Abstract
Given the increasing incidence of yeast infections and the presence of drug-resistant isolates, accurate identification of the pathogenic yeasts is essential for the management of yeast infections. In this review, we tried to introduce the routine and novel techniques applied for yeast identification. Laboratory identification methods of pathogenic yeast are classified into three categories; I. conventional methods, including microscopical and culture-base methods II. biochemical/physiological-processes methods III. molecular methods. While conventional and biochemical methods require more precautions and are not specific in some cases, molecular diagnostic methods are the optimum tools for diagnosing pathogenic yeasts in a short time with high accuracy and specificity, and having various methods that cover different purposes, and affordable costs for researchers. Nucleotide sequencing is a reference or gold standard for identifying pathogenic yeasts. Since it is an expensive method, it is not widely used in developing countries. However, novel identification techniques are constantly updated, and we recommend further studies in this field. The results of this study will guide researchers in finding more accurate diagnostic method(s) for their studies in a short period of time.
Keywords: Candidiasis, diagnostic equipment, diagnostic techniques and procedures, early diagnosis, molecular diagnostic techniques, yeasts
INTRODUCTION
The incidence of invasive fungal infections (IFIs) is increasing in patients with underlying diseases.[1,2,3,4] Most of these infections are caused by Candida, Trichosporon, Geotrichum, Cryptococcus, and Rhodotorula.[5,6,7] Candida species are the main leading cause of IFIs in patients with various underlying diseases such as diabetes, cancer, immunodeficiency, corticosteroids, broad-spectrum antibiotic users, and organ transplants.[8,9] Trichosporon species are the second-leading cause of yeast infections in patients with blood disorders.[10,11] Rhodotorula is another pathogenic yeast associated with infections caused by intravenous catheters,[12] while Cryptococcus species are more commonly observed in HIV-positive patients.[13,14] Geotrichum species are also associated with IFIs in patients with severe immunodeficiency.[15] Depending on the yeast type, the mortality rate ranged from 15–80%.[16] In addition, the incidence of drug-resistant non-albicans yeasts rapidly rises.[17,18,19] Therefore, identifying pathogenic yeasts is the first step in controlling and managing IFIs.
Nucleotide sequencing of the ribosomal region of DNA and other known genomic regions for yeasts is considered the gold standard method for identifying yeast species.[20,21,22] But these methods are expensive and, in many cases, in developing countries such as Iran, are not affordable. Although biochemical and morphological methods are time-consuming and have lower accuracy, they are routinely used in developing countries.[20] These methods are not suggested for identifying emerging fungal species such as C. auris and C. haemulonii complex.[23,24,25,26,27] Manual biochemical techniques such as API 20C AUX or automatic biochemical methods are relatively more comprehensive, sensitive, and time-consuming.[28,29,30]
Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS), a novel method with high sensitivity and specificity, has also been introduced to identify yeasts; however, it is not available in many diagnostic centers.[20,31] Molecular methods such as conventional polymerase chain reaction (PCR), nested PCR, semi-nested PCR, multiplex PCR, and restriction fragment length polymorphism (RFLP) PCR are still routine diagnostic methods in Iran.[30,32,33,34,35] PCR-RFLP is a user-friendly, cost-benefit, and reliable method widely used in many studies.[36,37] Recently, multiplex PCR has been applied to identify yeasts with higher accuracy rates.[28,29,30] Here, we attempted to take a comprehensive look and review the advantages and disadvantages of the status of yeast diagnostic methods, especially in Iranian laboratories.
MORPHOLOGICAL DIAGNOSTIC METHODS
Various morphological methods have been proposed to identify the yeast species, including the morphology of yeast colonies, physiological tests such as germ tube production and chlamydoconidia formation, and specific and non-specific staining methods for yeasts.[20,38]
Germ tube production test
The germ tube production test has been popular for several decades to identify C. albicans as the most common human pathogenic yeast. This test can differentiate C. albicans within 2–4 hours of incubation in a special serum or protein-rich medium. Germ tubes are closely similar to pseudohyphae (in the early stages of their formation); therefore, an specialized technician is needed.[39,40] It is important to differentiate germ tubes from filaments that are constricted and represent pseudohyphae. False-negative results have been reported in C. albicans and can be caused by heavy inoculum. This test is also positive in C. dubliniensis; therefore, an additional test, such as beta-galactosaminidase or L-proline, is recommended to differentiate these two species.[41] The germ tube along with the chlamydoconidia test was used in previous studies but was replaced by newer methods due to low reliability in yeast identification.[42]
Chlamydospore formation
Although many fungi produce chlamydospores, they are also used to identify Candida species. More than 90% of the C. albicans and C. dubliniensis species produce chlamydospores in corn meal agar containing twine 80.[43] Similar to the germ tube production test, this test differentiates C. albicans and C. dubliniensis from the rest of the Candida species.[43]
Stains used for early detection and identification of yeasts
Yeast staining follows two main perspectives; first, to observe them in direct testing of clinical specimens such as fluids, tissues, skin samples, vaginal swabs, and stools. Second, direct smears from colonies grown in a culture medium.[44,45] Different staining methods are applied to identify yeasts, including saline solutions, potassium hydroxide 10%, lactophenol, methylene blue, and rarely white calcofluor. Yeasts can also be stained with Wright and Giemsa stains.[46] In cases suspected of Cryptococcus spp., Indian ink staining is recommended. This dye targets fungal capsule structure and has been used routinely in most reports of cryptococcosis from Iran.[47] It should be noted that some species of Rhodotrolla, Candida, and Trichosporon have been capsulated and may be confused with Cryptococcus spp. In these cases, serological tests such as glucuronoxylomannan are helpful for the differentiation of yeast infections.[48] Currently, the serological test is not available in most laboratories, so molecular methods are used for accurate identification.[49,50] The most common staining methods used for histopathological investigations are hematoxylin and eosin (H and E), Grocott-methenamine silver, and periodic acid-Schiff.[51,52]
BIOCHEMICAL TESTS
Wickerham method
The Wickerham method is based on the absorption and fermentation of sugars. Absorption activity shows the yeast's ability to use carbon or nitrogen. This method has been a powerful tool for yeast characterization and taxonomic studies in recent decades. The Wickerham media are not commercially available and require homemade preparation. Due to the possibility of contamination, the negative control should be used along with other samples. Application of this test is tedious, time-consuming, and often difficult to interpret; hence, it is not recommended unless other additional tests support the results of this method.[53]
Exanographic method
The dye-pour-plate auxanogram method is a practical approach to phenotypic identification. Carbohydrate assimilation reactions are among the primary tests used to differentiate genera and species of yeasts. This is a modified method of the Wickerham test, which needs agar media and employs a heavy inoculum of yeasts with a pH detector such as bromocresol purple in a carbohydrate medium tube. The application of color detectors and agar-based media makes it much easier to interpret the results.[54] Same as the Wickerham test, this method is not routine. Although these modifications appear to be satisfactory, they possess several disadvantages. They use pH indicators to detect the assimilation of carbohydrates. This requires buffering, modification, and follow-up of the pH of the medium. Misinterpreting test results may occur with the reversion of the pH indicator or from acid diffusion in the medium. In addition, some methods require the preparation of sterile carbohydrate or yeast nitrogen base carbohydrate-impregnated disks, which is time-consuming.[54]
DIFFERENTIAL AND SELECTIVE CULTURE MEDIA
A variety of differential culture media have been designed for the identification of yeasts. Differential media are used to identify and differentiate microorganisms from a closely related group with the help of unique growth patterns. Still, selective media are used to isolate a particular type of microorganism by giving a specific condition for the growth of that particular microorganism. Generally, selective media only allow the growth of a single microorganism, but differential media allow several closely related microorganisms to grow. Urea agar is the most common differential medium to identify basidiomycete yeasts (Cryptococcus spp., Malassezia spp., Rhodotorula spp., and Trichosporon spp.), especially Cryptococcus spp.[55] Niger seed agar, canavanine-glycine-boron thymol blue media (CGB), and L-dopa glucose media are applied to differentiate Cryptococcus spp.[56,57,58] Sabouraud dextrose agar with chloramphenicol and cycloheximide (SCC) is recommended for the primary isolation and cultivation of pathogenic yeasts and molds from clinical specimens.[46] However, differential and selective media are generally used for primary isolation and identification of pathogenic yeasts and are not recommended to identify yeast species. In the first step, these culture media have been used for yeast isolation from clinical and environmental samples to purify colonies. Then advanced methods are used for final confirmation.[47,59,60]
CHROMOGENIC CULTURE MEDIA
A wide range of chromogens are used for their enzymatic activities. Most of them have hydrolytic activities leading to color production. In addition, they have been used for identifying drug-resistant strains.[61] Albicans ID2, CHROMagar™ Candida, and Fluoroplate Candida are well-known chromogenic media.[39] CHROMagar™ Candida is the most common commercial chromogenic medium. It accurately identifies common Candida spp., such as C. albicans, C. parapsilosis, C. krusei, C. tropicalis, and C. glabrata, according to their different color patterns [Figure 1].[62,63] The application of chromogenic culture media in Iran has increased due to their cost and time-consuming aspects. Another application of these media is their ability to detect multiple yeast infections[64] Chromogenic media show better detection ability of mixed yeast cultures than traditional media and allow the direct identification of C. albicans by means of colony color. Although chromogenic media are more expensive than conventional mycological media, they are used for reaching rapid results.[39] However, they have several limitations, such as detecting limited species and similar color patterns for several species.[65] For example, C. lipolytica, C. norvegensis, and C. inconspicua are indistinguishable from C. krusei in the CHROMagar™ Candida.[63,66] However, chromogenic media are a suitable choice for laboratories that do not have a better option for pathogenic yeast identification [Table 1].
