Abstract
Unraveling molecular mechanisms of adaptation to complex environments is crucial to understanding tolerance of abiotic pressures and responses to climatic change. Epigenetic variation is increasingly recognized as a mechanism that can facilitate rapid responses to changing environmental cues. To investigate variation in genetic and epigenetic diversity at spatial and thermal extremes, we use whole genome and methylome sequencing to generate a high-resolution map of DNA methylation in the bumble bee Bombus vosnesenskii. We sample two populations representing spatial and environmental range extremes (a warm southern low-elevation site and a cold northern high-elevation site) previously shown to exhibit differences in thermal tolerance and determine positions in the genome that are consistently and variably methylated across samples. Bisulfite sequencing reveals methylation characteristics similar to other arthropods, with low global CpG methylation but high methylation concentrated in gene bodies and in genome regions with low nucleotide diversity. Differentially methylated sites (n = 2066) were largely hypomethylated in the northern high-elevation population but not related to local sequence differentiation. The concentration of methylated and differentially methylated sites in exons and putative promoter regions suggests a possible role in gene regulation, and this high-resolution analysis of intraspecific epigenetic variation in wild Bombus suggests that the function of methylation in niche adaptation would be worth further investigation.
Subject terms: Epigenomics, Ecological genetics
Introduction
Understanding the ecological and evolutionary mechanisms of adaptation to complex ecological niches is a central goal of evolutionary genomics1–3. Species with large geographic distributions face diverse pressures from environmental heterogeneity across populations4, 5, and genotypic and phenotypic variation among dissimilar environments can provide the raw material for local adaptation6. Species in mountainous regions, in particular, can experience extreme variations in abiotic conditions such as temperature, precipitation, or air density2, 7, 8. Population-level genomic changes at the spatial-environmental extremes in widespread montane species could thus improve our current understanding of how species tolerate diverse bioclimatic conditions and provide insights into potential mechanisms of adaptability and robustness under global climate change6, 9, 10.
DNA sequence-based variation has been the most commonly examined form of genomic adaptation in wild populations, however, epigenetic variation, such as DNA methylation, histone modifications, and regulatory non-coding RNAs, is increasingly recognized as a potential mechanism of rapid environmental adaptation or plasticity11–13. Epigenetic mechanisms can generate flexible responses to various environmental stimuli without modifying genome sequences, and they are potentially important for species that occupy diverse bioclimatic niches14. Cytosine (CpG context) methylation is the most prevalent form of epigenetic methylation15, however, the extent of CpG methylation and its functional significance varies substantially across lineages16. For example, mammals exhibit higher (70–80%) of global CpG methylation17 compared to plants (4–40%)18 and arthropods (< 1% to 14%)19. While in plants, DNA methylation primarily occurs in repetitive regions, especially in transposon elements (TEs)20, in mammalian genomes, cytosine methylation is consistent except in the CpG islands (i.e., CG motif-rich genomic regions) near promoters and transcription start sites (TSS)21. Mammalian CpG methylation has been linked to various molecular functions17, such as gene silencing, genomic imprinting, and stabilization of regulation of gene expression22–25. In arthropods, methylation functionality has been attributed to varied biological processes such as reproduction26, caste determination27–29, and regulation of gene expression via differential exon usage30. Arthropod CpG methylation is most prominent in gene bodies compared to intrageneric and intergenic regions, but levels vary widely across lineages31. For example, model organism Drosophila melanogaster has very low amount of CpG methylation which is often not detected by bisulfite-sequencing19, 32 due to the absence of a key methyltransferase gene (Dnmt3)33. Characterizing genome-wide patterns of DNA methylation across a wide range of taxa34 will be important in understanding the distribution of consistent CpG methylation patterns across multiple lineages and identifying the extent of intraspecific epigenomic variability. The function of such variable epigenetic changes may be especially relevant in the context of adaptation to anthropogenic climate change.
Bumble bees are among the most economically and ecologically important pollinating insects35, 36 that primarily inhabit cool temperate, alpine, and arctic climates37. Bumble bees exhibit remarkable phenotypic and physiological adaptations for thermal regulation38, such as an insulated pile, generating heat through shivering of flight muscles, and shunting mechanisms that prevent overheating39–41. Such thermal adaptations allow bumble bees to fly and forage in diverse thermal niches than many other insects42, 43. Like many insects44, many bumble bee species have declined in geographic range and abundance45, seemingly driven at least in part from anthropogenic climate change46, 47. In North America, while several bumble bees have recently declined dramatically, many species remain common48–50, and species-specific responses to global climate change46 indicate that some species may tolerate warming temperatures better than others51. The nature of genomic and epigenomic variation within species that occupy diverse environments will be valuable for understanding why species may be vulnerable or resistant to climate change.
Bombus vosnesenskii is a common bumble bee species that is distributed across latitudinal and altitudinal gradients in Western North America, principally in California, Oregon, and Washington, USA (Fig. 1). Population genetic studies have found low levels of intra-specific genetic differentiation and weak population structures across the B. vosnesenskii range5, 52, and B. vosnesenskii is one of two North American bumble bee species projected to expand its range under future climate change scenarios51. Therefore, studying environment-associated genomic variation may provide insights into species-specific responses that may mitigate the negative impacts of climate change. As a widely distributed and ecologically crucial native pollinator, B. vosnesenskii has gained substantial attention as a research subject for population genetics52, 53, pollination biology54, and abiotic adaptation55, 56 studies. Genome scans across a broad latitudinal and altitudinal range using restriction site-associated DNA sequencing (RAD-Seq) and environmental association analysis identified relatively few potential genomic regions associated with thermal and desiccation tolerance55. However, analysis of thermal tolerance across latitude and altitude extremes of the B. vosnesenskii range provided some evidence for local adaptation, with population-level variation in lower thermal tolerance (CTMIN) of laboratory-reared bees that matched the annual temperature of respective source populations56. Moreover, transcriptional differences among populations were detected at these lower CTMIN thresholds. In contrast, there was no evidence of intrapopulation variation in responses to upper thermal limit (CTMAX), suggesting evolutionary conservation of physiological and molecular responses under heat stress56. Results from these studies provide a foundation for investigating other types of variation that may contribute to molecular responses, including epigenetics, which will contribute to a greater understanding of potentially adaptive thermal tolerance mechanisms in this species.
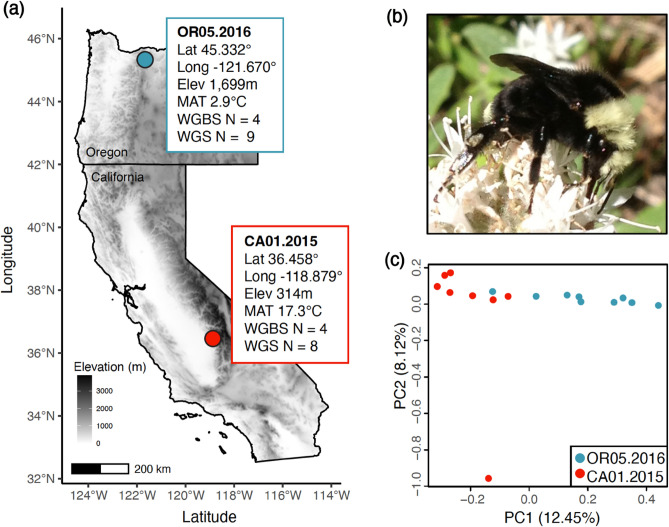
Figure 1.
(a) Map, spatial information [latitude (Lat), longitude (Long), elevation (Elev), and mean annual temperature (MAT) from the WorldClim v.257], and sample sizes for Whole Genome Bisulfite Sequencing (WGBS) and Whole Genome Sequencing (WGS) for the two B. vosnesenskii study populations. (b) Photograph of B. vosnesenskii, (c) Genome-wide Principal Components Analysis (PCA) from covariances estimated by PCAngsd for the B. vosnesenskii populations sampled for WGBS.
The majority of epigenetics and DNA methylation studies in bumble bees have centered around determining its role in sex/caste determination58, 59, reproduction60 and development61 using lab-reared individuals of two commonly used model species, B. terrestris, and B. impatiens62. However, little is known regarding the role of epigenetics in shaping niche-specific thermal adaptation in wild bumble bees, which might provide insights into the adaptive variation that could allow responses to environmental variation2, 63, 64. The availability of reference genomes for multiple bumble bee species65, 66 now facilitates expanding the phylogenetic scope of methylation research in bumble bees. In this study, we use very high-coverage whole genome bisulfite sequencing (WGBS) data to map epigenetic variation in B. vosnesenskii. We also evaluate the potential for intraspecific epigenomic variation by sequencing populations representing the spatial and thermal range extremes, focusing on wild-caught samples taken from two extreme populations: a southern low elevation population from California, USA (warm extreme) and a northern high elevation population from Oregon, USA (cold extreme) (Fig. 1). In addition to detailed characterization of the methylome of the species overall and testing for intraspecific epigenetic differentiation, we also assess possible relationships between methylation with population genetic diversity or structure using single nucleotide polymorphisms (SNPs) from whole genome sequencing (WGS). Specifically, we aim to: (i) characterize the major trends in consistent methylation patterns in B. vosnesenskii and identify putative major functions related to genome-wide CpG methylation; (ii) compare and contrast epigenetic profiles from populations at latitude and altitude extremes to assess variability in the methylome and characterize the genomic location and potential functional roles of differentially methylated CpGs; and (iii) investigate the potential relationship between population genetic diversity and genome-wide CpG methylation levels in B. vosnesenskii. Our research provides insights into the distinct nature of consistent and variable DNA methylation in populations from the spatial-environmental range of B. vosnesenskii, and it also highlights the existence of intraspecific epigenetic variation that may aid in generating regional variation in genotypes and phenotypes to adapt the species across a range of intricate biological niches.
