Abstract
A multiplex PCR was developed to identify enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains by amplifying genes encoding K99 and F41 fimbriae, heat-stable enterotoxin a, intimin, and Shiga toxins 1 and 2. This multiplex PCR was specific and sensitive. It will be useful for identification of E. coli strains which cause diarrhea in calves.
Diarrhea in calves is commonly caused by enterotoxigenic Escherichia coli (ETEC). More recently, attaching and effacing E. coli (AEEC) and Shiga toxin-producing E. coli (STEC) have also been identified as causes of diarrhea and dysentery in calves (2, 15).
ETEC has two groups of virulence factors: fimbriae (pili) and enterotoxins. K99 and/or F41 fimbriae mediate adherence to the ileum and are found on most calf ETEC (1, 9, 12). Calf ETEC produces heat-stable enterotoxin a (STa), which causes hypersecretion into the gut lumen (2).
Intestinal lesions caused by AEEC are termed attaching and effacing (AE) because of their intimate attachment to the enterocyte and effacement of the microvillus border (18). A chromosomal gene, eaeA, encodes the protein intimin, which is involved in AE activity (10). AEEC which causes disease and does not produce enterotoxins or Shiga toxins is referred to as enteropathogenic E. coli (EPEC).
STEC produces two types of E. coli Shiga toxins, those that are immunologically similar to the Shiga toxin produced by Shigella dysenteriae (Stx1) and those that are immunologically distinct from Shigella dysenteriae Shiga toxin (Stx2) (11). Bovine STEC produces either Stx1, Stx2, or both. These toxins act by inhibiting protein synthesis and are lethal for in vitro cultured Vero cells (8). Some E. coli strains, such as the zoonotic pathogen E. coli O157:H7, which produces Shiga toxins and has AE activity, are often termed enterohemorrhagic E. coli (EHEC) because of their association with hemorrhagic colitis in humans.
Diagnosis of E. coli infection currently relies on the phenotypic differentiation of pathogenic strains from nonpathogenic normal flora E. coli via bioassays or immunoassays for toxins and fimbriae. These tests can be time-consuming and complicated and are not routinely used in many clinical laboratories. Histological examination of the intestine may indicate involvement of ETEC or AEEC, but this is a postmortem diagnosis. Genotypic diagnosis may be accomplished by DNA colony blot hybridization to identify genes encoding virulence factors (17). However, the use of radioactive isotopes and the time required make this method unsuitable for many diagnostic laboratories. PCR is a useful diagnostic tool because it is quick, specific, sensitive, and relatively inexpensive. A multiplex PCR which detects genes encoding intimin and Stx1 and Stx2 has been developed (4). This paper reports the identification and differentiation of ETEC, AEEC, and STEC via multiplex PCR amplification of virulence-associated genes commonly found in these E. coli strains. Primers specific for genes encoding the fimbrial subunits of K99 and F41, STa enterotoxin, intimin, and the A subunits of Shiga toxins Stx1 and Stx2 were incorporated into a multiplex PCR format. This multiplex PCR was then tested on 99 E. coli strains obtained from the E. coli collection at the National Animal Disease Center in Ames, Iowa, and from the Center for Vaccine Development, University of Maryland. Strains were previously probed by colony blot hybridization for genes encoding these virulence factors.
Primers were chosen from published sequences with the aid of the Primer Select software (DNASTAR Inc, Madison, Wis.). Table 1 includes the primer sequences, the position of the primer in the open reading frame of the gene, and the predicted sizes of amplified products. Primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). Because the EHEC and EPEC genes which encode intimin have considerable heterogeneity on the 3′ end, the eaeA primers were made to amplify a constant 5′ region of the EHEC and EPEC genes (14). Other primers were chosen from the open reading frames of the published sequences. The primers were designed to have similar annealing temperatures and minimal interactions and resulted in different-sized products.
TABLE 1.
