Abstract
Metazoans are known to contain a limited, yet highly conserved, set of signal transduction pathways that instruct early developmental patterning mechanisms. Genomic surveys that have compared gene conservation in signal transduction pathways between various insects and Drosophila support the conclusion that these pathways are conserved in evolution. However, the degree to which individual components of signal transduction pathways vary among more divergent arthropods is not known. Here, we report our results of a survey of the genome of the two-spotted spider mite Tetranychus urticae, using a set of 294 Drosophila orthologs of genes that function in signal transduction. We find a third of all genes surveyed absent from the spider mite genome. We also identify several novel duplications that have not been previously reported for a chelicerate. In comparison with previous insect surveys, Tetranychus contains a decrease in overall gene conservation, as well as an unusual ratio of ligands to receptors and other modifiers. These findings suggest that gene loss and duplication among components of signal transduction pathways are common among arthropods and suggest that signal transduction pathways in arthropods are more evolutionarily labile than previously hypothesized.
Keywords: Arthropod comparative genomics, Chelicerate, Evolution, Development, Signal transduction
Introduction
A limited set of signal transduction pathways are known to pattern developing embryos throughout the metazoans. In model systems, where signal transduction pathways have been extensively studied, any given signaling pathway is composed of extracellular ligands, modifiers of these ligands, receptors and co-receptors, and varying numbers of cytoplasmic signal transducers. Ligand diversification for some pathways has been well studied. Surprisingly, little effort has been made toward a comparison of gene conservation within these signal transduction pathways across diverse taxa despite an increased number of metazoan genome sequences. The most comprehensive genomic surveys of signal transduction pathways are attributed to arthropods, with a focus on higher insects and conservation of key genes known to pattern Drosophila melanogaster during embryogenesis (Dearden et al. 2006; Shigenobu et al. 2010; Behura et al. 2011). As might be expected, based on the phylogenetic relationship between Drosophila and the species sampled, the majority of genes surveyed are conserved. Instances of lineage-specific gene duplications and losses were identified, demonstrating a perhaps unexpected degree of variation among signaling pathways. These results suggest that examination of the degree of conservation of Drosophila orthologs in the other major arthropod clades (Crustacea, Myriapoda, and Chelicerata) might reveal additional variation, as well as trends within signal transduction pathways during arthropod evolution.
The two-spotted spider mite Tetranychus urticae is one of several noninsect arthropods to have a recently completed genome project (Grbić et al. 2011). As a member of the phylogenetically basal group of arthropods, Chelicerata, Tetranychus is of taxonomic interest for comparative study. For example, Tetranychus forms its body segments sequentially like most arthropods, unlike the derived mode of development utilized by Drosophila in which all body segments form at once. In addition, Tetranychus contains several derived morphological features, including a reduced body plan achieved through a loss of posterior segments, a developmental delay of the fourth pair of walking legs, and silk spinnerets located on the anterior body tagma. With a sequenced genome (Grbić et al. 2011) and available methods for testing gene expression (Dearden et al. 2003) and embryonic gene silencing through RNA interference (RNAi) (Khila and Grbić 2007), Tetranychus represents a novel system to study evolution of developmental regulatory networks, including signal transduction pathways. As estimates place the Pancrustacea-Chelicerata time of divergence at ~725 million years ago (Pisani et al. 2004), it might be expected that there will be key differences in conservation of signal transduction pathways and other developmental processes between Tetranychus, Drosophila, and other insects.
Here, we present the first comprehensive characterization of six major signal transduction pathways (e.g., transforming growth factor-β (TGF-β), Wnt, Notch, Janus kinase/signal transducers and activators of transcription (JAK-STAT), receptor tyrosine kinase, and Hedgehog) in a chelicerate with a sequenced genome. The patterns of conservation of Drosophila orthologs within the insects, the unique morphology of Tetranychus, and the time of divergence between insects and chelicerates led us to hypothesize that conservation of similar pathway components in Tetranychus would differ from previous surveys in insects. Our results show that Tetranychus contains a reduced number of Drosophila orthologs, an expansion of lineage-specific gene duplications, and suggest that conservation of signal transduction pathway genes used during development in arthropods may be more plastic than previously understood.
Materials and methods
Identification and annotation of T. urticae signal transduction pathway genes
Using the genome of T. urticae (Sanger sequencing, 8.05× coverage) (Grbić et al. 2011), BLAST (blastn, blastp) analysis was performed using the bioinformatics suite located at online resource for community annotation of eukaryotes (OrcAE) (Sterck et al. 2012). For the list of genes, GI sequences, BLAST scores, and e-values, please see Supplemental Table 1. Signal transduction pathway genes were determined from previous surveys (Dearden et al. 2006; Shigenobu et al. 2010), gene ontology annotations from FlyBase (http://www.flybase.org) (St Pierre et al. 2014), the Kyoto Encyclopedia of Gene and Genomes (http://www.genome.jp/kegg/pathway.html) (Kanehisa and Goto 2000), and the Interactive Fly (http://www.sdbonline.org/fly/aimain/1aahome.htm) (Brody 1999). Sequences were then curated from the National Center for Biotechnology Information (NCBI) database. We used an e-value requirement of ≤0.001 for our initial BLAST. All gene orthology calls were corroborated by reciprocal BLAST analyses against the NCBI nr and D. melanogaster protein databases, to identify the top D. melanogaster sequence. Protein sequences for phylogenies were initially aligned with MUSCLE, followed by Gblocks to eliminate poorly aligned regions (except where noted), with the following parameters allowing for smaller final blocks, gap positions in final alignment, and less strict flanking positions (Castresana 2000; Talavera and Castresana 2007). Phylogenetic trees were constructed with PhyML (Guindon et al. 2010). The following parameters were used: amino acid substitution model=WAG; proportion of invariable sites—estimated; number of categories of substitution rate=4. Statistical support for phylogenetic grouping was assessed by approximate likelihood ratio tests based on a Shimodaira-Hasegawa-like procedure (SH-aLRT) with scores shown in the tree. In the case where no homologous sequences could be found, e-values were decreased and/or tblastn or tblastx was used to confirm absences from the genomic database. Gene annotations were entered manually using OrcAE. Accession numbers for D. melanogaster and T. urticae genes are provided in Supplemental Table 1.
Results
Gene and major signal transduction pathways that are expected conserved
Many cellular signal transduction pathways are expected conserved in Tetranychus based on their fundamental role in development in arthropods and throughout the Metazoa. However, little is known about the variation within signaling pathways in arthropods. To test this, we examined the Tetranychus genome for the presence and absence of genes from six major Drosophila signal transduction pathways (TGF-β, Wnt, Notch, receptor tyrosine kinase (RTK), JAK-STAT, and Hedgehog) and compared their conservation to the honeybee Apis mellifera and the pea aphid Acyrthosiphon pisum. In addition, in specific cases, we compared gene conservation in Tetranychus to the red flour beetle Tribolium castaneum and the deer tick Ixodes scapularis.
TGF-β
The TGF-β signaling pathway acts through a concentration gradient of secreted morphogens to regulate cell growth, proliferation, and differentiation during embryogenesis. Among the higher ordered developmental processes that TGF-β signaling regulates are dorsal-ventral polarity, patterning and growth of developing organs, and neuronal patterning.
