Abstract
Background:
Bacteria are ubiquitous in the marine environment. Increasing concern for human health has led to growing interest in contamination on public beaches. The presence of pathogenic microorganisms originating from anthropogenic activities such as defecation and disposal of sewage on beaches are of special concern. In this study, presence of pathogenic bacteria and bacterial load in beach zones and point sources were investigated.
Methods:
Sand core samples from the subtidal zone, intertidal zone, supratidal zone and point sources from 5 beaches in Accra, Ghana, were collected and analysed. Total aerobic, coliform and Escherichia (E. coli) counts were determined for each zone in the respective beaches. Bacteria isolates were presumptively identified using biochemical tests and confirmed with MALDI-TOF MS.
Results:
Mean total aerobic count and total coliform counts ranged from 2.10 to 3.01 log CFU/g and 0.29 to 2.18 log CFU/g respectively while E. coli counts ranged from 0.12 to 1.71 log CFU/g for the beaches. Total aerobic count from point sources was 2.4-folds higher than the subtidal zone while total coliform counts were 5-folds higher in the point sources compared to the supratidal zone. Point sources had 10 times (P = .0016) more E. coli counts as compared to the subtidal zone. Isolates recovered (n = 35) belonged to 10 bacteria genera. These were Bacillus spp. (25.7%), Acinetobacter spp. (14.3%), Aeromonas spp. (14.3%), Klebsiella pneumoniae (14.3%), Aerococcus viridans (8.6%), Staphylococcus spp. (8.6%), Shewanella profunda (5.7%), Rheinheimera soli (2.9%), Pseudomonas aeruginosa (2.9%), and Exiguobacterium aurantiacum (2.9%).
Conclusion:
Point sources are major contributors to contamination on beaches. The presence of potentially pathogenic bacteria in beach sand could be a public health risk. Sensitization on cleanliness in the marine environment including beaches in Ghana is needed to enhance public health and safety.
Keywords: Beach zones, point sources, bacteria counts, contamination, MALDITOF, Accra, Ghana
Significance Statement
This study reports presence of potential pathogenic bacteria on beaches of Accra, Ghana.
Bacillus species (25.7%) were the most frequently recovered bacteria from beach sand
Point sources are major contributors to contamination on beaches.
Sensitization on beach cleanliness need to be sustained to enhance public health and safety.
Introduction
Beach sand serves as a habitat to many microbes, including viruses, bacteria, fungi and protozoa. These microbes are introduced into the coastal waters and beach sand through sewage effluents, agricultural runoff, fecal droppings from birds and local animal populations, runoff from heavy rainfall, and the beachgoers. 1 In the sand, the survival of microbes is facilitated by biotic and abiotic factors such as moisture, nutrient availability and composition, physical habitat, and nature of the microbial community. 2
Most beach monitoring programs have mainly focused on water quality but in contrast, beach visitors spent more time in contact with the sand rather than seawater.3,4 Previous studies3,5 indicate that prolonged contact with contaminated beach sand might expose beach visitors to more pathogens than exposure to water. This is because of potentially higher concentrations of pathogens in the beach sand. Various studies have reported the presence of pathogenic bacteria in beach sands including Vibrio vulnificus, Salmonella and Campylobacter spp., Pseudomonas aeruginosa, and Staphylococcus aureus including methicillin resistant strains. 6 A study conducted by Weiskerger et al 2 suggests that bacterial contamination of coastal beach sand has increased globally and this is adversely impacting tourism.
Generally, infectious disease agents within the beach environments are transmitted to humans by direct exposure to the sand, water or both within the intertidal zone.7,8 Direct dermal contact with infectious agents such as Staphylococcus and Vibrio, contact with the eyes and ears and through inhalation and fecal-oral routes in cases of Pseudomonas, Salmonella, Shigella, Campylobacter have been reported.7,8 Although the success of infectious agents largely depend on the virulence of the strain and the immune status of the host, low numbers of these microbes are required to cause disease in a host. 9 The Health Canada MSDS for Salmonella species gives a human oral infectious dose of 102to 103 cfu, 10 enterohemorrhagic strains of E. coli require an infective dose of only about ten cells while adults can be infected with 102 cfu of Shigella. 11
The beaches of Ghana over the past decade have been confronted with the issues of poor sanitation. In Accra, several efforts have been made to ensure cleanliness at the beaches. However, these efforts have not been successful since most of the beaches have been converted into sites where solid and liquid wastes are disposed of by the people living in the communities in close proximity to these beaches. 12 Open defecation along the beaches is a major problem as most residential facilities along the Accra coast lack adequate places of convenience. Inhabitants of these communities therefore resort to the beaches as a place of convenience. 13
While pollution of the coastal waters from these activities has received considerable attention, the same cannot be said of contamination of the sandy beaches. Given that many beach goers spend more time on the beach sand than in the water, this study sought to investigate the bacterial burden in the beach sand at some popular beaches in Accra, Ghana. The specific aim was to determine the presence of potentially pathogenic bacteria in beach sand and the distribution of bacteria in the different beach zones.