Figure 1.
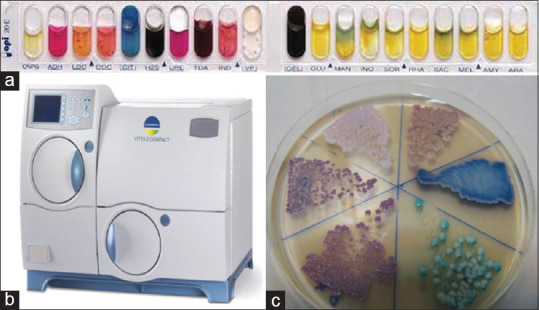
Some common biochemical methods for the identification of yeasts. (a) Biochemical media API 20C. A commercial system for the identification of yeasts from clinical specimens (b) The automated VITEK 2 YST ID Card system is a new yeast identification and susceptibility testing system that uses fluorescence-based technology (c) CHROMagar™ Candida Media: C. albicans, C. glabrata, C. tropicalis, and C. krusei have differed in green, purple, blue, and pink colors, respectively
Table 1.
Summary of some identification/detection methods for yeast species
| Author (year) | Identification/detection method | Source of yeast isolates | No. of isolates | Yeast species | Cost | Time | Sensitivity, specificity, PPV1, and NPV2 | Ref |
|---|---|---|---|---|---|---|---|---|
| Süleyha (2007) | Germ tube production | Clinical isolates | 157 | C. albicans | Low | 2–4 h | 98%, 100%, 100% and 92.3% | [124] |
| Perera (2004) | API ID 32C | Clinical isolates | 50 | C. dubliniensis | High | 24–72 h | Sensitivity: 96% | [125] |
| Bernal (1998) | API 20C | Clinical isolates | 198 | Candida species | High | 24–72 h | Sensitivity: With additional test: 96.3% Without additional test: 71.7% | [126] |
| Letscher (2002) | CHROMagar | Clinical isolates | 786 | C. albicans | Low | 24–4 h | Sensitivity: 97.7% | [127] |
| Yucesoy (2003) | CHROMagar | Various clinical specimens | 238 | C. albicans, C. glabrata, C. krusei and C. tropicalis | Low | 24–48 h | 98.3%, 100%, 100%, and 97.5 | [128] |
| Jafari (2017) | CHROMagar and PCR-RFLP | Standard strain and clinical isolates | 107 | Clinical Candida strains | Low | 24–48 h | Sensitivity for CHROMagar and PCR-RFLP was 33.3% and 95.4%, respectively | [66] |
| Kord (2021) | Multiplex PCR and PCR-RFLP | Clinical isolates | 173 | Candida species, Cryptococcus, Trichosporon and Saccharomyces | High | 4–5 h | Sensitivity: PCR-RFLP: 87.3% 21-plex PCR: 96.5%, | [28] |
| Romeo (2009) | Multiplex PCR | Clinical isolates | 81 | C. glabrata, C. nivariensis, and C. bracarensis | High | 4–5 h | Sensitivity: 100% Specificity: 100% | [129] |
| Kord (2020) | VITEK 2 | Blood culture | 137 | Candida species, Cryptococcus, Trichosporon | High | 16–24 h | Sensitivity: 87.6% | [30] |
| Arastehfar (2019) | rDNA sequencing, 21-plex PCR, API 20C AUX and MALDI-TOF | Clinical yeast isolates | 301 | Candida, Cryptococcus, Trichosporon, Saccharomyces, Rhodotorula | High | - | The sensitivity for rDNA sequencing, 21-plex PCR, API 20C AUX and MALDI-TOF was 100%, 88.7%, 83.7 and 98.3%, respectively | [130] |
| Wang (2014) | Real-Time PCR (18S rRNA) | Blood | 125 | C. albicans, C. parapsilosis, C. tropicalis, C. glabrata | High | 4–5 h | Sensitivity: 100% Specificity: 90% | [131] |
| Nieto (2019) | PCR (ITS1/2 rDNA) | Serum | 181 | C. albicans, C. parapsilosis, C. tropicalis, C. glabrata, C. krusei and C. guiliermondii | High | 4–5 h | Sensitivity: 16% Specificity: 93% | [132] |
| White (2005) | Real-Time PCR (18S rRNA) | Blood | 105 | Candida species | High | 4–5 h | Sensitivity: 95% Specificity: 97% PPV: 90.5% NPV: 98.5% | [133] |
| Klingspor (2006) | Real-Time PCR | Clinical specimens | 1650 | Candida species | High | 4–5 h | Specificity: 100% | [134] |
| Mark (2011) | Lateral Flow Antigen (LFA) | Serum | 704 | Cryptococcus | High | 30–60 min | Sensitivity: 100% | [135] |
| Tay (2022) | Real-Time PCR (cytochrome b gene) | Fresh frozen paraffin embedded specimens | 25 | Cryptococcus | High | 4–6 h | Sensitivity: 96.4% Specificity: 100% | [136] |
| Odabasi (2004) | β-D-glucan (BG) | Serum | 283 | Candida | High | 2–3 h | NPV: 100% Specificity: 90% | [137] |
| Fontana (2012) | Galactomannan + β-D-glucan (BG) | Serum | 46 | IFI (Candidiasis) | High | 2–3 h | 95.8%, 54.5%, 69.7% and 92.3%. | [138] |
| Ashrafi (2015) | Real-Time PCR | Blood | 72 | Candida | High | 4–5 h | Specificity: 100% | [139] |
| Shepard (2008) | FISH3 | Blood isolates | 197 | C. albicans and C. glabrata | High | 4–5 h | Sensitivity for C. albicans and C. glabrata were 98.7% and 100%. | [140] |
| Spanu (2012) | MALDI-TOF | Blood sample | 340 | Candida species | High | 30–60 min | Sensitivity: 95.9% Specificity: 100% | [141] |
1Positive predictive value. 2Negative predictive value. 3Fluorescence in situ hybridization
COMMERCIAL BIOCHEMICAL KITS
These systems should have the following characteristics; I. they should be fast, accurate, and user-friendly. II. applicable for detecting different types of yeasts in various clinical specimens. III. Identification of some rare yeast species. One of these systems is the commercial kit ID 32C (bioMe’rieux, France), commonly used in European countries. Newer methods, including the API 20C and the VITEK automated system [Figure 1], more widely used in the US.[67] The abilities of the API 20C and ID 32C yeast identification systems to identify 123 common and 120 rare clinical yeast isolates were compared. API 20C facilitated the correct identification of 97% common and 88% rare isolates, while ID 32C facilitated the correct identification of 92% common and 85% rare isolates.[67] Commercial biochemical kits have not been welcomed in Iran due to their high cost. However, some researchers applied these methods to their studies[68,69] [Table 1].