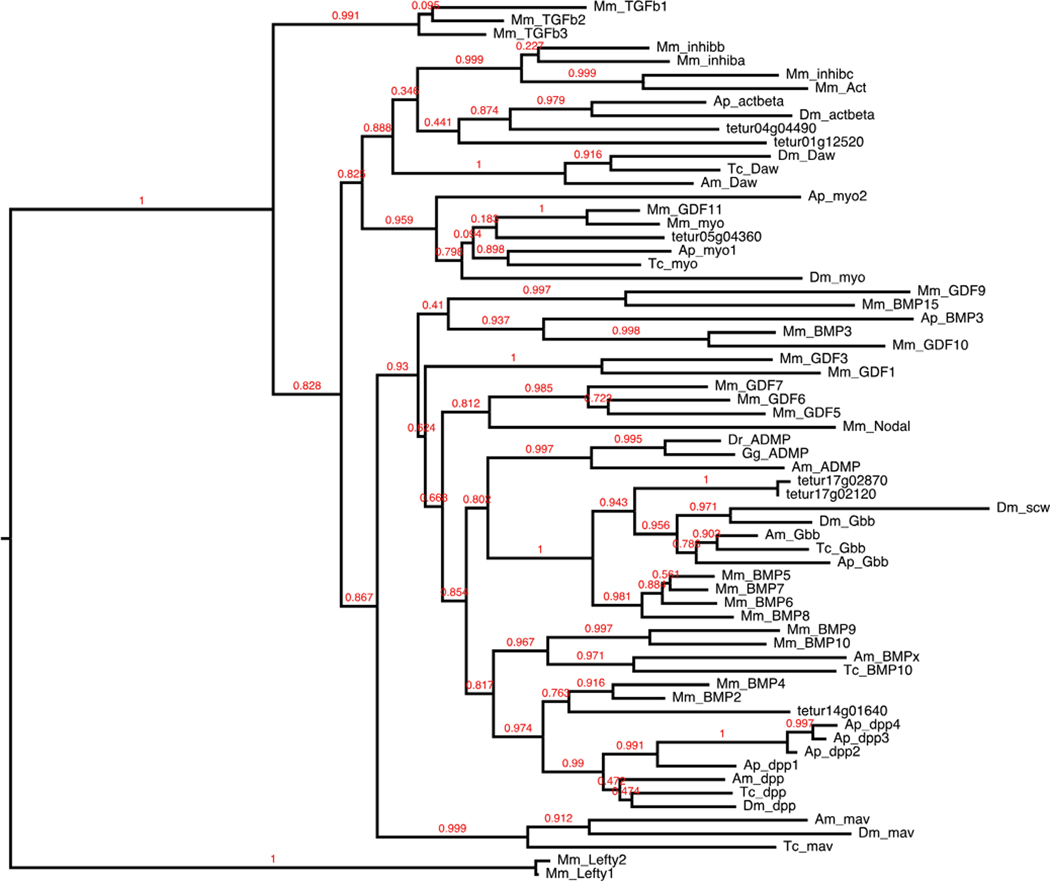
In total, we identified 45 of 63 Drosophila TGF-β signaling pathway components in the Tetranychus genome (Table 1). We identified four of seven Drosophila TGF-β ligands, including decapentaplegic (dpp), myoglianin (myo), activin-beta, and glass bottom boat, with duplications of the latter two (Fig. 1). The ligands maverick, Dawdle (Daw/ALP23), and screw are missing, with the absence of screw expected as it is derived in the lineage leading to Drosophila (Van der Zee et al. 2008). Two additional TGF-β ligands (BMP-10 and anti-dorsalizing morphogenetic protein) found in Tribolium, Apis, and vertebrates (Shigenobu et al. 2010; Van der Zee et al. 2008) are not conserved in Tetranychus. Four of five Drosophila TGF-β receptors were identified: baboon (type I), saxophone (type I), and duplications of thickveins (type I) and punt (type II). We were unable to find the type II receptor wishful thinking (wit), also absent from the Acyrthosiphon genome (Shigenobu et al. 2010). Orthologs of wit are found in Ixodes, Apis, Tribolium, and Drosophila, where they cluster most closely with mouse BMP type II receptor (Van der Zee et al. 2008) (Fig. 2), suggesting wit has been independently lost in Tetranychus and Acyrthosiphon. We found Smad family cytoplasmic transducers Mothers against dpp, Smad on X, and Medea, but not the anti-Smad daughters against dpp. We found several extracellular modifiers known to modulate TGF-β signal activity through interactions with both ligand receptors, including the dpp-inhibitor short gastrulation (sog) (Holley et al. 1996; reviewed in Massagué and Chen 2000) and the activin-inhibitor follistatin. Additional extracellular modifiers that counteract ligand inhibitors and enhance the TGF-β signal were also searched for including, crossveinless (cv1/tsg2), the sog cleavage protein tolloid (tld), and its relative tolloid-related (tlk). While cv1 and tlk are present, we did not find a homolog for tld.
Table 1.
Inventory list of key components from signal transduction pathways in Tetranychus
| Class/function | Name | Duplication |
|---|---|---|
|
| ||
| TGF-beta | ||
| Ligand | dpp | |
| actbeta | Duplication (2) | |
|
Gbb
myo |
Duplication (2) | |
| Extracellular modifiers | tlk | Duplication (4) |
|
sog
fs |
Duplication (2) | |
| Type I receptor | babo | |
|
tkv
Sax |
Duplication (2) | |
| Type II receptor | put | Duplication (2) |
| R-SMAD | Mad | |
| R-SMAD | Smox | |
| Co-SMAD (common) | Med | |
| Transcription factor |
Rbf
nej shn snoN |
|
| Smad binding |
fuss (CORL)
Rocla cul/linl9 Sara |
Duplication (2) |
| lack | Duplication (2) | |
| (absences) |
DAW,
ADMP, scw, mav, BMP-10, wit, tld |
|
| Wnt | ||
| Ligand |
Wnt4
Wnt5 |
|
|
Wnt6
Wnt8 Wnt 16 WntA |
Duplication (3) | |
| Transmcmbranc receptor | Vang | Duplication (2) |
| fz | Duplication (2) | |
|
fz2
fz4 arr |
Duplication (2) | |
| Activated by frizzled | dsh | Duplication (2) |
| Arm destruction complex |
APC
sgg |
Duplication (2) |
| Downstream effector | arm | Duplication (3) |
| (β-Catenin binding | pont | |
| Antagonist of β-catenin | Cby | |
| pan | Duplication (3) | |
| Transcription factor | CtBP | |
| Scaffolding protein | ebi | Duplication (2) |
| Ligand modifier | wls | |
| por | ||
| shf | ||
| Wnt-protein binding | dlp | |
| (absences) | wg, wnt2, wnt3, wnt7, wnt9, wnt 10, wnt11, Axn, pygo, Igs, nkd | |
| Notch | ||
| Ligand |
Dl
Ser |
|
| Transmcmbranc receptor | N | Duplication (4) |
| Receptor modifier |
O-fut
fng |
|
| Protease | Fur1 | Duplication (2) |
| Mctalloprotcasc | kuz | |
| γ-Secrctasc complex |
Psn
pen-2 nct |
|
| Aph01 | Duplication (2) | |
| Transcription factor | Su(H) | Duplication (3) |
| gro | Duplication (2) | |
| nub | Duplication (2) | |
| (absences) | Tace, dx, H, mam, E(spl) | |
| RTK | ||
| EGF | ||
| Ligand |
spi
vn |
|
| Transmcmbranc receptor | EGFR | |
| kek-1 | ||
| kek-3 | Duplication (2) | |
| EGFR inhibitor | aos | Duplication (2) |
| (absences) | grk, Km, Yan | |
| FGF | ||
| Transmcmbranc receptor | htl | Duplication (2) |
|
sgl
sfl |
||
| ECM component | trol | |
| (absences) | bnl, btl, pyr, ths, sty | |
| Scvenless/MAPK | ||
| Transmcmbranc receptor | sev | |
|
drk
Sos |
||
| Rasl | Ras85D | |
| MAPKKK (RAF) | phi | |
| MAPKK (MEK) |
Dsor1
hep lic |
|
| MAPK |
rl
p38b |
|
| (absences) | boss | |
| JAK-STAT | ||
| Receptor | Dome | |
| JAK | hop | |
| STAT | Stat92E (mrl) | Duplication (2) |
| (absences) | os, upd2, upd3 | |
| Hh | ||
| Ligand | Hh | |
| Transmembrane receptor |
ptc
smo mgl |
|
| Transcription factor | ci | |
| Repressor | Su(fu) | Duplication (2) |
| (absences) | cos, fu | |
TGF-beta transforming growth factor-β, RTK receptor tyrosine kinase, EGF epidermal growth factor, EGFR epidermal growth factor receptor, FGF fibroblast growth factor, ECM extracellular matrix, MAPK mitogen-activated protein kinase, JAKSTAT Janus kinase/signal transducers and activators of transcription, Hh hedgehog
Fig. 1.
