Abstract
Background
The carcinogenesis of hepatocellular carcinoma is complicated, and genetic factor may have the role in the malignant transformation of liver cells. IL-10 gene polymorphisms have been investigated for their potential roles in hepatocellular carcinoma This study aimed to investigate the relationship between polymorphisms of IL-10 (-1082 A/G, -819 T/C, -592 A/C), and hepatocellular carcinoma by performing a meta-analysis with eligible individual studies.
Methods
This study followed the PRISMA 2020 Checklist. Relevant studies were searched in health-related databases. The Newcastle-Ottawa Scale criteria were used to evaluate the studies quality. Pooled odds ratio (OR) and its 95% confidence interval (CI) were used to determine the strength of association between each polymorphism and hepatocellular carcinoma using five genetic models. Stratification was done by ethnic groups. Trial sequential analysis (TSA) was performed to determine the required information size.
Results
Fifteen case-control studies (n = 8182) were identified. Overall, the heterozygous model showed a marginal significant association only between IL-10 (-1082 A/G) and hepatocellular carcinoma risk (OR: 0.82, 95% CI: 0.67-1.00, 9 studies). On stratification, IL-10 (-1082 A/G) was significantly associated with hepatocellular carcinoma risk in the non-Asian population under dominant (OR: 0.62, 95% CI: 0.45–0.86, 4 studies), heterozygous (OR: 0.60, 95% CI: 0.43–0.85) and allelic models (OR: 0.79, 95% CI: 0.64–0.99). IL-10 (-819 T/C) was significantly associated with hepatocellular carcinoma risk only among non-Asians under the dominant (OR: 1.47, 95% CI: 1.02–2.13, 8 studies), recessive (OR: 1.99, 95% CI: 1.03–3.86, and homozygous models (OR: 2.18, 95% CI: 1.13–4.23). For IL-10 (-592 A/C) with 11 studies, there was no significant association with hepatocellular carcinoma in all five genetic models (P values > 0.5). TSA plots indicated that the information size for firm evidence of effect was sufficient only for the analysis of IL-10 (-592 A/C), but not for the − 1082 A/G or -819 T/C.
Conclusions
Findings suggest that IL-10 (-1082 A/G and − 819 T/C) polymorphisms are associated with hepatocellular carcinoma in ethnic-specific manner. However, this evidence is not conclusive because the sample size was insufficient. IL-10 (-592 A/C) polymorphism was not associated with hepatocellular carcinoma albeit with sufficient information size. Future well-designed large case-control studies on IL-10 (-1082 A/G and − 819 T/C) with different ethnicities are recommended.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-023-11323-1.
Keywords: IL-10 gene polymorphisms, Hepatocellular carcinoma, Genetic association, meta-analysis, Trial sequential analysis
Background
Hepatocellular carcinoma is the most common primary liver cancer, accounting 70–80% of total primary liver cancers [1]. Worldwide, it is the sixth most common type of cancer [2], and the third leading cause of cancer related deaths in 2020 [3, 4], albeit with variation in sex and geographic distribution. According to the GLOBOCAN estimate in 2020, the age-standardised incidences were highest in Eastern Asia, followed by South-Eastern Asia and Northern Africa [3], with male predominance in most countries [5–8].
Due to nonspecific symptoms at the early stages, the mortality of hepatocellular carcinoma is very high [9], and the mean survival time for end-stage hepatocellular carcinoma patients is around four to six months [10]. Numerous studies reported risk factors for the development of hepatocellular carcinoma, including chronic hepatitis B virus (HBV) and hepatitis C (HCV) infections [11], aflatoxin-contaminated food consumption [12], and alcohol consumption [4, 11] As a matter of fact, the carcinogenesis of hepatocellular carcinoma is complicated, and both epigenetic and genetic factors may have their roles in the malignant transformation of liver cells [10]. Since hepatocellular carcinoma is attributed to prolonged inflammation of liver cells, inflammatory markers like cytokines may have the roles in hepatocellular carcinoma development [13].
Among the genetic factors, Interleukin-10 (IL-10) is one of the anti-inflammatory cytokines [14] as well as a multifunctional cytokine, which can inhibit development of tumour and disease progression. Studies have reported that a lack of IL-10 may stimulate the secretion of pro-inflammatory cytokines that inhibit anti-tumor immune responses and enhance growth of tumor [15–17]. However, the exact mechanisms of these polymorphism in cancer development and growth is not fully understood [18]. Moreover, polymorphisms of various genes control and alter the cytokines production (IL-10 in this case), and individual variations exit [19]. Hence, it is crucial to understand the role of particular single nucleotide polymorphisms (SNPs). Although chronic HBV and HCV infections are the common risk factors of hepatocellular carcinoma, only a few chronic cases with HBV and HCV develop hepatocellular carcinoma later in their lives [9]. Therefore, it has hypothesized that host genetic factors may play a part in malignant transformation of liver cells [10]. Understanding of IL-10 genetic polymorphisms may help to estimate the influence of genetic alteration on the development of hepatocellular carcinoma.
There is a surge of individual studies that investigated the roles of IL-10 in patients with hepatocellular carcinoma. These studies varied in racial decent of participants, sample sizes, and the quality of study design, and these could contribute to heterogenous findings. Meta-analysis is a method that combines results from data collected from all eligible studies. There are published meta-analyses that assessed IL-10 on hepatocellular carcinoma [20, 21]. However, these published reviews did not provide evidence on adequate information size, and hence the results were inconclusive. As(single nucleotide polymorphisms (SNPs) in the cytokine genes are known to affect cytokine production levels, we focused on three SNPs of IL-10 (-1082 A/G, -819 T/C, -592 A/C) in this study. Taken together, the objective was to investigate the relationship between polymorphisms of IL-10 (-1082 A/G, -819 T/C, -592 A/C), and hepatocellular carcinoma by performing a meta-analysis.
Methods
We conducted the current study in adherence to the PRISMA 2020 checklist for reporting our meta-analysis (Additional File 1). A protocol of this study was approved by the Ethics Review Committee of the International Medical University in Malaysia (ID: BMS I/2021(10)). This study only used data from published studies. The need for consent from participants was waived by the Ethics Review Committee of the International Medical University in Malaysia.
Search strategy
Relevant studies were searched in electronic databases of PubMed, Ovid Medline, Cochrane library, EBSCOHOST, Science Direct, Latin American and Caribbean Health Sciences Literature (LILACS), and Google scholar. Keywords and MeSH terms were used with Boolean operators: [“Interleukin-10” or “IL-10”] AND [“polymorphism” or “gene polymorphisms”] AND [“hepatocellular carcinoma” or “liver cancer”]. To capture any additional studies, we performed a snowball method of manual cross-referencing of the retrieved studies and relevant systematic reviews. The search was restricted to studies published in English until June 2022. Search strategies are provided in Additional File 2.
Inclusion and exclusion criteria
Human studies that assessed hepatocellular carcinoma were included, if they.
-
i)
assessed IL-10 gene polymorphisms, − 1082 A/G (rs1800870; rs 1,800,896), -819 T/C (rs1800871; rs 3,021,097), and/or -592 A/C (rs1800872);
-
ii)
conducted case-control or nested case-control studies, irrespective of the method of DNA analysis;
-
iii)
compared hepatocellular carcinoma patients with the controls (healthy controls or non-hepatocellular carcinoma participants);
-
iv)
provided sufficient information to extract genotype frequencies in both cases and controls; and.