Materials and Methods
Study area
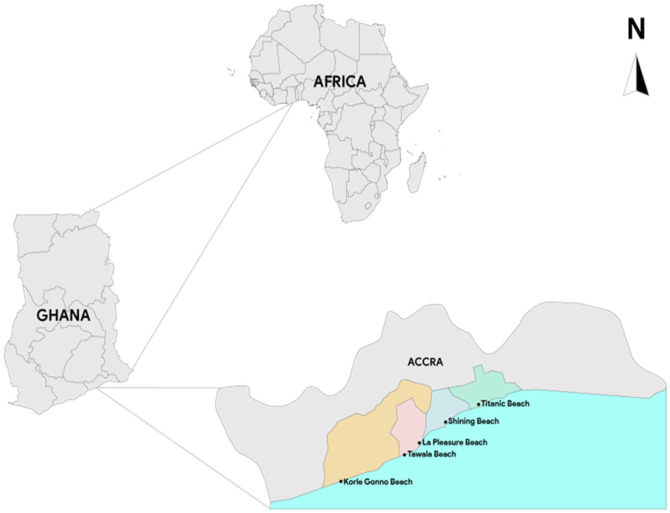
The study was conducted at 5 beaches (Figure 1); the La Pleasure beach, Korle beach, Tawala beach, Shining beach and Titanic beach all of which are located within the Greater- Accra region of Ghana. La Pleasure beach is located off the Accra-Teshie/Nungua Road (Lat. 5°33′48″N and 0°8′18″W). This beach which is the most popular in Accra is sandwiched by a 4-Star and a 5-Star hotel patronized by both foreign and local tourists. This beach also serves as the main landing site for local fishing activities. At La beach, point source sand samples were taken from water channels directed toward the beach sand and believed to be effluents from the restaurants and some small settlements that were present on the beaches. Sand samples were also taken from a rubbish dump site close to the water channel. The Korle beach (Lat 5°31′43″N and 0°13′36″W) serves as a site for local recreational and sporting activities with open defecation being a common sight. The beach also shares an open boundary with the main sewage disposal site for the Accra metropolitan assembly. Sand samples from point sources in this site were collected from big gutters that were channeled toward the beach sand in the supratidal zone. The Tawala beach (5°33′8″N and 0°9′54″W) which is located along the coast of the South La Estates is patronized mainly by the residents, mostly on weekends and holidays. The beach has largely organized residential facilities in close proximity with some few squatters. Effluent from the homes of these squatters is directly channeled onto the beach with open defecation and free ranging livestock being a regular phenomenon. Some residents were also seen throwing waste water from their houses onto the beach sand. Big gutters believed to carry effluents from the community were channeled across the sand. Sand samples from these areas which were considered point sources were taken. Shining (5°35′7″N and 0°5′17″W) and Titanic (5°36′41″N and 0°2′40″W) beaches which are located approximately 16 and 10 km from the port city of Tema also serves mainly for recreational activities with sparsely organized human settlements dotted some few meters away from the beaches. No point sources were observed in Shining beach. Like the other beaches, the Shining and the Titanic beaches serve as landing sites for local fishermen as well as fish market for trading activities. Sakumono lagoon which is a designated Ramsar site is located some 100 m away.
Figure 1.
Study sampling sites.
Field sampling
Sand samples (n = 53) were collected in the month of July 2020 with the aid of a hand-operated sampler (length- 5 cm, inner diameter- 1.8 cm). A transect, 100 m long and perpendicular to the shoreline was set up. The sand samples were taken 30 m apart along the transect from 3 sites (A, B, and C). Site A was the inundated wet sand and was approximately 1.5 m from the shoreline and in the water. The samples of sand from site B were collected approximately 10 m from the shoreline whiles samples collected from site C was mostly dry and located at a 30 m distance from the shores along the transect. Two other transects were set up 100 m on either side of the initial transect and samples were taken as described above. The sand samples taken at depths of not more than 5 cm were immediately placed in sterile labeled 50 mL falcon tubes. The samples were placed in cold boxes and transported to the Advanced Research laboratories (ARL) of the Noguchi Memorial Institute for Medical Research (NMIMR), University of Ghana for bacterial analysis. Sand samples were either processed within 12 hours or stored at −20°C overnight.
Point sources
In addition to sampling from 3 locations described above, sand samples were also taken from point sources on the beaches. In Ghana, only about 10% of urban wastewater emanating from domestic and municipal sources is disposed off through sewage networks connected to treatment plants. 14 In communities close to beaches, these point sources are final disposal sites for untreated wastewater (blackwater) emanating from domestic sources. Waste matter flushed from toilets and washoffs from domestic livestock panels are the major effluent disposed directly onto the beach sand.
Laboratory analysis
Assessment of bacteria load
Five grams (5 g) of each sand sample was weighed into different 50 mL falcon tubes containing 45 mL of sterile phosphate-buffered saline (pH 7.2). These were then homogenized by handshaking for 1 minute to make stock solutions from the supernatant. One milliliter (1 mL) of the stock solutions were pipetted into 15 mL centrifuge tubes containing 9 mL phosphate-buffered saline. Serial dilutions of 101 to 103 were prepared for each sample for plating on the appropriate media using the pour plate method.
