Abstract
We report here the development of a freeze-drying procedure allowing stabilization at ambient temperature of preoptimized, premixed, and predispensed PCR mixes aimed at the detection of mycobacteria in clinical materials. The freeze-dried mixes retained activity at 4°C and at 20°C for 1 year and for 3 months at 37°C, as judged by their performance with 50 and 500 fg of purified Mycobacterium bovis BCG target DNA.
PCR has brought new opportunities for the rapid, sensitive, and specific detection of mycobacteria. PCR has been shown to have a clear, although confined, value in the diagnosis of tuberculosis (2). The widespread use of PCR for the detection of mycobacteria, especially in countries with the greatest burden of disease, is limited because of, among other factors such as cost, the complexity of the method. One aspect of the PCR procedure which hampers its introduction is the need for standardized, quality-controlled frozen or refrigerated ingredients. The availability of ready-to-use freeze-dried PCR mixes of proven quality would greatly facilitate implementation in laboratories with relatively limited facilities.
Previously, we have described a sensitive multiplex PCR assay for the immediate identification of the infecting mycobacterial species in clinical samples (5). This technique is based on the simultaneous detection of insertion element IS6110 of Mycobacterium tuberculosis and DNA coding for 16S rRNA (3, 4, 5). Here, we report the development of a freeze-drying procedure allowing the stabilization of the PCR mix for up to 1 year at ambient temperature.
The composition of the PCR mix was the same as that described previously (5) and included buffer, primers, Thermus aquaticus DNA polymerase (1 U of AmpliTaq from Perkin-Elmer Cetus, Norwalk, Conn., or 0.2 U of SuperTaq from HT Biotechnology, Cambridge, United Kingdom, as indicated below), deoxynucleoside triphosphates dATP, dCTP, dGTP, and dUTP (Pharmacia, Uppsala, Sweden), and 0.2 U of uracil-DNA-glycosylase (UDG) (Gibco BRL, Gaithersburg, Md.). When SuperTaq was used, Triton X-100 was added after reconstitution (see below). Trehalose, needed for optimal freeze-drying (1), was added to the PCR mix at different concentrations up to 10% (wt/vol), as indicated below. Lyophilization of batches of PCR mixes (each for 15 reactions) was performed in a Klee pilot freeze-dryer (Marburg, Germany).
The freeze-dried PCR mixes were stored for up to 1 year at temperatures of 4, 20, 37, and 56°C and reconstituted to their original volume with distilled water and, for mixes with SuperTaq, also with Triton X-100 to a final concentration of 0.1% (vol/vol).
In all experiments DNA isolated from cultured Mycobacterium bovis BCG (ATCC 35733) was used (3). This mycobacterial species was chosen because it contains only one copy of the IS6110 element, facilitating the monitoring of any variations in the PCR performance due to the addition of stabilizers or to the freeze-drying process.
PCR was performed exactly as described before (5) and analyzed by agarose gel electrophoresis and reverse cross-blot hybridization with probe Pt3 (5′-GAACGGCTGATGACCAAACT) to capture the IS6110 PCR products and pTub1 (5′-ACCACAAGACATGCATCCCG) to capture the 16S rDNA PCR products of mycobacteria belonging to the M. tuberculosis complex (4, 5).
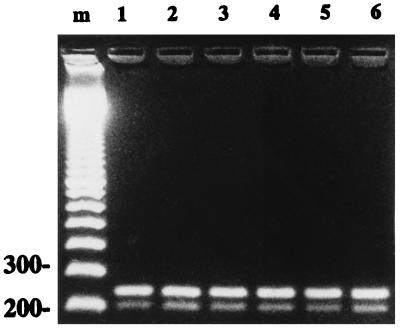
As shown in Fig. 1, an analysis of gel electrophoresis showed that the addition of up to 10% (wt/vol) trehalose to the PCR mix had no visible effect on the performance of the PCR, irrespective of the brand of Taq polymerase used. Both the 208-bp (16S PCR product) and 249-bp (IS6110 PCR product) bands could be clearly seen, and the intensities of the bands were only dependent on the amount of target DNA and were independent of the concentration of trehalose added.
FIG. 1.
Effect of trehalose on the performance of PCR with M. bovis BCG DNA. A gel electrophoresis analysis of PCR performed with the addition of trehalose at concentrations (% [wt/vol]) of 0 (lane 1), 2 (lane 2), 4 (lane 3), 6 (lane 4), 8 (lane 5), and 10 (lane 6) to the PCR mix shows the 208-bp (16S PCR product) and 249-bp (IS6110 PCR product) bands. Lane m contains a 100-bp ladder (Pharmacia) as a molecular weight marker; the 200- and 300-bp fragments are indicated.
