Abstract
Background: The prognostic assessment and therapeutic interventions of esophageal cancer (ESCA) require novel molecular targets. The prognostic value of necroptosis, a specific mode of programmed cell death strongly linked to cancer progression, remains largely unexplored in ESCA. The primary goal of this research is to develop a necroptosis-based prognostic signature, which will represent the microenvironmental characteristics and prognosis of individuals diagnosed with ESCA. Methods: Transcriptome data of ESCA samples from The Cancer Genome Atlas were utilized to screen for necroptosis-related long non-coding RNAs (NR-lncRNAs) and genes (NRGs). The research employed the least absolute shrinkage and selection operator (LASSO) regression and univariate Cox regression analysis to identify prognostic candidates. Based on these analyses, a signature was developed in the training set and subsequently verified in the testing and entire sets. A clinicopathologic relevance assessment was carried out, after which a nomogram was established. The features of the immune microenvironment, functional pathways, mutational burden, checkpoint expression, and stemness of tumors were analyzed. Moreover, the sensitivity of individuals to immunotherapy and chemotherapy was compared for therapeutic guidance. Results: A necroptosis-associated signature comprising two genes and eleven lncRNAs was constructed. High-risk patients showed worse prognosis and clinicopathologic features, with more tumor-infiltrating naïve B cells, CD4+ memory resting T cells, and regulatory T cells. Furthermore, stromal and ESTIMATE scores were decreased along with increased stemness scores and tumor mutational burden in high-risk individuals. For the quantitative prediction of the outcomes of individuals, a nomogram was established. High-risk individuals showed greater sensitivity to immunotherapy while low-risk individuals benefited more from conventional chemotherapeutic or targeted therapy. Conclusion: A necroptosis-related prognostic signature was developed to study the tumor microenvironment, mutational burden, clinical features, and the treatment response of ESCA patients. This may contribute to precision medicine for ESCA.
Keywords: Necroptosis, esophageal cancer, prognosis, lncRNA, immune microenvironment
Introduction
Esophageal cancer (ESCA) is the seventh most prevalent malignancy globally, with 604,100 new cases and 544,076 fatalities [1]. Despite employing chemotherapy, radiotherapy, and targeted molecular therapy as essential components for the extensive treatment of inoperable ESCA, the 5-year survival rate of the individuals remains unfavorable [2]. Immunotherapy, particularly immune checkpoint blockade (ICB) therapy, has shown remarkable clinical efficacy in ESCA. Several recent clinical studies have demonstrated that combining ICB therapy and chemotherapy improves the overall survival (OS) of individuals with advanced-stage ESCA [3-5]. However, the application of immunotherapy still faces limitations such as low response rate, drug resistance, and few targets [6]. This observation suggests a need to look for new biomarkers to improve prognosis prediction, risk stratification, and treatment, to achieve better efficacy of immunotherapy.
Cell death is one of the most important events in cellular physiologic activity related to tumorigenesis, metastasis, treatment resistance, and cancer immunity [7]. Necroptosis is non-caspase-dependent programmed cell death, triggering inflammatory responses by damage-associated molecular patterns (DAMPs), which subsequently stimulate the release of cytokines and chemokines to induce innate and adaptive immunity to kill tumor cells [8]. Another study further revealed how tumor cells undergo necroptosis and release IL-1α to activate dendritic cells (DCs), thereby eliciting anti-tumor immunity through the generation of IL-12 and activation of CD8+ T cells [9]. Furthermore, it was noted that activation of necroptosis is efficacious in avoiding apoptosis resistance of tumors [10]. Therefore, necroptosis and its related molecules are promising candidates for identifying molecular targets with prognostic and therapeutic value in ESCA.
Long non-coding RNAs (lncRNAs) are functional RNAs consisting of around 200 nucleotides that do not contribute to protein-encoding processes. LncRNA H19-derived miR-675 induces necroptosis of hepatocellular carcinoma (HCC) cells by raising pMLKL and RIP3 levels and inhibiting the expression of FADD, cleaved-caspase-3, and cleaved caspase-8 [11]. LncRNA SNHG1 has been implicated in inhibiting the acceleration and proliferation of necroptosis of stomach adenocarcinoma cells by downregulating miR-21-5p and TLR4 [12]. It was also reported that a lncRNA termed TRINGS (TP53-Regulated Inhibitor of Necrosis under Glucose Hunger) binds to serine-threonine kinase receptor-associated protein (STRAP) and impedes GSK3β-NF-κB necrotic signal, thus protecting malignant cells from death [13]. The functional roles and prognostic values of necroptosis-related genes (NRGs) and necroptosis-related lncRNAs (NR-lncRNAs) have not been fully demonstrated in ESCA. Thus, our objective was to identify novel therapeutic targets in NRGs or NR-lncRNAs with prognostic value in ESCA.
In this study, NRGs and NR-lncRNAs with prognostic value in ESCA were analyzed and newly proposed. A novel necroptosis-associated signature was subsequently constructed to uncover its prognostic role and association with the immune microenvironment along with its therapeutic potential in ESCA. The acquired data are expected to facilitate prognostic assessment and precision medicine for individuals with ESCA.
Materials and methods
Data acquisition and preprocessing
The RNA sequencing (RNA-seq) datasets of esophageal cancer (n = 160) and adjacent normal tissue (n = 11) were provided by The Cancer Genome Atlas (TCGA) (https://gdc.cancer.gov/). The limited number of normal esophagus samples led us to download the data of 150 normal samples from the database, GTEx (www.gtexportal.org/). Clinical data, including survival time, status, and stage, were also acquired from TCGA. After excluding samples with OS of less than 60 days, a total of 152 ESCA cases were obtained in this research. Those samples were further classified randomly into two sets, the training (n = 76) and testing (n = 76) sets. A total of 67 necroptosis-related genes (NRGs) were provided by the Molecular Signatures Database (MsigDB) (www.gsea-msigdb.org) and previous literature [14]. Figure 1 exhibits the general workflow of this study.
Figure 1.

Workflow of this study
Selection of NRGs and NR-lncRNAs
Differential analysis was carried out by employing the Limma package with the parameters set as false discovery rate (FDR) < 0.05 & |log2Fold Change (FC)| > 0.585 [15]. Afterward, the differentially expressed lncRNAs (DE-lncRNAs) and the differentially expressed necroptosis-related genes (DE-NRGs) were extracted, along with an evaluation of their expression correlation. LncRNAs showing considerable positive correlations (|R| > 0.3 & P < 0.001) with DE-NRGs were defined as NR-lncRNAs.