TGF-beta ligands. Full-length protein sequences were used to construct this maximum likelihood tree of TGF-beta ligands. Protein sequences for mouse (Mm), Drosophila melanogaster (Dm), Tribolium castaneum (Tc), and Apis mellifera (Am) were identified from the previous phylogenetic reconstruction of Van der Zee et al. (2008). Acyrthosiphon pisum (Ap) sequences were curated from Shigenobu et al. (2010) and from NCBI GenBank
Fig. 2.
TGF-beta receptors. Full-length protein sequences were used to construct this maximum likelihood tree of TGF-beta receptors. Tetranychus thickveins paralogs group away from the thickveins cluster but with the type I receptors. Mm, Dm, Tc, and Am protein sequences were identified from references in Fig. 1. Ixodes scapularis (Is) sequences were curated from NCBI GenBank
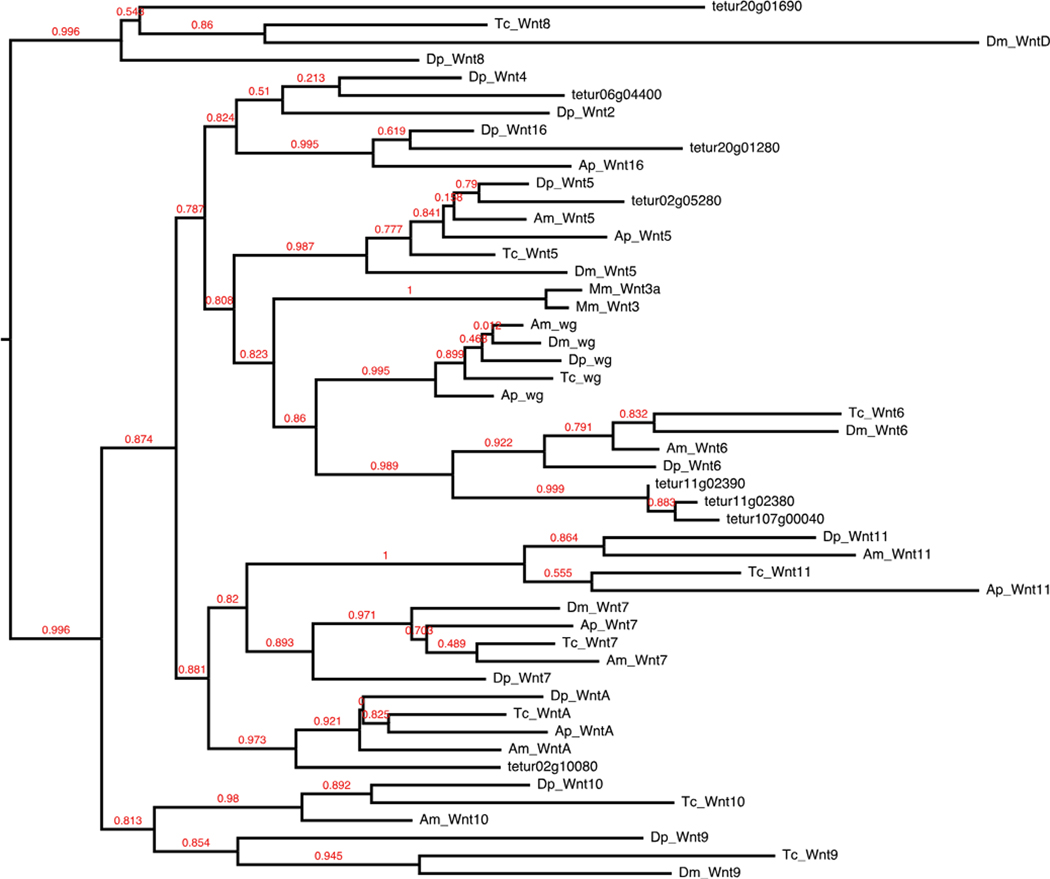
Wnt
The Wnts are a highly conserved family of secreted growth factors involved in early developmental patterning, including embryonic induction, cell fate, polarity, and death. Wnt ligands have been implicated in arthropods such as Tribolium and the spider Achaearanea in the establishment of posterior segments (Bolognesi et al. 2008; McGregor et al. 2008).
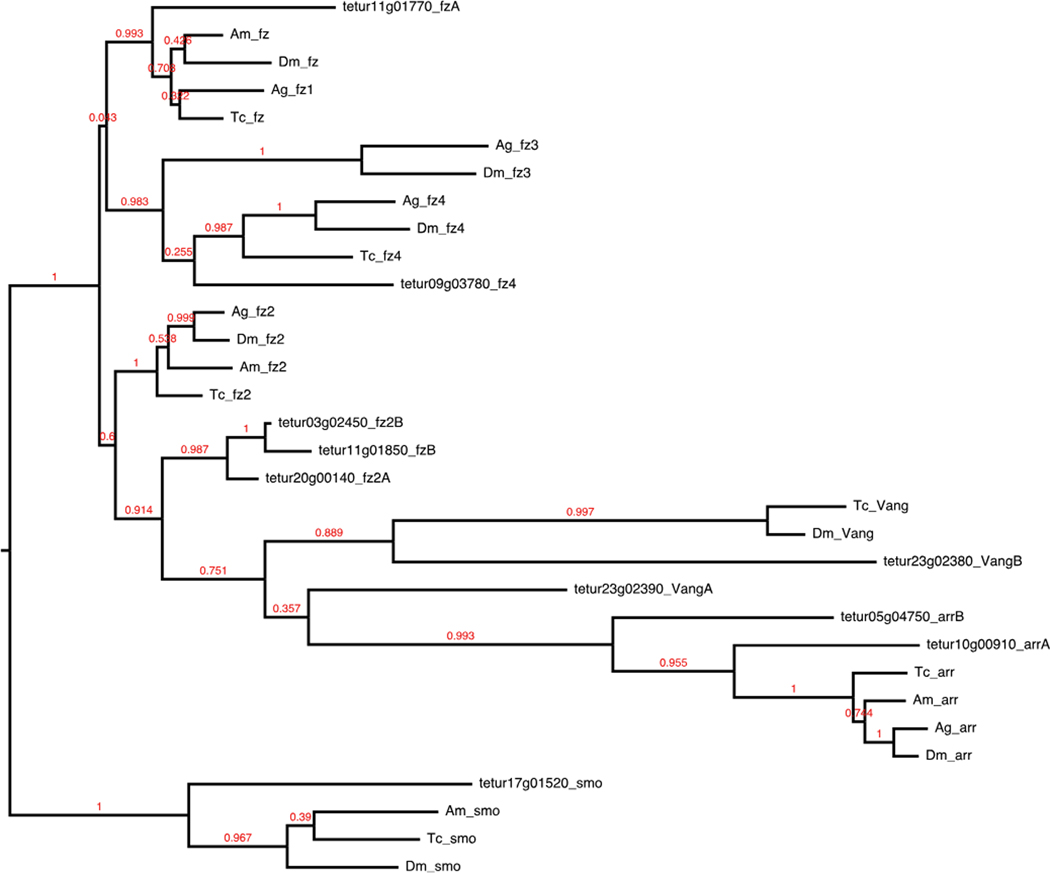
We identified 60 of 84 Drosophila Wnt signal transduction pathway components (Table 1). In Wnt-producing cells, the growth factors are glycosylated and lipid-modified for proper secretion and signal activity (Willert et al. 2003; Zhai 2004). Except for wnt8/D (Ching et al. 2008), palmitoylation by the ER membrane-bound protein porcupine is required for recognition of Drosophila Wnts by Wntless, a second membrane-bound protein essential for further transport and secretion (Bartscherer et al. 2006; Bänziger et al. 2006; Herr and Basler 2012). Both ligand modifiers required for proper Wnt secretion were found. Conservation of Wnt ligand subfamilies in arthropods varies. Insects have been found to contain as few as six in Anopheles and Acyrthosiphon, on up to nine in Tribolium. Both crustaceans and chelicerates have been found to contain 12 (Janssen et al. 2010). We identified eight sequences in Tetranychus with conserved Wnt domains, belonging to six subfamilies—Wnt4, Wnt5, Wnt6, Wnt8, Wnt16, and WntA, with three copies of Wnt6 (Fig. 3). Surprisingly, we did not find an ortholog of the wingless (wg/Wnt1) ligand that is conserved throughout Arthropoda and thus represents the first reported absence in an arthropod. Duplications of Wnt ligands are rare, but lineage-specific duplications have been reported in the cnidarian Nematostella, vertebrates, and the spider Achaearanea (Janssen et al. 2010). The three copies of Wnt6 present in Tetranychus represent the largest paralog group identified in an arthropod genome. Once secreted, Wnts bind to members from the frizzled family of transmembrane receptors and the LDL-receptor-related protein arrow. We found five of six Drosophila Wnt receptors, with additional duplications of the receptors Van gogh, frizzled, frizzled-2, and arrow (Fig. 4). Absence of ligand-receptor activity results in a complex of proteins (Axin, adenomatous polyposis coli (APC), and shaggy) phosphorylating and thus targeting the cytoplasmic transducer/transcription factor armadillo/β-catenin (arm) for degradation. Once the receptor binds to its respective ligand, the intracellular effector Disheveled is activated and blocks shaggy from phosphorylating arm. This allows hypophosphorylated arm to accumulate in the cytoplasm and promotes its translocation to the nucleus to regulate transcription with co-regulators, principally from the TCF/ LEF family of transcription factors. We found two copies of disheveled (Dsh), and three copies of armadillo/β-catenin. Of the three proteins that form the core β-catenin destruction complex, APC and shaggy homologs were present, but an Axin homolog is missing.