Plate count agar (PCA) from Oxoid and Brilliance E. coli Coliform Selective Medium Agar (Oxoid) for total aerobic count and total coliform count respectively were prepared according to the manufacturer’s instructions and allowed to cool to 50°C in a water-bath.
One milliliter (1 mL)of each dilution was pipetted onto a labeled duplicate sterile petri dish. Twenty-five (25 mL) of the prepared agar was transferred onto the inoculated petri dish, swirled and allow to set. The plates were inverted and incubated for 24 hours at 37°C.
Bacteria colonies
After incubation, all plates with 30 to 300 colonies were counted and the number of colonies per dilution recorded. Faint pink colonies for coliforms and purple colonies for E. coli on coliform selective media dishes containing between 30 and 300 colonies per dilution were counted and recorded. All counts were normalized to colony forming units (CFU) per 100 g of dry weight of sand.
Colony counts
For plates with 30 to 300 colonies, actual numbers were counted in the plates of each dilution per the formula given below 15 :
Where:
ΣC = the sum of colonies counted on all the dishes retained
N1 = the number of dishes retained in the first dilution
N2 = the number of dishes retained in the second dilution
D = the dilution factor corresponding to the first dilution
Microbial detection
Sample preparation
Five grams (5 g) each of the sand samples were weighed into sterile 50 mL falcon tubes and 45 mL of phosphate-buffered saline added. These were then shaken for 1 minute to dislodge any bacteria present. 16 With the aid of inoculating loops, the inocula were transferred unto petri dishes containing the respective media. Oxoid UTI Chromogenic Agar (clear) and Uriselect 4 were used for presumptive identification of bacteria such as E. coli, Enterococcus faecalis, Pseudomonas aeruginosa, and Staphylococcus aureus. Thiosulphate Citrate Bile Source Agar (TCBS) was used for presumptive identification of Vibrio sp. and Aeromonas sp., For Campylobacter, Mueller Hinton Agar with 5% sheep blood was used. Plates containing the various agar were placed in microaerophilic jars along with CampyGen. All streaked plates were incubated at 37°C overnight.
Bacteria bio-typing
Presumptive colonies identified were confirmed using Matrix-Assisted Laser-Desorption Ionization-Time of Flight (MALDI-TOF) biotyping. Well isolated and pure bacteria colonies were carefully picked from each of the culture plates and transferred unto spots on the MALDI-TOF target plate. One microliter(1 μL) of 70% formic acid was added to each spot and allowed to air dry. Extraction matrix (1 μL) was also added to each spot and allowed to air dry. The inoculated targets were placed in Bruker Biotyper instrument and analyzed to obtain bacterial profiles.
Data analysis
Bacteria counts were log transformed and statistical analysis was done using GraphPad prism version 9.0.1. Shapiro-Wilk test was done to test the normality of the data. In the present study, Kruskal-Wallis test followed by Dunn’s multiple comparison posthoc was used to determine differences in the bacterial counts from different beaches and between counts from the different beach zones.
Results
Bacterial counts
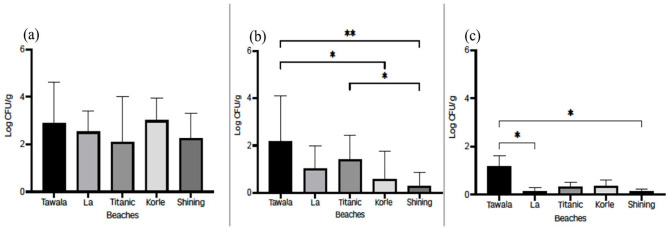
Overall, Korle beach had the highest mean total aerobic count (3.01 log CFU/g) while Titanic beach recorded the lowest (2.10 log CFU/g). The total coliform count for Tawala beach (2.18 log CFU/g), was 7.5-folds higher (P = .0055) than that recorded for Shining beach (0.29 log CFU/g) which had the lowest count. Tawala beach was 14-fold higher (P = .0340) in E. coli counts (1.71 log CFU/g), compared to the lowest Shining beach (0.12 log CFU/g) as shown in Figure 2.
Figure 2.
(a) Total aerobic count, (b) total coliform count and (c) E. coli counts of sand from different beaches.
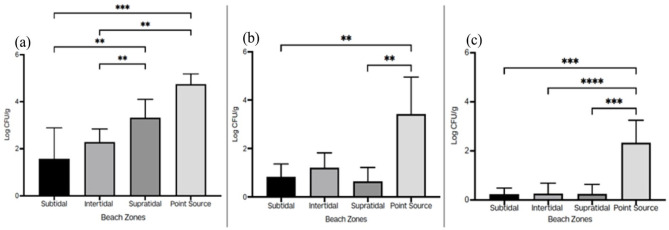
Total aerobic count from point sources was 2.4-folds higher (P = .0005) than sand from the subtidal zone which recorded the lowest (1.82 log CFU/g). Sand from the supratidal zone counts were also 1.8-folds higher (P = .0011) than that of the subtidal zone and 1.6-folds higher (P = .0064) than that of the intertidal zone. The intertidal zone counts were also 1.2 times higher (P = .5632) than that of the subtidal zone. Total coliform counts were 5-folds higher (P = .2552) in the point sources compared to the supratidal zone which had the lowest count (0.63 log CFU/g). The intertidal zone counts were 1.4-folds higher (P = .5632) than the subtidal zone and 1.9-folds higher (P = .0064) than the supratidal zone. Also, sand from the subtidal zone had a 1.3-fold higher (P = .6138) total coliform count as compared to sand from the supratidal zone. Point sources had 10 times (P = .0016) more E. coli counts as compared to the subtidal zone counts (0.22 log CFU/g). The intertidal zone had a 1-fold higher E. coli count compared to the supratidal zone (0.24 log CFU/g) (P = .1833) and the subtidal zone (P = .3848). Sand from the supratidal zone counts were also 1-fold higher compared to subtidal zone counts (P = .6138) for E. coli as shown in Figure 3.