Development and verification of a necroptosis-associated signature
Based on the clinical data of ESCA cases, expression of the DE-NRGs and NR-lncRNAs was combined with survival information, including OS time and status. Univariate Cox regression analysis was carried out to select NRGs and NR-lncRNAs with prognostic value (P < 0.05). Afterward, the LASSO regression was conducted utilizing the glmnet and survival packages. In total, 1000 random stimulations were set for each cycle to prevent overfitting. Hitherto, a model was established. The risk score was demonstrated utilizing the formula below:
The “X” represents the NRG or NR-lncRNA used for signature construction. The coefficient of the NRG or NR-lncRNA was denoted as coef (X), and the expression of the NRG or NR-lncRNA was denoted as expr (X). The samples were subdivided into high- and low-risk categories according to their median risk score. The OS difference was analyzed utilizing the Kaplan-Meier curve. Subsequently, heatmaps were generated to show expression profiles of NRGs and NR-lncRNAs.
Signature evaluation and nomogram establishment
For confirmation of the independence of the risk signatures, univariate and multivariate analyses were performed. Clinical characteristics comprising gender, risk score, and TNM staging parameters, were incorporated to evaluate the independent effects of the risk signature and other clinical variables. Additionally, the link between risk scores and clinical variables was revealed by correlation analysis. The receiver operating characteristics (ROC) curves of the model were plotted, then the area under the curve (AUC) of different factors was quantified. A nomogram for quantitative prediction of survival prognosis was developed utilizing the rms package.
Gene set enrichment analysis (GSEA)
GSEA was executed to identify the considerably enriched pathways and terms in each group. Two gene sets (go.v7.4.symbols.gmt and cp.kegg.v7.4.symbols.gmt) were acquired from the MsigDB. The process was conducted using R packages (limma, org.Hs.eg.db, clusterProfiler, etc.) [16].
Evaluation of immune function and infiltration
The infiltrative abundance of multiple immune cells was determined by xCELL, CIBERSORT, ESTIMATE, and MCPcounter [17,18]. Outcomes of CIBERSORT led us to determine the link between the proportion of immune cells, the risk score, and the gene expression used for model construction. In addition, tumor-associated stem cells are vital in the tumor immune microenvironment (TIME). Hence, the tumor stemness score was subjected to further analysis. R packages, such as ggplot2, ggpubr, tidyverse, and reshape2, were adopted in the analysis and visualization process. Furthermore, single sample Gene Set Enrichment Analysis (ssGSEA) was conducted, and the expression of checkpoint molecules was comparatively assessed to discover the variation in immune functional phenotypes between tumors with different risks.
Analysis of tumor mutational burden (TMB)
Somatic mutation data of ESCA were provided by the TCGA. The TMB for each individual was computed and compared across the two risk categories by the maftools package [19]. The visualization method was the same as mentioned above.
Evaluation of sensitivity to immunotherapy and chemotherapy
Sensitivity to chemotherapy and the immunotherapy response were analyzed to understand the effectiveness of this signature in evaluating the direction of clinical treatment. The R package pRRophetic was employed for the prediction of tumor sensitivity to commonly used chemotherapeutic agents in clinical practice [20]. The efficacy of drugs was evaluated by the half-maximal inhibitory concentration (IC50). The tumor Immune Dysfunction and Exclusion (TIDE) database (https:// http://tide.dfci.harvard.edu/) was used to analyze immunogenicity and immunoreactivity characteristics of the tumors; thus, TIDE scores can be used to assess whether patients may benefit from ICB therapy [21].
Statistical analysis
Statistical analyses were conducted using R software (4.0.3). Cox regression analyses were conducted to screen the prognostic molecules and test the independence of the risk signatures. Student’s t-test was employed for the comparative analysis of the variations in continuous variables. The Chi-square test was employed for the comparative assessment of the proportions of categorical variables. Multiple R packages used for data visualization have been listed in individual sections. For all analyses, the statistical significance level was set at P < 0.05.
Results
Mining for differentially expressed NRGs and NR-lncRNAs in ESCA
To find molecules upregulated or downregulated in ESCA, a total of 150 normal samples from GTEx, 11 paracancerous samples, and 160 ESCA samples from TCGA were acquired. Following intersections with the previously reported 67 NRGs, 55 NRGs in the expression data were identified. According to the criteria set for the expression difference (|Log2(FC)| > 0.585 & FDR < 0.05), 45 DE-NRGs and 1084 DE-lncRNAs were identified (Figure 2A, 2B). Afterward, 496 NR-lncRNAs were selected from 1084 DE-lncRNAs by correlation analysis (correlation coefficients > 0.3 and P < 0.001). Subsequent screening of the prognostic members was conducted from these 45 DE-NRGs and 496 NR-lncRNAs. GO and KEGG enrichment analysis demonstrated that these DE-NRGs were enriched in pathways correlated with cell apoptosis, necroptosis, and TNF signaling. Moreover, the enriched biologic processes included binding to ubiquitin ligase, programmed cell death, and necroptotic cell death (Figure 2C, 2D).
Figure 2.
Identification of the differentially expressed NRGs and lncRNAs with prognostic value in patients with ESCA. A, B. Differential expression of NRGs and NR-lncRNAs; C, D. GO and KEGG functional analysis of NRGs with differential expression; E. 34 genes preliminarily filtered by univariate Cox proportional hazard regression analysis; F, G. LASSO analysis selected ideal candidates and their coefficients for signature construction; H. Correlation network between NRGs and NR-lncRNAs (correlation coefficients > 0.3 and P < 0.001). The red ellipses depict prognostic NRGs, and the blue triangles depict lncRNAs. Positive and negative correlations are depicted by red and blue lines, respectively.
Construction and verification of the necroptosis-associated risk signature
A total of 152 individuals with ESCA were randomly classified into two categories (training and testing sets) with a ratio of 1:1. For the preliminary screening of prognostic NRGs and NR-lncRNAs, a univariate Cox analysis revealed 34 candidates (4 NRGs and 30 NR-lncRNAs) (Figure 2E). Then the Lasso regression was utilized, resulting in the identification of 13 candidates that were incorporated for signature construction (Figure 2F, 2G). The network figure revealing the correlation between prognostic NRGs, such as PANX1, SIRT1, STUB1, ATRX and NR-lncRNAs was developed to visualize the correlation better (Figure 2H). The mentioned formula evaluated the risk score for all the individuals under study: Risk score = Expr (ELFN1-AS1) × (0.0286) + Expr (PRRT3-AS1) × (0.0315) + Expr (AL035461.2) × (0.1070) + Expr (AC012467.2) × (0.2458) + Expr (AC073896.4) × (0.0272) + Expr (AC006026.3) × (0.1054) + Expr (MEG3) × (-0.1582) + Expr (AC079949.2) × (0.0547) + Expr (AC087741.1) × (0.0678) + Expr (UBR5-AS1) × (0.0729) + Expr (JPX) × (0.0109) + Expr (ATRX) × (0.0567) + Expr (STUB1) × (-0.0660).
