Abstract
Quantitative PCR (qPCR) has become a widely used technique for bacterial quantification. The affordability, ease of experimental design, reproducibility, and robustness of qPCR experiments contribute to its success. The establishment of guidelines for minimum information for publication of qPCR experiments, now more than 10 years ago, aimed to mitigate the publication of contradictory data. Unfortunately, there are still a significant number of recent research articles that do not consider the main pitfalls of qPCR for quantification of biological samples, which undoubtedly leads to biased experimental conclusions. qPCR experiments have two main issues that need to be properly tackled: those related to the extraction and purification of genomic DNA and those related to the thermal amplification process. This mini-review provides an updated literature survey that critically analyzes the following key aspects of bacterial quantification by qPCR: (i) the normalization of qPCR results by using exogenous controls, (ii) the construction of adequate calibration curves, and (iii) the determination of qPCR reaction efficiency. It is primarily focused on original papers published last year, where qPCR was applied to quantify bacterial species in different types of biological samples, including multi-species biofilms, human fluids, and water and soil samples.
Keywords: Bacterial load quantification, gDNA extraction efficiency, qPCR, gDNA yield, Exogenous control, Calibration curve, qPCR reaction efficiency
Introduction
Quantitative PCR (qPCR) has become a widely used technique for gene expression assessment, as well as for bacterial quantification (Kralik and Ricchi, 2017). The affordability, ease of experimental design, reproducibility, and robustness of qPCR experiments (Smith and Osborn, 2009) contribute to its success. In fact, qPCR is used in many other research fields beyond bacteriology and is even considered the gold standard in many applications (Pfaffl, 2010). While qPCR can be used to quantify DNA, RNA and even proteins, in this mini-review, the focus will be given to bacterial quantification in biological samples through genomic DNA (gDNA) amplification and quantification. Despite its success and technological potential, qPCR is not without its caveats, and the MIQE guidelines (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) were pivotal to improve reproducibility, data analysis, interpretation, and overall transparency of qPCR experimental reports (Bustin et al., 2009). As we will show in this mini-review, there are still a significant number of research articles that do not consider the main pitfalls of qPCR for bacterial quantification, which undoubtedly leads to biased conclusions (Bustin, 2010). The main objective of this mini-review is to perform an updated survey of recent literature to critically analyze how researchers conduct and report qPCR experiments for the quantification of bacteria in biological samples.
The general concept of qPCR beyond bacterial load quantification
When taking into consideration all required controls, the high reproducibility of a qPCR run, associated with a very low limit of detection, allows the accurate quantification of bacterial gDNA present in the initial biological sample (O’Connell et al., 2017a). As depicted in Fig. 1A, by performing a calibration curve relating initial bacterial concentration (before gDNA extraction) with the detection cycle threshold of a specific gene marker (present in the processed gDNA sample), qPCR data interpretation allows accurate determination of the initial bacterial load. However, the operative sentence here is “all the required controls.”
Fig. 1.
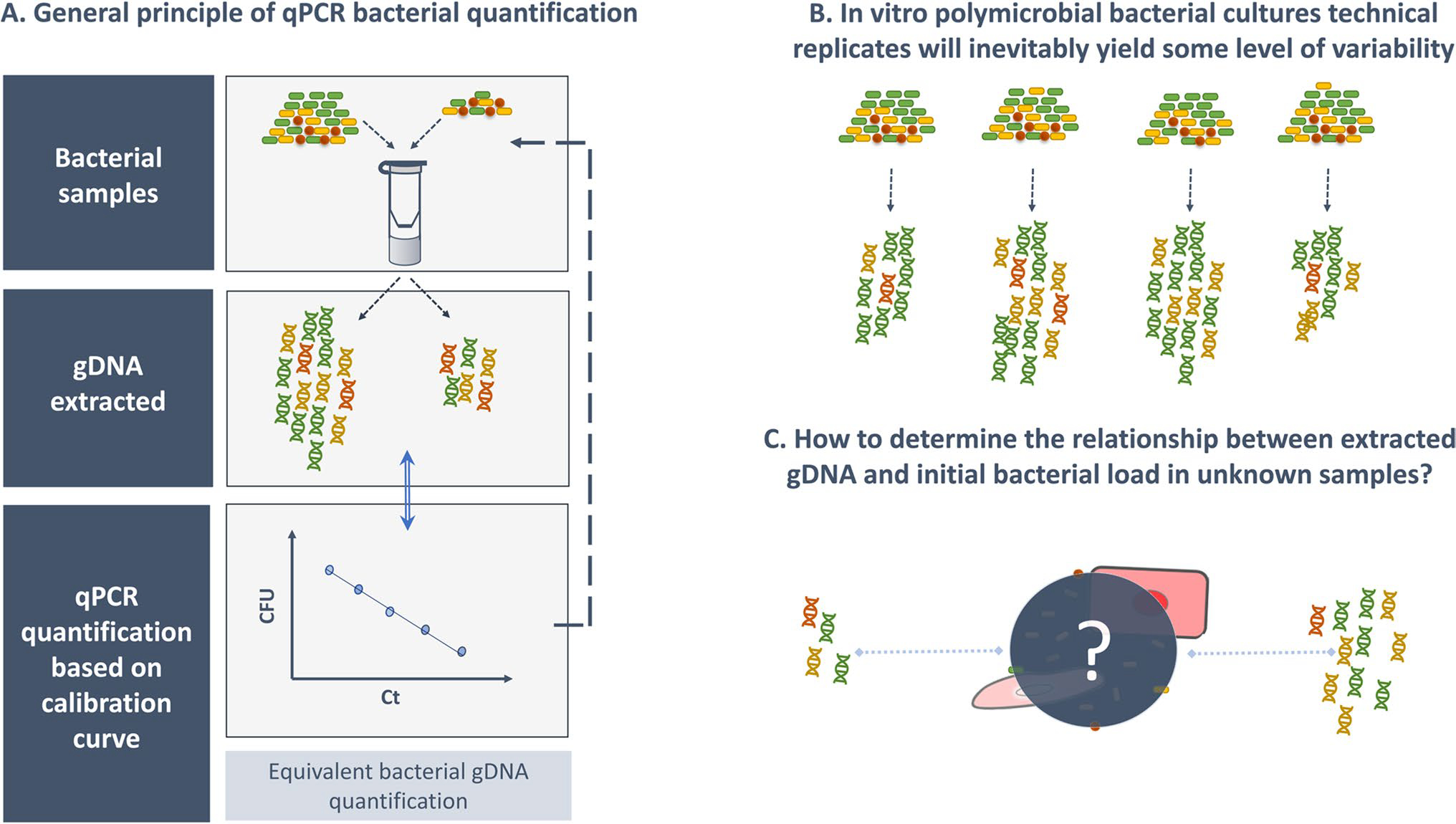
General concept and pitfalls behind bacterial load quantification by qPCR. Bacterial quantification by qPCR requires the preparation of a calibration curve, which includes gDNA isolated from samples with different concentrations of the bacterial species under study (A). However, due to the variable efficiency of the gDNA extraction procedure, each extraction, even from technical replicates, has inherent variability that can lead to biased bacterial load quantification (B). When the starting bacterial amount is known, gDNA loss, due to technical issues, can be easily detected. However, when the initial quantity of bacteria is unknown, it cannot be differentiated whether the variation detected was introduced by technical issues or if it was due to the initial bacterial load (C). As such, the addition of an exogenous control is imperative
As detailed in the MIQE guidelines (Bustin et al., 2009), qPCR experiments have two main issues that need to be properly addressed: those related to the extraction and purification of gDNA (McKee et al., 2015) and those related to the thermal amplification process (Ruiz-Villalba et al., 2017). While a lot of attention has been given to the latter, gDNA extraction troubleshooting is often oversimplified, mainly focusing on the gDNA yield and quality. However, it has been demonstrated by many studies that gDNA extraction efficiencies can vary significantly between experiments (Greathouse et al., 2019; Marotz et al., 2017; O’Connell et al., 2017b; Rezzonico et al., 2003). This can lead to biased quantification, as we have recently shown (Cerca et al., 2022). In addition to the inherent biological variability that occurs when growing in vitro cultures or collecting in vivo specimens, sample processing for qPCR experiments involves several steps, all of which can significantly contribute to variability (Caporaso et al., 2012; Greathouse et al., 2019; Sousa et al., 2014). It has been pointed out that the nucleic acid extraction step is the most important source of post-processing variability (Costea et al., 2017) as variation in the efficiency of this process will be further amplified during subsequent qPCR steps.
On one hand, if using in vitro technical replicates, with known bacterial concentrations, the variability in the gDNA extraction process can easily be observed by the differences in the total yield of gDNA obtained, as depicted in Fig. 1B. For example, after repeated extractions, if the user can anticipate a 1 × 108 CFU/mL bacterial pure culture yielding ~ 100 ng/μL of total DNA, then the user might exclude for downstream processing one extraction where only 5 ng/μL of total DNA was obtained and attribute this variation to some experimental flaw. However, if gDNA is extracted from an uncharacterized sample, which is often the goal of a qPCR experiment, DNA yield alone will not allow correct assessment of the initial bacterial load. Without the introduction of key experimental controls, it will not be possible to differentiate between a lower gDNA extraction efficiency and a lower initial bacterial load, since the same sample can yield two different gDNA concentrations, as depicted in Fig. 1C. This is even more relevant if human DNA is added to the mixture (Greathouse et al., 2019), as total DNA quantification will not allow inference of bacterial gDNA extraction yield. To tackle this issue, the addition of an exogenous DNA control before DNA extraction procedures, at a known concentration, has been identified as the best practice to normalize the inherent variations associated with gDNA extraction between samples (O’Connell et al., 2017a).
