Abstract
Understanding the specificity and complexity of the tumor microenvironment (TME) of Ewing sarcoma (ES) is essential for identifying the immune characteristics of ES, improving the prediction of immunotherapeutic response, and facilitating therapeutic target discovery. In this study, we not only evaluated the gene sets associated with TME in ES using ESTIMATE and WGCNA algorithms based on the transcriptome data of ES, but also constructed a prognostic model (ES Score) using univariate Cox regression and Lasso regression and assessed its predictive ability on immune cell infiltration. Subsequently, we identified ARAP3 as a key gene affecting the TME of ES. In addition, bioinformatic analyses and in vitro experiments proved that the high expression of ARAP3 regulated ES cell proliferation, migration, as well as apoptosis via the p53 signaling pathway and affected macrophage infiltration and osteoclast differentiation through regulating IL1B and IL11 secretion of tumor cells.
Keywords: Ewing sarcoma, ARAP3, tumor microenvironment, prognosis, macrophage, osteoclast
Introduction
Ewing Sarcoma (ES) is the second most common primary soft tissue and bone tumor of mesenchymal stem cell (MSC) origin in children and adolescents [1,2]. With the advances in the treatment strategy for early and local ES, the 5-year survival rate of these patients is more than 70%. However, about 20-30% ES patients with distant metastasis at the time of diagnosis are unresponsive to traditional treatment or develop relapses, resulting in a 5-year survival rate of less than 30% [2,3]. As one of the most significant features of ES, EWS-FLI1 chimeric protein has been proven to be crucial for the initiation and progression of ES, and thus may be used as a therapeutic target, which is an important breakthrough to improve the therapeutic effect for recurrent and metastatic ES [4,5]. In addition, EWS-FLI1 deletion has been reported to inhibit the growth of ES cells [6,7]; however, no drug targeting EWS-FLI1 has been reported so far [8]. Therefore, it is necessary to explore novel therapeutic strategies to improve the patient survival.
Although immunotherapies have achieved remarkable therapeutic efficacy in hematologic cancer, it is less impressive in other cancers, especially in solid tumors. Complex factors produced from the tumor microenvironment (TME) during the process of occurrence and progression of solid tumors may weaken or even undermine the effectiveness of immunotherapies [9,10]. The composition of TME includes not only tumor cells but also various immune cells and stromal cells, which are heterogeneous in different tumor types. These cells form a complex cellular communication network by secreting various cytokines and chemokines, which further regulate the tumor progression [11,12]. Tumor infiltrating immune and stromal cells can profoundly influence the efficacy of anti-tumor therapy by playing pro-tumor and anti-tumor roles [13,14]. A better understanding of TME in specific tumors is imperative to improve the targeting and immunotherapy strategies. In addition, the functional coordination of the characteristic osteoclasts (OCs) and osteoblasts (OBs) in the bone microenvironment of ES is the premise to maintain the stability of bone ecological niche, which affects tumor development and progression. The activation of OCs and the subsequent vertebral collapse and spinal compression caused by bone resorption are important clinical features of bone tumors. At the same time, ES has the characteristics of “moth eaten” in clinical imaging, namely multiple, fusion, and osteolytic lesions. Subperiosteal bone growth can be described by Codman triangle and “onion peel” appearances, representing the new periosteal layers on the tumors [2,15]. Such clinical information suggests the disturbance of OCs and OBs in the ES-TME.
With the continuous development of sequencing and bioinformatics technology, the gene profiles of specific diseases have been revealed by bioinformatics analysis of big data [16,17]. A variety of transcriptome analysis tools based on TME has provided the theoretical and methodological basis for evaluating the characteristics of ES-TME and mining the key factors affecting the occurrence and progression of ES [18,19]. Hence, the present study aimed to identify key prognostic molecules through a comprehensive analysis of the transcriptome characteristics of ES-TME to shed light on the clinical treatment of ES.
Materials and methods
Data source
The gene expression profiles of two independent cohorts of clinically annotated ES tumors (GSE63155 and GSE63156) from the NCBI Gene Expression Omnibus (GEO) database were used as the training cohort. The two datasets, containing 46 and 39 samples, respectively, were merged after removing the batch effects by using the ComBat function from “sva” R package. For the validation cohort, 57 ES samples with transcriptome and clinical data from the International Cancer Genome Consortium (ICGC) database were used.
Screening of TME-related genes
An ESTIMATE algorithm, which is developed to predict the TME phenotypes of tumor purity and stromal and immune cell infiltration in malignant tumor tissues based on gene expression data, was applied to evaluate the TME status in the ES samples. In addition, the WGCNA algorithm, which can calculate the correlation among genes from different samples and aggregate the highly correlated genes into different modules, was used to construct the co-expression network and identify different gene modules. Furthermore, association analysis was conducted between the modules and the TME phenotypes to obtain the gene set with the highest correlation with TME. The inclusion criteria were |correlation coefficient| ≥0.4 and p-value ≤0.05. R packages “ESTIMATE” and “WGCNA” were used for relevant analyses in the training cohort.
Prognostic analysis and construction of a TME-related risk model (ES score)
Univariate Cox analysis was performed for all TME-related gene sets to stratify genes that were significantly associated with prognosis. The Least Absolute Shrinkage and Selection Operator (LASSO) regression analysis was applied to further compress the gene variables and construct the prognostic model, namely, ES Score. Risk scores were calculated using the formula: Risk score = ∑coef(i) × Exp(i), where coef(i) and Exp(i) are the coefficient and the expression level of each gene in the model, respectively. Samples were divided into high- and low-ES Score groups based on the optimal cutoff value. Kaplan-Meier survival curves and receiver operating characteristic time-dependent (ROC) curves were employed to evaluate the model performance in both training and validation cohorts. R packages “glmnet”, “survival”, “survivalROC” and “timeROC” were used for prognostic analyses.
Exploration of the potential biological function of the ES score model
The single-sample Gene Set Enrichment Analysis (ssGSEA) algorithm implemented in R package “GSVA” was utilized to calculate the enrichment score of several biological processes and signaling pathways in each sample. Related gene sets were obtained from Molecular Signatures Database (MsigDB). Correlation analysis was performed between the ES Score and other scores to explore the potential biological functions. Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analyses were applied based on differentially expressed genes (DEGs), which were determined by using R package “limma” between the high- and the low-ES Score groups.
Estimation of TME cells infiltration and immune functions
The abundance of tumor infiltrating cells and immune functions was evaluated by ssGSEA algorithm. Related gene sets were obtained from previous studies, which included 28 types of immune cells and 13 types of immune functions [20,21]. Gene sets of OCs and OBs were downloaded from CellMarker and PanglaoDB databases. The gene list of osteoclastogenic cytokines was obtained from the study of Amarasekara et al. [22].
Cell lines and clinical samples
Human ES cell lines RD-ES, SK-ES-1, A673 and SK-NM-C and mouse cell line RAW264.7 (American Type Culture Collection (Manassas, VA, US)) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Human bone marrow-derived mesenchymal stem cells (hBMSCs; Cyagen Biosciences Inc., Suzhou, China) were cultured in Minimum Essential Medium alpha modification (α-MEM) supplemented with 10% FBS. Murine bone marrow-derived monocytes (BMMs) were isolated from C57BL/6 mice as previous described [23]. A total of 31 ES tumor and 9 peri-tumor samples were obtained from patients who underwent surgeries at Changzheng Hospital (Shanghai, China). Informed consents were obtained from all participants, and the research procedures involving human subjects were approved by the ethical committee of Changzheng Hospital (Shanghai, China).
RNA isolation and real-time quantitative PCR (RT-qPCR)
Total RNA was isolated by TRIZOL (Takara, Otsu, Shiga, Japan) methods. Reverse transcription and RT-qPCR assays were respectively conducted using HiScript III RT SuperMix and SYBR qPCR Master Mix (Vazyme, Nanjing, Jiangsu, China). Primer sequences are listed in Table S1.
Western blot
Total protein was extracted by RIPA lysis containing protease inhibitor cocktail (P1005; Beyotime, Shanghai, China). Protein samples were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were then blocked in PBST containing 5% bovine serum albumin (BSA) for 1 h and incubated with the indicated primary antibody at 4°C overnight. After extensive washing, the membranes were incubated with horse radish peroxidase (HRP) conjugated secondary antibody (BS13278; Bioworld, Bloomington, Indiana, USA) for 2 h, and the protein signals were detected by ECL and analyzed by ChemiDoc XRS+ Gel System (Bio-Rad, Hercules, CA, USA). The primary antibodies used in this study were as follows: ARAP3 (HPA042887; ATLAS ANTIBODIES, Stockholm, Sweden), IL1β (12703; Cell Signaling Technology, Danvers, MA, USA), IL11 (A1902; ABclonal, Wuhan, China), and GAPDH (AP0063; Bioworld, Bloomington, Indiana, USA).