Figure 3.
The total aerobic count (a), total coliform count (b) and E. coli counts (c) of sand from different beach zones.
Bacteria detection and distribution
A total of 35 bacterial isolates were detected in the sand from all the 5 beaches (Table 1). Bacteria species isolated include Bacillus spp. (25.7%), Klebsiella pneumoniae (14.3%), Acinetobacter spp. (14.3%), Aeromonas spp. (14.3%), Aerococcus viridans (8.6%), Staphylococcus spp. (8.6%), Shewanella profunda (5.7%), Rheinheimera soli (2.9%), Pseudomonas aeruginosa (2.9%), and Exiguobacterium aurantiacum (2.9%). A significant number of the isolates 17 were obtained from the Tawala beach which represented 48.6% of the total. This was followed by Shining beach (25.7%), Korle beach (11.4%), Titanic beach (8.6%) and La beach (5.7%). The most commonly occurring organism in the beach sand was from the genus Bacillus (25.7%). Four of Bacillus spp. were obtained from the Tawala beach whilst one species was obtained from La, Shining and Titanic beach. No Bacillus strains were isolated from Korle Beach (Table 1). Species of Bacillus that were isolated include B. pumilus, B. cereus, B. megaterium, and B. jeotgali.
Table 1.
Distribution of different species of bacteria isolates from the beach sand.
| Bacteria | Beaches | |||||
|---|---|---|---|---|---|---|
| Tawala beach | Shining beach | Korle beach | Titanic beach | La beach | Total | |
| Bacillus spp. | 4 | 3 | 0 | 1 | 1 | 9 (25.7%) |
| Acinetobacter spp. | 2 | 2 | 1 | 0 | 0 | 5 (14.3%) |
| Aeromonas spp. | 4 | 0 | 0 | 1 | 0 | 5 (14.3%) |
| Klebsiella pneumoniae | 4 | 0 | 1 | 0 | 0 | 5 (14.3%) |
| Aerococcus viridans | 0 | 3 | 0 | 0 | 0 | 3 (8.6%) |
| Staphylococcus spp. | 2 | 0 | 0 | 0 | 1 | 3 (8.6%) |
| Shewanella profunda | 0 | 0 | 1 | 1 | 0 | 2 (5.7%) |
| Pseudomonas aeruginosa | 0 | 1 | 0 | 0 | 0 | 1 (2.9%) |
| Rheinheimera soli | 1 | 0 | 0 | 0 | 0 | 1 (2.9%) |
| Exigobacterium aurantiacum | 0 | 0 | 1 | 0 | 0 | 1 (2.9%) |
| 17 (48.6%) | 9 (25.7%) | 4 (11.4%) | 3 (8.6%) | 2 (5.7%) | 35 (100%) | |
After Bacillus spp., Aeromonas spp., Klebsiella pneumoniae, and Acinetobacter spp. were the most isolated bacteria and represented 14.3% each. Species of Aeromonas isolated were Aeromonas bestarium, A. media, A. jandiae, A. ichthiosoma, and A. veronii. Most of the Aeromonas spp. were isolated from the Tawala beach (4) and the least from Titanic beach (1). The other beaches recorded no isolates for Aeromonas spp.
Klebsiella pneumoniae (4) was mostly isolated from Tawala beach and one isolate from Korle beach. Acinetobacter spp. were obtained from Tawala beach (2), Shining beach (2) and Korle beach (1). Two species of Acinetobacter were identified: Acinetobacter towneri and Acinetobacter tandoii.
Aerococcus viridans and Staphylococcus spp. were next most abundant with 8.6% of the bacteria isolated. All isolated Aerococcus viridans (3) were obtained from Shining beach and Staphylococcus species were obtained from Tawala beach (2) and La beach (1) only. These Staphylococci were characterized as Staphylococcus hominis, S. haemolyticus, and S. epidermidis.
Two isolates of Shewanella profunda were obtained from Korle beach (1) and Titanic beach (1) only. Rheinheimera soli was obtained in only Tawala beach. One Pseudomonas aeruginosa was isolated from Shining beach and one of Exiguobacterium aurantiacum from Korle beach. Each strain represented 2.9% of the total bacterial isolation of the study. No strain of Campylobacter spp. was identified during the study.
Among the 3 beach zones and point sources, the intertidal zone (B) (34.3%) recorded the highest, followed by the subtidal zone (A) (25.7%), then the supratidal zone (C) (22.9%) and then finally the point source (PS) number of bacterial isolates (17.1%) as shown in Table 2.
