Abstract
Tumor metastasis is a leading cause of death in nasopharyngeal carcinoma (NPC) patients. Previous research has identified that transcription factor Yin Yang 1 (YY1) acts as a tumor suppressor that inhibits cell proliferation and tumor growth in NPC; however, the role and the molecular mechanisms of YY1 in NPC invasion and metastasis remain unclear. In this study, we discovered that YY1 could inhibit the migration and invasion of NPC cells in vitro as well as NPC xenograft tumor metastasis in vivo. Furthermore, we identified eIF4E as a direct downstream target of YY1, and YY1 could negatively regulate the expression of eIF4E at transcriptional level. Moreover, we found that eIF4E promoted the migration and invasion of NPC cells as well as NPC lung metastasis, suggesting its potential as a pro-metastatic mediator in NPC. Importantly, restoring eIF4E expression could partially reverse the inhibitory effects of YY1 on NPC malignancy. In consistent with these findings, the expression of YY1 was downregulated while eIF4E was upregulated in NPC patients with metastasis, and there was a negative correlation between YY1 and eIF4E expression. Collectively, our results indicate that YY1 suppresses the invasion and metastasis of NPC by negatively regulating eIF4E transcription. Therefore, targeting the YY1/eIF4E transcriptional axis could be a potential therapeutic strategy for the treatment of patients with NPC.
Keywords: Nasopharyngeal carcinoma, YY1, eIF4E, transcription activity, tumor metastasis
Introduction
Nasopharyngeal carcinoma (NPC) is the most prevalent head and neck malignancy in Southeast Asia, especially in southern China [1,2]. The location of NPC is concealed, and many patients present advanced NPC accompanied with distant metastasis at the time of diagnosis [3,4]. Tumor cell migration and invasion are critical steps in the early stage of cancer metastasis, in which tumor cells disseminate from the primary tumor, invade the blood vessels, and enter the bloodstream, leading to the formation of new metastatic tumors [5]. The occurrence of tumor metastasis is thought to be associated with the process of epithelial-mesenchymal transition (EMT). Tumor metastasis is the major cause of poor therapeutic efficacy and death in patients with NPC, yet effective treatments remain limited. Therefore, it is essential to understand the molecular mechanism underlying the invasion and metastasis of NPC to develop effective therapeutic strategy for NPC.
Yin Yang 1 (YY1) is a member of the multifunctional zinc finger transcription factor family [6,7], which is involved in many critical biological processes, such as chromatin remodeling, cell proliferation, differentiation, apoptosis, and embryonic development [8,9]. As a transcription factor, YY1 activates or represses a wide spectrum of target genes depending on its associated proteins, the promoter context, and the chromatin structure [10]. Moreover, YY1 can function as a cofactor or scaffold protein by interacting with other proteins [11]. YY1 is abnormally expressed in most human tumors and plays dual biological roles as either a tumor suppressor or oncogene by regulating different target genes and signaling pathways. Although YY1 promotes tumor metastasis in most cancers, such as breast cancer and lung cancer [12,13], some studies have also shown that YY1 exerts an inhibitory effect on tumor metastasis by regulating the CDKN3/MdM2/P53/P21, MUC4/ErbB2/p38/MEF2C/MMP10, p38/MAPK and PI3K/AKT pathways in pancreatic ductal adenocarcinoma (PDAC) [14-16]. Our previous study has also shown that YY1 inhibits cell proliferation, cell cycle progression, and xenograft tumor growth in NPC by negatively regulating the c-Myc/miR-141 transcriptional axis, thus playing a tumor-suppressive role in NPC [17]. However, the molecular mechanism by which YY1 is involved in the invasion and metastasis of NPC remains to be elucidated.
Eukaryotic translation initiation factor 4E (eIF4E) is a critical rate-limiting factor in cap-dependent protein synthesis [18] and is upregulated in a variety of cancers as well as correlated with the poor prognosis of cancer patients [19-21]. Increasing evidence shows that eIF4E can promote the proliferation, invasion, and metastasis of cancer cells by initiating the translation of a series of oncogenes, thus playing an oncogenic role in multiple cancers [22]. Moreover, phosphorylated eIF4E is not only correlated with lymph node metastasis and poor prognosis of NPC [23,24], but also promotes NPC cell EMT and invasion by positively regulating the translation of Snail mRNA [25], therefore, eIF4E is a potential pro-invasive factor in NPC.
In this study, we demonstrated the suppressive role of YY1 in NPC invasion and metastasis and found that YY1 inhibited the transcription of eIF4E by binding to the promoter region of eIF4E gene. Importantly, restoring eIF4E expression in YY1-overexpressing NPC cells reversed the inhibitory effect of YY1 on cell invasion and tumor metastasis. Furthermore, we investigated the clinical significance of YY1 and eIF4E expression in NPC patients. In summary, our data suggest that eIF4E may mediate the function of YY1 in NPC invasion and metastasis, suggesting that the regulatory axis of YY1/eIF4E could potentially serve as a therapeutic target for NPC treatment.
Material and methods
Patients and samples
A total of 140 samples, including 113 NPC and 27 noncancerous NP tissues, were collected from patients at the Second Xiangya Hospital of Central South University. All patients provided informed consent, and this study was approved by the Central South University Ethics Committee.
Antibodies
Anti-YY1 (1:2000) and anti-p-eIF4E (S209) (1:2000) were purchased from Abcam (Cambridge, UK). Anti-GAPDH (1:10000) and anti-eIF4E (1:1000) were purchased from Proteintech Group Inc. (Wuhan, China). Anti-E-cadherin (1:1000) and anti-Snail (1:1000) were purchased from Cell Signaling Technology, Inc. (Beverly, MA). Anti-Vimentin (1:1000) was purchased from Arigo, Inc. (Taiwan, China), and anti-N-cadherin (1:1000) was purchased from Affinity Biosciences, Inc. (Jiangsu, China).
Cell culture
Human NPC cell lines 5-8F and HNE2 were obtained from The Central South University Cell Center (Changsha, China) and maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (BI, Jerusalem, Israel) at 37°C with 5% CO2.
Plasmid construction and stable cell lines
Cells stably overexpressing YY1 were generated as previously described [17]. Briefly, cDNAs encoding wildtype human YY1 with 3x FLAG tag was cloned into the lentiviral expression vector pCDH-copGFP. Then, the lentiviral particles were produced in 293T cells and were used to infect 5-8F and HNE2 cells to generate YY1-overexpressing cell lines. To generate stable eIF4E-overexpressing cells, the PIRESneo2-EGFP-eIF4E plasmid was constructed by cloning the full-length eIF4E ORF into the PIRESneo2-EGFP vector and then was transfected into 5-8F and HNE2 cells. After puromycin selection for 14-21 days, cells stably expressing elF4E were obtained and further analyzed. For the luciferase reporter assay, the promoter sequence of eIF4E was inserted into the pGL3 reporter vector (Promega), and the recombinant construct was named as the pGL3 enhancer/eIF4E promoter, which was confirmed by sequencing.
RNA interference and transfection
Specific small interfering RNAs (siRNAs) against YY1 or eIF4E were purchased from RiboBio (Guangzhou, China). The siRNA sequences were as follows (5’-3’): siRNA YY1#1, CGACGACTACATTGAACAA; siRNA YY1#2, GCACAAAGATGTTCAGGGA; siRNA eIF4E#1, TGGCGCTGTTGTTAATGTT; siRNA eIF4E#2, AGAGCTAAAGGTGATAAGA. To knockdown YY1 or eIF4E expression in cells, exponentially growing cells were transfected with siRNAs using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from 5-8F and HNE2 cells using TRIzol reagent (Invitrogen, Carlsbad, USA) and was reverse transcribed into complementary DNA (cDNA) using a reverse transcription kit (Accurate Biotechnology, Hunan, China). Then, qPCR analysis was performed using SYBR Green qPCR reagent (Accurate Biotechnology, Hunan, China) on a Bio-Rad CFX96 Touch Sequence Detection System (Bio-Rad Laboratories Inc.). eIF4E qPCR Forward Primer: 5’-CTGCGGCTGATCTCCAAG-3’; eIF4E qPCR Reverse Primer: 5’-TTCCCACATAGGCTCAATACC-3’. The relative gene expression was calculated by the 2-ΔΔCT method with triplicates for each sample.
RNA sequencing and data analysis
RNA sequencing assay was carried out as reported with some modifications [26]. Briefly, total RNA was isolated from control and YY1-overexpressing 5-8F and HNE2 cells, and the RNA quality was confirmed with Agilent 2100 system to meet the requirements for Illumina HiSeq™ 4000 sequencing (Lnc-seq). The sequencing data retrieval, quality assessment, comparative analysis, and gene annotation were completed by Genedenovo Biotechnology Co., Ltd.
