SUMMARY
In a healthy gut, microbes are often aggregated with host mucus, yet the molecular basis for this organization and its impact on intestinal health are unclear. Mucus is a viscous physical barrier separating resident microbes from epithelia, but also provides glycan cues that regulate microbial behaviors. Here, we describe a mucin-sensing pathway in an Aeromonas symbiont of zebrafish, Aer01. In response to the mucin-associated glycan N-acetylglucosamine, a sensor kinase regulates expression of an aggregation-promoting adhesin we named MbpA. Upon MbpA disruption, Aer01 colonizes to normal levels, but is largely planktonic and more pro-inflammatory. Increasing cell surface MbpA rescues these traits. MbpA-like adhesins are common in human-associated bacteria and expression of an Akkermansia muciniphila MbpA-like adhesin in MbpA-deficient Aer01 restores lumenal aggregation and reverses its pro-inflammatory character. Our work demonstrates how resident bacteria use mucin glycans to modulate behaviors congruent with host health.
Graphical Abstract
eTOC Blurb
The mechanisms resident microbes use to influence host health are underexplored. Smith et al. report a zebrafish symbiont, Aeromonas, that uses a mucin-regulated adhesin to control its inflammatory character and spatial organization in the gut. Functionally analogous Akkermansia adhesins suggest similar microbial mechanisms underly human gut health.
INTRODUCTION
Vertebrate intestinal mucus is vital for maintaining host-microbe mutualism. Mucus can modulate symbiosis by encapsulating and segregating resident microbes away from the gut epithelium 1,2. In a healthy digestive tract, microbes are often found associated with host mucus in the lumen, a distribution that has been described across vertebrates including humans, mice, and zebrafish 3–8. This lumenal microbial aggregation is commonly attributed to host processes such as active mucus secretion and peristalsis that encapsulate and displace lumenal contents, with the microbiota members being passive players within system. In this view, loss of gut health is attributed to host defects such as impaired mucus secretion or gut motility. Microbes however, can actively regulate their own aggregation and biofilm formation in response to environmental cues and recent studies indicate glycans associated with mucin can act as potent mediators of bacterial behaviors 9–15. The dual roles of mucin as a physical barrier and molecular signal are inseparable since the sugars that decorate mucins contribute to both its biophysical properties and its chemical signature. Defining the ways in which microbiota members sense and respond to mucins to regulate their own structural organization in the gut will refine our understanding of microbiota roles on gut ecosystem health and disease.
Mucin is secreted by intestinal goblet cells, with MUC2 being the dominant mucin in the human, mouse, and zebrafish gut 16–18. Mucin is heavily glycosylated with O-linked N-acetylgalactosamine (GalNAc), galactose (Gal), and N-acetylglucosamine (GlcNAc) glycan chains that are usually capped with fucose (Fuc) and/or sialic acid (SA) 19. The glycan composition of mucin varies along the intestinal tract 20 and various microbes encode adhesins that bind glycan epitopes O-linked to mucin 21,22. Resident microbes alter mucin’s glycan composition and stimulate its secretion 5,23–25. Additionally, microbes encode glycoside hydrolase (GH) enzymes that process glycan O-linkages on mucin, releasing energy-rich monomeric and multimeric glycan products that can also serve as molecular signals to microbiota 9,26–29. The mechanisms through which mucin glycans may signal to microbiota members and whether these processes impact symbiosis remain to be explored.
Most studies exploring the relationship between mucin’s barrier function and microbial symbiosis use mouse models with complete loss or severe defects in intestinal mucin secretion and thus dramatic changes to the gut’s glycan landscape. Loss of mucin often results in increased microbial proximity to the epithelium associated with intestinal inflammation that can progress to various intestinal diseases such as colitis and colorectal cancer 30,31. Another feature of the mucus-depleted gut is elevated levels of bacterial flagellin, indicative of motile bacteria and also a driver of intestinal inflammation 30,32. In these studies, intestinal mucus clearly promotes host health but the extent to which it does so as a barrier or through glycan-mediated regulation of microbial activities is difficult to disentangle.
The larval zebrafish (Danio rerio) has emerged as a powerful model system for dissecting mechanistic principles of vertebrate host-symbiont interactions due to its optical transparency, transgenic capabilities, relative ease of gnotobiology, and conservation of many mammalian innate immune processes 33,34. Live imaging of the larva zebrafish intestine demonstrates bacteria in the gut lumen adopt diverse aggregation states and localize to distinct regions of the gut 3,4, similar to observations in fixed mouse intestinal tissue 6,8,35. For example, the zebrafish symbiont Aeromonas ZOR0001 (Aer01) forms dense aggregates in the anterior midgut with few planktonic cells, while the closely related Aeromonas ZOR0002 is more planktonic and located more anteriorly. Recent live imaging analysis and modeling of bacterial population dynamics has highlighted the importance of host processes 36 as well as bacterial traits like motility 37, aggregate assembly, and fragmentation as critical determinants of bacterial spatial organization in the zebrafish gut 38,39.
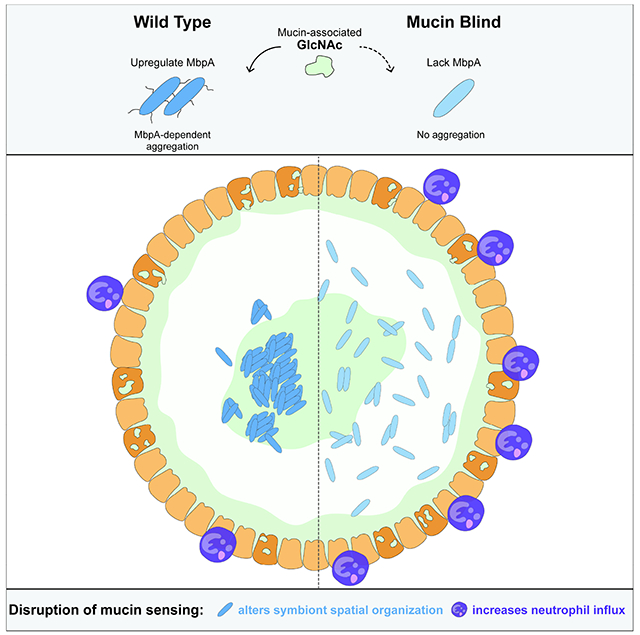
Here, we use the Aer01-zebrafish mutualistic relationship as a model to investigate the relative contributions of mucin’s barrier and signaling roles in shaping symbiosis in a healthy host. We found Aer01 actively promotes host intestinal health by self-limiting its intestinal distribution and inflammatory character in response to the mucin-associated glycan GlcNAc. Experimental evolution that selected for “mucin blind” (MB) Aer01 isolates that do not aggregate in response to mucin or GlcNAc revealed that a two-component system (TCS) sensor histidine kinase (MbpS) and regulatory protein (MbpD) coordinate to control the transcription and function of an adhesin we called MbpA. Inhibition of MbpA function is sufficient to enhance intestinal inflammation and alter Aer01 population structure in the gut, indicating regulation of this adhesin in response to GlcNAc signals is crucial for modulating the inflammatory tone of the intestine. MbpA analogs are distributed in various human-associated microbiota including Oxalobacter, Fusobacterium, and Akkermansia. Similar to MbpA, the MbpA analog in Akkermansia (Amuc_1620, here called MucA) is upregulated in the presence of mucin 14. Expression of MucA in an Aer01 strain with a non-functional MbpA (MB1) rescued aggregation and suppressed MB1’s pro-inflammatory activity, suggesting MucA may play a similar role in beneficial Akkermansia-human interactions. Together, these data indicate Aer01 actively promotes its hosts’ intestinal health in response to the glycan landscape of the gut environment and suggests this principle may shape beneficial interactions in other host-microbe systems.
RESULTS
Aer01 is associated with host mucus in the intestinal lumen.
To determine the spatial relationship between Aer01 and intestinal mucus, we mono-associated germfree larval zebrafish with dTomato (dTom)-tagged Aer01 four days post fertilization (dpf) and fixed whole larvae 24 hours later for confocal imaging. Aer01 distribution was visualized using anti-dTom. Mucus and goblet cell distribution were visualized using FITC-conjugated wheat germ agglutinin (WGA-FITC), which binds the mucin-associated glycan GlcNAc (Fig. 1A). Consistent with previous histological studies of goblet cell distribution in the zebrafish intestine, we found two anatomically separated populations of mucus-producing goblet cells in the pharynx/esophagus (Fig. 1B) and mid/distal gut (Fig. 1C) indicated by WGA-positive vesicles 40–42.
Figure 1. Aer01 associates with host intestinal mucus and aggregates in response to its mucolytic activity.
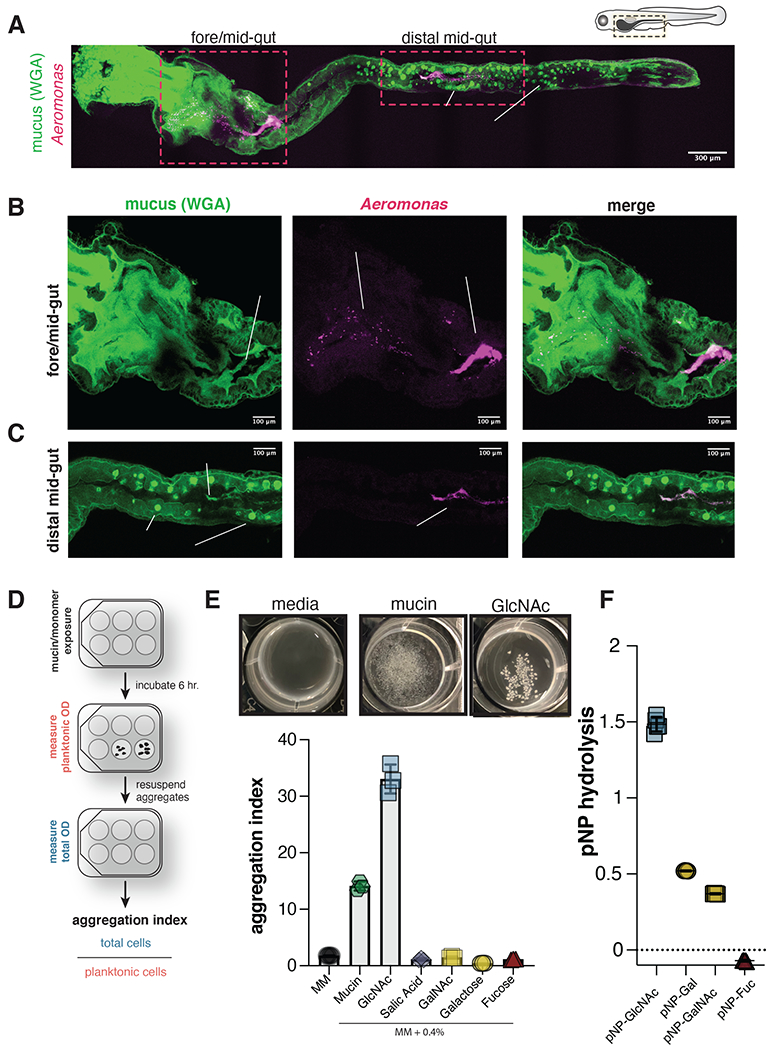
(A-C) Confocal microscopy of fixed 5 dpf larval zebrafish intestine indicating Aer01 (anti-dTom, magenta) and mucus (WGA-488, green) distribution (a) throughout the entire larval zebrafish gut, (b) the fore/mid-gut, and (c) distal mid-gut region. See also Figure S1.
(D) Experimental design of culture-based aggregation assay and calculation of aggregation index.
(E) Top: representative images of mucin-and GlcNAc-mediated Aer01 aggregation after 6 hr exposure. Bottom: mean aggregation index of Aer01 exposed to PGM or its monomeric glycan components (N=3).
(F) Aer01 hydrolysis of 4-Nitrophenyl (pNP) substrates. (N=5). The mean values are indicated in all figures.
WGA-stained mucus accumulates in the lumen of the anterior and proximal end of the midgut in fixed (Fig. 1B, left) and live (Fig. S1, Movie S1) larval zebrafish. The majority of Aer01 cells were aggregated in the same proximal midgut region as mucus (Fig. 1B, right and Movie S2), with a small number of planktonic Aer01 present in the foregut (Fig. 1B, middle). This spatial distribution in fixed tissue mirrors previous live imaging of Aer01 colonization dynamics 36,38. Planktonic Aer01 in the foregut may represent recently immigrated bacteria, as motility is critical for Aer01 immigration 43,44. Additionally, we observed small Aer01 aggregates in the same region as mucus in the distal midgut lumen (Fig. 1C, right and Movie S3), which are likely mucus-encased Aer01 undergoing excretion.
These data demonstrate a strong spatial relationship between Aer01 aggregates and GlcNAc-containing host gut mucus. Since Aer01 are motile in the aqueous environment but form aggregates in the gut environment, we next sought to determine if Aer01’s aggregation and spatial relationship with host gut mucus is due to mucus’ natively viscous features or if Aer01 actively aggregates in response to molecular cues associated with mucus.
Mucin and mucolytic byproducts drive Aer01 aggregation in culture.
We next tested if mucin, the primary component of mucus, can promote Aer01 aggregation in culture. We developed a culture-based approach outlined in Fig 1D to visualize and measure Aer01 aggregation upon exposure to mucin and various host-associated cues. In minimal medium (MM) supplemented with 0.4% commercially available porcine gastric mucin (mucin or PGM), Aer01 forms dense mucin-associated aggregates within 6 hours that are not formed in MM alone (Fig. 1E, top and Fig. S2). Further, Aer01 aggregated in mucin concentrations as low as 0.02%, suggesting aggregation was not due to polymer-driven depletion aggregation in the viscous mucin medium 45,46 (data not shown). We measured aggregation as the ratio of the total cell density to the planktonic cell density, called the aggregation index. Consistent with our visual assessment, we measured a higher aggregation index for Aer01 in mucin than in MM alone (Fig. 1E, bottom), suggesting Aer01 aggregation is an active response to mucin that may play a role in Aer01’s spatial organization and distribution in the intestinal lumen.