Table 2.
Distribution of different species of bacteria isolates in the different beach zones.
| Bacteria | Beach zones | |||
|---|---|---|---|---|
| Intertidal zone sand | Subtidal zone sand | Supratidal zone sand | Point source | |
| Aerococcus viridans | 1 | 2 | 0 | 0 |
| Acinetobacter spp. | 4 | 0 | 1 | 0 |
| Aeromonas spp. | 1 | 1 | 2 | 1 |
| Shewanella profunda | 1 | 0 | 0 | 1 |
| Rheinheimera soli | 0 | 1 | 0 | 0 |
| Klebsiella pneumoniae | 0 | 2 | 2 | 1 |
| Pseudomonas aeruginosa | 1 | 0 | 0 | 0 |
| Staphylococcus spp. | 1 | 1 | 0 | 1 |
| Bacillus spp. | 3 | 2 | 3 | 1 |
| Exiguobacterium aurantiacum | 0 | 0 | 0 | 1 |
| Total | 12 (34.3%) | 9 (25.7%) | 8 (22.9%) | 6 (17.1%) |
Discussion
Although total aerobic bacteria are an index of bacterial load, coliforms, fecal coliforms and E. coli are major indicators of the likelihood of fecal contamination in beaches in marine environments. 17 The presence of these indicators expresses the possibility of the presence of pathogenic bacteria and as such, is very useful for predicting the public health risks associated with the use of marine beaches. 17 The beach is often a dumping site for treated and untreated waste from anthropogenic activities. These wastes often contain very high levels of microorganisms including antibiotic resistant bacteria which are then spread to other environments. 18 This may account for the high total aerobic, total coliform and E. coli counts in this study from the point sources compared to the other beach zones. There was no point source at the Shining beach, and this could account for the low coliform and E. coli. counts recorded on the beach. Similar studies conducted by Cardonha et al 19 reported high E. coli levels from storm drains that were channeled toward the sea while Metcalf et al 20 found that E. coli levels in beach sand were different between sites. Previous studies on beach sand have established that the supratidal zone harbors more bacteria, 21 compared to the subtidal zone. The reason for this observation as stated by Abdool-Ghany et al 21 is that it offers favorable circumstances for growth (such ideal temperatures and moisture levels). The abundance of more bacteria in the supratidal zone has also been attributed to the movement of humans on the beach sand. 22 Bonilla et al 23 stated that fecal bacteria may be transferred from one place to another by people walking on the dry sand. Bonilla et al 23 further opines that there are lower rates of predation in the supratidal zone as compared to the subtidal zone. Sand from the supratidal zone have less water content as compared to the intertidal zone sand and the subtidal zone which consequently results in a decreased water film around the sand grains. Also, bacterivores may not be active in the supratidal zone. 23 In this study, total aerobic bacteria counts were 1.8-folds higher in the supratidal zone from all the 5 beaches compared to the subtidal zone and 1.6-folds higher as compared to the intertidal sand. However, there was no significant difference between the total coliform and E. coli counts in the supratidal zone, subtidal zone, and intertidal zone.
The marine environment is made up of a wide variety of bacteria; some of which are non-pathogenic and some, potentially pathogenic. 24 This study showed that some potentially pathogenic bacteria, some of which belong to the ESKAPE bacterial group of pathogens as well as the GLASS (Global Antimicrobial Resistance and Use Surveillance System) priority pathogens such as Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus spp. Acinetobacter are present in beach sand.
Staphylococcus spp. are opportunistic pathogens which may be found on the skin, hair and mucous membranes of humans and cause such as septicemia, wound sepsis, and pneumonia. 25 As pathogens of humans, Staphylococcus spp. are etiological agents of infection spread between humans. 25 S. hominis, S. epidermidis, and S. haemolyticus were the species of Staphylococcus isolated and identified in this study, and these results corroborate the findings of a study carried out in the Red Sea coast by Ullah et al 26 which found S. haemolyticus, S. epidermidis, and S. hominis in the beach samples.
In this study, 5.7% of the total isolates recovered were Aerococcus viridans. Aerococcus spp. have been isolated from clinical samples- A. urinae, A. sanguinocol, and A. viridans have been described as the most common cause of urinary tract infection among the several different species. 27
The genus Aeromonas are ubiquitous in the aquatic environment. 28 It is the main agent of infection in fishes. 29 A. hydrophila, A. bestiarum, A. caviae, A. sobria, and A. veronii species are responsible for causing motile aeromonad septicemia (MAS) as well as gastroenteritis, tissue infections and wounds to their hosts. 30 Aeromonas may be isolated from water sources such as sewage and wastewaters, freshwater, drinking water, and estuarine environments. 30 This agrees with this study in which A. bestarium was isolated from a point source. The range of Aeromonas associated with diarrheal diseases in humans and infections in wounds are A. hydrophila, A. jandiae, A. caviae, and A. veronii. 31 Majority of Bacillus spp. with the exception of a few, are non-pathogenic with no association with etiological causes of diseases in humans or animals. 32 Bacillus species are predominantly found in the soil although they have a wide distribution in the environment. 33 Although some species are frequently isolated as contaminants in cultures in the lab, recent reports indicate that the bacteria may be an etiological agent in local and systemic infections in humans. 33 The few species among this genus that are considered medically relevant are B. anthracis which causes anthrax and B. cereus which causes food-borne illness and also has been reported to cause bacteremia in patients with compromised immune systems. 34 B. pumilus has also been documented to cause sepsis in neonatal infants. 35 In the marine environment, B. pumilus, B. cereus, and B. subtilis form major components of the bacterial communities. B. pumilus has the ability to withstand severe environmental conditions such as reduced nutrient availability, irradiation from the sun and desiccation. 36 In this study, B. pumilus, B. megaterium, and B. cereus were isolated and identified from the beach sand with B. pumilus being the most predominant, similar to results by Parvathi et al 37 in the marine ecosystem.