Immunohistochemistry (IHC) staining
Clinical samples were prepared for tissue arrays and processed for IHC staining using the standard procedures. Images were scanned with a digital slide scanner PANNORAMIC 250 and analyzed with CaseViewer, Version 2.4.0 (3D HISTECH, Budapest, Hungary). IHC scores were measured to assess the expression of the indicated proteins as previously described [24].
Small interference RNA
RD-ES and SK-ES-1 cells were transfected with small interfering RNA (siRNA) specifically targeting ARAP3 (si1: 5’-GCAGAAAUGUGCGGCUCUAAATT-3’; si2: 5’-AGAGGCCUGGGUGAUGUUAAA-3’) or non-specific scramble siRNA as negative control (siNC: 5’-UUCUCCGAACGUGUCACGUTT-3’) for ARAP3 knockdown. siRNAs were purchased from Genomeditech Inc. (Shanghai, China).
Cell proliferation assays
The Cell Counting Kit-8 (CCK-8, C0042; Beyotime, Shanghai, China) and Colony formation assays were carried out to examine the proliferation of ES cells expressing ARAP3 siRNA or control siRNA. For CCK-8 assay, the cells were seeded into 96-well plates (1 × 103/well) and allowed to grow for 5 days. CCK-8 solution was added to cells every two days for 2 h before the optical density at a wavelength of 450 nm was recorded. For colony formation assay, cells were seeded into 6-well plates (1 × 103/well) and cultured for 7-10 days. Then, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution. The number of colonies were counted under microscope.
Flow cytometry
Cell cycle and the apoptotic cells were analyzed by flow cytometry (CyAnADP; BECKMAN, Indianapolis, IN, USA). For cell cycle analysis, cells were stained with propidium iodide (PI) using Cell cycle staining Kit (MULTI SCIENCES, Hangzhou, Zhejiang, China). For apoptosis assay, cells were stained with APC Annexin V and 7-Aminoactinomycin D (7-AAD) using Annexin V Apoptosis Detection Kit (BD Biosciences, NJ, USA).
Transwell migration assay
The impact of ARAP3 silencing on ES cell migration and the recruitment of monocytes was assessed by transwell migration assay. To determine ES cell migration, cells (5 × 104-1 × 105) were seeded in the upper chamber in FBS-free medium, while the bottom well was filled with complete medium. The RD-ES cells were cultured for 6 h (SK-ES-1 cells for 24 h), and the cells migrated to the lower side of the filter were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet solution, and counted under microscope. To detect monocyte recruitment, BMMs or RAW264.7 cells were seeded in the upper chamber in complete medium, while ES cells expressing either ARAP3 silencing or control siRNA were seeded in the bottom well in complete medium with 10 ng/mL M-CSF (R&D, Minneapolis, MN, USA). After incubation with BMMs for 8 h or incubation with RAW264.7 for 14 h, migrated cells were determined as described above.
Osteoclastic differentiation assay
To induce osteoclastic differentiation, isolated BMMs were seeded into 96-well plates (1.5 × 104/well) and cultured in OC differentiation medium containing 10 ng/ml M-CSF and 50 ng/ml RANKL (Novoprotein, Suzhou, Jiangsu, China) for 7-10 days. Alternatively, RAW264.7 cells were seeded into 48-well plates (5 × 103/well) and cultured in OC differentiation medium containing 30 ng/ml RANKL for 3-5 days. The medium was refreshed every two days. Then, the cells were fixed with 4% paraformaldehyde and stained with tartrate resistant acid phosphatase (TRAP) staining kit (Sigma-Aldrich, St. Louis, MO, USA).
Statistical analysis
All statistics analyses were conducted by using GraphPad Prism, version 8.0.1 (San Diego, CA, USA) and R project version 4.1.1 (http://www.r-project.org/). Student’s t-test, nonparametric Mann-Whitney test, and two-way ANOVA were performed accordingly. Error bar represented standard error ofmean (SEM) or standard deviation (SD). The relationship among variables was evaluated via Pearson’s correlation coefficients. Overall survival (OS) and event free survival (EFS) were generated by Kaplan-Meier method and compared using Log rank tests. The optimal cutoff values were determined by “surv_cutpoint” function in R package “survminer”. Area under the curve (AUC) was quantified to assess the accuracy of the ROC curves. P value (two-sided) of <0.05 was considered statistically significant.
Results
Identification of TME-related gene sets in ES patients
Batch effects of gene expression data from GSE63155 and GSE63156 were removed. The gene expression profile and principal component analysis (PCA) were visualized to evaluate the removal of batch effects. The expression and distribution of these two datasets were similar after data correction (Figure S1A-D). The data were further filtered according to the criterion that the average expression level of each gene was greater than 2, and a total of 17,133 genes were finally obtained.
The TME score (immune score and stromal score) of each sample was calculated using the ESTIMATE algorithm. Subsequently, Kaplan-Meier curves were used to analyze the impact of the TME scores on patients’ prognosis. We found that both immune and stromal scores were prognostic factors and negatively associated with OS (Figure 1A, 1B).
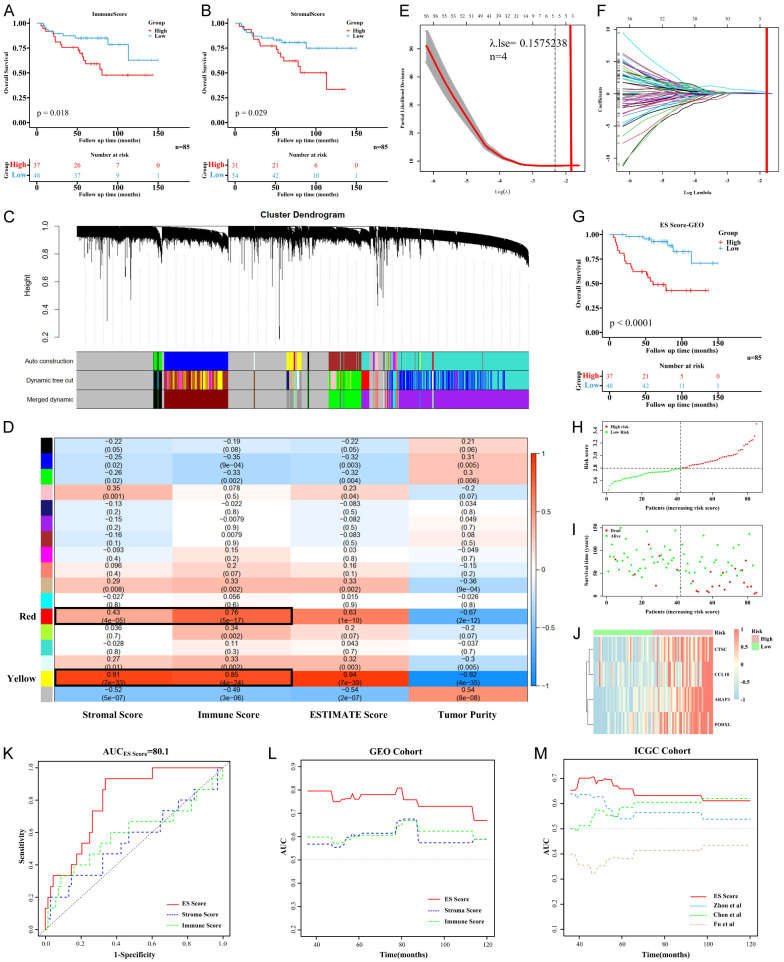
Figure 1.
Construction and validation of ES Score. A, B. Kaplan-Meier curves showed the overall survival between high and low immune or stromal scores of ES patients in GEO cohort. C. Gene modules identified by WGCNA. D. Heatmap of correlation between TME score phenotypes and gene modules. E, F. Lasso regression for variables compression and model construction. λ.1se was selected for further analysis. G. Kaplan-Meier curves showed the overall survival between high and low ES Score of ES patients in GEO cohort. H, I. The distribution diagram of risk score and survival status for each ES patient. J. Heatmap of the four candidate genes of ES patients in GEO cohort. Patients were arranged by ES Score from low risk to high risk. K, L. ROC and time-dependent ROC curves of ES Score, immune score and stromal score in GEO cohort. M. Time-dependent ROC curves of ES Score and other published prognostic models in ICGC cohort.