Bioinformatic analysis indicates Aer01 encodes several glycoside hydrolase families (GH2, GH18, GH20, GH84, and GH129) employed by bacteria to hydrolyze intestinal mucin at N-acetyl-glucosamine (GlcNAc), galactose (Gal), and N-acetyl-galactosamine (GalNAc) linkages 27,47. To test the idea that Aer01 aggregation may be driven in part by sensing the activity of its own mucin degrading activities, we first assayed if Aer01 can cleave common sugar linkages found in mucin. Consistent with Aer01’s predicted GH profile, Aer01 cleaved 4-Nitrophenyl (pNP) substrates containing GlcNAc, Gal, and GalNAc, but not Fucose (Fuc) (Fig. 1F), showing the highest hydrolytic activity against pNP-GlcNAc.
Next, we assayed Aer01 aggregation in MM containing 0.4% of each major O-linked glycan found on mucins: GlcNAc, Gal, GalNAc, Fuc, or sialic acid (SA). While several glycans supported Aer01 growth, only GlcNAc promoted Aer01 aggregation (Fig. 1E). Together, these data suggest Aer01 aggregation in response to mucin may be driven by release of GlcNAc glycans produced Aer01’s mucolytic activity.
Aer01 mucin-responses determine its spatial organization in the intestine.
To identify genetic pathways involved in Aer01 responses to mucin, we performed serial selections for Aer01 populations that remain planktonic in culture when exposed to GlcNAc, the key mucin glycan sensed by Aer01. For the serial selection, we exposed Aer01 to 0.4% GlcNAc and selected unaggregated Aer01 from the planktonic fraction after 6 hr (Fig. 2A, left). This planktonic sample was expanded by growing overnight in LB and again transferred back into GlcNAc medium for a total of 7 passages. Each population was cryopreserved and individual isolates were also selected from each passage by plating on LB agar. By the 5th passage, no Aer01 aggregates were observed in GlcNAc (Fig. 2A, right) or mucin. Thus, we named the non-responsive isolates from these populations “mucin-blind” (MB).
Figure 2. Defects in mucin responses alter Aer01 spatial organization in the intestine.
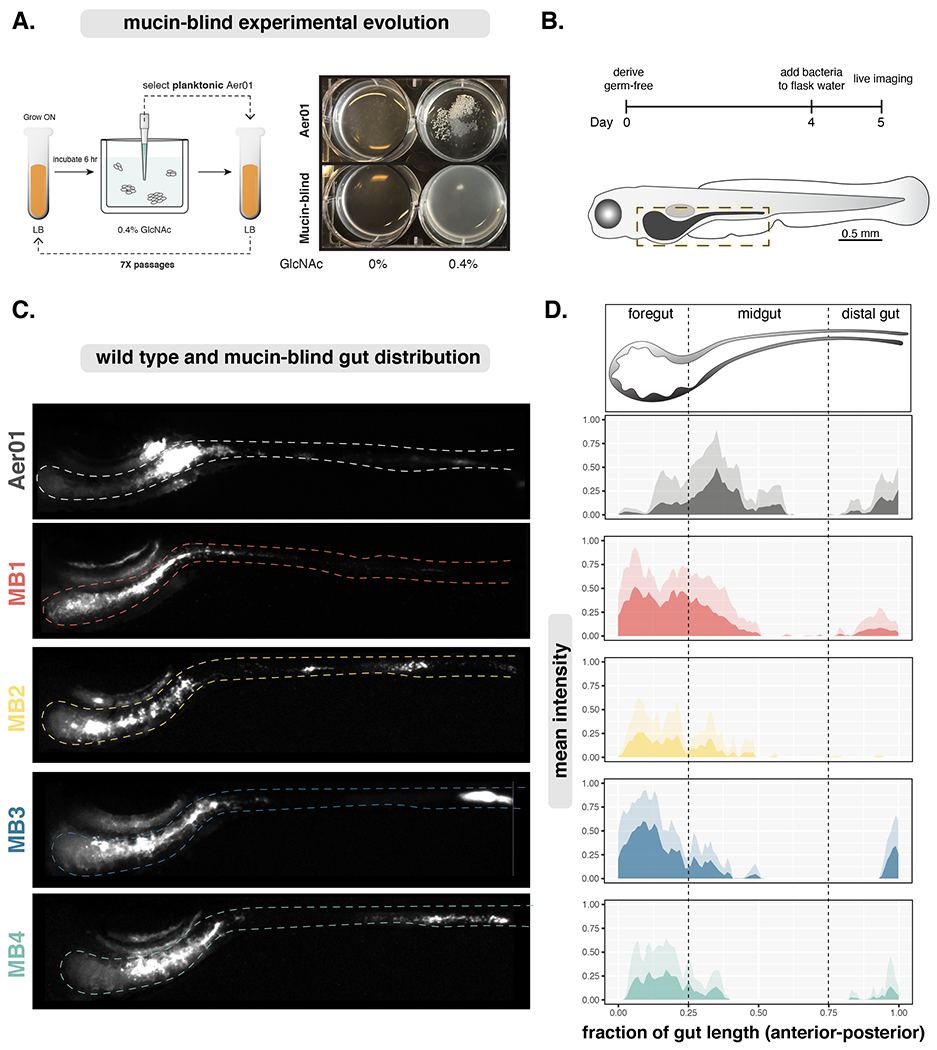
(A) Left: Experimental evolution schematic. Right: Evolved Aer01 are non-responsive to GlcNAc.
(B) Inoculation and imaging schedule for Aer01 and MB population structure analysis.
(C) Live stereoscope microscopy of population-level Aer01 and MB spatial organizations. The approximate intestinal boundary is outlined and corresponds the region measured for distribution analysis in (D).
(D) Quantification of bacterial spatial organization from lives images in (c). Mean pixel intensities (microbial abundance) are plotted along the normalized gut length and standard deviation at each point plotted in the corresponding lighter color. Aer01 (N=7), MB1 (N=7), MB2 (N=7), MB3 (N=5), MB4 (N=7).
Next, we tested if the genetic pathways disrupted in our culture-based experimental evolution screen contribute Aer01’s aggregation and population structure in the intestine. We randomly selected four MB isolates, two from passage 5 (MB1 and MB3) and two from passage 6 (MB2 and MB4), and assayed their spatial organization throughout the entire larval zebrafish gut using live fluorescence stereomicroscopy. Each MB isolate was labeled with dTom for live imaging using molecular genetic tools developed by our lab 4. Germfree larval zebrafish were mono-associated with an individual MB isolate on four dpf and their distribution throughout the gut imaged 24 hours later (Fig. 2B). The mean normalized pixel intensity corresponding to microbial abundance was plotted across the normalized gut length along with the standard deviation at each position for WT and our MB isolates of Aer01. Strikingly, all four MB isolates exhibited altered distributions with an anterior displacement into the foregut region while WT Aer01 formed large aggregates in the anterior midgut as expected (Fig. 2C and D). Consistent with the idea that the shift in MB spatial organization was due to defects in mucus sensing, the parental Aer01 strain exhibits a shifted spatial organization when mono-associated in a larval zebrafish mutant with decreased numbers of mucus-producing goblet cells (myd88−/−, Fig. S3A) 40 or upon colonization of wild type larval zebrafish co-incubated with WGA to mask free GlcNAc (Fig. S3B). Together, these data suggest Aer01 genetic pathways play a dominant role in its aggregation and spatial distribution in the intestinal environment.
MB mutations converge on a secondary messenger-regulated adhesin.
To uncover the genetic mutations responsible for disrupting aggregation in our four MB isolates, their genomes were sequenced and compared to the parental strain using breseq using the default settings48. In total, four single nucleotide polymorphisms (SNPs) and one indel were identified. The locus ID, putative functions, and mutations for each isolate are shown in Fig. 3A. This analysis identified mutations in genes associated with the Lap system or metabolism of the bacterial secondary messenger c-di-GMP (cdG).
Figure 3. Genomic analysis of experimentally evolved isolates reveals molecular basis for mucin-mediated aggregation defects.
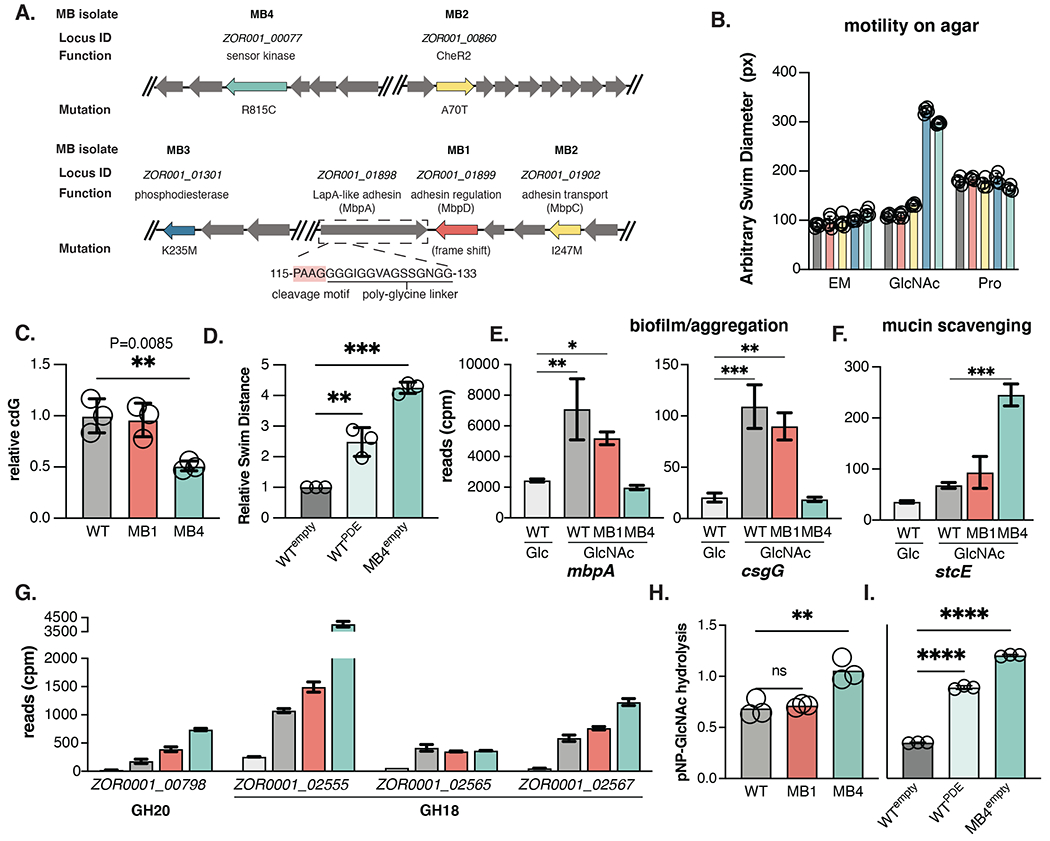
(A) Chromosomal organization of MB1-MB4 mutations (red: MB1; yellow: MB2; blue: MB3; teal: MB4). The locus ID, putative function, nucleotide substation, and corresponding amino acid change listed when applicable. The ORF encoding mbpA is boxed and the amino acids corresponding to the MbpG cleavage motif in the N-terminal retention module are indicated.
(B) Swim diameter of WT and MB isolates in MM alone (N=5 replicates) or supplemented with 0.4% GlcNAc (N= 5) or 1 mM proline (N=4) after 20 hr incubation at 30°C.
(C) Quantification of c-di-GMP in WT, MB1, and MB4 exposed to GlcNAc.
(D) Relative swim distance of Aer01 engineered to express the PDE ZOR0001_03617 upon aTc induction compared to Aer01 and MB4 carrying an empty induction construct.
(E-G) RNAseq analysis of indicated genes involved in biofilm formation/aggregation and mucin scavenging following exposure to 0.4% Glucose (WT, N=3) or 0.4% GlcNAc (WT, N=3; MB1, N=3; MB4, N=2). Ordinary one-way ANOVA with multiple comparisons. See also Table S1.
(H) Hydrolytic activity of WT, MB1, and MB4 on pNP-GlcNAc (N=3).
(I) Suppressing c-di-GMP accumulation increases Aer01 pNP-GlcNAc hydrolysis (N=3). Mean and standard deviations are indicated in all graphs.
The Lap system (LapABCDEG) has been best studied in Pseudomonas fluorescens 49. Here, the biofilm-promoting adhesin, LapA, is transported to the cell surface via its cognate Type 1 Secretion System (T1SS) secretion apparatus (LapBCE). Cell surface-associated levels of LapA are regulated in response cdG by the activity of a cdG-sensing regulatory protein (LapD) that suppresses the activity of a LapA-targeting protease (LapG) when cdG levels are high. Lap adhesins share little sequence similarity, even among closely related strains, but can be identified via a structurally conserved N-terminal retention module (NCBI HMM accession: NF033682.1) that tethers the adhesin to the outer membrane and contains the dialanine LapG cleavage motif required for cleavage-induced release from the cell surface 49. Because Lap adhesins share little sequence similarity and exhibit strain-specific substrate-binding properties, characterized Lap adhesins are often named after the condition or phenotype in which they were discovered (for instance the flagellum-regulated hemagglutinin adhesin FrhA of Vibrio cholerae). MB1 and MB2 harbor mutations in the Lap system and we thus named the Aer01 Lap system components according to their association as Mucin-blind proteins (mbpABCDEG).