-
v)
measured the outcome with odds ratio (OR) along with 95% confidence intervals (CI) (at least, provided enough data to estimate these).
We assessed whether the distribution of genotypes in the control group of the studies included was consistent with Hardy-Weinberg equilibrium (HWE) [22].
Hepatocellular carcinoma in this analysis was as defined in the primary studies. When more than one publication used the same participants, a publication with more comprehensive information was considered.
Studies that did not fit to the inclusion criteria (e.g., not case-control study, not genetic association studies, genetic studies with no genotype frequency) were not considered. Studies that assessed treatment response, disease severity, drug efficacy, and pre-clinical studies were excluded.
Data extraction
The two investigators (TYM and HHA) independently extracted information from each study using a piloted data extraction sheet. Information collected were: first author, publication year, study country, number of cases/controls, age group, male%, polymorphism frequencies in cases/controls, the racial descent (Asian or non-Asian), and minor allele frequency (MAF) in the controls, and HWE status (if MAF and HWE were not provided, we calculated it). Any discrepancy between the two investigators was resolved by discussion with the third investigator (CN).
Study quality assessment
The two investigators (TYM and HHA) independently evaluated the methodological quality of eligible studies using the Newcastle–Ottawa scale (NOS). The NOS checklist covers three main domains (selection, exposure, comparability) in eight items, and each item was awarded 1 or 2 stars in maximum for high quality, and a final score obtained was between 0 and 9 stars [23]. We considered studies with ≥ 7 stars as high quality. Any variations between the two investigators were settled through a discussion with the third investigator (WST).
Statistical analysis
The genotype frequencies in the control were checked for consistency using the HWE, and the exact test for goodness-of-fit was applied (p > 0.05) [24]. Described elsewhere [25], for individual studies, the strength of the association between IL-10 (-1082 A/G as an example) and hepatocellular carcinoma was estimated using OR and its 95%CI. For pooling of the estimates across studies, we calculated summary ORs and corresponding 95% Cis. We used random-effect model (Der Simonian and Laird method), accounting statistical heterogeneity of the studies. Otherwise, fixed-effect model (the Mental-Haenszel method) was used. Heterogeneity was evaluated with the I2 statistics (the percentage of total variation across studies), reflecting the heterogeneity rather than chance. I2 values greater than 50% is regarded as a substantial heterogeneity [26]. We calculated the pooled ORs and its 95% CIs under five genetic models (i.e., dominant, recessive, homozygous, heterozygous, and allelic models). The generic formula used for these calculations [27] is presented in Additional File 3.
To detect the sources of heterogeneity, we planned to perform meta-regression with covariates of personal factors (e.g. age, gender), and environmental factors (e.g. alcohol consumption, smoking, HBsAg status). Due to paucity or inconsistent reporting of these data, it was not able to do a meta-regression.
In small studies, a statistically significant finding would be actually a false-positive report probability (FPRP) [28]. We performed the FPRP test with the use of a pre-set FPRP < 0.2 and assigned prior probabilities of 0.25, 0.1, 0.01 or 0,001 to examine an OR of 1.5 (or its reciprocal 0.67 = 1/1.5 for ORs less than 1) of associated with the hepatocellular carcinoma risk. To evaluate whether an association is “noteworthy”, we used a FPRP cut-off value of 0.2. Statistical power and FPRP were computed by the Excel spreadsheet provided elsewhere [28].
To investigate the stability of results, we performed a sensitivity analysis with leave-one-out meta-analysis by removal of one study at a time. We planned to assess the publication bias by visual inspection of funnel plots [29]. This was done only for the − 592 A/C, where a minimum of 10 studies were available for this assessment.
Trial sequential analysis
Trial sequential analysis (TSA), an approach that adjusts for random error risk, was done for estimation of the required information size [30]. For dichotomous outcomes in this case, we calculated the information size adjusted for heterogeneity (diversity, D²) between trials (studies in this case) using the parameters described elsewhere [31]. Proportion of events in the control group estimated from the included studies (overall mean value), anticipated intervention effect (relative risk reduction) of 15%, alpha of 5% (one main outcome), and beta of 20% [32, 33]. It is classified as ‘potentially spurious evidence of effect’ (if the cumulative Z-curve did not cross the monitoring boundaries), or as ‘firm evidence of effect’ (if the cumulative Z-curve crossed the monitoring boundaries) [31].
Meta-analysis was done with Stata 16 (StataCorp TX), while TSA was with TSA version 0.9 beta (Copenhagen Trial Unit, Centre for Clinical Intervention Research, Copenhagen).
Results
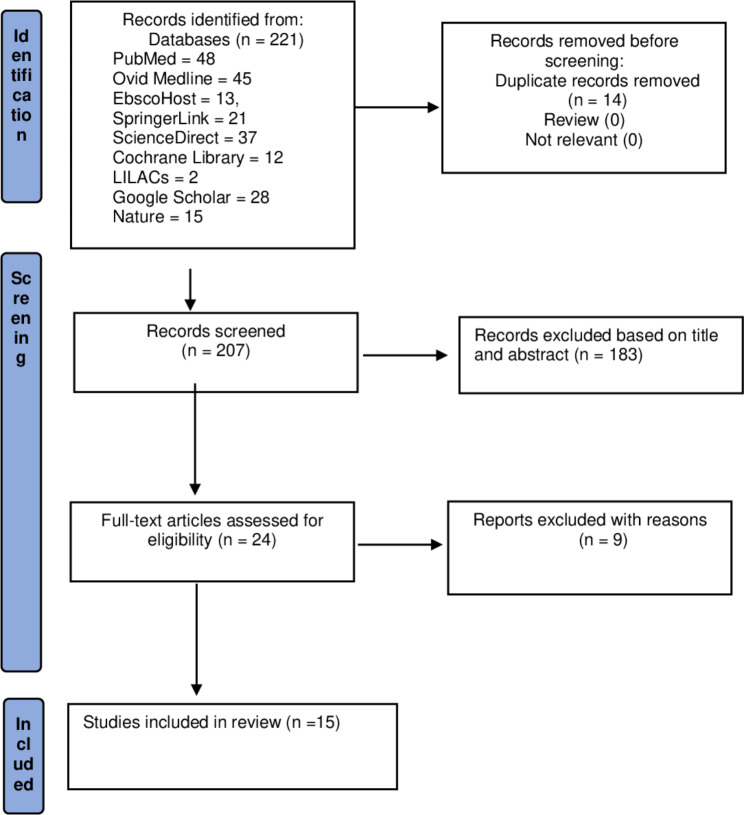
Figure 1 shows a study selection process. Initially, 221 studies were yielded from the database searches. After removal of 14 duplicates, 207 studies were further screened for the title and abstract. Twenty-four full-text studies were evaluated for eligibility, and finally, a total of 15 studies were identified for this meta-analysis [32, 34–48]. The reasons for nine excluded studies were presented in Additional File 4.
Fig. 1.