The VITEK (VITEK-2 YST ID) automatic system, currently used in a limited number of centers in Iran, is one of the biochemical systems for identifying more than 50 yeast species [Figure 1]. Also, the VITEK-2 is a popular system in developed countries, especially the US.[70] One limitation of this system is the possibility of misdiagnosing some emerging and/or rare species following a false interpretation of the results based on data recorded in the system's database. In a study conducted by Kord et al. on yeast agents isolated from blood infections in two large hospitals in Tehran using the VITEK-2 system, this method detected 92% of common species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei) and 50% of uncommon species.[30] However, with the passage of time and the updating of information in this system, we see more sensitivity and accuracy in identifying yeasts, especially emerging yeast pathogens.[71] Since this system is able to detect and identify both yeasts and types of bacteria, performing drug susceptibility tests on yeasts and bacteria is a proper choice for diagnostic centers in Iran. However, the high device price and ID cards limited its application [Table 1].
MOLECULAR METHODS IN THE IDENTIFICATION OF YEASTS
Recent developments in molecular biology, for example, PCR and DNA sequencing, have provided rapid and accurate tools for the identification of yeasts. We will introduce molecular methods and mechanisms underlying their activities in the following.
Identification of yeasts based on DNA sequencing
The most important genetic markers that are used for the identity of yeasts include: the internal transcribed spacer (ITS)—the primary fungal barcode,[21] translational elongation factor 1α (TEF1α)—the secondary fungal barcode,[72] the D1/D2 domain of the LSU ribosomal RNA,[73] the RNA polymerase II subunit RPB1, and the intergenic spacer (IGS) of ribosomal RNA genes.[74] Ribosomal DNA subunits contain protected domains separated by variable domains representing species-specific sequences, which are the primary targets for designing species-specific primer pairs for differentiation. rDNA contains genes encoded by small ribosomal subunits, including 16S and 5.8S, and the large ribosomal subunit, 26S. ITS separates these three regions.[75] One significant advantage of the rDNA genes in the molecular diagnosis of fungi is the high number of copies of this region (50–100 copies) in the fungal genome, which increases the sensitivity of the molecular methods. Moreover, data from the National Center for Biotechnology Information (NCBI) and Central Bureau Voor Schimmel cultures (CBS) gene banks allow for comparison and accurate identification of the results of the molecular marker. Panfungal primers have been designed for this aim. In Iran, the ITS1-5.8S-ITS2 and D1/D2 primer pairs are the most useful.[29,76,77,78] Although sequencing is the reference and/or gold standard method for fungal identification, it requires an advanced laboratory instrument (a sequencer) and an experienced technician. But considering all the limitations of other methods, it seems necessary to widely use DNA sequencing for emerging multidrug resistance yeast species in the future[79] [Table 1].
Species-specific primers
Some of the primer pairs are specific to a single species; however, different species can be identified by the same primer pairs.[80] These primers are also used to identify yeasts with similar characteristics. Nikmanesh et al.[81] used hyphal wall protein 1 (HWP1)-specific primers to differentiate C. albicans, C. africana, and C. dubliniensis species. Shirani et al. identified C. albicans, C. glabrata, C. parapsilosis, and C. krusei in patients with vulvovaginitis.[82] Species-specific primers can also identify yeasts directly from clinical specimens.[39] A significant limitation of this method is identifying a limited number of yeast species that cause the disease. Therefore, they are accommodating in identifying more common species. It is suggested to apply for specific purposes, such as tracking a particular species or detecting specific genes in yeast species isolated from the patient. Using specific primers is a cost-benefit and requires less time than genome sequencing. Specific primers are potent tools with high sensitivity for proving the fungal infection in a clinical specimen, but depending on the target probe, they may have different specificities. This method can also identify emerging species, for example, C. auris, in clinical samples.[83,84,85]
Multiplex PCR
This method uses different primers simultaneously to identify several yeast species.[86] The first step is knowing the target species in the target specimen, for example, yeasts that cause bloodstream infections. The main advantages of this method are fastness, cost-beneficent, high accuracy, and ease of interpreting the results. The main limitation is that it can be designed for a limited range of yeasts. However, multiplex PCR has been used in some studies to identify a wide range of pathogenic yeasts. Arastehfar et al. developed a hybrid multiplex PCR to identify 21 yeast species without sequencing as a reference method.[29] Although multiplex PCR is less efficient than the methods that have the ability to identify a wide range of yeasts; however, sometimes the result is the opposite. Kord et al. compared the results of multiplex PCR and the biochemical VITEK-2 system and concluded that the multiplex is more accurate in identifying yeast agents isolated from bloodstream fungal infections.[30] It is recommended that multiplex PCR can be an alternative method for DNA sequencing in Iran. New markers can be included in the multiplex panels according to the epidemiology of the geographical region. Multiplex PCR is also useful for identifying and differentiating the fungal agents that cause the fungal mycotic infection with the same clinical manifestations[87,88] [Table 1].
PCR-RFLP
During PCR-RFLP, the targeted gene locus is amplified via a conventional PCR tool, and then the PCR products are exposed to restriction enzymes. The resulting genomic fragments have specific patterns for each species on the electrophoretic results. Depending on the target gene region for amplification with PCR and the restriction enzyme used for RFLP, almost more than 20 yeast species can be identified.[37,89] Although it has some limitations, such as the similarity of the electrophoretic patterns of some species, PCR-RFLP has been widely used in Iran.[90,91] Another limitation is that it is more time-consuming than other molecular methods. This method can also not be used to identify all pathogenic yeasts. Mirhendi et al. typed six common species of Candida yeasts using the MspI restriction enzyme.[37] PCR-RFLP is also used for yeasts that are difficult to detect by conventional mycological methods, for example, Malassezia spp. Didehdar et al. identified Malassezia yeasts isolated from pityriasis versicolor by PCR-RFLP.[36] Sometimes this method is also helpful in identifying and differentiating species with the same characteristics using other identical methods. For example, Mirhendi et al. used the BlnI enzyme to distinguish between C. albicans and C. dubliniensis, which have common morphological, biochemical, and molecular properties.[89] This technique, along with multiplex PCR, is a suitable option for the molecular identification of yeasts in medical diagnostic or research laboratories with limited facilities. The existence of similar electrophoretic patterns in some species and the misidentification of some yeast-related species are other limitations of the PCR-RFLP method[28] [Table 1].
Real-Time PCR
In this method, the quantitative value (curve threshold, or CT) of the amplified target gene is measured and reported after each PCR cycle [Figure 2]. Despite the conventional PCR, which is reported at the end of the reaction, a fluorescent substrate previously added to the PCR master mix acts as the reaction reporter. As the reaction progresses and the amount of amplified target gene is increased, the photodynamic reaction is intensified by the fluorescence substrate and detected by the device's sensors, eventually being reported as specific curves. Results in real-time PCR can be qualitative (presence or absence of a sequence) or quantitative (number of target sequence copies). In other words, real-time PCR is a quantitative PCR (qPCR). Real-time platforms enable quantification of fungal load in the clinical specimen, which may provide information about the burden or progression of the disease. Compared to different types of PCR, the main advantage of this method is its higher sensitivity (especially in negative culture results) and shorter reaction time. Badiei et al. detected Candida and Aspergillus spp. in CSF samples via real-time PCR, although 80% of these specimens were culture-negative.[92] However, this method is costly and requires experienced technicians[93,94] [Table 1].
Figure 2.