MB1 contains a 5 nucleotide deletion in the cdG-sensing protease regulator mbpD (ZOR0001_01899) at nucleotides 76-80 of the 1926 nucleotide gene (Fig. 3A, red arrow). This indel generates a premature stop codon prior to translation of any regulatory domains, likely generating non-functional MbpD protein, allowing the protease MbpG to constitutively cleave its target adhesin, as has been observed in other Lap systems 50. Consistent with this idea, complementation with wild-type mbpD rescued MB1 aggregation in GlcNAc (Fig. S4), indicating the adhesin regulated by MbpDG activity is a key output in GlcNAc-mediated aggregation. The gene adjacent to mbpD, ZOR0001_01898, is a large ~500 kDa LapA-like adhesin containing a N-terminal retention module and putative di-alanine MbpG cleaveage site (Fig. 3A boxed region and insert) and was thus named mbpA.
A SNP in the MB2 isolate generates a I247M mutation in ZOR0001_0192, the membrane fusion protein (MFP) T1SS component, MbpC (Fig. 3A, yellow). In other MFPs, mutations in this region can diminish protein secretion by disrupting interactions between the MFP and its target outer membrane protein 51, suggesting this mutation may block transport or inhibit retention of MbpA. Since MB2 also contains a mutation in CheR2, we focused further analysis on MB1.
MB3 and MB4 contain mutations in predicted cdG metabolizing and sensory signaling. MB3 has a K235M mutation in the HD-related output domain (HDOD) of ZOR001_01301 (Fig. 3A, blue), a predicted phosphodiesterase (PDE) enzyme which are known to hydrolyze and degrade cdG. This protein has 64% amino acid similarity to CdgJ, a PDE involved in regulating Vibrio cholerae motility 52. HDODs are often associated with PDE and DGC enzymes, but their role in modulating cdG metabolism is poorly understood.
MB4 has a R815C mutation in an atypical two-component system (TCS) histidine kinase (HK), ZOR0001_00077 (Fig. 3A, teal). TCSs are regulatory phosphor-relay hubs comprised of an environment-sensing HK and a cognate response regulator (RR). The MB4 HK mutation sits in the dimerization and phosphor-acceptor domain, which is critical for TCS signaling. TCS HK-RR pairs are typically cotranslated, and the start codon for ZOR0001_00078, the likely RR, is within the ZOR0001_00077 ORF. The protein encoded by ZOR0001_00078 contains a PDE and receiver domain for HK-mediated phosphorylation, suggesting this HK is involved in relaying environmental information such as GlcNAc availability to modulate Aer01 cdG metabolism and thus Aer01 aggregation or motility. We thus named this sensor kinase and response regulator “mucin blind kinase sensor and regulator”, MbkS and MbkR. Given the close association with the Lap system and cdG signaling, the genetic analysis of our MB isolates suggests a model whereby Aer01 increase cdG levels in the presence of GlcNAc using a TCS (MbkSR) and PDE (ZOR001_01301), leading to MbpD-mediated inhibition of MbpG and increased cell surface levels of MbpA.
Aer01 uses distinct molecular signaling pathways for adapting to extra-host and host-associated lifestyles.
Increased levels of cdG stimulate biofilm formation/aggregation and inhibit motility, while depletion of cdG stimulates motility and inhibits aggregation. Since MB1 and MB2 mutations are not in cdG metabolizing enzymes, we predicted MB1 and MB2 would fail to aggregate despite increasing cdG levels, while MB3 and MB4 would have cdG accumulation defects and remain motile. In support of this idea, swim plate analysis on MM or MM + 0.4% GlcNAc indicate MB3 and MB4 are motile in the presence of GlcNAc (Fig. 3B; GlcNAc), which in MB4 corresponds to decreased cdG levels (Fig. 3C). Notably, all MB isolates swim similarly to WT in the presence of proline, a known stimulant of Aer01 motility 44 and on TSA plates (Fig. 3B, Fig. S5), suggesting MB3 and MB4 signaling defects are specific to GlcNAc. Further, induced the expression of the cdG-degrading PDE ZOR0001_03617 stimulated WT motility to MB4 levels in the presence on MM + 0.4% GlcNAc swim plates (Fig. 3D).
Cellular levels of cdG can directly and indirectly impact gene regulation 53,54. To understand how GlcNAc signaling shapes Aer01 transcription, RNA-seq was performed on Aer01 exposed to non-aggregating (Glucose, Glc) or aggregating (GlcNAc) conditions for 6 hr. Differential expression analysis identified 1268 genes differentially regulated in GlcNAc compared to Glucose. Additionally, RNA-seq was performed on MB isolates MB1 and MB4 exposed to GlcNAc, with differential expression analysis identifying 134 and 396 differentially regulated genes respectively compared to WT in GlcNAc (Supplementary Table 1). Consistent with the idea that GlcNAc stimulates aggregation through MbkS signaling, GlcNAc stimulated mbpA expression in WT and MB1, while mbpA expression in MB4 exposed to GlcNAc was similar to WT exposed to glucose (Fig. 3E). A similar transcriptional trend was observed for csgG, which is involved in curli biogenesis and aids in cdG-regulated biofilm formation in species such as E. coli, Aeromonas, and Vibrio 55. Importantly, mbkS levels were similar in the conditions tested.
GlcNAc signaling defects stimulate mucin degradation pathways.
Unexpectedly, MB4 exhibits marked up regulation of genes involved in mucin degradation and GlcNAc scavenging. Both stcE (Fig. 3F) and gbpA (not shown) are upregulated in MB4 compared to WT. StcE is a mucin-selective metalloprotease from E. coli that cleaves mucins as well as O-glycosylated cell surface glycoproteins 56–58. GbpA is a copper-dependent lytic polysaccharide monooxygenases (LPMO) first identified as a GlcNAc-binding protein from Vibrio cholerae that recognizes GlcNAc moieties on mucin and has since been shown to degrade GlcNAc-containing polymers such as chitin 59,60 and stimulate intestinal epithelial proliferation in the gnotobiotic zebrafish intestine 61. Additionally, while GlcNAc stimulated expression of several GH enzymes in WT and MB1, MB4 levels were nearly two fold higher (Fig. 3G). Together, these data suggest MbkS typically tunes Aer01 cdG levels in response to GlcNAc availability in the intestine, adapting Aer01’s physiological strategies to take advantage of available GlcNAc polymers through transcriptional regulation of mucin-degrading secreted factors. Consistent with this idea the MB4 isolate hydrolyzes pNP-GlcNAc more efficiently than WT and MB1 (Fig. 3H) and suppression of GlcNAc-mediated cdG accumulation by overexpressing the PDE ZOR0001_03617 increased WT pNP-GlcNAc activity (Fig. 3I).
MbpA function drives Aer01 aggregation in the intestine.
At the population level, MB isolates exhibit altered spatial organization, being displaced from their typical location in the proximal midgut into the foregut (Fig. 2C). However, fluorescence stereoscope microscopy is unable to discern single-cell resolution. Thus we next characterized the intestinal distributions and colonization dynamics, such as motility and aggregate size, of MB4 and MB1 using complementary confocal and light sheet microscopy approaches. First, using fluorescence confocal microscopy on fixed intestines as previously (Fig. 1A), we found MB1 and MB4 were largely anteriorly displaced into the foregut (Fig. 4A), supporting our fluorescence stereoscope microscopy analysis (Fig. 2CD). Both MB appeared to be more planktonic, seeming to form smaller aggregates than their wild type counterpart isolates and their localization overlapped to some extent with mucin in the midgut (Fig. 4 B and C). We did not observe alterations in the abundance or location WGA-positive goblet cells when larvae were mono-associated with WT, MB1, or MB4 (Fig 1A and Fig 4A), suggesting the planktonic traits of MB1 and MB4 do not impact host mucus production.
Figure 4. MB isolates are more planktonic in the mucus-rich lumen.
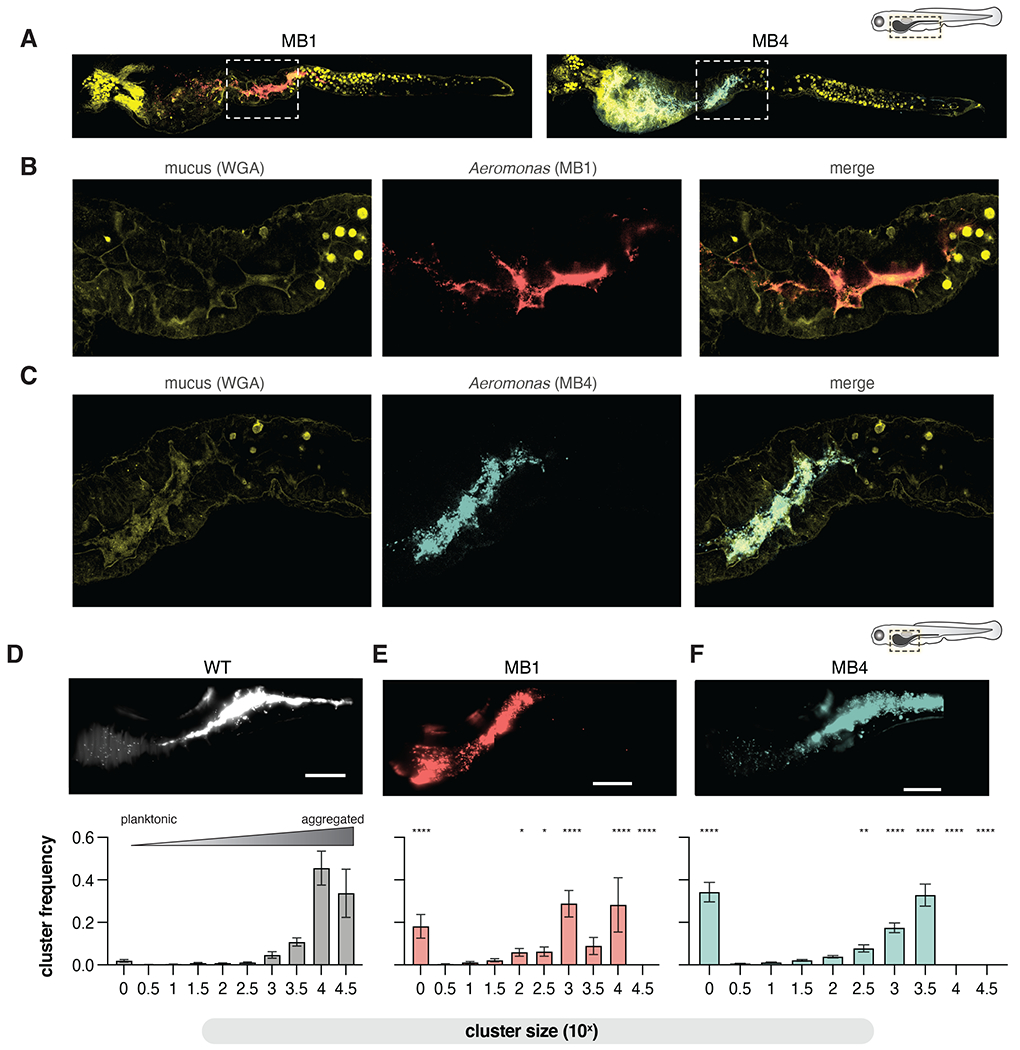
(A) Confocal microscopy showing MB1 (anti-dTom, red), MB4 (anti-dTom, blue), and mucus (WGA-488, yellow) distribution throughout the entire larval zebrafish gut.
(B-C) zoom in of the fore/mid-gut indicated in A and B boxed regions.
(D-F) Top: maximum intensity projections from live light sheet fluorescence microscopy of the distal foregut to proximal mid gut region of the larval zebrafish intestine showing population structures of WT, MB1, and MB4. Bottom: The probability of being in an n-cell cluster for WT, MB1, and MB4. The mean and standard deviation are indicated (WT, N=11; MB1, N=7, MB4, N=6). Two-way ANOVA with multiple comparisons to WT (*p <0.05, **p<0.01, ***p<0.0001). Scale bars=100 μm.
Given the motility differences between MB1 and MB4 motility in GlcNAc-rich culture conditions, we next sought to investigate MB1 and MB4 motility in the larval zebrafish gut using live light sheet fluorescence microscopy. Our analysis revealed striking differences between MB1 and MB4 behaviors in the intestinal foregut environment. While MB1 was predominately found as small aggregates and single non-motile planktonic cells (Movie S4), MB4 was quite motile, with planktonic cells frequently swimming into the epithelium (Movie S5).
To quantify the population structure of Aer01, MB1, and MB4, we again used live light sheet fluorescence microscopy to image a 800 μm region of interest corresponding to the anterior bulb and distal midgut of mono-associated larval zebrafish. The three-dimensional volume of the intestinal lumen was analyzed and Aer01 cells identified and categorized using custom software described previously 62. Here we compare the population structure of MB1 to our previously reported distributions of WT and MB4 62. Representative maximum intensity projections along this area are shown in Fig. 4 D–F. We found WT Aer01 are most often assembled into large multi-cellular aggregates of 104 and 104.5 cells (Fig. 4D), with planktonic cells (100) rarely observed. The cluster size distribution is shifted in MB1 (Fig. 4E) and MB4 (Fig. 4F). MB1 forms smaller (103-104cell) clusters with non-motile planktonic cells (100) dispersed throughout the intestine. The motile MB4 is predominantly planktonic (100) with some smaller cell clusters (up to about 103.5 cells), consistent with our confocal imaging. Together, these data highlight the crucial role of a microbial sensory pathway in determining a symbionts intestinal aggregation, population structure, and behavior along the length of the intestine.
Mucin regulatory pathways tune symbiont inflammation potential independently of motility.