Primers used in multiplex PCR of ETEC, AEEC, and STEC
| Virulence factor | Accession number | Primer sequence 5′-3′ | Position in open reading frame | Size of product (bp) | Reference |
|---|---|---|---|---|---|
| Stx1 | Z36899 | TTCGCTCTGCAATAGGTA | 125-142 of A subunit | 555 | 21 |
| TTCCCCAGTTCAATGTAAGAT | 659-679 of A subunit | ||||
| Intimin | Z11541 | ATATCCGTTTTAATGGCTATCT | 992-1013 of eaeA | 425 | 10, 28 |
| S90827 | AATCTTCTGCGTACTGTGTTCA | 1395-1416 of aeaA | |||
| F41 | X14354 | GCATCAGCGGCAGTATCT | 34-51 of fimbrial subunit | 380 | 7 |
| GTCCCTAGCTCAGTATTATCACCT | 390-413 of fimbrial subunit | ||||
| K99 | M35282 | TATTATCTTAGGTGGTATGG | 21-40 of fimbrial subunit | 314 | 23 |
| GGTATCCTTTAGCAGCAGTATTTC | 311-334 of fimbrial subunit | ||||
| Sta | M25607 | GCTAATGTTGGCAATTTTTATTTCTGTA | 9-36 | 190 | 25 |
| AGGATTACAACAAAGTTCACAGCAGTAA | 171-198 | ||||
| Stx2 | L11078 | GTGCCTGTTACTGGGTTTTTCTTC | 30-53 of A subunit | 118 | 22 |
| AGGGGTCGATATCTCTGTCC | 128-147 of A subunit |
The 20-μl PCR mixture contained 0.5 μM concentrations of each primer, 0.2 mM concentrations of each dNTP (di-deoxynucleotides), 1× AmpliTaq Gold buffer, 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Branchburg, N.J.), and bacterial DNA. All reagents except AmpliTaq Gold and template DNA were added to PCR tubes, lyophilized under centrifugation in a DNA Speed Vac (Savant Instruments, Farmingdale, N.Y.), and stored at −20°C. The use of tubes containing lyophilized reagents reduced the time for setup of the assay and decreased the possibility of false positives due to contamination. Bacterial DNA was obtained by suspending a colony of bacteria grown overnight on trypticase soy agar in 50 μl of H2O and boiling at 100°C for 10 min. After addition of 19.5 μl of bacterial DNA and 0.5 μl of AmpliTaq Gold to tubes containing the lyophilized reaction mixture, samples were amplified in a GeneAmp PCR 2400 thermal cycler (Perkin-Elmer) under the following conditions: 25 cycles beginning with a 30-s denaturation at 94°C, primer annealing at 50°C for 45 s, followed by extension for 1 min 30 s at 70°C. The extension time was ramped for an additional 3 s per cycle, and a final extension for 10 min at 70°C was performed. Products were electrophoresed in a 3% NuSieve 3:1 agarose gel (FMC Bioproducts, Rockland, Maine) for 1 h at 100 V, stained with ethidium bromide, and photographed under UV light. Each experiment contained negative controls with all reagents except template DNA. Also, a positive control was included with bacterial DNA from two strains containing all six genes of interest.
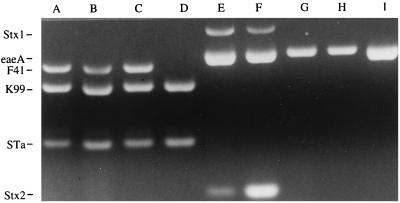
The multiplex PCR was tested on several reference strains with different phenotypes as illustrated in Fig. 1 and Table 2. These strains had been characterized phenotypically and by colony blot hybridization previously by others. The multiplex PCR assay correctly determined the presence or absence of the genes of interest in all of the reference strains.
FIG. 1.
Multiplex PCR of reference strains. DNA from reference strains was used as template DNA for PCR. Products were separated on a 3% agarose gel and compared to a positive control. Bands corresponding to Stx1, eaeA, F41, K99, STa, and Stx2 are indicated. Lane A, strain B41; lane B, strain B44; lane C, strain 431; lane D, strain 637; lane E, strain 3081; lane F, strain 933; lane G, strain CHMC6; lane H, strain E2348-69; lane I, RDEC-1.
TABLE 2.
Reference E. coli strains tested by multiplex PCR
| Literature no. | Phenotypic characterization | Genotypic characterizationa | PCR result | Refer- ence(s) |
|---|---|---|---|---|
| B41 | Calf ETEC | K99, F41, STa | K99, F41, STa | 19, 20 |
| B44 | Calf ETEC | K99, F41, STa | K99, F41, STa | 19, 20 |
| 431 | Pig ETEC | K99, F41, STa | K99, F41, STa | 16 |
| 637 | Pig ETEC | K99, STa | K99, STa | 26 |
| 3081 | Calf STEC O157: H7 | Intimin, Stx1, Stx2 | Intimin, Stx1, Stx2 | 5 |
| 933 | Human STEC O157: H7 | Intimin, Stx1, Stx2 | Intimin, Stx1, Stx2 | 6, 27 |
| CHMC#6 | Human EPEC | Intimin | Intimin | 24 |
| E2348-69 | Human EPEC | Intimin | Intimin | 13 |
| RDEC-1 | Rabbit EPEC | Intimin | Intimin | 3 |
Genes encoding the indicated virulence factors were determined previously by DNA colony blot hybridization.