Patients were then classified into two groups (high- and low-risk) as previously described. The risk scores of individuals in the training, testing, and entire sets are shown in Figure 3A-C, respectively. Moreover, the survival status of individuals in the above-mentioned three sets is depicted in Figure 3D-F, respectively. Figure 3G-I show the expression profiles of the NRGs and NR-lncRNAs in tumors with different risks. Notably, hazardous NR-lncRNAs such as ELFN1-AS1 were significantly upregulated in high-risk tumors, while the opposite was true for low-risk NRGs such as STUB1 (Figure 3G-I). The OS curves showed the high-risk patients to have poor prognoses in all three sets (Figure 3J-L, P < 0.05). The above findings implied that the necroptosis-based molecular signature can significantly identify the risk and prognosis of ESCA patients.
Figure 3.
Construction, validation, and independent evaluation of the necroptosis-associated signature. (A-C) Patient risk scores, calculated by the prognostic signature in the training, testing, and entire sets; (D-F) Survival status of individuals in the aforementioned three sets; (G-I) Heatmaps depicting the expression of the two NRGs and 11 NR-lncRNAs in these three sets, respectively; (J-L) Survival differences between low- and high-risk individuals in the three sets, respectively; (M-R) Univariate and multivariate independence analyses of the necroptosis-associated signature in the training (M, N), testing (O, P), and entire (Q, R) sets.
Univariate Cox regression revealed that the risk score had a hazard ratio (HR) of 9.668 and a 95% confidence interval (CI) of 3.360-27.818, in the training set. Furthermore, HR = 3.718 and CI = 1.840-7.515 were recorded in the testing set, and HR = 4.893 and CI = 2.858-8.376 in the entire set (P < 0.001). In multivariate Cox regression, the HR in the training set was recorded to be 8.391 and CI = 2.657-26.504 in the training set. HR = 3.959 and CI = 1.729-9.062 were recorded in the testing set with HR = 4.436 and CI = 2.508-7.846 in the entire set (P < 0.001) (Figure 3M-R). The above findings implied that the risk score can be considered an independent prognostic factor effective for ESCA patients.
Assessment of the clinicopathologic relevance of the necroptosis-related risk signature
ROC was utilized to evaluate the true and false positive rates of the prognostic model in a time-dependent manner. The results of the ROC were evaluated by the AUC of the ROC. The respective AUCs of the 1-, 2- and 3-year ROC in the training set were 0.753, 0.774, 0.703; whereas the corresponding values in the testing set were 0.879, 0.665, 0.574, respectively, and in the entire set were 0.806, 0.743, 0.665, respectively (Figure 4A-C). Moreover, a nomogram was developed, containing the gender, tumor stage, and risk score to anticipate the 1-, 2-, and 3-year OS of individuals with ESCA (Figure 4D). Additionally, heatmaps were established to demonstrate the distribution of individuals with different clinicopathologic features in each group, to measure the clinicopathologic relevance of risk scores. A larger distribution of patients with stages 3-4 was noted in the high-risk group (P < 0.05) (Figure 4E, 4F). The number of T2 patients was more in the low-risk group, while there were more T3 ones in the high-risk group (P < 0.01) (Figure 4E, 4G). Patients with N stage were also distributed differently between the two groups (P < 0.05) (Figure 4E, 4H). The above findings demonstrate that the necroptosis-associated signature correlated with the clinicopathologic features of tumors to a certain extent.
Figure 4.
Signature evaluation and clinical relevance analyses. A-C. The respective AUC values of the 1-, 2- and 3-year ROC in the training, testing, and entire sets; D. The nomogram that predicts the 1-, 3-, and 5-year OS by risk score, gender, and tumor stage; E. Distribution of individuals with various clinicopathologic features in two groups; F-H. Relevance of risk groups between clinical stage, T-stage, and N-stage, respectively.
GSEA
GSEA further investigated the enrichment or depletion of different biologic or functional features across high- and low-risk tumors. Pathways correlated with glutamate metabolism, nitrogen metabolism, and PPAR signaling were enriched in high-risk tumors (Figure 5B). The digestive system and its developmental processes, as well as lipoproteins and their receptor signaling processes, were also significantly enriched (Figure 5D). For low-risk tumors, enriched pathways were linked to cell growth and development, like focal adhesion and extracellular matrix (ECM) receptors. Moreover, skin development and differentiation pathways were also annotated (Figure 5A, 5C). The above analysis suggests that risk subgroups based on the necroptosis-associated signature may also differ significantly in functional phenotypes.
Figure 5.
Gene set enrichment analysis. A, C. Enriched KEGG pathways and GO terms in low-risk tumors; B, D. Enriched KEGG pathways and GO terms in high-risk tumors.