We previously demonstrated that bacterial motility is necessary and sufficient for a pro-inflammatory Vibrio species to elicit intestinal inflammation in the zebrafish intestine 37. We thus anticipated that the highly motile MB4 would elicit more intestinal inflammation than its non-motile WT or MB1 counterparts. To test this hypothesis, we enumerated intestinal neutrophil abundance using transgenic zebrafish Tg(mpx:gfp) with GFP labeled neutrophils (Fig. 5A) 24 hours after mono-association with either WT, MB1, or MB4 (Fig. 5B). Surprisingly, we found both MB1 and MB4 increase intestinal inflammation compared to WT (Fig. 5C) despite their clear motility differences in the host, suggesting motility is not necessary for Aer01-induced inflammation. The increase in intestinal inflammation was apparent when neutrophils were quantified along the entire length of the intestine, as shown, or if neutrophil enumeration was restricted to the mid and distal intestine (data not shown). For subsequent experiments, neutrophil quantification was restricted to the mid and distal intestine. We found no differences in intestinal colonization between WT, MB1, or MB4 (Fig. 5D), indicating heightened inflammation was not due to MB1 or MB4 intestinal overgrowth but rather to alterations in symbiont-innate immune system interactions.
Figure 5. Mucin-blind isolates are more proinflammatory.
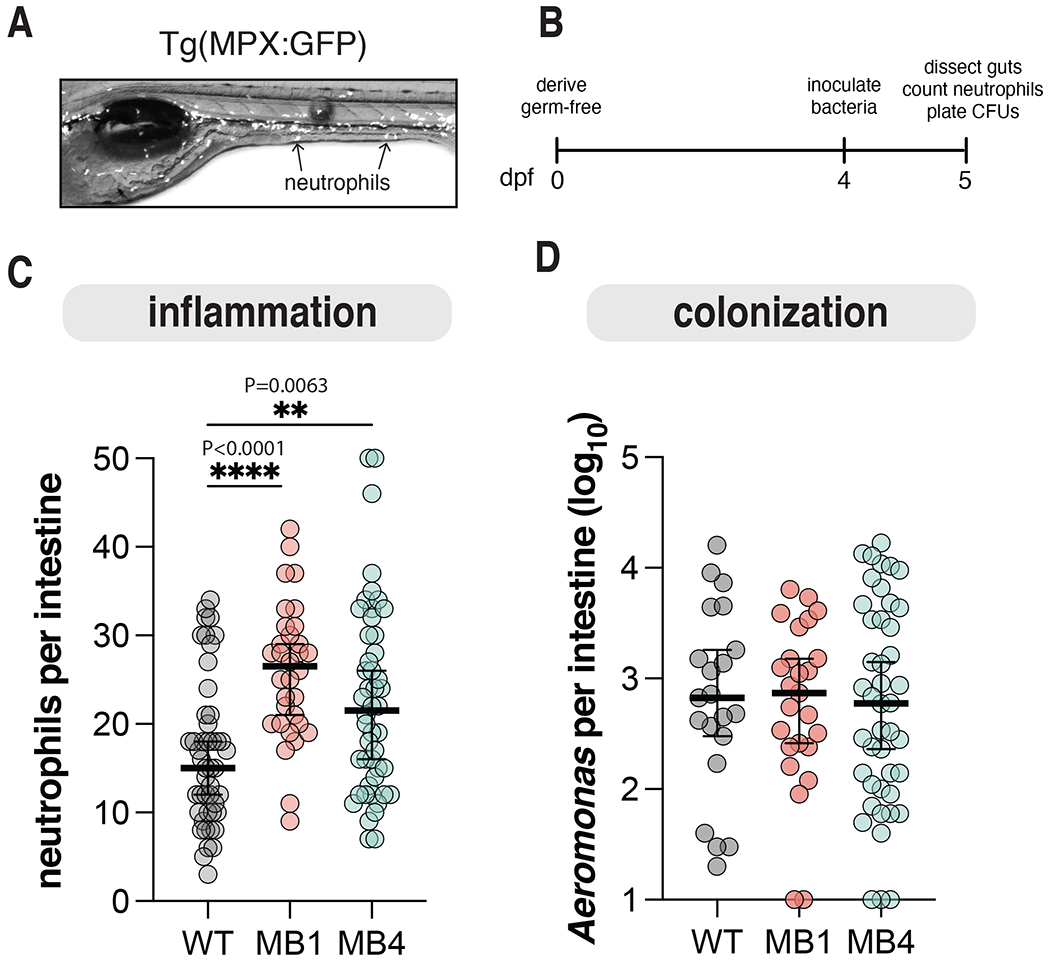
(A) Representative live fluorescence stereoscope microscopy image of Tg(MPX:GFP) larval zebrafish at 5 dpf. Arrows highlight neutrophils.
(B) Schedule for assaying Aer01 abundance and host intestinal inflammation.
(C) Neutrophil abundance from intestines of larval zebrafish mono-associated with the indicated Aer01 strain. Line and error bars represent median with 95% CI For all graphs each dot represents one fish; n ≥ 32 from at least 3 independent experiments. (WT, N=47; MB1, N=32; MB4, N=48). Ordinary one-way ANOVA was performed for Aeromonas abundance and neutrophils (P values: ****>0.0001, **=0.0063).
(D) Intestinal abundance of WT, MB1, and MB4. Line and error bars represent median with 95% CI. WT, N=23; MB1, N=26; MB4, N=46.
MbpA regulation limits Aer01’s inflammation character.
Our unexpected observation that non-motile MB1 cause similar gut inflammation to the hyper-motile MB4 isolate motivated us to explore the idea that Aer01’s inflammatory character is limited largely by proper MbpA regulation inside the gut environment. To accomplish this, we leveraged previous studies 63,64 on LapD-LapG regulation of LapA (MbpD-MbpG and MbpA in Aer01) to design a synthetic construct that can convert WT Aer01 to an MB1 phenotype upon addition of the inducer anhydrotetracycline (aTc). In the Lap system, over-expression of the LapA-targeting LapG protease phenocopies a genetic loss of lapD. Since MB1 contains a LOF in mbpD (Fig. 6A; left, MB1), we reasoned that induced expression of mbpG in WT Aer01 would result in loss of MbpA function and aggregation similarly to the MB1 isolate (Fig. 6A; right, WTmbpG). We engineered an inducible system as shown in Fig 6B: addition of the inducer aTc drives mbpG and mNeonGreen co-expression (mbpG) or only mNeonGreen as a control (empty). These constructs also encode constitutively expressed dTom for tracking live Aer01 cells in the larval zebrafish.
Figure 6. Disruption of MbpA function alters Aer01 spatial organization and increases intestinal inflammation.
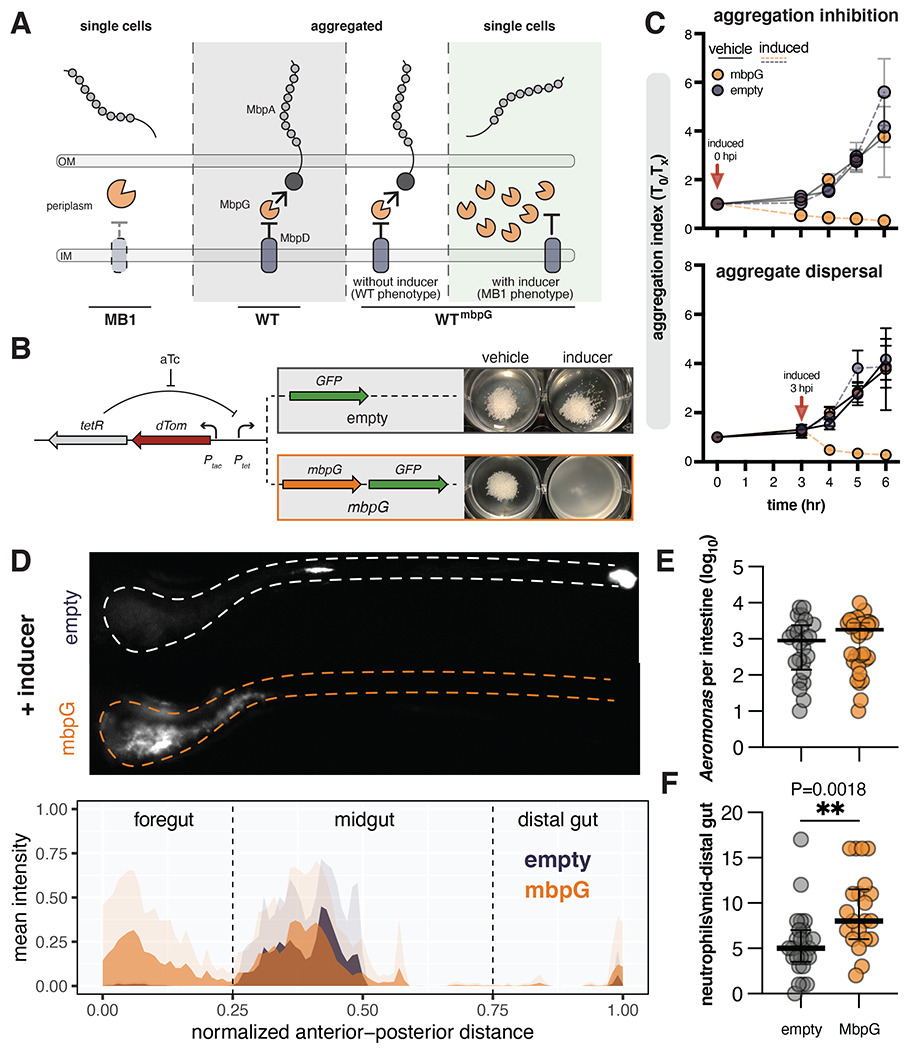
(A) Model of MbpA regulation. Middle (WT) and left (MB1): loss of the protease regulator MbpD in the MB1 mutant leads to constitutive protease activity and MbpA inactivation. Over expression of the MbpA-targeting protease MbpG in WT (WTmbpG) can overcome MbpD regulation, leading to MbpA inactivation.
(B) Left: design of engineered construct to inactivate MbpA activity in WT Aer01. The addition of aTc activates expression of GFP control (empty) or mbpG and GFP (mbpG). Right: mbpG induction is sufficient to suppress GlcNAc-mediated WT Aer01 aggregation in culture.
(C) Induction of mbpG inhibits GlcNAc-mediated aggregation (top) and disperses formed aggregates (bottom). N=3.
(D) Inhibition of MbpA activity via mbpG induction alters WT Aer01 population structure in the intestine, leading to displacement into the foregut region. Top: representative image WT Aer01 carrying either the empty or mbpG constructs outlined in (B). Bottom: population structure analysis across the entire gut. Solid colors indicate the mean value at the normalized distance with transparent colors indicating the standard deviation. WTempty N=5; WTmbpG, N=11.
(E) Intestinal abundance of WTempty and WTmbpG. Line and error bars represent median with 95% CI Unpaired t test. WTempty N=24; WTmbpG, N=34.
(F) Inactivation of MbpA function increases the inflammation character of WT Aer01. Intestinal inflammation is indicated by neutrophil abundance in the mid to distal gut. Line and error bars represent median with 95% CI. Unpaired t test. WTempty, N=25; WTmbpG, N=21.
Consistent with our model outlined in Fig. 6A, mbpG induction inhibited aggregation of Aer01 in the presence of GlcNAc (Fig. 6C, top), phenocopying MB1. Additionally, inducing mbpG expression following Aer01 aggregation dispersed the preformed aggregates (Fig. 6C, bottom), revealing that MbpA regulation is also involved in aggregate dispersion. Next, to test the functionality of our synthetic construct inside the host, we assayed Aer01 spatial organization, colonization, and gut inflammation by mono-associating larval zebrafish 4 dpf with WT Aer01 carrying either the mbpG or empty synthetic cassette and inducer. We found mpbG induction for 24 hours caused similar anterior displacement into the intestinal bulb region as seen in the MB1 isolate (Fig. 6D). The empty cassette control strain formed mid-gut aggregates similar to those previously observed with WT Aer01 (Fig. 6D). As expected, mbpG induction did not impact colonization levels (Fig. 6E). Phenocopying MB1, mbpG induction led to increased intestinal neutrophil abundance in the mid and distal gut (Fig. 6F). Together, these data indicate that symbiont adhesin regulation in response to host-associated environmental cues limits the inflammatory character of Aer01.
A human gut symbiont MbpA analog reduces MB1 induced gut inflammation.
SMART analysis 65 of MbpA’s domain architecture revealed it contains 20 repeats of a putative glycanbinding parallel beta-helix (PbH1) domain, 19 of which are identical in sequence (Fig. 7A, MbpA). To determine if other host-associated microbes encode putative adhesins with similar architecture, we again utilized the SMART database to identify likely adhesins containing PbH1 domains. We focused on adhesins belonging to the Type 5a Secretion System (T5aSS) in Gram-negative bacteria because adhesins of this group are easily identified by the presence of a C-terminal autotransporter domain. Our search for proteins with a C-terminal Autotransporter domain and at least 1 PbH1 domain returned >2,700 adhesins from various Gram-negative bacteria strains that ranged from 559 to 16,945 amino acids in length. Approximately 75% of the bacterial species encoding these adhesins have been identified in human ileum, colon, or stool samples via 16S sequencing according to the IBD TaMMA database 66. We provide a table with the uniport code, species, putative function, and amino acid sequences of MbpA-like adhesins identified (Table S2).
Figure 7. Expression of the Akkermansia adhesin MucA rescues MB1 population structure and reduces MB1 inflammation.