Results
CpG methylation across the B. vosnesenskii genome is broadly consistent among samples
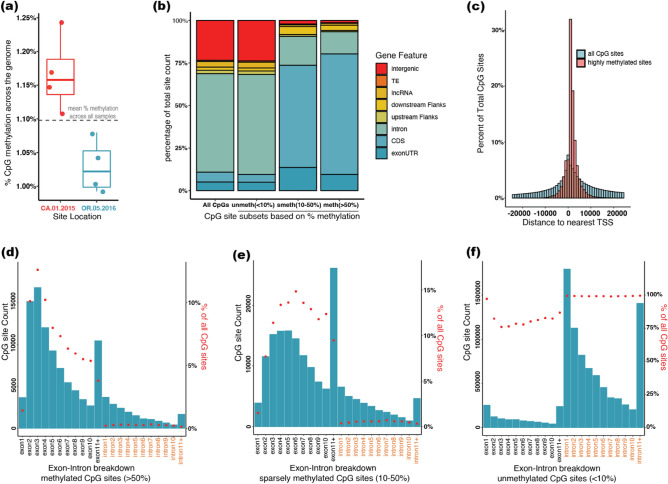
Overall CpG methylation across the genome was 1.1% ± 0.9% SD which was calculated from the percent methylation per CpG cytosine values across all samples (Fig. 2a). The low-elevation California (CA) population had slightly higher percent methylation (1.17% ± 0.06% SD) than the high-elevation Oregon (OR) population (1.03% ± 0.04% SD) (Fig. 2a). Most sequenced CpGs (methylated + unmethylated) were located in introns (57.90%) and intergenic (23.42%) regions, with 5.73% in coding sequences (CDS) and 5.09% in untranslated regions of exons (exon UTRs) (Fig. 2b). The distribution of methylated CpGs varied substantially from the overall distribution of CpGs, with both highly methylated (> 50% average methylation; n = 112,996, ~ 0.78% of all CpGs) and sparsely methylated (10–50% methylation, n = 186,846, ~ 1.28% of all CpGs) sites predominantly found in CDS (Fig. 2b). Specifically, 70.85% of sites that were classified as highly methylated in all samples were in CDS, 13.02% were in introns, 9.50% in exon UTRs, and much lower percentages in the remaining annotation feature categories (0.76–3.13%) (Fig. 2b). Although highly methylated CpGs are only ~ 0.78% of all CpG positions in the genome, the proportion of highly methylated CpGs per total sequenced CpGs in CDS is even more extreme (9.36% of all CpGs in CDS) compared to introns (0.17%) and intergenic (0.04%) regions. Annotation feature-specific distributions of highly methylated CpGs is significantly different from distribution of all CpGs (Pearson's Chi-squared test with Yates' continuity correction; FDR corrected P < 0.05) for all eight annotation features [i.e., exon UTR, CDS, intron, upstream flank, downstream Flank, long non-coding RNA, transposable elements (TE), intergenic; detailed results are available in Supplementary data repository].
Figure 2.
General Pattern of genome-wide methylation in B. vosnesenskii study samples. (a) Box plot exhibiting sample specific per base percent methylation for the CpGs present in every sample. (b) Bar plots of genomic feature-based annotation proportions for all CpGs, unmethylated sites, sparsely methylated sites, and highly methylated sites. (c) Histogram of distances to nearest TSS for all CpGs and highly methylated sites. (d–f) Exon intron breakdown of gene body methylation for highly methylated (d), sparsely methylated (e), and unmethylated CpGs (f). Y-axis blue bars represent the actual count, and red dots depict the proportion of individual genomic features (i.e., exons and introns) relative to similar annotation feature counts for all CpGs.
Consistent with the overabundance of methylated sites in CDS, a greater number of highly methylated sites clustered near the transcription start site (TSS) than predicted from the genome-wide distribution of TSS distances for all CpGs (Fig. 2c), with the absolute mean distance from TSS for highly methylated CpGs was much shorter (2438.78 bp) compared to the absolute mean distance from TSS for all CpGs (27,981.11 bp). There were ~ 4.5 times more CpGs in downstream (gene bodies and 5′ UTR) of TSS (n = 92,403) compared to the number of CpGs in upstream (e.g., likely promoter regions) of TSS (n = 20,561), which is substantially higher than for all CpGs [~ 1.51 × more sites in downstream of TSS (n = 8,708,196) compared to the CpGs in upstream (n = 5,774,550)]. The distribution of distances to the TSS for highly methylated sites was significantly different than that for all CpGs (two-sided two-sample Kolmogorov–Smirnov test, D = 0.35738, P < 2.2e−16). The distribution of sparsely methylated CpGs was similar to that of highly methylated CpGs (Fig. 2b). As expected, unmethylated CpGs (< 10% methylation average methylation; n = 14,283,650, ~ 97.94% of all CpGs) largely matched that of the genome-wide distribution of CpGs except for a slightly smaller proportion in CDS (due to the greater methylation presence in CDS).
To examine the distribution of methylation levels relative to CpG background within genes, we examined the frequencies of highly methylated, sparsely methylated, and unmethylated CpGs for exons, introns and other annotation features (Fig. 2d–f). The first clear pattern is that exons have much greater levels of methylation, both in absolute numbers of methylated CpGs and even more clearly apparent when visualized as a percentage of available CpGs per feature (Fig. 2d,e). For exons, exon 2–4 harbored substantially more highly methylated sites (10.1%, 12.6% and 10.2% relative percentages compared to all CpGs, respectively) than the first exon (1.4%), and generally decreased from exon 3 through the remaining exons (Fig. 2d). A similar pattern was apparent in the sparsely methylated sites, although the distribution was not as sharply biased toward exons 2 and 3 (Fig. 2e). In contrast, the exon-specific distribution pattern is reversed in unmethylated sites (Fig. 2f), as the first exon has more unmethylated sites (97%) than exon 2 (82%), exon 3 (76%) or rest of the exons, although as discussed above the number and proportion of unmethylated CpGs is reduced in exons relative to introns overall. For introns, there was a downward trend in raw counts from upstream to downstream intron locations across the gene body for all three (methylated, sparsely methylated and unmethylated) categories, however, the trend is absent when considered as percentages of available CpGs (Fig. 2d–f). We separately evaluated patterns in long non-coding RNAs, which showed a similar exon–intron breakdown (Supplementary Fig. 1).
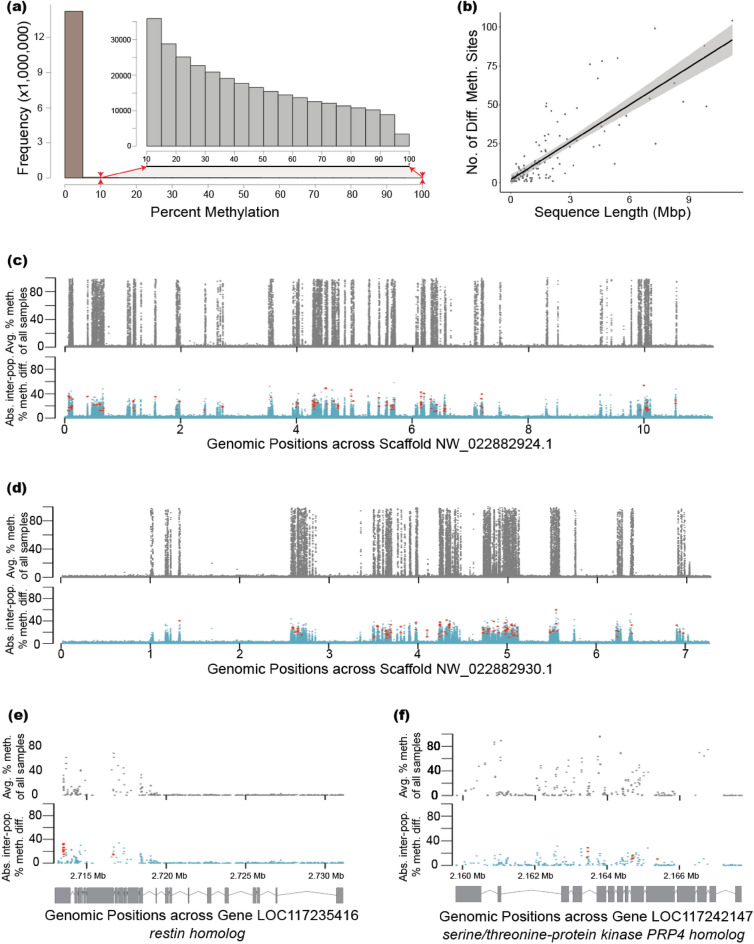
We also visualized the chromosomal distribution of CpGs across the genome (Fig. 3). Most of the CpGs across the genome have low methylation (< 10%) and highly methylated sites are relatively scarce (Fig. 3a), however, plotting the average per base percent methylation across the genome shows their distribution is non-random as we discovered the large regions of very low methylation in genomic scaffolds punctuated with peaks of methylation heavy regions (Fig. 3c–d); gene-specific visualization of CpGs (Fig. 3e,f) exhibits that this distinct pattern of clustering of highly methylated CpGs are predominantly located in gene bodies.
Figure 3.
Genome-wide distribution of CpG methylation in B. vosnesenskii. (a) Frequency histogram of percent methylation of all CpGs with the distribution of CpGs with more than 10% methylation zoomed-in inset plot. (b) Scatter plot of correlation between scaffold length (Mbp) and the number of differentially methylated sited harboured in the individual scaffolds. (c–f) Manhattan plots of the genomic landscape of average percent methylation (top panel) across all samples and absolute inter-population percent methylation difference (bottom panel) across scaffold NW_022882924.1 (c), scaffold NW_022882930.1 (d), restin homolog (e) and serine/threonine-protein kinase PRP4 homolog (f) with their exon–intron gene structures. Manhattan plots were drawn using fastman67 R package.
Patterns of differentially methylated CpGs between populations from spatial-environmental range extremes of B. vosnesenskii
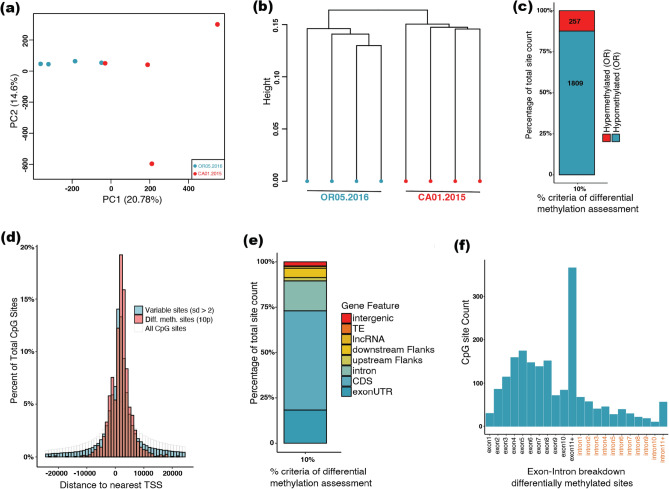
The principal component analysis (PCA) of all methylated CpGs showed that 31.44% of the variation was explained by first two principal components with weak separation of OR and CA samples, and greater variation within CA (Supplementary Fig. 2), although population-specific clustering was more prominent in a clustering dendrogram (Supplementary Fig. 2). When analyses were repeated using CpGs that were variably methylated among all samples (excluding sites within 2 SD of average per base percent methylation, n = 901,868 CpGs) there was more evident population-specific clustering (Fig. 4a), and hierarchical clustering also exhibited distinct population-specific clusters (Fig. 4b). PCA and hierarchical clustering analysis using only differentially methylated sites, obviously indicated clear distinction between two populations (Supplementary Fig. 2).
Figure 4.
General patterns of clustering and distribution of variable (SD > 2) and differentially methylated CpGs in B. vosnesenskii. (a) Principal Component Analysis (PCA) of methylation profiles of variable CpGs (SD > 2). (b) Hierarchical clustering of methylation patterns of variable CpGs using Ward.D2 algorithm (c) Bar plot of counts and percentages of hypermethylated and hypermethylated CpGs in high elevation (Oregon) samples assessed at 10% methylation difference. (d) Histogram of distances to nearest TSS for variable (SD > 2), differentially methylated, and all CpGs. (e) Genomic feature-based bar plots depicting annotation proportions for differentially methylated sites assessed at 10% methylation difference. (f) Exon-intron breakdown of gene body methylation for differentially methylated CpGs.
We identified 2066 significantly differentially methylated sites (≥ 10% methylation difference, FDR corrected q ≤ 0.01) between OR and CA. Most (n = 1809; 87.6%) were hypomethylated in OR relative to CA (Fig. 4c). This result is consistent with the sample-specific methylation frequencies (Fig. 2a) that shows slightly lower overall percent average methylation across the genome in OR.