Characteristics of TIME
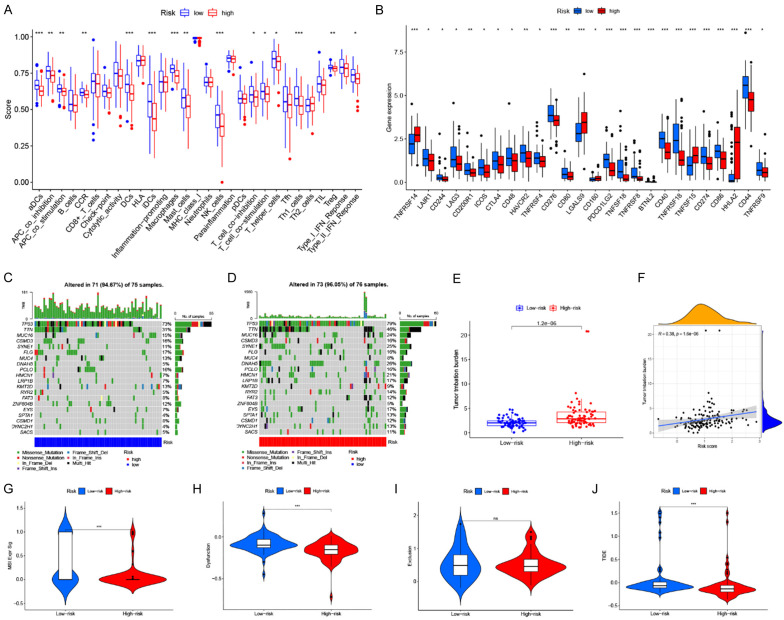
Immune infiltration analysis was carried out utilizing four algorithms (xCELL, MCPcounter, CIBERSORT, and ESTIMATE) and the infiltration level of different immune cells was quantified. CIBERSORT analysis revealed a greater proportion of naïve B cells, CD4+ memory resting T cells, and regulatory T cells (Tregs) in high-risk tumors while resting NK cells and M0 macrophages were higher in the low-risk tumors (Figure 6A). The TIME was also characterized and scored by ESTIMATE, which demonstrated that the low-risk individuals had increased stromal and ESTIMATE scores (Figure 6B). The xCELL algorithm analysis showed a positive correlation between risk scores and the infiltrating abundance of immune cells, including common lymphoid progenitors, and plasma B cells (Figure 6C). A positive correlation with the proportion of infiltrating naïve B cells, CD4+ memory resting T cells, follicular helper T cells, and regulatory T cells was observed using the CIBERSORT algorithm (Cor > 0.2 & P < 0.05) (Figure 6C). Moreover, a high proportion of tumor-infiltrating Tregs and CD4+ memory resting T cells also indicated poor prognosis (Figure S1). A negative correlation was observed between the risk scores and the infiltration of activated myeloid dendritic cells, myeloid dendritic cells, monocytes, CD4+ Th1 cells, cancer-related fibroblasts and M0 macrophages (Cor < -0.2) (all P < 0.05) (Figure 6C). The above findings demonstrate that there were considerable variations in the TIME of tumors with different risks.
Figure 6.
Tumor microenvironment characteristic analysis. A. Variation in the proportion of tumor-infiltrating immune cells determined by CIBERSORT across the two risk groups; B. TIME scores derived through ESTIMATE; C. Association of risk scores with the abundance of tumor-infiltrating immune cells; D. Association of expression of the prognostic genes with the abundance of tumor-infiltrating immune cells; E. Stemness correlation analysis.
Additionally, this research also aimed at the identification of NRGs or NR-lncRNAs closely linked to tumor-infiltrating immune cells. Among the signature NRGs and NR-lncRNAs, it was found that STUB1, ELFN1-AS1, AC012467.2, and AC006026.3 are closely linked to the tumor-infiltrating immune cells (Figure 6D). A positive correlation was found between tumor-infiltrating CD4+ memory resting T cells, naïve B cells, and AC012467.2 expression; naïve B cells and AC006026.3 expression; activated dendritic cells and AC079949.2 expression; neutrophils, naïve B cells and the expression of ELFN1-AS1. However, a negative correlation was observed between M0 macrophages and AC006026.3 expression, resting mast cells, and PRRT3-AS1 expression, CD4+ memory resting T cells, and STUB1 expression (Figure 6D). Stemness scores were also positively linked to the risk scores, implying that the tumors in the high-risk category have more robust tumor-associated stem cell properties (Figure 6E).
Evaluation of immune function status and immunotherapy response
The research subsequently asked whether this signature affected anti-tumor immunity and immunotherapy responsiveness. The characteristics of various immunomodulatory factors including immune checkpoint expression, TMB, and immune function scores were analyzed. Moreover, TIDE was utilized to predict the immunotherapy response of individuals. Initially, ssGSEA was carried out, which illustrated that most immune functions differed between the two groups. Among these, the most significant difference was harbored by the aDCs, APC-co-stimulation, DCs, iDCs, macrophages, and Th1-cells, suggesting these components to be functionally active in low-risk tumors (Figure 7A). The expression assessment of immune checkpoints illustrated that several immune checkpoints including CD274 (PD-L1), CTLA4, CD276 (B7-H3), PDCD1LG2, and HAVCR2 had remarkably increased expression in the low-risk group. At the same time, a substantially higher expression of TNFRSF14, LGALS9, TNFSF15, and HHLA2 was found in high-risk tumors (Figure 7B). Furthermore, the necroptosis-related risk score was negatively linked to the expression of most checkpoint molecules, such as PD-L1, CTLA4, and LAG3 (Figure S2).
Figure 7.
Immune function status and immunotherapy response. A. Variance in immune function scores between the two risk groups; B. Differences in the expression of immune checkpoints; C. Somatic mutation landscape of low-risk tumors; D. Somatic mutation landscape of high-risk tumors; E. Comparative assessment of TMB; F. The positive relationship between risk scores and TMB; G-J. Comparative analyses of MSI, T cell dysfunction score, T cell exclusion score, and TIDE scores.
The mutational landscapes are shown in Figure 7C, 7D, which revealed that the most frequently occurring somatic mutations in the low-risk category were as follows: TP53 > TTN > FLG > CSMD3 > PCLO > MUC16 > MUC4 > KMT2D > ZNF804B > SYNE1 > FAT3 > CSMD1 (Figure 7C). In contrast, the most frequently occurring somatic mutations in the high-risk category followed the order: TP53 > TTN > DNAH5 > SYNE1 > MUC16 > HMCN1 > LRP1B > EYS > FLG > CSMD3 > PCLO > RYR2 > SPTA1 > DYNC2H1 (Figure 7D). A comparative analysis of the TMB implied that high-risk patients had a remarkably elevated TMB in comparison to those low-risk ones (P = 4.2e-06) (Figure 7E). Furthermore, a positive relationship was observed between the risk score and TMB (R = 0.38, P = 1.6e-06) (Figure 7F). Patients with a high TMB showed poorer OS, while high-risk patients with an elevated TMB depicted a poor prognosis (Figure S1).
Increased microsatellite instability (MSI) and T-cell dysfunction scores were also evaluated in low-risk patients (Figure 7G, 7H). However, no difference was observed in T-cell exclusion scores (Figure 7I). The TIDE score system predicts the potential development of immune escape or response to immunotherapy of tumors, which can be applied to assess the efficacy of immunotherapy. The results showed a lower TIDE score in the high-risk group, implying that high-risk individuals could gain more benefit from ICB immunotherapy (Figure 7J). Studies also showed that individuals with ESCA depicting greater TMB tend to benefit more from immunotherapy, which is in line with the prediction of the TIDE score [22]. These findings suggest that the signature can accurately differentiate the immune function of patients with different risks. Moreover, high-risk individuals showed better responses to immunotherapy.