Impact of not assessing gDNA loss in bacterial quantification
Using a mock in vitro triple-species bacterial consortium, we recently demonstrated that the impact of not quantifying gDNA extraction efficiency in each reaction can significantly impact bacterial quantification (Cerca et al., 2022). As shown by others (Barton et al., 2006; Davis et al., 2019; Greathouse et al., 2019), at lower bacterial concentrations, the quantification error was significantly higher. Without considering gDNA extraction efficiency, we could find up to 46-fold under-representation of a particular species in the triple-species consortia. At higher bacterial concentrations, the quantification error was significantly lower, with no more than sixfold under-representation observed (Cerca et al., 2022). However, when we factored in the efficiency of gDNA extraction of each sample, as measured by the recovery rate of the exogenous control added before extraction, we were able to accurately (with less than 10% error) quantify each triple-species consortium with ~ 3 × 108 CFU/mL total bacterial load. When testing a consortium with only ~ 3 × 106 CFU/mL of total bacterial load, assessing gDNA extraction efficiency allowed no more than twofold under- or threefold over-representation of each species, which was significantly more accurate than the calculations excluding gDNA losses. Interestingly, different bacterial species incurred distinct gDNA losses.
Why determining qPCR reaction efficiency is not enough for accurate bacterial quantification?
One of the most basic rules of any qPCR experiment is the need to determine the qPCR reaction efficiency (Bustin et al., 2009). This is achieved by performing serial dilutions of a specific DNA sample (Svec et al., 2015). However, this is often mistakenly considered as a calibration curve for bacterial concentration determination. It is incorrectly assumed that the same linear response observed in the qPCR reaction efficiency mimics a linear calibration curve for total bacterial load and qPCR cycle threshold of a specific bacterial species. As depicted in Fig. 1A, the relationship between a known DNA concentration and qPCR cycle threshold is only able to accurately assess the initial concentration of the DNA present in the processed sample, but this is hardly the same as assessing the initial bacterial concentration of the unprocessed sample (before gDNA extraction) to be analyzed. So, while determining qPCR reaction efficiency is mandatory for accurate bacterial quantification, as described in the MIQE guidelines, this does not replace the need to perform a proper bacterial gDNA extraction calibration curve.
In this regard, Longin and co-workers pointed out important recommendations to perform accurate bacteria quantification using qPCR (Longin et al., 2016). One of the recommendations included (i) the use of a bacteria as a spike in control added to the unprocessed sample before starting the DNA extraction. There are already a few studies that have successfully used this strategy as exogenous controls (Scarsella et al., 2021; Stoeckel et al., 2009). In these studies, bacteria were incorporated into the culture right before the centrifugation step to account for biomass losses during centrifugation. It is important to highlight that the selected exogenous bacterial species should be absent from the target samples under study. In most cases, this is possible to predict. However, if not, an alternative strategy could be the utilization of a genetically manipulated strain containing a synthetic sequence. Furthermore, attention should be given to differences in cell wall composition, since Gram-positive and Gram-negative bacteria can have different lysis efficiency (Ketchum et al., 2018; Mahalanabis et al., 2009; Wang et al., 2020), mainly due to a thicker peptidoglycan layer in Gram-positive bacteria (Auer and Weibel, 2017). Furthermore, when studying complex samples that might include both Gram-positive and Gram-negative bacteria, a mixture of Gram-positive and Gram-negative exogenous bacterial controls should be used, as recently proposed (Scarsella et al., 2021).
Other important recommendations indicated by Login et al. was (ii) the use of calibration curves constructed with gDNA isolated form pure cultures prepared at different bacterial concentrations, as gDNA extraction efficiency depends on initial bacterial concentration. In addition, (iii) dilution of pure cultures to construct calibration curves should include the biological matrix of the samples.
Is bacterial quantification by qPCR being properly performed and reported? A recent literature review
To perform this review, we focused on the analysis of the Materials and Methods sections of original papers published last year, from a wide range of research fields, by performing a PUBMED search with the key words “bacterial qPCR quantification,” “qPCR exogenous control,” and “bacterial qPCR normalization”. We found 95 research papers (Table 1). We centered our analyses on (i) the normalization of qPCR results by using exogenous controls, (ii) the construction of adequate calibration curves, and (iii) the determination of reaction efficiency, which are key steps for both DNA extraction and thermal amplification normalization steps. Surprisingly, (82%) of the surveyed manuscripts did not include a normalization strategy to account for bacterial loss and to tackle gDNA extraction variability (Fig. 2). Among the studies where controls were included (8%), only 1% used proper exogenous controls, and the remaining 9% performed a normalization of gDNA concentration among samples. Regarding reaction efficiency, 53% of the studies did not consider this critical aspect, 25% did not determine the efficiency correctly, and thus, only 22% of the reports determined reaction efficiency properly. In the last aspect that we analyzed, the construction of calibration curves, we verified that only 4% of the studies performed proper calibration curves. In 45% of the studies analyzed, the calibration curves were constructed using dilutions of gDNA obtained from a single sample, a strategy routinely used to determine primer efficiency, instead of using gDNA obtained from samples with different bacteria concentrations, as discussed above. A large number (37%) of studies constructed calibration curves by cloning a gene into a plasmid, in order to determine absolute copy numbers. However, again, the calibration curve was constructed by performing dilutions of the same DNA sample. The most common errors found for each critical aspect of the qPCR are described in Fig. 3. It is important to highlight that none of the studies analyzed performed well in the three critical aspects evaluated. In this sense, we constructed a flowchart, Fig. 4, that summarizes the critical steps for absolute bacterial quantification by qPCR.
Table 1.
Literature survey of the three critical aspects for proper bacterial quantification by qPCR. The papers analyzed were all published in 2021
| Scientific field | Purpose | Normalization strategy # | Calibration curve ### | Reaction efficiency ## | Reference |
|---|---|---|---|---|---|
|
| |||||
| Vaginal infections | To quantify G. vaginalis before and after exposure to Lactobacillus spp. | F | 2 | vi) | (He et al., 2021a, b) |
| To quantify Candida spp. and bacterial load in vaginal swabs | F | 2 | vi) | (McKloud et al., 2021) | |
| To quantify Gardnerella spp., Lactobacillus spp. and total bacterial population in vaginal swabs | E | 2 | i) | (Turner et al., 2021) | |
| To quantify the bacterial load and G. vaginalis in cervical fluid | D | 3 | vi) | (Kacerovsky et al., 2021) | |
| To quantify G. vaginalis and Lactobacillus spp. in mouse vaginal tissue | B | 4 | iv) | (Selis et al., 2021) | |
| To quantify Enterobacteriaceae, Staphylococcus spp., and Streptococcus spp. in vaginal swabs | F | 2 | vi) | (Oh et al., 2021) | |
| Wound infections | To quantify different bacteria in a polymicrobial biofilm | D | 2 | vi) | (Li and Wu, 2021) |
| To quantify Staphylococcus aureus in mice tissues | F | 4 | vi) | (Do Pham et al., 2021) | |
| Gastrointestinal infections/microbiota | To quantify Clostridioides difficile in a polymicrobial biofilm | B | 3 | vi) | (Normington et al., 2021) |
| To quantify bacterial load and nine bacteria in feces samples | B | 3 | vi) | (Tonon et al., 2021) | |
| To quantify Prevotella histicola in human duodenal biopsies | F | 2 | i) | (Balakrishnan et al., 2021) | |
| To quantify bacterial load in cecum content and feces | F | 3 | vi) | (Taibi et al., 2021) | |
| To quantify enterotoxigenic E. coli F4 in colon and ileal mucosal samples of piglets | F | 2 | vi) | (Rodríguez-Sorrento et al., 2021) | |
| To quantify Bacteroides vulgatus in fecal samples | F | 3 | ii) | (Maier et al., 2021a, b) | |
| To quantify fungal load in stool samples | B | 2 | vi) | (Boutin et al., 2021) | |
| To quantify bacteria and fungi in honeybee gut samples | F | 3 | vi) | (Callegari et al., 2021) | |
| Pulmonary infections | To quantify Mycoplasma pneumoniae and Chlamydia pneumoniae in respiratory samples from cystic fibrosis patients | F | 3 | vi) | (Pittet et al., 2021) |
| To quantify three bacteria associated with ventilator-associated pneumonia in dual-species biofilms with Candida albicans | F | 2 | i) | (Luo et al., 2021) | |
| To quantify bacteria in bronchoalveolar fluid samples | F | 3 | vi) | (Invernizzi et al., 2021) | |
| Oral biofilms | To determine the composition of polymicrobial biofilms | F | 3 | vi) | (Redanz et al., 2021) |
| To determine and quantify bacteria in dental caries | F | 2 | vi) | (Chen et al., 2021) | |
| To quantify bacteria in a polymicrobial biofilm | F | 3 | vi) | (Verspecht et al., 2021a) | |
