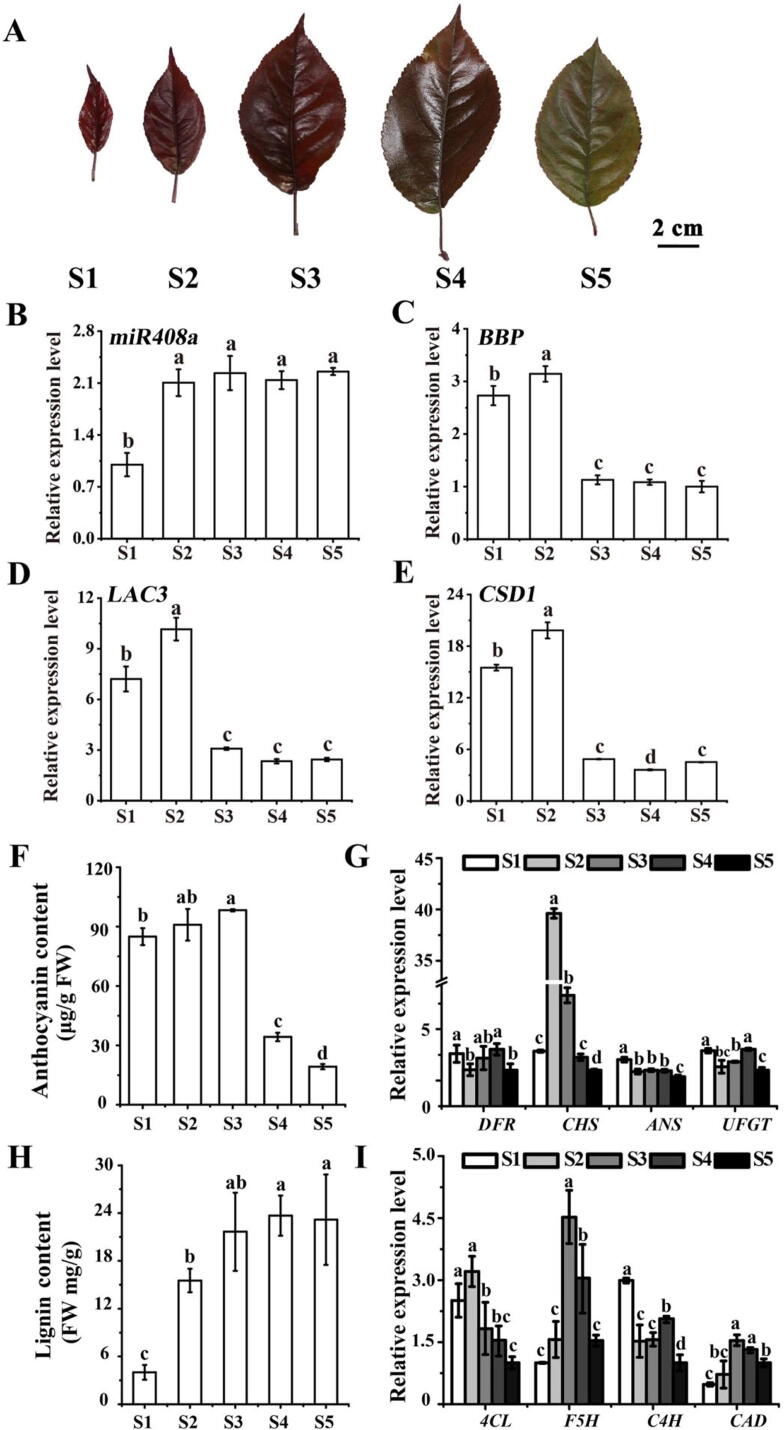
To determine whether miR408a, BBP, LAC3, and CSD1 are involved in the leaf colour transformation phenotype in Malus plants, we examined their expression levels, anthocyanin and lignin contents, and expression levels of genes involved in the anthocyanin and flavonoid pathways in Malus crabapple ‘Royalty’ leaves at five developmental stages (S1, S2, S3, S4, and S5). As shown in Fig. 6A, with the growth and development of leaves, the red colour of ‘Royalty’ leaves gradually became lighter, but the green background deepened. As shown in Fig. 6B, the abundance of miR408a was significantly upregulated from S2. Consistently, BBP expression was substantially downregulated from S2, which was contrary to the tendencies of the anthocyanin content and miR408a expression level (Fig. 6C). Interestingly, the expression patterns of both LAC3 and CSD1 were consistent with those of BBP (Fig. 6D and 6E), indicating that they might work together to synergistically regulate anthocyanin biosynthesis due to fluctuations in Cu homeostasis.
Fig. 6.
Analysis of ‘Royalty’ leaf coloration in five stages of leaf development in Malus crabapple ‘Royalty’. S1, 3 days after budding; S2, 9 days after budding; S3, 15 days after budding; S4, 21 days after budding; S5, 30 days after budding. (A) Leaf phenotypes in ‘Royalty’ at different stages (S1 to S5). (B) Analysis of the relative expression of miR408a in the leaves. (C) Relative expression of BBP in the leaves. (D) Relative expression of LAC3 in the leaves. (E) Relative expression of CSD1 in the leaves. (F) Total anthocyanin contents in ‘Royalty’ leaves at five developmental stages. (G) Expression analysis of anthocyanin pathway genes in ‘Royalty’ leaves at five developmental stages. (H) Quantification of total lignin content in ‘Royalty’ leaves at five developmental stages. (I) qRT–PCR analysis showing the relative expression of genes involved in the lignin pathway. Error bars indicate SD. Lowercase letters indicate significant differences.
Moreover, the red colour of ‘Royalty’ leaves was diluted in correlation with a sharp and significant decrease in the anthocyanin content from S3 (Fig. 6F). Interestingly, as suggested in Fig. 7F, the lignin content significantly increased throughout the periods of leaf development. Among anthocyanin biosynthesis genes, as shown in Fig. 6G, downward trends were observed for the gene expression of DFR, CHS, ANS, and UFGT. Concerning the common genes involved in anthocyanin and lignin biosynthesis, the expression levels of C4H and 4CL showed a downward trend. In the lignin biosynthesis pathway, the transcriptional levels of both F5H and CAD tended to be substantially up-regulated, resulting in increased lignin content (Fig. 6H and 6I). These results suggested a competitive relationship between lignin and anthocyanin accumulation, which might be related to leaf colour transformation and leaf senescence. Taken together, these results suggested that the miR408a-BBP-LAC3-CSD1 module might participate in regulating anthocyanin and lignin metabolism mediated by Cu homeostasis during the transformation of leaf colour in Malus plants.