Combined with TME score, the WGCNA algorithm was used to conduct cluster analysis of genes with immune score, stromal score, and tumor purity as sample phenotypes. According to the distribution evaluation criteria of Scale-free Topology Model, R2=0.89 was determined when the soft threshold weight parameter β was set to 22 (Figure S1E-G). Seventeen gene modules were then obtained, among which the genes in gray module had no co-expression characteristics. By comparing the correlation coefficients between the TME score and the gene modules, we found that yellow and red modules had the highest correlation with the immune score, stromal score and tumor purity. A total of 598 genes in yellow and red modules were selected for subsequent analysis (Figure 1C, 1D).
Construction and validation of ES score
Univariate Cox analysis was performed on the 598 genes, and 119 TME-related genes were identified to be associated with the prognosis of ES patients. Lasso regression was used to further compress the variables, and λ.1se was selected to construct the model using the optimal variables. Finally, a set of 4 genes, ARAP3, PODXL, CTSC and CCL18, were identified as the ES-TME genes and were used to construct the TME-related risk model (Figure 1E, 1F).
Kaplan-Meier analysis showed that ES score had a significant effect on the OS and EFS of ES patients (Figures 1G, S2). In terms of distribution, mortality events were relatively concentrated in patients in the high-ES Score group (Figure 1H, 1I). Also, the 4 ES-TME-related genes showed a consistent degree of clustering and were highly expressed in patients in the high-ES Score group (Figure 1J). Given that the TME scores had good prognostic discrimination, we analyzed their predictive efficacy together with ES Score. ROC results suggested that ES Score had a stronger OS prediction ability, and the AUC of predicting the 3-year survival was 80.1% (Figure 1K, 1L). Although both the immune and stromal scores were correlated with survival, the immune or stromal score alone could not predict the prognosis of ES patients.
Similarly, ES Score also showed a significant predictive effect on the OS and EFS of ES patients in the validation cohort (Figure S2). Since several studies have reported the prognostic model for ES, and some of them provided formulas for their models, we compared the prognostic prediction efficacy between our ES Score and these models and found that ES Score had the best performance in predicting OS (Figure 1M) [25-27]. Kaplan-Meier analysis also demonstrated that ES Score had a more significant predictive value on the OS and EFS of ES patients than other models did (Figure S2).
Functional annotations and immune traits of ES score
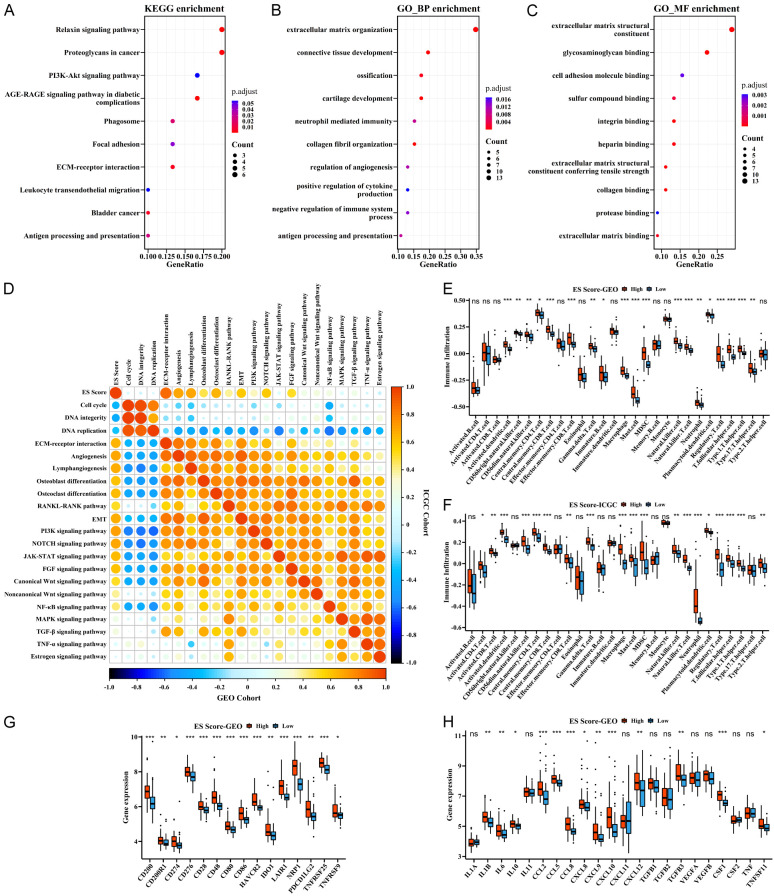
To determine the differences in functional phenotypes between the high- and low-ES Score groups, we identified DEGs between the two groups and performed GO and KEGG functional annotations. KEGG analysis identified several significantly enriched pathways that were related to tumor progression, stromal and immune activation (Figure 2A). For example, the classical PI3K-Akt signaling pathway played an important role in tumors. Relaxin signaling pathway exhibited pleiotropic functions of cell growth, angiogenesis, cell migration and extracellular matrix remodeling in the context of cancer progression [28-30]. Also, antigen processing and presentation, leukocyte transendothelial migration and ECM-receptor interaction were directly related to TME. Consistently, Go analysis also showed the enrichment of many biological functions that were related to stromal and immune regulation (Figure 2B, 2C).
Figure 2.
Functional annotations and immune traits of ES Score. A-C. KEGG and GO enrichment between high- and low-ES Score groups. D. Correlations between ES Score and other biological processes and signaling pathways in both cohorts. Negative correlation is marked in blue and positive correlation in orange. E, F. Immune cell infiltration characteristics of high- and low-ES Score groups in both cohorts. G, H. Expression of checkpoints and cytokines of high- and low-ES Score groups in GEO cohort. ***P<0.001, **P<0.01, *P<0.05 by Mann-Whitney U test.
Next, we explored the role of ES Score in ES progression and found that ES Score was highly correlated with a variety of tumor biological processes and signaling pathways, including cell cycle, angiogenesis, EMT, PI3K and NOTCH signaling pathways (Figure 2D). Notably, the bone tumor characteristics of ES suggested that ES score may correlated with OB and OC differentiation, as well as the activation of the RANKL-RANK signaling pathway (Figure 2D). Importantly, these functional phenotypes of ES Score showed good consistency in both training and validation cohorts.
Since the ES Score was derived from immune and stromal scores and associated with immune functions, we predicted that there were differences in immune characteristics between the high- and the low-ES Score groups. Indeed, immune cell infiltration analysis showed that high-ES Score group was prominently enriched with various immune cells including macrophages, mast cells, MDSCs, natural killer cells, neutrophils, and regulatory-T cells in both training and validation cohorts (Figure 2E, 2F). Due to the significant clinical application of immune checkpoint inhibitors, we examined the expression of several immune checkpoint proteins including PD-L1 (CD274) and revealed an upregulation of these immune checkpoint proteins in the high-ES Score group (Figures 2G, S3A). Furthermore, since cytokines are one of the main components of the TME and often act as the mediators of different cells in the microenvironment, thereby promoting or inhibiting the recruitment and the proliferation of these cells, we analyzed the expression of several important cytokines and found that IL6, CCL5, CXCL12, TGFB3, CSF1 and RANKL (TNFSF11) were highly expressed in the high-ES Score group (Figures 2H, S3B).
ARAP3 was a key ES-TME gene
To further explore the independent predictive value of these four ES-TME genes on patient survival, the OS and EFS of patients in both training and validation cohorts were analyzed by Kaplan-Meier curves. Our results illustrated that these four genes were associated with the OS and EFS of patients in the training cohort (Figures 3A, S4A); nevertheless, only ARAP3 showed its predictive value on both OS and EFS in the validation cohort, while ARAP3 and PODXL were associated with OS only (Figures 3B, S4B). Univariate analysis further showed that among the four genes, ARAP3 was the most influential factor on OS and EFS, with the largest HR value of 4.836 and 2.871, respectively (Figure S4C, S4D). These results suggested ARAP3 as the key ES-TME gene; therefore, we focused our experimental validation on ARAP3.