MALDI-TOF and real-time PCR methods known as two valid molecular methods for yeast identification. (a) MALDI-TOF instrument. A rapid microbiological method that can be used to identify a wide variety of microorganisms (b) Development of Candida-Specific real-time PCR assays for the detection and identification of eight medically important Candida species
Random amplified polymorphic DNA (RAPD)-PCR
RAPD-PCR, also known as arbitrarily primed PCR (AP-PCR), is a rapid method of DNA fingerprinting. RAPD-PCR is a technique that allows the study of gene polymorphisms in a target population of organisms. This method is based on the production of amplitudes by oligonucleotides that are randomly attached to the target fragment. The fragments are randomly amplified and scanned. The resulting pattern provides a unique profile of the RAPD reaction based on the organism's characteristics under study. This tool is an excellent way to analyze genetic polymorphisms in bacteria, fungi, plants, and humans. The advantages of RAPD include its appropriateness to analyze unknown genomes, its efficiency, affordability, and quickness, and the fact that it requires less expertise in conducting analysis.[95] Gene polymorphism assay tools such as RAPD-PCR, amplified fragment length polymorphism (AFLP)-PCR, and microsatellites provide a powerful option in the classification and study of genotypic variants among species. Some studies have been conducted using RAPD-PCR in Iran. Fahami et al. analyzed the primary origin of the yeast isolated from hospitalized patients with cancer using this RAPD-PCR.[96] The accuracy of species identification with RAPD-PCR can be improved by combining RAPD with other genotypic methods, such as RFLP. Poor reproducibility of fingerprints and the requirement for strict standardization of reaction parameters are the limitations of RAPD.[97]
Multilocus sequence typing (MLST)
MLST is a genome sequencing method for several gene regions simultaneously. Sequencing results obtained from each gene locus are known as an allele in a yeast species, and the alleles obtained at each locus are collectively called an allelic profile or a sequence type. Therefore, each isolate contained several allelic characters. MLST is a method based on nucleotide sequences, and its underlying technology has acceptable efficiency and reliability. This method facilitates the exchange of data obtained from molecular typing between research laboratories via a database and leads to the development of epidemiological studies worldwide. In this regard, databases for several organisms are now available online at www.mlst.net, allowing the typing results of species management and integration anywhere.[98] Afsarian et al. studied seven different gene regions (2883 nucleotides per species) and detected 71 nucleotide positions (2.5%) of variations in C. albicans isolates.[98] They recorded 18 sequence types from seven loci of thirty C. albicans isolates. This technique facilitates epidemiological studies worldwide. The patterns of transfer and evolution of yeasts are revealed.
MALDI-TOF MS
MS is an analytical method based on the ionization of crystallized materials by short laser pulses. The ions accelerate, and their escape time is measured in a vacuum tube [Figure 2]. Various detection methods have been designed and developed based on ionization techniques. The most common biomolecule analysis method is laser adsorption/ionization-time using a flight or escape MS matrix. MALDI-TOF has been used to determine the mass of proteins and peptides, identify unknown proteins, and diagnose tumors, rheumatoid arthritis, Alzheimer's, and allergies.[99] In recent years, MALDI-TOF has also been used in research laboratories and has proposed a new approach to identifying bacteria and yeasts. This tool is a quick, accurate, and easy method of identifying filamentous and yeast fungi.[100] Another advantage of this method is the possibility of using it as a powerful tool and an alternative to gold standard methods for less common and rare yeasts. Aslani et al. identified rare yeasts such as Saccharomyces cerevisiae, C. lusitaniae, and Pichia coliuri from oral lesions of cancer patients with the MALDI-TOF method.[101] This method has been used in several studies to identify common and rare yeasts.[102,103,104,105] This system has been used in a limited number of research laboratories in the country but is limited with less access to databases and high prices [Table 1].
CONCLUDING REMARKS AND PERSPECTIVES
The prevalence of yeast infections increases with the escalation of leading risk factors. The main risk factors are immunosuppression and/or immunosuppressive treatments, long-term and severe neutropenia, broad-spectrum antibiotics, corticosteroids, failure of the mechanical and/or mucosal barriers, underlying diseases, for example, diabetes, and epidemics, for example, COVID-19.[1,2,77,106] The most prevalent yeast infection is candidiasis.[107] Candidiasis is a spectrum of opportunistic infections with various clinical forms caused by Candida species.[108,109] It is reported that systemic candidiasis (SC) involves more than 700,0000 cases per year worldwide.[110] Also, SC is the fourth most common bloodstream infection in the US and contains almost 55% of all fungal infections.[111,112,113,114,115] Moreover, Candida species are considered the most frequent fungi isolated from patients with malignancy,[110,116,117] the second causative agent of catheter-associated urinary tract infections, and the third pathogenic organism responsible for catheter-associated and pediatric sepsis.[118,119,120] Although C. albicans is the most prevalent causative agent,[107] other species include C. glabrata, C. krusei, C. parapsilosis, C. tropicalis, C. kefyr, C. dubliniensis, C. lusitaniae, and emerging multidrug-resistant C. auris are also important. Infections by non-Candida yeasts such as Cryptococcus, Geotrichum, and Trichosporon are also increasing.[121,122] Moreover, emerging multidrug-resistant (MDR) species are becoming a global concern. Identification of the MDR isolates is a great challenge. These problems increase the need to level up and apply diagnostic laboratory tests.[1]
Conventional methods need experienced medical mycologists in addition to their low accuracy. Biochemical methods require more precautions and are not specific in some cases. Identification using chromogenic media is easy and accessible and does not require an experienced technician. However, they are designed for a limited number of yeasts, and in some cases, the results are similar. Despite these limitations, chromogenic detection methods are a good choice for diagnostic centers without advanced molecular facilities and devices. Molecular diagnostic methods are the optimum tool for diagnosing pathogenic yeasts quickly, with high accuracy and specificity.[123] In addition to various methods that cover different purposes, they have affordable costs for Iranian researchers.
This review tried to introduce the techniques applied for yeast identification. The data of this study will guide researchers in finding more accurate diagnostic method(s) for their studied isolates in a short period of time.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors are grateful to kindly support of the Medical Mycology laboratories of Tehran, Shiraz, Zanjan, and Sanandaj Universities of Medical Sciences.
REFERENCES
- 1.Enoch DA, Ludlam HA, Brown NM. Invasive fungal infections: A review of epidemiology and management options. J Med Microbiol. 2006;55:809–18. doi: 10.1099/jmm.0.46548-0. [DOI] [PubMed] [Google Scholar]
- 2.Gullo A. Invasive fungal infections: The challenge continues. Drugs. 2009;69((Suppl 1)):65–73. doi: 10.2165/11315530-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Jenks JD, Cornely OA, Chen SCA, Thompson GR, 3rd, Hoenigl M. Breakthrough invasive fungal infections: Who is at risk? Mycoses. 2020;63:1021–32. doi: 10.1111/myc.13148. [DOI] [PubMed] [Google Scholar]
- 4.Ramana KV, Kandi S, Venkata BP, Sharada CHV, Rao R, Mani R, et al. Invasive fungal infections: A comprehensive review. Am J Infect Dis. 2013;1:64–9. [Google Scholar]
- 5.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, et al. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010;48:1366–77. doi: 10.1128/JCM.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–51. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 7.Richardson M, Lass-Flörl C. Changing epidemiology of systemic fungal infections. Clin Microbiol Infect. 2008;14((Suppl 4)):5–24. doi: 10.1111/j.1469-0691.2008.01978.x. [DOI] [PubMed] [Google Scholar]