Wound healing assay
Briefly, cells were grown in a 6-well plate to confluence. Then, a scratch was created using a 10 μl pipette tip. Images of the scratch were captured at 0, 24, and 48 h after scratching at the same spot, and the reduction of the wound area was measured to assess the cell migration capacity.
Transwell assay
A transwell assay was used to evaluate the cell invasion ability. Briefly, transwell inserts in a 24-well plate (Corning, New York, USA) were first coated with 30 µl Matrigel (BD Biosciences, New York, USA) for 2-4 hours. Then, 3×104 cells/well in 200 μl serum-free medium were seeded in the upper chamber, while 600 μl medium containing 20% FBS was added to the lower compartment. After 36-48 hours of incubation, cells on the top side of the upper chamber were gently removed with a cotton swab, while cells that had invaded into the lower surface of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet for 5 minutes (Beyotime). Images were acquired, and the invaded cells were counted under an optical microscope (IX71, OLYMPUS, Japan) in three random fields of view.
Three-dimensional culture
First, the wells in 24-well plate were coated with 100 μl of Matrigel (reduced growth factors) at 37°C for 2 hours. Then, 1×104 cells in 200 μl of serum-free medium containing 10% Matrigel and 100 µl of complete growth medium were seeded in the wells and cultured for 10 days. Experiments were done in triplicate. Images were acquired, and the number of invaded cells was determined under microscope.
Luciferase reporter assay
A luciferase reporter assay was used to determine the effect of YY1 on eIF4E promoter activity. Briefly, the reporter plasmid and the pRL-TK internal reference vector were co-transfected into cells using Lipofectamine 3000 reagent. A dual-luciferase reporter assay system (Promega) was used to quantify the activity of firefly and Renilla luciferase. Experiments were done in triplicate.
Chromatin immunoprecipitation (ChIP) assay
A ChIP assay was performed to detect YY1 binding to eIF4E promoter by immunoprecipitation with IgG (Proteintech, Wuhan, China) or an anti-YY1 antibody (Abcam, Cambridge, UK). To evaluate the enrichment of H3K27ac in the eIF4E promoter, immunoprecipitation with an anti-H3K27ac antibody (ABclonal, Wuhan, China) was carried out using the Protein A/G Magnetic Beads system (Selleck Chemicals, Houston, TX, USA) as previously described [27]. qPCR was used to quantify the amount of precipitated DNA. Table 1 lists the primers used in ChIP assay.
Table 1.
The primers sequences for ChIP assay
| PCR primer name | Sequence (5’-3’) | Length (bp) |
|---|---|---|
| ChIP-eIF4E-promoter-F1 | ATCTCCCAGTAGCTGGGACTA | 21 |
| ChIP-eIF4E-promoter-R1 | TCTATTTGGGCTGGGTACGG | 20 |
| ChIP-eIF4E-promoter-F2 | GTTTATGGGGAGGTAGTGTACA | 22 |
| ChIP-eIF4E-promoter-R2 | CAAACCTGTATGGCATGTTACC | 22 |
Protein extraction and western blot analysis
Briefly, cells were lysed with cell lysis reagent (NCM Biotech, Suzhou, China), and the protein concentration was determined by a BCA protein assay kit (Thermo Fisher Scientific, Inc.). A total of 20-30 µg of total protein from each sample was separated on a 4-15% SDS gel and then transferred onto 0.22 µM or 0.45 µM PVDF membranes (Millipore, Billerica, USA). Then, the membranes were blocked with 5% skim milk for 60-120 minutes at room temperature and incubated with primary antibody overnight at 4°C followed by incubation with the corresponding secondary antibody at 37°C for 1 hour. The protein signals were detected with a modified chemiluminescence detection system (MiniChemi™ I, SAGECREATION, China) using western blotting substrate (Accurate Biotechnology, Hunan, China).
In vivo lung metastasis models
All animal studies were approved by the Institutional Animal Care and Use Committee of Central South University (Changsha, China) and were carried out following the National Research Council guidelines for the care and use of laboratory animals. Female BALB/c nude mice aged 4-6 weeks were purchased from Hunan Slake Jingda Experimental Animal Co. and were housed in a specific pathogen-free (SPF) environment. To generate lung metastasis models, 2×106 cells/100 µl 0.9% saline solution were injected into the tail veins of the nude mice, and the mice were weighed every 4 days. At 8 weeks post injection, the mice were sacrificed, and the lung tissues were excised, photographed, and paraffin embedded. Subsequently, H&E staining was performed, and the number of lung metastatic nodules was quantified.
Immunohistochemistry (IHC) staining
The expression of eIF4E and YY1 in patient tissue samples was examined by IHC staining. Briefly, slices were boiled in citrate buffer solution (pH 6.0) for 10 minutes for antigen retrieval, preincubated with 0.3% hydrogen peroxide for 10 minutes to block endogenous peroxidase activity, incubated with 5% goat serum to block nonspecific antigens, and incubated with the primary antibody followed by incubation with the corresponding secondary antibody. Then, the tissue sections were stained with diaminobenzidine (DAB) for 3-5 minutes, counterstained with hematoxylin, dehydrated in a graded ethanol series, rehydrated, mounted, and examined. The representative images were photographed. Staining results was independently assessed by two pathologists blinded to the sample diagnosis. IHC score was calculated based on the staining intensity and the percentage of stained cells [28-30]. Briefly, the staining intensity was graded as 0 (negative) to 3 (strong). The percentage of positive cells was scored as 0 (0%), 1 (1-25%), 2 (26-50%), 3 (51-75%), or 4 (76-100%).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7.0 software (San Diego, CA, USA). All quantified data were presented as the mean ± standard deviation (SD). The χ2 test was used to evaluate the correlation between the expression levels of YY1 and eIF4E as well as the correlation between their expression levels and the clinicopathological features of patients with NPC. Statistical significance was assessed using Student’s t test. A two-sided P value <0.05 was considered statistically significant. All experiments were repeated at least three times.
Results
YY1 inhibits the migration, invasion and EMT process of NPC cells
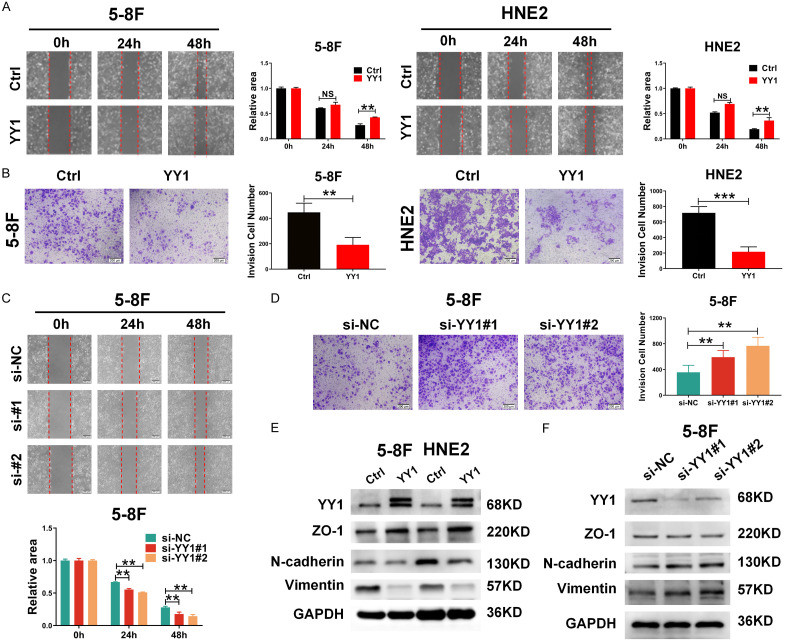
Our previous study has demonstrated that YY1 is downregulated in NPC and exerts a tumor-suppressive effect by inhibiting cell proliferation and tumor growth [17]; however, the role of YY1 in the invasion and metastasis of NPC is remains unclear. Hence, in this study, we first examined the influence of altered YY1 expression on the migration and invasion of NPC 5-8F and HNE2 cells. The successful overexpression and knockdown of YY1 were confirmed by western blot (Figure S1A). Subsequently, wound healing and transwell assays respectively demonstrated that YY1 overexpression inhibited NPC cell migration and invasion (Figure 1A, 1B), while YY1 silencing showed the opposite effects (Figures 1C, 1D, S1B, S1C). In line with these findings, compared to the control cells, YY1-overexpressing NPC cells showed a more epithelial cell-like morphology (Figure S1D), suggesting that YY1 might affect the mesenchymal-epithelial transition (EMT) process. To direct prove this, we examined the protein levels of EMT-related molecular markers by western blot and found that the ectopic expression of YY1 led to reduced levels of N-cadherin and Vimentin, while an upregulated expression of ZO-1 (Figure 1E). Conversely, silencing YY1 resulted in the opposite changes (Figures 1F, S1E), indicating that YY1 plays a crucial role in maintaining an epithelial phenotype in NPC cells. Collectively, these results demonstrate that YY1 inhibits cell migration and invasion by suppressing the EMT process in NPC cells.