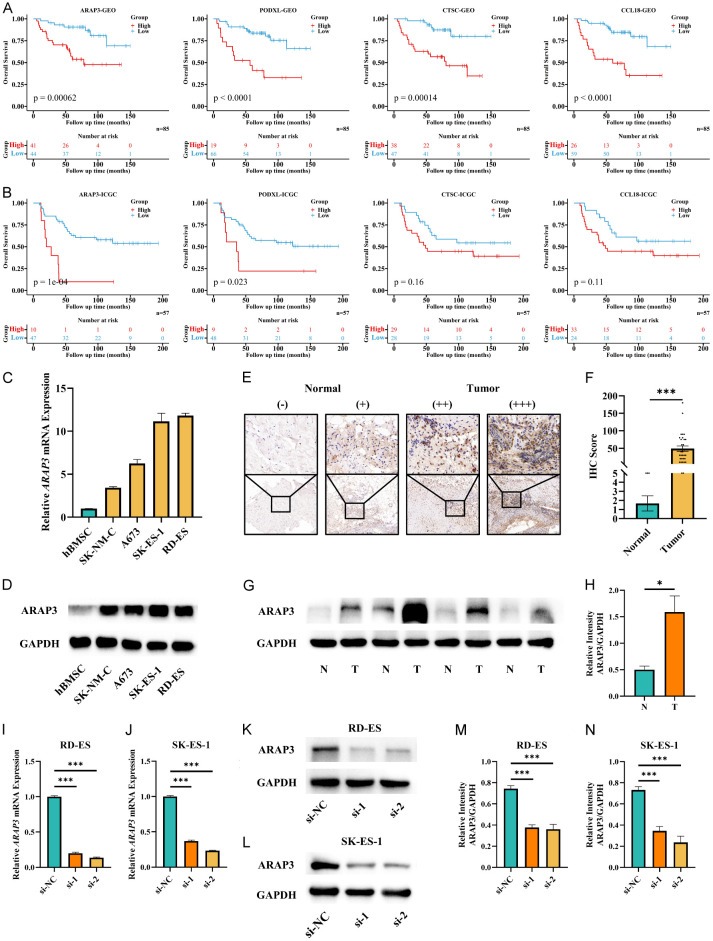
Figure 3.
ARAP3 was a key ES-TME gene. A, B. Kaplan-Meier curves showed the overall survival between high- and low-expressed groups of the four candidate genes of ES patients in both cohorts. C, D. Expression of ARAP3 in hBMSCs and ES cell lines detecting by RT-qPCR and WB. E-H. Expression of ARAP3 in normal and tumor tissues detecting by IHC and WB. The IHC images were captured and magnified at 10× and 40×. ***P<0.001 by Mann-Whitney U test for IHC. Mean ± SEM. *P<0.05 by unpaired t test for WB. Mean ± SD. I-N. Knockdown of ARAP3 in RD-ES and SK-ES-1 cell lines. ***P<0.001 by unpaired t test. Mean ± SD. Experiments were performed in triplicate.
We first assessed the expression of ARAP3 in normal hBMSCs and four human ES cell lines by RT-qPCR as well as western blot and discovered that ARAP3 was upregulated in all ES cell lines examined (Figure 3C, 3D). Furthermore, the expression of ARAP3 in ES tumor and normal peri-tumor samples was detected by IHC and western blot, and the results demonstrated that ARAP3 was highly expressed in tumor tissues with heterogeneity (Figure 3E-H). Subsequently, RD-ES and SK-ES-1 cell lines were selected for ARAP3 knockdown experiments to evaluate the effect of altered ARAP3 expression on cell behavior (Figure 3I-N).
ARAP3 knockdown inhibited the malignant behavior of ES cells via activating p53 signaling pathway
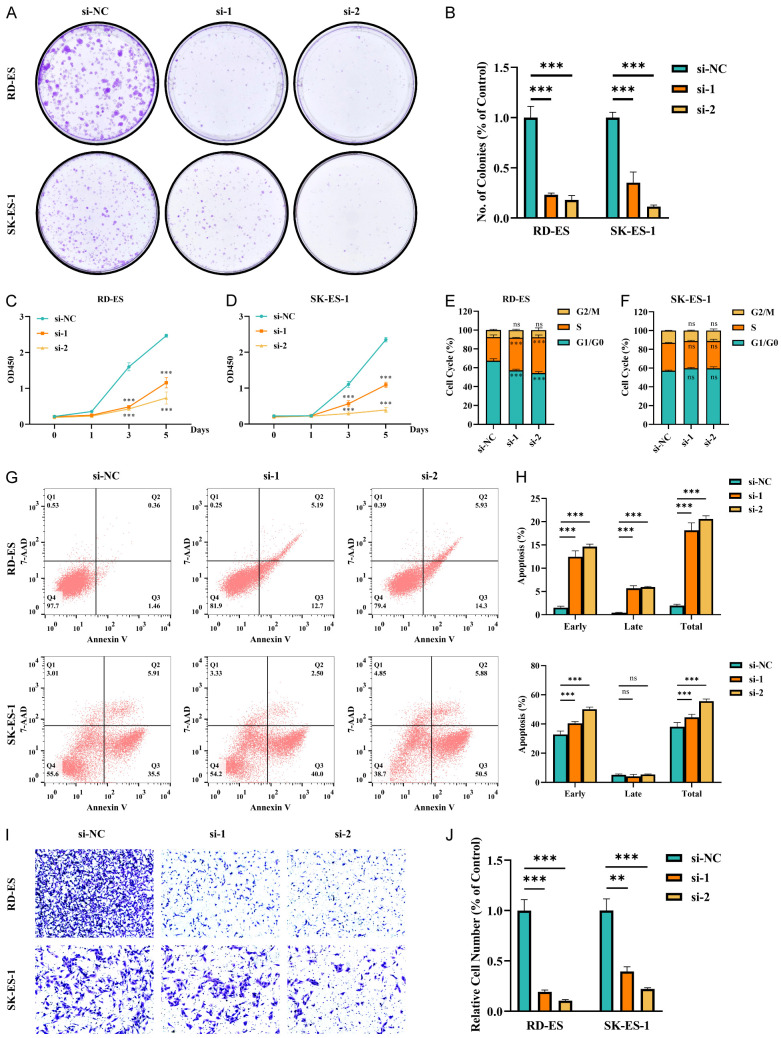
First, we discovered that the colony formation ability of ES cells was attenuated by ARAP3 knockdown (Figure 4A, 4B). Next, CCK-8 assay showed the proliferation of ES cells was significantly decreased in both ARAP3 knockdown cell lines (Figure 4C, 4D). Flow cytometry further showed that there was a significant increase in the S phase of ARAP3-knockdown RD-ES cells, while cells in G1/G0 phase were significantly decreased; nevertheless, there was no difference in the cell cycle distribution of SK-ES-1 cells (Figures 4E, 4F and S5A). Accordingly, ARAP3 knockdown significantly promoted early apoptosis in both ES cell lines and promoted the late apoptosis in RD-ES cells (Figure 4G, 4H). Moreover, transwell assay demonstrated that ARAP3 knockdown significantly inhibited ES cell migration (Figure 4I, 4J).
Figure 4.
ARAP3 knockdown inhibited the malignant behavior of ES cells. A, B. ARAP3 knockdown significantly depressed tumor colony formation in ES cell lines. ***P<0.001 by unpaired t test. C, D. ARAP3 knockdown significantly inhibited proliferation of ES cells by CCK8 assays. ***P<0.001 by Two-way ANOVA. E, F. Cell cycle analysis showed that ARAP3 knockdown significantly increased cell numbers in S phase and decreased cell numbers in G0/G1 phase in RD-ES cells. No differences were noted in SK-ES-1 cells. ***P<0.001 by Two-way ANOVA. G, H. Apoptosis analysis showed that ARAP3 knockdown significantly promoted early apoptosis in both ES cell lines, and also exhibited a promotional effect on late apoptosis in RD-ES cells. ***P<0.001 by Two-way ANOVA. I, J. ARAP3 knockdown significantly reduced cell migration by transwell assay. ***P<0.001 by unpaired t test. Experiments were performed in triplicate. Mean ± SD.
To further explore the functional mechanism of ARAP3 in ES progression, we collected three pairs of ARAP3 knockdown and control RD-ES cells for RNA sequencing. KEGG analysis of the RNA-sequencing data showed that PI3K-Akt signaling pathway, cytokine-cytokine receptor interaction, cell adhesion molecules, and p53 signaling pathway were significantly enriched (Figure S5B), consistent with previous reports that ARAP3 plays an important role in PI3K signaling pathway [31,32].