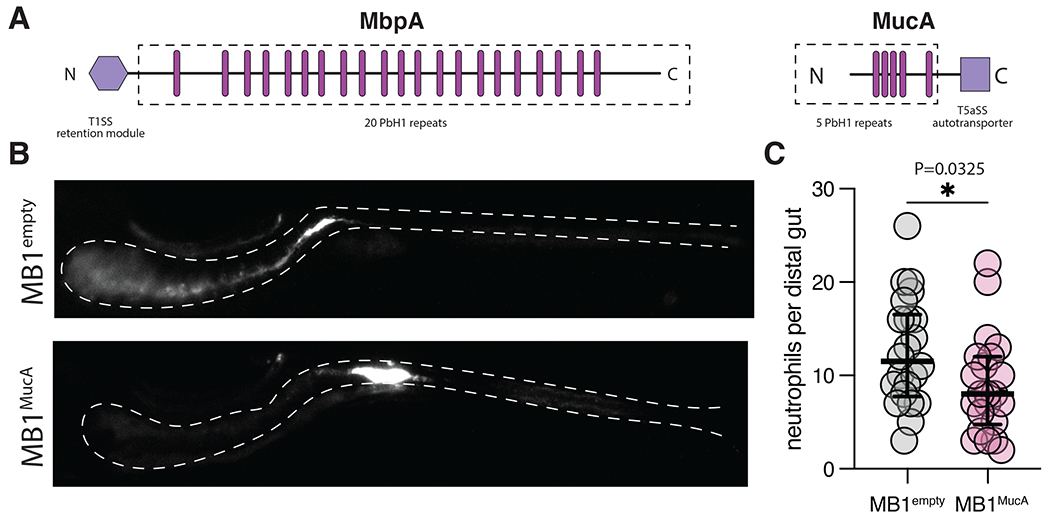
(A) Aeromonas MbpA and Akkermansia muciniphila MucA contain similar domain architectures but are secreted and regulated via distinct mechanisms (T1SS and T5aSS). The dashed boxed region indicates putative surface exposed domains, the pink bars approximate the location of each PbH1 domain as indicated by SMART analysis, and purple hexagons are the domains involved in retaining the respective adhesins at the cell surface. The size of each adhesin is scaled according to the amino acid length.
(B) Expression of MucA rescues population structure defects associated with MB1 in the intestine.
(D) Expression of MucA reduces intestinal inflammation associated with the MB1 evolved isolate. Neutrophil abundance was compared using an unpaired t test (N=22, each). Line and error bars represent median with 95% CI.
We hypothesized adhesins of this group likely function in microbial aggregation or binding glycan-rich substrates such as mucus. To test this idea, we sought to determine if an uncharacterized MbpA analog from Akkermansia muciniphilia--Amuc_1620 (Fig. 7A, MucA for muciniphilia adhesin)—could rebalance MB1 population structure and inflammatory potential. Previous proteomic analysis indicated MucA is ~20 fold more abundant in the outer membrane of Akkermansia grown in mucin-rich growth conditions 14, suggesting Akkermansia and Aer01 deploy functionally analogous adhesins to their cell surface in response to host-associated signals. We reasoned Aer01 can display MucA at its cell surface because T5aSS adhesins are broadly encoded by Gram-negative bacteria (including Aeromonas species) and their transport and insertion into the outer membrane rely on the highly conserved Sec and Bam machinery. Importantly, unlike MbpA, MucA is not a substrate for the Lap system and its function is regulated independently of MbpG activity. This feature allows us to express MucA in the MB1 background (where MbpG is constitutively active) without impacting MucA function.
We used a similar cloning and induction strategy as detailed previously (Fig. 6B) to express the mucA open reading frame from A. muciniphilia ATCC BAA-835 under regulation of the tet operator, such that MucA protein would be induced upon addition of aTc. In support of the idea that E. coli and Aeromonas can translocate and display MucA, we observed the lab strain of E. coli (S17) carrying the mbpA construct formed filament-like aggregate structures upon mbpA induction (data not shown), further suggesting a role for MucA and MbpA-like adhesins in aggregation.
We next tested whether expression of the MbpA analog from Akkermansia in MB1 could repair the altered intestinal population structure and pro-inflammatory properties associated with the MB1 isolate. When induced in MB1 colonizing the zebrafish intestine, MucA largely restored MB1’s population organization and distribution, reducing the population of cells in the foregut and leading to the formation of large aggregates in the anterior midgut region (Fig. 7C), characteristic of the parental Aer01 (Fig. 2C). Concomitant with this rescue of population structure, mucA expression in MB1 significantly reduced the capacity of this strain to elicit intestinal inflammation. Together, these data suggest a broad role for MbpA-like adhesins in bacterial aggregation and host immunomodulation in response to mucin.
Discussion
Mucin’s physical barrier function has long been appreciated for its ability to modulate the structure and function of microbiota 67. In this study, we demonstrate the active role bacterial play in determining their population structure and host interactions within the gut. We show that the zebrafish mutualist Aer01 governs its gut distribution and inflammatory character in response to host mucin signals by regulating the activity of a large ~500 kDa adhesin we named MbpA. Microbial adhesins are well know for their role in biofilm formation and aggregation in culture, but their contribution to the organization and distribution of microbial communities in the intestine and corresponding impacts on host health are poorly understood. Our work demonstrates that Aer01 detects molecular cues from the healthy gut mucus ecosystem (GlcNAc) using a TCS sensor kinase (MbkS) to control various aspects of its physiology inside the host including tuning the transcription and cell surface localization of MbpA--an adhesin required for aggregation in response to GlcNAc.
Bacterial sensing of environmental cues such as mucin glycans through TCSs can rapidly alter transcription and physiological behaviors to change their interactions with the host immune system. For example, the TCS RprXY of Enterotoxigenic Bacteroides fragilis represses production of the metalloprotease B. fragilis toxin (BFT) in response to yet to be determined mucus signals, and induces BTF expression when the mucus ecosystem is compromised 11. Similarly, in Pseudomonas aeruginosa mucin glycans act through the sensor kinase RetS to inhibit GacS and Type-6 secretion system (T6SS) activity and toxin secretion 9. RNA-seq analysis of WT Aer01 and the MB4 isolate exposed to GlcNAc in culture indicate MbkS activity modulates the transcription of mbpA, motility-associated genes (ie,fliD and fliE), and several mucin-degrading enzyme genes (stcE and gbpA), the latter of which induces proliferative responses in the zebrafish intestinal epithelium 61. In the gut, the motile MB4 isolate resists formation of large aggregates characteristic of its WT counterpart and elicits more intestinal inflammation. We initially hypothesized that MB4’s increased motility would render it especially pro-inflammatory, possibly through elevated flagellin-TLR5 mediated NF-κB signaling 32,68. Additionally, MB4’s increased mucus degrading capacity is a microbial attribute associated with increased intestinal inflammation 69. Unexpectedly, the MB1 isolate--which is non-motile and has WT levels of mucin degrading gene expression and activity—is equally as pro-inflammatory as MB4.
A trait that is common to these two pro-inflammatory isolates is defective MbpA cell-surface regulation, indicating a crucial role for this GlcNAc-responsive adhesin in limiting the inflammatory capacity of Aer01. Consistent with this idea, controlled inhibition of MbpA function in WT Aer01 is sufficient to stimulate intestinal inflammation and displaces Aer01 from the anterior midgut region into the foregut, mimicking traits of the evolved MB1 isolate. We found the PbH1 domain repeat architecture of MbpA in various adhesins encoded by vertebrate intestinal symbionts, including human-associated Akkermansia species. Like Aer01, Akkermansia increases production of a PbH1-containing adhesin (Amuc_1620) in the presence of mucin 70, suggesting a similar role in mucus-mediated aggregation. Consistent with this idea, ectopic Amuc_1620 expression is sufficient to rescue aggregation defects of MB1 isolate in culture and in the intestine and to reduce its pro-inflammatory activity.
We speculate that MbpA activity could limit Aer01’s inflammatory potential through two potential mechanisms. Aer01 interactions with mucin could shield potentially immunogenic epitopes from host detection, as has been shown for some mucin absorbed bacteria 71. Additionally, MbpA could act on host cells directly to inhibit inflammatory behaviors, as has been shown for a PbH1-containing adhesin from Streptococcus pneumoniae that inhibits phagocytosis 72. The impacts of PbH1 proteins are likely context-dependent and contingent on local mucin-derived cues and bacterial behaviors that they regulate. One PbH1 adhesin identified in our bioinformatic analysis, Fap2 of Fusobacterium nucleatum, regulates co-aggregation between F. nucleatum and Porphyromonas gingivalis 73 and binds Gal-GalNAc epitopes which are overexpressed in colon rectal cancer (CRC). Fap2 was found to direct F. nucleatum localization to mouse colon tumors 74 and promote invasion of cultured HCT116 CRC cells and stimulation of IL-8 and CXCL1, which are associated with CRC progression 75.
Similar to Fabp2’s role in F. nucleatum and P. gingivalis co-aggregation, we have shown that MbpA plays a role in bacterial-bacterial interactions in the larval zebrafish intestine. In model bacterial communities in gnotobiotic zebrafish, we found that Aer01 and a highly aggregated zebrafish isolate Enterobacter ZOR0014 (Ent14) have strong antagonistic relationships 76. In combination with the MB4 variant of Aer01, Ent14 was at an even greater colonization disadvantage than with WT Aer01. The presence of MB4 dramatically alters Ent14’s population structure, causing Ent14 to rapidly disaggregate. This disaggregation behavior may be due to MB4’s altered cell surface chemistry or its increased production of carbohydrate-active enzymes, as many mucin-associated glycans, including GlcNAc, are also components of bacterial extracellular polysaccharides 77. MB4 could also indirectly act on Ent14 aggregates through its pro-inflammatory capacity in the host, with disaggregation being triggered by MB4-induced host inflammation.
As microbiome research transitions from descriptive to mechanistic studies, it will be important to identify the most tractable features of host-microbe systems that can be tuned to promote system health. Our study reveals how bacteria sense and respond to a dominant feature of the intestinal environment, mucin glucans, to regulate their cell wall properties and aggregation behavior. In a recent study, we show the importance of this process in determing the community structure of a 5-member synthetic community 62, as MB4 is able to disaggregate and displace community members such as Ent14, likely due to its increased production of secreted factors capable of degrading carbohyrdate matricies such as bacterial EPS (Fig. 3G). Invasion experiments conducted in this previous study show MB4 also disaggregates Aer01, allowing MB4 to overtake Aer01. Cell surface adhesion expression impacts not only immediate properties of the bacterial cells such as the size of cellular aggregates in which they reside, but also organ-wide features such as bacterial distribution along the length of the gut, bacterial-bacterial competitive interactions, and host inflammatory tone. These system-wide features are altered both by impairment of bacterial mucin sensing and even by disruption of a single feature of their mucin response program, cell surface retention of a mucin-responsive adhesin. These findings point to multiple avenues for manipulating microbiota-member behaviors. As a proof of concept, we demonstrate that expression of a MbpA-like Akkeromansia mucin-responsive adhesin in Aer01 is sufficient to reverse its proinflammatory activity. Engineered mucin-responsive cell surface adhesins may be useful in other context to mask proinflammatory bacterial cell surface molecules, aggregate proinflammatory community members, or modulate host immune responses.
STAR methods
RESOURCE AVAILABILITY
Lead contact
Further information and requestis for resources should be directed to the corresponding author, Karen Guillemin (kguillem@uoregon.edu).
Materials availability
Bacterial strains and plasmids generated by this study are available through the lead contact.
Data and code availability
The raw sequence reads can be accessed using the BioProject Accession: PRJNA988826.
Code for identifying bacterial single cells and clusters has been deposited at GitHub (https://github.com/rplab/Bacterial_Automated_Pipeline).
Any additional information to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECTS DETAILS
Ethics statement
All experiments with zebrafish (Danio rerio) were performed according to protocols approved by the University of Oregon Institutional Animal Care and Use Committee (IACUC) and following standard protocols. The animal protocol number for this work is 20-16.
Zebrafish (Danio rerio) studies
All experiments with zebrafish (Danio rerio) were performed according to protocols approved by the University of Oregon Institutional Animal Care and Use Committee (IACUC). All animal experiments were conducted with larval zebrafish aged 4-7 dpf that have yet undergone sexual differentiation, which occurs 20-25 dpf. The zebrafish strains employed in this study were wild type (ABxTU), myd88−/− 40, and Tg(BACmpx:GFP)i114 (referred as MPX:GFP) 78. Fish were not fed in any of the experiments described here.
Gnotobiotic (germfree, GF) larval zebrafish were derived as previously described 79. Briefly, 0 days post fertilization (dpf) embryos were washed with sterile embryo medium (SEM) containing antibiotics (100 μg/ml Ampicillin, 250 ng/ml Amphotericin B, 10 μg/ml Gentamycin, 1 μg/ml Tetracycline, 1 μg/ml Chloramphenicol), then sequentially submerged in bleach (0.003%) and iodine (0.1%) solutions. SEM contains: NaCl (0.8 g/L), KCl (40 mg/L), Na2HPO4 (4 mg/L), KH2PO4 (6 mg/L), CaCl2,2H2O (1.2 mM), MgSO4,7H2O (1 mM), NaHCO3 (4.2 mM). Sterilized embryos were transferred to tissue culture flasks containing SEM at densities of 1 embryo/mL (10 mL per flask and 10 embryos). The flasks were stored in a temperature controlled room at 28°C on a 14 h/10 h light/dark cycle.
Prior to all imaging experiments, larvae were anesthetized in SEM including 20 μL/mL MS-222 (Tricaine). Larvae were euthanized using SEM with 40 μL/mL MS-222.
Bacterial strains.
This study uses the bacterial zebrafish isolate Aer01 (Aeromonas veronii ZOR0001; BioProject Accession: PRJNA205571). Variants of Aer01 or evolved mucin blind isolates were generated using yeast cloning derivatives of previously described genetic tools4 that are detailed in “Generation of pYTn7xTS and aTc-inducible plasmids”. These tools allow chromosomal insertion of a DNA cassette of interest at the attTn7 site. For all experiments, bacterial strains were cultured overnight at 30°C in lysogeny broth (LB) medium with shaking. Gentamicin (10μg/mL), kanamycin (50μg/mL), or ampicillin (100 μg/mL) were used when noted. Tn7-tagged Aer01 were maintained in gentamicin. When selecting for recombinant Aer01 tagged using pTn7xTS or pYTn7xTS, the triparental conjugation mixture was plated on tryptic soy agar (TSA) containing 25 μg/mL gentamicin and 50 μg/mL kanamycin and grown for 18 hr at 37°C. For anhydrotetracycline (aTc) induction, aTc was added at 50 ng/mL in culture or 200 ng/mL to flask water. For bacterial experiments, the minimal medium recipe (MM) is identical to SEM detailed above, but supplemented as indicated in the experimental design. All experiments were conducted under traditional sterile technique. All strains described here are cryopreserved in 25% glycerol at −80°C.