Figure 1.
YY1 inhibits the migration and invasion of NPC cell lines. A. Wound-healing assays were used to compare the motility of 5-8F and HNE2 cells infected with YY1-overexpression lentiviruses (left), quantification of the wound recovery rate of the two groups (right), scale bar, 200 μm. B. The cell migration assay (transwell assay) was used to compare the migration of 5-8F and HNE2 cells infected with YY1-overexpression lentiviruses. C and D. The scratch wound healing analysis and Matrigel invasion analysis of cell invasive capabilities in 5-8F cells with transfection of YY1 siRNAs or negative control. E and F. Significantly differently expressed proteins involved in EMT progression in 5-8F and HNE2 cells stably with YY1 overexpression or siRNAs, respectively. GAPDH served as an internal control. Error bars represent the mean ± SD. **P<0.01; ***P<0.001; NS, no significance.
EIF4E is a downstream target gene of YY1 and is negatively regulated by YY1 at transcriptional level
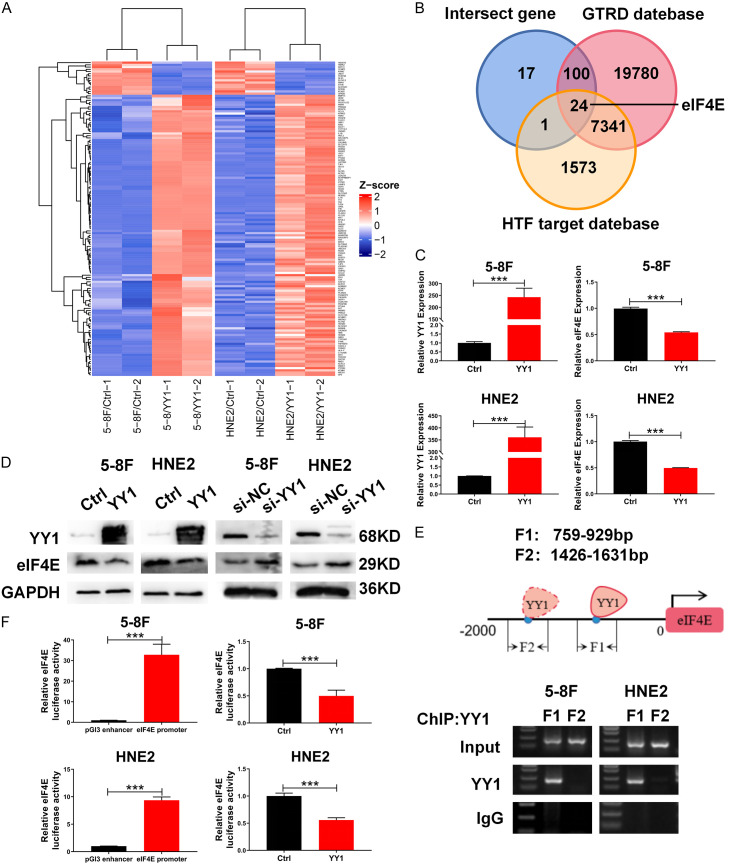
To investigate the molecular mechanisms by which YY1 inhibits NPC cell migration and invasion, we performed high-throughput RNA sequencing and bioinformatic analyses in YY1-overexpressing 5-8F and HNE2 cells and identified a total of 1791 genes that were regulated by YY1 (1513 upregulated genes and 278 downregulated genes) in 5-8F cells and 2555 genes (1337 upregulated genes and 1218 downregulated genes) regulated by YY1 in HNE2 cells (Figure S2A). Among these genes, 142 genes showed a consistent expression pattern in both 5-8F and HNE2 cells (Figures 2A, S2B). Notably, the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that adherens junctions and cell adhesions were among the top 10 signaling pathways associated with YY1 regulation (Figure S2C), reinforcing the potential association between YY1 and EMT. In addition, we further identified the direct downstream target genes of YY1. Using the GTRD web interface (http://gtrd20-06.biouml.org/) and HTF target web interface (http://bioinfo.life.hust.edu.cn/hTFtarget/), the Venn diagram showed that 24 genes were simultaneously annotated in these two databases and were differentially expressed in both 5-8F and HNE2 cells (Figure 2B; Table S1). Among these genes, we focused on eukaryotic translation initiation factor 4E (eIF4E), the top downregulated gene. To validate our findings, we first confirmed the YY1-regulated eIF4E expression by qPCR and western blot, and our results showed that the overexpression of YY1 led to a significant decrease in both the mRNA and protein levels of eIF4E, whereas silencing YY1 resulted in an upregulated expression of eIF4E (Figure 2C, 2D), indicating that eIF4E is a downstream target of YY1.
Figure 2.
EIF4E is a downstream target gene of YY1 that negatively regulates its promoter activity and expression. A. Heatmap visualization of the cluster signatures of the differentially expressed genes regulated by YY1. B. The intersection Venn diagram shows the potential downstream target genes of YY1 predicted by GTRD, HTF target and RNA-seq differentially expressed genes. C and D. qRT-PCR and western blotting assays were used to confirm the expression of YY1 and eIF4E in mRNA and protein levels, respectively. GAPDH served as an internal control. E. ChIP-PCR assays using antibodies specific for YY1 validated the YY1-binding sites in the eIF4E potential promoter region. F. The dual-luciferase reporter assays determined the effect of YY1 on the promoter activity of eIF4E. Error bars represent the mean ± SD. ***P<0.001.
We then investigated the molecular mechanism by which YY1 regulates eIF4E expression. As YY1 is a transcription factor, we explored whether YY1 binds to the promoter region of eIF4E gene and thereby regulates its expression at the transcriptional level. We first predicted two YY1 binding sites (named Site 1 and Site 2) in the eIF4E promoter region through JASPAR (http://jaspar.genereg.net/) bioinformatics software (Figure S2D). Next, a ChIP assay was performed to verify whether YY1 binds to either of these two putative binding sites in the eIF4E promoter. The results showed that YY1 bound to site 1 but not site 2 (Figure 2E). In addition, we functionally investigated the effect of YY1 on eIF4E promoter activity by using a luciferase reporter assay and determined that YY1 overexpression significantly suppressed the luciferase activity driven by eIF4E promoter (Figure 2F), suggesting that YY1 inhibited the transcription of eIF4E gene in NPC cells. Although the UCSC bioinformatics software revealed that the eIF4E promoter region was enriched with H3K27ac modification (Figure S2E), our ChIP-PCR results show that overexpression of YY1 did not change the enrichment of H3K27ac at eIF4E promoter (Figure S2F). Taken together, we revealed that eIF4E is a direct downstream target gene of YY1 and that YY1 negatively regulates the transcription of eIF4E gene by directly binding to its promoter region, regardless of the chromatin state of the eIF4E promoter.