There was a significant positive correlation between the number of differentially methylated sites and the sequence length of the scaffolds (Pearson’s r = 0.82, P < 0.001; Spearman’s rho = 0.8, P < 0.001) (Fig. 3b), however, as for consistently methylated CpGs, the distribution within scaffolds was clearly not random (Fig. 3c-f). Differentially methylated sites were distributed even more closely to the TSS (mean = 3622.9 bp) than all CpGs (27,981.1 bp, two-sided Two-sample Kolmogorov–Smirnov test, D = 0.329, P < 2.2 × 10–16) or variably methylated CpGs (absolute mean distance 16,304.33 bp; two-sided Two-sample Kolmogorov–Smirnov test, D = 0.174, P < 2.2 × 10–16) (Fig. 4d). Also similar to consistently methylated CpGs, differentially methylated CpGs are also more numerous downstream of the TSS (n = 1540) than upstream (n = 524), indicating greater abundance in gene bodies compared to the promoters. Differentially methylated CpGs (10% methylation difference threshold) were mostly found in coding sequences (54.72%) and exon UTRs (18.32%) while relatively few were in introns (16.53%) and intergenic regions (2.22%) (Fig. 4e). Again, the first exon had fewer differentially methylated CpGs compared to downstream exons (Fig. 4f), and differentially methylated CpGs declined from upstream to downstream across the gene body (Fig. 4f). Long non-coding RNAs also showed more differentially methylated CpGs in exons (n = 92) compared to introns (n = 50) (Supplementary Fig. 1). Annotation-specific distributions of differentially methylated CpGs were significantly different from the distributions of all sequenced CpGs (Pearson’s Chi-squared test with Yates’ continuity correction, FDR-corrected P < 0.05) for seven out of the eight annotation features (exon UTR, CDS, intron, downstream Flank, long non-coding RNA, TE, intergenic); only the “upstream” feature was not significant (P = 0.509) (Detailed results are available in Supplementary data repository).
Genome-wide population structure, genetic diversity and the relationship with CpG methylation
Population structure was weak (FST = 0.025, 95% CI: 0.025–0.026). Some separation of samples by population was apparent along the first PC axis, which explained only 12.45% of variance (percent variance explained largely plateaued for remaining PC axes), consistent with the low FST (Fig. 1c). Nucleotide diversity (π) per site was similar between the populations, with global π = 0.00197 (95% CI: 0.00196–0.00198), OR π = 0.00191 (95% CI: 0.00191–0.00192), and CA = 0.00193 (95% CI: 0.00192–0.00194), suggesting that differences in genetic diversity do not drive differences in observed methylation levels between populations.
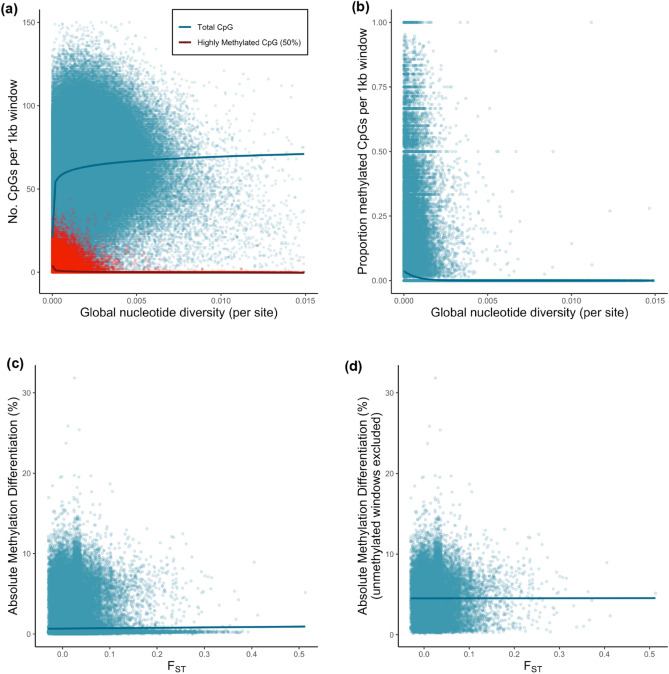
We tested the relationship between general methylation patterns and population genetic diversity across 1 kb regions within the B. vosnesenskii genome. There was a strong correlation between the mean methylation proportion per CpG per 1 kb window (n = 232,788 windows) and both the raw number (Pearson’s r = 0.84, t232786 = 737.97, P < 0.001; Spearman’s rho = 0.46, S = 1 × 1015, P < 0.001) and proportion (r = 0.96, t232786 = 1642.3, P < 0.001; rho = 0.46, S = 1 × 1015, P < 0.001) of highly methylated CpGs per window. We thus performed analysis only on counts of highly methylated CpGs. The number of highly methylated sites per 1 kb window was negatively correlated with π (Fig. 5a) (Pearson’s r = − 0.22, t232786 = 110.59, P < 0.001; Spearman’s rho = − 0.29, S = 2.7 × 1015, P < 0.001). This relationship was not seemingly driven by the number of available CpGs per window, as low diversity windows had fewer CpGs in general (Fig. 5a), so the proportion of CpGs methylated per window thus also declined significantly with π (Fig. 5b; Supplementary Table 1) (Pearson’s r = − 0.24, t232786 = -118.88, P < 0.001; Spearman’s rho = − 0.29, S = 2.7 × 1015, P < 0.001). There was a weak relationship for FST and the mean percent methylation difference per CpG per 1 kb window between populations (Pearson’s r = 0.01; t229624 = 4.97, P < 0.001), although this was not significant for Spearman’s rank correlation (Spearman’s rho = 0.004; P > 0.05) (Fig. 5c). Because above data suggested that certain sites were likely to never be methylated in B. vosnesenskii and thus would not differ among populations, it is possible that such regions could affect patterns of differentiation within methylated regions. We thus also evaluated the FST-methylation difference relationships after excluding windows with no methylated CpGs (< 10% threshold; n = 22,542 1 kb windows retained) and there was no correlation (Pearson’s r = 0.000, Spearman’s rho = 0.000; both P > 0.05; Fig. 5d), suggesting that the weak positive correlation above was likely driven by intragenic or intronic windows with both weak divergence and no methylation.
Figure 5.
Relationships between genetic diversity from whole genome re-sequencing and methylation patterns. (a) Relationship between global nucleotide diversity across samples and the number of methylated CpGs (50% threshold, red) and all sequenced CpGs (blue). Lines fit with a log relationship for visualization (see Supplementary Table 1 for statistical model). (b) Relationship between global nucleotide diversity across samples (log-transformed) and the proportion of methylated CpGs (50% threshold). Line fit with a binomial model for visualization (see Supplementary Table 1 for statistical results from the zero-inflated beta-binomial model). (c) Relationship between FST and absolute percent methylation differences between populations, (d) Relationship between population-level FST and absolute percent methylation differences excluding 1 kb windows with no methylated CpGs. Line fit with a linear relationship for visualization, as no substantial relationship between the variables was detected (see Supplementary Table 1 for detailed statistics).
Gene ontology analysis of genes harboring highly methylated and differentially methylated CpGs
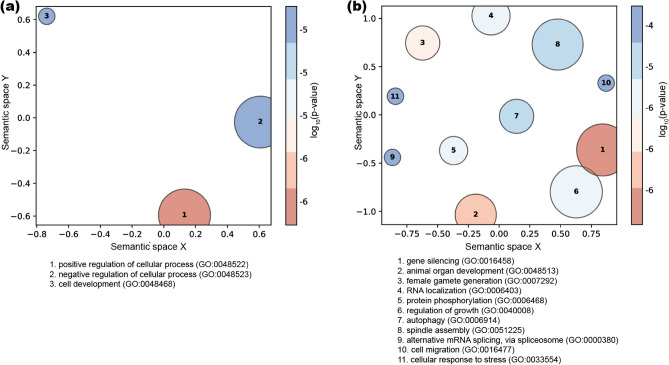
Analyses of unique genes (n = 44) containing ≥ 100 highly methylated sites provided 18 statistically significant [family-wise error rate (FWER) ≤ 0.1] GO terms and a total of seven summarized GO term clusters. Overall, these GO terms and summarized GO clusters were linked to fundamental cellular activities, such as metabolism, binding, regulation of biological processes, neuronal activities, gene expression machinery and cell development (Fig. 6a; Supplementary Table 2 and 3). Gene Ontology (GO) analyses of genes (n = 1272) harboring differentially methylated sites (10% difference threshold) produced 89 significantly enriched terms (Supplementary Table 4) that grouped into 31 clusters that were likewise associated with diverse biological processes, including binding, various enzymatic activities, reproduction, cell cycle, development, metabolism, response to stress and cell communication, and signaling activities (Fig. 6b; Supplementary Table 5). Five overlapping GO terms from the general and differential methylation enrichment analyses included two overlapping biological process-related terms [positive regulation of cellular process (GO:0048522), regulation of cellular component organization (GO:0051128), two molecular function related terms [mRNA binding (GO:0003729), RNA binding (GO:0003723)] and one cellular component related term [nuclear speck (GO:0016607)] (Supplementary Table 6). Comparison of GO terms from this study with two previous studies55, 56 indicates a functional-level convergence regarding population-specific thermal/environmental adaptation as we noticed several overlapping GO terms, such as, cell signaling and communication, development, reproduction, metabolic functions, neuronal activities and stress response (Supplementary Table 7).
Figure 6.
Summarized visualization of biological process-related GO terms from unique genes (n = 44) harboring a minimum of 100 highly methylated (> 50% methylation) sites (a) and unique genes (n = 1272) harboring differentially methylated sites assessed at 10% methylation difference (b). The top ten biological process-related clusters with their corresponding GO IDs were listed at the bottom of each plot.
Discussion
This study presents a high-coverage methylome analysis for the North American bumble bee B. vosnesenskii and it is the first to provide initial insights into CpG methylation patterns in wild-caught bumble bees from climatically distinct locations. Genome-wide methylation patterns in B. vosnesenskii are similar to those observed in other arthropods and hymenopterans, with a preponderance of highly and sparsely methylated sites found in gene bodies and unmethylated sites disproportionately represented in introns and intragenic regions. We also identified multiple (n = 2066) differentially methylated CpGs between the two sampled populations, predominantly in exons and putative promoter regions, suggesting that epigenetic marks can vary across bumble bee species’ ranges. Our study also reconfirmed previous findings of low genetic diversity and genome-wide genetic homogeneity in B. vosnesenskii and showed that while highly methylated regions tended to occur in genome regions with relatively low nucleotide diversity, there was no clear relationship between methylation differentiation and genetic differentiation across genome regions. This in-depth high-coverage analysis of epigenetic variations in B. vosnesenskii offers novel biological insights into the factors that may shape the genome-wide distribution of DNA methylation in bumble bees and provides a valuable starting point for more detailed studies of epigenetic mechanisms that may be involved in environmental adaptation or plasticity in this species.