Methods Details
Generation of pYTn7xTS and aTc-inducible plasmids.
pYTn7xTS
To generate pYTn7xTS, yeast cloning was used to replace the AmpR cassette of pTn7xTS with an autonomously replicating yeast centromere and URA3 gene cassette for yeast cloning compatibility from the plasmid pMQ30 80. Primers JP27/JP28 were used to amplify the yeast centromere and URA gene cassette from pMQ30 and JP29/JP30 were used to amplify pTn7xTS lacking the AmpR cassette. To facilitate homologous recombination in yeast, the JP27 and JP30 contain 42 base pairs of homology and JP28 and JP29 share 20. The PCR products were recombined to engineer pYTn7xTS using yeast cloning as previously described80. Briefly, URA- auxotrophic yeast were grown in YPD at 30°C for 24 hours. A 0.5 mL aliquot of overnight yeast culture growth was gently pelleted at 8K rcf for 30 sec and washed 2X in sterile T.E. buffer. Yeast cells were resuspended in 0.5 mL Lazy Bones Solution (40% polyethylene glycol (MW 3350), 0.1M Lithium acetate, 10 mM Tris-HCl pH7.5, 1 mM EDTA). The JP27/JP28 and JP29/JP30 PCR products were added to the solution with 40 μg of boiled salmon sperm DNA (carrier DNA). The yeast cloning reaction was allowed to incubate for 24 hours at room temperature and the reaction was plated on URA-plates (6.7 YBN, 0.76 g CSM-URA, 15 g Dextrose, 20 g Agar per liter) after washing 2X in 1 mL T.E. buffer. The plates were incubated at 30°C for 3-5 days until colonies appeared. The assembled pYTn7xTS plasmid was purified using the Zymoprep Yeast Plasmid Miniprep II Kit and transformed into E. coli S17.
aTc-inducible mbpG, mbpD, ZOR0001_03617, and mucA plasmids
The pYTn7xTS tagging constructs for inducing mbpG, mbpD, ZOR0001_03617 (PDE), and mucA expression were modeled after aTc-inducible genetic switches recently engineered by our lab 37. The Tet operator and Ptac::dTomato-TetR regions were first amplified from pTW32437 using the primer pairs JP85/JP86 (TetO elements) and JP83/JP84 (constitutive dTomato, TetR elements). The mbpG ORF was amplified from Aer01 genomic DNA using primer pairs JP165/JP88. The mbpD ORF was amplified using primers JP175/JP175. The mucA ORF was amplified from Akkermansia muciniphilia BAA-835 (MucT) genomic DNA using primers JP179/JP180. All PCR products were purified using the QIAGEN PCR Purification Kit (Cat # 28106). For yeast cloning, the JP85/JP86 and JP83/JP84 reactions were added with mbpG, mbpD, ZOR0001_03617, or mucA reactions and SmaI-digested pYTn7xTS following the yeast cloning protocol detailed above. A. mucinphilia MucT was a gift from Dr. Raphale Valdivia’s lab. Oligonucleotides used in this study are provided in Table S3.
Triparental mating.
Aer01 and MB isolates were tagged with the pTn7xTS or pYTn7xTS constructs as recently described 4. Briefly, triparental conjugation was performed with the Aeromonas target strain, an E. coli SM10 donor strain carrying the transposase-containing pTNS2 helper plasmid, and an E. coli S17 donor strain carrying the pTn7xTS or pYTn7xTS-based tagging construct. All strains were grown in LB supplemented with the appropriate antibiotic. To set up the mating reaction, overnight growth from each strain was diluted 1:50 in fresh LB and grown to 0.5 OD600. A 750 μl aliquot of each strain was washed 2X in PBS, mixed, resuspended in 25 μl PBS and then the entire reaction spotted on a LB agar plate and incubated for 6 hr at 30°C. The cell mass was transferred to 1 mL of fresh PBS. To select for Aer01 recombinants, the triparental mating was plated on TSA plates containing gentamycin and kanamycin, as Aer01 is naturally KanR but both donor strains are KanS.
Aggregation assay.
Overnight cultures of Aer01 were washed 3 times in MM, then diluted 1:10 in 2 ml MM containing 0.4% GlcNAc (wt/vol.) in a 12-well plate and incubated at 30°C for 6 hr with gentle rotation (120 RPM). To quantify aggregation, a sample of the planktonic fraction containing unaggregated Aer01 cells was taken. The Aer01 aggregates were gently resuspended by pipetting and a sample of the resuspension representing the total bacteria taken. The OD600 for the total and planktonic samples were taken and the aggregation index calculated as the ratio of total/planktonic cells.
Experimental evolution of Aer01.
To select for Aer01 that do not aggregate when exposed to GlcNAc, the aggregation assay was performed as detailed above, but after 6 hr, a 250 μl sample of the planktonic (non-aggregated) fraction was inoculated in LB to enrich for isolates that remain unaggregated in GlcNAc. The following day, a sample of the overnight growth was cryopreserved and the aggregation assay was performed on the passaged sample. A total of 7 passages were performed.
Evolved isolates were purified by plating cryopreserved stocks of whole populations from passages 5 and 6 on TSA plates. After incubating for 18 hr. at 30°C, isolates were randomly picked and grown overnight in TSB with shaking in a 96-well plate. Isolates were cryopreserved in 96-well plates in 25% glycerol and stored at −80°C.
Preparation of genomic DNA for sequencing.
Genomic DNA was extracted from overnight cultures grown in LB using the QIAGEN DNeasy Blood & Tissue Kit (QIAGEN Cat#69506) according to the manufacture’s specifications. Library preparation and sequencing were conducted at the Microbial Genome Sequencing Center (MiGS, https://www.migscenter.com/).
Preparation of RNA for sequencing.
RNA was harvested from wild type Aer01 grown for 6 hr. in MM supplemented with 0.4% glucose or 0.4% GlcNAc, and MB1 and MB4 grown in MM supplemented with 0.4% GlcNAc using the RNeasy Qiagen kit (Cat. #74104). Sequencing and transcript analysis (transcript level quantification, count normalization, and differential expression analysis) was performed at the Microbial Genome Sequencing Center (MiGS, https://www.migscenter.com/).
Quantification of pNP hydrolysis.
Stock solutions of 0.1% pNP-GlcNAc, pNP-Gal, pNP-Fuc, or pNP-GalNAc were diluted 1:1 in buffer solution (MM) containing stationary Aer01 cells diluted to OD600~0.05. pNP hydrolysis (indicated by OD405) was monitored for 5 hours in a 96-well plate (100μl total volume) at 30°C with 120 rpm gentle rotation. Readings were taken every 5 minutes on BMG LABTECH FLUOstar Omega.
Swim plate assay
Overnight cultures were grown in LB at 30°C. Each strain normalized to an OD600 of 5, washed in 1 ml MM, then concentrated 10 fold in 100 μl MM. The cell concentrate (5 μl) was spotted onto freshly prepared swim agar plates (0.2% agar) as detailed below. MM swim plates supplemented with 0.4% GlcNAc or 1 mM Proline were made by heat-dissolving 100 mg agar in 20 ml MM, cooling slightly, then adding 30 ml MM supplemented with GlcNAc, Proline, no supplement. The media was added to 100 mm petri dishes at 20 ml per plate, cooled for 2 hrs then inoculated with the parental Aer01 and the MB isolates. The swim plates were incubated for 24 hr at 30 °C and the swim diameter was measured. The plates were imaged from a fixed distance and the swim diameter measured as the pixel length in Fiji.
Quantification of intracellular c-di-GMP
Levels of intracellular c-di-GMP were measured using the Luceran cyclic-di-GMP assay kit. Briefly, the aggregation assay was performed with Aer01, MB1 and MB4. After 6 hr each strain was normalized to OD600 of 0.5 in RNase-free water in a total volume of 70 μl. The assay reagents (50 μl CA buffer, 50 μl of BC reagent, 20 μl 10X DFHBI-1T, and 10 μl 20X c-di-GMP sensor) were mixed with the bacterial cell sample and incubated for 30 min at 25°C. GFP readings, which correspond to c-di-GMP levels, were read at ex/em of 485/520. Values are reported as relative to the parental Aer01 strain.
Confocal microscopy of Aer01 and mucus distribution in fixed larvae.
Germfree larval zebrafish inoculated on 4 dpf with dTomato-tagged WT, MB1 or MB4 Aer01 and colonized larvae were fixed 24 hr later in PBS containing 4% paraformaldehyde (PFA). Larvae were rinsed in PBS, washed in PBS + 0.5% Triton-X, and permeabilized in deionized water. Larvae were blocked in PBS containing 5% NGS, 2% BSA, 1% DMSO (blocking solution). Aer01 tagged with dTomato were labeled by incubating larvae in blocking solution containing Rabbit anti-dsRed (Takara Bio #6324960) at 1:500 dilution. The primary anti-body was washed away in PBS containing 1% DMSO (wash buffer) and larvae were incubated in blocking solution containing Goat anti-Rabbit546 (ThermoFisher #A11010) at 1:1000 dilution. Secondary antibody was washed away in wash buffer. Host mucus was labeled by adding WGA-FITC (Vector Laboratories, #FL-1021) at 1:100 in wash buffer. Samples were mounted in Prolong Diamond antifade (Cat # P36966). Image acquisition was performed using a Leica SPE laser scanning confocal microscope.
Analysis of Aer01 population structure by stereomicroscopy.
Germfree larval zebrafish were mono-associated on 4 dpf and imaged 24 hr later on 5 dpf. Before imaging, larvae were anesthetized in MS-222 as detailed above and mounted onto glass slides containing a layer of 4% methylcellulose (wt/vol.). A Leica M165 FC Fluorescent Stereo Microscope was used to image of the spatial organization of dTomato labeled wild type, MB1, MB2, MB3, and MB4 Aeromonas throughout the larval zebrafish gut. The full length of the intestine was manually outlined from the images and the pixel intensity across this region measured using plot profile function in Fiji. The gut length for each fish was normalized to the total pixel distance and the mean intensity at every 1/100 normalized distance calculated. The standard deviation at each position was calculated for each group and plotted with the mean values using the ggplot2 package in R.
Live light sheet fluorescence imaging of MB isolates
3D imaging of the larval zebrafish gut was conducted using a previously described home-built set-up 81. GF larvae were inoculated with dTomato-labeled Aer01, MB1 or MB4 5 dpf and imaged 7dpf using a custom-built light sheet microscope described in detail elsewhere81. Larvae are anesthetized by placing in MS-222 (Syndel) solution in sterile embryo medium (20μL/mL) for approximately 2 minutes and mounted in 0.6% agarose gel using glass capillaries. The larvae are extruded from the capillaries within a temperature controlled (30°C) sample chamber for imaging. The approximately 1mm larval zebrafish gut is imaged in 3D in 4 sub-regions in ~45s. Lasers at 488 nm and 568nm were used to excite for mNeonGreen and dTomato when indicated. After imaging, larvae are euthanized by placing in MS-222 solution in sterile embryo medium (40μL/mL).
Identification of single cells and clusters in 3D light sheet fluorescence images.
Single cells and cell clusters were identified as recently described62. Briefly, a convolutional neural network was used to classify bacteria and noise blobs 82. Potential objects were classified by the network and the output was curated manually. For aggregate segmentation, the zebrafish gut was segmented in 2D, images were threshold and the total number of cells in an aggregate determined by dividing the total fluorescence intensity from the segmented object by the median intensity of individuals in the image. Once the single cell and aggregate net populations were determined, they were used to calculate the planktonic fraction. The code is available at https://github.com/rplab/Bacterial_Automated_Pipeline.
Aer01 and mucin distribution in culture.
Aer01 aggregates were developed as detailed in aggregation assay but in buffer containing WGAFITC-stained mucin and imaged using fluorescence stereomicroscopy. To stain mucin, WGAFITC was added to buffer supplemented with 0.4% Type III Mucin (Sigma, cat #M1778) at 5 μg/mL. The solution was incubated for 15 min at room temperature with gentle rotation in the dark then pelleted and washed three times in sterile MM to remove unbound WGAFITC. The 12-well plate was covered in aluminum foil during incubation.
Quantification of bacterial colonization.
Flasks containing germfree larval zebrafish were inoculated with ~106 CFU/ml of Aer01 or the indicated mucus blind isolate on 4 dpf and colonization measured 24 hr later by dissecting and homogenizing individual intestines following euthanasia with Tricaine. For each measurement, an individual intestine was added to a 1.6 ml tube containing 1 mL SEM and 100 μl of 0.5 mm zirconium oxide beads and homogenized using a bullet blender tissue homogenizer (Next Advance, Averill park, NY, US) for 30 seconds at the power 4 setting. The mixture was plated (100 μl) on TSA and incubated for 18 hrs at 30°C.
Quantification of intestinal neutrophils.
The intestines of Tricaine-euthanized 5 dfp Tg(MPX:GFP) larval zebrafish were dissected using previously described methods44. Fluorescent myeloperoxidase positive (MPX+) cells were visualized using fluorescent stereomicroscopy and manually enumerated throughout the entire gut or in the mid and distal intestine. Prior to counting, the researcher was blinded to the treatment groups.
Identification of human-associated microbiota with PbH1 adhesins.
The SMART database (http://smart.embl-heidelberg.de/) was queried using the term “PbH1 AND Autotransporter”. The species retrieved from this search were cross-referenced with a list of bacteria species in the IBD TaMMA (https://github.com/Humanitas-Danese-s-omics/ibd-meta-analysis-data/blob/main/manual/bacteria_species_list.tsv).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analyses were performed using Prism9 (GraphPad Software). The statistical test used and number of samples are included and described in each figure legend. For fish experiments N represents the number of animals. For bacterial experiments N represents the number of biological replicates. Group statistics (e.g., mean or median) are indicated in the figure legends when plotted. P values of p≥0.05 not significant (ns), p<0.05 (*), p<0.01 (**), p<0.001 (***), and p<0.0001 (****).