Fig. 3.
Wnt ligands. Mm, Dm, Tc, Am, Ap, and Daphnia pulex (Dp) protein sequences were identified from Janssen et al. (2010)
Fig. 4.
Wnt receptors. Dm, Tc, Am, and Anopheles gambiae (Ag) protein sequences were curated from NCBI GenBank
Notch
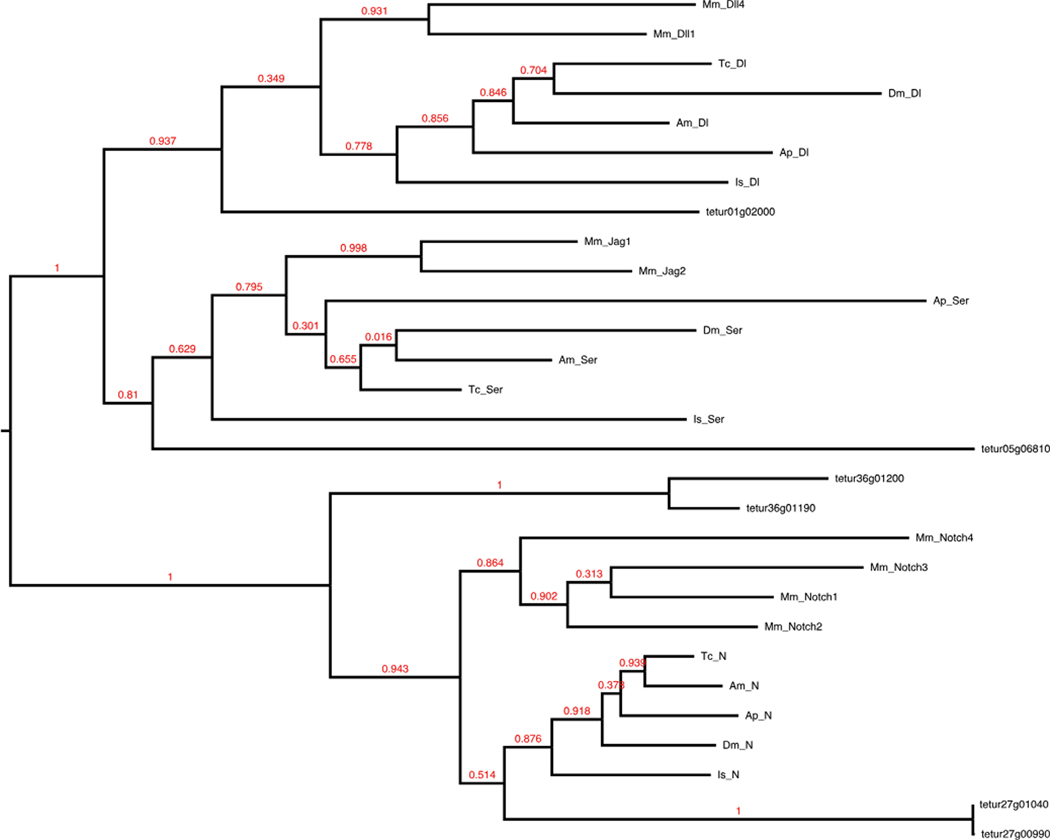
Notch and its ligands Delta and Serrate are transmembrane proteins that provide direct cell-cell communication. Notch signaling is required during neurogenesis in vertebrates and invertebrates and likely has an ancestral role in the establishment and maintenance of posterior segments in arthropods (Chesebro et al. 2013; Chipman and Akam 2008; Pueyo et al. 2008; Schoppmeier and Damen 2005; Stollewerk et al. 2003), a function similar to its role during vertebrate somitogenesis (Dequeant et al. 2006; Palmeirim et al. 1997).
A majority of the core Drosophila Notch signaling pathway is present in the Tetranychus genome (42/66) (Table 1). There are four copies of the Notch receptor and single copies of the ligands Delta and Serrate (Fig. 5). During trafficking to the cell membrane and upon ligand binding, Notch undergoes a tripartite series of cleavages (S1, S2, and S3) that results in activation and release from the cell membrane to the cytosol. The cleaved portion, or Notch intracellular domain, then translocates into the nucleus to regulate transcription. Among the proteases that participate in Notch cleavage, we found duplications of the S1 protease furin-1 and the S3 γ-secretase component anterior pharynx defective 1 (APH-1). Of the metalloproteases that participate in S2 extracellular cleavage of Notch, Kuzbanian (Kuz) is present, but not TNF-α converting enzyme (TACE). The loss of TACE appears to be lineage-specific as a homolog is present in I. scapularis (XP_002405453) and may be attributed to its redundancy in canonical Notch signaling. For example, Kuz and TACE have been reported both genetically and biochemically to have partially redundant functions in ecdysozoans (Brou et al. 2000; Mumm et al. 2000). In addition, Drosophila TACE does not appear to be required for most Notch-mediated cell decisions as loss of Kuz leads to associated Notch phenotypes (Pan and Rubin 1997; Rooke et al. 1996), and overexpression of TACE is insufficient for restoring appropriate Notch S2 cleavage (Lieber 2002). Several other Notch protein regulators such as deltex and mastermind are also missing and have been reported absent in several other metazoan genomes suggesting Notch signaling functions in their absence (Gazave et al. 2009; Maier 2006; Shigenobu et al. 2010). The absence of deltex appears to be lineage-specific, as a homolog has been recorded for Ixodes (IscW_ISCW003605), and also suggests that Tetranychus lacks this noncanonical Notch signaling pathway. Additional missing regulators (Table 1) appear to be derived in Drosophila and other higher insects and have been expected absent in more basally branching arthropods. For example, the Notch antagonist hairless to date has only been found within dipterans and hymenopterans (Maier 2006).
Fig. 5.
Notch receptors and ligands. Full-length protein sequences were used to construct this maximum likelihood tree of Notch receptors and ligands
We also identified several well-known transcriptional targets of Notch, including genes from the achaete-scute (AS-C) and enhancer of split complexes (E(spl)-C) (Schlatter and Maier 2005). The Drosophila AS-C is a 40-kb complex of four basic helix-loop-helix (bHLH) transcription factors, achaete, scute, lethal of scute, and asense (reviewed in García-Bellido and de Celis 2009). We found apparent duplications of both achaete and scute on multiple contigs (Supplemental Table 1) but did not find homologs of lethal of scute and asense. The E(spl)-C of Drosophila spans a 45-kb region and contains a group of four bearded family and seven bHLH transcription factors thought to have been duplicated from a pair of ancestral genes (reviewed in Dearden and Duncan 2010). We found two classes of bHLH members in tandem (tetur01g03010 and tetur01g03020; tetur08g02740 and tetur08g02780). Our phylogenetic analysis suggests that these may be part of an ancestral E(spl)-C or that these genes likely represent a lineage-specific duplication of a single ancestral bHLH gene (Supplemental Fig. 1). These findings parallel previous pancrustacean surveys that revealed similar reductions in the number of E(spl)-C genes, and none have been found in Ixodes (Dearden and Duncan 2010; Dearden et al. 2006; Schlatter and Maier 2005).