Our first research objective was to characterize consistent patterns of methylation observed across B. vosnesenskii workers collected from distinct climatic regions within the species’ range to identify features that were highly or rarely methylated in all individuals. The low genome-wide CpG methylation (~ 1.1%) is similar to other Hymenoptera68, including other bumble bees59–61, the honey bee Apis mellifera69, the wasp Nasonia vitripennis70. Such trends are generally common in holometabolous insects31 apart from a few unusual instances31. Despite low overall methylation, sparsely distributed peaks of high CpG methylation were non randomly distributed across scaffolds owing to a concentration of methylation in gene bodies, especially exon sequences. This intragenic CpG methylation is also a classic characteristic in insects19, 28, 31, 60, 69–72, and gene body methylation is likely ancestral73. Thus, our results add to the growing body of evidence across the multiple insect orders where the prevalence of gene body methylation was observed irrespective of substantial variability in global methylation levels31.
Within genes, methylation was substantially biased towards the 5′ region, with a higher concentration of CpG methylation near the TSS (Fig. 2c) and a relatively gradual decrease of CpG methylation across (5′ to 3′) the transcription unit (Fig. 2d–f). At a more granular level, exon sequences have substantially more methylated sites than introns (Fig. 2d), with a disproportionate distribution of highly methylated sites in exon 2–4, with fewer in exon 1 (Fig. 2d). Similar 5′ biased methylation is observed in bees69, wasps70, ants71 and more generally in holometabolous insects. In contrast, 3′ bias is more prominent in many hemimetabolous insects74, 75 and mammals with much higher global methylation72. The disproportionate exon–intron breakdown patterns across genes and depleted methylation in the first exon, are also common in Hymenoptera31, 72 and other arthropods, such as Crustaceans72, 76. In several Hymenoptera species, clusters of CpG methylation are found across the exon–intron boundaries, as we tend to observe here68, which may contribute to alternative splicing via its presumed role of exon–intron “tagging”28, 70. Several studies in arthropods indicate a potential role of gene body methylation in transcription elongation and alternative splicing19, based on the apparent correlation of CpG methylation with alternative splicing found in honeybees30, 69, 77 and ants71. However, evidence from multiple insect orders suggests that CpG methylation is not directly correlated to differential exon usage31, 59, 70. The mixed evidence on the potential involvement of gene body methylation on alternative splicing indicates the need for future methylation studies in bumble bees that explore the possible link between CpG methylation and differential exon usage by utilizing complementary gene expression and methylation datasets.
One consistent pattern in insects is that gene body methylation is believed to be associated with unimodal expression of highly expressed “housekeeping” genes19, 31, 60, 70, 78. These highly expressed “housekeeping” genes are uniformly (i.e., not developmental stage- or tissue-specific) expressed31, exhibiting low variability in their expression pattern70, 79. Gene ontology analysis results from our study also support this as we noticed functional enrichment of many important essential activities in our list of GO terms, such as biological processes related to the regulation of gene expression, alternative splicing, metabolism, development, neuronal activities and other fundamental aspects of cell machinery (Supplementary Table 2). Highly methylated genes in other arthropods26, 69–72, 79–81 also exhibit functional level enrichment of essential cellular functions such as metabolism, mRNA processing, organelle function and transport related terms. Thus, the extent and the functional properties of gene body methylation in B. vosnesenskii complement the similar patterns observed in other holometabolous insects exhibiting overall low global methylation and clustered exon-biased gene body methylation, in contrast to the relatively higher global methylation and higher methylation levels extending to other genomic features (e.g., promoters, introns, and transposable elements) in hemimetabolous insects19, 31.
Several insect studies also suggest a link between gene body methylation and other epigenetic mechanisms82. For example, nucleosome dynamics, histone post-translation modifications, and associated changes in local chromatin state83 have been hypothesized to act in concert with CpG methylation to mediate the extent and timing of access to the transcriptional machinery and, thus, regulate subsequent gene expression levels84. Our data support potential cooperation among these epigenetic mechanisms as GO analysis of highly methylated CpGs also included a histone modification-related term [negative regulation of histone H2A K63-linked ubiquitination (GO:1901315); Supplementary Table 2]. In insects, CpG methylation is strongly associated with histone post-translational modification and transcriptionally active chromatin marks82, 85. It may play a critical role in ensuring the consistent expression of highly methylated genes across insect lineages via the exclusion of a chemically modified TSS-associated histone variant (H2A.Z) that exhibits a negative correlation to gene expression activity28. Thus, high CpG methylation concentration patterns of near TSS and subsequent 5′ bias could be potentially linked to CpG methylation-mediated chromatin remodeling near TSS82. Methylation levels in arthropods can also be related to nucleosome occupancy around the TSS, with nucleosome occupancy exhibiting positive correlations with CpG methylation31. No nucleosome positioning data is available for bumble bees yet; however, we hypothesize that distinct distribution pattern of distance to TSS for both highly methylated sites and differentially methylated sites observed in B. vosnesenskii could be potentially linked to nucleosome occupancy, especially given differences in methylation levels observed between the populations. Future multi-omics studies examining the multiple components of individual-specific epigenomes would be especially advantageous to address knowledge gaps relating to the total epigenetic landscape and regulation of context-dependent gene expression.
The second objective of this study was to evaluate the potential for differences in methylation levels among B. vosnesenskii from the spatial-environmental extremes of its broad distribution. We identified 2,066 differentially methylated sites between the two populations and the genomic distribution of these differentially methylated CpGs matched the trends of general CpG methylation, and were similarly overrepresented in gene bodies, especially in exons, consistent with the distribution of differentially methylated sites between sexes and castes in the bumble bee B. terrestris59. The colder high-elevation Oregon site exhibited lower percent methylation (1.03% ± 0.04% SD) than warmer southern low-elevation sites in California (1.17% ± 0.06% SD), and 87.6% of differentially methylated sites were hypomethylated in the northern high-elevation samples. Although our results must be evaluated in additional populations for robust conclusions, several insect studies have reported a propensity for hypomethylation at low temperatures, including reduced CpG methylation under low-temperature stress in the tick Haemaphysalis longicornis86 and under relatively low rearing temperatures in the cockroach Diploptera punctata87. Interestingly, while highly methylated genes are evolutionary conserved, hypomethylated genes are often faster evolving, and can be order-, genus- or species specific70, 72 and exhibit tissue73 or developmental stage specific70 expression. Thus, hypomethylated genes may be more plastic, exhibiting more variability and flexibility regarding their adaptability towards environmental cues88. The reduced methylation observed in the high-elevation Oregon B. vosnesenskii population is intriguing given that this population was also found to have the broadest range in critical thermal limits in laboratory experiments (CTMIN vs CTMAX), and also exhibited the most unique gene expression patterns, especially at CTMIN56. Although we could compare GO terms from prior gene expression and coding sequence variation datasets to identify shared biological functions or cellular components with our methylation data, we cannot link our detected methylation levels directly to thermal tolerance with the current dataset as we currently lack corresponding gene expression data at the sample-level. Establishing causal links with direct comparisons between transcription and methylation/coding sequence variance will be needed to formulate insights into niche adaptation. Given differences observed between the latitude-altitude extremes in this study, future studies involving CpG methylation and complementary gene expression data from specimens sampled across the altitudinal and latitudinal gradients of its wide species range would be advantageous2.
Genes harboring at least one differentially methylated CpG were enriched for GO terms related to several biological processes such as metabolism, reproduction, cell cycle process, and fundamental cellular activities and molecular functions including binding, transmembrane transport, and various enzymatic functions (Supplementary Table 4 and 5). These results are broadly consistent with gene ontology analysis of differentially methylation between reproductive states60 or during colony development61 in B. terrestris. Similar functional enrichment results have also been reported in differentially methylated gene sets from abiotic stress response-related studies involving silkworm89, water fleas90, and ticks86. Numerous GO terms overlap with previous population genomic and thermal stress studies in B. vosnesenskii55, 56, including cellular communication/signaling and functions related to neuronal activities, gene expression regulation, metabolism and reproduction (see Results and Supplementary Table 7). Of particular interest from the perspective of thermal tolerance, we observe GO term related to “cellular response to stress” (GO:0033554) within the summarized biological function-related GO term clusters for differentially methylated gene sets (Supplementary Table 5). At the gene level, at least one differentially methylated CpG was observed in ion channel and membrane transport-related genes [sodium/calcium exchanger regulatory protein 1-like (LOC117234134), TWiK family of potassium channels protein 7 (LOC117238582), chloride channel CLIC-like protein 1 (LOC117236045), calcium homeostasis endoplasmic reticulum protein (LOC117242823)] and heat shock protein-related genes [97kDa heat shock protein (LOC117234768), heat shock protein 83-like (LOC117235089)]. Heat shock protein machinery91–93 and ion channel/transmembrane transport mechanisms94, especially those linked to calcium regulation95 are widely recognized for their essential role in mediating molecular responses to thermal stress95, 96, and have been previously observed in B. vosnesenskii55, 56. The presence of chromatin-related GO terms (i.e., GO:0043044, GO:0003682) in the GO term lists of differentially methylated genes (Supplementary Table 4) is consistent with the potential involvement of CpG methylation in mediating access transcription machinery and particularly with a previously reported case of enrichment of chromatin related GO terms for differentially methylated genes related to caste determination in bumble bees59. Although the potential link between differential methylation and differential expression is still unclear in insects as there is mixed evidence if the differential methylation is positively correlated to differential expression97, 98 (but see99–103) or differential exon usage60, 74, these reported genes from our study could serve as promising candidates to more closely examine in future studies of thermal stress or other niche specific gene expression regulation in bumble bees.
Finally, our third objective was to explore the potential link between genomic and epigenetic variability in B. vosnesenskii. Interestingly, B. vosnesenskii appears to exhibit variation in thermal tolerance among populations with minimal genome-wide population structure56. Although we observe weak differentiation in both genome-wide SNP polymorphisms and CpG methylation, there is substantial range-wide genetic connectivity between the populations selected for WGBS (FST = 0.025). There was also no substantial correlation between methylation differences and FST in 1 kb windows across the genome, especially once methylation-free regions were removed, indicating that regions with variable methylation are not located in high- or low-differentiation regions. This is consistent with a recent study in another insect, Diploptera punctata, which also failed to find any correlation between genetic and epigenetic variability87. We did observe a significant negative correlation between nucleotide diversity (π) and methylation levels across genomic windows, which is consistent with the elevated levels of methylation in gene bodies, as protein-coding regions tend to have lower levels of variation, including reduced nonsynonymous π in B. vosnesenskii5. Methylation analysis in lab-reared bumble bees also reported a potential relationship between evolutionary sequence conservation and CpG methylation59. While CpG methylation can potentially act as a mutagen on individual cytosines104, 105, paradoxically, CpG methylation in insects is enriched in evolutionary conserved “housekeeping” genes31 where it may play a counterintuitive role as a stabilizing factor59. The potential complex relationship between underlying genomic diversity and epigenetic variability in bumble bees should be further investigated, ideally including other species with more variable heterozygosity or population structure5.