Further isolates from the National Animal Disease Center and the Center for Vaccine Development were chosen to verify that the multiplex PCR could detect genes with a high level of agreement with colony blot hybridization (Table 3). The isolates tested included the following groups: a, 20 ETEC isolates from calves, pigs, or humans; b, 16 EPEC isolates from humans; c, 29 STEC isolates from calves or humans; d, 14 isolates representing various combinations of the six genes of interest; and e, 11 nonpathogenic E. coli isolates containing none of the six genes of interest. The multiplex PCR correctly detected the presence or absence of all genes of interest in all 90 isolates from groups a to e.
TABLE 3.
Results of multiplex PCR test on groups of E. coli representing different phenotypes
| Group | Phenotypic characterization | Serogroup | Host species | Genotypic characterizationa | PCR results (no. in agreement/no. tested) |
|---|---|---|---|---|---|
| a | ETEC | O101, O9 | Calf and pig | K99, F41, STa | 9/9 |
| O8 | Calf | K99, STa | 2/2 | ||
| O9 | Pig | F41, STa | 1/1 | ||
| O9, O20, O141, O148, O149, O78 | Pig and human | STa | 8/8 | ||
| b | EPEC | O55, O111, O119, O125, O126, O127, O128, O142 | Human | Intimin | 16/16 |
| c | STEC | O157:H7, O157 NM, O15, O17 | Calf and human | Intimin, Stx1, Stx2 | 19/19 |
| O5, O84, O103, O133 | Calf | Intimin, Stx1 | 10/10 | ||
| d | Various | O101 | Calf | F41 | 1/1 |
| O9 | Calf | F41, K99 | 1/1 | ||
| O133, OX3, O22 | Calf | Stx1, Stx2 | 3/3 | ||
| O98 | Pig | Stx1 | 1/1 | ||
| O5, O8, O171 | Calf | Stx2 | 3/3 | ||
| O139 | Pig | Stx2e | 1/1 | ||
| O8, O138 | Pig | Stx2e, STa | 3/3 | ||
| O119 | Calf and pig | Stx2, STa | 1/1 | ||
| e | Nonpathogens | Calf and pig | No genes of interest | 11/11 |
Genes encoding the indicated virulence factors were determined previously by DNA colony blot hybridization.
This multiplex PCR was highly specific, because it did not detect any genes which were determined to be absent by probing. The multiplex method was also highly sensitive, as it correctly detected all genes of interest in 100% (88 of 88) of the strains and isolates containing them. This assay will be useful in diagnostic situations for identification and characterization of E. coli isolated from calves with diarrhea. This multiplex PCR may also prove useful in detecting any of the genes of interest in E. coli from other host species.
Acknowledgments
This research was funded in part by Merck and Company, Inc., through the Merck Scholars program at Iowa State University and by the Frank K. Ramsey endowment.
We would like to thank Rob Schneider and James Kaper for providing the genotypically characterized strains.
