Abstract
Noncovalent adsorption of DNA on nanoparticles has led to their widespread implementation as gene delivery tools and optical probes. Yet, the behavior and stability of DNA-nanoparticle complexes once applied in biomolecule-rich, in vivo environments remains unpredictable, whereby biocompatibility testing usually occurs in serum. Here, we demonstrate time-resolved measurements of exchange dynamics between solution-phase and adsorbed corona-phase DNA and protein biomolecules on single-walled carbon nanotubes (SWCNTs). We capture real-time binding of fluorophore-labeled biomolecules, utilizing the SWCNT surface as a fluorescence quencher, and apply this corona exchange assay to study protein corona dynamics on ssDNA-SWCNT-based dopamine sensors. We study exchange of two blood proteins, albumin and fibrinogen, adsorbing to and competitively displacing (GT)6 vs (GT)15 ssDNA from ssDNA-SWCNTs. We find that (GT)15 binds to SWCNTs with a higher affinity than (GT)6 and that fibrinogen interacts with ssDNA-SWCNTs more strongly than albumin. Albumin and fibrinogen cause a 52.2% and 78.2% attenuation of the dopamine nanosensor response, coinciding with 0.5% and 3.7% desorption of (GT)6, respectively. Concurrently, the total surface-adsorbed fibrinogen mass is 168% greater than that of albumin. Binding profiles are fit to a competitive surface exchange model which recapitulates the experimental observation that fibrinogen has a higher affinity for SWCNTs than albumin, with a fibrinogen on-rate constant 1.61-fold greater and an off-rate constant 0.563-fold smaller than that of albumin. Our methodology presents a generic route to assess real-time corona exchange on nanoparticles in solution phase and more broadly motivates testing of nanoparticle-based technologies in blood plasma rather than the more ubiquitously tested serum conditions.
Graphical abstract
1. INTRODUCTION
Adsorption of polymers on single-walled carbon nanotubes (SWCNTs) has enabled developments in molecular sensing,1 in vivo imaging,2 genetic cargo delivery,3 and chirality sorting.4 Noncovalent SWCNT functionalization offers a route that preserves the pristine atomic structure, thus retaining the intrinsic near-infrared (nIR) fluorescence of the SWCNTs for the aforementioned applications. However, noncovalent adsorption is an inherently dynamic process, where exchange occurs between molecules in the bulk solution and molecules on the surface, into what is known as the “corona phase”. In the case of polymers on SWCNTs, the nature, strength, and kinetics of both the polymer binding and unbinding processes are key contributors to the success of polymer-SWCNT based technologies.5 Understanding this exchange process is especially critical for intended uses of functionalized SWCNTs to probe biological environments. When a nanoparticle is injected into a biological system, the nanoparticle surface is spontaneously and rapidly coated with proteins to form the “protein corona”.6 For noncovalent polymer–SWCNT complexes, we hypothesize that native biomolecules compete with the original polymer to occupy the nanoparticle surface. Binding of proteins and other biomolecules to the SWCNT can disrupt the intended functionality of the nanoparticle and causes potentially adverse biocompatibility outcomes.7,8 This phenomenon of protein corona formation leads to challenges in translating in vitro sensing or biomolecule delivery platforms to in vivo application. Moreover, the generally accepted method of simulating in vivo biological conditions involves testing nanotechnology performance in blood serum.2,9 Yet, the absence of blood coagulation proteins from serum could yield a false outcome in assessing robustness of the nanotechnology and accordingly result in unpredicted failure when applied in vivo.
To clarify how nanoparticle–polymer conjugates behave in biologically relevant environments, it is pivotal to understand the kinetics describing molecular exchange on nanoparticle surfaces. Hence, we aim to gain a mechanistic understanding of how SWCNT-based neuromodulator sensors behave in protein-rich milieus. These sensors are based on noncovalent functionalization of (GT)6 single-stranded DNA (ssDNA) on SWCNTs, resulting in a complex that exhibits ultrasensitive ΔF/F0 = 2400% and 3500% fluorescence “turn-on” responses in the presence of neuromodulators dopamine and norepinephrine, respectively.10–13 However, the drastic enhancement of SWCNT fluorescence experienced upon in vitro exposure to dopamine is attenuated to ΔF/F0 ≈ 20% once the sensors are applied in brain tissue,12 presumably due to protein adsorption and/or disruption of the ssDNA corona phase originally on the SWCNT surface.