Fig. 7.
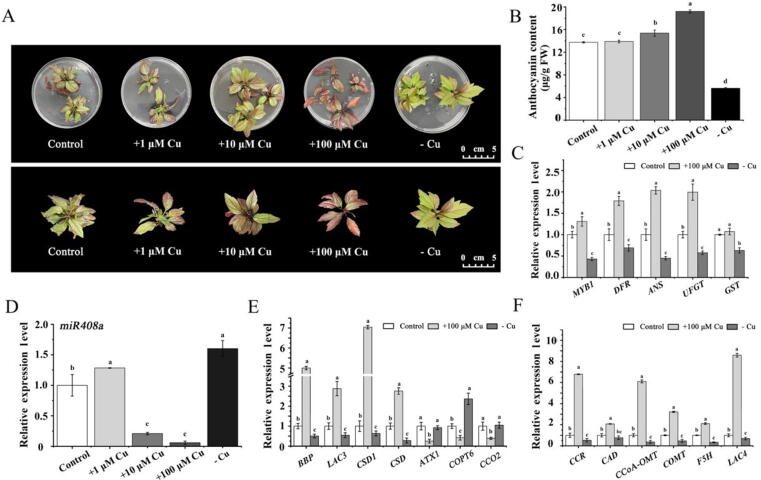
Effects of different Cu conditions on anthocyanin accumulation in ‘Royalty’ plantlets. (A) Phenotypic comparison of ‘Royalty’ plantlets after treatment with a lack of copper (-Cu), normal copper (control), and 100 μM copper (+100 μM Cu). (B) The total anthocyanin content in ‘Royalty’ plantlets under different Cu treatments. (C) Expression of anthocyanin pathway-related genes in leaves under different Cu treatments. (D) Expression levels of miR408a. (E) Expression levels of BBP, LAC3, CSD1, ATX1, COPT6, and CCO2 under different Cu conditions. (F) Expression of lignin pathway-related genes in leaves under different Cu treatments. Error bars denote the standard deviations of three replicate measurements. Different lowercase letters denote significant differences (ANOVA, P < 0.05).
The expression of miR408a and its target BBP is modulated under Cu stress
To determine whether anthocyanin biosynthesis regulated by miR408a and BBP is mediated by Cu homeostasis, ‘Royalty’ seedlings were subjected to treatment with copper at different concentrations. As shown in Fig. 8A, as the concentration of Cu increased, the leaves of ‘Royalty’ seedlings exhibited a deepened red color on the 15th day of treatment. Consistently, the anthocyanin contents in the leaves increased significantly (Fig. 7B). Excessive Cu might facilitate the transportation or stability of anthocyanins due to the protective and stabilizing effects of Cu ions on pigments. The expression levels of genes involved in anthocyanin biosynthesis were increased significantly, including DFR, ANS, UFGT, and GST, under 100 μM Cu conditions (Fig. 7C). The expression of miR408a decreased significantly with increasing concentrations of Cu, especially under the condition of 100 μM Cu (Fig. 7D). However, the transcript levels of BBP, LAC3, CSD1, and CSD, were increased significantly under 100 μM Cu conditions (Fig. 7E). The expression of the Cu transporter genes ATX1, COPT6, and CCO2 decreased significantly under 100 μM Cu conditions. These results indicated that the miR408a-BBP module might avoid the toxic effects of excessive Cu by reducing the transport of Cu. In addition, the expression of CCR, CAD, CCoA-OMT, COMT, F5H, and LAC4 in the lignin pathway also increased significantly under 100 μM Cu conditions (Fig. 7J).
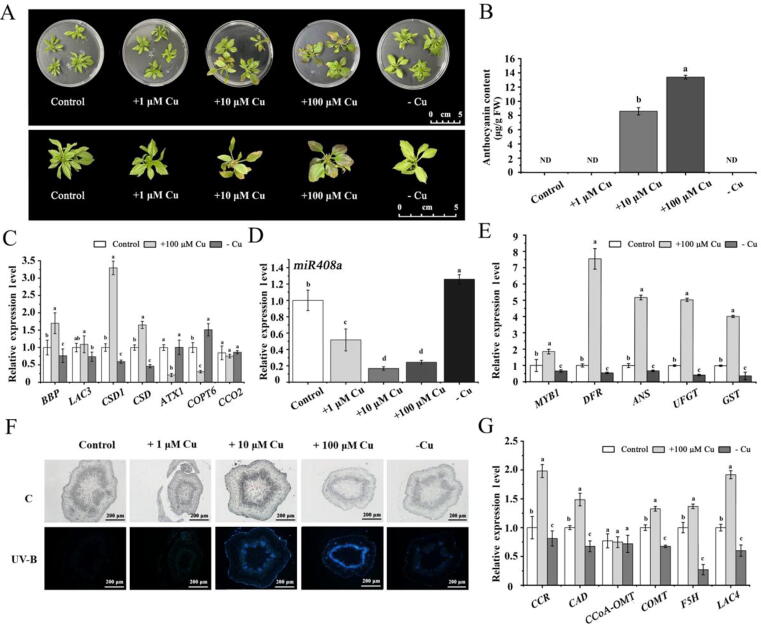
Fig. 8.
Effects of different Cu conditions on ‘Golden Delicious’ plantlets. (A) Phenotypic comparison of ‘Golden Delicious’ plantlets under different Cu concentrations. (B) The total anthocyanin content in ‘Golden Delicious’ plantlets under different Cu treatments. (C) Relative expression levels of BBP, LAC3, CSD1, ATX1, COPT6, and CCO2 in plantlet leaves under different Cu conditions. (D) Relative fold changes in the abundance of miR408a. (E) Expression of anthocyanin pathway-related genes in leaves under different Cu treatments. (F) Microscopic observation of stem tissue in ‘Golden Delicious’. (G) Expression of lignin pathway-related genes in leaves under different Cu treatments. Error bars denote the standard deviations of three replicate measurements. Different lowercase letters denote significant differences (ANOVA, P < 0.05).
In contrast, the leaf colour of ‘Royalty’ seedlings was green in the absence of Cu, and the anthocyanin content decreased significantly (Fig. 8A and 8B). The expression levels of anthocyanin biosynthesis genes decreased significantly, including CHS, DFR, UFGT, and GST (Fig. 7C). The expression of miR408a significantly increased under Cu deficiency, accompanied by a decrease in the BBP, LAC3, CSD1, and CSD expression levels (Fig. 7D). The expression of the Cu transporter gene COPT6 increased significantly under Cu deficiency (Fig. 7E), indicating that the miR408a-BBP module might maintain cellular Cu homeostasis by accelerating Cu transport. Moreover, the transcriptional levels of CCR, CAD, CCoA-OMT, COMT, F5H, and LAC4 in the lignin pathway markedly decreased under Cu deficiency conditions (Fig. 7F).