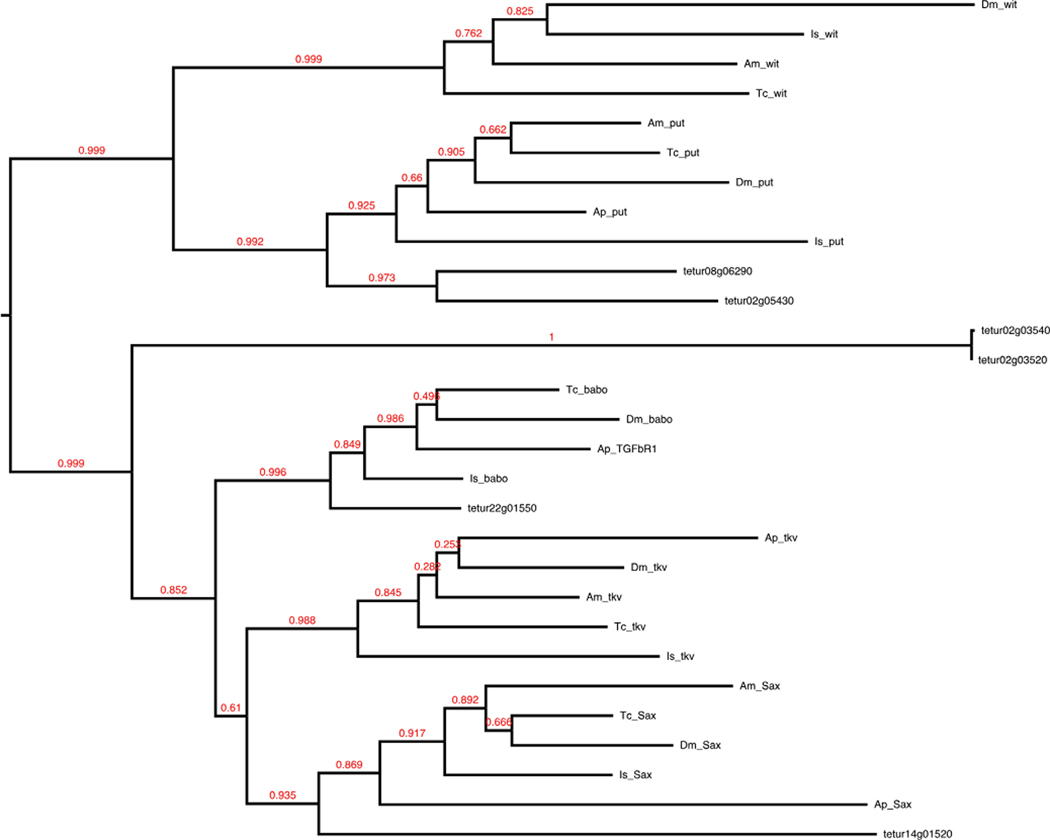
RTK (EGF/FGF/Sevenless)
RTK signaling contributes to changes in cell shape and terminal and dorsal patterning, among several other developmental mechanisms that all rely on intracellular MAP kinase activity. We searched for 50 Drosophila genes among three well-known RTK pathways: epidermal growth factor (EGF), fibroblast growth factor (FGF), and Sevenless. These pathways share a MAPK signal cascade that includes Sos, Ras1, Raf1, Dsor1, and rolled; all of which are conserved in Tetranychus (Table 1).
EGF signaling
Fourteen of 30 EGF signaling components were identified, with the core pathway conserved (Table 1). We found two of four ligands: spitz and vein. Of the four receptors searched for, we found single copies of epidermal growth factor receptor and kekkon-1, along with two kekkon-3 orthologs. We found two copies of the extracellular EGFR inhibitor argos (giant lens), also found with four copies in Acyrthosiphon.
FGF signaling
The FGF signaling pathway appears highly modified in Tetranychus (Table 1). Three FGF ligands (branchless, pyramus, and thisbe) and two FGF receptors (breathless and heartless) have been identified in Drosophila. We did not find any ligands and only found a single FGF receptor, heartless. Similar modifications of FGF signaling has been reported for the genomes of three mosquito species, with Aedes aegypti missing the ligand branchless, and all three mosquitoes missing the receptor breathless, and the heartless ligands pyramus and thisbe.
Sevenless signaling
Except for the absence of the canonical ligand boss of Sevenless (boss), the Sevenless signaling pathway is conserved in Tetranychus (Table 1). A review of the Ixodes genome also shows boss to be missing. The absence of boss is striking due to its role in determining cell fate during fly retina development (Krämer et al. 1991). Further inquiry into the developmental genetic mechanisms that underlie chelicerate eye development may provide further insight into whether Tetranychus is likely to have a functioning Sevenless pathway in the absence of this canonical ligand.
JAK/STAT
The JAK/STAT signaling pathway is a conserved, pleiotropic cell signaling pathway involved in cytokine transduction and growth factor signaling (Behura et al. 2011; Darnell 1997; Rawlings et al. 2004; Shigenobu et al. 2010; Zeidler et al. 2000) (Table 1). The pathway involves stimulation of a variety of receptors, each having a characteristic JAK tyrosine kinase attached. This results in the transphosphorylation of JAKs, which in turn phosphorylate STATs, a family of otherwise inactive transcription factors that reside in the cytoplasm. STATs then enter the nucleus and control expression of JAK/STAT target genes. We were unable to identify any previously identified JAK/STAT ligands (os, upd2, upd3). Interestingly, Tetranychus contains duplications of both hopscotch and Stat92E, which are classified as canonical JAK- and STAT-like proteins in Drosophila (Zeidler et al. 2000). Stat92E duplications are not unique to Tetranychus as three copies have been reported in the mosquito Culex quinquefasciatus (Behura:2011dv). Additional effectors of the JAK/STAT signaling pathway include protein families such as STAMs and SOCs, which activate and repress different pathway components, respectively (Rawlings et al. 2004). Orthologs of Socs36E and Socs44A were not found in our search of the genome, while Socs16D has duplicated. The JAK/STAT inhibitor phosphatidylinositol 3-kinase was also found present in multiple copies.
Hedgehog
The hedgehog (Hh) protein functions as a morphogen during development. The hedgehog ligand is highly lipid-modified; a cholesterol moiety is attached to the C-terminus, and the N-terminus undergoes palmitoylation. The covalent coupling of cholesterol to Hh acts to tether the ligand to the plasma membrane and regulate long-range signaling. High conservation of hedgehog signaling components has been observed in several higher insects, with each genome yielding a complete pathway (Dearden et al. 2006; Shigenobu et al. 2010). However, this trend does not hold for Tetranychus.
Eleven of 14 pathway components were found; the genome lacks dispatched, costa, and fused (Table 1). The absence of dispatched is striking as it is required for release of cholesterol anchored Hh from Hh-secreting cells (Burke et al. 1999). Curiously, costa and fused are essential components of the protein complex that, in unstimulated cells, collects the transcription factor cubitus interruptus (ci) in the cytoplasm (Alves et al. 1998; Farzana and Brown 2008; Sisson et al. 1997). This action encourages cleavage of the ci N-terminus before entering the nucleus, which conversely produces a repressive effect on targeted genes (Aza-Blanc et al. 1997). In stimulated cells, smoothened inhibits cleavage of ci and allows the full protein to enter the nucleus and activate target genes (Alexandre et al. 1996). Without costa and fused, the ci-sequestering complex is incomplete, making the canonical cleavage of ci impossible in this system and allowing the constant transcription of Hh target genes. Reports of lineage-specific gene duplications are rare in this pathway. However, two copies of supernumerary limbs (Slmb) have been reported in the vector mosquito, C. quinquefasciatus (Behura:2011dv). Tetranychus also contains a duplication of Slmb, as well as a duplication of Suppressor of fused.