Pseudomonas aeruginosa has been described as an opportunistic bacterium that causes infections of the skin, eye and ear via swimming in seawater that has been contaminated and the use of sandy beaches for recreation. 38 Studies done by Mohammed et al 39 on establishing survival trends of P. aeruginosa concluded that they were able to thrive and replicate in sterile beach sand and as such could be used to assess sanitary conditions of beaches and also recurrent health risk assessments of beach sand.
Acinetobacter spp. is naturally found in the environment, in soil and in animals. It is of research interest in recent years due to its ability to cause nosocomial infections and their ability to develop multidrug resistance and extreme drug resistance. These bacteria have developed distinguishing capabilities from other bacteria which allows them to colonize most surfaces and acquire resistance to antibiotics. 40
The Shewanella genus is mostly found in deep sediments in marine environments. 41 Toffin et al 42 first discovered Shewanella profunda from a sediment sample that was 4.15 m below the sea floor contrary to this study in which it was isolated from the intertidal zone and from a point source. It is ubiquitous in nature and is usually isolated from the soil and the marine environments. 43 Shewanella spp. are deemed to be opportunistic pathogens of animals and humans. It has also been linked to food spoilage. 43
Klebsiella spp. are found in different habitats and commonly in soil, sewage, and on mucosal surfaces in humans and animals. 44 They are said to be opportunistic pathogens and can cause serious infections such as pneumonia, sepsis and urinary tract infections. 45
The association of Exiguobacterium aurantiacum with human infections is very rare. 46 It is an environmental bacterium which is saprophytic in nature. Studies from Pitt et al 47 show that immunocompromised individuals may be susceptible to infections from E. aurantiacum.
Rheinheimera spp. are gammaproteobacteria mostly isolated from aquatic environments and its associated habitats. In a study conducted in Korea, R. soli was isolated from a soil sample in a playground. 48 Little is known about their pathogenicity to humans.
Conclusion
This study is the first to record the bacterial concentrations and presence of potential pathogenic bacteria in various sands across and along sandy beaches in Accra, Ghana. Results from this study reveal that point sources are major contributors to contamination on the beaches. Also, the presence of potential pathogenic bacteria in the beach sands may be a potential public health risk to beach goers. Beaches are treasured natural resources that must be preserved for future generations. Reducing bacteria contamination of beaches should therefore be a shared responsibility of government, corporate institutions and individuals. Short term solutions such as regular clean up campaigns, provision of public toilets and waste bins at beaches as well as long term solutions such as conservation will ameliorate the waste and pollution that leads to bacteria contamination. While it is necessary to sensitize the general public on the need to keep the marine environments clean to enhance public health and safety, enactment and enforcement of regulatory policies to safeguard sanitation at the beach may yield better results.
Acknowledgments
The laboratory analysis was carried out at the Advanced Research Laboratories (ARL) of the Noguchi Memorial Institute for Medical Research (NMIMR), University of Ghana and we gratefully acknowledge the support of Mr. Christian Bonsu, Mr. Lorenzo Akyeh, Mr. Christian Owusu and staff of the bacteriology department of NMIMR.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Study was supported by funds from authors.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: D Oduro: Conceptualization, funds provision, samples acquisition, writing/review/editing, data analysis and supervision. S Darko: Conceptualization, samples acquisition, experimentation and writing of first draft of manuscript. ER Blankson: Expert support, mapping out sampling points at beaches and review/editing. GI Mensah: Technical/expert support for bacteriological analysis, funds provision, supervision of experiments, statistical analysis and review/editing. All authors read and approved the final manuscript.
Ethics Approval and Consent to Participate: Not applicable.
Consent for Publication: Not applicable.