| To detect and quantify bacteria in polymicrobial biofilms from saliva and dental plaque | B | 4 | vi) | (Oliveira et al., 2021) | |
| To quantify pathogenic bacteria in subgingival plaque and saliva samples | F | 2 | vi) | (Sereti et al., 2021) | |
| To quantify bacteria and fungi in a tooth model | F | 4 | i) | (Leelapornpisid et al., 2021) | |
| To quantify bacteria in a dual-species biofilm | B | 5 | vi) | (Millones-Gómez et al., 2021) | |
| To quantify bacteria in a polymicrobial biofilm | F | 3 | vi) | (Verspecht et al., 2021b) | |
| To quantify bacteria in a multi-species community and bacteria present in ligatures placed around teeth of mice | C | 2 | vi) | (Hoare et al., 2021) | |
| To quantify bacteria in a polymicrobial biofilm | F | 2 | vi) | (Chathoth et al., 2021) | |
| To quantify Porphyromonas gengivalis in oral samples | F | 2 | vi) | (Franciotti et al., 2021) | |
| To quantify two bacteria in oral samples and colon tissue | F | 2 | vi) | (Pignatelli et al., 2021) | |
| To quantify microbial load in saliva samples | F | 2 | vi) | (Marotz et al., 2021) | |
| To quantify four bacteria in subgingival samples | F | 2 | v) | (Cuenca et al., 2021) | |
| To quantify bacterial load, and two different bacteria in periapical tissue | F | 2 | vi) | (Bordagaray et al., 2021) | |
| To quantify Streptococcus mutans and Candida albicans in oral samples | F | 2 | vi) | (Yang et al., 2022) | |
| Soil microbiota | To quantify bacterial and fungal load and Fusarium oxisporum in soil samples | F | 6 | v) | (Zhu et al., 2021a, b) |
| To quantify bacterial and fungal load | F | 2 | i) | (Ammitzboll et al., 2021) | |
| To quantify ammonia-oxidizing archaea and bacteria in soil samples | F | 3 | v) | (Dai et al., 2021) | |
| To quantify Streptomyces bottropensis and Brevibacillus laterosporus in soil samples | F | 3 | v) | (Li et al., 2021a, b, c) | |
| To quantify bacterial load in wheat root samples | F | 3 | v) | (Usyskin-Tonne et al., 2021) | |
| To quantify ammonia-oxidizing archaea and bacteria in soil samples | F | 3 | v) | (Samaddar et al., 2021) | |
| To quantify bacterial and fungal load in soybean soil samples | F | 3 | v) | (Gao et al., 2021a, b) | |
| To quantify Calonectria ilicicola in soybean soil samples | F | 2 | vi) | (Ochi and Kuroda, 2021) | |
| To quantify ammonia-oxidizing archaea and bacteria in soil samples | F | 3 | vi) | (He et al., 2021a, b) | |
| To quantify F. oxysporum, bacterial and fungal load, ammonia-oxidizing archaea and bacteria in soil samples | F | 3 | ii) | (Liu and Zhang, 2021) | |
| To quantify bacterial load, ammonia-oxidizing archaea and bacteria in soil samples | F | 3 | vi) | (Li et al., 2021a, b, c) | |
| To quantify bacterial load in soil samples | F | 3 | vi) | (Nogrado et al., 2021) | |
| To quantify bacterial load in soil samples | F | 2 | v) | (Han et al., 2021) | |
| To quantify bacterial load in dry soil samples | F | 3 | ii) | (Zhao et al., 2021) | |
| To quantify bacterial load in soil samples | F | 3 | ii) | (Wang et al., 2021a, b) | |
| To quantify bacterial load in soil samples | F | 3 | ii) | (Zhu et al., 2021a, b) | |
| To quantify bacterial load in soil samples | F | 3 | vi) | (Zhang et al., 2021a, b) | |
| To quantify Brevibacillus laterosporus and S. bottropensis in soil samples | F | 3 | v) | (Li et al., 2021a, b, c) | |
| To quantify ammonia-oxidizing archaea and bacteria in soil samples | F | 3 | vi) | (Wei et al., 2021) | |
| To quantify ammonia-oxidizing archaea, ammonia-oxidizing bacteria, Nitrobacter and Nitrospira spp. in soil samples | F | 3 | vi) | (Yin et al., 2021) | |
| To quantify Rhizoctonia solani and Rhizoctonia solani AG1-IB in lettuce and soil samples | D | 2 | i) | (Wallon et al., 2021) | |
| To quantify bacterial load in soil samples | B | 2 | vi) | (Alvarez et al., 2021) | |
| To quantify F. oxysporum f. sp. cubense, Fusobacterium solani and Aspergillus fumigatus in soil samples | F | 3 | vi) | (Yuan et al., 2021) | |
| To quantify bacterial and fungal load, Ralstonia solanacearum and F. oxysporum f. sp. Lycopersici in soil samples | F | 6 | vi) | (Deng et al., 2021) | |
| To quantify Azospirillum brazilense in soil samples | F | 1 | iii) | (Urrea-Valencia et al., 2021) | |
| To quantify Pseudomonas protegens and Bacillus subtilis in soil samples | F | 3 | ii) | (Zhang et al., 2021a, b) | |
| Bacterial contamination on surfaces | To quantify Lactobacillus delbrueckii subsp. Bulgaricus in surface samples | F | 2 | i) | (Wang et al., 2021a, b) |
| To quantify four Staphylococcus spp. in hospital surfaces | F | 2 | vi) | (Gismondi et al., 2021) | |
| To quantify Candida auris in skin swabs and hospital surfaces | F | 4 | vi) | (Sexton et al., 2021) | |
| Food quality control / Foodborne pathogens | To quantify three Lactobacillus spp. in sourdough bread | F | 2 | i) | (Baek et al., 2021) |
| To quantify Lactobacillus spp. in fermented dairy products samples | F | 2 | i) | (Yang et al., 2021) | |
| To quantify Brucella spp. in milk and cheese samples | F | 2 | vi) | (Marouf et al., 2021) | |
| To quantify seven proteolytic Pseudomonas spp. in raw milk samples | F | 1 | iii) | (Maier et al., 2021a, b) | |
| To quantify Nucleospora cyclopteri in fish and blood samples | F | 2 | i) | (Naung et al., 2021) | |
| To quantify different bacteria in meat samples | F | 2 | i) | (Bahlinger et al., 2021) | |
| To quantify Colletotrichum acutatum in olive fruit samples | F | 3 | v) | (Azevedo-Nogueira et al., 2021) | |
| To quantify Lysteria monocytogenes in meat samples | F | 1 | iii) | (Labrador et al., 2021) | |
| To quantify Shiga toxin-producing Escherichia coli in meat samples | F | 2 | vi) | (Rey et al., 2021) | |
| To quantify Campylobacter coli in meat samples | F | 2 | i) | (Lazou et al., 2021) | |
| To quantify Brochothrix thermosphacta in fish samples | F | 2 | i) | (Bouju-Albert et al., 2021) | |
| To quantify Streptococcus iniae in fish samples | F | 2 | i) | (Torres-Corral and Santos, 2021) | |
| To quantify Cronobacter sakazakii in spiked powdered infant formula samples | F | 2 | i) | (Gao et al., 2021a, b) | |
| To quantify four Campylobacter spp. in meat samples | F | 2 | i) | (Vizzini et al., 2021) | |
| To quantify Salmonella spp. in poultry floor dust | F | 1 | i) | (Ahaduzzaman et al., 2021) | |
| To quantify six bacteria genera and four bacteria phyla in milk samples | F | 2 | vi) | (Sanjulián et al., 2021) | |
| Waterborne pathogens | To quantify bacteria in polymicrobial biofilms present in a drinking water distribution system | C | 3 | i) | (Fu et al., 2021) |
| To quantify several enteric opportunistic pathogens in influent and recycled water | F | 2 | i) | (Drigo et al., 2021) | |
| To quantify Enterococcus spp. and Salmonella spp. in water, sand, and sediments samples | F | 3 | ii) | (Li et al., 2021a, b, c) | |
| To quantify Helicobacter pylori and Legionella spp. in filtered water samples | F | 5 | vi) | (Ribes et al., 2021) | |
| To quantify pathogenic fungi, enteric and opportunistic pathogens | F | 3 | ii) | (Hu et al., 2021) | |
| To quantify several bacteria in water samples | F | 1 | iii) | (Ambili and Sebastian, 2021) | |
| Other fields | To detect and quantify bacteria in breast implant samples | E | 2 | vi) | (Crowe et al., 2021) |
| To quantify bacteria in bovine digital dermatitis | B | 3 | v) | (Caddey et al., 2021) | |
| To quantify bacterial load in livestock fecal manure samples | F | 3 | vi) | (Wongsaroj et al., 2021) | |
| To quantify Zobellia genus in macroalgae surface | F | 2 | i) | (Brunet et al., 2021) | |
| To quantify Campylobacter fetus subsp. fetus and Salmonella enterica subsp. enterica serovar Typhimurium in bovine endometrial cells | F | 4 | vi) | (Muzquiz et al., 2021) | |
| To quantify five bacteria in cosmetic cream samples | F | 2 | i) | (Bermond et al., 2021) | |
| To quantify bacterial and fungal load in dust samples | F | 4 | vi) | (Haines et al., 2021) | |
| To quantify bacterial load in skin swabs, and skin samples from patients with atopic dermatitis | F | 2 | vi) | (Edslev et al., 2021) | |
| To quantify bacterial load in milk, feces and blood samples from healthy cows and cows suffering from bovine mastitis | A & B | 6 | vi) | (Scarsella et al., 2021) | |
Normalization strategy—A—An exogenous control was used; B- Initial gDNA concentration normalization among samples; C—gene copy numbers were normalized by the filtrate membrane surface area / ligature length; D-16 s rRNA gene (internal control) was used; E—an exogenous control was used only in the qPCR run. F- No normalization strategy was used
Calibration curve—1—Calibration curve performed using gDNA isolated from pure cultures with different initial concentrations, or pure cultures mixed with sample background of interest; 2—calibration curve performed with gDNA dilutions from one sample; 3—calibration curve performed by cloning a gene into a plasmid to determine absolute copy numbers by performing dilutions of the sample; 4-it is not clear whether the calibration curve was performed using dilutions of one extraction or different extractions from different bacterial concentrations; 5—standard curve was mentioned but it was not explained how it was constructed. 6 -No standard curve was mentioned
Reaction efficiency—i—gDNA from one sample; ii—dilution of one sample with a plasmid containing the gene of interest; iii—gDNA extracted from samples with different concentrations of bacteria; iv—it is not clear whether qPCR primer efficiency was determined by using dilutions of one gDNA sample or from gDNA samples isolated from samples with different initial bacteria concentration; v—considered but the procedure to determine the efficiency was not mentioned; vi—not mentioned
Fig. 2.