EIF4E promotes NPC cell migration and invasion
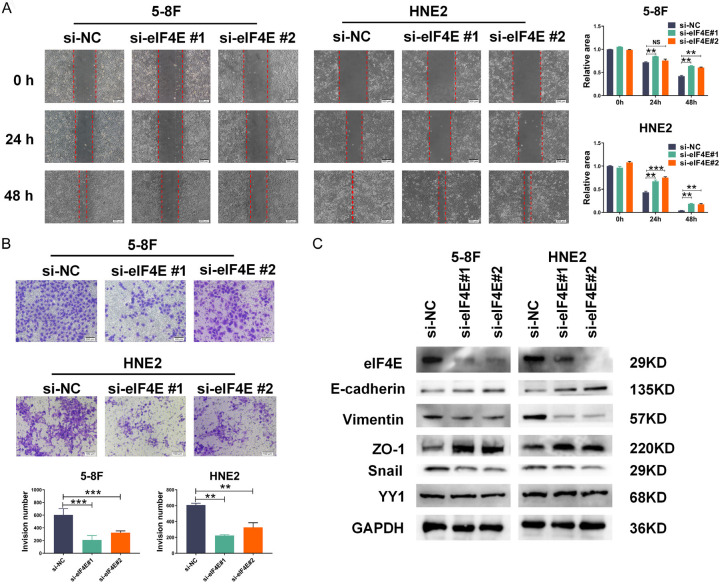
Although eIF4E has been reported to be associated with the invasion and metastasis of a variety of tumors, such as breast cancer and gastric cancer [31,32], the roles of eIF4E in NPC invasion and metastasis have yet to be determined. In this study, we first examined the effect of altered eIF4E expression on NPC cell migration and invasion in vitro by respective wound healing and transwell assays. Silencing efficiency was evaluated using western blotting and the results showed that eIF4E was successfully knocked down in 5-8F and HNE2 cells (Figure S3). Notably, the scratch wound healing assay results suggested that eIF4E knockdown significantly suppressed the NPC cell migration and invasion ability (Figure 3A, 3B). To investigate the functional mechanisms of eIF4E, we conducted an analysis on the impact of eIF4E knockdown on Snail. Snail is a crucial transcription factor involved in the EMT process, where it suppresses the transcription of E-cadherin. Our findings revealed that eIF4E knockdown resulted in a decrease in Snail expression and an increase in E-cadherin expression (Figure 3C). In addition, we evaluated the expression of EMT-related markers by western blot and found that the expression of the mesenchymal marker ZO-1 increased, while the expression of the epithelial marker vimentin decreased in eIF4E knockdown cells (Figure 3C). These results provide evidences supporting the hypothesis that eIF4E plays a negative regulatory role in the snail/E-cadherin axis and is crucial for NPC cell migration and invasion.
Figure 3.
Depletion of eIF4E by siRNA inhibits cell migration and metastasis in NPC cell lines. A. Cell migration assay was performed with the scratch wound healing analysis (left), quantification of the wound recovery rate of the three groups (right). B. Matrigel invasion analysis of cell invasive capabilities in 5-8F and HNE2 cells with transfection of eIF4E siRNAs or negative control. C. Significantly differently expressed proteins involved in EMT progression in 5-8F and HNE2 cells with eIF4E siRNAs or negative control, respectively. GAPDH served as an internal control. Error bars represent the mean ± SD. **P<0.01; ***P<0.001; NS, no significance.
Restoring the expression of eIF4E reverses the inhibitory effect of YY1 on the migration and invasion of NPC cells
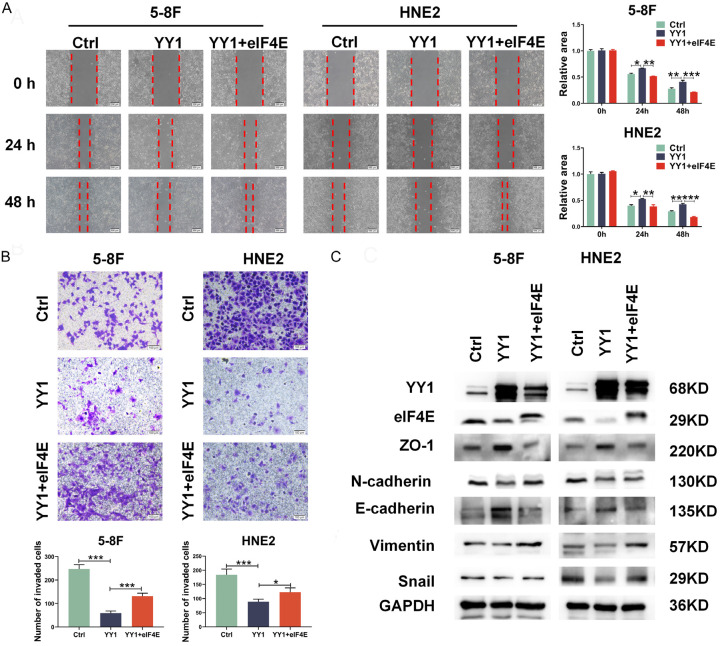
Given our results above showing the opposite roles of YY1 and eIF4E in the migration and invasion of NPC cells and the inhibition of eIF4E by YY1, we sought to determine whether YY1 suppressed NPC cell mobility through inhibiting eIF4E activity. Therefore, we performed rescue experiments by overexpressing eIF4E in YY1-overexpressing cells. The wound healing and transwell assays demonstrated that the restoration of eIF4E expression effectively reversed the suppressive effect of YY1 overexpression on NPC cell migration and invasion (Figure 4A, 4B). Previous research has shown that the role of eIF4E as an oncogene is dependent on the phosphorylation of serine 209. Our analysis using western blot assay demonstrated that the overexpression of YY1 reduced eIF4E phosphorylation at S209, while the knockdown of YY1 increased its expression in 5-8F and HNE2 cells (Figure S4A). To further investigate the impact of eIF4E phosphorylation at serine 209 on YY1’s regulation of eIF4E-mediated migration and invasion of NPC cells, we constructed a recombinant with a mutation in eIF4E at serine 209 (eIF4E S209A) [33]. Through scratch healing and invasion assays, we verified that restoring the expression of eIF4E mutant failed to reverse the inhibitory effect of YY1 overexpression on the invasion of NPC cells (Figure S4B, S4C). Furthermore, western blot analysis revealed that the restoration of eIF4E expression at least partially reversed eIF4E phosphorylation at S209 regulated by YY1 as well as the expression of EMT-related markers, including Vimentin, N-cadherin, Snail, E-cadherin, and ZO-1 (Figure 4C). However, eIF4E mutant with S209A failed to rescue the expression of the above molecules (Figure S4D). Moreover, we performed invasion assays with a reconstituted three-dimensional basement membrane. Strikingly, the overexpression of YY1 significantly reduced the number of spherical colonies and protrusions at the edges of invading cells, while the restoration of eIF4E expression at least partially reversed these effects (Figure S5). Collectively, these findings demonstrate that YY1 inhibits the EMT process and the mobility of NPC cells by negatively regulating eIF4E expression.
Figure 4.
Restoration of eIF4E expression reverses the inhibitory effect of YY1 on cell migration and invasion. A. Scratch wound healing analysis of cell migration in 5-8F and HNE2 cells stably with YY1 overexpression, YY1 and eIF4E simultaneous overexpression or control (left), quantification of the wound recovery rate of the three groups (right). B. Matrigel invasion analysis of cell invasive capabilities in 5-8F and HNE2 cells stably with YY1 overexpression, YY1 and eIF4E simultaneous overexpression or control. C. Significantly differently expressed proteins involved in EMT progression (E-cadherin, N-cadherin, Vimentin, ZO-1, Snail) in YY1 overexpression and eIF4E restoration cells, respectively. GAPDH served as an internal control. Error bars represent the mean ± SD. *P<0.05; **P<0.01; ***P<0.001.
YY1 inhibits tumor metastasis through negatively regulating eIF4E expression in vivo
Importantly, to further study the effect of the YY1/eIF4E axis on the metastasis of NPC tumors in vivo, we injected 5-8F stable cell lines expressing either vector plasmid (5-8F/Ctrl), YY1 (5-8F/YY1), eIF4E (5-8F/eIF4E), or YY1 plus eIF4E (5-8F/YY1 plus eIF4E) into mice via the tail vein to establish a lung metastatic colonization model. After eight weeks, we examined the lung metastatic tumor lesions which were quantified by histopathological analysis. As expected, the overexpression of YY1 reduced, whereas the overexpression of eIF4E increased the number of tumor nodules; however, the restoration of eIF4E expression in YY1-overexpressing cells significantly reversed the suppressive effect of YY1 (Figure 5A). Furthermore, H&E staining of tissue sections confirmed the presence of lung metastatic nodules. Consistently, compared to the control group, eIF4E overexpression increased lung metastasis, while YY1 expression significantly attenuated lung colonization, which could be reversed by eIF4E overexpression (Figure 5B, 5C). Taken together, these results further support the notion that YY1 suppresses tumor metastasis at least partly by downregulating the expression of eIF4E.
Figure 5.