These results revealed that the expression levels of both miR408a and BBP were sensitive to fluctuations in the external Cu status. Both excessive Cu and Cu deficiency affected the expression of miR408a and BBP, leading to variations in anthocyanin accumulation. Taken together, these results suggest that the miR408a-BBP module is critical for the regulation of Cu homeostasis, as it affects the anthocyanin accumulation in Malus plants.
The miR408a-BBP-LAC3/CSD1 module is critical for the regulation of Cu homeostasis
To further investigate the roles of miR408a, BBP, LAC3, and CSD1 in regulating Cu homeostasis, ‘Golden Delicious’ plantlets were treated with Cu under various conditions. As shown in Fig. S1A and 1B, under conditions of both Cu deficiency and excessive Cu (100 μM Cu), the plantlets showed a dwarfing phenotype with a decreased stem length, indicating that both excessive Cu and Cu deficiency are not conducive to the nutritional growth of Malus plants. Interestingly, in the case of excessive Cu (10 μM Cu and 100 μM Cu), the leaves of ‘Golden Delicious’ seedlings exhibited a red colour at the blade edge. Consistently, the anthocyanin contents increased significantly (Fig. 8A and 8B). Consistent with the ‘Royalty’, the expression of miR408a decreased significantly in ‘Golden Delicious’ with increasing concentrations of Cu (Fig. 8D). However, the expression of BBP, LAC3, and CSD1 was increased significantly under 100 μM Cu (Fig. 8C). The expression of the Cu transporter genes ATX1 and COPT6 was decreased significantly under 100 μM Cu conditions (Fig. 8C). The transcriptional levels of CCR, CAD, COMT, F5H, and LAC4 in the lignin pathway markedly increased under 100 μM Cu conditions (Fig. 8G).
Cu deficiency significantly downregulated the expression of BBP, LAC3, CSD1, and CSD, and the pattern was opposite that of miR408a (Fig. 8C). However, the expression of the Cu transporter gene COPT6 increased significantly under Cu deficiency (Fig. 8C). Moreover, lignin accumulation was decreased under Cu deficiency conditions (Fig. 8F), whereas lignin accumulation increased significantly as the concentration of copper increased. Consistently, the transcriptional levels of CCR, CAD, COMT, F5H, and LAC4 in the lignin pathway markedly decreased under Cu deficiency conditions (Fig. 8G). Overall, these results further revealed that the miR408a-BBP-LAC3/CSD1 module is critical for the regulation of Cu homeostasis, thus regulating anthocyanin biosynthesis in Malus plants.
Discussion
Secondary metabolism plays extensive roles in plant growth, development, and responses to various biotic and abiotic stresses, and methods for coping with extracellular Cu changes and maintaining cellular Cu homeostasis are crucial for plant growth and development.
MiR408a and its target BBP regulate Cu homeostasis and anthocyanin biosynthesis in Malus plants
In Arabidopsis, miR408 was reported to promote anthocyanin biosynthesis [36]. Consistently, the differential expression of miR408a and its target BBP was shown to be involved in leaf colour transformation and apple fruit colouration in the current study. Overexpression of miR408a positively regulated anthocyanin biosynthesis and Cu content by increasing the expression of the Cu-binding genes LAC3, CSD1, and CSD, as well as the Cu transporter genes ATX1, COPT6, and CCO2. Contrasting results were obtained when miR408a was transiently silenced. Moreover, overexpression of BBP significantly reduced anthocyanin biosynthesis, coinciding with the downregulation of the Cu content, accompanied by the downregulation of Cu-binding genes LAC3, CSD1, and CSD, as well as transporter gene expression, including COPT6 and CCO2. These results demonstrated that miR408a targets BBP to regulate Cu homeostasis by regulating the binding and transportation of Cu, leading to anthocyanin accumulation in Malus plants. The latest research findings suggest that the intracellular Cu homeostasis mediated by the PCY-SAG14 phytocyanin module repressed by miR408 promotes dark-induced leaf senescence in Arabidopsis [68]. Further study needs to be performed to understand the Cu homeostasis regulation mechanism that would provide new insights into apple fruit quality formation.
Cu homeostasis affects anthocyanin accumulation and growth in Malus plants
Cu homeostasis is critical for the plant growth and development. Under conditions of excess Cu and Cu starvation, the ‘Golden Delicious’ plantlets showed a dwarfing phenotype, and the plant height was decreased significantly, suggesting that cellular Cu homeostasis is associated with alteration of the extracellular Cu status and that plant growth is sensitive to the undulation of cellular Cu homeostasis. In the current study, we found that BBP is bound directly to LAC3 and CSD1.
Cu can stabilize chlorophyll and other pigments. In grape berries, the comparative transcriptomic analysis revealed that Cu amine oxidase and 4-coumarate-CoA ligase played essential roles in total anthocyanin variations induced by bud sport [69]. In barley, transcriptome analysis disclosed that tolerance to cobalt and Cu is associated with anthocyanin biosynthesis pathways, as well as the MAPK signalling, glutathione biosynthesis, phenylalanine metabolism, and photosynthesis [5]. Consistently, 100 μM Cu significantly enhanced leaf colour and promoted anthocyanin accumulation in Malus ‘Royalty’ and ‘Golden Delicious’ plantlets, indicating that Cu plays an important role in the biosynthesis of anthocyanins. These findings are consistent with those of a previous study reporting that anthocyanins accumulate at high levels under sufficient Cu and high-light conditions in Arabidopsis [36]. However, anthocyanins accumulation was decreased significantly under Cu deficiency. AtACS8 is elicited by Cu ions and plays a critical role in the early biosynthesis of ethylene in Arabidopsis [69]. Thus, we hypothesized that the promotion of anthocyanin accumulation induced by Cu might be associated with the induction of ethylene.