Comparison to previous insect surveys
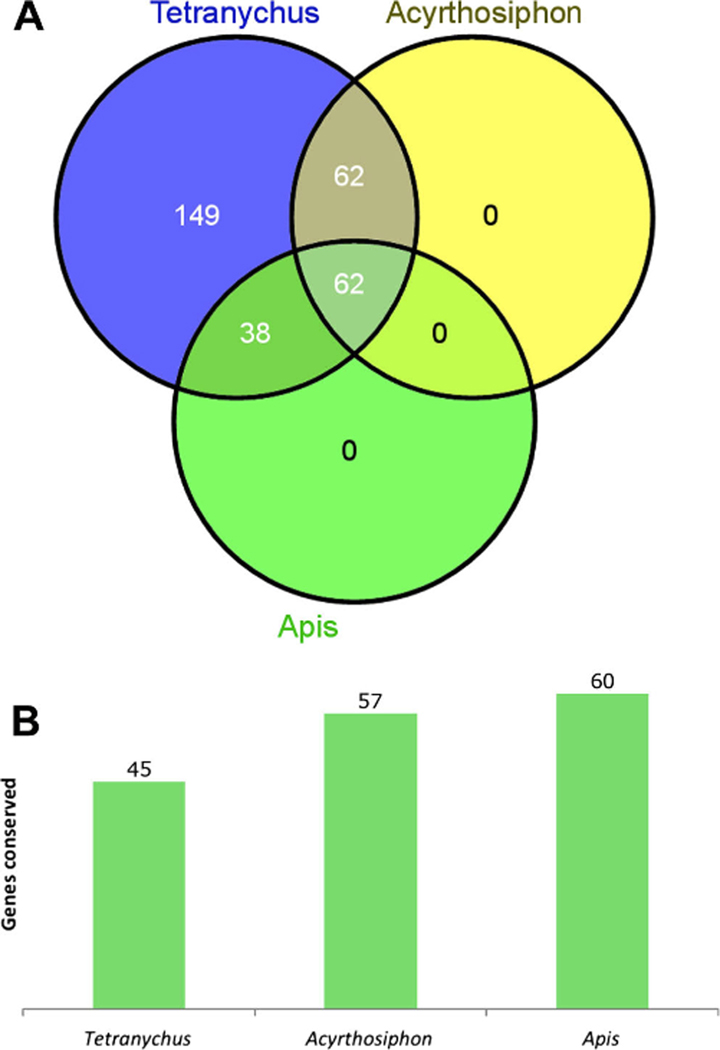
Unique sets of genes from the six signaling pathways surveyed in Tetranychus have been previously surveyed in insects. Of the 294 Drosophila genes that we surveyed in the Tetranychus genome, 100 were surveyed in the hymenopteran honey bee Apis mellifera genome (Dearden et al. 2006), and 124 were surveyed in the more distantly related hemipteran pea aphid Acyrthosiphon pisum genome (Shigenobu et al. 2010) (Fig. 6). Sixty-two genes were found to overlap within all three sets (Fig. 6a, Supplemental Table 2). Of these genes, there appears to be a phylogenetic trend of conservation, with 73, 92, and 97 % conserved in Tetranychus (45/62), Acyrthosiphon (57/62), and Apis (60/62), respectively (Fig. 6b). Only two genes, screw and gurken, are missing from all three genomes, although these are expected absences as they are believed to have originated within Diptera (Dearden et al. 2006). Acyrthosiphon is missing three additional genes, of which one is shared with Tetranychus (wnt10). Tetranychus contains an additional 14 unique gene absences from the set of 62 genes shared among the three surveys. These results show that gene absences appear far more common in Tetranychus, both among the set of 62 genes shared among the three surveys and in the total number of genes surveyed.
Fig. 6.
Number of genes conserved among genes shared in arthropod surveys. a Venn diagram of Drosophila genes surveyed for in Tetranychus, Acyrthosiphon, and Apis. b Number of shared Drosophila orthologs conserved among 62 genes surveyed for in the three arthropod surveys. Tetranychus (44/62), Acyrthosiphon (57/62), and Apis (60/62)
In contrast, 33 of 124 genes surveyed in both Acyrthosiphon and Tetranychus genomes are found duplicated in Acyrthosiphon and Tetranychus, Acyrthosiphon alone, or Tetranychus alone (Table 2). Within this set of duplicated genes, Tetranychus contains almost twice the number of duplications as Acyrthosiphon. In total, however, these duplications represent less than half the number of duplications in signaling pathways found in the Tetranychus genome.
Table 2.
Lineage-specific and shared gene duplications found in the 124 Drosophila orthologs surveyed for in the Tetranychus and Acyrthosiphon genomes
| Tetranychus | Acyrthosiphon | |
|---|---|---|
|
| ||
| activin-beta | 2 | |
| anterior pharynx defective 1 | 2 | |
| APC-like | 2 | |
| canoe | 2 | |
| dishevelled | 2 | |
| frizzled | 2 | |
| frizzled 3 | 3 | |
| glass bottom boat | 2 | |
| groucho | 2 | |
| heartless | 3 | |
| kekkon-3 | 2 | |
| Notch | 4 | |
| nubbin | 2 | |
| pangolin/TCF-LEF1 | 3 | |
| punt | 2 | |
| short gastrulation | 2 | |
| supernumerary limbs | 2 | |
| Suppressor of fused | 2 | |
| thickveins | 2 | |
| Wnt6 | 3 | |
| argos | 2 | 4 |
| armadillo/β-catenin | 2 | 2 |
| Stat | 2 | 2 |
| Suppressor of Hairless | 3 | 2 |
| decapentaplegic | 4 | |
| Domeless | 5 | |
| kekkon-1 | 2 | |
| kekkon-2 | 2 | |
| Medea | 5 | |
| Mothers against dpp | 2 | |
| myoglianin | 2 | |
| Rhomboid-4 | 2 | |
| shaggy/GSK-3 | 2 | |
| Number of duplicated orthologs | 24 | 13 |
| Total number of paralogs | 55 | 36 |
To determine whether the duplications found in Tetranychus represent lineage-specific duplications or are more broadly representative of duplications within acarid chelicerates, we searched the tick I. scapularis draft genome for similar sets of paralogs. The gene duplications Tetranychus that shares with Acyrthosiphon (Su(H), argos, arm, and Stat) appear to have arisen independently in both lineages as only single orthologs were found in Ixodes. Of the remaining Tetranychus duplications, only two were found duplicated in the Ixodes genome: gro and kek-3. Our findings suggest that gene duplication and losses within signal transduction pathways are prominent features of the Tetranychus genome.
Discussion
We have completed a genomic survey in the two-spotted spider mite T. urticae to determine the degree of conservation of the genes involved in several major signal transduction pathways in arthropods. We used a sequence homology-based approach to identify genes as present or absent in the Tetranychus genome. In specific cases, our approach identifies genes missing in Drosophila but present in other arthropods but does not identify genes specific to Tetranychus. Additionally, as with all genome sequencing projects, gene absences should be treated as putative due to the incomplete nature of current genome sequencing, assembly methods, and limitations using sequence homology to identify orthology. However, genuine gene absences in Tetranychus may arise from several scenarios. The first being the gene is derived only within Drosophila and/or higher insects. The second being the gene is present in other chelicerates but was subsequently lost in Tetranychus. We have identified genes that fall into both categories.
While it is understood that signal transduction pathways are conserved throughout the Metazoa, explorations of how much variation characterizes signal transduction pathways in arthropods that group more phylogenetically basal has not been previously explored. Conservation of Drosophila signal transduction pathway orthologs in insects has been found to range from 82 % in Acyrthosiphon (109/124) (Shigenobu et al. 2010) to 99 % in Apis (99/100) (Dearden et al. 2006). This conservation is higher than the 66 % conservation we report here for Tetranychus (195/294). Although the sample size is small, these datasets suggest that as evolutionary distance from Drosophila increases, gene conservation decreases, with gene conservation following a phylogenetic trend. The observed loss of conservation is not due to an increase in the sample size, as limiting the comparison to only the 62 genes shared among the three surveys follows a similar trend. We expect that analyses of the recently sequenced genomes from the crustacean Daphnia pulex and myriapod Strigamia maritima will provide support for this trend.