- 8.Mavor AL, Thewes S, Hube B. Systemic fungal infections caused by Candida species: Epidemiology, infection process and virulence attributes. Curr Drug Targets. 2005;6:863–74. doi: 10.2174/138945005774912735. [DOI] [PubMed] [Google Scholar]
- 9.Zarrin M, Mahmoudabadi AZ. Invasive candidiasis; A review article. Jundishapur Journal of Microbiology. 2009;2:1–6. [Google Scholar]
- 10.Colombo AL, Padovan ACB, Chaves GM. Current knowledge of Trichosporon spp.and Trichosporonosis. Clin Microbiol Rev. 2011;24:682–700. doi: 10.1128/CMR.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Almeida Júnior JN, Hennequin C. Invasive Trichosporon infection: A systematic review on a re-emerging fungal pathogen. Front Microbiol. 2016;7:1629. doi: 10.3389/fmicb.2016.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuon FF, Duboc de Almeida GM, Costa SF. Central venous catheter-associated fungemia due to Rhodotorula spp.–A systematic review. Med Mycol. 2007;45:441–7. doi: 10.1080/13693780701381289. [DOI] [PubMed] [Google Scholar]
- 13.Chayakulkeeree M, Perfect JR. Cryptococcosis. Infect Dis Clin North Am. 2006;20:507–44. doi: 10.1016/j.idc.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Antinori S. New insights into HIV/AIDS-associated cryptococcosis. ISRN AIDS. 2013;2013:471363. doi: 10.1155/2013/471363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao G-X, Tang H-L, Zhang X, Xin X-L, Feng J, Chen X-Q. Invasive fungal infection caused by Geotrichum capitatum in patients with acute lymphoblastic leukemia: A case study and literature review. Int J Clin Exp Med. 2015;8:14228–35. [PMC free article] [PubMed] [Google Scholar]
- 16.Cleveland AA, Harrison LH, Farley MM, Hollick R, Stein B, Chiller TM, et al. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008–2013: Results from population-based surveillance. PLoS One. 2015;10:e0120452. doi: 10.1371/journal.pone.0120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kołaczkowska A, Kołaczkowski M. Drug resistance mechanisms and their regulation in non-albicans Candida species. J Antimicrob Chemother. 2016;71:1438–50. doi: 10.1093/jac/dkv445. [DOI] [PubMed] [Google Scholar]
- 18.Hii I-M, Liu C-E, Lee Y-L, Liu W-L, Wu P-F, Hsieh M-H, et al. Resistance rates of non-albicans Candida infections in Taiwan after the revision of 2012 Clinical and Laboratory Standards Institute breakpoints. Infect Drug Resist. 2019;12:235–40. doi: 10.2147/IDR.S184884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadeghi G, Ebrahimi-Rad M, Mousavi SF, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. Emergence of non-Candida albicans species: Epidemiology, phylogeny and fluconazole susceptibility profile. J Mycol Med. 2018;28:51–8. doi: 10.1016/j.mycmed.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Arastehfar A, Wickes BL, Ilkit M, Pincus DH, Daneshnia F, Pan W, et al. Identification of mycoses in developing countries. J Fungi (Basel) 2019;5:90. doi: 10.3390/jof5040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaw SN, Chang HC, Sun HF, Barton R, Bouchara J-P, Chang TC. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J Clin Microbiol. 2006;44:693–9. doi: 10.1128/JCM.44.3.693-699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linton CJ, Borman AM, Cheung G, Holmes AD, Szekely A, Palmer MD, et al. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom Mycology Reference Laboratory. J Clin Microbiol. 2007;45:1152–8. doi: 10.1128/JCM.02061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol. 2017;55:2445–52. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Černáková L, Roudbary M, Brás S, Tafaj S, Rodrigues CF. Candida auris: A quick review on identification, current treatments, and challenges. Int J Mol Sci. 2021;22:4470. doi: 10.3390/ijms22094470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vatanshenassan M, Boekhout T, Meis JF, Berman J, Chowdhary A, Ben-Ami R, et al. Candida auris identification and rapid antifungal susceptibility testing against echinocandins by MALDI-TOF MS. Front Cell Infect Microbiol. 2019;9:20. doi: 10.3389/fcimb.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, et al. Multidrug-resistant candida haemulonii and C.Auris, tel aviv, Israel. Emerg Infect Dis. 2017;23:195–203. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arastehfar A, Fang W, Badali H, Vaezi A, Jiang W, Liao W, et al. Low-cost tetraplex PCR for the global spreading multi-drug resistant fungus, Candida auris and its phylogenetic relatives. Front Microbiol. 2018;9:1119. doi: 10.3389/fmicb.2018.01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kord M, Elmimoghaddam A, Hashemi SJ, Reziae S, Daie Ghazvini R, Salehi M, et al. Comparison of PCR-RFLP with 21-plex PCR and rDNA sequencing for identification of clinical yeast isolates. Mycopathologia. 2021;186:213–20. doi: 10.1007/s11046-020-00522-0. [DOI] [PubMed] [Google Scholar]
- 29.Arastehfar A, Daneshnia F, Kord M, Roudbary M, Zarrinfar H, Fang W, et al. Corrigendum: Comparison of 21-Plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA sequencing for a wide range of clinically isolated yeast species: Improved identification by combining 21-Plex PCR and API 20C AUX as an alternative strategy for developing countries. Front Cell Infect Microbiol. 2019;9:176. doi: 10.3389/fcimb.2019.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kord M, Salehi M, Khodavaisy S, Hashemi SJ, Ghazvini RD, Rezaei S, et al. Epidemiology of yeast species causing bloodstream infection in Tehran, Iran (2015–2017); superiority of 21-plex PCR over the Vitek 2 system for yeast identification. J Med Microbiol. 2020;69:712–20. doi: 10.1099/jmm.0.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader O. MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics. 2013;13:788–99. doi: 10.1002/pmic.201200468. [DOI] [PubMed] [Google Scholar]
- 32.Hasanvand S, Qomi HA, Kord M, Didehdar M. Molecular epidemiology and in vitro antifungal susceptibility of candida isolates from women with vulvovaginal candidiasis in northern cities of Khuzestan province, Iran. Jundishapur Journal of Microbiology. 2017;10:e12804. [Google Scholar]
- 33.Ashraf MJ, Kord M, Morovati H, Ansari S, Shekarkhar G, Badali H, et al. Evaluating a semi-nested PCR to support histopathology reports of fungal rhinosinusitis in formalin-fixed paraffin-embedded tissue samples. J Clin Lab Anal. 2022;36:e24209. doi: 10.1002/jcla.24209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005;61:281–4. doi: 10.1016/j.mimet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Mahmoudabadi AZ, Zarrin M, Azish M. Detection of Malassezia species isolated from patients with pityriasis versicolor and seborrheic dermatitis using Nested-PCR. Jentashapir Journal of Health Research. 2014;5:e26683. [Google Scholar]
- 36.Didehdar M, Mehbod ASA, Eslamirad Z, Mosayebi M, Hajihossein R, Ghorbanzade B, et al. Identification of Malassezia species isolated from patients with pityriasis versicolor using PCR-RFLP method in Markazi Province, Central Iran. Iran J Public Health. 2014;43:682–6. [PMC free article] [PubMed] [Google Scholar]
- 37.Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H. A one-enzyme PCR-RFLP assay for identification of six medically important Candida species. Nihon Ishinkin Gakkai Zasshi. 2006;47:225–9. doi: 10.3314/jjmm.47.225. [DOI] [PubMed] [Google Scholar]
- 38.Lin CC, Fung DY. Conventional and rapid methods for yeast identification. Crit Rev Microbiol. 1987;14:273–89. doi: 10.3109/10408418709104441. [DOI] [PubMed] [Google Scholar]
- 39.Pincus DH, Orenga S, Chatellier S. Yeast identification—past, present, and future methods. Med Mycol. 2007;45:97–121. doi: 10.1080/13693780601059936. [DOI] [PubMed] [Google Scholar]
- 40.Gow NA. Germ tube growth of Candida albicans. Curr Top Med Mycol. 1997;8:43–55. [PubMed] [Google Scholar]
- 41.Heelan JS, Siliezar D, Coon K. Comparison of rapid testing methods for enzyme production with the germ tube method for presumptive identification of Candida albicans. J Clin Microbiol. 1996;34:2847–9. doi: 10.1128/jcm.34.11.2847-2849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liguori G, Di Onofrio V, Gallé F, Lucariello A, Albano L, Catania MR, et al. Candida albicans identification: Comparison among nine phenotypic systems and a multiplex PCR. J Prev Med Hyg. 2010;51:121–4. [PubMed] [Google Scholar]