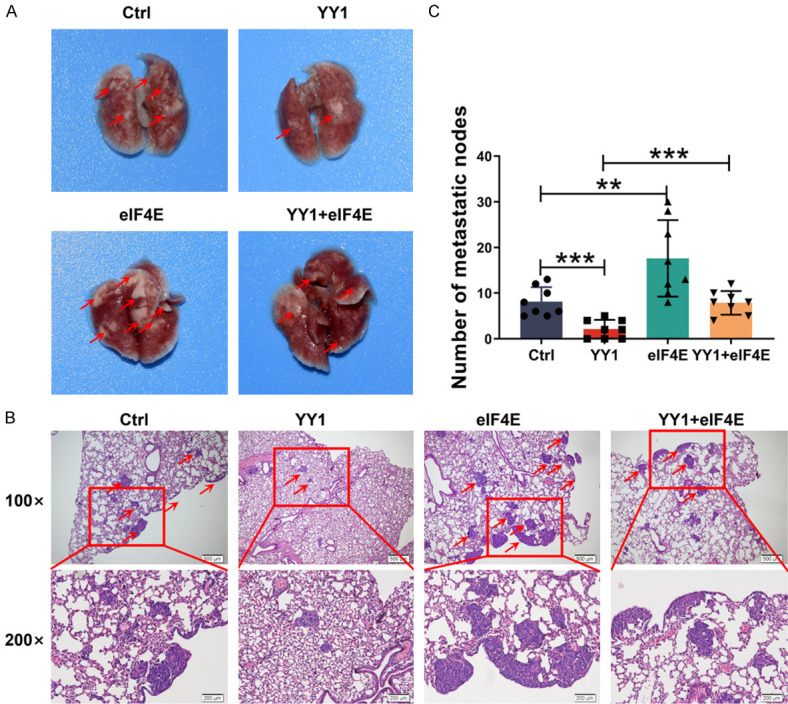
YY1 inhibits tumor metastasis in vivo through regulation of eIF4E expression. A. Representative image of macroscopic mouse lung tissue in the metastatic tumor model, n = 8 mice per group. B. H&E staining is shown in control, YY1 overexpression, eIF4E overexpression and eIF4E restoration group. Red arrows indicate metastatic tumors. C. The number of metastatic lung nodules of every mouse per group was counted in microscopy. Error bars represent the mean ± SD. **P<0.01; ***P<0.001.
The abnormal expression of YY1 and eIF4E is closely related to the metastasis and poor prognosis in NPC patients
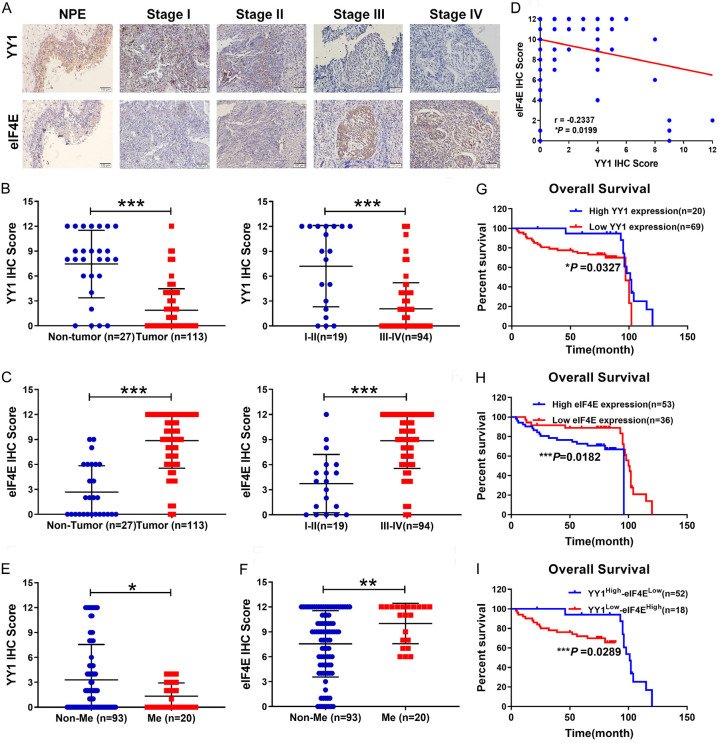
Based on the function and mechanism of YY1/eIF4E axis identified above, we further investigated the expression and clinical relevance of YY1 and eIF4E in NPC and normal NP tissue samples by IHC staining of 27 noncancerous tissues and 113 NPC tissues from patients with different ages, sexes, lymph node metastasis statuses, distant metastasis statuses, and clinical stages. The results showed that YY1 expression was lower in NPC tissues than in normal NP tissues and was significantly lower in stage III and IV tissues than in stage I and II NPC tissues (Figure 6A, 6B). In contrast, eIF4E expression showed an opposite pattern as it was higher in NPC tissues than in normal NP tissues and was significantly higher in clinical stage III and IV tissues than in stage I and II tissues (Figure 6A-C; Table 2). In addition, we found a strong negative correlation between the expression of YY1 and eIF4E in NPC tissues (Pearson correlation coefficient, r = -0.2337, P = 0.0199) (Figure 6D). We further analyzed the correlation between the YY1 expression level and six clinical variables: tumor size, lymph node metastasis status, distant metastasis status, pathological tumor stage, gender, and age. The results showed that the expression of YY1 was negatively correlated with lymph node metastasis (Table 2) and distant metastasis in patients with NPC (Figure 6E), and that high YY1 expression was a favorable factor for patient survival and prognosis (Figure 6G). Conversely, eIF4E expression was positively correlated with lymph node metastasis (Table 2) and distant metastasis in NPC patients (Figure 6F); its high expression predicted poor survival and prognosis (Figure 6H). Consistently, the combination of high YY1 and low eIF4E expression predicted a better prognosis in patients with NPC (Figure 6I). Similarly, the combined analysis showed that high YY1 expression combined with low eIF4E expression and low YY1 expression combined with high eIF4E expression were associated with tumor size, lymph node metastasis, distant metastasis and TNM stage (Table 2). Therefore, our data suggest that both YY1 and eIF4E may be involved in the metastasis of NPC and that the YY1/eIF4E axis may be a new prognostic biomarker and therapeutic target for NPC.
Figure 6.
YY1 expression was negatively correlated with eIF4E in clinical NP and NPC tissues. A. Representative image of YY1 and eIF4E expression in noncancerous nasopharyngeal tissues and different clinical TNM stages of NPC detected by IHC. B and C. Statistical diagram of YY1 and eIF4E expression in noncancerous nasopharyngeal tissues and different clinical TNM stages of NPC. D. Correlation between YY1 expression and eIF4E expression. E and F. Statistical diagram of YY1 and eIF4E expression with distant metastases [28] of NPC patients. G. Kaplan-Meier analyses comparing overall survival between high- and low-YY1 expression groups. H. Kaplan-Meier analyses comparing overall survival between high- and low-eIF4E expression groups. I. Kaplan-Meier analyses comparing overall survival between high YY1 and low eIF4E or high eIF4E and low YY1 expression groups. Error bars represent the mean ± SD. *P<0.05; **P<0.01; ***P<0.001.
Table 2.
Association between the expression of YY1, eIF4E and NPC clinical pathological features (n = 113)
| Variables features | YY1 expression | eIF4E expression | YY1/eIF4E | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Low | High | P | Low | High | P | L-H | H-L | P | |
| Gender | |||||||||
| Male (n = 76) | 64 (84%) | 12 (16%) | 0.6764 | 30 (39%) | 46 (61%) | 0.0289 | 46 (61%) | 11 (14%) | 0.6200 |
| Famale (n = 37) | 30 (81%) | 7 (19%) | 7 (19%) | 30 (81%) | 28 (76%) | 5 (14%) | |||
| Age | |||||||||
| ≤44 (n = 54) | 44 (81%) | 10 (19%) | 0.6430 | 17 (31%) | 37 (69%) | 0.7845 | 35 (65%) | 8 (15%) | 0.8444 |
| >44 (n = 59) | 50 (85%) | 9 (15%) | 20 (34%) | 39 (66%) | 39 (66%) | 8 (14%) | |||
| Tumor size | |||||||||
| I (n = 22) | 12 (55%) | 10 (45%) | 0.0006*** | 15 (68%) | 7 (32%) | 0.0009*** | 7 (32%) | 10 (45%) | 0.0001*** |
| II (n = 37) | 34 (92%) | 3 (8%) | 11 (30%) | 26 (70%) | 26 (70%) | 2 (5%) | |||
| III (n = 31) | 29 (94%) | 2 (6%) | 6 (19%) | 25 (81%) | 24 (77%) | 1 (3%) | |||
| IV (n = 23) | 19 (83%) | 4 (17%) | 5 (22%) | 18 (78%) | 17 (74%) | 3 (13%) | |||
| LNM | |||||||||
| I (n = 17) | 11 (65%) | 6 (35%) | 0.0055** | 10 (59%) | 7 (41%) | 0.0088** | 7 (41%) | 6 (35%) | 0.0001*** |
| II (n = 72) | 67 (93%) | 5 (7%) | 16 (22%) | 56 (78%) | 56 (78%) | 2 (3%) | |||
| III (n = 10) | 9 (90%) | 1 (10%) | 2 (20%) | 8 (80%) | 8 (80%) | 1 (10%) | |||
| Metastatic | |||||||||
| No-DM (n = 93) | 68 (73%) | 25 (27%) | 0.0086** | 35 (38%) | 58 (62%) | 0.0180* | 55 (59%) | 17 (18%) | 0.0221* |
| DM (n = 20) | 20 (100%) | 0 (0%) | 2 (10%) | 18 (90%) | 18 (90%) | 0 (0%) | |||
| Clinical stages | |||||||||
| I (n = 7) | 1 (14%) | 6 (86%) | 0.0001*** | 7 (100%) | 0 (0%) | 0.0001*** | 0 (0%) | 6 (86%) | 0.0001*** |
| II (n = 12) | 7 (58%) | 5 (42%) | 9 (75%) | 3 (25%) | 5 (42%) | 3 (25%) | |||
| III (n = 63) | 56 (89%) | 7 (11%) | 17 (27%) | 46 (73%) | 46 (73%) | 4 (6%) | |||
| IV (n = 31) | 28 (90%) | 3 (10%) | 5 (16%) | 26 (84%) | 25 (81%) | 1 (3%) | |||
The following abbreviations were used: H, high expression; L, low expression; LNM, lymph node metastasis, Statistical analysis was performed using the Chi-squared test.