On the other hand, Cu ions might have a stable protective effect on anthocyanins, which is conducive to their transport from the cytoplasm into vacuoles. BBP could be exactly localized in tonoplast. Thus, we speculated that the significant upregulation of BBP, LAC3, CSD1, and CSD expression under excessive Cu might be beneficial to the transfer of Cu ions from the cytoplasm into the vacuole, resulting in more sufficient Cu ions being stored in the vacuole and enhancing the stability of anthocyanins in the vacuole. Another possibility is the existence of an unknown cotransport mechanism by which anthocyanins are transferred into vacuoles, accompanied by Cu ions acting as cofactors of GST. Therefore, we hypothesize that Cu homeostasis plays a role in temporal leaf colour or fruit colouration with growth and development in Malus plants.
The miR408a-BBP module might play a central role in the regulation of Cu homeostasis
In Arabidopsis, plants accumulate miR397, miR408, and miR857 in response to low Cu availability, which mediates the systemic downregulation of Cu protein expression, including plantacyanin and members of the LAC Cu-containing protein family, indicating a general mechanism by which miRNAs negatively regulate those nonessential Cu proteins at the posttranscriptional level [41]. It was reported that miR408 and its targets are crucial to the HY5-SPL7 module in regulating Cu homeostasis-related genes in Arabidopsis [36], [70], [71].
Moreover, the differential expression of miR408 and its target genes in response to changing light and copper conditions is associated with the allocation of Cu in chloroplasts and with the PC levels in Arabidopsis [36]. In oceanic diatoms, the majority of differentially regulated genes are associated with photosynthetic metabolism, suggesting that chloroplasts are the primary target of low Cu availability [72]. In Arabidopsis, under Cu exposure, the expression of Cu transporter (COPT) genes was shown to be down-regulated, indicating that the relocation and redistribution of Cu ions may be an important mechanism for regulating Cu homeostasis in plants under Cu stress [73]. In our study, under Cu starvation conditions, the expression levels of miR408a and its target BBP were upregulated and downregulated in both ‘Royalty’ and ‘Golden Delicious’ plant leaves, respectively. When exposed to excess Cu, the opposite results were observed, indicating that miR408a and BBP are very sensitive to the extracellular Cu environment; this phenomenon could be a hallmark of Cu homeostasis in Malus plants. Moreover, overexpression of miR408a significantly downregulated the abundance of BBP in Malus plants, whereas transiently silencing miR408a substantially increased BBP expression. When BBP expression was undermined, the physiological Cu content was increased significantly, and the anthocyanin content was increased in Malus plants, accompanied by a reduced expression of the Cu transporter genes ATX1, COPT6, and CCO2. However, the opposite results were observed under conditions of BBP overexpression. These results indicated that BBP negatively regulates cellular Cu homeostasis in plants as a target of miR408.
In addition, the interaction among BBP, CSD1, and LAC3 might occur on the cell membrane and tonoplast, which may coordinately participate in the binding of Cu ions and affect the transport of extracellular Cu ions into the cell to maintain intracellular Cu homeostasis under Cu deficiency or excess Cu conditions. Interestingly, bioinformatic analysis showed that the BBP protein has a transmembrane structure, and subcellular localization experiments revealed that it was localized at the cell membrane and vacuolar membranes, suggesting that BBP recruits or cooperates with other Cu transporters or copper-containing proteins to regulate Cu homeostasis. Thus, BBP protein might play an important role in the shuttling of Cu ions between vacuoles and the cytoplasm, and transmitting extracellular and intracellular Cu signals. Our results suggested that silencing BBP significantly decreased the expression levels of other Cu-containing proteins, including CSD1 and LAC3, and increased the cellular Cu content, indicating that BBP might be located upstream of these proteins and thus play a more central role in regulating Cu homeostasis.
The expression of the Cu chaperone COPT2 was upregulated in leaves under the copper and zinc deficiency. Under Cu-deficient conditions, the expression levels of ATOPT3 and three nicotinamide synthase genes were up-regulated in roots [74]. Consistently, in our study, the expression levels of the Cu transporter COPT6 were upregulated significantly under Cu-deficient conditions in Malus plants. Recently, MdWRKY11 was reported to enhance the Cu tolerance by directly promoting the expression of Cu transporter MdHMA5 in apples [75]. Nevertheless, whether and how BBP works together with Cu transport proteins or Cu chaperones to precisely regulate Cu transmembrane transport and thereby maintain cellular Cu homeostasis still needs to be further studied. Taken together, these results suggested that the miR408a-BBP module plays an important role in regulating the intracellular Cu ion concentration and homeostasis by modulating the cellular localization, transport, and storage of Cu ions in the cell.
The module consisting of miR408a, BBP, CSD1, and LAC3 coordinately regulates the crosstalk between Cu homeostasis and ROS homeostasis
Due to the disturbance or destruction of Cu homeostasis, Cu deficiency prevents photosynthesis in plants, and excessive Cu triggers the production and diffusion of harmful ROS, resulting in oxidative damage to plant cells [30]. ROS homeostasis is known to be widely involved in the responses of plants to adverse stresses. Transcriptomic analysis revealed that the expression levels of genes related to oxidative stress defence systems were up-regulated in oceanic diatom cells, indicating that increased ROS triggers cellular damage under Cu-deficient conditions [53]. In Hematococcus pluvialis, the application of Cu increased the transcriptional expression of antioxidant enzyme-related genes and led to a decrease in ROS levels [76]. These studies indicated that the Cu content affects the levels of ROS. Under high Cu concentrations, plants are exposed to excessive Cu-derived cellular toxicity, and excessive Cu damages cellular membrane systems and various organelles and inhibits electron transport in the photosynthesis chain. In our study, the interaction between BBP and CSD1 occurred on the cell membrane, implying that the regulation of Cu homeostasis might be coupled with ROS homeostasis.
The most abundant Cu proteins are PC in thylakoids and CSD, of which the major isoforms are found in the cytosol and the chloroplast stroma. In Arabidopsis, CSD1 and CSD2 were shown to be targeted by miR398 and to be important for oxidative stress tolerance [77], [78]. In Arabidopsis thaliana, excess Cu resulted in a reduced protein level and reduced enzymatic activity of CSD1 in siz1, indicating that the expression of CSD1 was influenced by Cu stress [31]. In our study, the application of excess Cu (10 μM, 100 μM) significantly upregulated the expression of BBP, LAC3, and CSD1 in ‘Golden Delicious’ plantlets, resulting in increased binding of Cu ions and quick movement into vacuoles to maintain cellular Cu homeostasis. As expected, the application of exogenous 100 μM Cu promoted anthocyanin accumulation in ‘Golden Delicious’ and ‘Royalty’ leaves, indicating that excessive Cu induced relatively high ROS accumulation and ROS signalling, leading to the biosynthesis and accumulation of antioxidants, such as anthocyanins; this result supports the existence of a relationship between Cu stress and antioxidant metabolism [79], [80]. Anthocyanins are beneficial to the removal of ROS, further denoting the crosstalk between Cu homeostasis and ROS homeostasis.