- 43.Staib P, Morschhäuser J. Chlamydospore formation in Candida albicans and Candida dubliniensis–an enigmatic developmental programme. Mycoses. 2007;50:1–12. doi: 10.1111/j.1439-0507.2006.01308.x. [DOI] [PubMed] [Google Scholar]
- 44.Wilson GS, Miles AA. Topley and Wilson's principles of bacteriology and immunity. London: Edward Arnold Publishers; 1955. [Google Scholar]
- 45.Harrigan WF. Topley and Wilson's Microbiology and Microbial Infections, 9th edn. International Journal of Food Science and Technology. 1998;33:191–7. [Google Scholar]
- 46.Topley W. Topley and Wilson's Microbiology and Microbial Infections, 8 Volume Set [Google Scholar]
- 47.Bandalizadeh Z, Javidnia J, Hosseini SA, Moosazadeh M, Amouei A, Kermani F, et al. Cryptococcus and cryptococcosis in Iran during 1969–2019: A systematic review and meta-analysis. J Mycol Med. 2020;30:100917. doi: 10.1016/j.mycmed.2019.100917. [DOI] [PubMed] [Google Scholar]
- 48.Probert M, Zhou X, Goodall M, Johnston SA, Bielska E, Ballou ER, et al. A glucuronoxylomannan epitope exhibits serotype-specific accessibility and redistributes towards the capsule surface during titanization of the fungal pathogen Cryptococcus neoformans. Infect Immun. 2019;87:e00731–18. doi: 10.1128/IAI.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moslem M, Fatahinia M, Kiasat N, Mahmoudabadi AZ. Predominance of Cryptococcus neoformans var.grubii in Ahvaz, molecular identification and evaluation of virulence factors. Jundishapur Journal of Microbiology. 2020;13:e112408. [Google Scholar]
- 50.Bandalizadeh Z, Shokohi T, Badali H, Abastabar M, Babamahmoudi F, Davoodi L, et al. Molecular epidemiology and antifungal susceptibility profiles of clinical Cryptococcus neoformans/Cryptococcus gattii species complex. J Med Microbiol. 2020;69:72–81. doi: 10.1099/jmm.0.001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Principles C. CLSI Document M54-A Wayne. PA, USA: CLSI; 2012. Procedures for Detection of Fungi in Clinical Specimens-Direct Examination and Culture; Approved Guideline. [Google Scholar]
- 52.Guarner J, Brandt ME. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev. 2011;24:247–80. doi: 10.1128/CMR.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickerham LJ. A critical evaluation of the nitrogen assimilation tests commonly used in the classification of yeasts. J Bacteriol. 1946;52:293–301. doi: 10.1128/JB.52.3.293-301.1946. [DOI] [PubMed] [Google Scholar]
- 54.Mickelsen PA, McCarthy LR, Propst MA. Further modifications of the auxanographic method for identification of yeasts. J Clin Microbiol. 1977;5:297–301. doi: 10.1128/jcm.5.3.297-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts GD, Horstmeier CD, Land GA, Foxworth JH. Rapid urea broth test for yeasts. J Clin Microbiol. 1978;7:584–8. doi: 10.1128/jcm.7.6.584-588.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein KR, Hall L, Deml SM, Rysavy JM, Wohlfiel SL, Wengenack NL. Identification of Cryptococcus gattii by use of L-canavanine glycine bromothymol blue medium and DNA sequencing. J Clin Microbiol. 2009;47:3669–72. doi: 10.1128/JCM.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min KH, Kwon-Chung KJ. The biochemical basis for the distinction between the two Cryptococcus neoformans varieties with CGB medium. Zentralbl Bakteriol Mikrobiol Hyg A. 1986;261:471–80. doi: 10.1016/s0176-6724(86)80079-7. [DOI] [PubMed] [Google Scholar]
- 58.Nishikawa MM, Sant’Anna OD, Lazera MS, Wanke B. Use of D-proline assimilation and CGB medium for screening Brazilian Cryptococcus neoformans isolates. J Med Vet Mycol. 1996;34:365–6. [PubMed] [Google Scholar]
- 59.Pakshir K, Fakhim H, Vaezi A, Meis JF, Mahmoodi M, Zomorodian K, et al. Molecular epidemiology of environmental Cryptococcus species isolates based on amplified fragment length polymorphism. J Mycol Med. 2018;28:599–605. doi: 10.1016/j.mycmed.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Hedayati MT, Mayahi S, Fakhar M, Shokohi T, Majidi M. Cryptococcus neoformans isolation from swallow (Hirundo rustica) excreta in Iran. Rev Inst Med Trop Sao Paulo. 2011;53:125–7. doi: 10.1590/s0036-46652011000300002. [DOI] [PubMed] [Google Scholar]
- 61.Perry JD, Freydiere AM. The application of chromogenic media in clinical microbiology. J Appl Microbiol. 2007;103:2046–55. doi: 10.1111/j.1365-2672.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- 62.Kooshki P, Rezaei-Matehkolaei A, Mahmoudabadi AZ. The patterns of colonization and antifungal susceptibility of Candida, isolated from preterm neonates in Khorramabad, South West of Iran. J Mycol Med. 2018;28:340–4. doi: 10.1016/j.mycmed.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 63.Razzaghi-Abyaneh M, Sadeghi G, Zeinali E, Alirezaee M, Shams-Ghahfarokhi M, Amani A, et al. Species distribution and antifungal susceptibility of Candida spp.isolated from superficial candidiasis in outpatients in Iran. J Mycol Med. 2014;24:e43–50. doi: 10.1016/j.mycmed.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Beighton D, Ludford R, Clark DT, Brailsford SR, Pankhurst CL, Tinsley GF, et al. Use of CHROMagar Candida medium for isolation of yeasts from dental samples. J Clin Microbiol. 1995;33:3025–7. doi: 10.1128/jcm.33.11.3025-3027.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yücesoy M, Marol S. Performance of CHROMAGAR candida and BIGGY agar for identification of yeast species. Ann Clin Microbiol Antimicrob. 2003;2:8. doi: 10.1186/1476-0711-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jafari Z, Motamedi M, Jalalizand N, Shokoohi GR, Charsizadeh A, Mirhendi H. Comparison of CHROMagar, polymerase chain reaction-restriction fragment length polymorphism, and polymerase chain reaction-fragment size for the identification of Candida species. Curr Med Mycol. 2017;3:10–5. doi: 10.29252/cmm.3.3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramani R, Gromadzki S, Pincus DH, Salkin IF, Chaturvedi V. Efficacy of API 20C and ID 32C systems for identification of common and rare clinical yeast isolates. J Clin Microbiol. 1998;36:3396–8. doi: 10.1128/jcm.36.11.3396-3398.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falahati M, Farahyar S, Akhlaghi L, Mahmoudi S, Sabzian K, Yarahmadi M, et al. Characterization and identification of candiduria due to Candida species in diabetic patients. Curr Med Mycol. 2016;2:10–4. doi: 10.18869/acadpub.cmm.2.3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jahanshiri Z, Razzaghi-Abyaneh M. Comparison of conventional and molecular methods in identification of Candida species isolated from oropharyngeal candidiasis. Pathobiology Research. 2020;23:1–9. [Google Scholar]
- 70.Shawky SM, Gaballah AH, Abdallah A, Fadel S, El Kholy MA. Automated identification and antifungal susceptibility testing of Candida species using vitek 2 compact system in ICUs and pediatric oncology unit, Alexandria, Egypt. Egyptian Journal of Medical Microbiology. 2017;26:101–9. [Google Scholar]
- 71.Tan YE, Teo JQ-M, Rahman NBA, Ng OT, Kalisvar M, Tan AL, et al. Candida auris in Singapore: Genomic epidemiology, antifungal drug resistance, and identification using the updated 8.01 VITEKⓇ 2 system. Int J Antimicrob Agents. 2019;54:709–15. doi: 10.1016/j.ijantimicag.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 72.Meyer W, Irinyi L, Hoang MTV, Robert V, Garcia-Hermoso D, Desnos-Ollivier M, et al. Database establishment for the secondary fungal DNA barcode translational elongation factor 1α (TEF1α) Genome. 2019;62:160–9. doi: 10.1139/gen-2018-0083. [DOI] [PubMed] [Google Scholar]
- 73.Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50:1351–71. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- 74.Diaz MR, Boekhout T, Theelen B, Fell JW. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Syst Appl Microbiol. 2000;23:535–45. doi: 10.1016/S0723-2020(00)80028-4. [DOI] [PubMed] [Google Scholar]
- 75.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics.PCR protocols: A guide to methods and applications. 1990;18:315–22. [Google Scholar]