REFERENCES
- 1.Braaten B A, Myers L L. Biochemical characteristics of enterotoxigenic and nonenterotoxigenic Escherichia coli isolated from calves with diarrhoea. Am J Vet Res. 1977;38:1989–1991. [PubMed] [Google Scholar]
- 2.Butler D G, Clarke R C. Diarrhoea and dysentery in calves. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 91–116. [Google Scholar]
- 3.Cantey J R, Inman L R. Diarrhea due to Escherichia coli strain RDEC-1 in the rabbit: the Peyer’s patch as the initial site of attachment and colonization. J Infect Dis. 1981;143:440–446. doi: 10.1093/infdis/143.3.440. [DOI] [PubMed] [Google Scholar]
- 4.China B, Pirson V, Mainil J. Typing of bovine attaching and effacing Escherichia coli by multiplex in vitro amplification of virulence-associated genes. Appl Environ Microbiol. 1996;62:3462–3465. doi: 10.1128/aem.62.9.3462-3465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cray W C, Jr, Moon H W. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean-Nystrom, E. A., B. T. Bosworth, W. C. Cray, Jr., and H. W. Moon. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. 65:1842–1848. [DOI] [PMC free article] [PubMed]
- 7.Fidock D A, McNicholas P A, Lehrbach P R. Nucleotide sequence of the F41 fimbriae subunit gene in Escherichia coli B41. Nucleic Acids Res. 1989;17:2849. doi: 10.1093/nar/17.7.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyles C L. Escherichia coli verotoxins and other cytotoxins. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 365–398. [Google Scholar]
- 9.Isaacson R E, Moon H W, Schneider R A. Distribution and virulence of Escherichia coli in the small intestines of calves with and without diarrhea. Am J Vet Res. 1978;39:1750–1755. [PubMed] [Google Scholar]
- 10.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7943. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karmali M A, Petric M, Louie S, Cheung R. Antigenic heterogenicity of Escherichia coli verotoxins. Lancet. 1986;i:164–165. doi: 10.1016/s0140-6736(86)92307-x. [DOI] [PubMed] [Google Scholar]
- 12.Krogh H V. Occurrence of enterotoxigenic Escherichia coli in calves with acute neonatal diarrhoea. Nord Vetmed. 1983;35:346–352. [PubMed] [Google Scholar]
- 13.Levine M M, Nalin D R, Hornick R B, Bergquist E J, Waterman D H, Young C R, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 14.Louie M, De Azavedo J, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mainil J G, Jacquemin E, Kaeckenbeeck A, Pohl P. Association between the effacing gene (eae) and the Shiga-like toxin-encoding genes in Escherichia coli isolates from cattle. Am J Vet Res. 1993;54:1064–1068. [PubMed] [Google Scholar]
- 16.Mainil J G, Sadowski P L, Tarsio M, Moon H W. In vivo emergence of enterotoxigenic Escherichia coli variants lacking genes for K99 fimbriae and heat-stable enterotoxin. Infect Immun. 1987;55:3111–3116. doi: 10.1128/iai.55.12.3111-3116.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon H W, Schneider R A, Mosley S C. Comparative prevalence of four enterotoxin genes among Escherichia coli isolated from swine. Am J Vet Res. 1986;47:210–212. [PubMed] [Google Scholar]
- 18.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris J A, Thorns C, Scott A C, Sojka W J, Wells G A. Adhesion in vitro and in vivo associated with an adhesive antigen (F41) produced by a K99 mutant of the reference strain Escherichia coli B41. Infect Immun. 1982;36:1146–1153. doi: 10.1128/iai.36.3.1146-1153.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orskov I, Orskov F, Smith H W, Sojka W J. The establishment of K99, a thermolabile, transmissible, Escherichia coli K antigen, previously called “Kco,” possessed by calf and lamb enterotoxigenic strains. Acta Pathol Microbiol Scand Sect B. 1975;83:31–36. doi: 10.1111/j.1699-0463.1975.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 21.Paton A W, Beutin L, Paton J C. Heterogeneity of the amino-acid sequences of Escherichia coli Shiga-like toxin type-I operons. Gene. 1995;153:71–74. doi: 10.1016/0378-1119(94)00777-p. [DOI] [PubMed] [Google Scholar]
- 22.Paton A W, Paton J C, Manning P A. Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb Pathog. 1993;15:77–82. doi: 10.1006/mpat.1993.1058. [DOI] [PubMed] [Google Scholar]
- 23.Roosendaal B, Gaastra W, de Graaf F K. The nucleotide sequence of the gene encoding the K99 subunit of enterotoxigenic Escherichia coli. FEMS Microbiol Lett. 1984;22:253–258. [Google Scholar]
- 24.Rothbaum R, McAdams A J, Gianella R, Partin J C. A clinicopathologic study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology. 1982;83:441–454. [PubMed] [Google Scholar]
- 25.Sekizaki T, Akashi H, Terakado N. Nucleotide sequences of the genes for Escherichia coli heat-stable enterotoxin I of bovine, avian, and porcine origins. Am J Vet Res. 1985;46:909–912. [PubMed] [Google Scholar]
- 26.Smith H W, Linggood M A. Further observations on Escherichia coli enterotoxins with particular regard to those produced by atypical piglet strains and by calf and lamb strains: the transmissible nature of these enterotoxins and of a K antigen possessed by calf and lamb strains. J Med Microbiol. 1972;5:243–250. doi: 10.1099/00222615-5-2-243. [DOI] [PubMed] [Google Scholar]
- 27.Strockbine N A, Marques L R M, Newland J W, Smith H W, Holmes R K, O’Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]