This study provides the first look at the potential for ecologically associated epigenetic variation across the B. vosnesenskii range, however there are several limitations which should be considered when interpreting our reported results and must be addressed with future research. First, methylation status may be influenced by developmental age of the bumble bees and other associated ecological and environmental variables106 which are common caveats in ecological epigenetic studies. Although, the collection of wild bees prohibited any control for many variables, prior studies suggest that most such variation is driven by sex, tissue, and developmental stages58–60 so sampling of all adult female workers may minimize such concerns. A second concerns is that the challenges of collecting populations from range extremes necessitated sampling populations on different dates, which could introduce biases due to different local conditions experienced by samples prior to collection (as opposed to more bioclimatic divergence associated at range extremes). Thus, we cannot fully exclude the possibility that some differential methylation could be from variable specimen age or recent environmental experience. Increasing sample size beyond our small initial sample size (n = 8) may also help improve statistical power of future analyses to detect important but subtle population-specific methylation changes, while reducing error introduced by factors like sample age or prior individual experiences107.
In summary, our study provides the first high-coverage methylation profiling in a widespread North American bumble bee, B. vosnesenskii, and unravels the key characteristics and trends of CpG methylation in this montane species. B. vosnesenskii is a crucial pollinator and one of two species available commercially to be used for greenhouse crop pollination in North America54 and is also one of few North American bumble bees that may benefit from projected future climate change scenarios51. Although more work is needed, understanding region-specific genomic and epigenomic variation, particularly their connection to thermal adaptation, may hold considerable economic and practical conservation value. Epigenetic variation is only recently beginning to be evaluated in bumble bees, nevertheless, given the substantial colony-specific variation in bumble bee methylomes60, it is also possible that environmentally associated colony-specific “epi-alleles” at the population level108 may exist and play a role in niche-specific adaptation or may contribute to phenotypic plasticity. Further, our study only evaluated one tissue type which, while relevant for thermoregulation and flight, should be expanded to incorporate additional tissues to fully understand variation in the methylation landscape in B. vosnesenskii. Overall, this study provides baseline data for future studies that will include integrative multi-omics approaches (e.g., genomics, transcriptomics, epigenomics, metabolomics) from field and laboratory experiments to build a conceptual framework on the interplay between multiple modes of non-genomic epigenetic variations and its influence across multi-level molecular processes that are mediating tolerance to a broad set of environmental conditions in this species2, 109.
Methods
Samples
DNA was extracted using Qiagen DNeasy kits from the thoracic tissue of worker bees from a previous study5 which were collected from southern California at low elevations and from northern Oregon at high elevation (see Table 1 for detailed information). These sites generally represent warm and cold extremes of the species range56 (Fig. 1a). All B. vosnesenskii workers (Fig. 1b) represent unique colonies based on inferences of relatedness from reduced representation SNP data5.
Table 1.
Detailed information about the samples used in Whole Genome Sequencing (WGS) and Whole Genome Bisulfite Sequencing (WGBS) approach.
| Population | Latitude | Longitude | Elevation (m) | Date | Sample Name | Sequencing data type |
|---|---|---|---|---|---|---|
| Northern high elevation (Oregon) | 45.332 | − 121.670 | 1699 | 3-Aug-16 | JDL3148-OR052016 | WGBS + WGS |
| JDL3150-OR052016 | WGBS + WGS | |||||
| JDL3152-OR052016 | WGBS + WGS | |||||
| JDL3147-OR052016 | WGBS | |||||
| JDL3144-OR052016 | WGS | |||||
| JDL3145-OR052016 | WGS | |||||
| JDL3146-OR052016 | WGS | |||||
| JDL3153-OR052016 | WGS | |||||
| JDL3154-OR052016 | WGS | |||||
| JDL3155-OR052016 | WGS | |||||
| Southern low elevation (California) | 36.458 | − 118.879 | 314 | 12-May-15 | JDL928-CA012015 | WGBS + WGS |
| JDL929-CA012015 | WGBS + WGS | |||||
| JDL931-CA012015 | WGBS + WGS | |||||
| JDL940-CA012015 | WGBS | |||||
| JDL930-CA012015 | WGS | |||||
| JDL932-CA012015 | WGS | |||||
| JDL933-CA012015 | WGS | |||||
| JDL937-CA012015 | WGS | |||||
| JDL938-CA012015 | WGS |
Whole genome methylation sequencing and WGBS data analysis
Whole genome methylation libraries were prepared using the Swift AccelNGS Methyl-Seq DNA library approach for bisulfite-converted DNA (with lambda control genome spike-in) and they were sequenced on an Illumina Hiseq X sequencer by HudsonAlpha Institute for Biotechnology Genome Services Lab (Huntsville, Alabama, USA). Samples (n = 8) were run in individual lanes to generate 2 × 151 bp paired-end libraries. 3.6 × 107 raw read pairs and 1,088Gbp in total were sequenced in the raw WGBS data set (per sample mean = 450.19 × 106 ± 50.21 × 106 SD read pairs and 135.96Gbp ± 15.16Gbp SD of sequence). Quality assessment of the sequenced specimens was conducted using FastQC v.0.11.9110. Based on the generated sequence quality assessment and a large amount of sequence data, we conducted stringent quality filtering, including adapter removal, quality trimming, removal of short sequences (< 50 bp) and removed specific fixed lengths from both 5′ and 3′ ends to minimize bisulfite conversion bias using Trim Galore! v.0.6.6111; custom command line parameters:–illumina –q 20 –clip_R1 20 –clip_R2 20 –three_prime_clip_R1 20 –three_prime_clip_R2 60 –length 50). After stringent trimming and quality filtering of these high coverage data, we discarded ~ 34.27% of raw reads, resulting in 295.90 × 106 ± 107.65 × 106 SD trimmed read pairs and 52.33 Gbp ± 19.32 Gbp SD per sample. We generated post-trimming sequence quality reports and sample-specific statistics using FastQC v.0.11.9110 and SeqKit v.0.15.0112. All samples were sequenced to very high coverage, but the total number of reads varied among sample, so to reduce possible biases in methylation calling and subsequent analyses due to sequencing depth we normalized read coverage by random subsampling with SeqKit v.0.15.0112 to match the smallest number of read pairs in any sample (n = 187,618,210 read pairs per sample). After performing the read-pair subsampling across samples, 187.62 million read pairs for each sample were aligned to the B. vosnesenskii genome assembly, which resulted in 75.70 ± 3.04 SD fold sequencing depth per sample.
Subsampled read pairs were aligned to the B. vosnesenskii genome assembly (RefSeq accession GCF_011952255.165) using bwa-meth v.0.2.2113 and alignment files were sorted using SAMtools v.1.9114. PCR duplicates were removed using MarkDuplicates from Picard tools v.2.23.9115. Methylation extraction in the CpG context from sorted post-processed BAM files was conducted using MethylDackel v.0.6.1116 by setting an absolute minimum coverage and employing bioinformatic removal of CpGs that were potentially C-to-T variant sites using the following parameters (–minDepth 10 –maxVariantFrac 0.5 –minOppositeDepth 10 –methylKit). Bioinformatic removal of probable C > T variants by MethylDackel resulted in the exclusion of 64,847.63 ± 5138.26 SD CpGs per sample and resulted in a methylation call dataset containing 22,189,312.75 ± 1,919,429.19 SD CpG locations per sample. Further processing was conducted in R v.4.1.3117 utilizing methylKit v.1.20118 (analysis summary is available in Supplementary Table 8). We removed CpGs with < 10 × coverage and with unusually high coverage (> 99th percentile) to minimize the effects of paralogs or repetitive regions, which excluded 1.04% ± 0.01% SD sites from the samples (Supplementary Table 8) and resulted in 21,961,863.38 ± 1,898,407.22 SD CpGs per sample. We calculated the per base read coverage and per base methylation statistics for each sample before and after filtering using the getCoverageStats and getMethylationStats functions in methylKit, respectively, and utilized the average percent methylation per CpG site matrix for tabulating genome-level sample-specific and population-specific mean percent CpG methylation. There remained some dissimilarity of per base coverage within and across the samples even after read normalization, so we also normalized the coverage of the CpGs per sample using the normalizeCoverage function (method = “median”) in methylKit. We then obtained a united methylation call dataset for all samples using the unite function in methylKit that included all CpGs present in every sample at ≥ 10 × coverage (n = 14,627,533 CpGs). As the presence of C > T SNPs can impact the accuracy of detected methylation levels in CpGs119, in addition to using a built-in bioinformatic detection in MethylDackel v.0.6.1, we also filtered sites using SNP data from whole genome sequencing in these populations (see the following section: “Whole genome resequencing and variant calling”). We excluded 44,041 CpGs that overlapped SNP positions so that we could focus on sites that should only be affected by methylation. After filtering, the final dataset used for general and differential methylation analysis contained 14,583,492 CpGs containing no missing data (i.e., sites that are present in every sample). Although the consistent patterns of low and similarly distributed methylation in all samples indicated successful WGBS (see Results), we repeated bioinformatic analyses by mapping reads to Escherichia phage Lambda (NCBI GenBank accession J02459.1) to assess bisulfite conversion efficiency. We found an average of 99.80% ± 0.01% SD successful raw conversion rate, and when applying a liberal 10% threshold to call a site as methylated, we found that a mean of 0.01% ± 0.01% SD of sites were called as C and thus would be considered erroneously methylated. Upon further investigation, all these calls (a single base each in four of eight samples) were at the same genomic location near the start of the genome (J02459.1—base location 8), suggesting a possible technical or bioinformatic artifact rather than any issues in the WGBS conversion (See details in Supplementary data).
To investigate the general differences in methylation among samples, we conducted principal component Analysis (PCA) using the PCASamples function in methylKit by utilizing all CpGs (n = 14,627,533) sequenced in at least 10 × coverage. We also used the same CpG dataset to conduct hierarchical clustering analysis by calculating a correlation matrix from per base percent methylation data utilizing Ward’s minimum variance method with the clusterSamples function in methylKit.
Analysis of consistent patterns genome-wide CpG methylation in B. vosnesenskii
We calculated the percent methylation per CpG site (percentage reads at each CpG cytosine with a C or T) for each sample using percMethylation function of methylKit. Based on the average percent methylation for each CpG site, we categorized these sites into three categories; methylated (with ≥ 50% methylation); sparsely methylated (10–50% percent methylation), and unmethylated (≤ 10% percent methylation). We first calculated the distance from the nearest transcription start site (TSS) for all CpGs (getAssociationWithTSS function of methylKit from the NCBI B. vosnesenskii RefSeq annotation65). We used a two-sided two-sample Kolmogorov–Smirnov test to compare distributions of the distances from TSS of highly methylated sites and all CpGs using ks.test function in R v.4.1.3. We then used the NCBI B. vosnesenskii RefSeq annotation65 to generate feature-specific custom annotation genome tracks [i.e., Untranslated Regions of exon (exon UTR), Coding Sequences (CDS), Intron, Upstream flanking regions (Upstream Flank), Downstream flanking regions (Downstream Flank), long non-coding RNA (lncRNA), Transposable elements (TE), intergenic] following previously described methods120 and publicly available codes (available at: https://github.com/RobertsLab/project-gigas-oa-meth). We produced feature-specific breakdowns for all three CpG subsets (i.e., highly methylated, sparsely methylated, and unmethylated CpGs) and all sequenced CpGs. To test for statistically significant enrichment of highly methylated CpGs and the overall abundance of sequenced CpGs in each genomic annotation feature, for each feature class, we compared all CpGs against methylated sites using a Pearson’s Chi-squared test with Yates’ continuity correction implemented by chisq.test function in R.