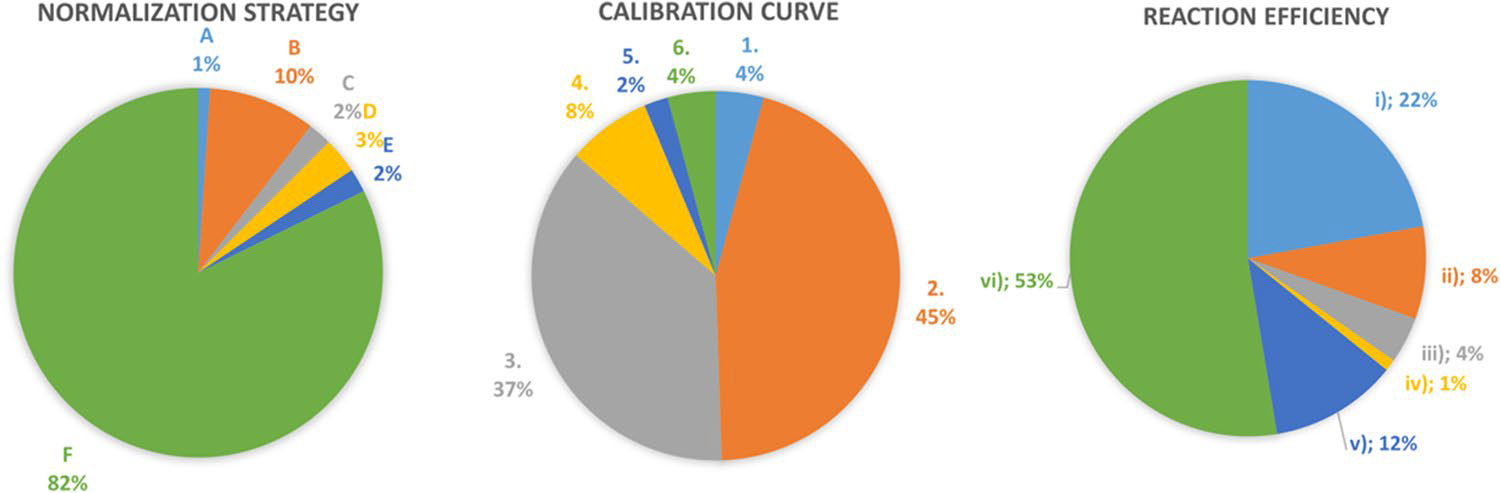
Graphical representation of the percentage (%) of studies presented in Table 1 that considered the three critical aspects for bacterial quantification by qPCR. The caption is the same as described in the Table 1 footer section
Fig. 3.
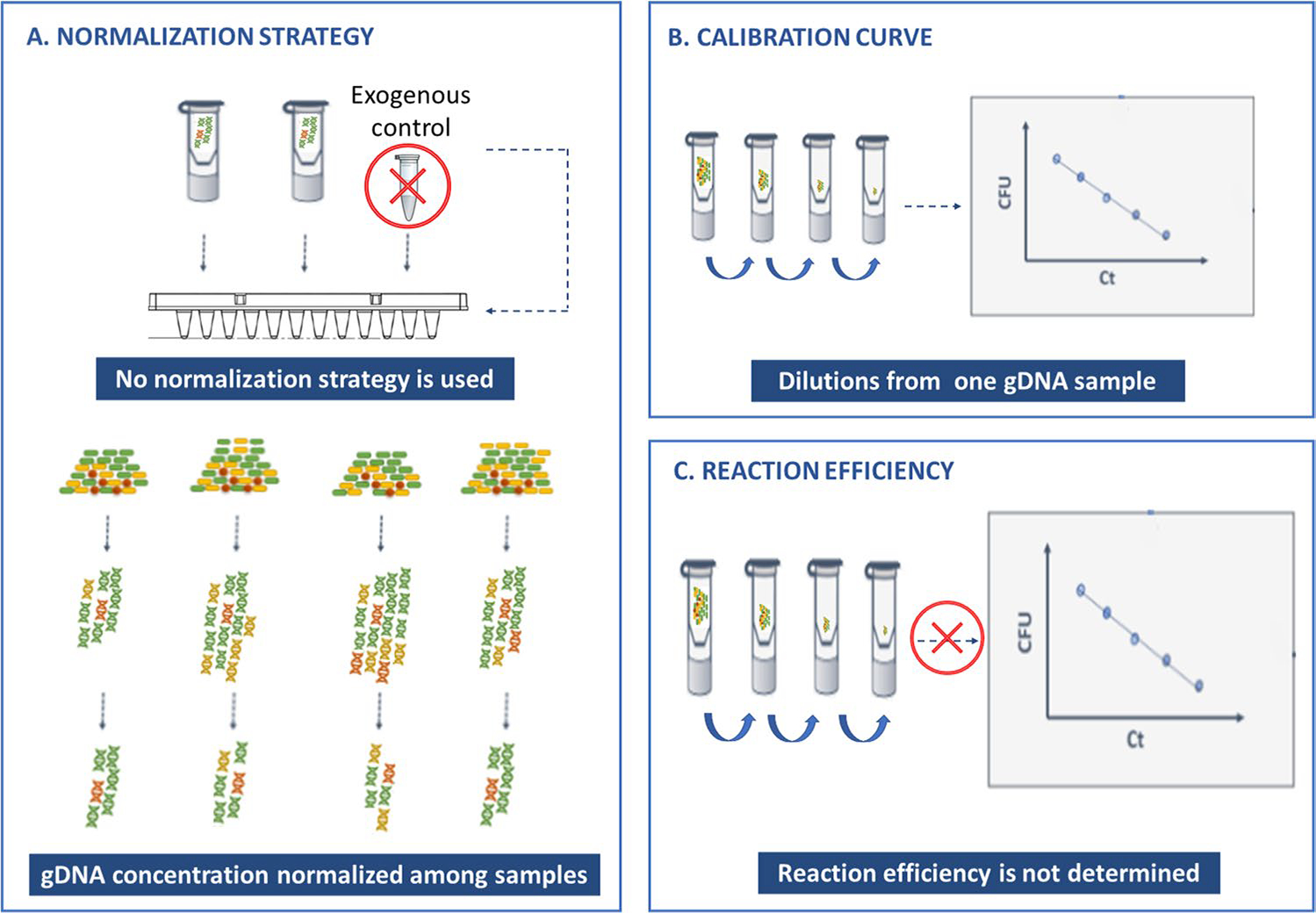
Common flaws when quantifying bacterial load using qPCR. In most of the studies surveyed, (A) DNA extraction normalization was not performed (or described) or was normalized by total gDNA concentration, which can bias the results. Often (B), the qPCR/bacterial load calibration curve is performed by diluting a known gDNA sample, but this fails to consider the different extraction efficiencies at different bacterial concentrations. Also (C) some studies failed to consider the qPCR reaction efficiency that needs to be determined for each primer set and varies according to reagent and equipment used
Fig. 4.
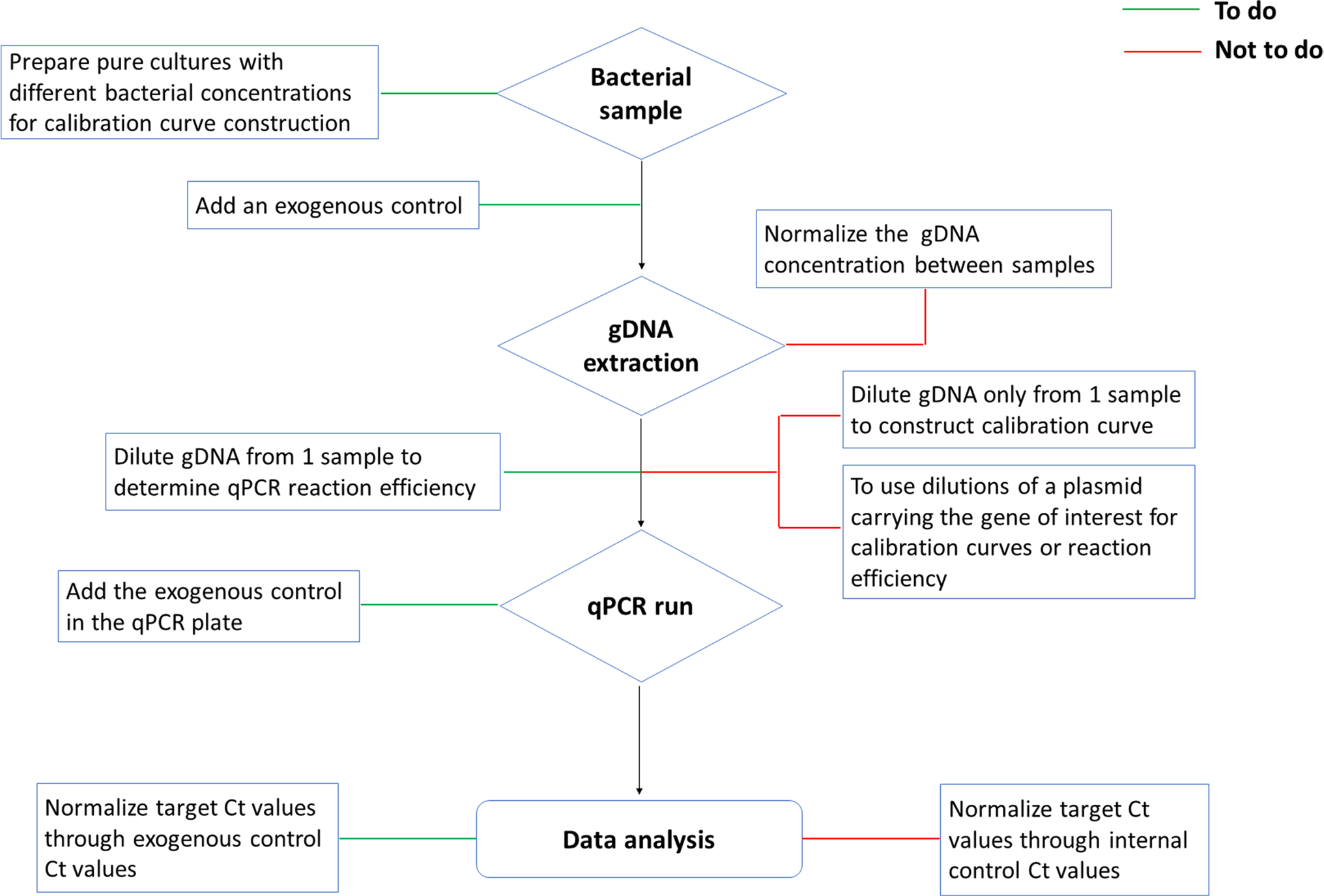
Flowchart highlighting the “do’s” and “don’ts” of procedures in bacterial quantification by qPCR. This scheme represents our recommended actions to perform a proper bacterial quantification by qPCR (green lines). It also includes common errors that are generally made (red lines) and should be avoided
Concluding remarks
Our review of recent literature clearly shows that the use of qPCR for bacteria quantification is not properly described, with a lack of clarity to what concerns the description on how quantification was performed. Although the publication of MIQE guidelines were pivotal to improve scientific literature on the use of qPCR for a multitude of applications, they have focused on RNA extraction experiments and fail to mention the need to include the utilization of an exogenous control to determine gDNA extraction efficiency. However, this is of utmost importance to guarantee proper bacterial quantification by gDNA amplification. The inadequate use of qPCR for bacterial quantification will bias the results obtained, leading to the publication of inconsistent data and, consequently, misleading and erroneous conclusions. As such, it is vital to alert the scientific community of the pitfalls associated with the quantification of bacteria by qPCR and to establish guidelines on how to proceed to properly address the limitations of these assays and, this way, obtain reproducible, reliable, and meaningful data.