Assessment of chemotherapeutic sensitivity
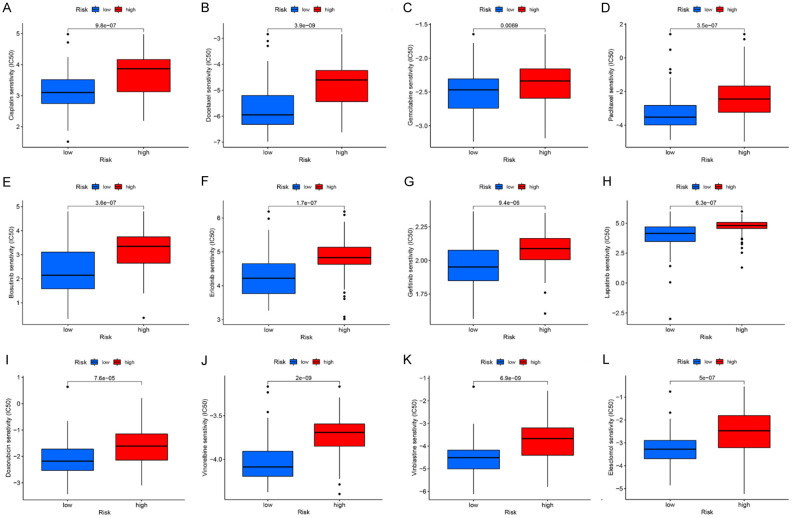
Although the likelihood of the high-risk individuals benefiting from immunotherapy was high, it was crucial to investigate the potential of the signature in differentiating patients in terms of sensitivity to chemotherapeutic agents. This analysis showed a higher estimated IC50 of common chemotherapeutic drugs like Cisplatin, Docetaxel, Paclitaxel, Gemcitabine, Bosutinib, Erlotinib, Gefitinib, Lapatinib, Doxorubicin, Vinorelbine, Vinblastine, and Elesclomol for high-risk individuals (P < 0.01), suggesting an increased sensitivity of the low-risk individuals to these chemotherapeutic drugs and targeted drugs (Figure 8A-L). Targeted drugs showed the same propensity. Src/Ab1 dual inhibitor, Bosutinib, showed decreased IC50 in the low-risk when compared to the high-risk group. A similar trend for EGFR inhibitors Erlotinib, Gefitinib, and Lapatinib was also found. Low-risk patients also demonstrated greater sensitivity to the copper carrier Elesclomol. Taken together, common chemotherapeutic and targeted drugs were more effective for low-risk patients. Hence, this necroptosis-associated signature holds promise for providing more accurate advice on clinical medication regimens for ESCA patients.
Figure 8.
Chemotherapeutic sensitivity assessment. High-risk patients possessed greater estimated IC50 to 12 kinds of drugs, in contrast to low-risk patients.
Discussion
As a novel anti-cancer strategy, immunotherapy enhances anti-tumor immunity in the body, presenting broad application prospects in the treatment of ESCA. Recently, emerging ICB agents such as Tislelizumab and Sintilimab have shown remarkable therapeutic efficacy in treating advanced ESCA [3-5]. However, factors such as insufficient effector cells in the TIME and low response rate still limit the further application of immunotherapy [23]. Therefore, there is an urgent need to establish molecular markers to improve the survival of individuals with ESCA and to develop new therapeutic targets.
Several studies have documented the function of necroptosis in regulating tumorigenesis and progression and influencing the efficiency of immunotherapy and chemotherapy, so necroptosis is considered a promising target in treating ESCA [7,8,10,24,25]. However, the prognostic value and potential immunomodulatory functions of necroptosis-related molecules, especially the NR-lncRNAs, in ESCA have not been fully explored. Furthermore, reliable prognostic models for ESCA based on necroptosis-related molecules are still rare. Therefore, establishing a prognostic signature based on NRGs and NR-lncRNAs in ESCA to comprehensively characterize the prognosis, TIME, and therapeutic sensitivity of ESCA patients is of great significance.
In this study, a total of 34 necroptosis-related molecules with prognostic significance were identified. Among these, two NR-NRGs and 11 NR-lncRNAs were further selected to establish a prognostic signature. This necroptosis-based molecular signature enabled risk stratification of ESCA patients, and high-risk individuals experienced a worse prognosis. Moreover, this signature was capable of representing the clinicopathologic, tumor immunological, chemotherapeutic, and immunotherapy sensitivity characteristics of patients in different risk groups. This 13-molecule signature is anticipated to offer novel insight for the advancement of precision medicine in ESCA.
It is noteworthy that certain members of the 13 signature genes have been reported to affect prognosis and regulate biologic functions in tumors. STUB1, known as STIP homologous and U box-containing protein 1, possesses E3 ligase activity and can bind to Hsp70, Hsp90, or other molecular chaperones, which crucially regulate protein folding, assembly, transport, and degradation, thus regulating the physiologic or pathologic processes in cells [26]. It was shown that STUB1-mediated proteasomal degradation of METTL3 inhibited the metastasis of ESCA [27]. Consistently, this study identified it as a low-risk NRG. Unlike STUB1, ATRX (ATRX Chromatin Remodeler), a chromatin remodeling protein whose mutations are associated with alpha thalassemia X-linked intellectual disability (ATR-X) syndrome, was identified as a poor prognostic marker in this study. ATRX was found to be targeted by miR-1269a as a downstream gene and was linked to adverse outcome among individuals with ESCA, further validating our findings [28]. Furthermore, one of the identified adversely prognostic NR-LncRNAs in this study, ELFN1-AS1, has been reported to act as a competing endogenous RNA that promotes the proliferation, invasion, and migration of ESCA, achieved through the ELFN1-AS1/miR-183-3p/GFPT1 pathway [29]. In this study, a favorable NR-lncRNA MEG3 was also identified, which has been reported to inhibit Treg differentiation and immune escape in ESCA through mediation of the miR-149-3p/FOXP3 axis [30]. Downregulation of lncRNA MEG3 has been found to induce epithelial-to-mesenchymal transition (EMT), which subsequently promotes tumor progression and leads to poor prognosis in ESCA individuals [31]. Up-regulated lncRNA AC079949.2, which features in the cGMP-PKG signaling pathway, might be a poor prognostic factor in ESCA [32]. JPX was also reported to play a tumor-promoting role in ESCA, by interacting with miR-516b-5p and VEGFA [33]. Although some of the signature lncRNAs have been reported to have cancer-promoting or suppressing roles in ESCA, there are still several newly proposed prognostic NR-lncRNAs in this study and their functions require further exploration.