Study selection flowchart
Study characteristics
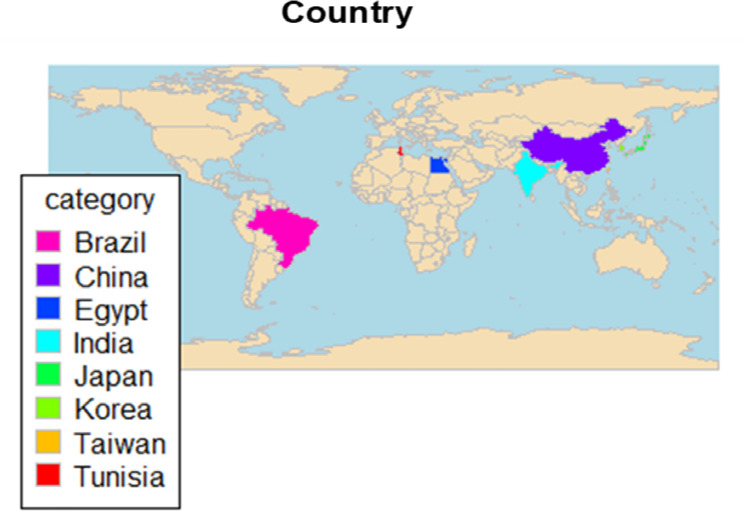
Table 1. presents the main characteristics of the included studies. A total of 15 studies involving 8182 participants (2923 cases and 5259 controls) were identified. These studies were conducted across eight countries most frequently in China [37, 40, 42, 47, 48] (Fig. 2). Study samples varied from 100 [35] to 1504 participants [37], and the years of publication spanned from 2003 [40, 45] to 2020 [36, 39, 43]. With regard to ethnic groups, ten studies (67%) were done with the Asians population [36, 37, 40–42, 44–48], while five studies (33%) were with the non-Asian population [34, 35, 38, 39, 43]. Ten studies were hospital-based studies [34, 35, 37–40, 42, 43, 45–48], while 5 studies were done on patients at the outpatient clinic/special clinic [35, 36, 39, 43, 44]. Eight studies (53%) examined more than one SNP [32, 34, 40–42, 44, 45, 45], while nine [32, 34–36, 38–42, 45], eight [34, 36, 38, 40–44, 47], and 11 studies [32, 36, 37, 40–42, 44–48] examined the single SNP − 1082 A/G, -819 T/C, and − 592 A/C, respectively. Several genotyping methods were used in these studies, and the most frequently used genotyping methods were AS-PCR, TaqMan, and PCR-RFLP.
Table 1.
Characteristics of study
| Author, yr [ref] | Yr of publication | Country | Setting | Ethnic group | Samples (case/ control) | Age in yr1 | Male%1 | Underlying cause | Polymorphism | Genotyping method | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Aroucha 2016 [34] |
2016 | Brazil | Hospital | Non-Asian | 108/280 | 62.7 ± 8.7 | 68.5 | HCV related HCC | -1082 A/G.-819 C/T, -592 A/C | TaqMan | Yes |
| Bahgat 2015 [35] | 2015 | Egypt | Community clinic | Non-Asian | 50/50 | 52.30 ± 4.41 | 82 | HCV related HCC | -1082 A/G | RT-PCR | Yes |
| Barooh 2020 [36] | 2020 | India | OPD | Asian | 60/306 | 54.8 ± 9.1 | 49 | HCV related HCC | -1082 A/G.-819 C/T, -592 A/C | PCR-RFLP | No |
| Bei 2014 [37] | 2014 | China | Hospital | Asian | 720/784 | 48.65 ± 11.03 | 86 | Newly diagnosed HCC | -592 A/C | TaqMan | Yes |
| Bouzgarrou 2009 [38] | 2009 | Tunisia | Hospital | Non-Asian | 58/145 | 61.6 ± 9.8 | 34.5 | HCV related HCC | -1082 A/G | AS- PCR- | Yes |
| El-Baky 2020 [39] | 2020 | Egypt | Hospital | Non-Asian | 54/92 | 64.16 ± 12.1 | 90.7 | HCV complicated with HCC | -1082 A/G | TaqMan | No |
|
Heneghan 2003 [40] |
2003 | China | Hospital | Asian | 98/175 | 55 year (range: 14–77 year) | 92.9 | HBV related HCC | -1082 A/G.-819 C/T, -592 A/C | PCR-SSCP | Yes |
| Migita 2003 [41] | 2003 | Japan | OPD | Asian | 48/188 | 62.5 ± 8.9 | 81.25 | HBV related HCC | -1082 A/G.-819 C/T, -592 A/C | PCR-SSP | No |
| Peng 2016 [42] | 2016 | China | Hospital | Asian | 173/182 | 56.32 ± 7.55 | 36.42 | HBV related HCC | -1082 A/G.-819 C/T, -592 A/C | PCR-RFLP | Yes |
| Saleh 2020 [43] | 2020 | Egypt | HCC clinic | Non-Asian | 73/85 | 56.21 ± 4.62 | 58.9 | HCV related HCC | -819 C/T | RT-PCR | No |
| Saxena 2014 [44] | 2014 | India | OPD | Asian | 59/331 | 55.31 ± 12.67 | 18.66 | HBV related HCC | -819 C/T, -592 A/C | AS-PCR | No |
| Shin 2003 [45] | 2003 | Korea | Hospital | Asian | 230/792 | Only cutoff age | HBV related HCC | -1082 A/G, -592 A/C | MAPA | No | |
| Tseng 2006 [46] | 2006 | Taiwan | Hospital | Asian | 208/528 | 55 (23–85) | HBV related HCC | -592 A/C | PCR/RFLP | Yes | |
| Wang 2019 [47] | 2019 | China | Hospital | Asian | 554/612 (277/306) | ≤ 35- >50 | 84.8 | HBV-related HCC2 | -819 C/T, -592 A/C | PCR-based | No |
| Zhou 2017 [48] | 2017 | China | Hospital | Asian | 430/709 | < 55-≥55 | 88.1 | HBV-related HCC | -592 A/C | MassARRAY | Yes |
1: values for the cases; 2: 87%of the cases;
AS: Allele-specific; HCC: hepatocellular carcinoma; HWE: Hardy–Weinberg equilibrium: MAPA: multiplex automated primer extension analysis; OPD: Outpatient Department/Outpatient clinic; PCR: Polymerase chain reaction; PCR/RFLP: mis- matched PCR/restriction fragment length polymorphism; RT-PCR: Reverse transcription polymerase chain reaction; PCR-SSCP polymerase chain reaction-single-strand conformation polymorphism
Fig. 2.
Geographic distribution of the included studies
Frequency of individual SNP is provided in Additional File 5. Seven studies (47%) did not follow HWE [36, 39, 41, 43–45, 47]. We retained them for an initial overall analysis.
Methodological quality assessment
Based on the NOS criteria, the studies included achieved the scores between 5 and 9. Less than half of these studies (6/15, 40%) obtained the high scores (i.e., > 7.0 score) [36, 37, 40–42, 45] (Additional File 6).
Genetic model assessments
IL-10 (-1082 A/G)
Overall, IL-10 (-1082 A/G) showed a tendency toward significant relationship with hepatocellular carcinoma under heterozygous model (OR: 0.82, 95%CI:0.67-1.0, I2 = 49%, fixed effect model, 9 studies), however other four genetic models did not (All p values > 0.05) (Table 2).