After initial analyses confirmed that most methylated CpGs were confined to gene bodies, we next investigated the breakdown of CpGs based on their location within the gene body. To avoid complications that may arise from the existence of multiple transcripts due to alternative splicing, we selected the annotation of the longest isoform for each gene using the AGAT genomic toolset v.0.8.0121 and tabulated the fine-scale gene-body feature annotation count for each exon and intron. CpG counts for each exon and intron for protein-coding genes and long non-coding RNAs were conducted using custom bash scripts.
Differential methylation analysis
To conduct the differential methylation analysis, we first calculated the mean and standard deviation of all CpGs using rowSds and rowMeans2 function of R package matrixStats v.0.62122. Because the majority of CpGs in the genome were found to be unmethylated, as is typical for insects27, we removed low-variability CpGs (i.e., within less than 2 standard deviations of per base percent methylation calculated for each CpG site location across all samples) as they are not informative for differential methylation and would increase the total number of comparisons for significance testing. Overall, 93.82% of CpGs were excluded in this process. The remaining variable (SD > 2) CpGs (n = 901,868) were used in differential methylation analysis in methylKit v.1.20118. We implemented Chi-square test to test for significance between two population groups with basic overdispersion correction123 along with a false discovery rate (FDR) correction using the Benjamini-Hochberg (BH) procedure. We considered a site as differentially methylated only if there was ≥ 10% methylation change between two populations with an FDR corrected q ≤ 0.01. We defined the CpGs as “hypomethylated” and “hypermethylated” when we found statistically significant lower and higher levels of percent methylation difference in OR samples compared to CA samples, respectively.
To compare the sample-specific methylation patterns, we also tabulated distances from the nearest transcription start site (TSS), compare distributions of the distances from TSS of differentially methylated sites with all CpGs, principal component analysis (PCA) and hierarchical clustering analysis for both variable (SD > 2) CpGs (n = 901,868) and differentially methylated sites (n = 2066; assessed at 10% methylation difference) using the methods described in “Analysis of general methylation patterns” section above. We then annotated the differentially methylated sites (n = 2066) and investigated the exon–intron breakdown of these differentially methylated sites using the methods described above and used the chi-square based contingency test as above to examine if the annotation-specific distribution of differentially methylated sites (assessed at 10% methylation difference) is significantly different than the distribution of all CpGs sequenced in the WGBS data set.
Whole genome resequencing and variant calling
We used additional samples from the two bisulfite sequencing localities for whole genome resequencing to characterize genome-wide diversity and differentiation and identify genome positions with SNPs that could be artifactually inferred as differential methylation. We selected B. vosnesenskii individuals from each locality (8 for CA01.2015, 9 for OR05.2016; Fig. 1) that represent unique colonies based on inferences of relatedness from reduced representation data5. DNA was extracted from thoracic muscle using DNeasy kits as above and provided to the University of Oregon Genomics & Cell Characterization Core Facility for library preparation and sequencing on an Illumina HiSeq 4000 instrument. Resulting sequence data were filtered for quality using bbduk v.37.32124 to remove adaptors, trim low-quality bases, and remove short reads (ktrim = r k = 23 mink = 11 hdist = 1 tpe tbo ftm = 5 qtrim = rl trimq = 10 minlen = 25). Reads were mapped to the B. vosnesenskii reference genome (RefSeq Accession GCF_011952255.1)65 using BWA v.0.7.15-r1140125. SAM files were converted to the BAM using SAMtools v1.10114 and Picard Tools v.2.23.9115 was then used to sort, mark duplicates, and index BAM files. To identify a SNP set for filtering methylation data (see above) we used freebayes v.1.3.2126. We filtered the resulting VCF with VCFtools v.0.1.13127 to remove indels and non-binary SNPs, scored genotypes with depth < 4 × as missing, and then retained sites with ≤ 10% missing data, Q ≥ 20, and a minor allele count of ≥ 2. Finally, we removed a SNPs with unusually high sequencing coverage (> 2 times mean coverage per site) and with significant heterozygosity excess using Bonferroni correction (–hardy flag in VCFtools) (following128). The resulting VCF included 1,162,015 SNPs after filtering, with a mean sequencing coverage of 9.97 ± 1.68 SD reads per SNP per individual and a mean missingness of 1.78% ± 1.40% per SNP per individual (98.2% complete data matrix).
The called SNP set was needed for filtering methylation calls, however genetic diversity and population structure analyses used a genotyping-free approach in the software ANGSD v.0.935-53-gf475f10129. ANGSD employs methods to estimate population genetic statistics from BAM files while accounting for genotype uncertainty associated with high throughput sequencing data130, 131. We estimated the folded site frequency spectrum (SFS)131 across 151 genome scaffolds of at least 100 kb in length (total genome size analysed = 241,826,154 bp). We estimated nucleotide diversity (π) for the two populations separately and combined using the angsd -doSfs command with minimum mapping and base quality of 20, mapping quality downgrading of C = 50, GATK132 genotype likelihoods (GL = 2), and base quality recalibration (baq = 1). We then ran the realSFS program with the-fold 1 option to produce a folded SFS and thetastat –do_stat to estimate diversity parameters per site and in stepping windows of 1 kb (window and step both 1,000 bp). Narrow windows were used due to the rapid breakdown of linkage disequilibrium in bumble bee genomes62 and to avoid dilution of signal in comparisons with bisulfite data due to the globally sparse but locally clustered methylation in the B. vosnesenskii genome (see Results). Weighted FST was determined for the two populations by estimating the folded 2D SFS using the realSFS program and was determined per site and for 1 kb windows (window and step both 1,000 bp). Confidence intervals around mean nucleotide diversities and FST were obtained by nonparametric bootstrapping (1,000 replicates across windows with 1,000 sequenced sites) in the R package boot v.1.3-28133. Population structure was visualized using PCA with the PCAngsd v.1.03 program134 from ANGSD genotype likelihoods.
For genomic window-based analyses, we retained windows with complete sequence data across 1 kb, and for comparison with methylation data, we only retained windows with at least one CpG. To test for a significant effect of methylation counts and π (log-transformed) per 1 kb window, we used the R package glmmTMB v.1.1.5135 to perform a zero-inflated generalized linear model (family = negative binomial 2 to account for overdispersion). We also tested the relationship for the proportion of highly methylated (> 50% category) CpGs and π (log-transformed) within each window using zero-inflated logistic regression.
Gene Ontology (GO) analysis of highly methylated and differentially methylated CpGs
To understand the putative functional roles of genes carrying CpGs, we conducted a gene ontology (GO) analysis of two different gene sets of highly methylated and differentially methylated sites, respectively. Because there are substantial numbers of unique genes (n = 6010) with at least one highly methylated CpG site represented in the gene set, we decided to set a predefined criterion (i.e., use the subset of unique genes harboring a minimum of 100 highly methylated sites) to conduct functional enrichment analysis. Based on this criterion, we selected a subset of unique genes (n = 44) which were subsequently used in our gene ontology analysis. We also conducted a separate functional enrichment analysis where we included all unique genes (n = 1272) harboring all differentially methylated sites (n = 2066) assessed at 10% methylation difference. We conducted functional enrichment analysis for both gene sets using R package GofuncR v.1.14136 and utilized the curated B. vosnesenskii GO annotations from Hymenoptera Genome Database137. We considered the GO terms significant using a stringent Familywise Error Rate cut-off, FWER ≤ 0.1 using the refine function implemented in R package GofuncR v.1.14. We used semantic similarity-based reduction of GO terms and visualized the enriched GO term list using GO-Figure!138. We independently compared the statistically significant GO terms from both gene sets with GO term lists from two previous studies55, 56. In one of these studies, Pimsler et al.56 identified 1786 enriched statistically significant GO terms (assessed at P ≤ 0.05) for seven different contrasts and directions of gene expressions). We combined these GO term lists into a single list representing the unique GO terms (n = 1398) found at least once in any of these contrasts to compare them with our study's two individual GO term lists. We also compared our gene ontology (GO) results with another study55 by Jackson et al. which provided two different enriched GO term lists from outlier gene lists detected from tested for associations with variable temperature (n = 151 GO terms) and precipitation (n = 86 GO terms). We combined these two GO term lists into a single list representing 221 unique GO terms from both categories and compared them with GO term lists from our study.
Supplementary Information
Acknowledgements
This work was supported by National Science Foundation for under grants no. DEB 1457645 and URoL 1921585 to awarded to J.D.L. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. We thank Jason M. Jackson for his assistance with specimen collection and sample processing. We also thank Lozier lab members for useful their helpful feedback and discussion.
Author contributions
J.D.L. conceptualized the study, managed funding support, performed population genetics/ WGS- bioinformatic and statistical analysis, and supervised the project. S.R.R. designed and conducted bioinformatic and statistical analyses for WGBS and functional genomic approaches. S.R.R. wrote the manuscript with contributions from J.D.L.. Both authors have contributed, read, and approved the final manuscript.
Data availability
Raw WGBS reads generated in this study has been deposited and is currently available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under NCBI BioProject PRJNA956115. Final methylation call set (n = 14,627,533), variant calling file for population genomics analyses, analysis codes/scripts and other associated files to reproduce the research in available from Zenodo data repository (https://doi.org/10.5281/zenodo.8327218).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-41896-7.