Supplementary Material
Movie S1. Mucus distribution in a live larval zebrafish, Related to Figure 1 and S1. Scan of z-stacks used to for the maximum projection image in Fig. S1. Green indicates WGA-FITC staining.
Movie S2. Association between Aer01 and mucus signals in the anterior midgut of fixed intestines, Related to Figure 1. Scan of z-stacks from region used for the maximum projection image in Fig. 1B. Aer01 and mucus are colored as described in main figure.
Movie S3. Association between Aer01 and mucus signals in the distal midgut of fixed intestines, Related to Figure 1. Scan of z-stacks from region used for the maximum projection image in Fig. 1C. Aer01 and mucus are colored as described in main figure.
Movie S4. MB1 are planktonic and non-motile in the zebrafish foregut, Related to Figure 4. Real-time video of MB1 physiology acquired using light sheet fluorescence microscopy on single oprical plane.
Movie S5. MB4 are planktonic and motile in the zebrafish foregut, Related to Figure 4. Real-time video of MB4 physiology acquired using light sheet fluorescence microscopy on single oprical plane.
Figure S1. Mucus distribution in live larval zebrafish, Related to Figure 1. Maximum intensity projection of WGA-FITC staining (green) of gut and skin mucus in live larval zebrafish aged 6 dpf acquired using light sheet fluorescence microscopy. Individual z-stacks are shown in Movie S1.
Figure S2. Aer01 associates with WGA-stained mucin in culture, Related to Figure 2. dTomato-tagged Aer01 were exposed to 0.4% mucin that was stained with WGA-FITC (left) or untreated (right) in the aggregation assay. The merged image shows Aer01 (red) associated with WGA-stained mucin (green). WGA-stained mucin does not aggregate unless Aer01 is present (not shown).
Figure S3. Aer01 distribution in two models of mucus depletion, Related to Figure 2. (A) Distribution of dTom-tagged Aer01 mono-associated with myd88−/− larval zebrafish using the experimental design highlighted in Fig 2B; n=7. (B) Distribution of dTom-tagged Aer01 mono-associated in wild type fish treated with WGA-FITC.
Figure S4. MB1 GlcNAc-mediated aggregation is rescued by mbpD expression, Related to Figure 3. Aggregation assay in GlcNAc with MB1 expressing empty vector or the wild-type mbpD allele. Expression of mbpD rescues GlcNAc-mediated aggregation of MB1 which contains a LOF mutation in mbpD.
Figure S5. Swim plate analysis on TSA, Related to Figure 3. WT and MB1-4 were inoculated onto TSA swim plates and incubated for 12 hr at 30°C. Plates were imaged and the swim distance calculated in FIJI. MB isolates were normalized to the plate-specific WT control.
Supplemental Table 1. RNA-seq analysis, Related to Figure 3.
Supplemental Table 2. MbpA-like adhesins from bacteria identified in IBD TaMMa, Related to Figure 7.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-dsRed | Takara Bio | Cat # 6324960 |
| Goat anti-Rabbit546 | ThermoFisher | Cat # A11010; RRID:AB_2534077 |
| Bacterial and virus strains | ||
| Aeromonas veronii ZOR0001 | Karen Guillemin lab | BioProject Accession: PRJNA205572 |
| Aeromonas veronii ZOR0001; dTomato | Karen Guillemin lab | NA |
| Aeromonas veronii ZOR0001; dTomato, aTc-mbpG | This study | NA |
| Aeromonas veronii ZOR0001; dTomato, aTc-mbpD | This study | NA |
| Aeromonas veronii ZOR0001; dTomato, aTc-empty | This study | NA |
| Aeromonas veronii ZOR0001 MB1 | This study | NA |
| Aeromonas veronii ZOR0001 MB1; dTomato | This study | NA |
| Aeromonas veronii ZOR0001 MB1; dTomato, aTc-mucA | This study | NA |
| Aeromonas veronii ZOR0001 MB2 | This study | NA |
| Aeromonas veronii ZOR0001 MB2; dTomato | This study | NA |
| Aeromonas veronii ZOR0001 MB3 | This study | NA |
| Aeromonas veronii ZOR0001 MB3; dTomato | This study | NA |
| Aeromonas veronii ZOR0001 MB4 | This study | NA |
| Aeromonas veronii ZOR0001 MB4; dTomato | This study | NA |
| Escherichia coli S17 | Karen Guillemin lab | NA |
| E. coli SM10 pTNS2 | Karen Guillemin lab | NA |
| E. coli SM10 pTn7xTS-dTomato | Karen Guillemin lab | Addgene #117391 |
| Chemicals, peptides, and recombinant proteins | ||
| Type III porcine gastric mucin | Sigma | Cat # M1778 |
| N-acetylglucosamine (GlcNAc) | Chem-Impex | Cat #1426 |
| D-Galactose (Gal) | Sigma-Aldrich | Cat # G0625 |
| N-acetylgalactosamine. (GalNAc) | Chem-Impex | Cat # 21629 |
| Sialic Acid (SA) | Sigma-Aldrich | Cat # 1612619 |
| L-Fucose (Fuc) | Chem-Impex | Cat # 736 |
| D-Glucose (Glu) | Acros | Cat # 170080010 |
| pNP-B-D-GlcNAc | GoldBio | Cat # N-230; CAS: 3459-18-5 |
| pNP-B-D-Gal | GoldBio | Cat # N-290; CAS: 3150-24-1 |
| pNP-B-D-GalNAc | GoldBio | Cat # N-200; CAS: 14948-96-0 |
| pNP-A-L-Fuc | GoldBio | Cat # N-250; CAS: 10231-84-2 |
| Anhydrotetracycline (aTc) | Takara | Cat # 631310 |
| Yeast Nitrogen Base without Amino Acids | Research Products International | Cat # Y20040 |
| CSM-URA | Sunrise Science | Cat # 1004-100 |
| Critical commercial assays | ||
| QIAquick PCR Purification | QIAGEN | Cat # 28106 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat # 27104 |
| QIAGEN DNeasy Blood & Tissue Kit | QIAGEN | Cat # 69506 |
| RNeasy | QIAGEN | Cat # 74104 |
| Zymoprep Yeast Plasmid Miniprep II | Zymo Research | Cat # D2004 |
| Cyclic-di-GMP Assay | Lucerna Technologies | Cat # 200-100 |
| Deposited data | ||
| Raw Sequence Reads | This study | SRA Accession: PRJNA988826 |
| Experimental models: Organisms/strains | ||
| AB X Tu wild type zebrafish; Danio rerio | Univ. of Oregon Zebrafish facility | NA |
| MPX:GFP reporter zebrafish; Tg(BACmpx:GFP)i114 | Guillemin lab (Renshaw et al, 2006) | NA |
| Myd88 mutant zebrafish, myd88−/−, Danio rerio | Guillemin lab (Troll et al, 2018) | NA |
| Oligonucleotides | ||
| See supplemental Table S3 | This study | NA |
| Recombinant DNA | ||
| pYTn7xTS | This study | NA |
| pYTn7xTS-dTomato, aTc-empty | This study | NA |
| pYTn7xTS-dTomato, aTc-mbpG | This study | NA |
| pYTn7xTS-dTomato, aTc-mbpD | This study | NA |
| pYTn7xTS-dTomato, aTc-mucA | This study | NA |
| pYTn7xTS-dTomato, aTc-PDE | This study | NA |
| Software and algorithms | ||
| Prism 9 | Dotmatics | https://www.graphpad.com |
| SnapGene | Dotmatics | https://www.snapgene.com |
| breseq | Deatherage and Barrick, 2014 | https://barricklab.org/twiki/bin/view/Lab/ToolsBacterialGenomeResequencing |
| Fiji | ImageJ | https://imagej.net/software/fiji/downloads |
| Identification of bacterial cells and clusters in 3D light sheet fluorescence images | Parthasarathy Lab University of Oregon | https://github.com/rplab/Bacterial_Automated_Pipeline |
| Other | ||
| WGA-FITC | Vector Laboratories | Cat #FL-1021; RRID:AB_2336866 |
| Prolong Diamond antifade | ThermoFisher | Cat #P36966 |
| MS-222 (Tricaine) | Syndel | NA |
Highlights.
Bacterial aggregates are a hallmark of healthy gut microbial communities
In Aeromonas, MbpA controls aggregation in response to mucus-associated GlcNAc
Strains lacking MbpA are proinflammatory and spatially reorganized within the gut
A MbpA-like Akkermansia adhesin restores Aeromonas distribution and gut health
Acknowledgments
We thank Cathy Robinson for her motility assay insight. We thank Rose Sockol and UO Zebrafish Facility staff for expert fish husbandry. We thank Caitlin Kowalski and members of the Guillemin lab for thoughtful discussion throughout this study. Research reported in this publication was supported by the National Institutes of Health under award numbers F32DK124033 and 1P01GM125576. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, and Hansson GC (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A 105, 15064–15069. 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaishnava S, Yamamoto M, Severson KM, Ruhn K. a, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV (2011). The Antibacterial Lectin RegIII-gamma Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science (80-.). 334, 255–258. 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlomann BH, Wiles TJ, Wall ES, Guillemin K, and Parthasarathy R (2018). Bacterial Cohesion Predicts Spatial Distribution in the Larval Zebrafish Intestine. Biophys. J 115, 2271–2277. 10.1016/j.bpj.2018.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiles TJ, Wall ES, Schlomann BH, Hay EA, Parthasarathy R, and Guillemin K (2018). Modernized Tools for Streamlined Genetic Manipulation and Comparative Study of Wild and Diverse Proteobacterial Lineages. MBio 9, 1–19. 10.1128/mbio.01877-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergstrom K, Shan X, Casero D, Batushansky A, Lagishetty V, Jacobs JP, Hoover C, Kondo Y, Shao B, Gao L, et al. (2020). Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science (80-.). 370, 467–472. 10.1126/science.aay7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frese SA, MacKenzie DA, Peterson DA, Schmaltz R, Fangman T, Zhou Y, Zhang C, Benson AK, Cody LA, Mulholland F, et al. (2013). Molecular Characterization of Host-Specific Biofilm Formation in a Vertebrate Gut Symbiont. PLoS Genet. 9. 10.1371/journal.pgen.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, and Gordon JI (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (80-.). 307, 1955–1959. 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 8.Welch JLM, Hasegawa Y, McNulty NP, Gordon JI, and Borisy GG (2017). Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. U. S. A 114, E9105–E9114. 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang BX, Wheeler KM, Cady KC, Lehoux S, Cummings RD, Laub MT, and Ribbeck K (2020). Mucin Glycans Signal through the Sensor Kinase RetS to Inhibit Virulence-Associated Traits in Pseudomonas aeruginosa. Curr. Biol, 1–13. 10.1016/j.cub.2020.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler KM, Cárcamo-Oyarce G, Turner BS, Dellos-Nolan S, Co JY, Lehoux S, Cummings RD, Wozniak DJ, and Ribbeck K (2019). Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat. Microbiol 4, 2146–2154. 10.1038/s41564-019-0581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecht AL, Casterline BW, Choi VM, and Bubeck Wardenburg J (2017). A Two-Component System Regulates Bacteroides fragilis Toxin to Maintain Intestinal Homeostasis and Prevent Lethal Disease. Cell Host Microbe 22, 443–448.e5. 10.1016/j.chom.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kavanaugh NL, Zhang AQ, Nobile CJ, Johnson AD, and Ribbeck K (2014). Mucins suppress virulence traits of Candida albicans. MBio 5, 1–8. 10.1128/mBio.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacheco AR, Munera D, Waldor MK, Sperandio V, and Ritchie JM (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ottman N, HuuskonenL L, Reunanen J, Boeren S, Klievink J, Smidt H, Belzer C, and De Vos WM (2016). Characterization of outer membrane proteome of akkermansia muciniphila reveals sets of novel proteins exposed to the human intestine. Front. Microbiol 7, 1–13. 10.3389/fmicb.2016.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Co JY, Cárcamo-Oyarce G, Billings N, Wheeler KM, Grindy SC, Holten-Andersen N, and Ribbeck K (2018). Mucins trigger dispersal of Pseudomonas aeruginosa biofilms. npj Biofilms Microbiomes 4, 23. 10.1038/s41522-018-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jevtov I, Samuelsson T, Yao G, Amsterdam A, and Ribbeck K (2014). Zebrafish as a model to study live mucus physiology. Sci. Rep 4, 1–6. 10.1038/srep06653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massaquoi MS, Kong G, Chilin D, Hamilton MK, and Melancon E (2022). GLOBAL HOST RESPONSES TO THE MICROBIOTA AT SINGLE CELL Keywords. 10.1101/2022.03.28.486083. [DOI] [Google Scholar]
- 18.Willms RJ, Ocampo Jones L, Hocking JC, and Foley E (2021). A Cell Atlas of Microbe-Responsive Processes in the Zebrafish Intestine. SSRN Electron. J 38, 110311. 10.2139/ssrn.3778935. [DOI] [PubMed] [Google Scholar]
- 19.Bergstrom K, and Xia L (2022). The barrier and beyond: Roles of intestinal mucus and mucin-type O-glycosylation in resistance and tolerance defense strategies guiding host-microbe symbiosis. Gut Microbes 14. 10.1080/19490976.2022.2052699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson JMH, Karlsson H, Sjövall H, and Hansson GC (2009). A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 21.Maresca M, Alatou R, Pujol A, Nicoletti C, Perrier J, Giardina T, Simon G, Méjean V, and Fons M (2021). RadA, a MSCRAMM adhesin of the dominant symbiote Ruminococcus gnavus e1, binds human immunoglobulins and intestinal mucins. Biomolecules 11. 10.3390/biom11111613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juge N (2012). Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 20, 30–39. 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arike L, Holmén-Larsson J, and Hansson GC (2017). Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 27, 318–328. 10.1093/glycob/cww134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, and Guillemin K (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol 297, 374–386. 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Wardman JF, Bains RK, Rahfeld P, and Withers SG (2022). Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol 0123456789. 10.1038/s41579-022-00712-1. [DOI] [PubMed] [Google Scholar]
- 27.Ndeh D, and Gilbert HJ (2018). Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol. Rev 42, 146–164. 10.1093/femsre/fuy002. [DOI] [PubMed] [Google Scholar]