Table 2.
Associations with the risk of hepatocellular carcinoma
| SNP | Number of studies included | Total participants | Genetic Model | Effect estimates, OR (95%CI); p value |
||
|---|---|---|---|---|---|---|
| -1082 A/G |
9 (5 Asian group, 4 non-Asian group) |
3089 | Overall | Asian group | Non-Asian group | |
| Dominant |
0.85 (0.70–1.03) [p:0.09] |
1.01 (0.79–1.29) [p:0.95] |
0.62 (0.45–0.86) [p:0.004] |
|||
| Recessive |
1.10 (0.79–1.54) [p:0.56] |
1.30 (0.80–2.13) [p:0.29] |
0.97 (0.62–1.52) [p:0.88] |
|||
| Heterozygous) |
0.82 (0.67–0.99) [p:0.04] |
0.97 (0.75–1.25) [p:0.65] |
0.60 (0.43–0.85) [p:0.007] |
|||
| Homozygous |
0.91 (0.64–1.30) [p:0.61] |
1.20 (0.72–1.99) [p:0.65] |
0.71 (0.43–1.17) [p:0.51] |
|||
| Allelic ( |
0.92 (0.80–1.07) [p:0.28] |
1.05 (0.86–1.29) [p:0.63] |
0.80 (0.64–0.99) [p:0.04] |
|||
| -819 T/C |
8 (6 Asian group, 2 non-Asian group) |
3332 | Overall | Asian group | Non-Asian group | |
| Dominant |
1.25 (1.05–1.48) [p:0.37] |
1.19 (0.98–1.45) [p:0.27] |
1.47 (1.02–2.13) [p:0.032] |
|||
| Recessive |
1.10 (0.81–1.50) [p:0.] |
0.96 (0.74–1.26) [p:0.] |
1.99 (1.03–3.86) [p:0.] |
|||
| Heterozygous | 1.20 (1.00-1.43) [p:0.73] | 1.19 (0.97–1.46) [p:0.49] | 1.23 (0.82–1.84) [p:0.93] | |||
| Homozygous | 1.17 (0.80–1.72) [p:0.37] | 0.97 (0.65–1.44) [p:0.12] | 2.18 (1.13–4.23) [p:0.03] | |||
| Allelic | 1.08 (0.89–1.32) [p:0.35] | 0.98 (0.81–1.19) [p:0.11] | 1.59 (0.96–2.63) [p:0.64] | |||
| -592 A/C |
11 (10 Asian group, 1 non-Asian group) |
7516 | Overall | Asian group | Non-Asian group | |
| Dominant | 0.90 (0.68–1.18) [p:0.44] |
0.86 (0.63–1.16) [p:0.32] |
1.29 (0.82–2.03) [p:0.27] |
|||
|
Recessive TT vs. CT |
0.89 (0.72–1.11) [p:0.31] |
0.86 (0.69–1.08) [p:0.2] |
1.45 (0.75–2.78) [p:0.26] |
|||
| Heterozygous | 0.96 (0.75–1.22) [p:0.31] | 0.93 (0.71–1.21) [p:0.57] |
1.21 (0.75–1.96) [p:0.43] |
|||
| Homozygous |
0.93 (0.77–1.12) [p:0.43] |
0.91 (0.75–1.10) [p:0.33] |
1.32 (0.66–2.63) [p:0.44] |
|||
| Allelic | 0.90 (0.76–1.08) [p:0.25] |
0.87 (0.72–1.05) [p:0.15] |
1.26 (0.91–1.74) [p:0.17] |
|||
Bold indicates significant at p value < 0.05. CI: Confidence interval; OR: Odds ratio:
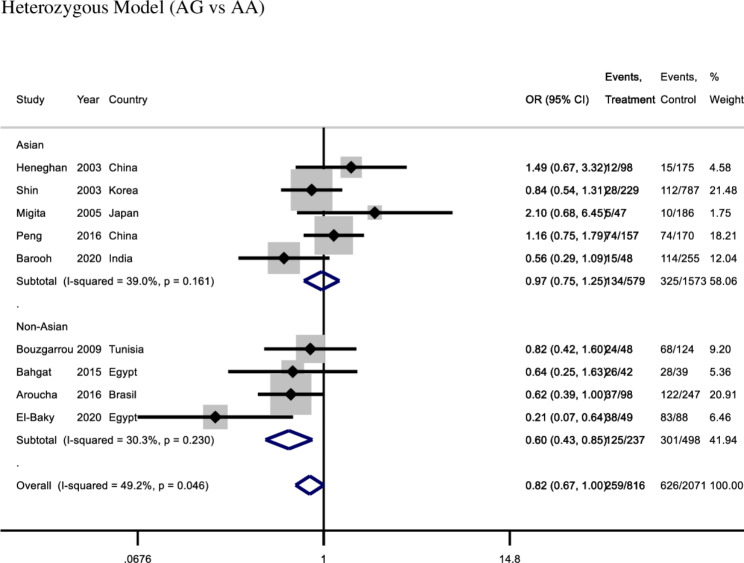
For a subgroup of non-Asian population, there was a significant association between the IL-10 (-1082 A/G) and hepatocellular carcinoma in protection using the heterozygous model (OR: 0.60, 95% CI: 0.43–0.85, I2 = 30.3%, fixed effect model, four studies). However, for the Asian population, there was no significant association between the IL-10 (-1082 A/G) and hepatocellular carcinoma in heterozygous model (OR:0.97, 95% CI: 0.75–1.25, I2 = 39%, fixed effect model, five studies) (Fig. 3). There was a significant association between the IL-10 (-1082 A/G) and hepatocellular carcinoma in protection using the dominant model (OR: 0.62, 95% CI: 0.45–0.86, I2 = 33.1%, fixed effect model) and allelic model (G vs. A) (OR: 0.79, 95% CI: 0.64–0.99, I2 = 0.0%, fixed effect model) with the non-Asian population. However, there was no significant association with hepatocellular carcinoma under the remaining two genetic models (recessive and homozygous models). For a subgroup of Asian population (5 studies), there was no significant association with hepatocellular carcinoma under any of the five genetic models (Table 2).
Fig. 3.
Forest plot of − 1082 A/ G under heterozygous model
IL-10 (-819 T/C)
Overall, Il-10 (-819 T/C) was associated with hepatocellular carcinoma susceptibility under dominant model (OR: 1.25, 95% CI:1.05–1.48, I2 = 15.7%, fixed effect model, 8 studies), and heterozygous model (OR: 1.20, 95% CI: 1.00-1.43, I2 = 0.0%, fixed effect model, 8 studies (Additional File 7). There was no significant association with hepatocellular carcinoma under other three genetic models (Table 2).
On a subgroup of the non-Asian population (two studies), There was significant association with hepatocellular carcinoma susceptibility under dominant model (OR: 1.47, 95% CI: 1.02–2.13, I2 = 0.0%, fixed effect model, two studies) and recessive model (OR:1.99, 95% CI: 1.03–3.86, I2 = 46.1%, fixed effect model), and homozygous model (OR: 2.18, 95% CI: 1.13–4.23, I2 = 36.5%, fixed effect model, 111 participants) (Table 2). However, there was no significant association with hepatocellular carcinoma under the remaining two genetic models (heterozygous and allelic models). The subgroup Asian population (six studies) had no significant association with hepatocellular carcinoma under any five genetic models (Table 2).