It has been well known that p53 signaling affects various tumor phenotypes such as cell cycle and apoptosis. Our analysis showed the enrichment of p53 signaling pathway by ARAP3 knockdown, suggesting that it may be an important mediator of ARAP3 in regulating ES progression. In the p53 signal transduction cascade, MDM2 plays a major negative role in regulating p53 level, while p53 controls cell cycle and apoptosis mainly through the activation of downstream target genes. Hence, we examined the expression of MDM2, p21 and Bax in ARAP3 knockdown and control cells and found that ARAP3 knockdown downregulated the expression of MDM2 and significantly upregulated the expression of p53 as well as its downstream target genes p21 and Bax, indicating the activation of p53 signaling (Figure S5C). Moreover, we examined the proliferation, migration, and apoptosis of RD-ES cells that were co-transfected with si-ARAP3 and si-TP53. The results revealed that, compared to ARAP3 knockdown alone, double knockdown of ARAP3 and TP53 enhanced cell proliferation and migration, along with a decreased cell apoptosis, demonstrating that combined ARAP3 and TP53 knockdown could partially reversed the anti-tumor effect by ARAP3 knockdown (Figure S6).
ARAP3 regulated macrophage infiltration and osteoclast differentiation
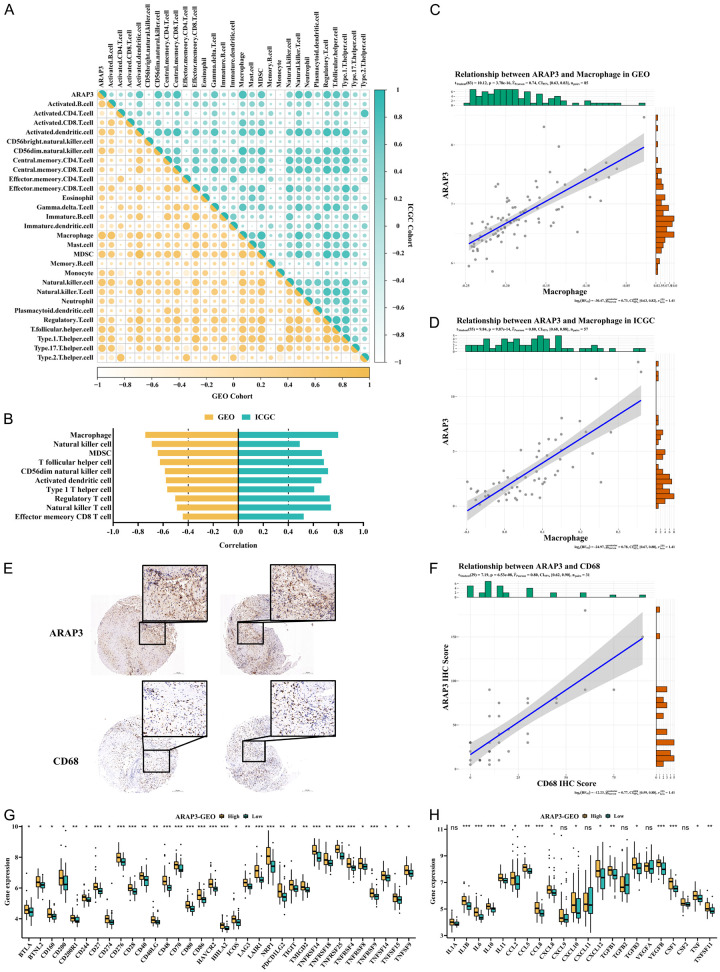
Our above bioinformatics analysis on immune cell infiltration showed a correlation between ARAP3 expression and the composition of immune infiltrating cell in both training and validation cohorts. Specifically, macrophages, MDSCs, dendritic cells, natural killer cells, and regulatory-T cells were enriched in ARAP3-high expression group (Figures 5A, S7A, S7B). To further confirm the immune cell types that were highly correlated with ARAP3 level, we extracted immune cells that had a correlation coefficient greater than 0.4 with ARAP3 in both cohorts and found that macrophages had the highest correlation with ARAP3 (Figure 5B-D). Then, IHC was performed to assess the expression of CD68, a main marker of monocyte-derived macrophages, in ES samples. Combined with the IHC score of ARAP3, we found that CD68 expression was highly correlated with ARAP3 level (Figure 5E, 5F). In addition, we also compared the expression of cytokines and checkpoint proteins between the low- and the high-ARAP3 expression groups (Figures 5G, 5H, S7C, S7D). Notably, the differences in the expression of a large number of cytokines and checkpoint proteins suggested the potential role of ARAP3 in immune cell activation and immunotherapeutic monitoring.
Figure 5.
Regulation of ARAP3 in immune traits. A. Correlations between ARAP3 and immune cells in both cohorts. B-D. Macrophages had the highest correlation with ARAP3 in both cohorts. r GEO=0.74, r ICGC=0.80, P<0.001 by Pearson’s correlation. E, F. CD68 was highly correlated with ARAP3 as detected by IHC. The IHC images were captured and magnified at 5× and 20×. r=0.80, P<0.001 by Pearson’s correlation. G, H. Expression of checkpoints and cytokines of high- and low-expressed ARAP3 groups in GEO cohort. ***P<0.001, **P<0.01, *P<0.05 by Mann-Whitney U test.
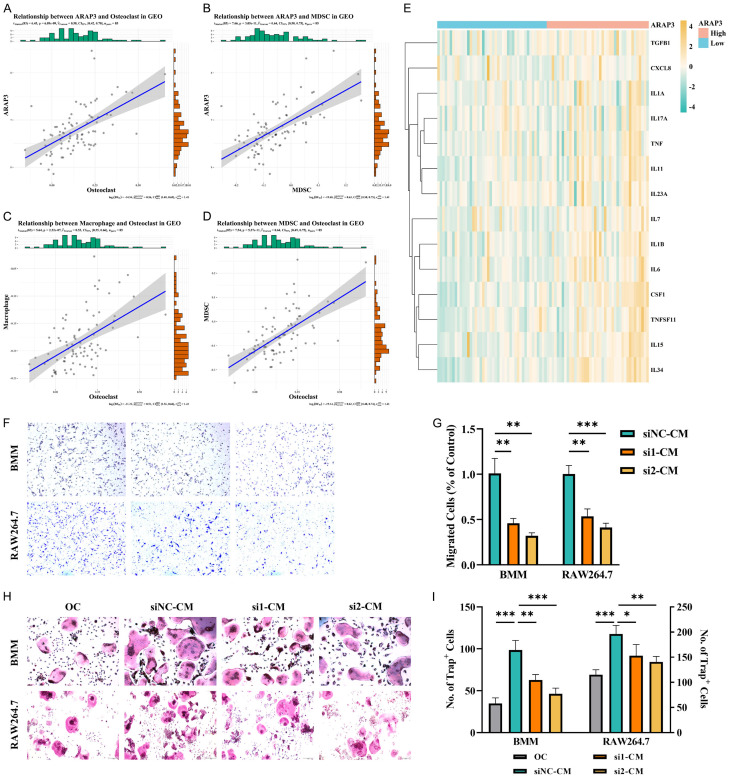
Furthermore, we found a strong correlation of ARAP3 with OCs and OBs, further demonstrating the role of ARAP3 in the ES microenvironment, in line with the ES Score result above (Figures 6A, S8A, S8E, S8F). In addition, previous studies have shown that macrophages and MDSCs are closely related to OCs, which is consistent with the findings in our study (Figures 6B-D, S8B-D). In fact, the expression of osteoclastogenic cytokines was higher in the ARAP3-high expression group (Figures 6E, S8G). It is known that monocytes are often recruited by tumor cells in TME where they are induced into mature macrophages or OCs by a variety of cytokines secreted by tumor cells. Similar to the findings in the monocyte recruitment experiments, we also observed that ARAP3 knockdown decreased the recruitment of BMMs and Raw264.7 cells (Figure 6F, 6G). Moreover, we used the conditioned medium (CM) from tumor cells to induce the OC differentiation of BMMs and Raw264.7 cells and found that ARAP3 knockdown attenuated the CM-induced osteoclast differentiation (Figure 6H, 6I). Consistently, the CM-induced expression of OC markers in Raw264.7 cells was also decreased by ARAP3 knockdown (Figure S8H-J).
Figure 6.