Key points.
qPCR is a widely used technique used for absolute bacterial quantification.
Recently published papers lack proper qPCR methodologies.
Not including proper qPCR controls significantly affect experimental conclusions.
Acknowledgements
Christina A. Muzny, MD, MSPH, Christopher M. Taylor, PhD, Nuno Cerca, PhD and Ângela Lima, BSc, are currently funded by the National Institute of Allergy and Infectious Diseases (R01AI146065–01A1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation.
Footnotes
Conflict of interest The authors declare no competing interests.
Ethical approval This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Ahaduzzaman M, Groves PJ, Walkden-Brown SW, Gerber PF (2021) A molecular based method for rapid detection of Salmonella spp in poultry dust samples. MethodsX 8:101356. 10.1016/j.mex.2021.101356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D, Mendes KF, Tosi M, Fonseca de Souza L, Campos Cedano JC, de Souza Falcão NP, Dunfield K, Tsai SM, Tornisielo VL (2021) Sorption-desorption and biodegradation of sulfometuron-methyl and its effects on the bacterial communities in Amazonian soils amended with aged biochar. Ecotoxicol Environ Saf 207 10.1016/j.ecoenv.2020.111222 [DOI] [PubMed] [Google Scholar]
- Ambili A, Sebastian D (2021) Evaluation of sensitivity and cost-effectiveness of molecular methods for the co-detection of waterborne pathogens in India. Mar Biotechnol 23:955–963. 10.1007/s10126-021-10078-9 [DOI] [PubMed] [Google Scholar]
- Ammitzboll H, Jordan GJ, Baker SC, Freeman J, Bissett A (2021) Diversity and abundance of soil microbial communities decline, and community compositions change with severity of post-logging fire. Mol Ecol 30:2434–2448. 10.1111/mec.15900 [DOI] [PubMed] [Google Scholar]
- Auer G, Weibel D (2017) Bacterial cell mechanics. Biochem 56:3710–3724. 10.1021/acs.biochem.7b00346 (Bacterial) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo-Nogueira F, Gomes S, Lino A, Carvalho T, Martins-Lopes P (2021) Real-time PCR assay for Colletotrichum acutatum sensu stricto quantification in olive fruit samples. Food Chem 339:127858. 10.1016/j.foodchem.2020.127858 [DOI] [PubMed] [Google Scholar]
- Baek HW, Kim SA, Min WK, Kang SD, Shim S, Han NS, Seo JH (2021) A Species-Specific qPCR Method for Enumeration of Lactobacillus sanfranciscensis, Lactobacillus brevis, and Lactobacillus curvatus During Cocultivation in Sourdough. Food Anal Methods 14:750–760. 10.1007/s12161-020-01920-2 [DOI] [Google Scholar]
- Bahlinger E, Dorn-In S, Beindorf PM, Mang S, Kaltner F, Gottschalk C, Gareis M, Schwaiger K (2021) Development of two specific multiplex qPCRs to determine amounts of Pseudomonas, Enterobacteriaceae, Brochothrix thermosphacta and Staphylococcus in meat and heat-treated meat products. Int J Food Microbiol 337:108932. 10.1016/j.ijfoodmicro.2020.108932 [DOI] [PubMed] [Google Scholar]
- Balakrishnan B, Luckey D, Bodhke R, Chen J, Marietta E, Jeraldo P, Murray J, Taneja V (2021)Prevotellahisticola Protects From Arthritis by Expansion of Allobaculum and Augmenting Butyrate Production in Humanized Mice. Front Immunol 12:1–14. 10.3389/fimmu.2021.609644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton HA, Taylor NM, Lubbers BR, Pemberton AC (2006) DNA extraction from low-biomass carbonate rock: an improved method with reduced contamination and the low-biomass contaminant database. J Microbiol Methods 66:21–31. 10.1016/j.mimet.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Bermond C, Cherrad S, Trainoy A, Ngari C, Poulet V (2021) Real-time qPCR to evaluate bacterial contamination of cosmetic cream and the efficiency of protective ingredients. J Appl Microbiol 10.1111/jam.15310 [DOI] [PubMed] [Google Scholar]
- Bordagaray MJ, Fernández A, Garrido M, Astorga J, Hoare A, Hernández M (2021) Systemic and extraradicular bacterial translocation in apical periodontitis. Front Cell Infect Microbiol 11:1–9. 10.3389/fcimb.2021.649925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouju-Albert A, Saltaji S, Dousset X, Prévost H, Jaffrès E (2021) Quantification of viable Brochothrix thermosphacta in cold-smoked salmon using PMA/PMAxx-qPCR. Front Microbiol 12 10.3389/fmicb.2021.654178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin RCT, Sbihi H, McLaughlin RJ, Hahn AS, Konwar KM, Loo RS, Dai D, Petersen C, Brinkman FSL, Winsor GL, Sears MR, Moraes TJ, Becker AB, Azad MB, Mandhane PJ, Subbarao P, Turvey SE, Finlay BB (2021) Composition and associations of the infant gut fungal microbiota with environmental factors and childhood allergic outcomes. MBio 12 10.1128/mBio.03396-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet M, Le Duff N, Fuchs BM, Amann R, Barbeyron T, Thomas F (2021) Specific detection and quantification of the marine flavobacterial genus Zobellia on macroalgae using novel qPCR and CARD-FISH assays. Syst Appl Microbiol 44 10.1016/j.syapm.2021.126269 [DOI] [PubMed] [Google Scholar]
- Bustin SA (2010) Why the need for qPCR publication guidelines?— The case for MIQE. Methods 50:217–226. 10.1016/j.ymeth.2009.12.006 [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. 10.1373/CLINCHEM.2008.112797 [DOI] [PubMed] [Google Scholar]
- Caddey B, Orsel K, Naushad S, Derakhshani H, De Buck J (2021) Identification and quantification of bovine digital dermatitis-associated microbiota across lesion stages in feedlot beef cattle. mSystems 6:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari M, Crotti E, Fusi M, Marasco R, Gonella E, De Noni I, Romano D, Borin S, Tsiamis G, Cherif A, Alma A, Daffonchio D (2021) Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. npj Biofilms Microbiomes 7:42. 10.1038/s41522-021-00212-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerca N, Lima Â, França A (2022) Accurate qPCR quantification in polymicrobial communities requires assessment of gDNA extraction efficiency. J Microbiol Methods 194:1–4. 10.1016/j.mimet.2022.106421 [DOI] [PubMed] [Google Scholar]
- Chathoth K, Martin B, Bonnaure-Mallet M, Baysse C (2021) Method for screening antimicrobial gels against multi-species oral biofilms. J Microbiol Methods 187:1–4. 10.1016/j.mimet.2021.106253 [DOI] [PubMed] [Google Scholar]
- Chen J, Kong L, Peng X, Chen Y, Ren B, Li M, Li J, Zhou X, Cheng L (2021) Core microbiota promotes the development of dental caries. Appl Sci 11 10.3390/app11083638 [DOI] [Google Scholar]
- Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung FE, Kultima JR, Hayward MR, Coelho LP, Allen-Vercoe E, Bertrand L, Blaut M, Brown JRM, Carton T, Cools-Portier S, Daigneault M, Derrien M, Druesne A, De Vos WM, Finlay BB, Flint HJ, Guarner F, Hattori M, Heilig H, Luna RA, Van HylckamaVlieg J, Junick J, Klymiuk I, Langella P, Le Chatelier E, Mai V, Manichanh C, Martin JC, Mery C, Morita H, O’toole PW, Orvain C, Patil KR, Penders J, Persson S, Pons N, Popova M, Salonen A, Saulnier D, Scott KP, Singh B, Slezak K, Veiga P, Versalovic J, Zhao L, Zoetendal EG, Ehrlich SD, Dore J, Bork P (2017) Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 35:1069–1076. 10.1038/NBT.3960 [DOI] [PubMed] [Google Scholar]
- Crowe SA, Simister RL, Spence JS, Kenward PA, van Slyke AC, Lennox P, Carr N (2021) Microbial community compositions in breast implant biofilms associated with contracted capsules. PLoS One 16:1–19. 10.1371/journal.pone.0249261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca M, Marín MJ, Nóvoa L, O’Connor A, del Sánchez MC, Blanco J, Limeres J, Sanz M, Diz P, Herrera D (2021) Periodontal condition and subgingival microbiota characterization in subjects with down syndrome. Appl Sci 11:1–15. 10.3390/app11020778 [DOI] [Google Scholar]
- Dai X, Guo Q, Song D, Zhou W, Liu G, Liang G, He P, Sun G, Yuan F, Liu Z (2021) Long-term mineral fertilizer substitution by organic fertilizer and the effect on the abundance and community structure of ammonia-oxidizing archaea and bacteria in paddy soil of south China. Eur J Soil Biol 103:103288. 10.1016/j.ejsobi.2021.103288 [DOI] [Google Scholar]
- Davis A, Kohler C, Alsallaq R, Hayden R, Maron G, Margolis E (2019) Improved yield and accuracy for DNA extraction in microbiome studies with variation in microbial biomass. Biotechniques 66:285–289. 10.2144/btn-2019-0016 [DOI] [PubMed] [Google Scholar]