IL-10 (-592 A/C)
Overall, there was no association with hepatocellular carcinoma under all five genetic models. On stratification by ethnic groups (Asian in 10 studies and non-Asian in one study), the Asian groups as well as the non-Asan group showed no significant association with hepatocellular carcinoma under any five genetic models (Table 2).
FPRP test
Table 3 shows the FPRP of IL10 (-1082 A/G) and (-819 T/C) gene polymorphisms of significant association with hepatocellular carcinoma risk. At prior probability of 0.25 and 0.1 FPRP test results show that many associations of the non-Asian group remained “noteworthy”. At prior probability of 0.01 and 0.001, and statistical power to detect an OR of 1.5, none of the associations were considered “noteworthy” (FPRP < 0.2). This means at low level of prior probability of 0.001(1000: 1), none were “noteworthy” and may not be true association.
Table 3.
False positive report probability power and value
| Genetic model | OR (95%CI) |
FPRP | |||||
|---|---|---|---|---|---|---|---|
| P value | Statistical power | Prior probability | |||||
| 0.25 | 0.1 | 0.01 | 0.001 | ||||
| -1082 A/G | |||||||
| Dominant | |||||||
| Non-Asian | 0.62 (0.45–0.86) | 0.004 | 0.321 | 0.038 | 0.105 | 0.564 | 0.929 |
| Heterozygous) | |||||||
| Overall | 0.82 (0.67–0.99) | 0.039 | 0.982 | 0.106 | 0.263 | 0.797 | 0.975 |
| Non-Asian | 0.60 (0.43–0.85) | 0.004 | 0.267 | 0.043 | 0.12 | 0.6 | 0.938 |
| Allelic | |||||||
| Non-Asian | 0.80 (0.64–0.99) | 0.04 | 0.953 | 0.112 | 0.275 | 0.806 | 0.977 |
| -819 T/C | |||||||
| Dominant | |||||||
| Non-Asian | 1.47 (1.02–2.13) | 0.042 | 0.543 | 0.188 | 0.409 | 0.884 | 0.987 |
| Recessive | |||||||
| Non-Asian | 1.99 (1.03–3.86) | 0.042 | 0.202 | 0.383 | 0.651 | 0.954 | 0.995 |
| Homogenous | |||||||
| Non-Asian | 2.18 (1.13–4.23) | 0.021 | 0.134 | 0.321 | 0.587 | 0.940 | 0.994 |
FPRP: False positive report probability. Statistical power is the power to detect an odds ratio of 1.5 with the genetic variant (or, 0.67 = 1/1.5 for protective effect). The results in bold implies noteworthiness of association at 0.2 level by FPRP. Not performed FPRP test for IL10 (-592 A/C) as there was no significant association with hepatocellular carcinoma risk (See Table 2)
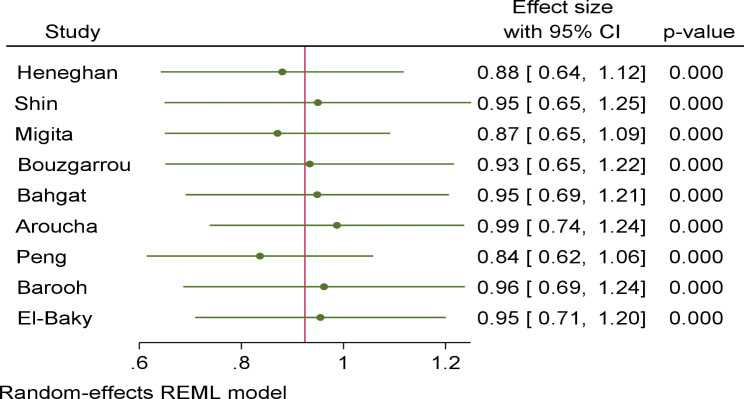
Sensitivity analysis
For robustness of the findings, we performed’ leave-one-out meta-analysis’ by sequentially removing each study. For the SNP − 1082 A/G (nine studies), the omission of any single study did not significantly change the direction of estimates (Fig. 4). This was also found for IL-10 (-819 T/C and − 592 A/C) under all five genetic models (data not shown). These implied the stability of the effect estimates.
Fig. 4.
Forest plot of leave-one-out meta-analysis in IL-10-1082 A/G under dominant
Publication bias
For the SNP (-592 A/C) (11 studies), publication bias was detected in the dominant model (p = 0.051), homozygous (p = 0.059), heterozygous (p = 0.066), and allelic (p = 0.050) models. Due to less than ten studies included, we could not investigate publication bias for IL-10 (-1082 A/G) and IL-10 ( -819 T/C).
Trial sequential analysis
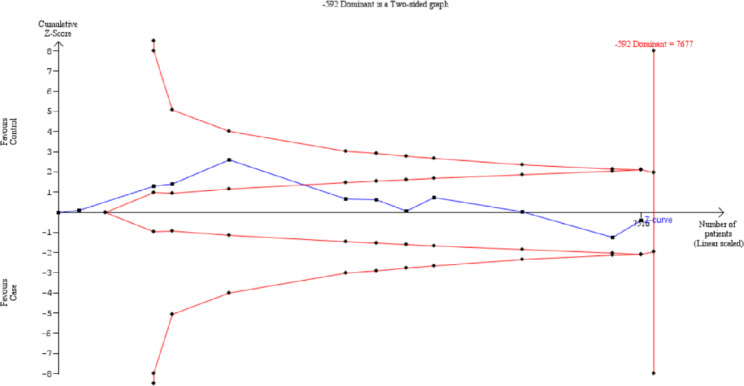
TSA approach was performed for all three SNPs using an overall 5% type I error and 80% power. For IL-10 (-592 A/C), the cumulative z-curve cut crossed the futility boundary, indicating the information size was adequate to establish a firm conclusion (Fig. 5).
Fig. 5.
Trial sequential analysis plot of IL-10 (-592 A/C)
In IL-10 -1082 A/G and − 819 T/C, the cumulative Z-curves did not pass the monitoring thresholds and was not within the futility area, indicating that the existing sample size for these SNPs were not adequate to establish a firm conclusion. This implied that the available information size in these two SNP (-1082 A/G and − 819 T/C) were not sufficient to provide firm evidence on the relationship with hepatocellular carcinoma. Therefore, further case-control studies investigating with IL-10 -1082 A/G and − 819 T/C are needed to provide a more robust conclusion.
Discussion
The candidate gene approach is increasingly attractive to distinguish susceptibility genes that may trigger the initiation and progression of various types of cancer. In the present meta-analysis, we assessed the role of three important SNPs in the IL-10 gene (-1082 A/G, -819 T/C and − 592 A/C) in the risk of hepatocellular carcinoma across eight countries.
The gene encoding IL-10 is located on chromosome 1q31-1q32, and has three common IL-10 promoter variants (-1082 A/G, -819 T/C, -592 A/C) that significantly affect the gene transcription and expression [49]. Cytokines play an important role on the pathogenesis of virus-associated hepatitis. Clearance of hepatitis viruses following acute infection is associated with a vigorous cytotoxic T-cell response, through inhibition of viral replication and gene expression by the proinflammatory and Th1 cytokines [46, 50]. IL-10 is a potent suppressor of proinflammatory, and Th1 cytokine production [50], subsequently attenuate these inflammatory responses and reduce liver injury [46].