ARAP3 regulated osteoclast differentiation. A, B. Correlation of ARAP3 with osteoclasts and MDSCs in GEO cohort. r ARAP3-OC=0.58, r ARAP3-MDSC=0.64, P<0.001 by Pearson’s correlation. C, D. Correlation of osteoclasts with macrophages and MDSCs in GEO cohort. r Macrophage-OC=0.53, r MDSC-OC=0.64, P<0.001 by Pearson’s correlation. E. Heatmap of osteoclastogenic cytokines of ES patients in GEO cohort. Patients were arranged by ARAP3 from low expression to high expression. F, G. ARAP3 knockdown significantly inhibited recruitment of BMMs and Raw264.7 cells by ES cells. ***P<0.001, **P<0.01 by unpaired t test. H, I. ARAP3 knockdown significantly attenuated ES-promoted osteoclast differentiation. ***P<0.001, **P<0.01, *P<0.05 by One-way ANOVA. Experiments were performed in triplicate. Mean ± SD.
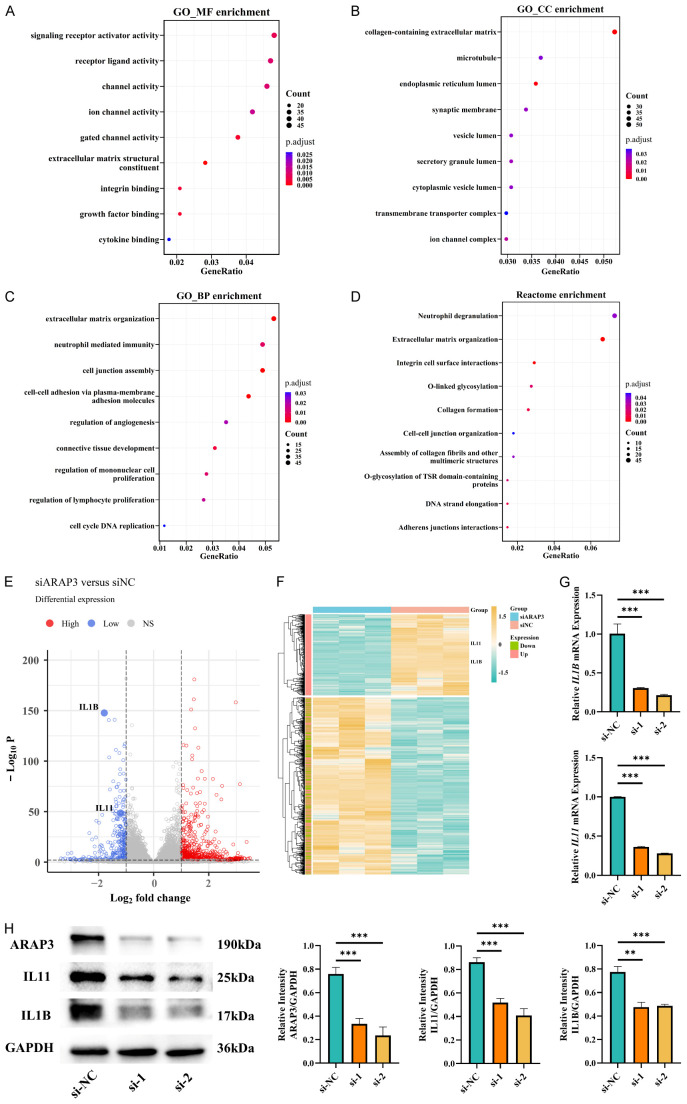
Based on the results above, we concluded that cytokines may play a major role in ARAP3 regulation of TME, which was confirmed by RNA sequencing. In addition, KEGG and GO analyses showed the enrichment in the interaction between cytokine and cytokine receptor, as well as the functions of receptor ligand, cytokine binding and types of channel activity (Figures 7A, S5B). Moreover, the function of extracellular matrix remodeling, cell cycle and immune response were also enriched, consistent with the phenotype analysis discussed above (Figure 7B-D). We also analyzed the differential expression of cytokines and found that IL1B and IL11 were significantly downregulated by ARAP3 knockdown, which was further verified by RT-qPCR and western blot (Figure 7E-H). Therefore, we proposed that ARAP3 may affect TME, especially macrophage infiltration and osteoclast differentiation, by regulating IL1B and IL11 secretion of tumor cells.
Figure 7.
ARAP3 affected TME by regulating IL1B and IL11 secretion of tumor cells. A-D. GO and REACTOME enrichment between ARAP3 knockdown and control cells. E, F. Volcano plot and heatmap of DEGs between ARAP3 knockdown and control cells. The location of IL1B and IL11 were marked. G, H. Expression of IL1B and IL11 in ARAP3 knockdown and control cells as detected by RT-qPCR and WB. ***P<0.001, **P<0.01 by unpaired t test. Experiments were performed in triplicate. Mean ± SD.
Discussion
During tumorigenesis, the co-evolution of malignant cells and their microenvironment facilitates tumor progression. The composition and the alteration of the microenvironment vary among different tumor types, but in general the TME consists of immune cells, stromal cells, and non-cellular components including blood vessels and extracellular matrix [33]. In addition, the crosstalk between malignant tumors and the bone microenvironment has been extensively studied, especially in the bone metastasis of breast and prostate cancers, based on the “seed and soil” theory proposed by Paget [34] which is also applicable to primary bone tumors such as osteosarcoma and chondrosarcoma, but is rarely studied in ES [35,36]. In this study, we not only assessed the TME properties of ES, but also included OCs and OBs in our assessment.
ES Score is a TME-related prognostic signature based on the clinical and transcriptome data from the GEO cohort, which was used to evaluate the TME characteristics of ES patients as well as to predict prognosis and guide immunotherapy strategies. We further compared the functions and pathways associated with the DEGs between the low- and high-ES Score groups and found an enrichment of immune cell-related functions, including antigen presentation, of immune system regulation and cytokine production, functions involving bone stromal cells such as ossification and cartilage development, and regulation of angiogenesis and ECM function. In addition, we analyzed the effect of ES Score on the differentiation of OCs and OBs and discovered a highly positive correlation between them.
Recent studies on tumor progression and therapies have also paid attention to TME central models, such as immune checkpoint blockade which targets T cell activation in TME by blocking receptor/ligand interactions (e.g., CTLA-4 and PD-1) [37,38]. However, the outcomes of therapeutic strategies targeting TME, especially targeting specific cells or pathways of TME, remain unsatisfactory [39,40]. Different TME phenotypes may represent different survival outcomes and beneficial effect from immunotherapy. Our results showed that patients in high-ES Score group presented more immune cell infiltration, including more immunosuppressive cells such as MDSCs, macrophages, Treg, and Th17. Immune checkpoint protein analysis showed that the expression of multiple checkpoint proteins, including PD-1, was elevated in patients with high ES Score. Previous studies have shown that PD-1/PD-L1 treatment does not produce a satisfactory therapeutic efficacy in ES; hence, further in vivo experiments and clinical trials are necessary to explore the benefits of checkpoint blockade in ES. Cytokines are the main mediators by which tumor cells shape their microenvironment, thus leading to the reprogramming of surrounding cells and tumor survival and progression [41]. In this study, we found that multiple cytokines were enriched in patients with high ES Score, thereby promoting directional cell migration and homing of the immune system. Specifically, pro-inflammatory cytokines such as IL1B, IL6, CXCL9 and CXCL10 are chemoattractant for T cells [42]. Among the cytokines, CXCL12 has been reported to play a key role in tumor progression by promoting tumor cell proliferation, survival, metastatic processes and angiogenesis [43], while IL1B, IL11, CSF1 and TNFSF11 can promote OC differentiation [22].
We identified four genes that can be used to generate ES Score, among which, CCL18 can promote cancer cell proliferation, invasion, EMT and angiogenesis, and may serve as a marker for M2-type macrophages that recruit regulatory T cells, CAFs and tumor-associated dendritic cells, thereby promoting the formation of tumor immunosuppressive microenvironment [44]. On the other hand, CTSC is mainly associated with inflammatory diseases. Notable, tumor derived CTSC can promote the lung metastasis of breast cancer by regulating neutrophil infiltration and the formation of neutrophil extracellular traps in early metastatic niche [45]. The third gene, PODXL, a membrane protein involved in tumor progression, is aberrantly expressed in several cancer types and is an indicator of poor prognosis. Notably, a novel and specific monoclonal antibody (PODO447) against PODXL, combined with a unique tumor-restricted glycan epitope, has been manufactured [46].