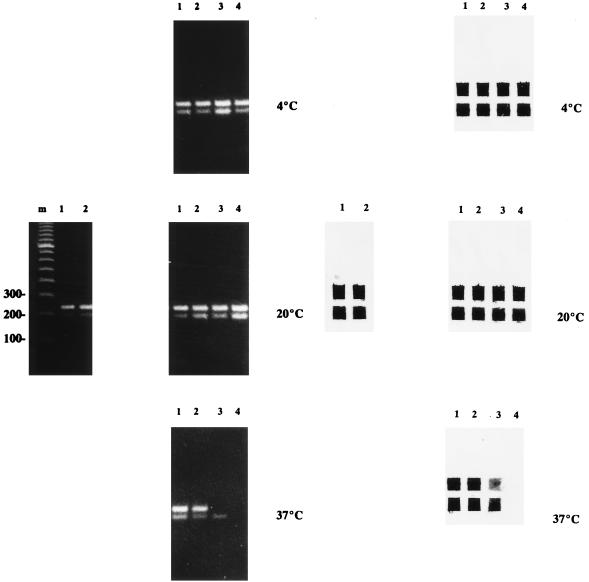
We chose a 5% (wt/vol) trehalose concentration for freeze-drying. The freeze-dried PCR mixes were stored for up to a year at different temperatures and, upon reconstitution, were tested with 50 and 500 fg of purified M. bovis BCG DNA (equivalent to 10 and 100 mycobacteria, respectively) to check their performance. If not freeze-dried, PCR reaction mixes retained activity for 1 year at −20°C and for 1 week at 4°C, but lost activity when stored for more than 24 h at 20 or 37°C (6). Figure 2 shows the gel electrophoresis and reverse cross-blot hybridization results for the reconstituted freeze-dried PCR mixes prepared with SuperTaq after storage. The freeze-dried mixes retained activity at 4 and at 20°C for 1 year and for 3 months at 37°C. The freeze-dried mixes were only active for 1 week at 56°C (results not shown). The activity of the freeze-dried mixes prepared with AmpliTaq was retained for 1 year at 4 and 20°C but was lost after 1 week at 37 and 56°C (results not shown). The UDG in the freeze-dried mixes was active for the same periods of time as those observed for the Taq polymerases; this was evidenced by the absence of any bands on gels after reamplification of 10−4 and 10−6 dilutions of 208- and 249-bp amplicons originating from 1 pg of purified M. bovis BCG DNA amplified in the reconstituted PCR mixes (results not shown).
FIG. 2.
Gel electrophoresis (left) and reverse cross-blot hybridization (right) results of the reconstituted PCR mixes after storage. The freeze-dried PCR mixes were stored for 3 months (lanes 1 and 2) and 1 year (lanes 3 and 4) at different temperatures as indicated and, upon reconstitution, were tested with 500 (lanes 1 and 3) and 50 (lanes 2 and 4) fg of purified M. bovis BCG DNA. The photographs at the left in each panel show the results for PCR mixes reconstituted directly after freeze-drying. Lane m contains a 100-bp ladder (Pharmacia) as a molecular weight marker; the 100-, 200-, and 300-bp fragments are indicated. Reverse cross-blot hybridization was done with probe Pt3 to capture the 249-bp IS6110 PCR products (top wells) and pTub1 to capture the 208-bp 16S rDNA PCR products (bottom wells).
The lesser stability of the AmpliTaq mix may be related to the difference in glycerol concentration in the final freeze-dried preparation between it and the SuperTaq mix: 0.48% (vol/vol) versus 0.28% (vol/vol). Both the Taq polymerases and the UDG used in this study were supplied in 50% (vol/vol) glycerol solutions. Glycerol could not be sublimated in our freeze-dryer, since that would require a very low vacuum. The glycerol concentration increases during the freeze-drying process as water disappears. Since glycerol is hygroscopic, its presence in the final freeze-dried product likely results in a high moisture content, which may affect the stability of the product. The commercial availability of glycerol-free Taq polymerases and UDG would help to prolong the shelf life of freeze-dried PCR mixes.
Our results show that freeze-drying allows the preparation of off-the-shelf, preoptimized, premixed, predispensed PCR reaction mixes for the detection of mycobacterial DNA which are stable for a year at 20°C and for up to 3 months at 37°C. All that is required is to reconstitute the mix and add template DNA. A commercial freeze-dried basic PCR mix recently became available (Pharmacia Biotech; Ready-To-Go PCR beads). Our system has the advantage that it includes a complete prevalidated PCR system, including primers and dUTP-UDG to prevent contamination, which has previously been shown to be of value in testing clinical samples from patients suspected of having a mycobacterial infection (5, 6). Such freeze-dried PCR mixes will be useful for both research and diagnostic purposes in laboratories with limited experience or limited facilities for PCR.
Acknowledgments
We thank the Q. M. Gastmann-Wichers Foundation and the Netherlands Leprosy Relief Association for financial support.
REFERENCES
- 1.Franks F. Freeze drying: from empiricism to predictability. Cryoletters. 1990;11:93–110. [PubMed] [Google Scholar]
- 2.Klatser P R. Amplification reactions in mycobacteriology. J Microbiol Methods. 1995;23:75–87. [Google Scholar]
- 3.Kolk A H J, Schuitema A R J, Kuijper S, van Leeuwen J, Hermans P W M, van Embden J D A, Hartskeerl R A. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a nonradioactive detection system. J Clin Microbiol. 1992;30:2567–2575. doi: 10.1128/jcm.30.10.2567-2575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kox L F F, van Leeuwen J, Kuijper S, Jansen H M, Kolk A H J. PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J Clin Microbiol. 1995;33:3225–3233. doi: 10.1128/jcm.33.12.3225-3233.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kox L F F, Jansen H M, Kuijper S, Kolk A H J. Multiplex PCR assay for immediate identification of the infecting species in patients with mycobacterial disease. J Clin Microbiol. 1997;35:1492–1498. doi: 10.1128/jcm.35.6.1492-1498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kox L F F, Rhientong D, Medo Miranda A, Udomsantisuk N, Ellis K, van Leeuwen J, van Heusden S, Kuijper S, Kolk A H J. A more reliable PCR for the detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1994;32:672–678. doi: 10.1128/jcm.32.3.672-678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]