- Deng X, Zhang N, Shen Z, Zhu C, Liu H, Xu Z, Li R, Shen Q, Salles JF (2021) Soil microbiome manipulation triggers direct and possible indirect suppression against Ralstonia solanacearum and Fusarium oxysporum. npj Biofilms Microbiomes 7. 10.1038/s41522-021-00204-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Pham DD, Jenčová V, Kaňuchová M, Bayram J, Grossová I, Šuca H, Urban L, Havlíčková K, Novotný V, Mikeš P, Mojr V, Asatiani N, Košťáková EK, Maixnerová M, Vlková A, Vítovská D, Šanderová H, Nemec A, Krásný L, Zajíček R, Lukáš D, Rejman D, Gál P (2021) Novel lipophosphonoxin-loaded polycaprolactone electrospun nanofiber dressing reduces Staphylococcus aureus induced wound infection in mice. Sci Rep 11:1–15. 10.1038/s41598-021-96980-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigo B, Brunetti G, Aleer SC, Bell JM, Short MD, Vasileiadis S, Turnidge J, Monis P, Cunliffe D, Donner E (2021) Inactivation, removal, and regrowth potential of opportunistic pathogens and antimicrobial resistance genes in recycled water systems. Water Res 201:117324. 10.1016/j.watres.2021.117324 [DOI] [PubMed] [Google Scholar]
- Edslev SM, Olesen CM, Nørreslet LB, Ingham AC, Iversen S, Lilje B, Clausen ML, Jensen JS, Stegger M, Agner T, Andersen PS (2021) Staphylococcal communities on skin are associated with atopic dermatitis and disease severity. Microorganisms 9:1–17. 10.3390/microorganisms9020432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciotti R, Pignatelli P, Carrarini C, Romei FM, Mastrippolito M, Gentile A, Mancinelli R, Fulle S, Piattelli A, Onofrj M, Curia MC (2021) Exploring the connection between Porphyromonas gingivalis and neurodegenerative diseases: a pilot quantitative study on the bacterium abundance in oral cavity and the amount of antibodies in serum. Biomolecules 11 10.3390/biom11060845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Peng H, Liu J, Nguyen TH, Hashmi MZ, Shen C (2021) Occurrence and quantification of culturable and viable but non-culturable (VBNC) pathogens in biofilm on different pipes from a metropolitan drinking water distribution system. Sci Total Environ 764 10.1016/j.scitotenv.2020.142851 [DOI] [PubMed] [Google Scholar]
- Gao S, Sun C, Hong H, Gooneratne R, Mutukumira A, Wu X (2021a) Rapid detection of viable Cronobacter sakazakii in powdered infant formula using improved propidium monoazide (PMAxx) and quantitative recombinase polymerase amplification (qRPA) assay. Food Control 124:107899. 10.1016/j.foodcont.2021.107899 [DOI] [Google Scholar]
- Gao W, Gao K, Guo Z, Liu Y, Jiang L, Liu C, Liu X, Wang G (2021b) Different Responses of Soil Bacterial and Fungal Communities to 3 Years of Biochar Amendment in an Alkaline Soybean Soil. Front Microbiol 12:1–11. 10.3389/fmicb.2021.630418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gismondi A, Di Marco G, Redi EL, Ferrucci L, Cantonetti M, Canini A (2021) The antimicrobial activity of Lavandula angustifolia Mill essential oil against Staphylococcus species in a hospital environment. J Herb Med 26:100426. 10.1016/j.hermed.2021.100426 [DOI] [Google Scholar]
- Greathouse K, Sinha R, Vogtmann E (2019) DNA extraction for human microbiome studies: The issue of standardization. Genome Biol 20:1–4. 10.1186/S13059-019-1843-8/TABLES/1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines SR, Hall EC, Marciniak K, Misztal PK, Goldstein AH, Adams RI, Dannemiller KC (2021) Microbial growth and volatile organic compound (VOC) emissions from carpet and drywall under elevated relative humidity conditions (Microbiome, (2021), 9, 1, (209), DOI: 10.1186/s40168-021-01158-y). Microbiome 9:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Bai M, Chen Y, Gong Y, Wu M, Yang H, Chen Q, Xu T, Wei Y, Ding G, Li J (2021) Dynamics of diversity and abundance of sulfonamide resistant bacteria in a silt loam soil fertilized by compost. Antibiotics 10 10.3390/antibiotics10060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Li Y, Mu H, Zhao Z, Wang J, Liu S, Sun Z, Zheng M (2021a) Ammonium concentration determines differential growth of comammox and canonical ammonia-oxidizing prokaryotes in soil microcosms. Appl Soil Ecol 157 10.1016/j.apsoil.2020.103776 [DOI] [Google Scholar]
- He Y, Na R, Niu X, Xiao B, Yang H (2021b) Lactobacillus rhamnosus and Lactobacillus casei affect various stages of gardnerella species biofilm formation. front. Cell Infect Microbiol 11:1–13. 10.3389/fcimb.2021.568178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare A, Wang H, Meethil A, Abusleme L, Hong BY, Moutsopoulos NM, Marsh PD, Hajishengallis G, Diaz PI (2021) A cross-species interaction with a symbiotic commensal enables cell-density-dependent growth and in vivo virulence of an oral pathogen. ISME J 15:1490–1504. 10.1038/s41396-020-00865-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Hong H, Rong B, Wei Y, Zeng J, Zhu J, Bai L, Guo F, Yu X (2021) A comprehensive investigation of the microbial risk of secondary water supply systems in residential neighborhoods in a large city. Water Res 205 10.1016/j.watres.2021.117690 [DOI] [PubMed] [Google Scholar]
- Invernizzi R, Wu BG, Barnett J, Ghai P, Kingston S, Hewitt RJ, Feary J, Li Y, Chua F, Wu Z, Wells AU, George PM, Renzoni EA, Nicholson AG, Rice A, Devaraj A, Segal LN, Byrne AJ, Maher TM, Lloyd CM, Molyneaux PL (2021) The respiratory microbiome in chronic hypersensitivity pneumonitis is distinct from that of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 203:339–347. 10.1164/rccm.202002-0460OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacerovsky M, Pliskova L, Bolehovska R, Lesko D, Gerychova R, Janku P, Matlak P, Simetka O, Stranik J, Faist T, Mls J, Vescicik P, Jacobsson B, Musilova I (2021) Cervical Gardnerella vaginalis in women with preterm prelabor rupture of membranes. PLoS ONE 16:1–19. 10.1371/journal.pone.0245937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum RN, Smith EG, Vaughan GO, Phippen BL, McParland D, Al-Mansoori N, Carrier TJ, Burt JA, Reitzel AM (2018) DNA extraction method plays a significant role when defining bacterial community composition in the marine invertebrate Echinometramathaei. Front Mar Sci 5:1–13. 10.3389/fmars.2018.0025529552559 [DOI] [Google Scholar]
- Kralik P, Ricchi M (2017) A basic guide to real time PCR in microbial diagnostics : definitions, parameters, and everything. Front Microbiol 8:1–9. 10.3389/fmicb.2017.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Giménez-Rota C, Rota C (2021) Real-time pcr method combined with a matrix lysis procedure for the quantification of Listeria monocytogenes in meat products. Foods 10 10.3390/foods10040735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazou TP, Gelasakis AI, Chaintoutis SC, Iossifidou EG, Dovas CI (2021) Method-dependent implications in foodborne pathogen quantification: the case of Campylobacter coli survival on meat as comparatively assessed by colony count and viability PCR. Front Microbiol 12. 10.3389/fmicb.2021.604933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelapornpisid W, Novak-Frazer L, Qualtrough A, Rautemaa-Richardson R (2021) Effectiveness of D, L-2-hydroxyisocaproic acid (HICA) and alpha-mangostin against endodontopathogenic microorganisms in a multispecies bacterial–fungal biofilm in an ex vivo tooth model. Int Endod J 54:2243–2255. 10.1111/iej.13623 [DOI] [PubMed] [Google Scholar]
- Li C, Shi W, Wu D, Tian R, Wang B, Lin R, Zhou B, Gao Z (2021a) Biocontrol of potato common scab by Brevibacillus laterosporus BL12 is related to the reduction of pathogen and changes in soil bacterial community. Biol Control 153 10.1016/j.biocontrol.2020.104496 [DOI] [Google Scholar]
- Li D, Van De Werfhorst LC, Steets B, Ervin J, Murray JLS, Blackwell A, Devarajan N, Holden PA (2021b) Sources of low level human fecal markers in recreational waters of two Santa Barbara, CA beaches: roles of WWTP outfalls and swimmers. Water Res 202:117378. 10.1016/j.watres.2021.117378 [DOI] [PubMed] [Google Scholar]
- Li M, Zhang J, Yang X, Zhou Y, Zhang L, Yang Y, Luo L, Yan Q (2021c) Responses of ammonia-oxidizing microorganisms to biochar and compost amendments of heavy metals-polluted soil. J Environ Sci (china) 102:263–272. 10.1016/j.jes.2020.09.029 [DOI] [PubMed] [Google Scholar]
- Li Y, Wu MX (2021) Reversal of polymicrobial biofilm tolerance to ciprofloxacin by blue light plus carvacrol. Microorganisms 9:2074. 10.3390/microorganisms9102074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Y (2021) Exploring the communities of bacteria, fungi and ammonia oxidizers in rhizosphere of Fusarium-diseased greenhouse cucumber. Appl Soil Ecol 161 10.1016/j.apsoil.2020.103832 [DOI] [Google Scholar]