Immune infiltration analyses revealed an increased fraction of naïve B cells, Tregs, and CD4+ memory resting T cells, and a decreased fraction of resting NK cells and dendritic cells in high-risk tumors. Investigations conducted in the past have corroborated that naïve B cells and Tregs were strongly linked to unfavorable prognosis in ESCA [34,35]. The prognostic model of gastric cancer also depicted that patients with poor prognoses had an elevated proportion of resting memory CD4+ T cells [36]. These cells were also associated with poor prognosis in melanoma and urothelial carcinoma [37]. The prognostic value of resting NK and dendritic cells in esophageal cancer has been documented [38,39]. Moreover, M0 and M1 macrophages were found increased dramatically in ESCC tissues [40]. In our study, we also explored the association between tumor-infiltrating immune cells and expression of NRGs. STUB1, AC006026.3, AC006026.3 and ELFN1. AS1 were both NRGs or NR-lncRNAs with prognostic value and high relevance to the TIME in ESCA. This should be of value for further experiments.
To better explore the differential response to immunotherapy, we compared the somatic mutation status, immune checkpoint expression, and TIDE scores between groups. Although the high-risk group had lower PD-L1 expression, they had a higher TMB, which is emerging as an indicator to evaluate the prognostic outcome in individuals with cancer and predict the efficacy of immunotherapy [41]. For advanced gastroesophageal adenocarcinoma, patients with TMB > 12 mut/Mb indicated a higher benefit from the anti-PD-1 immunotherapy [42]. At the same time, it is noteworthy that one study indicated that the characteristics of TMB and PD-L1 expression had a certain degree of temporal and spatial heterogeneity in gastroesophageal cancer, which is crucial when considering immunotherapy [43]. This may partly explain the lower PD-L1 expression in the high-risk group. Moreover, lower TIDE scores of high-risk patients also indicated a lower possibility of immune evasion. This observation further suggests greater sensitivity of the high-risk group to immunotherapy, despite the worse prognosis.
Interestingly, the low-risk group showed a higher sensitivity to all 12 chemotherapeutic or targeted agents. Anticancer chemotherapeutic drugs commonly used in clinical practice, including Cisplatin, Docetaxel, Gemcitabine, Paclitaxel, Doxorubicin, Vinorelbine, and Vinblastine were more effective in treating low-risk tumors. Erlotinib, Gefitinib, and Lapatinib are targeted molecular drugs that have demonstrated good therapeutic efficacy in gastric and esophageal tumors [44-46]. Bosutinib was primarily used for leukemia but could significantly induce apoptosis of esophageal cancer cells in vitro [47]. Elesclomol, a copper ion binding agent, could induce oxidative stress and promotes apoptosis of cancer cells [48]. The stemness characteristics of tumors may partially explain the differences in chemosensitivity of tumors in different groups. Cancer stem cells (CSCs), also called tumor-initiating cells, regulate treatment resistance and recurrence of various cancers [49]. As expected, high-risk tumors presented higher stemness scores, suggesting stronger CSC traits, accompanied by greater resistance to multiple chemotherapeutic agents.
In summary, a prognostic signature for ESCA patients was developed based on two NRGs and 11 NR-lncRNAs. The signature accurately performed risk stratification of ESCA patients and showed good relevance to clinicopathologic factors. There was considerable variation in functional features, TIME landscapes, TMB, CSC characteristics, and therapeutic sensitivity between the high- and low-risk groups.
This study had some limitations. First, this is a retrospective analysis solely based on bioinformatics data, warranting in vivo or in vitro experimental validation. Second, the prognostic role of these signature genes and lncRNAs requires further exploration through more prospective studies. Nevertheless, this signature and its involved molecules are expected to bring new insight into the discovery of novel therapeutic targets and precision medicine for ESCA.
Acknowledgements
This research was supported by the Medical Education Collaborative Innovation Fund of Jiangsu University (Grant No. JDY2022016). The authors also thank Yixing hospital affiliated to Jiangsu University, Dongtai hospital of traditional Chinese medicine and the first affiliated hospital of Naval Medical University for providing research support.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Eyck BM, van Lanschot JJB, Hulshof MCCM, van der Wilk BJ, Shapiro J, van Hagen P, van Berge Henegouwen MI, Wijnhoven BPL, van Laarhoven HWM, Nieuwenhuijzen GAP, Hospers GAP, Bonenkamp JJ, Cuesta MA, Blaisse RJB, Busch OR, Creemers GM, Punt CJA, Plukker JTM, Verheul HMW, Spillenaar Bilgen EJ, van der Sangen MJC, Rozema T, Ten Kate FJW, Beukema JC, Piet AHM, van Rij CM, Reinders JG, Tilanus HW, Steyerberg EW, van der Gaast A CROSS Study Group. Ten-year outcome of neoadjuvant chemoradiotherapy plus surgery for esophageal cancer: the randomized controlled CROSS trial. J. Clin. Oncol. 2021;39:1995–2004. doi: 10.1200/JCO.20.03614. [DOI] [PubMed] [Google Scholar]