- 28.Crouch LI, Liberato MV, Urbanowicz PA, Baslé A, Lamb CA, Stewart CJ, Cooke K, Doona M, Needham S, Brady RR, et al. (2020). Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown. Nat. Commun 11. 10.1038/s41467-020-17847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koropatkin NM, Cameron EA, and Martens EC (2012). How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol 10, 323–335. 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, et al. (2013). Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14, 571–581. 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson MEV, Holmén Larsson JM, and Hansson GC (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U. S. A 108, 4659–4665. 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, and Aderem A (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 33.Burns AR, and Guillemin K (2017). The scales of the zebrafish: host–microbiota interactions from proteins to populations. Curr. Opin. Microbiol 38, 137–141. 10.1016/j.mib.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flores EM, Nguyen AT, Odem MA, Eisenhoffer GT, and Krachler AM (2020). The zebrafish as a model for gastrointestinal tract–microbe interactions. Cell. Microbiol 22. 10.1111/cmi.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, and Sonnenburg JL (2015). Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe 18, 478–488. 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, Melancon E, Eisen JS, Guillemin K, and Parthasarathy R (2016). Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLoS Biol. 14, 1–24. 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiles TJ, Schlomann BH, Wall ES, Betancourt R, Parthasarathy R, and Guillemin K (2020). Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation 10.1371/journal.pbio.3000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlomann BH, Wiles TJ, Wall ES, Guillemin K, and Parthasarathy R (2019). Sublethal antibiotics collapse gut bacterial populations by enhancing aggregation and expulsion. Proc. Natl. Acad. Sci. U. S. A 116, 21392–21400. 10.1073/pnas.1907567116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlomann BH, and Parthasarathy R (2021). Gut bacterial aggregates as living gels. Elife 10, 1–22. 10.7554/eLife.71105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troll JV, Hamilton MK, Abel ML, Ganz J, Bates JM, Stephens WZ, Melancon E, van der Vaart M, Meijer AH, Distel M, et al. (2018). Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development 145, dev155317. 10.1242/dev.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace KN, Akhter S, Smith EM, Lorent K, and Pack M (2005). Intestinal growth and differentiation in zebrafish. Mech. Dev 122, 157–173. 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Ng ANY, De Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PDS, Stainier DYR, and Heath JK (2005). Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev. Biol 286, 114–135. 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Robinson CD, Klein HS, Murphy KD, Parthasarathy R, Guillemin K, and Bohannan BJM (2018). Experimental bacterial adaptation to the zebrafish gut reveals a primary role for immigration. PLoS Biol. 16, 1–26. 10.1371/journal.pbio.2006893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson CD, Sweeney EG, Ngo J, Ma E, Perkins A, Smith TJ, Fernandez NL, Waters CM, Remington SJ, Bohannan BJM, et al. (2021). Host-emitted amino acid cues regulate bacterial chemokinesis to enhance colonization. Cell Host Microbe 29, 1221–1234.e8. 10.1016/j.chom.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaels LA, Singh PK, Jennings LK, Secor PR, and Ratjen A (2018). Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci 115, 10780–10785. 10.1073/pnas.1806005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azimi S, Thomas J, Cleland SE, Curtis JE, Goldberg JB, and Diggle SP (2021). Cell surface hydrophobicity determines aggregate assembly type in Pseudomonas aeruginosa. MBio 12. 10.1128/mBio.00860-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mondal M, Nag D, Koley H, Saha DR, and Chatterjee NS (2014). The Vibrio cholerae extracellular chitinase ChiA2 is important for survival and pathogenesis in the host intestine. PLoS One 9. 10.1371/journal.pone.0103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deatherage DE, and Barrick JE (2014). Identification of mutations in laboratory evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol, 165–188. 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins AJ, Jarrod Smith T, Sondermann H, and O’Toole GA (2020). From Input to Output: The Lap/c-di-GMP Biofilm Regulatory Circuit. Amur. Rev. Microbiol 74, 607–631. 10.1146/annurev-micro-011520-094214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newell PD, Monds RD, and O’Toole GA (2009). LapD is a bis-(3’,5’)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A 106, 3461–3466. 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JS, Song S, Lee M, Lee S, Lee K, and Ha NC (2016). Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type i Secretion System of Gram-Negative Bacteria. Strudure 24, 477–485. 10.1016/j.str.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GCL, and Yildiz FH (2015). C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in Vibrio cholerae. PLoS Pathog. 11, 1–27. 10.1371/journal.ppat.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJF, Yildiz F, and Klose KE (2009). The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol 191, 6555–6570. 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanco-Romero E, Redondo-Nieto M, Martínez-Granero F, Garrido-Sanz D, Ramos-Gonzalez MI, Martín M, and Rivilla R (2018). Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci. Rep 8, 1–13. 10.1038/s41598-018-31371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dueholm MS, Albertsen M, Otzen D, and Nielsen PH (2012). Curli Functional Amyloid Systems Are Phylogenetically Widespread and Display Large Diversity in Operon and Protein Structure. PLoS One 7, 1–10. 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grys TE, Walters LL, and Welch RA (2006). Characterization of the StcE protease activity of Escherichia coli O157:H7. J. Bacteriol 188, 4646–4653. 10.1128/JB.01806-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hews CL, Tran SL, Wegmann U, Brett B, Walsham ADS, Kavanaugh D, Ward NJ, Juge N, and Schuller S (2017). The StcE metalloprotease of enterohaemorrhagic Escherichia coli reduces the inner mucus layer and promotes adherence to human colonic epithelium ex vivo. Cell. Microbiol 19, 1–10. 10.1111/cmi.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malaker SA, Pedram K, Ferracane MJ, Bensing BA, Krishnan V, Pett C, Yu J, Woods EC, Kramer JR, Westerlind U, et al. (2019). The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sd. U. S. A 116, 7278–7287. 10.1073/pnas.1813020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, and Chatterjee NS (2008). Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun 76, 4968–4977. 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loose JSM, Forsberg Z, Fraaije MW, Eijsink VGH, and Vaaje-Kolstad G (2014). A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett. 588, 3435–3440. 10.1016/j.febslet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 61.Banse AV, Vanbeuge S, Smith TJ, Logan SL, and Guillemin K (2022). Secreted Aeromonas GlcNAc binding protein GbpA stimulates epithelial cell proliferation in the zebrafish intestine. bioRxiv. 10.1101/2022.06.27.497793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sundarraman D, Smith TJ, Kast JVZ, Guillemin K, and Parthasarathy R (2022). Disaggregation as an interaction mechanism among intestinal bacteria. Biophys. J 121, 3458–3473. 10.1016/j.bpj.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newell PD, Boyd CD, Sondermann H, and O’Toole GA (2011). A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9. 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyd CD, Chatterjee D, Sondermann H, and O’Toole GA (2012). LapG, required for modulating biofilm formation by pseudomonas fluorescens Pf0-1, is a calcium-dependent protease. J. Bacteriol 194, 4406–4414. 10.1128/JB.00642-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letunic I, Khedkar S, and Bork P (2021). SMART: Recent updates, new developments and status in 2020. Nucleic Adds Res. 49, D458–D460. 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massimino L, Lamparelli LA, Houshyar Y, D’Alessio S, Peyrin-Biroulet L, Vetrano S, Danese S, and Ungaro F (2021). The Inflammatory Bowel Disease Transcriptome and Metatranscriptome Meta-Analysis (IBD TaMMA) framework. Nat. Comput. Sci 1, 511–515. 10.1038/s43588-021-00114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hansson GC (2012). Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol 15, 57–62. 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voogdt CGP, Wagenaar JA, and van Putten JPM (2018). Duplicated TLR5 of zebrafish functions as a heterodimeric receptor. Proc. Natl. Acad. Sci 115, E3221–E3229. 10.1073/pnas.1719245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353.e21. 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ottman N, Davis M, Suarez-Diez M, Boeren S, Schaap PJ, Martins dos Santos VAP, Smidt H, Belzer C, and de Vos WM (2017). Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin- Degrading Lifestyle. Applies Environ. Microbiol 83, e01014–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mikhalchik E, Balabushevich N, Vakhrusheva T, Sokolov A, Baykova J, Rakitina D, Scherbakov P, Gusev S, Gusev A, Kharaeva Z, et al. (2020). Mucin adsorbed by E. coli can affect neutrophil activation in vitro. FEBS Open Bio 10, 180–196. 10.1002/2211-5463.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamaguchi M, Hirose Y, Takemura M, Ono M, Sumitomo T, Nakata M, Terao Y, and Kawabata S (2019). Streptococcus pneumoniae Evades Host Cell Phagocytosis and Limits Host Mortality Through Its Cell Wall Anchoring Protein PfbA. Front. Cell. Infect. Microbiol 9, 1–14. 10.3389/fcimb.2019.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, and Bachrach G (2015). Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun 83, 1104–1113. 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. (2016). Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 20, 215–225. 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umanã A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, et al. (2020). Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sd. Signal 13, 1–13. 10.1126/SCISIGNAL.ABA9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sundarraman D, Hay EA, Martins DM, Shields DS, Pettinari NL, and Parthasarathy R (2020). Higher-order interactions dampen pairwise competition in the zebrafish gut microbiome. MBio 11, 1–15. 10.1128/mBio.01667-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bales PM, Renke EM, May SL, Shen Y, and Nelson DC (2013). Purification and Characterization of Biofilm-Associated EPS Exopolysaccharides from ESKAPE Organisms and Other Pathogens. PLoS One 8. 10.1371/journal.pone.0067950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, and Whyte MKB (2006). A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976–3978. 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 79.Melancon E, Gomez De La Torre Canny S, Sichel S, Kelly M, Wiles TJ, Rawls JF, Eisen J, and Guillemin K (2017). Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 138, 61–100. 10.1016/bs.mcb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, and O’Toole G a (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol 72, 5027–5036. 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K, and Parthasarathy R (2014). Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. MBio 5, 1–8. 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hay EA, and Parthasarathy R (2018). Performance of convolutional neural networks for identification of bacteria in 3D microscopy datasets. PLoS Comput. Biol 14, 1–17. 10.1371/journal.pcbi.1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Mucus distribution in a live larval zebrafish, Related to Figure 1 and S1. Scan of z-stacks used to for the maximum projection image in Fig. S1. Green indicates WGA-FITC staining.
Movie S2. Association between Aer01 and mucus signals in the anterior midgut of fixed intestines, Related to Figure 1. Scan of z-stacks from region used for the maximum projection image in Fig. 1B. Aer01 and mucus are colored as described in main figure.
Movie S3. Association between Aer01 and mucus signals in the distal midgut of fixed intestines, Related to Figure 1. Scan of z-stacks from region used for the maximum projection image in Fig. 1C. Aer01 and mucus are colored as described in main figure.
Movie S4. MB1 are planktonic and non-motile in the zebrafish foregut, Related to Figure 4. Real-time video of MB1 physiology acquired using light sheet fluorescence microscopy on single oprical plane.
Movie S5. MB4 are planktonic and motile in the zebrafish foregut, Related to Figure 4. Real-time video of MB4 physiology acquired using light sheet fluorescence microscopy on single oprical plane.
Figure S1. Mucus distribution in live larval zebrafish, Related to Figure 1. Maximum intensity projection of WGA-FITC staining (green) of gut and skin mucus in live larval zebrafish aged 6 dpf acquired using light sheet fluorescence microscopy. Individual z-stacks are shown in Movie S1.
Figure S2. Aer01 associates with WGA-stained mucin in culture, Related to Figure 2. dTomato-tagged Aer01 were exposed to 0.4% mucin that was stained with WGA-FITC (left) or untreated (right) in the aggregation assay. The merged image shows Aer01 (red) associated with WGA-stained mucin (green). WGA-stained mucin does not aggregate unless Aer01 is present (not shown).
Figure S3. Aer01 distribution in two models of mucus depletion, Related to Figure 2. (A) Distribution of dTom-tagged Aer01 mono-associated with myd88−/− larval zebrafish using the experimental design highlighted in Fig 2B; n=7. (B) Distribution of dTom-tagged Aer01 mono-associated in wild type fish treated with WGA-FITC.
Figure S4. MB1 GlcNAc-mediated aggregation is rescued by mbpD expression, Related to Figure 3. Aggregation assay in GlcNAc with MB1 expressing empty vector or the wild-type mbpD allele. Expression of mbpD rescues GlcNAc-mediated aggregation of MB1 which contains a LOF mutation in mbpD.
Figure S5. Swim plate analysis on TSA, Related to Figure 3. WT and MB1-4 were inoculated onto TSA swim plates and incubated for 12 hr at 30°C. Plates were imaged and the swim distance calculated in FIJI. MB isolates were normalized to the plate-specific WT control.
Supplemental Table 1. RNA-seq analysis, Related to Figure 3.
Supplemental Table 2. MbpA-like adhesins from bacteria identified in IBD TaMMa, Related to Figure 7.
Data Availability Statement
The raw sequence reads can be accessed using the BioProject Accession: PRJNA988826.
Code for identifying bacterial single cells and clusters has been deposited at GitHub (https://github.com/rplab/Bacterial_Automated_Pipeline).
Any additional information to reanalyze the data reported in this work paper is available from the lead contact upon request.