- Longin C, Guilloux-benatier M, Alexandre H (2016) Design and Performance Testing of a DNA Extraction Assay for Sensitive and Reliable Quantification of Acetic Acid Bacteria Directly in Red Wine Using Real Time PCR. Front Microbiol 7:1–9. 10.3389/fmicb.2016.00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, McAuley DF, Fulton CR, Pessoa JS, McMullan R, Lundy FT (2021) Targeting Candida albicans in dual-species biofilms with antifungal treatment reduces Staphylococcus aureus and MRSA in vitro. PLoS One 16:1–14. 10.1371/journal.pone.0249547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D, Klapperich CM (2009) Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip 9:2811–2817. 10.1039/b905065p [DOI] [PubMed] [Google Scholar]
- Maier C, Hofmann K, Huptas C, Scherer S, Wenning M, Lücking G (2021a) Simultaneous quantification of the most common and proteolytic Pseudomonas species in raw milk by multiplex qPCR. Appl Microbiol Biotechnol 105:1693–1708. 10.1007/s00253-021-11109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L, Goemans CV, Wirbel J, Kuhn M, Eberl C, Pruteanu M, Müller P, Garcia-Santamarina S, Cacace E, Zhang B, Gekeler C, Banerjee T, Anderson EE, Milanese A, Löber U, Forslund SK, Patil KR, Zimmermann M, Stecher B, Zeller G, Bork P, Typas A (2021b) Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599:120–124. 10.1038/s41586-021-03986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotz C, Amir A, Humphrey G, Gogul G, Knight R (2017) DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques 62:290–293. 10.2144/000114559 [DOI] [PubMed] [Google Scholar]
- Marotz C, Morton JT, Navarro P, Coker J, Belda-Ferre P, Knight R, Zengler K (2021) Quantifying Live Microbial Load in Human Saliva Samples over Time Reveals Stable Composition and Dynamic Load. mSystems 6 10.1128/msystems.01182-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouf A, Hanifian S, Shayegh J (2021) Prevalence of Brucella spp in raw milk and artisanal cheese tested via real-time qPCR and culture assay. Int J Food Microbiol 347:109192. 10.1016/j.ijfoodmicro.2021.109192 [DOI] [PubMed] [Google Scholar]
- McKee AM, Spear SF, Pierson TW (2015) The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biol Conserv 183:70–76. 10.1016/j.biocon.2014.11.031 [DOI] [Google Scholar]
- McKloud E, Delaney C, Sherry L, Kean R, Williams S, Metcalfe R, Thomas R, Richardson R, Gerasimidis K, Nile CJ, Williams C, Ramage G (2021) Recurrent Vulvovaginal Candidiasis: a Dynamic Interkingdom Biofilm Disease of Candida and Lactobacillus. mSystems 6 10.1128/msystems.00622-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millones-Gómez PA, Amaranto REB, Torres DJM, Calla-Poma RD, Requena-Mendizabal MF, Alvino-Vales MI, Calla-Poma R (2021) Identification of proteins associated with the formation of oral biofilms. Pesqui Bras Odontopediatria Clin Integr 21:1–10. 10.1590/pboci.2021.084 [DOI] [Google Scholar]
- Muzquiz LG, Gómez D, Cruz T, Olvera ET (2021) Evaluation of intracellular survival of Campylobacter fetus subsp. fetus in bovine endometrial cells by qPCR. Iran J Vet Res 22:94–99. 10.22099/IJVR.2021.38693.5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naung M, Uren Webster TM, Lloyd R, Garcia de Leaniz C, Consuegra S (2021) A novel qPCR assay for the rapid detection and quantification of the lumpfish (Cyclopterus lumpus) microsporidian parasite Nucleospora cyclopteri. Aquaculture 531 10.1016/j.aquaculture.2020.735779 [DOI] [Google Scholar]
- Nogrado K, Unno T, Hur HG, Lee JH (2021) Tetracycline-resistant bacteria and ribosomal protection protein genes in soils from selected agricultural fields and livestock farms. Appl Biol Chem 64 10.1186/s13765-021-00613-6 [DOI] [Google Scholar]
- Normington C, Moura IB, Bryant JA, Ewin DJ, Clark EV, Kettle MJ, Harris HC, Spittal W, Davis G, Henn MR, Ford CB, Wilcox MH, Buckley AM (2021) Biofilms harbour Clostridioides difficile, serving as a reservoir for recurrent infection. npj Biofilms Microbiomes 7 10.1038/s41522-021-00184-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell GC, Chantler PD, Barr TL (2017a) High interspecimen variability in nucleic acid extraction efficiency necessitates the use of spike-in control for accurate qPCR-based measurement of plasma cell-free DNA levels. Lab Med 48:332–338. 10.1093/LABMED/LMX043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi S, Kuroda T (2021) Developing a qPCR assay for the quantification of Calonectriailicicola in soil of soybean field. Trop Plant Pathol 46:186–194. 10.1007/s40858-020-00399-w [DOI] [Google Scholar]
- Oh KY, Lee S, Lee MS, Lee MJ, Shim E, Hwang YH, Ha JG, Yang YS, Hwang IT, Park JS (2021) Composition of vaginal microbiota in pregnant women with Aerobic Vaginitis. Front Cell Infect Microbiol 11:1–12. 10.3389/fcimb.2021.677648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G, Sant’Anna C, Lamarão L, Guimarães A, Da Rocha C, Bahia M, De Souza C, Calcagno D, De Assumpção P, Burbano R (2021) Quantitative difference of oral pathogen between individuals with gastric cancer and individuals without cancer. Oncotarget 12:1677–1686. 10.18632/ONCOTARGET.28034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2010) The ongoing evolution of qPCR. Methods 50:215–216. 10.1016/J.YMETH.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Pignatelli P, Iezzi L, Pennese M, Raimondi P, Cichella A, Bondi D, Grande R, Cotellese R, Di Bartolomeo N, Innocenti P, Piattelli A, Curia MC (2021) The potential of colonic tumor tissue Fusobacterium nucleatum to predict staging and its interplay with oral abundance in colon cancer patients. Cancers (basel) 13:1–19. 10.3390/cancers13051032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet LF, Bertelli C, Scherz V, Rochat I, Mardegan C, Brouillet R, Jaton K, Mornand A, Kaiser L, Posfay-Barbe K, Asner SA, Greub G (2021) Chlamydia pneumoniae and Mycoplasma pneumoniae in children with cystic fibrosis: Impact on bacterial respiratory microbiota diversity. Pathog Dis 79:1–7. 10.1093/FEMSPD/FTAA074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redanz U, Redanz S, Treerat P, Prakasam S, Lin LJ, Merritt J, Kreth J (2021) Differential response of oral mucosal and gingival cells to Corynebacterium durum, Streptococcus sanguinis, and Porphyromonas gingivalis Multispecies Biofilms. Front Cell Infect Microbiol 11:1–12. 10.3389/fcimb.2021.686479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey MÁ, Cap M, Favre LC, Rodríguez Racca A, Dus Santos MJ, Vaudagna SR, Mozgovoj M (2021) Evaluation of PMA-qPCR methodology to detect and quantify viable Shiga toxin-producing Escherichia coli in beef burgers. J Food Process Preserv 45:1–8. 10.1111/jfpp.15338 [DOI] [Google Scholar]
- Rezzonico F, Moe Y, Zu C, Lyon CB, Villeurbanne F (2003) Effect of stress on the ability of a phlA -based quantitative competitive PCR assay to monitor biocontrol strain Pseudomonas fluorescens CHA0. Appl Env Microbiol 69:686–690. 10.1128/AEM.69.1.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes S, Ruiz-Rico M, Moreno-Mesonero L, Moreno Y, Barat JM (2021) Natural antimicrobial compounds immobilised on silica microparticles as filtering materials: Impact on the metabolic activity and bacterial viability of waterborne microorganisms. Environ Technol Innov 21:101219. 10.1016/j.eti.2020.101219 [DOI] [Google Scholar]
- Rodríguez-Sorrento A, Castillejos L, López-Colom P, Cifuentes-Orjuela G, Rodríguez-Palmero M, Moreno-Muñoz JA, Luise D, Trevisi P, Martín-Orúe SM (2021) Effects of the Administration of Bifidobacterium longum subsp. infantis CECT 7210 and Lactobacillus rhamnosus HN001 and Their Synbiotic Combination With Galacto-Oligosaccharides Against Enterotoxigenic Escherichia coli F4 in an Early Weaned Piglet Model Front Microbiol 12 10.3389/fmicb.2021.642549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Villalba A, van Pelt-Verkuil E, Gunst QD, Ruijter JM, van den Hoff MJ (2017) Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR). Biomol Detect Quantif 14:7–18. 10.1016/j.bdq.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaddar S, Truu J, Chatterjee P, Oopkaup K, Truu M, Kim K, Kim S, Schmidt R, Roy Choudhury A, Choi J, Sa T (2021) Long-term inorganic nitrogen application changes the ammonia-oxidizing archaeal community composition in paddy soils. Eur J Soil Sci 72:2246–2260. 10.1111/ejss.13112 [DOI] [Google Scholar]
- Sanjulián L, Lamas A, Barreiro R, Cepeda A, Fente CA, Regal P (2021) Bacterial diversity of breast milk in healthy Spanish women: Evolution from birth to five years postpartum. Nutrients 13:1–22. 10.3390/nu13072414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsella E, Zecconi A, Cintio M, Stefanon B (2021) Characterization of microbiome on feces, blood and milk in dairy cows with different milk leucocyte pattern. Animals 11:1–14. 10.3390/ani11051463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selis NN, Oliveira HBM, Souza CLS, Almeida JB, Andrade YMFS, Silva LSC, Romano CC, Rezende RP, Yatsuda R, Uetanabaro APT, Marques LM (2021) Lactobacillus plantarum Lp62 exerts probiotic effects against Gardnerella vaginalis ATCC 49154 in bacterial vaginosis. Lett Appl Microbiol 73:579–589. 10.1111/lam.13547 [DOI] [PubMed] [Google Scholar]