ARAP3 has been less studied in tumors and exhibits different properties in different tumors. Yagi et al. revealed that ARAP3 inhibited peritoneal dissemination of scirrhous gastric carcinoma cells. In contrast, ARAP3 was reported to promote tumor progression in both breast and thyroid cancers [47-50]. Our study suggested that ARAP3 depletion might activate p53 signaling pathway, thereby inhibiting the proliferation, migration, and invasion, as well as promoting the apoptosis of ES cells. The functional heterogeneity of ARAP3 in tumor progression may result from its specific structural features. ARAP3 is characterized by the existence of 5 unique domains: two GAP domains (Arf GAP and Rho GAP domains), one RA domain, one SAM domain and five PH domains. Accumulating evidence has indicated that ARAP3 plays an important role in PI3K signaling. Upon PI3K activation, ARAP3 can be recruited to the plasma membrane, where its main substrates RhoA-GTP and Arf6-GTP are located after PIP3 binding to the PH domains of ARAP3 [32,51,52]. Thus, ARAP3 regulates not only the activation of p53 signaling pathway but also the activities of PI3K and GTPase signaling pathways. In consistent with these studies, our analysis also revealed the co-enrichment of PI3K-Akt pathway and p53 pathway; nevertheless, further validation is required.
In this study, ARAP3 also showed strong potential to regulate TME. On one hand, high expression of ARAP3 can affect the infiltration of immune cells, especially macrophages, as well as the expression of a large number of checkpoint proteins and cytokines; on the other hand, ARAP3 can regulate ES cells to induce OC differentiation. Our GO and KEGG enrichment analyses and in vitro experiments suggest that ARAP3 modulated microenvironment through cytokine secretion. Indeed, we found that the expression of IL1B and IL11 was significantly decreased by ARAP3 knockdown. Previous studies have demonstrated that IL1B regulates tumor progression and TME crosstalk. Specifically, IL1B secreted by tumor cells can induce chemotaxis of macrophages, while in the modified bone microenvironment, IL1B can promote the induction of OCs and regulate the expression of RANKL/OPG in OBs [53,54]. As for IL-11, a member of the IL-6 family, IL-11 can stimulate hematopoiesis and platelet production, regulate macrophage differentiation, and induce osteoclast formation [55,56]. However, the specific signaling pathway by which ARAP3 regulates the expression of IL1B and IL11 remains to be elucidated.
In conclusion, we constructed a TME-related prognostic model to predict the prognosis of ES patients and guide immunotherapy strategy. ARAP3 could regulate tumor progression via p53 signaling pathway and inducing macrophage infiltration and OC differentiation.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81874180, 81902732, 81972505).
Disclosure of conflict of interest
None.
Abbreviations
- ES
Ewing Sarcoma
- MSC
Mesenchymal stem cell
- TME
Tumor microenvironment
- OCs
Osteoclasts
- OBs
Osteoblasts
- hBMSCs
Human bone marrow-derived mesenchymal stem cells
- BMMs
Bone marrow-derived monocytes
- MDSCs
Myeloid-derived suppressor cells
- PCA
Principal component analysis
- OS
Overall survival
- EFS
Event-free survival
- ICGC
International Cancer Genome Consortium
- GEO
The Gene Expression Omnibus
- GO
Gene ontology
- ssGSEA
The single-sample Gene Set Enrichment Analysis
- MsigDB
Molecular Signatures Database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ESTIMATE
Estimation of STromal and Immune cells in MAlignant Tumour tissues using Expression data
- WGCNA
Weighted gene co-expression network analysis
- LASSO
The Least Absolute Shrinkage and Selection Operator
- ROC
the receiver operating characteristic curve
- HR
Hazard ratio
- DEGs
Differentially expressed genes
- IHC
Immunohistochemical
- CM
Conditional medium
- ECM
Extracellular matrix
- EMT
Epithelial-mesenchymal transition
Supporting Information
References
- 1.Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, Sorensen PH, Delattre O, Dirksen U. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 2.Riggi N, Suva ML, Stamenkovic I. Ewing’s sarcoma. N Engl J Med. 2021;384:154–164. doi: 10.1056/NEJMra2028910. [DOI] [PubMed] [Google Scholar]
- 3.Zollner SK, Amatruda JF, Bauer S, Collaud S, de Alava E, DuBois SG, Hardes J, Hartmann W, Kovar H, Metzler M, Shulman DS, Streitburger A, Timmermann B, Toretsky JA, Uhlenbruch Y, Vieth V, Grunewald TGP, Dirksen U. Ewing sarcoma-diagnosis, treatment, clinical challenges and future perspectives. J Clin Med. 2021;10:1685. doi: 10.3390/jcm10081685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 5.Thangaretnam KP, Gopisetty G, Ramanathan P, Rajkumar T. A polypeptide from the junction region sequence of EWS-FLI1 inhibits Ewing’s sarcoma cells, interacts with the EWS-FLI1 and partner proteins. Sci Rep. 2017;7:7172. doi: 10.1038/s41598-017-07482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka K, Iwakuma T, Harimaya K, Sato H, Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing’s sarcoma and primitive neuroectodermal tumor cells. J Clin Invest. 1997;99:239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casey DL, Lin TY, Cheung NV. Exploiting signaling pathways and immune targets beyond the standard of care for ewing sarcoma. Front Oncol. 2019;9:537. doi: 10.3389/fonc.2019.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, Chew A, Gonzalez VE, Zheng Z, Lacey SF, Mahnke YD, Melenhorst JJ, Rheingold SR, Shen A, Teachey DT, Levine BL, June CH, Porter DL, Grupp SA. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 11.Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, Osokin N, Kozlov I, Frenkel F, Gancharova O, Almog N, Tsiper M, Ataullakhanov R, Fowler N. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39:845–865. e7. doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 13.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 14.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Zhang T, Chen M, Yao K, Huang X, Zhang B, Li Y, Liu J, Wang Y, Zhao Z. Bone physiological microenvironment and healing mechanism: basis for future bone-tissue engineering scaffolds. Bioact Mater. 2021;6:4110–4140. doi: 10.1016/j.bioactmat.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu H, Zhu Y, Wang Y, Liu Z, Zhang J, Xie H, Fu Q, Dai B, Ye D, Xu J. Identification and validation of stromal immunotype predict survival and benefit from adjuvant chemotherapy in patients with muscle-invasive bladder cancer. Clin Cancer Res. 2018;24:3069–3078. doi: 10.1158/1078-0432.CCR-17-2687. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H, Wang L, Lu T, Zhang Y, Sun Z, Han X. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat Commun. 2022;13:816. doi: 10.1038/s41467-022-28421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Liang JY, Wang DS, Lin HC, Chen XX, Yang H, Zheng Y, Li YH. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int J Biol Sci. 2020;16:2430–2441. doi: 10.7150/ijbs.45050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amarasekara DS, Yun H, Kim S, Lee N, Kim H, Rho J. Regulation of osteoclast differentiation by cytokine networks. Immune Netw. 2018;18:e8. doi: 10.4110/in.2018.18.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Huang J, Wang F, Li W, Wu X, Zhao C, Zhao J, Wei H, Wu Z, Qian M, Sun P, He L, Jin Y, Tang J, Qiu W, Siwko S, Liu M, Luo J, Xiao J. Dual targeting of bile acid receptor-1 (TGR5) and farnesoid X receptor (FXR) prevents estrogen-dependent bone loss in mice. J Bone Miner Res. 2019;34:765–776. doi: 10.1002/jbmr.3652. [DOI] [PubMed] [Google Scholar]