References
- 1. WHO. WHO Guidelines on Recreational Water Quality: Volume 1: Coastal and Fresh Waters. World Health Organization; 2021. [PubMed] [Google Scholar]
- 2. Weiskerger CJ, Brandão J, Ahmed W, et al. Impacts of a changing earth on microbial dynamics and human health risks in the continuum between beach water and sand. Water Res. 2019;162:456-470. [DOI] [PubMed] [Google Scholar]
- 3. WHO. Guidelines for Safe Recreational Water Environments: Coastal and Fresh Waters. World Health Organization; 2003. [PubMed] [Google Scholar]
- 4. Buzzi NS, Menéndez MC, Truchet DM, Delgado AL, Severini MD. An overview on metal pollution on touristic sandy beaches: is the COVID-19 pandemic an opportunity to improve coastal management? Mar Pollut Bull. 2021;174:113275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young I, Sanchez JJ, Desta BN, Heasley C, Tustin J. Recreational water exposures and illness outcomes at a freshwater beach in Toronto, Canada: a prospective cohort pilot study. PLoS One. 2023;18:e0286584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Plano LRW, Shibata T, Garza AC, et al. Human-associated methicillin-resistant Staphylococcus aureus from a subtropical recreational marine beach. Microb Ecol. 2013;65:1039-1051. [DOI] [PubMed] [Google Scholar]
- 7. Solo-Gabriele HM, Harwood VJ, Kay D, et al. Beach sand and the potential for infectious disease transmission: observations and recommendations. J Mar Biol Assoc UK. 2016;96:101-120. [Google Scholar]
- 8. Sabino R, Rodrigues R, Costa I, et al. Routine screening of harmful microorganisms in beach sands: implications to public health. Sci Total Environ. 2014;472:1062-1069. [DOI] [PubMed] [Google Scholar]
- 9. Haas CN, Rose JB, Gerba CP. Quantitative Microbial Risk Assessment. John Wiley & Sons; 2014. [Google Scholar]
- 10. Public Health Agency of Canada (PHA), Pathogen Regulation Directorate: Material Safety Data Sheet: Health Canada. 2003. Accessed July 4, 2023. http://www.phac-aspc.gc.ca/msds-ftss/index.html.
- 11. Schmid-Hempel P, Frank SA. Pathogenesis, virulence, and infective dose. PLoS Pathog. 2007;3:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marfo AA. Environmental effects of sewage pollution and waste water effluent quality assessment. University bof Ghana digital collections (UG Space); 2014. https://ugspace.ug.edu.gh/handle/123456789/8121. [Google Scholar]
- 13. Peprah D, Baker KK, Moe C, et al. Public toilets and their customers in low-income Accra, Ghana. Environ Urban. 2015;27:589-604. [Google Scholar]
- 14. Gyampo MA, ed. Wastewater production, treatment, and use in Ghana. Third regional workshop of the project ‘safe use of wastewater in agriculture; 2012. [Google Scholar]
- 15. Maturin L, Peeler J. Aerobic plate count. In: Bacteriological Analytical Manual (BAM), Chapter 3, US Food and Drug Administration. Silver Spring; 2001. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm063346.htm. [Google Scholar]
- 16. Mudryk Z, Gackowska J, Skórczewski P, Perliński P, Zdanowicz M. Occurrence of potentially human pathogenic bacteria in the seawater and in the sand of the recreational coastal beach in the southern Baltic Sea. Oceanol Hydrobiol Stud. 2014;43:366-373. [Google Scholar]
- 17. Nana PA, Ebonji Seth R, Ndjuissi Tamko NA, et al. Tidal effect on the dispersion of fecal pollution indicator bacteria and associated health risks along the Kribi beaches (southern Atlantic coast, Cameroon). Reg Stud Mar Sci. 2023;60:102831. [Google Scholar]
- 18. Vaz-Moreira I, Nunes OC, Manaia CM. Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol Rev. 2014;38:761-778. [DOI] [PubMed] [Google Scholar]
- 19. Cardonha AM, Cardonha AM, Vieira RH, Rodrigues DP, Macrae A, Peirano G, et al. Fecal pollution in water from storm sewers and adjacent seashores in Natal, Rio Grande do Norte, Brazil. Int Microbiol. 2004;7:213-218. [PubMed] [Google Scholar]
- 20. Metcalf R, White HL, Moresco V, Ormsby MJ, Oliver DM, Quilliam RS. Sewage-associated plastic waste washed up on beaches can act as a reservoir for faecal bacteria, potential human pathogens, and genes for antimicrobial resistance. Mar Pollut Bull. 2022;180:113766. [DOI] [PubMed] [Google Scholar]
- 21. Abdool-Ghany AA, Sahwell PJ, Klaus J, Gidley ML, Sinigalliano CD, Solo-Gabriele HM. Fecal indicator bacteria levels at a marine beach before, during, and after the COVID-19 shutdown period and associations with decomposing seaweed and human presence. Sci Total Environ. 2022;851:158349. [DOI] [PubMed] [Google Scholar]
- 22. Skórczewski P, Mudryk Z, Gackowska J, Perlinski P. Abundance and distribution of fecal indicator bacteria in recreational beach sand in the southern Baltic Sea. Rev Med Biol Mar y Oceanogr. 2012;47:503-512. [Google Scholar]
- 23. Bonilla TD, Nowosielski K, Cuvelier M, et al. Prevalence and distribution of fecal indicator organisms in South Florida beach sand and preliminary assessment of health effects associated with beach sand exposure. Mar Pollut Bull. 2007;54:1472-1482. [DOI] [PubMed] [Google Scholar]