Our findings suggest IL-10 pertinent to AA and GG genotypes in -1082 A/G would be a protective role in hepatocellular carcinoma. A published study reported that high levels of IL-10 reduced the risk of hepatocellular carcinoma by reducing hepatic inflammation, which in turn prevents the malignant transformation of liver cells [51]. The significant association with subgroup of the non-Asian population implied that even the same polymorphism, there might have different effects on individuals from different ethnic backgrounds. Ethnicity plays a role in giving the genetic diversities among two different ethnic groups, leading to different susceptibilities to hepatocellular carcinoma [52]. With regards to -819 T/C, there was significant association with hepatocellular carcinoma under dominant model in the non-Asian population. This might be due to a prominent role of TT homozygous and CT heterozygous genotypes, which were linked to hepatocellular carcinoma. Among different ethnicities, the same gene polymorphism may influence differently among various ethnic populations. There were only four studies in IL-10 (-1082 A/G) and two studies in IL-10 (-819 T/C) for the non-Asian group. This reflects that non-Asian populations do not share the common genetic basis. A large-scale study in non-Asian populations is thereby require to substantiate the current findings.
It has been established that hepatocellular carcinoma is a multifactorial process, involving genetic factors and other factors such as lifestyle and environment factors, for the carcinogenesis [12]. Therefore, variations in lifestyle factors such as alcohol consumption and diet intake among participants in these ethnic groups may also contribute to the different susceptibilities to hepatocellular carcinoma even for the same polymorphism [53].
For IL-10 (-592 A/C), no significant association in both Asians and non-Asians might be explored that IL-10 − 592 A/C had neither the causal nor inhibitory effect in hepatocellular carcinoma development in these study population. Preclinical studies reported that − 592 A/C did not play a part in altering the level of production of IL-10 [54, 55].
Comparisons with published reviews
A review including ten studies [56], assessed only allelic and dominant models, and reported no significant associations with any of these three SNPs and hepatocellular carcinoma. Another published review including 12 observational studies [21] reported a marginal association between IL-10 (-1082 A/G) and hepatocellular carcinoma for overall population, while significant association with − 592 A/C under dominant and allelic models for overall population, but no significant association with − 819 T/C under any genetic models for overall population or subgroups. IL-10 (-1082 A/G) and hepatocellular carcinoma were significantly associated in the non-Asian population under three genetic models (dominant, heterozygous, and allelic models), and IL-10 (-819 T/C) was significantly associated in the non-Asian population under three genetic models (dominant, recessive, and homozygous models), according to our review of 15 studies that evaluated five genetic models. However, TSA indicated that the significant relationships reported in our review may be spurious effects due to inadequate information (sample) size. Another review including seven case-control studies reported no significant associations between IL10 (-819 T/C) and hepatocellular carcinoma risk [20]. The published reviews did not assess adequate information size using the TSA method. This review applied TSA for the required information size, which is important to classify the effect estimates as ‘firm evidence of effect’ or ‘potentially spurious evidence of effect’ [30, 31]. Based on the TSA report, more sample size was required for the assessment of -1082 A/G and − 819 T/C in hepatocellular carcinoma risk for confirmatory evidence.
The difference in the number of included studies along with different total participants between the published reviews [20, 21, 56] and the current study may be the reason of the discrepancy in the findings. With SNPs (-819 T/C, -592 A/C) that had fewer studies than the recommended minimum limit of ten studies [26], the Zhang review [21] still performed publication bias. The current assessment, which is supported by TSA plots, as well as the previous reviews [20, 21, 56], concurred that findings should be interpreted with caution.
Study limitations
There are several limitations that should be acknowledged. First, the sample sizes in the current study were small. For example, less than ten studies were recruited in the overall meta-analyses of -1082 A/G and − 819 T/C, and only two studies included for subgroup analysis of -819 T/C. Hence, there may be a type II error, regarding the small number of studies.
It is possible for specific environmental and lifestyle factors to alter those associations between gene polymorphisms and hepatocellular carcinoma risk [21]. However, we could not assess adjusted estimates with these potential confounding variables due to a lack of data or inconsistent reporting of these data. TtThe FRPP, which is the probability of no association between a genetic variant and disease (hepatocellular carcinoma in this case) given a statistically significant results, depends on the observed p value, the prior probability that the association between the genetic variant and the disease is real, and the statistical power of the test [28,57]. In this analysis, the FPRP approach did not support the associations as noteworthy of true associations at the 0.001 level. The FPRP approach is essentially Bayesian in that it formally integrates data from direct observation of study results with other information about the likelihood of a true association [28]. TSA plots also demonstrated the issue of inadequate samples in the two SNPs (-1082 A/G and − 819 T/C). In some studies, the genotypic distribution of controls did not conform to the HWE. Nevertheless, sensitivity analysis showed the stability of the estimates. Moreover, we included only published studies in English. Hence, there might be relevant studies in other languages or non-published studies, which could contribute to selection bias. Lastly, due to the complexity of the etiology of hepatocellular carcinoma, there might be a contextual interaction between polymorphisms of IL-10 gene and other genes together with environmental/lifestyle related factors, which were beyond the scope of our study [5–60]. Taken together, the current findings are limited guidance for screening susceptible populations in the real world.
Nevertheless, the current meta-analysis has strengths. We could identify a greater number of case- control studies that could enhance the power of the current meta-analysis, and additional TSA plot. The earlier reviews did not address adequate information size using the TSA method. The present study performed TSA as the required information size is important to classify the effect estimates as ‘firm evidence of effect’ or ‘potentially spurious evidence of effect’ [30, 31].
Implications for clinical practice
Based on limited data presented in this review, we still do not know whether IL10 gene polymorphisms (-1082 A/G and − 819 T/C) increases or reduces the risk of hepatocellular carcinoma. Our analysis highlights the need for additional studies on the role of the IL10 gene in the development of hepatocellular carcinoma, and the findings should be used to inform healthcare providers considering screening of genetic risk factors. Such research could aid in identifying patients who have considerably higher risks of disease progression and may guide the development of customized treatment plans for chronic HCV infection. Since IL10 can diminishes the antiviral response, a practical approach could be the indication of earlier HCV treatment for those carrying low expression of IL-10. This would halt the inflammation induced by HCV infection and potentially inhibit the development of hepatocellular carcinoma [34].
Conclusion
The findings suggested that IL-10 -1082 A/G and − 819 T/C have some roles in associated risk of hepatocellular carcinoma in the non-Asians group. The information size for confirmatory evidence was adequate only in the IL-10 (-592 A/C) to reach a conclusion. Future well-designed case-control studies with adequate number of participants in multi-ethnic groups are recommended to substantiate the evidence on the relationship between these two polymorphisms (IL10- 1082 A/G and − 819 T/C) and hepatocellular carcinoma.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: PRISMA 2020 Checklist
Supplementary Material 2: Search strategies in databases
Supplementary Material 3: Generic formula
Supplementary Material 4: Excluded nine studies
Supplementary Material 5: Frequency of genetic polymorphisms
Supplementary Material 6: Methodological quality of studies assessed via NOS criteria
Supplementary Material 7: Forest plot of ? 819 C/T (a) dominant model (b) heterozygous model
Acknowledgements
The authors thank the patients and researchers of the primary studies. We thank our institutions for allowing us to perform this study.