- 3.Lu Z, Wang J, Shu Y, Liu L, Kong L, Yang L, Wang B, Sun G, Ji Y, Cao G, Liu H, Cui T, Li N, Qiu W, Li G, Hou X, Luo H, Xue L, Zhang Y, Yue W, Liu Z, Wang X, Gao S, Pan Y, Galais MP, Zaanan A, Ma Z, Li H, Wang Y, Shen L ORIENT-15 study group. Sintilimab versus placebo in combination with chemotherapy as first line treatment for locally advanced or metastatic oesophageal squamous cell carcinoma (ORIENT-15): multicentre, randomised, double blind, phase 3 trial. BMJ. 2022;377:e068714. doi: 10.1136/bmj-2021-068714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen L, Kato K, Kim SB, Ajani JA, Zhao K, He Z, Yu X, Shu Y, Luo Q, Wang J, Chen Z, Niu Z, Zhang L, Yi T, Sun JM, Chen J, Yu G, Lin CY, Hara H, Bi Q, Satoh T, Pazo-Cid R, Arkenau HT, Borg C, Lordick F, Li L, Ding N, Tao A, Shi J, Van Cutsem E RATIONALE-302 Investigators. Tislelizumab versus chemotherapy as second-line treatment for advanced or metastatic esophageal squamous cell carcinoma (RATIONALE-302): a randomized phase III study. J. Clin. Oncol. 2022;40:3065–3076. doi: 10.1200/JCO.21.01926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Li Y, Fan Q, Shu Y, Yang L, Cui T, Gu K, Tao M, Wang X, Cui C, Xu N, Xiao J, Gao Q, Liu Y, Zhang T, Bai Y, Li W, Zhang Y, Dai G, Ma D, Zhang J, Bai C, Huang Y, Liao W, Wu L, Chen X, Yang Y, Wang J, Ji S, Zhou H, Wang Y, Ma Z, Wang Y, Peng B, Sun J, Mancao C. Clinical and biomarker analyses of sintilimab versus chemotherapy as second-line therapy for advanced or metastatic esophageal squamous cell carcinoma: a randomized, open-label phase 2 study (ORIENT-2) Nat Commun. 2022;13:857. doi: 10.1038/s41467-022-28408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo H, Lu J, Bai Y, Mao T, Wang J, Fan Q, Zhang Y, Zhao K, Chen Z, Gao S, Li J, Fu Z, Gu K, Liu Z, Wu L, Zhang X, Feng J, Niu Z, Ba Y, Zhang H, Liu Y, Zhang L, Min X, Huang J, Cheng Y, Wang D, Shen Y, Yang Q, Zou J, Xu RH ESCORT-1st Investigators. Effect of camrelizumab vs placebo added to chemotherapy on survival and progression-free survival in patients with advanced or metastatic esophageal squamous cell carcinoma: the ESCORT-1st randomized clinical trial. JAMA. 2021;326:916–925. doi: 10.1001/jama.2021.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer. 2022;8:21–27. doi: 10.1016/j.trecan.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SY, Hsieh SY, Fan YT, Wei WC, Hsiao PW, Tsai DH, Wu TS, Yang NS. Necroptosis promotes autophagy-dependent upregulation of DAMP and results in immunosurveillance. Autophagy. 2018;14:778–795. doi: 10.1080/15548627.2017.1386359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt SV, Seibert S, Walch-Ruckheim B, Vicinus B, Kamionka EM, Pahne-Zeppenfeld J, Solomayer EF, Kim YJ, Bohle RM, Smola S. RIPK3 expression in cervical cancer cells is required for PolyIC-induced necroptosis, IL-1alpha release, and efficient paracrine dendritic cell activation. Oncotarget. 2015;6:8635–8647. doi: 10.18632/oncotarget.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarcognato S, Jong IEM, Fabris L, Cadamuro M, Guido M. Necroptosis in cholangiocarcinoma. Cells. 2020;9:982. doi: 10.3390/cells9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harari-Steinfeld R, Gefen M, Simerzin A, Zorde-Khvalevsky E, Rivkin M, Ella E, Friehmann T, Gerlic M, Zucman-Rossi J, Caruso S, Leveille M, Estall JL, Goldenberg DS, Giladi H, Galun E, Bromberg Z. The lncRNA H19-derived microRNA-675 promotes liver necroptosis by targeting FADD. Cancers (Basel) 2021;13:411. doi: 10.3390/cancers13030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N, Liu D. Identification and validation a necroptosis-related prognostic signature and associated regulatory axis in stomach adenocarcinoma. Onco Targets Ther. 2021;14:5373–5383. doi: 10.2147/OTT.S342613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan MR, Xiang S, Song Z, Wu M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017;36:3483–3500. doi: 10.15252/embj.201696239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z, Liu H, Zhou X, Fang D, Ou X, Ye J, Peng J, Xu J. Necroptosis-related lncRNAs: predicting prognosis and the distinction between the cold and hot tumors in gastric cancer. J Oncol. 2021;2021:6718443. doi: 10.1155/2021/6718443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Xu X, Wang D, Liu J, Sun J, Lu M, Wang R, Hui B, Li X, Zhou C, Wang M, Qiu T, Cui S, Sun N, Li Y, Wang F, Liu C, Shao Y, Luo J, Gu Y. Neoadjuvant sintilimab and chemotherapy in patients with potentially resectable esophageal squamous cell carcinoma (KEEP-G 03): an open-label, single-arm, phase 2 trial. J Immunother Cancer. 2023;11:e005830. doi: 10.1136/jitc-2022-005830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang TX, Fu L. The immune landscape of esophageal cancer. Cancer Commun (Lond) 2019;39:79. doi: 10.1186/s40880-019-0427-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan K, Cheng H, Jin K, Ni Q, Yu X, Liu C. The role of necroptosis in cancer biology and therapy. Mol Cancer. 2019;18:100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aaes TL, Kaczmarek A, Delvaeye T, De Craene B, De Koker S, Heyndrickx L, Delrue I, Taminau J, Wiernicki B, De Groote P, Garg AD, Leybaert L, Grooten J, Bertrand MJ, Agostinis P, Berx G, Declercq W, Vandenabeele P, Krysko DV. Vaccination with necroptotic cancer cells induces efficient anti-tumor immunity. Cell Rep. 2016;15:274–287. doi: 10.1016/j.celrep.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Ranek MJ, Stachowski MJ, Kirk JA, Willis MS. The role of heat shock proteins and co-chaperones in heart failure. Philos Trans R Soc Lond B Biol Sci. 2018;373:20160530. doi: 10.1098/rstb.2016.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao L, He Y, Li SJ, Zhang GG, Yu W, Yang J, Huang ZJ, Zheng CC, He QY, Li Y, Li B. Anti-HIV drug elvitegravir suppresses cancer metastasis via increased proteasomal degradation of m6A methyltransferase METTL3. Cancer Res. 2022;82:2444–2457. doi: 10.1158/0008-5472.CAN-21-4124. [DOI] [PubMed] [Google Scholar]
- 28.Xie Z, Zhong C, Duan S. miR-1269a and miR-1269b: emerging carcinogenic genes of the miR-1269 family. Front Cell Dev Biol. 2022;10:809132. doi: 10.3389/fcell.2022.809132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Lian H, Xie L, Yin N, Cui Y. LncRNA ELFN1-AS1 promotes esophageal cancer progression by up-regulating GFPT1 via sponging miR-183-3p. Biol Chem. 2020;401:1053–1061. doi: 10.1515/hsz-2019-0430. [DOI] [PubMed] [Google Scholar]