- 24.Dong K, Chen W, Pan X, Wang H, Sun Y, Qian C, Chen W, Wang C, Yang F, Cui X. FCER1G positively relates to macrophage infiltration in clear cell renal cell carcinoma and contributes to unfavorable prognosis by regulating tumor immunity. BMC Cancer. 2022;22:140. doi: 10.1186/s12885-022-09251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Wang X, Wang G, Xiao B, Ma Z, Huo H, Li W. A seven-lncRNA signature for predicting Ewing’s sarcoma. PeerJ. 2021;9:e11599. doi: 10.7717/peerj.11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Z, Yu B, Liu M, Wu B, Hou Y, Wang H, Jiang Y, Zhu D. Construction of a prognostic signature in Ewing’s sarcoma: based on metabolism-related genes. Transl Oncol. 2021;14:101225. doi: 10.1016/j.tranon.2021.101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Xu B, Wu S, Liu Y. Prognostic immune-related genes of patients with Ewing’s sarcoma. Front Genet. 2021;12:669549. doi: 10.3389/fgene.2021.669549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neschadim A, Summerlee AJ, Silvertown JD. Targeting the relaxin hormonal pathway in prostate cancer. Int J Cancer. 2015;137:2287–2295. doi: 10.1002/ijc.29079. [DOI] [PubMed] [Google Scholar]
- 29.Ng HH, Shen M, Samuel CS, Schlossmann J, Bennett RG. Relaxin and extracellular matrix remodeling: mechanisms and signaling pathways. Mol Cell Endocrinol. 2019;487:59–65. doi: 10.1016/j.mce.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valkovic AL, Bathgate RA, Samuel CS, Kocan M. Understanding relaxin signalling at the cellular level. Mol Cell Endocrinol. 2019;487:24–33. doi: 10.1016/j.mce.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 31.Gambardella L, Hemberger M, Hughes B, Zudaire E, Andrews S, Vermeren S. PI3K signaling through the dual GTPase-activating protein ARAP3 is essential for developmental angiogenesis. Sci Signal. 2010;3:ra76. doi: 10.1126/scisignal.2001026. [DOI] [PubMed] [Google Scholar]
- 32.Kartopawiro J, Bower NI, Karnezis T, Kazenwadel J, Betterman KL, Lesieur E, Koltowska K, Astin J, Crosier P, Vermeren S, Achen MG, Stacker SA, Smith KA, Harvey NL, Francois M, Hogan BM. Arap3 is dysregulated in a mouse model of hypotrichosis-lymphedema-telangiectasia and regulates lymphatic vascular development. Hum Mol Genet. 2014;23:1286–1297. doi: 10.1093/hmg/ddt518. [DOI] [PubMed] [Google Scholar]
- 33.Tian X, Shen H, Li Z, Wang T, Wang S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12:84. doi: 10.1186/s13045-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 35.Alfranca A, Martinez-Cruzado L, Tornin J, Abarrategi A, Amaral T, de Alava E, Menendez P, Garcia-Castro J, Rodriguez R. Bone microenvironment signals in osteosarcoma development. Cell Mol Life Sci. 2015;72:3097–3113. doi: 10.1007/s00018-015-1918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buenrostro D, Mulcrone PL, Owens P, Sterling JA. The bone microenvironment: a fertile soil for tumor growth. Curr Osteoporos Rep. 2016;14:151–158. doi: 10.1007/s11914-016-0315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dovedi SJ, Elder MJ, Yang C, Sitnikova SI, Irving L, Hansen A, Hair J, Jones DC, Hasani S, Wang B, Im SA, Tran B, Subramaniam DS, Gainer SD, Vashisht K, Lewis A, Jin X, Kentner S, Mulgrew K, Wang Y, Overstreet MG, Dodgson J, Wu Y, Palazon A, Morrow M, Rainey GJ, Browne GJ, Neal F, Murray TV, Toloczko AD, Dall’Acqua W, Achour I, Freeman DJ, Wilkinson RW, Mazor Y. Design and efficacy of a monovalent bispecific PD-1/CTLA4 antibody that enhances CTLA4 blockade on PD-1(+) activated T cells. Cancer Discov. 2021;11:1100–1117. doi: 10.1158/2159-8290.CD-20-1445. [DOI] [PubMed] [Google Scholar]
- 38.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 39.Blando J, Sharma A, Higa MG, Zhao H, Vence L, Yadav SS, Kim J, Sepulveda AM, Sharp M, Maitra A, Wargo J, Tetzlaff M, Broaddus R, Katz MHG, Varadhachary GR, Overman M, Wang H, Yee C, Bernatchez C, Iacobuzio-Donahue C, Basu S, Allison JP, Sharma P. Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer. Proc Natl Acad Sci U S A. 2019;116:1692–1697. doi: 10.1073/pnas.1811067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei Q, Wang D, Sun K, Wang L, Zhang Y. Resistance mechanisms of anti-PD1/PDL1 therapy in solid tumors. Front Cell Dev Biol. 2020;8:672. doi: 10.3389/fcell.2020.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morales E, Olson M, Iglesias F, Dahiya S, Luetkens T, Atanackovic D. Role of immunotherapy in Ewing sarcoma. J Immunother Cancer. 2020;8:e000653. doi: 10.1136/jitc-2020-000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janssens R, Struyf S, Proost P. The unique structural and functional features of CXCL12. Cell Mol Immunol. 2018;15:299–311. doi: 10.1038/cmi.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korbecki J, Olbromski M, Dziegiel P. CCL18 in the progression of cancer. Int J Mol Sci. 2020;21:7955. doi: 10.3390/ijms21217955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Cong M, Li J, He D, Wu Q, Tian P, Wang Y, Yang S, Liang C, Liang Y, Wen J, Liu Y, Luo W, Lv X, He Y, Cheng DD, Zhou T, Zhao W, Zhang P, Zhang X, Xiao Y, Qian Y, Wang H, Gao Q, Yang QC, Yang Q, Hu G. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell. 2021;39:423–437. e7. doi: 10.1016/j.ccell.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Canals Hernaez D, Hughes MR, Dean P, Bergqvist P, Samudio I, Blixt O, Wiedemeyer K, Li Y, Bond C, Cruz E, Kobel M, Gilks B, Roskelley CD, McNagny KM. PODO447: a novel antibody to a tumor-restricted epitope on the cancer antigen podocalyxin. J Immunother Cancer. 2020;8:e001128. doi: 10.1136/jitc-2020-001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han JJ, Du BR, Zhang CH. Bioinformatic analysis of prognostic value of ARAP3 in breast cancer and the associated signaling pathways. Eur Rev Med Pharmacol Sci. 2017;21:2405–2412. [PubMed] [Google Scholar]
- 48.Loskutov YV, Kozyulina PY, Kozyreva VK, Ice RJ, Jones BC, Roston TJ, Smolkin MB, Ivanov AV, Wysolmerski RB, Pugacheva EN. NEDD9/Arf6-dependent endocytic trafficking of matrix metalloproteinase 14: a novel mechanism for blocking mesenchymal cell invasion and metastasis of breast cancer. Oncogene. 2015;34:3662–3675. doi: 10.1038/onc.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang QX, Chen ED, Cai YF, Zhou YL, Zheng ZC, Wang YH, Jin YX, Jin WX, Zhang XH, Wang OC. Next-generation sequence detects ARAP3 as a novel oncogene in papillary thyroid carcinoma. Onco Targets Ther. 2016;9:7161–7167. doi: 10.2147/OTT.S115668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yagi R, Tanaka M, Sasaki K, Kamata R, Nakanishi Y, Kanai Y, Sakai R. ARAP3 inhibits peritoneal dissemination of scirrhous gastric carcinoma cells by regulating cell adhesion and invasion. Oncogene. 2011;30:1413–1421. doi: 10.1038/onc.2010.522. [DOI] [PubMed] [Google Scholar]
- 51.Krugmann S, Anderson KE, Ridley SH, Risso N, McGregor A, Coadwell J, Davidson K, Eguinoa A, Ellson CD, Lipp P, Manifava M, Ktistakis N, Painter G, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Dove SK, Michell RH, Grewal A, Nazarian A, Erdjument-Bromage H, Tempst P, Stephens LR, Hawkins PT. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- 52.Krugmann S, Williams R, Stephens L, Hawkins PT. ARAP3 is a PI3K- and rap-regulated GAP for RhoA. Curr Biol. 2004;14:1380–1384. doi: 10.1016/j.cub.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 53.Levescot A, Chang MH, Schnell J, Nelson-Maney N, Yan J, Martinez-Bonet M, Grieshaber-Bouyer R, Lee PY, Wei K, Blaustein RB, Morris A, Wactor A, Iwakura Y, Lederer JA, Rao DA, Charles JF, Nigrovic PA. IL-1beta-driven osteoclastogenic Tregs accelerate bone erosion in arthritis. J Clin Invest. 2021;131:e141008. doi: 10.1172/JCI141008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao C, Cai X, Wang Y, Wang D, Wang T, Gong H, Sun H, Jia Q, Zhou W, Wu Z, Li Z, Xiao J. NAT1 promotes osteolytic metastasis in luminal breast cancer by regulating the bone metastatic niche via NF-kappaB/IL-1B signaling pathway. Am J Cancer Res. 2020;10:2464–2479. [PMC free article] [PubMed] [Google Scholar]
- 55.Cai WL, Huang WD, Li B, Chen TR, Li ZX, Zhao CL, Li HY, Wu YM, Yan WJ, Xiao JR. microRNA-124 inhibits bone metastasis of breast cancer by repressing interleukin-11. Mol Cancer. 2018;17:9. doi: 10.1186/s12943-017-0746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putoczki T, Ernst M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J Leukoc Biol. 2010;88:1109–1117. doi: 10.1189/jlb.0410226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.