List of abbreviations
- CI
Confidence interval
- FPRR
False positive report probability
- HWE
Hardy–Weinberg equilibrium
- IL-10
Interleukin-10
- MAF
Minimum Alley frequency
- NOS
Newcastle–Ottawa scale
- OR
Odds ratio
- PCR
Polymerase chain reaction
- PCR/RFLP
PCR/restriction fragment length polymorphism
- PCR-SSCP
Polymerase chain reaction-single-strand conformation polymorphism
- PRISMA
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- RT-PCR
Reverse transcription polymerase chain reaction
- SNP
Single nucleotide polymorphisms
Authors’ contributions
HHA, CN, TYM: designed; TYM, HHA: collected data; TYM, HHA, CN: Analysed; WST: assisted in analysis; HHA, CN, TYM, WST: interpreted; HHA: wrote and revised the manuscript together with CN. TYM, WST: Provided additional information; All authors approved the final version.
Funding
International Medical University Malaysia (project ID No: BMS I/2021(10).
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
A protocol of this study was approved by the Ethics Review Committee of the International Medical University in Malaysia (ID: BMS I/2021(10)). This study only used data from published studies. The need for consent from participants was waived by the Ethics Review Committee of the International Medical University in Malaysia.
Consent for publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Htar Htar Aung and Cho Naing contributed equally to this work.
References
- 1.Petrick JL, Braunlin Laversanne_M, Valery_PC, Bray_F, McGlynn_KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978–2007. Int J Cancer. 2016;139:1534–45. doi: 10.1002/ijc.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balogh J, Victor D, Asham EH, Burroughs SG, Boktour M, Saharia A, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Global Cancer Statistics. 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 Countries - Sung – 2021 - CA: a Cancer Journal for Clinicians - https://acsjournals.onlinelibrary.wiley.com/doi/10.3322/caac.21660. [DOI] [PubMed]
- 4.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, et al. Global, Regional, and National Cancer incidence, mortality, years of Life Lost, Years lived with disability, and disability-adjusted life-years for 29 Cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5(12):1749–68. doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–44. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Li H, Spitsbergen JM, Gong Z. Males develop faster and more severe hepatocellular carcinoma than females in krasV12 transgenic zebrafish. Sci Rep. 2017;7:41280. doi: 10.1038/srep41280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu I, editor. Preventive female sex factors against the development of chronic liver disease. Bentham Science Publishers; 2012.
- 8.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5 Suppl 1):5–16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Cartier V, Aube C. Diagnosis of hepatocellular carcinoma. Diagn Interv Imaging. 2014;95(7):709–19. doi: 10.1016/j.diii.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Ottaviano M, Palmieri G, Damiano V, Tortora M, Montella L. Rescue of sorafenib-pretreated advanced hepatocellular carcinoma with tamoxifen. Clin Res Trials. 2017;3(6).
- 11.Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019;70(1):151–71. doi: 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68(3):526–49. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Refolo MG, Messa C, Guerra V, Carr BI, D’Alessandro R. Inflammatory mechanisms of hepatocellular carcinoma development. Cancers. 2020;12(3):641. doi: 10.3390/cancers12030641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 15.Bijjiga E, Martino A. Interleukin 10 (IL-10) Regulatory cytokine and its clinical consequences. J Clin Cell Immunol. 2011;1:1. [Google Scholar]
- 16.Shi J, Li J, Guan H, Cai W, Bai X, Fang X, et al. Anti-fibrotic actions of interleukin-10 against hypertrophic scarring by activation of PI3K/AKT and STAT3 signaling pathways in scar-forming fibroblasts. PLoS ONE. 2014;9(5):e98228. doi: 10.1371/journal.pone.0098228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol. 2012;32(1):23–63. doi: 10.1615/CritRevImmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Z, Liu Q, Huang C, Wu M, Li G. The interleukin 10–819 C/T polymorphism and cancer risk: a HuGE review and meta-analysis of 73 studies including 15,942 cases and 22,336 controls. Omics J Integr Biol. 2013;17(4):200–14. doi: 10.1089/omi.2012.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karhukorpi J, Laitinen T, Karttunen R, Tiilikainen AS. The functionally important IL-10 promoter polymorphism (-1082 G–>A) is not a major genetic regulator in recurrent spontaneous abortions. Mol Hum Reprod. 2001;7(2):201–3. doi: 10.1093/molehr/7.2.201. [DOI] [PubMed] [Google Scholar]
- 20.Wei Y-G, Liu F, Li B, Chen X, Ma Y, Yan L-N, et al. Interleukin-10 gene polymorphisms and hepatocellular carcinoma susceptibility: a meta-analysis. World J Gastroenterol. 2011;17(34):3941–7. doi: 10.3748/wjg.v17.i34.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Wang Y, Liu C, Jiang J, Jiang H, Tang H, et al. Polymorphisms in the interleukin-10 gene and hepatocellular carcinoma: an updated meta-analysis of 12 case-control studies. Int J Clin Exp Med. 2016;9(9):17140–52. [Google Scholar]
- 22.Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy- Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13:840–8. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]
- 23.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 24.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–72. doi: 10.2307/2532296. [DOI] [PubMed] [Google Scholar]
- 25.Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291–306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 26.Page MJ, Higgins JP, Sterne JA et al. Chapter 13: Assessing risk of bias due to missing results in a synthesis. In: Higgins JP, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, editors. Cochrane Handbook for Systematic Reviews of Interventions version 6.3 (updated February 2022). Cochrane, 2022.