The downregulation of CSD under Cu limitation was observed in A. thaliana, Brassica juncea, Lycopersicum lycopersicum, Zea mays, and Oryza sativa [81], indicating that the expression of CSD was also sensitive to Cu availability. Consistently, under Cu-deficient conditions, BBP, LAC3, and CSD1 were downregulated significantly to decrease the binding of Cu ions, thereby increasing the cellular Cu content in plants. These results further corroborate that the interaction between BBP and CSD1 mediates the crosstalk between Cu homeostasis and ROS homeostasis and suggest that the expression levels of Cu/ZnSOD enzymes are good indicators of excessive Cu or Cu deficiency. In addition, miR408 was identified to be significantly induced during leaf senescence in Arabidopsis [52,82]. Our results demonstrated that the expression levels of miR408a and BBP gradually increased and decreased, respectively, during leaf growth and senescence. The expression of BBP, CSD1, and LAC3 was downregulated consistently during leaf development and senescence, accompanied by a reduction in anthocyanin accumulation and upregulation of lignin biosynthesis. This result indicates the downregulation of Cu availability, which has important biological significance for the recycling of Cu and serves as a good explanation for the transfer of Cu from old to young leaves in plants. Further investigation revealed strong interactions between BBP and LAC3 and between BBP and CSD1, indicating that BBP might be a hub protein linking Cu homeostasis with ROS homeostasis.
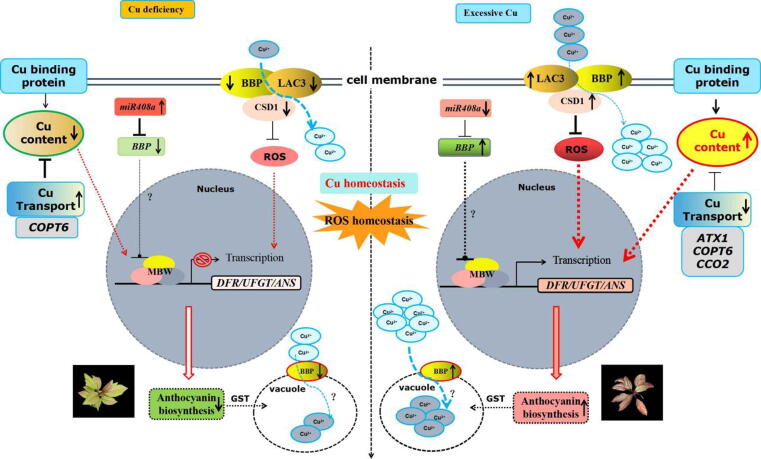
However, COPT6 expression increases and promotes transportation to Cu ions under Cu deficiency to maintain Cu homeostasis. Under conditions of Cu deficiency, the expression levels of miR408a and its target BBP were significantly upregulated and downregulated, respectively. Moreover, the levels of LAC3 and CSD1 are significantly decreased, leading to reductions in the absorption of extracellular Cu. Finally, the decreased abundance of BBP protein localized at the tonoplast weakens the binding of Cu ions, thereby reducing the transport of copper ions from the cytoplasm to the vacuole. Meanwhile, the expression levels of the Cu transport gene COPT6 promote Cu transportation, thus maintaining the intracellular Cu level to alleviate damage to plant growth and development induced by Cu deficiency. In a word, the miR408a-BBP-LAC3/CSD1 module that mediates the crosstalk between Cu homeostasis and ROS homeostasis to regulate anthocyanin biosynthesis in Malus during copper stress.
A model for miR408a-BBP-LAC3/CSD1-regulated Cu homeostasis and ROS homeostasis regulating anthocyanin accumulation under Cu stress in Malus plant.
Based on the results and observations described above, we proposed a working model including miR408a, BBP, LAC3, and CSD1 that perceives and integrates extracellular Cu to regulate Cu homeostasis and ROS homeostasis, thereby affecting anthocyanin accumulation in Malus plants (Fig. 9). Under conditions of excess Cu, the expression levels of miR408a and its target BBP were significantly downregulated and upregulated, respectively. At the same time, the expression levels of LAC3 and CSD1, which interact with and might recruited by BBP to synergistically increase the absorption of extracellular Cu, are increased significantly, thereby increasing the intracellular Cu content. At the same time, the expression levels of the Cu transport genes ATX1, COPT6, and CCO2 were decreased significantly to reduce Cu transportation to avoid the toxic effects of excessive Cu. Moreover, BBP localized to the tonoplast enhance the binding of Cu ions, thereby promoting the transport of Cu ions in the cytoplasm to the vacuole and restoring intracellular Cu homeostasis to supply the Cu needed for plant growth and development. The upregulation of CSD1 expression might clear the large amounts of ROS induced by excessive Cu, which also promotes antioxidant anthocyanin accumulation. Under conditions of Cu deficiency, the expression levels of miR408a and its target BBP were significantly upregulated and downregulated, respectively. Moreover, the levels of LAC3 and CSD1 are significantly decreased, leading to reductions in the absorption of extracellular Cu. Finally, the decreased abundance of BBP protein localized at the vacuolar membrane weakens the binding of copper ions, thereby reducing the transport of copper ions from the cytoplasm to the vacuole. Meanwhile, the expression levels of the Cu transport gene COPT6 promote Cu transportation, thus maintaining the intracellular Cu level to alleviate damage to plant growth and development induced by copper deficiency. Based on these results, we proposed a working model that the miR408a-BBP-LAC3/CSD1 module regulate the anthocyanin accumulation under conditions of Cu excess and Cu deficiency in Malus plant leaves.
Fig. 9.