While many of the gene absences appear to be specific to the acarid chelicerate lineage as half the genes absent from Tetranychus are also absent from the Ixodes genome (16/33) (Supplemental Table 3), Tetranychus contains unexpected gene losses in every pathway we investigated. In several cases, these absences are incongruous with finding the core pathway conserved. For example, a fifth of all absences are attributed to ligands. For JAK/STAT, FGF, and Sevenless, these pathways are left without canonical ligands. Several other absences are known to be essential to proper signal reception (e.g., breathless), processing (e.g., dispatched), or transduction (e.g., Axin). Functional and genomic data provide insight into some, but not all, of the putative gene absences. In general, absent genes fall into two categories: those with partially redundant function in Drosophila (e.g., TACE) and those believed derived or not essential for signal transduction (e.g., deltex, mastermind, hairless, and E(spl)-C homologs). Nonetheless, many missing genes are known to be essential for proper signaling in Drosophila. While no functional data exists for these pathways in Tetranychus, observations of developmental stage and feeding RNA-seq data confirm that core pathway components are expressed. This suggests that the pathways function in the absence of what we assume to be essential components. This could be a consequence of the co-option of novel components into the pathway. Based on the number of protein coding genes predicted in the Tetranychus genome (~16,000, compared to the ~14,000 in Drosophila), it is possible for novel genes to act in these signaling pathways that are unidentifiable by sequence homology alone. Further analyses using structural homology-based searches may be of value in the identification of additional pathway components.
Also of particular interest is the absence of the Wnt signaling ligand wg from the Tetranychus genome. In Drosophila, wg functions in multiple developmental events including limb development (Cohen et al. 1993; Simcox et al. 1989), midgut morphogenesis (Mathies et al. 1994), and segmentation where it acts as a segment polarity gene (Nüsslein-Volhard et al. 1984). Loss of wg could therefore have dramatic consequences on developmental patterning and may be reflected in the reduced posterior segmentation of Tetranychus. However, functional analyses of wg from several insects suggest that unlike the segment polarity gene engrailed, which has a conserved function throughout Arthropoda, the role of wg within insects is evolving despite retaining a segmental expression pattern (Kraft and Jäckle 1994; Nagy and Carroll 1994). For example, RNAi depletion of wg in several insects that form posterior segments from a growth zone have shown it to have little to no effect on posterior segmentation (Bolognesi et al. 2008; Miyawaki et al. 2004; Ober and Jockusch 2006). In these arthropods, another Wnt ligand, Wnt8, of which a single copy is found in Tetranychus, appears to be important for segmentation. Functional data from Tribolium and the spider Achaearanea tepidariorum have shown Wnt8 to be involved in establishing and maintaining segments as RNAi-treated samples lack a growth zone and posterior segments (Bolognesi et al. 2008; McGregor et al. 2008). While more diverse arthropod taxa await a functional analysis of Wnt8, it remains to be determined if this is merely a matter of functional convergence or if it is representative of an ancestral function in posterior segmentation. However, if Wnt8 is involved in posterior segmentation in more phylogenetically basal arthropods, it is expected that changes in its regulation may contribute to the reduced posterior segmentation in Tetranychus.
In contrast with the number of genes absent, we also found Tetranychus signaling pathways to be characterized by unique duplications of paralogs within a given gene family. In general, gene duplication has played a prominent role during the evolution of signal transduction pathways. Many components from the signal transduction pathways present in extant arthropods are the result of ancestral duplications (e.g., the 12 arthropod Wnt ligand subfamilies (Janssen et al. 2010)). There are also cases of lineage-specific duplications in arthropod signaling pathways (Shigenobu et al. 2010; Van der Zee et al. 2008). Both Tribolium and Drosophila contain lineage-specific duplications of the TGF-β ligand glass bottom boat. The 11 and 4 members of the E(spl)-C from Drosophila and Apis, respectively, are believed to have arisen from an ancestral pair of genes found conserved in Anopheles (Dearden and Duncan 2010; Schlatter and Maier 2005). Eighteen signal transduction pathway genes are believed to have arose from lineage-specific duplications in Acyrthosiphon (Shigenobu et al. 2010). However, Tetranychus contains an increased number of unique genes present in multiple copies when directly compared against duplications identified from surveys in Acyrthosiphon and Apis. In Tetranychus, 30 genes are present with at least two copies. An additional six genes are present with three copies, and three genes are present with four copies (Supplemental Table 1). For example, receptors from each pathway are observed to have undergone duplications, and several pathways have multiple receptors duplicated. Likewise, duplications can be found for ligand modifiers, transducers, and transcription factors in each pathway. Similar, albeit larger in number, expansions in plant defense and pesticide resistance genes have occurred in Tetranychus (Grbić et al. 2011). In addition, the unusual ratio of ligand to receptors and modifiers may function in pathway specificity. As signaling pathways rely on these proteins for specificity, gene duplication may provide a substrate for the expansion or refinement of pathway specificity through differences in ligand-receptor kinetics, combinatorial interactions, and novel targets of activity.
Conclusion
Our data provide additional evidence for the labile nature of arthropod signal transduction pathways and complement previous surveys by showing that gene loss and duplication are common themes during arthropod evolution. At the same time, our data is in contrast with the general view that the conservation of complete signal transduction pathways is fundamental to cellular and developmental processes in metazoans (Carroll et al. 2005; Pires-daSilva and Sommer 2003; Wilkins 2002). The lack of overall conservation of signal transduction pathway components and the amount of lineage-specific duplications suggests that novel genes may participate in these pathways. These components may not share sequence identity, instead sharing structural homologies, and are likely to be discovered as emerging arthropod models are better characterized at the developmental genetic level.
Supplementary Material
Acknowledgments
We thank Matt Stansbury and two anonymous reviewers for careful reading and helpful comments on earlier drafts. This research was carried out while R.M.P. and P.C.E was supported through NSF-IGERT grant DGE-0114420. R.M.P. was also supported through NIH Training Grant T32GM008659.
Footnotes
Conflict of interest The authors declare that there are no conflicts of interest in this report.
Electronic supplementary material The online version of this article (doi:10.1007/s00427-014-0479-7) contains supplementary material, which is available to authorized users.
Contributor Information
Ryan M. Pace, Department of Molecular and Cellular Biology, University of Arizona, Tucson, AZ 85721, USA
P. Cole Eskridge, Graduate Interdisciplinary Program in Entomology and Insect Science, University of Arizona, Tucson, AZ 85721, USA.
Miodrag Grbić, Department of Biology, University of Western Ontario, London, Ontario N6A 5B7, Canada; Instituto de Ciencias de la Vid y del Vino CSIC, Universidad de la Rioja, Logroño 26006, Spain.