P<0.05;
P<0.01;
P<0.001.
Discussion
As an important transcription factor, YY1 regulates the expression of different target genes involved in various signaling pathways and plays dual biological roles in tumorigenesis serving as an oncogene or a tumor suppressor. Our previous studies show that YY1 functions as a cofactor of c-MYC to negatively regulate the expression of miR-141, thereby inhibiting the proliferation, reducing the colony-forming ability, and suppressing the G1/S phase progression of NPC cells, which supports the role of YY1 as a tumor suppressor in NPC [17]. In recent years, the function of YY1 in tumor invasion and metastasis has been widely reported in various tumor types [34-36], except in NPC. In this study, we for the first time revealed that the overexpression of YY1 inhibited cell migration and invasion, the EMT process and tumor metastasis, supporting the hypothesis that YY1 plays an anti-metastatic role in NPC.
As a transcription factor, YY1 activates or represses the transcription of target genes by directly binding to promoter regions [37-39]. Therefore, we performed RNA-seq analysis to identify YY1 target genes that mediate the function of YY1 in suppressing cell mobility. The results revealed that YY1 overexpression significantly downregulated eIF4E mRNA level in 5-8F and HNE2 NPC cells. Mechanistically, we discovered that YY1 directly bound to the promoter region of eIF4E and exerted a negative regulatory effect on its promoter activity. It has been known that the transcriptional activation function of YY1 is often accompanied by the enrichment of H3K4me3 and H3K27ac on the promoters of its target genes [40,41]; interestingly, we didn’t observe an enrichment of H3K27ac in the promoter region of eIF4E, suggesting that YY1 regulates the transcription of eIF4E independent of its influence on the open chromatin state. Nevertheless, further investigation is required to elucidate the epigenetic mechanisms involved in YY1 function.
EIF4E is an mRNA cap-binding protein and acts as an initiation factor for mRNA-ribosome interactions and cap-dependent translation in human cells [42]. The activities of eIF4E are regulated at multiple levels. Firstly, eIF4E expression is regulated extensively at the transcriptional and mRNA stability levels. A series of studies have shown that c-Myc directly activates the transcription of eIF4E. Interestingly, multiple studies have indicated the existence of positive regulatory feedback loops between c-Myc and eIF4E at the transcriptional and translational levels. Specifically, eIF4E promotes the translation of c-Myc, and c-Myc promotes the transcription and expression of eIF4E [43-45]. In addition, the mechanism by which YY1 regulates gene expression is partially mediated through its interaction with other proteins to function as a molecular scaffold or transcriptional coactivator. Furthermore, our previous studies have confirmed that YY1 negatively regulates the transcription of c-Myc and its downstream target genes, consistent with the current findings. Another mechanism of eIF4E regulation is through its phosphorylation at serine 209 [46,47]. Recent studies have shown that eIF4E is widely overexpressed and hyperphosphorylated in human cancers [48,49]. Phosphorylated eIF4E activates several essential biological processes in cancer, especially tumor metastasis and angiogenesis [50]. Our previous studies have demonstrated that YY1 negatively regulates c-Myc transcriptional activity in NPC cells via Max/Mad and that eIF4E is the target gene of c-Myc. Therefore, we speculate that the transcriptional activity of eIF4E might depend on the YY1/c-Myc axis. Moreover, eIF4E phosphorylation at serine 209 is negatively regulated by YY1, suggesting the existence of more molecular mechanisms for the regulatory effect of YY1 on eIF4E.
EIF4E acts as an upstream regulator of several oncogenes as well as plays a crucial role in integrating and amplifying multiple oncogenic signaling pathways [51]. Yao et al. have reported that eIF4E level is positively correlated with Snail expression, the core transcription factor of EMT [52]. Snail is a zinc finger transcription factor that represses E-cadherin transcription by directly binding to the E-box motif within the E-cadherin promoter [53]. Similarly, our results showed that the overexpression of YY1 downregulated the protein level of Snail and upregulated the expression of E-cadherin, thereby inhibiting the EMT process in NPC cells. Importantly, restoring the expression of eIF4E at least partially rescued the expression of Snail/E-cadherin and reversed YY1-mediated inhibition of the EMT process in NPC cells, further indicating that YY1 suppressed the invasion and metastasis of NPC cells via inhibiting the transcription of eIF4E.
The expression of YY1 has been found to be upregulated in multiple cancers, making it a potential biomarker for predicting cancer metastasis and prognosis. YY1 regulates the expression of several genes, and in PDAC, its expression was significantly negatively correlated with the expression of CDKN3. Mechanistically, YY1 directly binds to the promoter region of CDKN3 and negatively regulates its expression [14]. In our study, we confirmed that eIF4E is a direct downstream target gene of YY1. We observed a negative correlation between the expression of YY1 and eIF4E in NPC patients. Additionally, we found a correlation between the expression of YY1 and eIF4E with the lymph node metastasis status, clinical stage, distant metastasis status, and overall survival rate of NPC patients. These findings provide strong evidence suggesting that YY1 acts as a suppressor of tumor metastasis by directly regulating the transcription of eIF4E. Furthermore, our results suggest that YY1 may serve as a potential target for predicting the prognosis of patients with NPC.
Conclusions
This study revealed that YY1 directly binds to the promoter region of eIF4E gene, thereby negatively regulating eIF4E transcription. Restoring the expression of eIF4E could reverse the inhibitory effect of YY1 on the invasive and metastatic potential of NPC cells both in vivo and in vitro. Moreover, eIF4E expression is significantly upregulated in NPC and is positively associated with advanced TNM stages, metastasis, and shorter survival times in patients. Therefore, these results suggest that YY1 inhibits tumor metastasis at least partly by negatively regulating the activity of eIF4E/Snail/E-cadherin signal axis (Figure 7). Targeting the YY1/eIF4E regulatory axis is a potential novel strategy for the treatment of NPC. However, the specific mechanism underlying YY1’s dual biological role in tumorigenesis needs further exploration.
Figure 7.
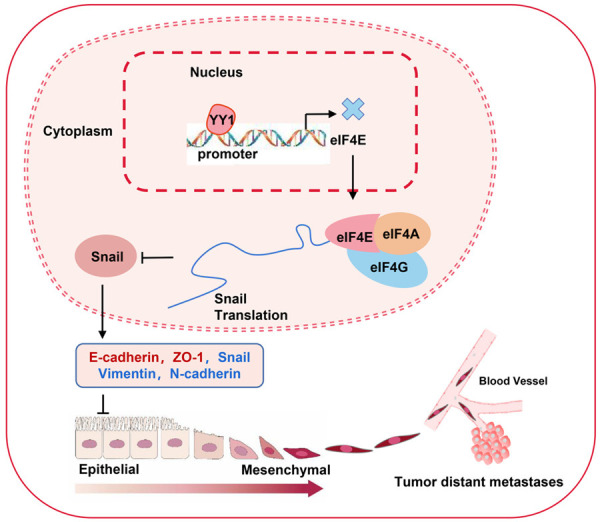
Schematic representation of the tumor-suppressive mechanism of YY1 that occurs by repressing the transcription activity and expression of eIF4E in NPC. YY1 negatively regulates the transcriptional activity of eIF4E by binding to the promoter region of eIF4E, thereby inhibiting eIF4E-mediated Snail protein translation, and then efficiently inhibits EMT progression and metastasis in nasopharyngeal carcinoma cells. Red represents upregulated genes, and blue represents downregulated genes.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (grant No. 82172592), the program of Introducing Talents of Discipline to Universities (grant No. 111-2-12), Project Funded by China Postdoctoral Science Foundation (grant No. 2022M711125), the Science and Technology Innovation Program of Hunan Province (grant No. 2021SK51110).