A working model for the mechanism by which the miR408a-BBP-LAC3/CSD1 module regulates Cu homeostasis and anthocyanin accumulation under Cu deficiency and excessive Cu in Malus Plant. Under conditions of excessive Cu, miR408a perceives an excessive Cu signal and shows a significant downregulation, resulting in the upregulation of its target BBP expression. The expression levels of other Cu-binding proteins CSD1 and LAC3 increased synergistically, increasing the intracellular Cu content and anthocyanin accumulation characterized by red color in leaves. At the same time, the expression levels of Cu transport genes were decreased significantly to inhibit the toxic effects of excessive Cu. Under Cu deficiency conditions, the expression levels of miR408a and its target BBP were significantly upregulated and downregulated, respectively. The expression levels of CSD1 and LAC3 decreased synergistically. Meanwhile, the increase in Cu transport protein expression restored cellular Cu homeostasis. A continuous decrease in intracellular Cu content reduces the anthocyanin biosynthesis characterized by a green color in leaves. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
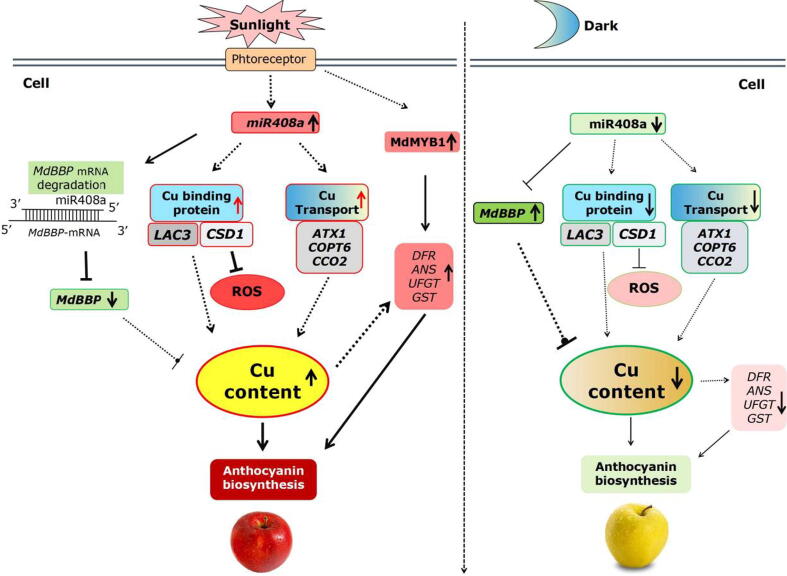
A model for miR408a-BBP-LAC3/CSD1-regulated Cu homeostasis and anthocyanin accumulation during light induction in apple fruit.
Based on the above results, we propose that the miR408a-BBP-LAC3/CSD1 module regulates Cu homeostasis and anthocyanin accumulation during light induction in apple fruit (Fig. 10). When apple fruit perceived light signals after bag removal, the expression of the miR408a and BBP was significantly upregulated and downregulated, respectively, leading to an increase in Cu content. Moreover, the levels of Cu-binding protein LAC3 and CSD1, as well as the Cu transport protein ATX1, COPT6, and CCO2 transcripts, increased significantly. Consequently, these changes eventually lead to a substantial increase in Cu content in the apple pericarp. In addition, the levels of CSD1 transcript increased significantly during light induction, leading to a reduction in ROS levels. Finally, relatively high Cu status facilitates the anthocyanin biosynthesis gene expression and apple colouration. In short, these results showed that the miR408a-BBP-LAC3/CSD1 module also regulates anthocyanin biosynthesis in apple fruit involved in the fluctuation in both Cu homeostasis and ROS homeostasis during light induction. This highlights the important role of Cu homeostasis play in apple colouration during light induction.
Fig. 10.
A working model for the mechanism by which the miR408a-BBP-LAC3/CSD1 module regulates anthocyanin accumulation during light induction in apple fruit. During light induction, miR408a and BBP expression are significantly upregulated and downregulated, respectively, resulting in the upregulation of Cu content. On the other hand, the upregulation of Cu-binding protein (LAC3, CSD1) and Cu transport protein (ATX1, COPT6, CCO2) also strengthened the promotion of the Cu level. In addition, the levels of CSD1 transcripts increased significantly during light induction, leading to a reduction in ROS levels. Eventually, the relatively high Cu status facilitates anthocyanin biosynthesis gene expression and apple coloration. Under dark conditions, the expression of miR408a and BBP is downregulated and upregulated, respectively, accompanied by the downregulation of Cu-binding protein and Cu transport protein expression, leading to a reduction in Cu homeostasis and anthocyanin accumulation.
Conclusion
This study reveals that BBP targeted by miR408a is the key regulator of Cu and ROS homeostasis through interacting with LAC3 and CSD1, which links the light signal and Cu signal involved in anthocyanin biosynthesis in Malus plants. In addition, this study elucidated the mechanism of the miR408a-BBP-LAC3/CSD1 module regulating anthocyanin biosynthesis under light induction and Cu stress and provide a new insights into the the crosstalk between Cu homeostasis and ROS homeostasis during plant growth and development.
Compliance with ethics requirements
All the authors state that this article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 31901997), the General Project of the Scientific Research Program of the Beijing Municipal Commission of Education (KM202010020013), the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032), the Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing University of Agriculture, and the Beijing Collaborative Innovation Center for Eco-Environmental Improvement with Forestry and Fruit Trees.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.11.005.
Contributor Information
Jie Zhang, Email: anyzj034@126.com.
Yuncong Yao, Email: yaoyc_20@126.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
The gene sequences listed in this article are available in the GenBank data libraries with the following accession numbers: miR408a (LOC103344813), MdBBP (LOC103402332), MdLAC3 (LOC103447291), MdCSD1 (LOC103413973), MdATX1 (LOC103408642), MdCOPT6 (103419433) and MdCCO2 (LOC103448680).