- 27.Liang B, Guo Y, Li Y, Kong H. Association between IL-10 gene polymorphisms and susceptibility of tuberculosis: evidence based on a meta-analysis. Plos One. 2014;9(2): e88448. [DOI] [PMC free article] [PubMed]
- 28.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434 − 42. [DOI] [PMC free article] [PubMed]
- 29.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorlund K, Wetterslev J, Brok J, Imberger G, Gluud G. User manual for trial sequential analysis (TSA) Copenhagen, Denmark: Copenhagen Trial Unit, Centre for Clinical Intervention Research; 2011. pp. 1–115. [Google Scholar]
- 31.Wetterslev J, Thorlund K, Brok J, Gluud C. Estimating required information size by quantifying diversity in random-effects model meta-analyses. BMC Med Res Methodol 2009;9(1):1–2 [DOI] [PMC free article] [PubMed]
- 32.Jakobsen J, Wetterslev J, Winkel P, Lange T, Gluud C. Thresholds for statistical and clinical significance in systematic reviews with meta-analytic methods. BMC Med Res Methodol 2014;14(1):120 [DOI] [PMC free article] [PubMed]
- 33.Wetterslev J, Jakobsen JC, Gluud C. Trial Sequential Analysis in systematic reviews with meta-analysis. BMC Med Res Methodol. 2017;17:39. doi: 10.1186/s12874-017-0315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aroucha DC, Carmo RF, Vasconcelos LRS, Lima RE, Mendonça TF, Arnez LE, et al. TNF-α and IL-10 polymorphisms increase the risk to hepatocellular carcinoma in HCV infected individuals. J Med Virol. 2016;88(9):1587–95. doi: 10.1002/jmv.24501. [DOI] [PubMed] [Google Scholar]
- 35.Bahgat NA, Kamal MM, Abdelaziz AO, Mohye MA, Shousha HI, Ahmed MM, et al. Interferon-γ and interleukin-10 gene polymorphisms are not predictors of chronic hepatitis C (Genotype-4) disease progression. Asian Pac J Cancer Prev APJCP. 2015;16(12):5025–30. doi: 10.7314/APJCP.2015.16.12.5025. [DOI] [PubMed] [Google Scholar]
- 36.Barooah P, Saikia S, Kalita MJ, Bharadwaj R, Sarmah P, Bhattacharyya M, et al. IL-10 polymorphisms and haplotypes predict susceptibility to hepatocellular carcinoma occurrence in patients with hepatitis c virus infection from Northeast India. Viral Immunol. 2020;33(6):457–67. doi: 10.1089/vim.2019.0170. [DOI] [PubMed] [Google Scholar]
- 37.Bei C-H, Bai H, Yu H-P, Yang Y, Liang Q-Q, Deng Y-Y, et al. Combined effects of six cytokine gene polymorphisms and SNP-SNP interactions on hepatocellular carcinoma risk in Southern Guangxi, China. Asian Pac J Cancer Prev. 2014;15(16):6961–7. doi: 10.7314/APJCP.2014.15.16.6961. [DOI] [PubMed] [Google Scholar]
- 38.Bouzgarrou N, Hassen E, Farhat K, Bahri O, Gabbouj S, Maamouri N, et al. Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis C severity. Hum Immunol. 2009;70(4):230–6. doi: 10.1016/j.humimm.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Abd El-Baky RM, Hetta HF, Koneru G, Ammar M, Shafik EA, Mohareb DA, et al. Impact of interleukin IL-6 rs-1474347 and IL-10 rs-1800896 genetic polymorphisms on the susceptibility of HCV-infected egyptian patients to hepatocellular carcinoma. Immunol Res. 2020;68(3):118–25. doi: 10.1007/s12026-020-09126-8. [DOI] [PubMed] [Google Scholar]
- 40.Heneghan MA, Johnson PJ, Clare M, Ho S, Harrison PM, Donaldson PT. Frequency and nature of cytokine gene polymorphisms in hepatocellular carcinoma in Hong Kong Chinese. Int J Gastrointest Cancer. 2003;34(1):19–26. doi: 10.1385/IJGC:34:1:19. [DOI] [PubMed] [Google Scholar]
- 41.Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, et al. Cytokine gene polymorphisms in japanese patients with hepatitis B virus infection–association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42(4):505–10. doi: 10.1016/j.jhep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 42.Peng MW, Lu SQ, Liu J, Dong CY. Role of IL-10 polymorphisms in susceptibility to hepatitis B virus-related hepatocellular carcinoma. Genet Mol Res GMR. 2016; 18;15(1). [DOI] [PubMed]
- 43.Saleh A, Saed AM, Mansour M. Association of IL-10 and TNF-α polymorphisms with risk and aggressiveness of hepatocellular carcinoma in patients with HCV-related cirrhosis. Egypt Liver J. 2020;10(1):43. doi: 10.1186/s43066-020-00052-w. [DOI] [Google Scholar]
- 44.Saxena R, Chawla YK, Verma I, Kaur J. Association of interleukin-10 with hepatitis B virus (HBV) mediated disease progression in indian population. Indian J Med Res. 2014;139(5):737–45. [PMC free article] [PubMed] [Google Scholar]
- 45.Shin HD, Park BL, Kim LH, Jung JH, Kim JY, Yoon JH, et al. Interleukin 10 haplotype associated with increased risk of hepatocellular carcinoma. Hum Mol Genet. 2003;12(8):901–6. doi: 10.1093/hmg/ddg104. [DOI] [PubMed] [Google Scholar]
- 46.Tseng L-H, Lin M-T, Shau W-Y, Lin W-C, Chang F-Y, Chien K-L, et al. Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens. 2006;67(2):127–33. doi: 10.1111/j.1399-0039.2006.00536.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang JJ, Wang ZB, Tan TC. Association of CTLA-4, TNF alpha and IL 10 polymorphisms with susceptibility to hepatocellular carcinoma. Scand J Immunol. 2019;90(6):e12819. doi: 10.1111/sji.12819. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J, Liao W, Zhao Y, Chen Y, Qin L, Zhang H, et al. IL-10 and IL-10RB gene polymorphisms are correlated with hepatitis B-related hepatocellular carcinoma in the chinese Han population. Transl Cancer Res. 2017;6(2):432–40. doi: 10.21037/tcr.2017.04.09. [DOI] [Google Scholar]
- 49.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 50.Napoli J, Bishop GA, McGuinness PH, Painter DM, McCaughan GW. Progressive liver injury in chronic hepatitis C infection correlates with increased intrahepatic expression of Th1-associated cytokines. Hepatology. 1996;24:759–65. doi: 10.1002/hep.510240402. [DOI] [PubMed] [Google Scholar]
- 51.Chow MT, Moller A, Smyth MJ. Inflammation and immune surveillance in cancer. Semin Cancer Biol. 2012;22(1):23–32. doi: 10.1016/j.semcancer.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Thylur RP, Roy SK, Shrivastava A, LaVeist TA, Shankar S, Srivastava RK. Assessment of risk factors, and racial and ethnic differences in hepatocellular carcinoma. JGH Open Access J Gastroenterol Hepatol. 2020;4(3):351–9. doi: 10.1002/jgh3.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M, Metanat M, Khosravi S, et al. Association between IL-10 gene promoter polymorphisms (-592 A/C, -819 T/C, -1082 A/G) and susceptibility to HBV infection in an iranian Population. Hepat Mon. 2016;16(2):e32427. doi: 10.5812/hepatmon.32427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 55.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–73. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi YH, Zhao DM, Wang YF, Li X, Ji MR, Jiang DN, et al. The association of three promoter polymorphisms in interleukin-10 gene with the risk for colorectal cancer and hepatocellular carcinoma: a meta-analysis. Sci Rep. 2016;6:30809. doi: 10.1038/srep30809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299(20):2423–36. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdallah E, Waked E, Abdelwahab MA. Evaluating the association of interleukin-10 gene promoter – 592 A/C polymorphism with lupus nephritis susceptibility. Kidney Res Clin Pract. 2016;35(1):29–34. doi: 10.1016/j.krcp.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu L, Liu Z, Zeng C, Chen Z, Yu C, Li L. Association of interleukin-10 gene-592 A/C polymorphism with the clinical and pathological diversity of lupus nephritis. Clin Exp Rheumatol. 2005;23(6):854. [PubMed] [Google Scholar]
- 60.McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453(7198):1112–6. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: PRISMA 2020 Checklist
Supplementary Material 2: Search strategies in databases
Supplementary Material 3: Generic formula
Supplementary Material 4: Excluded nine studies
Supplementary Material 5: Frequency of genetic polymorphisms
Supplementary Material 6: Methodological quality of studies assessed via NOS criteria
Supplementary Material 7: Forest plot of ? 819 C/T (a) dominant model (b) heterozygous model
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.