- 24. Skórczewski P, Mudryk Z, Miranowicz J, Perlinski P, Zdanowicz M. Antibiotic resistance of Staphylococcus-like organisms isolated from a recreational sea beach on the southern coast of the Baltic Sea as one of the consequences of anthropogenic pressure. Oceanol Hydrobiol Stud. 2014;43:41-48. [Google Scholar]
- 25. Efuntoye M. Study of antibiotic sensitivity pattern and enterotoxigenicity of staphylococci isolated from swimming, pools in Ibadan, Nigeria. World Appl Sci J. 2010;9:1324-1327. [Google Scholar]
- 26. Ullah R, Yasir M, Bibi F, et al. Taxonomic diversity of antimicrobial-resistant bacteria and genes in the Red Sea coast. Sci Total Environ. 2019;677:474-483. [DOI] [PubMed] [Google Scholar]
- 27. Senneby E, Petersson A-C, Rasmussen M. Epidemiology and antibiotic susceptibility of aerococci in urinary cultures. Diagn Microbiol Infect Dis. 2015;81:149-151. [DOI] [PubMed] [Google Scholar]
- 28. Baron S, Granier SA, Larvor E, et al. Aeromonas diversity and antimicrobial susceptibility in freshwater—an attempt to set generic epidemiological cut-off values. Front Microbiol. 2017;8:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Austin B, Adams C. Fish pathogens. In Austin B, Altwegg M, Gosling PJ, Joseph S. (eds). The Genus Aeromonas. John Wiley and Sons, Chichester; 1996:197-243. [Google Scholar]
- 30. Majeed S, De Silva LADS, Kumarage PM, Heo G-J. Occurrence of potential virulence determinants in Aeromonas spp. isolated from different aquatic environments. J Appl Microbiol. 2023;134:lxad031. [DOI] [PubMed] [Google Scholar]
- 31. Shin HB, Yoon J, Lee Y, Kim MS, Lee K. Comparison of MALDI-TOF MS, housekeeping gene sequencing, and 16S rRNA gene sequencing for identification of Aeromonas clinical isolates. Yonsei Med J. 2015;56:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turnbell P, Kramer J. Bacillus: manual of clinical microbiology 5th ed. In Balows A, Hauster JR, Herman KL, et al. (eds). American Society of Microbiology, Washington D.C.; 1991:296-303. [Google Scholar]
- 33. Lotte R, Chevalier A, Boyer L, Ruimy R. Bacillus cereus invasive infections in preterm neonates: an up-to-date review of the literature. Clin Microbiol Rev. 2022;35:e00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jeßberger N, Krey VM, Rademacher C, et al. From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front Microbiol. 2015;6:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimouli M, Vrioni G, Papadopoulou M, et al. Two cases of severe sepsis caused by Bacillus pumilus in neonatal infants. J Med Microbiol. 2012;61:596-599. [DOI] [PubMed] [Google Scholar]
- 36. Dobrzyński J, Jakubowska Z, Dybek B. Potential of Bacillus pumilus to directly promote plant growth. Front Microbiol. 2022;13:1069053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parvathi A, Krishna K, Jose J, Joseph N, Nair S. Biochemical and molecular characterization of Bacillus pumilus isolated from coastal environment in Cochin, India. Braz J Microbiol. 2009;40:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Januário AP, Afonso CN, Mendes S, Rodrigues MJ. Faecal indicator bacteria and Pseudomonas aeruginosa in marine coastal waters: is there a relationship? Pathogens. 2019;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mohammed RL, Echeverry A, Stinson CM, et al. Survival trends of Staphylococcus aureus, Pseudomonas aeruginosa, and Clostridium perfringens in a sandy South Florida beach. Mar Pollut Bull. 2012;64:1201-1209. [DOI] [PubMed] [Google Scholar]
- 40. Gupta N, Gandham N, Jadhav S, Mishra R. Isolation and identification of Acinetobacter species with special reference to antibiotic resistance. J Nat Sci Biol Med. 2015;6:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Delong EF, Franks DG, Yayanos AA. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl Environ Microbiol. 1997;63:2105-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Toffin L, Bidault A, Pignet P, et al. Shewanella profunda sp. nov., isolated from deep marine sediment of the Nankai Trough. Int J Syst Evol Microbiol. 2004;54:1943-1949. [DOI] [PubMed] [Google Scholar]
- 43. Janda JM, Abbott SL. The genus Shewanella: from the briny depths below to human pathogen. Crit Rev Microbiol. 2014;40:293-312. [DOI] [PubMed] [Google Scholar]
- 44. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen X, Wang L, Zhou J, et al. Exiguobacterium sp. A1b/GX59 isolated from a patient with community-acquired pneumonia and bacteremia: genomic characterization and literature review. BMC Infect Dis. 2017;17:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pitt TL, Malnick H, Shah J, et al. Characterisation of Exiguobacterium aurantiacum isolates from blood cultures of six patients. Clin Microbiol Infect. 2007;13:946-948. [DOI] [PubMed] [Google Scholar]
- 48. Ryu SH, Chung BS, Park M, Lee SS, Lee S-S, Jeon CO. Rheinheimera soli sp. nov., a gamma proteobacterium isolated from soil in Korea. Int J Syst Evol Microbiol. 2008;58:2271-2274. [DOI] [PubMed] [Google Scholar]