- 30.Xu QR, Tang J, Liao HY, Yu BT, He XY, Zheng YZ, Liu S. Long non-coding RNA MEG3 mediates the miR-149-3p/FOXP3 axis by reducing p53 ubiquitination to exert a suppressive effect on regulatory T cell differentiation and immune escape in esophageal cancer. J Transl Med. 2021;19:264. doi: 10.1186/s12967-021-02907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li MK, Liu LX, Zhang WY, Zhan HL, Chen RP, Feng JL, Wu LF. Long non-coding RNA MEG3 suppresses epithelial-to-mesenchymal transition by inhibiting the PSAT1-dependent GSK-3beta/Snail signaling pathway in esophageal squamous cell carcinoma. Oncol Rep. 2020;44:2130–2142. doi: 10.3892/or.2020.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Zhang Q, Lou Q, Zhang X, Cui Y, Wang P, Yang F, Wu F, Wang J, Fan T, Li S. Differential analysis of lncRNA, miRNA and mRNA expression profiles and the prognostic value of lncRNA in esophageal cancer. Pathol Oncol Res. 2020;26:1029–1039. doi: 10.1007/s12253-019-00655-8. [DOI] [PubMed] [Google Scholar]
- 33.He Y, Hua R, Yang Y, Li B, Guo X, Li Z. LncRNA JPX promotes esophageal squamous cell carcinoma progression by targeting miR-516b-5p/VEGFA axis. Cancers (Basel) 2022;14:2713. doi: 10.3390/cancers14112713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan F, Xu B, Li J. A low proportion of regulatory T cells before chemoradiotherapy predicts better overall survival in esophageal cancer. Ann Palliat Med. 2021;10:2195–2202. doi: 10.21037/apm-21-196. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Chen P, Zhao Y, Feng X, Gao S, Qi Y. Immune infiltration represents potential diagnostic and prognostic biomarkers for esophageal squamous cell carcinoma. Biomed Res Int. 2022;2022:9009269. doi: 10.1155/2022/9009269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng Z, Xie D, Gong J. Genome-wide identification of CpG island methylator phenotype related gene signature as a novel prognostic biomarker of gastric cancer. PeerJ. 2020;8:e9624. doi: 10.7717/peerj.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R, Yang F, Yin JY, Liu YZ, Zhang W, Zhou HH. Influence of tumor immune infiltration on immune checkpoint inhibitor therapeutic efficacy: a computational retrospective study. Front Immunol. 2021;12:685370. doi: 10.3389/fimmu.2021.685370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan X, Song J, Fan Y, Li J, Chen Y, Zhu H, Zhang Z. CSMD1 mutation related to immunity can be used as a marker to evaluate the clinical therapeutic effect and prognosis of patients with esophageal cancer. Int J Gen Med. 2021;14:8689–8710. doi: 10.2147/IJGM.S338284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Wang H, Wu H, Qiang G. The functionalities and clinical significance of tumor-infiltrating immune cells in esophageal squamous cell carcinoma. Biomed Res Int. 2021;2021:8635381. doi: 10.1155/2021/8635381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Feng Z, Qu J, Liu X, Liang J, Li Y, Jiang J, Zhang H, Tian H. Integrated bioinformatics analysis of differentially expressed genes and immune cell infiltration characteristics in esophageal squamous cell carcinoma. Sci Rep. 2021;11:16696. doi: 10.1038/s41598-021-96274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Wei XL, Wang FH, Xu N, Shen L, Dai GH, Yuan XL, Chen Y, Yang SJ, Shi JH, Hu XC, Lin XY, Zhang QY, Feng JF, Ba Y, Liu YP, Li W, Shu YQ, Jiang Y, Li Q, Wang JW, Wu H, Feng H, Yao S, Xu RH. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol. 2019;30:1479–1486. doi: 10.1093/annonc/mdz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou KI, Peterson B, Serritella A, Thomas J, Reizine N, Moya S, Tan C, Wang Y, Catenacci DVT. Spatial and temporal heterogeneity of PD-L1 expression and tumor mutational burden in gastroesophageal adenocarcinoma at baseline diagnosis and after chemotherapy. Clin Cancer Res. 2020;26:6453–6463. doi: 10.1158/1078-0432.CCR-20-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hecht JR, Bang YJ, Qin SK, Chung HC, Xu JM, Park JO, Jeziorski K, Shparyk Y, Hoff PM, Sobrero A, Salman P, Li J, Protsenko SA, Wainberg ZA, Buyse M, Afenjar K, Houe V, Garcia A, Kaneko T, Huang Y, Khan-Wasti S, Santillana S, Press MF, Slamon D. Lapatinib in combination with capecitabine plus oxaliplatin in human epidermal growth factor receptor 2-positive advanced or metastatic gastric, esophageal, or gastroesophageal adenocarcinoma: TRIO-013/LOGiC--a randomized phase III trial. J. Clin. Oncol. 2016;34:443–451. doi: 10.1200/JCO.2015.62.6598. [DOI] [PubMed] [Google Scholar]
- 45.Petty RD, Dahle-Smith A, Stevenson DAJ, Osborne A, Massie D, Clark C, Murray GI, Dutton SJ, Roberts C, Chong IY, Mansoor W, Thompson J, Harrison M, Chatterjee A, Falk SJ, Elyan S, Garcia-Alonso A, Fyfe DW, Wadsley J, Chau I, Ferry DR, Miedzybrodzka Z. Gefitinib and EGFR gene copy number aberrations in esophageal cancer. J. Clin. Oncol. 2017;35:2279–2287. doi: 10.1200/JCO.2016.70.3934. [DOI] [PubMed] [Google Scholar]
- 46.Xie C, Jing Z, Luo H, Jiang W, Ma L, Hu W, Zheng A, Li D, Ding L, Zhang H, Xie C, Lian X, Du D, Chen M, Bian X, Tan B, Xia B, Xie R, Liu Q, Wang L, Wu S. Chemoradiotherapy with extended nodal irradiation and/or erlotinib in locally advanced oesophageal squamous cell cancer: long-term update of a randomised phase 3 trial. Br J Cancer. 2020;123:1616–1624. doi: 10.1038/s41416-020-01054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ha YNE, Dai Y, Wufuer D, Pidayi M, Anasihan G, Chen L. Second-generation Src/Abl inhibitor bosutinib effectively induces apoptosis in human esophageal squamous cell carcinoma (ESCC) cells via inhibiting Src/Abl signaling. Neoplasma. 2020;67:54–60. doi: 10.4149/neo_2019_190131N94. [DOI] [PubMed] [Google Scholar]
- 48.Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R, Spangler RD, Eaton JK, Frenkel E, Kocak M, Corsello SM, Lutsenko S, Kanarek N, Santagata S, Golub TR. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu K, Zhao T, Wang J, Chen Y, Zhang R, Lan X, Que J. Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett. 2019;458:21–28. doi: 10.1016/j.canlet.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.