- 76.Rafat Z, Hashemi SJ, Ashrafi K, Nikokar I, Jafari A, Foroushani AR, et al. Epidemiology, laboratory diagnosis and clinical aspects of fungal pulmonary infections in 384 patients hospitalized in pulmonary units in Guilan province, Iran. Iran J Microbiol. 2020;12:353–63. doi: 10.18502/ijm.v12i4.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salehi M, Ahmadikia K, Mahmoudi S, Kalantari S, Jamalimoghadamsiahkali S, Izadi A, et al. Oropharyngeal candidiasis in hospitalised COVID-19 patients from Iran: Species identification and antifungal susceptibility pattern. Mycoses. 2020;63:771–8. doi: 10.1111/myc.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gholami M, Mokhtari F, Mohammadi R. Identification of Malassezia species using direct PCR-sequencing on clinical samples from patients with pityriasis versicolor and seborrheic dermatitis. Curr Med Mycol. 2020;6:21–6. doi: 10.18502/cmm.6.3.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colombo AL, de Almeida JN Júnior, Guinea J. Emerging multidrug-resistant Candida species. Curr Opin Infect Dis. 2017;30:528–38. doi: 10.1097/QCO.0000000000000411. [DOI] [PubMed] [Google Scholar]
- 80.Harrison E, Muir A, Stratford M, Wheals A. Species-specific PCR primers for the rapid identification of yeasts of the genus Zygosaccharomyces. FEMS Yeast Res. 2011;11:356–65. doi: 10.1111/j.1567-1364.2011.00724.x. [DOI] [PubMed] [Google Scholar]
- 81.Nikmanesh B, Ahmadikia K, Getso MI, Gharehbolagh SA, Aboutalebian S, Mirhendi H, et al. Candida africana and Candida dubliniensis as causes of pediatric candiduria: A study using HWP1 gene size polymorphism. AIMS Microbiol. 2020;6:272–9. doi: 10.3934/microbiol.2020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shirani K, Allameh Z, Moshtzan A. Identification of etiologic agents of recurrent vulvovaginal candidiasis in patients from Isfahan, Iran. Immunopathologia Persa. 2021;7:e18. [Google Scholar]
- 83.Safari F, Madani M, Kheirollahi M, Mirhendi H. Molecular investigation of the incidence of Candida auris infections at selected hospitals in Iran. Mycoses. 2022;65:1137–45. doi: 10.1111/myc.13504. [DOI] [PubMed] [Google Scholar]
- 84.Safari F, Madani M, Mirhendi H, Kheirollahi M. Evaluation of Candida auris Colonization using Clinical Skin Swabs: A Single-Center Study in Isfahan, Iran. Journal of Health Reports and Technology. 2022;8:e121844. [Google Scholar]
- 85.Armaki MT, Omran SM, Kiakojuri K, Khojasteh S, Jafarzadeh J, Tavakoli M, et al. First fluconazole-resistant Candida auris isolated from fungal otitis in Iran. Curr Med Mycol. 2021;7:51–4. doi: 10.18502/cmm.7.1.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luo G, Mitchell TG. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol. 2002;40:2860–5. doi: 10.1128/JCM.40.8.2860-2865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aboutalebian S, Ahmadikia K, Fakhim H, Chabavizadeh J, Okhovat A, Nikaeen M, et al. Direct detection and identification of the most common bacteria and fungi causing otitis externa by a stepwise multiplex PCR. Front Cell Infect Microbiol. 2021;11:644060. doi: 10.3389/fcimb.2021.644060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carvalho-Pereira J, Fernandes F, Araújo R, Springer J, Loeffler J, Buitrago MJ, et al. Multiplex PCR based strategy for detection of fungal pathogen DNA in patients with suspected invasive fungal infections. J Fungi (Basel) 2020;6:308. doi: 10.3390/jof6040308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mirhendi H, Makimura K, Zomorodian K, Maeda N, Ohshima T, Yamaguchi H. Differentiation of Candida albicans and Candida dubliniensis using a single-enzyme PCR-RFLP method. Jpn J Infect Dis. 2005;58:235–7. [PubMed] [Google Scholar]
- 90.Shokohi T, Hashemi Soteh MB, Saltanat Pouri Z, Hedayati MT, Mayahi S. Identification of Candida species using PCR-RFLP in cancer patients in Iran. Indian J Med Microbiol. 2010;28:147–51. doi: 10.4103/0255-0857.62493. [DOI] [PubMed] [Google Scholar]
- 91.Fallahi AA, Korbacheh P, Zaini F, Mirhendi H, Zeraati H, Noorbakhsh F, et al. Candida species in cutaneous candidiasis patients in the Guilan province in Iran; identified by PCR-RFLP method. Acta Med Iran. 2013;51:799–804. [PubMed] [Google Scholar]
- 92.Badiee P, Alborzi A. Assessment of a real-time PCR method to detect human non-cryptococcal fungal meningitis. Arch Iran Med. 2011;14:381–4. [PubMed] [Google Scholar]
- 93.Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 94.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hadrys H, Balick M, Schierwater B. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1992;1:55–63. doi: 10.1111/j.1365-294x.1992.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 96.Fahami S, Kordbacheh P, Moazeni M, Mahmoudi M, Mirhendi H. Species identification and strain typing of Candida isolates by PCR-RFLP and RAPD-PCR analysis for determining the probable sources of nosocomial infections. Iranian Red Crescent Medical Journal. 2010;12:539–47. [Google Scholar]
- 97.Tabit FT. Advantages and limitations of potential methods for the analysis of bacteria in milk: A review. J Food Sci Technol. 2016;53:42–9. doi: 10.1007/s13197-015-1993-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Afsarian MH, Badali H, Boekhout T, Shokohi T, Katiraee F. Multilocus sequence typing of Candida albicans isolates from a burn intensive care unit in Iran. J Med Microbiol. 2015;64:248–53. doi: 10.1099/jmm.0.000015. [DOI] [PubMed] [Google Scholar]
- 99.Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics—identification of microorganisms and beyond (mini review) Appl Microbiol Biotechnol. 2012;93:965–74. doi: 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- 100.Panda A, Ghosh AK, Mirdha BR, Xess I, Paul S, Samantaray JC, et al. MALDI-TOF mass spectrometry for rapid identification of clinical fungal isolates based on ribosomal protein biomarkers. J Microbiol Methods. 2015;109:93–105. doi: 10.1016/j.mimet.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 101.Aslani N, Janbabaei G, Abastabar M, Meis JF, Babaeian M, Khodavaisy S, et al. Identification of uncommon oral yeasts from cancer patients by MALDI-TOF mass spectrometry. BMC Infect Dis. 2018;18:24. doi: 10.1186/s12879-017-2916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahangarkani F, Ilkit M, Vaseghi N, Zahedi N, Zomorodian K, Khodavaisy S, et al. MALDI-TOF MS characterisation, genetic diversity and antifungal susceptibility of Trichosporon species from Iranian clinical samples. Mycoses. 2021;64:918–25. doi: 10.1111/myc.13306. [DOI] [PubMed] [Google Scholar]
- 103.Alizadeh M, Kolecka A, Boekhout T, Zarrinfar H, Ghanbari Nahzag MA, Badiee P, et al. Identification of Candida species isolated from vulvovaginitis in Mashhad, Iran by Use of MALDI-TOF MS. Curr Med Mycol. 2017;3:21–5. doi: 10.29252/cmm.3.4.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ahangarkani F, Shokohi T, Rezai MS, Ilkit M, Mahmoodi Nesheli HM, Karami H, et al. Epidemiological features of nosocomial candidaemia in neonates, infants and children: A multicentre study in Iran. Mycoses. 2020;63:382–94. doi: 10.1111/myc.13053. [DOI] [PubMed] [Google Scholar]
- 105.Arastehfar A, Khodavaisy S, Daneshnia F, Najafzadeh M-J, Mahmoudi S, Charsizadeh A, et al. Molecular identification, genotypic diversity, antifungal susceptibility, and clinical outcomes of infections caused by clinically underrated yeasts, Candida orthopsilosis, and Candida metapsilosis: An Iranian multicenter study (2014–2019) Front Cell Infect Microbiol. 2019;9:264. doi: 10.3389/fcimb.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cafardi J, Haas D, Lamarre T, Feinberg J. Opportunistic fungal infection associated with COVID-19. Open Forum Infect Dis. 2021;8:ofab016. doi: 10.1093/ofid/ofab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 108.Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers. 2018;4:18026. doi: 10.1038/nrdp.2018.26. [DOI] [PubMed] [Google Scholar]
- 109.Suleyman G, Alangaden GJ. Nosocomial Fungal Infections: Epidemiology, Infection Control, and Prevention. Infect Dis Clin North Am. 2021;35:1027–53. doi: 10.1016/j.idc.2021.08.002. [DOI] [PubMed] [Google Scholar]
- 110.Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel) 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: A three-year analysis. Clin Infect Dis. 1999;29:239–44. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 112.Rangel-Frausto MS, Wiblin T, Blumberg HM, Saiman L, Patterson J, Rinaldi M, et al. National epidemiology of mycoses survey (NEMIS): Variations in rates of bloodstream infections due to Candida species in seven surgical intensive care units and six neonatal intensive care units. Clin Infect Dis. 1999;29:253–8. doi: 10.1086/520194. [DOI] [PubMed] [Google Scholar]