References
- 1.Shang Y., Venail J., Mackay S., Bailey P.C., Schwinn K.E., Jameson P.E., et al. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011;189(2):602–615. doi: 10.1111/j.1469-8137.2010.03498.x. [DOI] [PubMed] [Google Scholar]
- 2.Yousuf B., Gul K., Wani A.A., Singh P. Health benefits of anthocyanins and their encapsulation for potential use in food systems: a review. Cri Rev in Food Sci. 2016;56:2223–2230. doi: 10.1080/10408398.2013.805316. [DOI] [PubMed] [Google Scholar]
- 3.Macar O., Kalefetoğlu Macar T., Çavuşoğlu K., Yalçın E. Protective effects of anthocyanin-rich bilberry (Vaccinium myrtillus L.) extract against copper(II) chloride toxicity. Environ Sci Pollut R. 2020;27:1428–1435. doi: 10.1007/s11356-019-06781-9. [DOI] [PubMed] [Google Scholar]
- 4.Peng Z., Tian J., Luo R., Kang Y., Lu Y., Hu Y., et al. MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ. 2019;43:1148–1159. doi: 10.1111/pce.13697. [DOI] [PubMed] [Google Scholar]
- 5.Lwalaba J.L.W., Zvobgo G., Gai Y., Issaka J.H., Mwamba T.M., Louis L.T., et al. Transcriptome analysis reveals the tolerant mechanisms to cobalt and copper in barley. Ecotoxicol Environ Saf. 2021;209 doi: 10.1016/j.ecoenv.2020.111761. [DOI] [PubMed] [Google Scholar]
- 6.Axtell M.J., Bowman J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Jones-Rhoades M.W., Bartel D.P., Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Bio. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 9.Ramachandran V., Chen X.M. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;2008(13):368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S., Liu L., Zhuang X., Yu Y., Liu X., Cui X., et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Sunkar R., Li Y.F., Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physio. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F., Jing C., Yi Y., Liu Z. Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tiss Org. 2013;115(2):159–167. [Google Scholar]
- 15.Jia X., Shen J., Liu H., Li F., Ding N., Gao C., et al. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta. 2015;242:283–293. doi: 10.1007/s00425-015-2305-5. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y., Cheng H., Zhang Y., Zhang J., Niu S., Wang X., et al. The MdMYB16/MdMYB1-miR7125-MdCCR module regulates the homeostasis between anthocyanin and lignin biosynthesis during light induction in apple. New Phytol. 2021;231:1105–1122. doi: 10.1111/nph.17431. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y.F., Tao Y.L., Zhu C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot. 2013;64:3077–3086. doi: 10.1093/jxb/ert164. [DOI] [PubMed] [Google Scholar]
- 18.Lin J.S., Lin C.C., Lin H.H., Chen Y.C., Jeng S.T. MicroR828 regulates lignin and H2O2 accumulation in sweet potato on wounding. New Phytol. 2012;196:427–440. doi: 10.1111/j.1469-8137.2012.04277.x. [DOI] [PubMed] [Google Scholar]
- 19.Chávez-Hernández E.C., Alejandri-Ramírez N.D., Juárez-González V.T., Dinkova T.D. Maize mirna and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front Plant Sci. 2015;6:555. doi: 10.3389/fpls.2015.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C., Li D., Mao D., Liu X., Ji C., Li X., et al. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.) Plant Cell Environ. 2013;36:2207–2218. doi: 10.1111/pce.12130. [DOI] [PubMed] [Google Scholar]
- 21.Zhou M., Li D., Li Z., Hu Q., Yang C., Zhu L., et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physio. 2013;161:1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y., Xue H., Zhang F., Jiang Q., Yang S., Yue P., et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol J. 2020;19:311–323. doi: 10.1111/pbi.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiou T.J. The role of micrornas in sensing nutrient stress. Plant Cell Environ. 2010;30:323–332. doi: 10.1111/j.1365-3040.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 24.Matthewman C.A., Kawashima C.G., Húska D., Csorba T., Dalmay T., Kopriva S. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 2012;586:3242–3248. doi: 10.1016/j.febslet.2012.06.044. [DOI] [PubMed] [Google Scholar]
- 25.Liu T.Y., Lin W.Y., Huang T.K., Chiou T.J. MicroRNA-mediated surveillance of phosphate transporters on the move. Trends Plant Sci. 2014;19:647–655. doi: 10.1016/j.tplants.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Zeng H., Wang G., Hu X., Wang H., Du L., Zhu Y. Role of microRNAs in plant responses to nutrient stress. Plant Soil. 2014;374:1005–1021. [Google Scholar]
- 27.Sun Q, Liu X, Yang J, Liu W, Du Q, Wang H. et al. microRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol Plant 2018:S167420521830100X. [DOI] [PubMed]
- 28.Marschner H. 2nd edn. Academic Press; London: 1995. Mineral Nutrition in Higher Plants; pp. 79–115. [Google Scholar]
- 29.Burkhead J.L., Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. Copper homeostasis. New Phytol. 2009;182:799–816. doi: 10.1111/j.1469-8137.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen C.C., Chen Y.Y., Yeh K.C. Effect of Cu content on the activity of Cu/ZnSOD1 in the Arabidopsis SUMO E3 ligase siz1 mutant. Plant Signal Behav. 2011;6(10):1428–1430. doi: 10.4161/psb.6.10.16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K.K., Pilon M. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell. 2003;15:1333–1346. doi: 10.1105/tpc.011817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Axtell M.J., Bowman J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;2011(39):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., He H., Wang X., Wang X., Yang X., Li L., et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65(3):346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H., Zhao X., Li J., Cai H., Deng X.W., Li L. MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell. 2014;26:4933–4953. doi: 10.1105/tpc.114.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nersissian A.M., Immoos C., Hill M.G., Hart P.J., Williams G., Herrmann R.G., et al. Uclacyanins, stellacyanins, and plantacyanins ape distinct subfamilies of phytocyanins: plant-specific mononuclear blue copper proteins. Protein Sci. 1998;7:1915–1929. doi: 10.1002/pro.5560070907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi M., Davidson V.L. Cupredoxins-A study of how proteins may evolve to use metals for bioenergetic processes. Metallomics. 2011;2011(3):140–151. doi: 10.1039/c0mt00061b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., et al. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuetz M., Benske A., Smith R.A., Watanabe Y., Tobimatsu Y., Ralph J., et al. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014;166:798–807. doi: 10.1104/pp.114.245597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Ghany S.E., Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Bio Chem. 2008;283(23):15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J.P., Yu Y., Feng Y.Z., Zhou Y.F., Zhang F., Yang Y.W., et al. MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 2017;2017(175):1175–1185. doi: 10.1104/pp.17.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan J., Huang D., Guo Z., Kuang Z., Zhang H., Xie X., et al. Overexpression of miR408 enhances photosynthesis, growth, and seed yield in diverse plants. JIPB. 2018;60:61–78. doi: 10.1111/jipb.12634. [DOI] [PubMed] [Google Scholar]
- 43.Ma C., Burd S., Lers A. MiR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84:169–187. doi: 10.1111/tpj.12999. [DOI] [PubMed] [Google Scholar]