Lisa M. Nagy, Department of Molecular and Cellular Biology, University of Arizona, Tucson, AZ 85721, USA
References
- Alexandre C, Jacinto A, Ingham PW (1996) Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev 10:2003–2013 [DOI] [PubMed] [Google Scholar]
- Alves G, Limbourg-Bouchon B, Tricoire H, Brissard-Zahraoui J, Lamour-Isnard C, Busson D (1998) Modulation of Hedgehog target gene expression by the Fused serine-threonine kinase in wing imaginal discs. Mech Develop 78:17–31 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramírez-Weber FA, Laget MP, Schwartz C, Kornberg TB (1997) Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89:1043–1053 [DOI] [PubMed] [Google Scholar]
- Bänziger C, Soldini D, Schütt C, Zipperlen P, Hausmann G, Basler K (2006) Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125:509–522 [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M (2006) Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125:523–533 [DOI] [PubMed] [Google Scholar]
- Behura SK, Haugen M, Flannery E, Sarro J, Tessier CR, Severson DW, Duman-Scheel M (2011) Comparative genomic analysis of Drosophila melanogaster and vector mosquito developmental genes. PLoS ONE 6:e21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi R, Farzana L, Fischer TD, Brown SJ (2008) Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr Biol 18:1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T (1999) The Interactive Fly: gene networks, development and the Internet. Trends Genet 15:333–334 [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A (2000) A novel proteolytic cleavage involved in Notch signaling—the role of the disintegrin-metalloprotease TACE. Mol Cell 5:10–10 [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K (1999) Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99:803–815 [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, and Weatherbee SD (2005). From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell, London [Google Scholar]
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552 [DOI] [PubMed] [Google Scholar]
- Chesebro JE, Pueyo JI, Couso JP (2013) Interplay between a Wnt-dependent organiser and the Notch segmentation clock regulates posterior development in Periplaneta americana. Biology Open 2: 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching W, Hang HC, Nusse R (2008) Lipid-independent secretion of a Drosophila Wnt protein. J Biol Chem 283:17092–17098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman AD, Akam M (2008) The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair-rule gene homologues. Dev Biol 319:160–169 [DOI] [PubMed] [Google Scholar]
- Cohen B, Simcox AA, Cohen SM (1993) Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development 117: 597–608 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr (1997) STATs and gene regulation. Science 277:1630–1635 [DOI] [PubMed] [Google Scholar]
- Dearden PK, Duncan EJ (2010) Evolution of a genomic regulatory domain: the role of gene co-option and gene duplication in the Enhancer of split complex. Genome Res 20:917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearden PK, Grbić M, Donly C (2003) Vasa expression and germ-cell specification in the spider mite Tetranychus urticae. Dev Genes Evol 212:599–603 [DOI] [PubMed] [Google Scholar]
- Dearden PK, Wilson MJ, Sablan L, Osborne PW, Havler M, McNaughton E, Kimura K, Milshina NV, Hasselmann M, Gempe T et al. (2006) Patterns of conservation and change in honey bee developmental genes. Genome Res 16:1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquie O (2006) A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 314:1595–1598 [DOI] [PubMed] [Google Scholar]
- Farzana L, Brown SJ (2008) Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev Genes Evol 218: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bellido A, de Celis JF (2009) The complex tale of the achaetescute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics 182: 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E (2009) Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol 9:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, Osborne EJ, Dermauw W, Ngoc PCT, Ortego F et al. (2011) The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479:487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biol 59:307–321 [DOI] [PubMed] [Google Scholar]
- Herr P, Basler K (2012) Developmental biology. Dev Biol 361:392–402 [DOI] [PubMed] [Google Scholar]
- Holley SA, Neul JL, Attisano L, Wrana JL, Sasai Y, O’Connor MB, De Robertis EM, Ferguson EL (1996) The Xenopus dorsalizing factor noggin ventralizes Drosophila embryos by preventing DPP from activating its receptor. Cell 86:11–11 [DOI] [PubMed] [Google Scholar]
- Janssen R, Brown SJ, Le Gouar M, Pechmann M, Poulin F, Bolognesi R, Schwager EE, Hopfen C, Colbourne JK, Budd GE et al. (2010) Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol Biol 10:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khila A, Grbić M (2007) Gene silencing in the spider mite Tetranychus urticae: dsRNA and siRNA parental silencing of the Distal-less gene. Dev Genes Evol 217:241–251 [DOI] [PubMed] [Google Scholar]
- Kraft R, Jäckle H (1994) Drosophila mode of metamerization in the embryogenesis of the lepidopteran insect Manduca sexta. Pnas 91(14):6634–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer H, Cagan RL, Zipursky SL (1991) Interaction of bride of Sevenless membrane-bound ligand and the Sevenless tyrosine-kinase receptor. Nature 352:207–212 [DOI] [PubMed] [Google Scholar]
- Lieber T (2002) kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev 16:209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D (2006) Hairless: the ignored antagonist of the Notch signalling pathway. Hereditas 143:212–221 [DOI] [PubMed] [Google Scholar]
- Massagué J, Chen Y-G (2000) Controlling TGF-β signaling. Genes Dev 14:627–644 [PubMed] [Google Scholar]
- Mathies LD, Kerridge S, Scott MP (1994) Role of the teashirt gene in Drosophila midgut morphogenesis: secreted proteins mediate the action of homeotic genes. Development 120:2799–2809 [DOI] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Feitosa NM, Kruck S, Aranda M, Damen WGM (2008) Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Curr Biol 18:1619–1623 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Mito T, Sarashina I, Zhang H, Shinmyo Y, Ohuchi H, Noji S (2004) Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech Develop 121: 119–130 [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R (2000) A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell 5:197–206 [DOI] [PubMed] [Google Scholar]
- Nagy LM, Carroll S (1994) Conservation of wingless patterning functions in the short-germ embryos of Tribolium castaneum. Nature 367(6462):460–463 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E, Kluding H (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. 1. Zygotic loci on the 2nd chromosome. Wilhelm Rouxs Arch Dev Biol 193:267–282 [DOI] [PubMed] [Google Scholar]
- Ober KA, Jockusch EL (2006) The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol 294:391–405 [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O (1997) Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91: 639–648 [DOI] [PubMed] [Google Scholar]
- Pan DJ, Rubin GM (1997) Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90:271–280 [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ (2003) The evolution of signalling pathways in animal development. Nat Rev Genet 4:39–49 [DOI] [PubMed] [Google Scholar]
- Pisani D, Poling LL, Lyons-Weiler M, Hedges S (2004) The colonization of land by animals: molecular phylogeny and divergence times among arthropods. BMC Biol 2:1. doi: 10.1186/1741-7007-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo JI, Lanfear R, Couso JP (2008) Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc Natl Acad Sci 105:16614–16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA (2004) The JAK/STAT signaling pathway. J Cell Sci 117:1281–1283 [DOI] [PubMed] [Google Scholar]
- Rooke J, Pan D, Xu T, Rubin GM (1996) KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science 273:1227–1231 [DOI] [PubMed] [Google Scholar]
- Schlatter R, Maier D (2005) The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee. BMC Evol Biol 5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppmeier M, Damen WGM (2005) Suppressor of Hairless and Presenilin phenotypes imply involvement of canonical Notch-signalling in segmentation of the spider Cupiennius salei. Dev Biol 280:211–224 [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Dearden PK, Bickel RD, Brisson JA, Butts T, Chang C-C, Christiaens O, Davis GK, Duncan EJ, Ferrier DEK et al. (2010) Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: frequent lineage-specific duplications and losses of developmental genes. Insect Mol Biol 19(Suppl 2):47–62 [DOI] [PubMed] [Google Scholar]
- Simcox AA, Roberts IJ, Hersperger E, Gribbin MC, Shearn A, Whittle JR (1989) Imaginal discs can be recovered from cultured embryos mutant for the segment-polarity genes engrailed, naked and patched but not from wingless. Development 107:715–722 [DOI] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP (1997) Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell 90:235–245 [DOI] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P, FlyBase Consortium (2014) FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res 42:D780–D788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck L, Billiau K, Abeel T, Rouze P, Van de Peer Y (2012) ORCAE: online resource for community annotation of eukaryotes. Nat Meth 9:1041–1041 [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Schoppmeier M, Damen WGM (2003) Involvement of Notch and Delta genes in spider segmentation. Nature 423:863–865 [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biol 56:564–577 [DOI] [PubMed] [Google Scholar]
- Van der Zee M, Fonseca RN, Roth S (2008) TGFβ signaling in Tribolium: vertebrate-like components in a beetle. Dev Genes Evol 218:203–213 [DOI] [PubMed] [Google Scholar]
- Wilkins AS (2002) The evolution of developmental pathways. [Google Scholar]
- Sinauer MA Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, Nusse R (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423:448–452 [DOI] [PubMed] [Google Scholar]
- Zeidler MP, Bach EA, Perrimon N (2000) The roles of the Drosophila JAK/STAT pathway. Oncogene 19:2598–2606 [DOI] [PubMed] [Google Scholar]
- Zhai L (2004) Drosophila Wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts, a process that requires porcupine. J Biol Chem 279:33220–33227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.