- Sereti M, Zekeridou A, Cancela J, Mombelli A, Giannopoulou C (2021) Microbiological testing of clinical samples before and after periodontal treatment. A comparative methodological study between real-time PCR and real-time-PCR associated to propidium monoazide. Clin Exp Dent Res 7:1069–1079. 10.1002/cre2.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton DJ, Bentz ML, Welsh RM, Derado G, Furin W, Rose LJ, Noble-Wang J, Pacilli M, McPherson TD, Black S, Kemble SK, Herzegh O, Ahmad A, Forsberg K, Jackson B, Litvintseva AP (2021) Positive Correlation Between Candida auris Skin-Colonization Burden and Environmental Contamination at a Ventilator-Capable Skilled Nursing Facility in Chicago. Clin Infect Dis 73:1142–1148. 10.1093/cid/ciab327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Osborn AM (2009) Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol 67:6–20. 10.1111/j.1574-6941.2008.00629.x [DOI] [PubMed] [Google Scholar]
- Sousa C, França A, Cerca N (2014) Assessing and reducing sources of gene expression variability in Staphylococcus epidermidis biofilms. Biotechniques 57:295–301. 10.2144/000114238 [DOI] [PubMed] [Google Scholar]
- Stoeckel DM, Stelzer EA, Dick LK (2009) Evaluation of two spike-and-recovery controls for assessment of extraction efficiency in microbial source tracking studies. Water Res 43:4820–4827. 10.1016/j.watres.2009.06.028 [DOI] [PubMed] [Google Scholar]
- Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M (2015) How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif 3:9–16. 10.1016/j.bdq.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taibi A, Ku M, Lin Z, Gargari G, Kubant A, Lepp D, Power KA, Guglielmetti S, Thompson LU, Comelli EM (2021) Data on cecal and fecal microbiota and predicted metagenomes profiles of female mice receiving whole flaxseed or its oil and secoisolariciresinol diglucoside components. Data Br 38:107409. 10.1016/j.dib.2021.107409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon KM, Morais TB, Taddei CR, Araújo-Filho HB, Abrão ACFV, Miranda A, De Morais MB (2021) Gut microbiota comparison of vaginally and cesarean born infants exclusively breastfed by mothers secreting a1–2 fucosylated oligosaccharides in breast milk. PLoS One 16:1–15. 10.1371/journal.pone.0246839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Corral Y, Santos Y (2021) Development of a real-time PCR assay for detection and quantification of Streptococcus iniae using the lactate permease gene. J Fish Dis 44:53–61. 10.1111/jfd.13267 [DOI] [PubMed] [Google Scholar]
- Turner E, Sobel JD, Akins RA (2021) Prognosis of recurrent bacterial vaginosis based on longitudinal changes in abundance of Lactobacillus and specific species of Gardnerella. PLoS One 16:1–18. 10.1371/journal.pone.0256445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrea-Valencia S, Etto RM, Takahashi WY, Caires EF, Bini AR, Ayub RA, Stets MI, Cruz LM, Galvão CW (2021) Detection of Azospirillum brasilense by qPCR throughout a maize field trial. Appl Soil Ecol 160 10.1016/j.apsoil.2020.103849 [DOI] [Google Scholar]
- Usyskin-Tonne A, Hadar Y, Yermiyahu U, Minz D (2021) Elevated CO2 and nitrate levels increase wheat root-associated bacterial abundance and impact rhizosphere microbial community composition and function. ISME J 15:1073–1084. 10.1038/s41396-020-00831-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspecht T, Van Holm W, Boon N, Bernaerts K, Daep CA, Masters JG, Zayed N, Quirynen M, Teughels W (2021a) Potential prebiotic substrates modulate composition, metabolism, virulence and inflammatory potential of an in vitro multi-species oral biofilm. J Oral Microbiol 13 10.1080/20002297.2021.1910462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspecht T, Van Holm W, Boon N, Bernaerts K, Daep CA, Zayed N, Quirynen M, Teughels W (2021b) Comparison of the modulatory effects of three structurally similar potential prebiotic substrates on an in vitro multi-species oral biofilm. Sci Rep 11:1–15. 10.1038/s41598-021-94510-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzini P, Vidic J, Manzano M (2021) Enrichment Free qPCR for Rapid Identification and Quantification of Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis in Chicken Meat Samples by a New Couple of Primers. Foods 10:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallon T, Sauvageau A, Van der Heyden H (2021) Detection and quantification of Rhizoctonia solani and Rhizoctonia solani ag1-ib causing the bottom rot of lettuce in tissues and soils by multiplex qpcr. Plants 10:1–17. 10.3390/plants10010057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Peng W, Zhang B, Cao Y, Zhao J, Cao H (2021a) Succession of bacterial community composition in coastal agricultural soils along a 1000-year reclamation chronosequence in Hangzhou Bay, China Ecol Indic 121 10.1016/j.ecolind.2020.106972 [DOI] [Google Scholar]
- Wang P, Zhang N, Lee PKH, Li Y (2021) Quantification of Lactobacillus delbrueckii subsp Bulgaricus and its applicability as a tracer for studying contamination spread on environmental surfaces. Build Environ 197:107869. 10.1016/j.buildenv.2021.107869 [DOI] [Google Scholar]
- Wang S, Zhu Y, Yang Y, Li J, Hoffmann MR (2020) Electrochemical cell lysis of gram-positive and gram-negative bacteria: DNA extraction from environmental water samples. Electrochim Acta 338:1–9. 10.1016/j.electacta.2020.135864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Zeng S, Hou D, Zhou R, Xing C, Deng X, Yu L, Wang H, Deng Z, Weng S, Huang Z, He J (2021) Community diversity and abundance of ammonia-oxidizing archaea and bacteria in shrimp pond sediment at different culture stages. J Appl Microbiol 130:1442–1455. 10.1111/jam.14846 [DOI] [PubMed] [Google Scholar]
- Wongsaroj L, Chanabun R, Tunsakul N, Prombutara P, Panha S, Somboonna N (2021) First reported quantitative microbiota in different livestock manures used as organic fertilizers in the Northeast of Thailand. Sci Rep 11:1–15. 10.1038/s41598-020-80543-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Dinis M, Haghighi F, He X, Shi W, Chaichanasakul Tran N (2022) Oral colonization of Candida albicans and Streptococcus mutans in children with or without fixed orthodontic appliances: A pilot study. J Dent Sci 17:451–458. 10.1016/j.jds.2021.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu Y, Shu Y, Xia W, Xu R, Chen Y (2021) Modified PMA-qPCR Method for Rapid Quantification of Viable Lactobacillus spp. in Fermented Dairy Products. Food Anal Methods 14:1908–1918. 10.1007/s12161-021-02022-3 [DOI] [Google Scholar]
- Yin C, Schlatter DC, Kroese DR, Paulitz TC, Hagerty CH (2021) Impacts of lime application on soil bacterial microbiome in dryland wheat soil in the Pacific Northwest. Appl Soil Ecol 168:104113. 10.1016/j.apsoil.2021.104113 [DOI] [Google Scholar]
- Yuan X, Hong S, Xiong W, Raza W, Shen Z, Wang B, Li R, Ruan Y, Shen Q, Dini-Andreote F (2021) Development of fungal-mediated soil suppressiveness against Fusarium wilt disease via plant residue manipulation. Microbiome 9:1–15. 10.1186/s40168-021-01133-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhong B, Zhao C, Wang E, Wang Y, Chen D, Shi F (2021a) Change of soil physicochemical properties, bacterial community and aggregation during desertification of grasslands in the Tibetan Plateau. Eur J Soil Sci 72:274–288. 10.1111/ejss.12939 [DOI] [Google Scholar]
- Zhang Q, Stummer BE, Guo Q, Zhang W, Zhang X, Zhang L, Harvey PR (2021b) Quantification of Pseudomonas protegens FD6 and Bacillus subtilis NCD-2 in soil and the wheat rhizosphere and suppression of root pathogenic Rhizoctoniasolani AG-8. Biol Control 154:104504. 10.1016/j.biocontrol.2020.104504 [DOI] [Google Scholar]
- Zhao J, Chen D, Gao W, Guo Z, Jia Z, Hernández M (2021) Resuscitation of soil microbiota after > 70-years of desiccation Eur J Soil Biol 103 10.1016/j.ejsobi.2021.103290 [DOI] [Google Scholar]
- Zhu H, Teng Y, Wang X, Zhao L, Ren W, Luo Y, Christie P (2021a) Changes in clover rhizosphere microbial community and diazotrophs in mercury-contaminated soils. Sci Total Environ 767:1–10. 10.1016/j.scitotenv.2021.145473 [DOI] [PubMed] [Google Scholar]
- Zhu J, Cao A, Wu J, Fang W, Huang B, Yan D, Wang Q, Li Y (2021b) Effects of chloropicrin fumigation combined with biochar on soil bacterial and fungal communities and Fusarium oxysporum. Ecotoxicol Environ Saf 220 10.1016/j.ecoenv.2021.112414 [DOI] [PubMed] [Google Scholar]