Clinical samples of nasopharyngeal carcinoma and normal nasopharyngeal epithelium samples were all collected from the Second Xiangya Hospital of Central South University. All patients involved in this article signed informed consent.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Zhao R, Wei Y, Li M, Wang H, Niu W, Zhou Y, Qiu Y, Fan S, Zhan Y, Xiong W, Zhou Y, Li X, Li Z, Li G, Zhou M. BRD7 expression and c-Myc activation forms a double-negative feedback loop that controls the cell proliferation and tumor growth of nasopharyngeal carcinoma by targeting oncogenic miR-141. J Exp Clin Cancer Res. 2018;37:64. doi: 10.1186/s13046-018-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan W, Xiong B, Tian T, Zou X, He Z, Zhang L. Radiomics in nasopharyngeal carcinoma. Clin Med Insights Oncol. 2022;16:11795549221079186. doi: 10.1177/11795549221079186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang LL, Chen YP, Chen CB, Chen MY, Chen NY, Chen XZ, Du XJ, Fang WF, Feng M, Gao J, Han F, He X, Hu CS, Hu DS, Hu GY, Jiang H, Jiang W, Jin F, Lang JY, Li JG, Lin SJ, Liu X, Liu QF, Ma L, Mai HQ, Qin JY, Shen LF, Sun Y, Wang PG, Wang RS, Wang RZ, Wang XS, Wang Y, Wu H, Xia YF, Xiao SW, Yang KY, Yi JL, Zhu XD, Ma J. The Chinese Society of Clinical Oncology (CSCO) clinical guidelines for the diagnosis and treatment of nasopharyngeal carcinoma. Cancer Commun (Lond) 2021;41:1195–1227. doi: 10.1002/cac2.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng WT, Corry J, Langendijk JA, Lee AWM, Mäkitie A, Mendenhall WM, Rinaldo A, Rodrigo JP, Saba NF, Smee R, Strojan P, Suárez C, Vermorken JB, Ferlito A. Current management of stage IV nasopharyngeal carcinoma without distant metastasis. Cancer Treat Rev. 2020;85:101995. doi: 10.1016/j.ctrv.2020.101995. [DOI] [PubMed] [Google Scholar]
- 6.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson S, Whitehouse H, Naidoo K, Byers RJ. Yin Yang 1 in human cancer. Crit Rev Oncog. 2011;16:245–260. doi: 10.1615/critrevoncog.v16.i3-4.80. [DOI] [PubMed] [Google Scholar]
- 9.Zaravinos A, Spandidos DA. Yin yang 1 expression in human tumors. Cell Cycle. 2010;9:512–522. doi: 10.4161/cc.9.3.10588. [DOI] [PubMed] [Google Scholar]
- 10.Figiel M, Gorecki A. Physical interaction of human Yin Yang 1 protein with DNA. Crit Rev Oncog. 2017;22:75–97. doi: 10.1615/CritRevOncog.2017020976. [DOI] [PubMed] [Google Scholar]
- 11.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 12.Xu P, Xiao H, Yang Q, Hu R, Jiang L, Bi R, Jiang X, Wang L, Mei J, Ding F, Huang J. The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp Mol Med. 2020;52:41–55. doi: 10.1038/s12276-019-0356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberthal JG, Kaminsky M, Parkhurst CN, Tanese N. The role of YY1 in reduced HP1alpha gene expression in invasive human breast cancer cells. Breast Cancer Res. 2009;11:R42. doi: 10.1186/bcr2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D, Zhang J, Wu Y, Shi G, Yuan H, Lu Z, Zhu Q, Wu P, Lu C, Guo F, Chen J, Jiang K, Miao Y. YY1 suppresses proliferation and migration of pancreatic ductal adenocarcinoma by regulating the CDKN3/MdM2/P53/P21 signaling pathway. Int J Cancer. 2018;142:1392–1404. doi: 10.1002/ijc.31173. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JJ, Zhu Y, Xie KL, Peng YP, Tao JQ, Tang J, Li Z, Xu ZK, Dai CC, Qian ZY, Jiang KR, Wu JL, Gao WT, Du Q, Miao Y. Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol Cancer. 2014;13:130. doi: 10.1186/1476-4598-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, Yang C, Chen L, Zhang JJ, Ge WL, Yuan H, Meng LD, Huang XM, Shen P, Miao Y, Jiang KR. YY1 targets tubulin polymerisation-promoting protein to inhibit migration, invasion and angiogenesis in pancreatic cancer via p38/MAPK and PI3K/AKT pathways. Br J Cancer. 2019;121:912–921. doi: 10.1038/s41416-019-0604-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Liu Y, Wei Y, Wu C, Meng H, Niu W, Zhou Y, Wang H, Wen Q, Fan S, Li Z, Li X, Zhou J, Cao K, Xiong W, Zeng Z, Li X, Qiu Y, Li G, Zhou M. Zinc-finger protein YY1 suppresses tumor growth of human nasopharyngeal carcinoma by inactivating c-Myc-mediated microRNA-141 transcription. J Biol Chem. 2019;294:6172–6187. doi: 10.1074/jbc.RA118.006281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 19.Li F, Sun H, Li Y, Bai X, Dong X, Zhao N, Meng J, Sun B, Zhang D. High expression of eIF4E is associated with tumor macrophage infiltration and leads to poor prognosis in breast cancer. BMC Cancer. 2021;21:1305. doi: 10.1186/s12885-021-09010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst BP, Mikstas C, Stöver T, Stauber R, Strieth S. Association of eIF4E and SPARC expression with lymphangiogenesis and lymph node metastasis in hypopharyngeal cancer. Anticancer Res. 2018;38:699–706. doi: 10.21873/anticanres.12275. [DOI] [PubMed] [Google Scholar]
- 21.Oblinger JL, Burns SS, Huang J, Pan L, Ren Y, Shen R, Kinghorn AD, Welling DB, Chang LS. Overexpression of eIF4F components in meningiomas and suppression of meningioma cell growth by inhibiting translation initiation. Exp Neurol. 2018;299:299–307. doi: 10.1016/j.expneurol.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang S, Guo R, Zhang Z, Liu D, Xu H, Xu Z, Wang X, Yang L. Upregulation of the eIF4E signaling pathway contributes to the progression of gastric cancer, and targeting eIF4E by perifosine inhibits cell growth. Oncol Rep. 2013;29:2422–2430. doi: 10.3892/or.2013.2397. [DOI] [PubMed] [Google Scholar]
- 23.Zheng J, Li J, Xu L, Xie G, Wen Q, Luo J, Li D, Huang D, Fan S. Phosphorylated Mnk1 and eIF4E are associated with lymph node metastasis and poor prognosis of nasopharyngeal carcinoma. PLoS One. 2014;9:e89220. doi: 10.1371/journal.pone.0089220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Pang TY, Wang Y, Wang S, Kang HX, Ding WB, Yong WW, Bie YH, Cheng XG, Zeng C, Yao YH, Li Q, Hu XR. LMP1 stimulates the transcription of eIF4E to promote the proliferation, migration and invasion of human nasopharyngeal carcinoma. FEBS J. 2014;281:3004–3018. doi: 10.1111/febs.12838. [DOI] [PubMed] [Google Scholar]
- 25.Yao Y, Pang T, Cheng Y, Yong W, Kang H, Zhao Y, Wang S, Hu X. Positive correlative over-expression between eIF4E and snail in nasopharyngeal carcinoma promotes its metastasis and resistance to cisplatin. Pathol Oncol Res. 2020;26:1639–1649. doi: 10.1007/s12253-019-00733-x. [DOI] [PubMed] [Google Scholar]