References
- 1.Orr HA. The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 2.Dillon ME, Lozier JD. Adaptation to the abiotic environment in insects: the influence of variability on ecophysiology and evolutionary genomics. Curr. Opin. Insect Sci. 2019;36:131–139. doi: 10.1016/j.cois.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Hoban S, et al. Finding the genomic basis of local adaptation: Pitfalls, practical solutions, and future directions. Am. Nat. 2016;188:379–397. doi: 10.1086/688018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- 5.Jackson JM, et al. Distance, elevation and environment as drivers of diversity and divergence in bumble bees across latitude and altitude. Mol. Ecol. 2018;27:2926–2942. doi: 10.1111/mec.14735. [DOI] [PubMed] [Google Scholar]
- 6.Fitzpatrick MC, Keller SR. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 2015;18:1–16. doi: 10.1111/ele.12376. [DOI] [PubMed] [Google Scholar]
- 7.Dillon ME, Frazier MR, Dudley R. Into thin air: Physiology and evolution of alpine insects. Integr. Comp. Biol. 2006;46:49–61. doi: 10.1093/icb/icj007. [DOI] [PubMed] [Google Scholar]
- 8.Keller I, Alexander JM, Holderegger R, Edwards PJ. Widespread phenotypic and genetic divergence along altitudinal gradients in animals. J. Evol. Biol. 2013;26:2527–2543. doi: 10.1111/jeb.12255. [DOI] [PubMed] [Google Scholar]
- 9.Franks SJ, Hoffmann AA. Genetics of climate change adaptation. Annu. Rev. Genet. 2012;46:185–208. doi: 10.1146/annurev-genet-110711-155511. [DOI] [PubMed] [Google Scholar]
- 10.Verheyen J, Tüzün N, Stoks R. Using natural laboratories to study evolution to global warming: Contrasting altitudinal, latitudinal, and urbanization gradients. Curr. Opin. Insect Sci. 2019;35:10–19. doi: 10.1016/j.cois.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Jeremias G, et al. Synthesizing the role of epigenetics in the response and adaptation of species to climate change in freshwater ecosystems. Mol. Ecol. 2018;27:2790–2806. doi: 10.1111/mec.14727. [DOI] [PubMed] [Google Scholar]
- 12.Liew YJ, et al. Intergenerational epigenetic inheritance in reef-building corals. Nat. Clim. Change. 2020;10:254–259. [Google Scholar]
- 13.McGuigan K, Hoffmann AA, Sgrò CM. How is epigenetics predicted to contribute to climate change adaptation? What evidence do we need? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021;376:20200119. doi: 10.1098/rstb.2020.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan EJ, Gluckman PD, Dearden PK. Epigenetics, plasticity, and evolution: How do we link epigenetic change to phenotype? J. Exp. Zool. B Mol. Dev. Evol. 2014;322:208–220. doi: 10.1002/jez.b.22571. [DOI] [PubMed] [Google Scholar]
- 15.Bird A, et al. Studies of DNA methylation in animals. J. Cell Sci. Suppl. 1995;19:37–39. doi: 10.1242/jcs.1995.supplement_19.5. [DOI] [PubMed] [Google Scholar]
- 16.He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards CL, et al. Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecol. Lett. 2017;20:1576–1590. doi: 10.1111/ele.12858. [DOI] [PubMed] [Google Scholar]
- 19.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. Evolution of DNA Methylation across Insects. Mol. Biol. Evol. 2016;34:654–665. doi: 10.1093/molbev/msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederhuth CE, et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016;17:194. doi: 10.1186/s13059-016-1059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharif J, Endo TA, Toyoda T, Koseki H. Divergence of CpG island promoters: a consequence or cause of evolution? Dev. Growth Differ. 2010;52:545–554. doi: 10.1111/j.1440-169X.2010.01193.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Mazzio EA, Soliman KFA. Basic concepts of epigenetics: Impact of environmental signals on gene expression. Epigenetics. 2012;7:119–130. doi: 10.4161/epi.7.2.18764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heard E, Chaumeil J, Masui O, Okamoto I. Mammalian X-chromosome inactivation: An epigenetics paradigm. Cold Spring Harb. Symp. Quant. Biol. 2004;69:89–102. doi: 10.1101/sqb.2004.69.89. [DOI] [PubMed] [Google Scholar]
- 26.Xu G, et al. Intragenic DNA methylation regulates insect gene expression and reproduction through the MBD/Tip60 complex. iScience. 2021;24:102040. doi: 10.1016/j.isci.2021.102040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glastad KM, Hunt BG, Yi SV, Goodisman MAD. DNA methylation in insects: On the brink of the epigenomic era. Insect Mol. Biol. 2011;20:553–565. doi: 10.1111/j.1365-2583.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- 28.Glastad KM, Hunt BG, Goodisman MA. Evolutionary insights into DNA methylation in insects. Curr. Opin. Insect Sci. 2014;1:25–30. doi: 10.1016/j.cois.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Weiner SA, Toth AL. Epigenetics in social insects: a new direction for understanding the evolution of castes. Genet. Res. Int. 2012;2012:609810. doi: 10.1155/2012/609810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li-Byarlay H, et al. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12750–12755. doi: 10.1073/pnas.1310735110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis SH, et al. Widespread conservation and lineage-specific diversification of genome-wide DNA methylation patterns across arthropods. PLoS Genet. 2020;16:e1008864. doi: 10.1371/journal.pgen.1008864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshmukh S, Ponnaluri VC, Dai N, Pradhan S, Deobagkar D. Levels of DNA cytosine methylation in the Drosophila genome. PeerJ. 2018;6:e5119. doi: 10.7717/peerj.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulz NKE, et al. Dnmt1 has an essential function despite the absence of CpG DNA methylation in the red flour beetle Tribolium castaneum. Sci. Rep. 2018;8:16462. doi: 10.1038/s41598-018-34701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonasio R. The expanding epigenetic landscape of non-model organisms. J. Exp. Biol. 2015;218:114–122. doi: 10.1242/jeb.110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalifa SAM, et al. Overview of bee pollination and its economic value for crop production. Insects. 2021;12:688. doi: 10.3390/insects12080688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velthuis HHW, van Doorn A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie. 2006;37:421–451. [Google Scholar]
- 37.Williams PH. An annotated checklist of bumble bees with an analysis of patterns of description (Hymenoptera: Apidae, Bombini) Bull. Nat. Hist. Mus. Entomol. Ser. 1998;67:79–152. [Google Scholar]
- 38.Heinrich B. Bumblebee Economics. Cambridge: Harvard University Press; 2004. [Google Scholar]
- 39.Heinrich B, Kammer AE. Activation of the fibrillar muscles in the bumblebee during warm-up, stabilization of thoracic temperature and flight. J. Exp. Biol. 1973;58:677–688. [Google Scholar]
- 40.Heinrich B. Thermoregulation in bumblebees. J. Comp. Physiol. B. 1975;96:155–166. [Google Scholar]
- 41.Heinrich B. Heat exchange in relation to blood flow between thorax and abdomen in bumblebees. J. Exp. Biol. 1976;64:561–585. doi: 10.1242/jeb.64.3.561. [DOI] [PubMed] [Google Scholar]
- 42.Maebe K, et al. Bumblebee resilience to climate change, through plastic and adaptive responses. Glob. Change Biol. 2021;27:4223–4237. doi: 10.1111/gcb.15751. [DOI] [PubMed] [Google Scholar]
- 43.Martinet B, et al. Global effects of extreme temperatures on wild bumblebees. Conserv. Biol. 2021;35:1507–1518. doi: 10.1111/cobi.13685. [DOI] [PubMed] [Google Scholar]
- 44.Harvey JA, et al. Scientists’ warning on climate change and insects. Ecol. Monogr. 2022;93:e1553. [Google Scholar]
- 45.Cameron SA, Sadd BM. Global trends in Bumble Bee health. Annu. Rev. Entomol. 2020;65:209–232. doi: 10.1146/annurev-ento-011118-111847. [DOI] [PubMed] [Google Scholar]
- 46.Kerr JT, et al. Climate change impacts on bumblebees converge across continents. Science. 2015;349:177–180. doi: 10.1126/science.aaa7031. [DOI] [PubMed] [Google Scholar]
- 47.Soroye P, Newbold T, Kerr J. Climate change contributes to widespread declines among bumble bees across continents. Science. 2020;367:685–688. doi: 10.1126/science.aax8591. [DOI] [PubMed] [Google Scholar]
- 48.Goulson D, Lye GC, Darvill B. Decline and conservation of bumble bees. Annu. Rev. Entomol. 2008;53:191–208. doi: 10.1146/annurev.ento.53.103106.093454. [DOI] [PubMed] [Google Scholar]
- 49.Cameron SA, et al. Patterns of widespread decline in North American bumble bees. Proc. Natl. Acad. Sci. U. S. A. 2011;108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arbetman MP, Gleiser G, Morales CL, Williams P, Aizen MA. Global decline of bumblebees is phylogenetically structured and inversely related to species range size and pathogen incidence. Proc. R. Soc. B. 2017;284:20170204. doi: 10.1098/rspb.2017.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson HM, et al. Climate change winners and losers among North American bumblebees. Biol. Lett. 2022;18:20210551. doi: 10.1098/rsbl.2021.0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lozier JD, Strange JP, Stewart IJ, Cameron SA. Patterns of range-wide genetic variation in six North American bumble bee (Apidae: Bombus) species. Mol. Ecol. 2011;20:4870–4888. doi: 10.1111/j.1365-294X.2011.05314.x. [DOI] [PubMed] [Google Scholar]
- 53.Lozier JD, et al. Divergence in body mass, wing loading, and population structure reveals species-specific and potentially adaptive trait variation across elevations in Montane bumble bees. Insect Syst. Divers. 2021;5:3. [Google Scholar]
- 54.Strange JP. Bombus huntii, Bombus impatiens, and Bombus vosnesenskii (Hymenoptera: Apidae) Pollinate Greenhouse-Grown Tomatoes in Western North America. J. Econ. Entomol. 2015;108:873–879. doi: 10.1093/jee/tov078. [DOI] [PubMed] [Google Scholar]
- 55.Jackson JM, et al. Local adaptation across a complex bioclimatic landscape in two montane bumble bee species. Mol. Ecol. 2020;29:920–939. doi: 10.1111/mec.15376. [DOI] [PubMed] [Google Scholar]
- 56.Pimsler ML, et al. Biogeographic parallels in thermal tolerance and gene expression variation under temperature stress in a widespread bumble bee. Sci. Rep. 2020;10:17063. doi: 10.1038/s41598-020-73391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. [Google Scholar]
- 58.Marshall H, Jones ARC, Lonsdale ZN, Mallon EB. Bumblebee workers show differences in allele-specific DNA methylation and allele-specific expression. Genome Biol. Evol. 2020;12:1471–1481. doi: 10.1093/gbe/evaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marshall H, et al. DNA methylation is associated with codon degeneracy in a species of bumblebee. Heredity. 2023;130:188–195. doi: 10.1038/s41437-023-00591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall H, Lonsdale ZN, Mallon EB. Methylation and gene expression differences between reproductive and sterile bumblebee workers. Evol. Lett. 2019;3:485–499. doi: 10.1002/evl3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pozo MI, et al. The effect of DNA methylation on bumblebee colony development. BMC Genomics. 2021;22:73. doi: 10.1186/s12864-021-07371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadd BM, et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCaw BA, Stevenson TJ, Lancaster LT. Epigenetic responses to temperature and climate. Integr. Comp. Biol. 2020;60:1469–1480. doi: 10.1093/icb/icaa049. [DOI] [PubMed] [Google Scholar]
- 64.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 65.Heraghty SD, et al. De novo genome assemblies for three North American bumble bee species: Bombus bifarius, Bombus vancouverensis, and Bombus vosnesenskii. G3 Genes|Genomes|Genet. 2020;10:2585–2592. doi: 10.1534/g3.120.401437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun C, et al. Genus-wide characterization of bumblebee genomes provides insights into their evolution and variation in ecological and behavioral traits. Mol. Biol. Evol. 2021;38:486–501. doi: 10.1093/molbev/msaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paria SS, Rahman SR, Adhikari K. fastman: A fast algorithm for visualizing GWAS results using Manhattan and Q-Q plots. bioRxiv. 2022 doi: 10.1101/2022.04.19.488738. [DOI] [Google Scholar]