- 44.Candar-Cakir B., Arican E., Zhang B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J. 2016;14:1727–1746. doi: 10.1111/pbi.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo X.R., Niu J.F., Cao X.Y. Heterologous expression of salvia miltiorrhiza microRNA408 enhances tolerance to salt stress in nicotiana benthamiana. Int J Mol Sci. 2018;19(12):3985. doi: 10.3390/ijms19123985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuo Y.W., Lin J.S., Li Y.C., Jhu M.Y., King Y.C., Jeng S.T. MicroR408 regulates defense response in sweet potato upon wounding. J Exp Bot. 2018;70(2):469–483. doi: 10.1093/jxb/ery381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinbothe S., Reinbothe C., Apel K., Lebedev N. Evolution of chlorophyll biosynthesis-The challenge to survive photooxidation. Cell. 1996;86(5):703–705. doi: 10.1016/s0092-8674(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 48.Yang L., Zhang J., He J., Qin Y., Hua D., Duan Y., et al. Aba-mediated ros in mitochondria regulate root meristem activity by controlling plethora expression in arabidopsis. PLoS Genet. 2014;10:e1004791. doi: 10.1371/journal.pgen.1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., et al. Antagonistic basic helix-loop-helix/bzip transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell. 2013;25:1657–1673. doi: 10.1105/tpc.112.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Q., Su N., Zhang X., Liu Y., Cui J., Liang Y. Hydrogen peroxide, nitric oxide and uv resistance locus8 interact to mediate UV-B-induced anthocyanin biosynthesis in radish sprouts. Sci Rep. 2016;6:29164. doi: 10.1038/srep29164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thatcher S.R., Burd S., Wright C., Lers A., Green P.J. Differential expression of miRNAs and their target genes in senescing leaves and siliques: insights from deep sequencing of small RNAs and cleaved target RNAs. Plant Cell Environ. 2015;38:188–200. doi: 10.1111/pce.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong L.L., Price N.M. Transcriptomes of an oceanic diatom reveal the initial and final stages of acclimation to copper deficiency. Environ Microbiol. 2022;24(2):951–966. doi: 10.1111/1462-2920.15609. [DOI] [PubMed] [Google Scholar]
- 53.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔ C T method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 55.Bruce R.J., West C.A. Elicitation of lignin biosynthesis and isoperoxidase activity by pectic fragments in suspension cultures of castor bean. Plant Physiol. 1989;1989(91):889–897. doi: 10.1104/pp.91.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Revilla E., Ryan J.M. Analysis of several phenolic compounds with potential antioxidant properties in grape extracts and wines by high-performance liquid chromatography-photodiode array detection without sample preparation. J Chromatogr A. 2000;881(1):461–469. doi: 10.1016/s0021-9673(00)00269-7. [DOI] [PubMed] [Google Scholar]
- 57.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. BLAST+: architecture and applications. BMC Bioinf. 2009;10(1):421–430. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hofmann K., Stoffel W. TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 61.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein Identification and Analysis Tools on the ExPASy Server; (In) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press. pp. 571-607.
- 62.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Bio. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 63.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 64.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. Edited in Evolving Genes and Proteins by V. Bryson and H.J. Vogel, pp. 97-166. Academic Press, New York. [DOI] [PubMed]
- 66.Chou K.C., Shen H.B. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE. 2010;5:e11335. doi: 10.1371/journal.pone.0011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hao C, Yang YZ, Du JM, Deng XW, Li Lei. The PCY-SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. Proc Natl Acad Sci U S A 2022;119(3). https://doi.org/10.1073/pnas.2116623119. [DOI] [PMC free article] [PubMed]
- 68.Zhang B, Liu H, Ding X, Qiu J, Zhang M, Chu Z. 2018. Arabidopsis thaliana ACS8 plays a crucial role in the early biosynthesis of ethylene elicited by Cu2+ ions. J Cell Sci 2018; 131(2): jcs.202424. [DOI] [PubMed]
- 69.Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. SQUAMOSA promoter binding protein-like 7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell. 2009;2009(21):347–361. doi: 10.1105/tpc.108.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Araki R., Mermod M., Yamasaki H., Kamiya T., Fujiwara T., Shikanai T. SPL7 locally regulates copper-homeostasis-related genes in Arabidopsis. J Plant Physiol. 2018:224–225. doi: 10.1016/j.jplph.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 71.Kong L.L., Price N.M. Transcriptomes of an oceanic diatom reveal the initial and final stages of acclimation to copper deficiency. Environ Microbiol. 2022;; 24(2):951–966 doi: 10.1111/1462-2920.15609. [DOI] [PubMed] [Google Scholar]
- 72.Amaral Dos Reis R., Hendrix S., Mourato M.P., Louro Martins L., Vangronsveld J., Cuypers A. Efficient regulation of copper homeostasis underlies accession-specific sensitivities to excess copper and cadmium in roots of Arabidopsis thaliana. J Plant Physiol. 2021;2021(261) doi: 10.1016/j.jplph.2021.153434. [DOI] [PubMed] [Google Scholar]
- 73.Utz M., Andrei A., Milanov M., Trasnea P.I., Marckmann D., Daldal F., et al. The Cu chaperone copz is required for cu homeostasis in rhodobacter capsulatus and influences cytochrome cbb3 oxidase assembly. Mol Microbiol. 2019;111:764–783. doi: 10.1111/mmi.14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi K., Liu X., Zhu Y., Bai Y., Shan D., Zheng X., et al. MdWRKY11 improves copper tolerance by directly promoting the expression of the copper transporter gene MdHMA5. Hortic Res. 2020;7:105. doi: 10.1038/s41438-020-0326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo H., Li T., Zhao Y.T., Yu X.Y. Role of copper in the enhancement of astaxanthin and lipid coaccumulation in Haematococcus pluvialis exposed to abiotic stress conditions. Bioresour Technol. 2021;335(5) doi: 10.1016/j.biortech.2021.125265. [DOI] [PubMed] [Google Scholar]
- 76.Sunkar R., Kapoor A., Zhu J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;; 18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagae M., Takahashi Y. Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 2008;146(4):1687–1696. doi: 10.1104/pp.107.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neill S.O., Gould K.S., Neil S.O., Gould K.S. Anthocyanins in leaves: Light attenuators or antioxidants? Funct Plant Biol. 2003;30(8):865–873. doi: 10.1071/FP03118. [DOI] [PubMed] [Google Scholar]
- 79.Merzlyak M.N., Chivkunova O.B., Solovchenko A.E., Naqvi K.R. Light absorption by anthocyanins in juvenile, stressed, and senescing leaves. J Exp Bot. 2008;59:3903–3911. doi: 10.1093/jxb/ern230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cohu C.M., Pilon M. Regulation of superoxide dismutase expression by copper availability. Physiol Plantarum. 2007;129:747–755. [Google Scholar]
- 81.Zhang H., Li L. SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 2013;74:98–109. doi: 10.1111/tpj.12107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene sequences listed in this article are available in the GenBank data libraries with the following accession numbers: miR408a (LOC103344813), MdBBP (LOC103402332), MdLAC3 (LOC103447291), MdCSD1 (LOC103413973), MdATX1 (LOC103408642), MdCOPT6 (103419433) and MdCCO2 (LOC103448680).