- 26.Niu W, Luo Y, Zhou Y, Li M, Wu C, Duan Y, Wang H, Fan S, Li Z, Xiong W, Li X, Li G, Ren C, Li H, Zhou M. BRD7 suppresses invasion and metastasis in breast cancer by negatively regulating YB1-induced epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2020;39:30. doi: 10.1186/s13046-019-1493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Wei Y, Liu Y, Wei J, Zhou X, Duan Y, Chen S, Xue C, Zhan Y, Zheng L, Deng H, Tang F, Fan S, Xiong W, Li G, Tan M, Zhou M. BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma. Cell Death Dis. 2023;14:121. doi: 10.1038/s41419-023-05632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen Q, Li J, Wang W, Xie G, Xu L, Luo J, Chu S, She L, Li D, Huang D, Fan S. Increased expression of flotillin-2 protein as a novel biomarker for lymph node metastasis in nasopharyngeal carcinoma. PLoS One. 2014;9:e101676. doi: 10.1371/journal.pone.0101676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan Y, Feng J, Lu J, Xu L, Wang W, Fan S. Expression of LEF1 and TCF1 (TCF7) proteins associates with clinical progression of nasopharyngeal carcinoma. J Clin Pathol. 2019;72:425–430. doi: 10.1136/jclinpath-2019-205698. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Wen Q, Xu L, Wang W, Luo J, Chu S, Xie G, Shi L, Huang D, Li J, Fan S. Fatty acid synthase-associated protein with death domain: a prognostic factor for survival in patients with nasopharyngeal carcinoma. Hum Pathol. 2014;45:2447–2452. doi: 10.1016/j.humpath.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Guo Q, Bartish M, Gonçalves C, Huang F, Smith-Voudouris J, Krisna SS, Preston SEJ, Emond A, Li VZ, Duerr CU, Gui Y, Cleret-Buhot A, Thebault P, Lefrère H, Lenaerts L, Plourde D, Su J, Mindt BC, Hewgill SA, Cotechini T, Hindmarch CCT, Yang W, Khoury E, Zhan Y, Narykina V, Wei Y, Floris G, Basik M, Amant F, Quail DF, Lapointe R, Fritz JH, Del Rincon SV, Miller WH Jr. The MNK1/2-eIF4E axis supports immune suppression and metastasis in postpartum breast cancer. Cancer Res. 2021;81:3876–3889. doi: 10.1158/0008-5472.CAN-20-3143. [DOI] [PubMed] [Google Scholar]
- 32.Wu S, Yang L, Wu D, Gao Z, Li P, Huang W, Wang X. AEG-1 induces gastric cancer metastasis by upregulation of eIF4E expression. J Cell Mol Med. 2017;21:3481–3493. doi: 10.1111/jcmm.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Abronzo LS, Bose S, Crapuchettes ME, Beggs RE, Vinall RL, Tepper CG, Siddiqui S, Mudryj M, Melgoza FU, Durbin-Johnson BP, deVere White RW, Ghosh PM. The androgen receptor is a negative regulator of eIF4E phosphorylation at S209: implications for the use of mTOR inhibitors in advanced prostate cancer. Oncogene. 2017;36:6359–6373. doi: 10.1038/onc.2017.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Li D, Sui G. YY1 is an inducer of cancer metastasis. Crit Rev Oncog. 2017;22:1–11. doi: 10.1615/CritRevOncog.2017021314. [DOI] [PubMed] [Google Scholar]
- 35.Zhou W, Feng Y, Lin C, Chao CK, He Z, Zhao S, Xue J, Zhao XY, Cao W. Yin Yang 1-induced long noncoding RNA DUXAP9 drives the progression of oral squamous cell carcinoma by blocking CDK1-mediated EZH2 degradation. Adv Sci (Weinh) 2023:e2207549. doi: 10.1002/advs.202207549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Wei J, Xue C, Zhou X, Chen S, Zheng L, Duan Y, Deng H, Xiong W, Tang F, Li G, Zhou M. Dissecting the roles and clinical potential of YY1 in the tumor microenvironment. Front Oncol. 2023;13:1122110. doi: 10.3389/fonc.2023.1122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao P, Li L, Yang L, Gui D, Zhang J, Han J, Wang J, Wang N, Lu J, Chen S, Hou L, Sun H, Xie L, Zhou J, Peng C, Lu Y, Peng X, Wang C, Miao J, Ozcan U, Huang Y, Jia W, Liu J. Yin Yang 1 protein ameliorates diabetic nephropathy pathology through transcriptional repression of TGFβ1. Sci Transl Med. 2019;11:eaaw2050. doi: 10.1126/scitranslmed.aaw2050. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal N, Theodorescu D. The role of transcription factor YY1 in the biology of cancer. Crit Rev Oncog. 2017;22:13–21. doi: 10.1615/CritRevOncog.2017021071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verheul TCJ, van Hijfte L, Perenthaler E, Barakat TS. The why of YY1: mechanisms of transcriptional regulation by Yin Yang 1. Front Cell Dev Biol. 2020;8:592164. doi: 10.3389/fcell.2020.592164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YC, Lin YH, Chi HC, Huang PS, Liao CJ, Liou YS, Lin CC, Yu CJ, Yeh CT, Huang YH, Lin KH. CRNDE acts as an epigenetic modulator of the p300/YY1 complex to promote HCC progression and therapeutic resistance. Clin Epigenetics. 2022;14:106. doi: 10.1186/s13148-022-01326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong X, Guo R, Ji T, Zhang J, Xu J, Li Y, Sheng Y, Wang Y, Fang K, Wen Y, Liu B, Hu G, Deng H, Yao H. YY1 safeguard multidimensional epigenetic landscape associated with extended pluripotency. Nucleic Acids Res. 2022;50:12019–12038. doi: 10.1093/nar/gkac230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan X, Yang T, Cuesta A, Pang X, Balius TE, Irwin JJ, Shoichet BK, Taunton J. Discovery of lysine-targeted eIF4E inhibitors through covalent docking. J Am Chem Soc. 2020;142:4960–4964. doi: 10.1021/jacs.9b10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen P, Reineke LC, Knutsen E, Chen M, Pichler M, Ling H, Calin GA. Metformin blocks MYC protein synthesis in colorectal cancer via mTOR-4EBP-eIF4E and MNK1-eIF4G-eIF4E signaling. Mol Oncol. 2018;12:1856–1870. doi: 10.1002/1878-0261.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 45.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–5334. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- 46.Knight JRP, Alexandrou C, Skalka GL, Vlahov N, Pennel K, Officer L, Teodosio A, Kanellos G, Gay DM, May-Wilson S, Smith EM, Najumudeen AK, Gilroy K, Ridgway RA, Flanagan DJ, Smith RCL, McDonald L, MacKay C, Cheasty A, McArthur K, Stanway E, Leach JD, Jackstadt R, Waldron JA, Campbell AD, Vlachogiannis G, Valeri N, Haigis KM, Sonenberg N, Proud CG, Jones NP, Swarbrick ME, McKinnon HJ, Faller WJ, Le Quesne J, Edwards J, Willis AE, Bushell M, Sansom OJ. MNK inhibition sensitizes KRAS-mutant colorectal cancer to mTORC1 inhibition by reducing eIF4E phosphorylation and c-MYC expression. Cancer Discov. 2021;11:1228–1247. doi: 10.1158/2159-8290.CD-20-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Yue P, Chan CB, Ye K, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Fu H, Khuri FR, Sun SY. Inhibition of mammalian target of rapamycin induces phosphatidylinositol 3-kinase-dependent and Mnk-mediated eukaryotic translation initiation factor 4E phosphorylation. Mol Cell Biol. 2007;27:7405–7413. doi: 10.1128/MCB.00760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi B, Cai AL, Keiper BD, Minich WB, Mendez R, Beach CM, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, Zhong W, Cao R. Phosphorylation of the mRNA cap-binding protein eIF4E and cancer. Cell Signal. 2020;73:109689. doi: 10.1016/j.cellsig.2020.109689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Fan S, Koo J, Yue P, Chen ZG, Owonikoko TK, Ramalingam SS, Khuri FR, Sun SY. Elevated expression of eukaryotic translation initiation factor 4E is associated with proliferation, invasion and acquired resistance to erlotinib in lung cancer. Cancer Biol Ther. 2012;13:272–280. doi: 10.4161/cbt.18923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16:4914–4920. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, Larsson O, Muller WJ, Miller WH, Sonenberg N. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34:2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JH, Park S, Chung H, Oh S. Wnt5a attenuates the pathogenic effects of the Wnt/β-catenin pathway in human retinal pigment epithelial cells via down-regulating β-catenin and snail. BMB Rep. 2015;48:525–530. doi: 10.5483/BMBRep.2015.48.9.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.