- 68.Jeong H, Wu X, Smith B, Yi SV. Genomic landscape of methylation islands in hymenopteran insects. Genome Biol. Evol. 2018;10:2766–2776. doi: 10.1093/gbe/evy203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, et al. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 2013;9:e1003872. doi: 10.1371/journal.pgen.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonasio R, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kvist J, et al. Pattern of DNA methylation in Daphnia: Evolutionary perspective. Genome Biol. Evol. 2018;10:1988–2007. doi: 10.1093/gbe/evy155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarda S, Zeng J, Hunt BG, Yi SV. The evolution of invertebrate gene body methylation. Mol. Biol. Evol. 2012;29:1907–1916. doi: 10.1093/molbev/mss062. [DOI] [PubMed] [Google Scholar]
- 74.Glastad KM, Gokhale K, Liebig J, Goodisman MAD. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci. Rep. 2016;6:37110. doi: 10.1038/srep37110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bewick AJ, et al. Dnmt1 is essential for egg production and embryo viability in the large milkweed bug, Oncopeltus fasciatus. Epigenetics Chromatin. 2019;12:6. doi: 10.1186/s13072-018-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hearn J, Plenderleith F, Little TJ. DNA methylation differs extensively between strains of the same geographical origin and changes with age in Daphnia magna. Epigenetics Chromatin. 2021;14:4. doi: 10.1186/s13072-020-00379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flores K, et al. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics. 2012;13:480. doi: 10.1186/1471-2164-13-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glastad KM, Hunt BG, Goodisman MAD. Epigenetics in insects: Genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 2019;64:185–203. doi: 10.1146/annurev-ento-011118-111914. [DOI] [PubMed] [Google Scholar]
- 79.Ventós-Alfonso A, Ylla G, Montañes J-C, Belles X. DNMT1 promotes genome methylation and early embryo development in cockroaches. iScience. 2020;23:101778. doi: 10.1016/j.isci.2020.101778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wurm Y, et al. The genome of the fire ant Solenopsis invicta. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5679–5684. doi: 10.1073/pnas.1009690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones CM, Lim KS, Chapman JW, Bass C. Genome-wide characterization of DNA methylation in an invasive lepidopteran pest, the cotton bollworm Helicoverpa armigera. G3 Genes|Genomes|Genetics. 2018;8:779–787. doi: 10.1534/g3.117.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glastad KM, Hunt BG, Goodisman MAD. DNA methylation and chromatin organization in insects: Insights from the Ant Camponotus floridanus. Genome Biol. Evol. 2015;7:931–942. doi: 10.1093/gbe/evv039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richard G, Jaquiéry J, Le Trionnaire G. Contribution of epigenetic mechanisms in the regulation of environmentally-induced polyphenism in insects. Insects. 2021;12:649. doi: 10.3390/insects12070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hunt BG, Glastad KM, Yi SV, Goodisman MAD. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol. Evol. 2013;5:591–598. doi: 10.1093/gbe/evt030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nwanade CF, et al. DNA methylation variation is a possible mechanism in the response of Haemaphysalis longicornis to low-temperature stress. Int. J. Mol. Sci. 2022;23:15207. doi: 10.3390/ijms232315207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Villalba de la Peña M, Piskobulu V, Murgatroyd C, Hager R. DNA methylation patterns respond to thermal stress in the viviparous cockroach Diploptera punctata. Epigenetics. 2021;16:313–326. doi: 10.1080/15592294.2020.1795603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Asselman J, De Coninck DIM, Pfrender ME, De Schamphelaere KAC. Gene body methylation patterns in Daphnia are associated with gene family size. Genome Biol. Evol. 2016;8:1185–1196. doi: 10.1093/gbe/evw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen P, et al. Comparative genome-wide DNA methylation analysis reveals epigenomic differences in response to heat-humidity stress in Bombyx mori. Int. J. Biol. Macromol. 2020;164:3771–3779. doi: 10.1016/j.ijbiomac.2020.08.251. [DOI] [PubMed] [Google Scholar]
- 90.Feiner N, et al. Environmentally induced DNA methylation is inherited across generations in an aquatic keystone species. iScience. 2022;25:104303. doi: 10.1016/j.isci.2022.104303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sørensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003;6:1025–1037. [Google Scholar]
- 92.Sørensen JG. Application of heat shock protein expression for detecting natural adaptation and exposure to stress in natural populations. Curr. Zool. 2010;56:703–713. [Google Scholar]
- 93.Chen S, Bawa D, Besshoh S, Gurd JW, Brown IR. Association of heat shock proteins and neuronal membrane components with lipid rafts from the rat brain. J. Neurosci. Res. 2005;81:522–529. doi: 10.1002/jnr.20575. [DOI] [PubMed] [Google Scholar]
- 94.Andersen JL, MacMillan HA, Overgaard J. Muscle membrane potential and insect chill coma. J. Exp. Biol. 2015;218:2492–2495. doi: 10.1242/jeb.123760. [DOI] [PubMed] [Google Scholar]
- 95.Overgaard J, MacMillan HA. The integrative physiology of insect chill tolerance. Annu. Rev. Physiol. 2017;79:187–208. doi: 10.1146/annurev-physiol-022516-034142. [DOI] [PubMed] [Google Scholar]
- 96.Robertson RM, Spong KE, Srithiphaphirom P. Chill coma in the locust, Locusta migratoria, is initiated by spreading depolarization in the central nervous system. Sci. Rep. 2017;7:10297. doi: 10.1038/s41598-017-10586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson KL, Tohidi-Esfahani D, Lo N, Simpson SJ, Sword GA. Evidence for widespread genomic methylation in the migratory locust, Locusta migratoria (Orthoptera: Acrididae) PLoS ONE. 2011;6:e28167. doi: 10.1371/journal.pone.0028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mashoodh R, Sarkies P, Westoby J, Kilner RM. Evolved changes in DNA methylation in response to the sustained loss of parental care in the burying beetle. bioRxiv. 2021 doi: 10.1101/2021.03.25.436923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu X, et al. Sex-specific transcription and DNA methylation landscapes of the Asian citrus psyllid, a vector of huanglongbing pathogens. Evolution. 2023;77:1203–1215. doi: 10.1093/evolut/qpad036. [DOI] [PubMed] [Google Scholar]
- 100.Cunningham CB, et al. Changes of gene expression but not cytosine methylation are associated with male parental care reflecting behavioural state, social context and individual flexibility. J. Exp. Biol. 2019;222:jeb188649. doi: 10.1242/jeb.188649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morandin C, Brendel VP, Sundström L, Helanterä H, Mikheyev AS. Changes in gene DNA methylation and expression networks accompany caste specialization and age-related physiological changes in a social insect. Mol. Ecol. 2019;28:1975–1993. doi: 10.1111/mec.15062. [DOI] [PubMed] [Google Scholar]
- 102.Bebane PSA, et al. The effects of the neonicotinoid imidacloprid on gene expression and DNA methylation in the buff-tailed bumblebee Bombus terrestris. Proc. Biol. Sci. 2019;286:20190718. doi: 10.1098/rspb.2019.0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cardoso-Júnior CAM, et al. DNA methylation is not a driver of gene expression reprogramming in young honey bee workers. Mol. Ecol. 2021;30:4804–4818. doi: 10.1111/mec.16098. [DOI] [PubMed] [Google Scholar]
- 104.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Drewell RA, et al. The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development. 2014;141:2702–2711. doi: 10.1242/dev.110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lamka GF, et al. Epigenetics in ecology, evolution, and conservation. Front. Ecol. Evol. 2022;10:871791. [Google Scholar]
- 107.Tsai P-C, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol. 2015;44:1429–1441. doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Flores KB, Wolschin F, Amdam GV. The role of methylation of DNA in environmental adaptation. Integr. Comp. Biol. 2013;53:359–372. doi: 10.1093/icb/ict019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lozier JD, Zayed A. Bee conservation in the age of genomics. Conserv. Genet. 2017;18:713–729. [Google Scholar]
- 110.Andrews, S. FastQC: a quality control tool for high throughput sequence data. Babraham Institute Webpage. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 111.Krueger, F. Trim Galore!: A wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data. GitHub Repository.https://github.com/FelixKrueger/TrimGalore (2015).
- 112.Shen W, Le S, Li Y, Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pedersen, B. S., Eyring, K., De, S., Yang, I. V. & Schwartz, D. A. Fast and accurate alignment of long bisulfite-seq reads. arXiv. https://arxiv.org/abs/1401.1129 (2014).
- 114.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Broad Institute. Picard Toolkit. GitHub Repository.https://github.com/broadinstitute/picard (2016).
- 116.Ryan, D. MethylDackel: A (mostly) universal methylation extractor for BS-seq experiments. GitHub Repository.https://github.com/dpryan79/MethylDackel (2021).
- 117.R Core Team. R: A language and environment for statistical computing. The R Project for Statistical Computinghttps://www.R-project.org (2021).
- 118.Akalin A, et al. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012;13:R61. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Venkataraman YR, White SJ, Roberts SB. Differential DNA methylation in Pacific oyster reproductive tissue in response to ocean acidification. BMC Genomics. 2022;23:556. doi: 10.1186/s12864-022-08781-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dainat, J. AGAT: AGAT: Another Gff Analysis Toolkit to handle annotations in any GTF/GFF format (v.0.8.0). GitHub Repository.https://github.com/NBISweden/AGAT/tree/v0.8.0 (2021).
- 122.Bengtsson, H. et al. matrixStats: Functions that Apply to Rows and Columns of Matrices (and to Vectors). GitHub Repository.https://github.com/HenrikBengtsson/matrixStats (2022).
- 123.McCullagh, P. & Nelder, J. A. Generalized Linear Models, Second Edition. (CRC Press, 1989).
- 124.Bushnell, B. BBMap: short read aligner, and other bioinformatic tools. SourceForge Webpage. https://sourceforge.net/projects/bbmap/ (2014).
- 125.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. arXivhttps://arxiv.org/abs/1207.3907 (2012).
- 127.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ghisbain G, et al. Substantial genetic divergence and lack of recent gene flow support cryptic speciation in a colour polymorphic bumble bee (Bombus bifarius) species complex. Syst. Entomol. 2020;45:635–652. [Google Scholar]
- 129.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinform. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Korneliussen TS, Moltke I, Albrechtsen A, Nielsen R. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinform. 2013;14:289. doi: 10.1186/1471-2105-14-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLoS ONE. 2012;7:e37558. doi: 10.1371/journal.pone.0037558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Canty, A. & Ripley, B. boot: Bootstrap R (S-Plus) functions. CRAN R Project.https://cran.r-project.org/web/packages/boot/index.html (2017).
- 134.Meisner J, Albrechtsen A. Inferring population structure and admixture proportions in low-depth NGS data. Genetics. 2018;210:719–731. doi: 10.1534/genetics.118.301336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brooks ME, et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 2017;9:378–400. [Google Scholar]
- 136.Grote S. GOfuncR: Gene ontology enrichment using FUNC. Bioconductor R Package. 2018 doi: 10.18129/B9.bioc.GOfuncR. [DOI] [Google Scholar]
- 137.Walsh AT, Triant DA, Le Tourneau JJ, Shamimuzzaman M, Elsik CG. Hymenoptera Genome Database: New genomes and annotation datasets for improved go enrichment and orthologue analyses. Nucleic Acids Res. 2022;50:D1032–D1039. doi: 10.1093/nar/gkab1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reijnders MJMF, Waterhouse RM. Summary visualizations of gene ontology terms with GO-Figure! Front. Bioinform. 2021;1:638255. doi: 10.3389/fbinf.2021.638255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw WGBS reads generated in this study has been deposited and is currently available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under NCBI BioProject PRJNA956115. Final methylation call set (n = 14,627,533), variant calling file for population genomics analyses, analysis codes/scripts and other associated files to reproduce the research in available from Zenodo data repository (https://doi.org/10.5281/zenodo.8327218).