- 113.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 114.Wisplinghoff H, Seifert H, Tallent SM, Bischoff T, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in pediatric patients in United States hospitals: Epidemiology, clinical features and susceptibilities. Pediatr Infect Dis J. 2003;22:686–91. doi: 10.1097/01.inf.0000078159.53132.40. [DOI] [PubMed] [Google Scholar]
- 115.Wisplinghoff H, Seifert H, Wenzel RP, Edmond MB. Current trends in the epidemiology of nosocomial bloodstream infections in patients with hematological malignancies and solid neoplasms in hospitals in the United States. Clin Infect Dis. 2003;36:1103–10. doi: 10.1086/374339. [DOI] [PubMed] [Google Scholar]
- 116.Arastehfar A, Daneshnia F, Farahyar S, Fang W, Salimi M, Salehi M, et al. Incidence and spectrum of yeast species isolated from the oral cavity of Iranian patients suffering from hematological malignancies. J Oral Microbiol. 2019;11:1601061. doi: 10.1080/20002297.2019.1601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maheronnaghsh M, Fatahinia M, Dehghan P, Teimoori A. Identification of Candida species and antifungal susceptibility in cancer patients with oral lesions in Ahvaz, Southern West of Iran. Adv Biomed Res. 2020;9:50. doi: 10.4103/abr.abr_214_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brissaud O, Guichoux J, Harambat J, Tandonnet O, Zaoutis T. Invasive fungal disease in PICU: Epidemiology and risk factors. Ann Intensive Care. 2012;2:6. doi: 10.1186/2110-5820-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Korula A, Abraham A, Abubacker FN, Viswabandya A, Lakshmi KM, Abraham OC, et al. Invasive fungal infection following chemotherapy for acute myeloid leukaemia—Experience from a developing country. Mycoses. 2017;60:686–91. doi: 10.1111/myc.12646. [DOI] [PubMed] [Google Scholar]
- 120.Mellinghoff SC, Panse J, Alakel N, Behre G, Buchheidt D, Christopeit M, et al. Primary prophylaxis of invasive fungal infections in patients with haematological malignancies: 2017 update of the recommendations of the Infectious Diseases Working Party (AGIHO) of the German Society for Haematology and Medical Oncology (DGHO) Ann Hematol. 2018;97:197–207. doi: 10.1007/s00277-017-3196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eriksen HM, Iversen BG, Aavitsland P. Prevalence of nosocomial infections in hospitals in Norway, 2002 and 2003. J Hosp Infect. 2005;60:40–5. doi: 10.1016/j.jhin.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 122.John GT, Shankar V, Talaulikar G, Mathews MS, Abraham MA, Thomas PP, et al. Epidemiology of systemic mycoses among renal-transplant recipients in India. Transplantation. 2003;75:1544–51. doi: 10.1097/01.TP.0000061610.34110.04. [DOI] [PubMed] [Google Scholar]
- 123.Borman AM, Linton CJ, Miles S-J, Johnson EM. Molecular identification of pathogenic fungi. J Antimicrob Chemother. 2008;61((Suppl 1)):i7–12. doi: 10.1093/jac/dkm425. [DOI] [PubMed] [Google Scholar]
- 124.Hilmioglu S, Ilkit M, Badak Z. Comparison of 12 liquid media for germ tube production of Candida albicans and C.tropicalis. Mycoses. 2007;50:282–5. doi: 10.1111/j.1439-0507.2007.01359.x. [DOI] [PubMed] [Google Scholar]
- 125.Cárdenes-Perera C-D, Torres-Lana A, Alonso-Vargas R, Moragues-Tosantas M-D, Pontón-San Emeterio J, Quindós-Andrés G, et al. Evaluation of API ID 32C® and VITEK-2® to identify Candida dubliniensis. Diagn Microbiol Infect Dis. 2004;50:219–21. doi: 10.1016/j.diagmicrobio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 126.Bernal S, Mazuelos EM, Chávez M, Coronilla J, Valverde A. Evaluation of the new API Candida system for identification of the most clinically important yeast species. Diagn Microbiol Infect Dis. 1998;32:217–21. doi: 10.1016/s0732-8893(98)00119-9. [DOI] [PubMed] [Google Scholar]
- 127.Letscher-Bru V, Meyer M-H, Galoisy A-C, Waller J, Candolfi E. Prospective evaluation of the new chromogenic medium Candida ID, in comparison with Candiselect, for isolation of molds and isolation and presumptive identification of yeast species. J Clin Microbiol. 2002;40:1508–10. doi: 10.1128/JCM.40.4.1508-1510.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yücesoy M, Marol S. Performance of CHROMAGAR candida and BIGGY agar for identification of yeast species. Ann Clin Microbiol Antimicrob. 2003;2:8. doi: 10.1186/1476-0711-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Romeo O, Scordino F, Pernice IDA, Lo Passo C, Criseo G. A multiplex PCR protocol for rapid identification of Candida glabrata and its phylogenetically related species Candida nivariensis and Candida bracarensis. J Microbiol Methods. 2009;79:117–20. doi: 10.1016/j.mimet.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 130.Arastehfar A, Daneshnia F, Kord M, Roudbary M, Zarrinfar H, Fang W, et al. Corrigendum: Comparison of 21-Plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA sequencing for a wide range of clinically isolated yeast species: Improved identification by combining 21-Plex PCR and API 20C AUX as an alternative strategy for developing countries. Front Cell Infect Microbiol. 2019;9:176. doi: 10.3389/fcimb.2019.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang H-Y, Kim S, Kim H, Kim J, Kim Y, Park S-D, et al. Real-time PCR TaqMan assay for rapid screening of bloodstream infection. Ann Clin Microbiol Antimicrob. 2014;13:3. doi: 10.1186/1476-0711-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nieto M, Robles JC, Causse M, Gutiérrez L, Cruz Perez M, Ferrer R, et al. Polymerase chain reaction versus blood culture to detect Candida species in high-risk patients with suspected invasive Candidiasis: The MICAFEM Study. Infect Dis Ther. 2019;8:429–44. doi: 10.1007/s40121-019-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.White PL, Archer AE, Barnes RA. Comparison of non-culture-based methods for detection of systemic fungal infections, with an emphasis on invasive Candida infections. J Clin Microbiol. 2005;43:2181–7. doi: 10.1128/JCM.43.5.2181-2187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Klingspor L, Jalal S. Molecular detection and identification of Candida and Aspergillus spp.from clinical samples using real-time PCR. Clin Microbiol Infect. 2006;12:745–53. doi: 10.1111/j.1469-0691.2006.01498.x. [DOI] [PubMed] [Google Scholar]
- 135.Lindsley MD, Mekha N, Baggett HC, Surinthong Y, Autthateinchai R, Sawatwong P, et al. Evaluation of a newly developed lateral flow immunoassay for the diagnosis of cryptococcosis. Clin Infect Dis. 2011;53:321–5. doi: 10.1093/cid/cir379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tay E, Chen SC-A, Green W, Lopez R, Halliday CL. Development of a real-time PCR assay to identify and distinguish between Cryptococcus neoformans and Cryptococcus gattii species complexes. J Fungi (Basel) 2022;8:462. doi: 10.3390/jof8050462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Odabasi Z, Mattiuzzi G, Estey E, Kantarjian H, Saeki F, Ridge RJ, et al. β-D-glucan as a diagnostic adjunct for invasive fungal infections: Validation, cutoff development, and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin Infect Dis. 2004;39:199–205. doi: 10.1086/421944. [DOI] [PubMed] [Google Scholar]
- 138.Fontana C, Gaziano R, Favaro M, Casalinuovo I, Pistoia E, Di Francesco P. (1-3)-β-D-glucan vs galactomannan antigen in diagnosing invasive fungal infections (IFIs) Open Microbiol J. 2012;6:70–3. doi: 10.2174/1874285801206010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ashrafi M, Nabili M, Shokohi T, Janbabaie G, Hedayati MT, Ali-Moghaddam K. A real time PCR assay on blood for diagnosis of invasive candidiasis in immunocompromised patient. Curr Med Mycol. 2015;1:35–41. doi: 10.18869/acadpub.cmm.1.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shepard JR, Addison RM, Alexander BD, Della-Latta P, Gherna M, Haase G, et al. Multicenter evaluation of the Candida albicans/Candida glabrata peptide nucleic acid fluorescent in situ hybridization method for simultaneous dual-color identification of C. albicans and C. glabrata directly from blood culture bottles. J Clin Microbiol. 2008;46:50–5. doi: 10.1128/JCM.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Spanu T, Posteraro B, Fiori B, D’Inzeo T, Campoli S, Ruggeri A, et al. Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: An observational study in two large microbiology laboratories. J Clin Microbiol. 2012;50:176–9. doi: 10.1128/JCM.05742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]