Current methods to measure dynamic, noncovalent exchange on nanoparticles exist but are limited in scope. Most research on protein–surface interactions involves characterizing macroscopic surfaces using a series of well-developed techniques that broadly entail an input signal modulated by changing adsorbate mass on the surface as a function of time, including total internal reflection fluorescence microscopy, surface plasmon resonance, biolayer interferometry, and quartz-crystal microbalance with dissipation monitoring. To apply these surface techniques to nanoparticles, the nanoparticles must be surface immobilized, thus introducing unrealistic topographical constraints that affect ligand exchange kinetics, lead to mass transport limitations,14 do not reproduce solution-phase nanosensor responses,15 and cause nonselective protein adsorption to any surface left exposed during the immobilization process.15
An alternative method that permits the study of SWCNTs in solution takes advantage of SWCNT sensitivity to their local dielectric environment16–18 by monitoring SWCNT fluorescence intensity changes and solvatochromic shifts upon corona exchange.19,20 This technique is applied to study polymer–surfactant exchange kinetics,21–24 whereby SWCNTs suspended with surfactant exhibit higher quantum yield and optical transition energy (i.e., blue-shifted spectra) compared to SWCNTs suspended with most biomolecules such as protein or ssDNA. Previous work has successfully applied measurable differences in SWCNT fluorescence spectra to study relative changes in corona surface composition.25,26 However, this approach cannot distinguish the exchange of two biomolecules (here, ssDNA to protein) nor can it distinguish between molecular rearrangement vs molecular desorption from the SWCNT surface. Despite the advantage of undertaking corona exchange studies in the solution phase with this approach, its low sensitivity, nonquantitative nature, and inability to distinguish between adsorbed biomolecules nullifies its potential for monitoring ssDNA–protein exchange.
In this work, we present an assay that overcomes the limitations of previous characterization methods to study corona exchange dynamics between solution-phase and corona-phase biopolymers on SWCNTs, specifically applied to ssDNA and protein. This assay exploits the quenching of fluorophores when in close proximity to the SWCNT surface to monitor ligand binding and unbinding events.27 While prior literature has similarly harnessed fluorophore quenching by SWCNTs to study the ssDNA-to-SWCNT binding process,8,19,28 far less is known regarding how preadsorbed ssDNA and biologically native proteins exchange on the SWCNTs. To our knowledge, this method is unique in enabling real-time monitoring of SWCNT surface exchange between ssDNA and proteins, tracing the fate of all biomolecules involved in the binding exchange. We conduct multiplexed fluorescence tracking of polymer adsorption and desorption events to/from the SWCNT surface. As a case study for this assay, we focus on comparing the sorption behavior of two specific blood proteins, human serum albumin and fibrinogen, chosen because (i) both are highly abundant in plasma, with albumin as ∼55% (w/v) of blood plasma, or 35–50 mg/mL29 and fibrinogen as ∼4% (w/v) of blood plasma, or 1.5–4.5 mg/mL,30 (ii) albumin is present in both blood plasma and serum, whereas fibrinogen is a key coagulation protein present in plasma but depleted from serum, and (iii) albumin and fibrinogen are known to be interfacially active proteins prone to surface-adsorption and are implicated in the formation of many other nanoparticle coronas.31–33 Binding profiles from the experimental assay in conjunction with a competitive-exchange model are used to extract kinetic parameters for each adsorbent species. Although this study specifically examines competitive adsorption of individual plasma proteins, albumin and fibrinogen, onto (GT)6- and (GT)15-SWCNTs, the assay is general to any molecules that can be fluorescently labeled and to any nanomaterial surface to which these species may adsorb and display quenched fluorescence. Binding is also compared to the orthogonal and more ubiquitously used platform monitoring solvatochromic shifting of the nIR SWCNT spectrum as a proxy for SWCNT corona coverage.25,26 The work presented herein develops an understanding of the fundamental corona exchange mechanism, contextualizes the nature of the ligand exchange process vs SWCNT solvatochromic shifting, and provides guidance for testing the performance of SWCNT-based systems in biologically relevant, protein-rich conditions.
2. RESULTS AND DISCUSSION
2.1. Proteins Attenuate Dopamine Sensor Response.
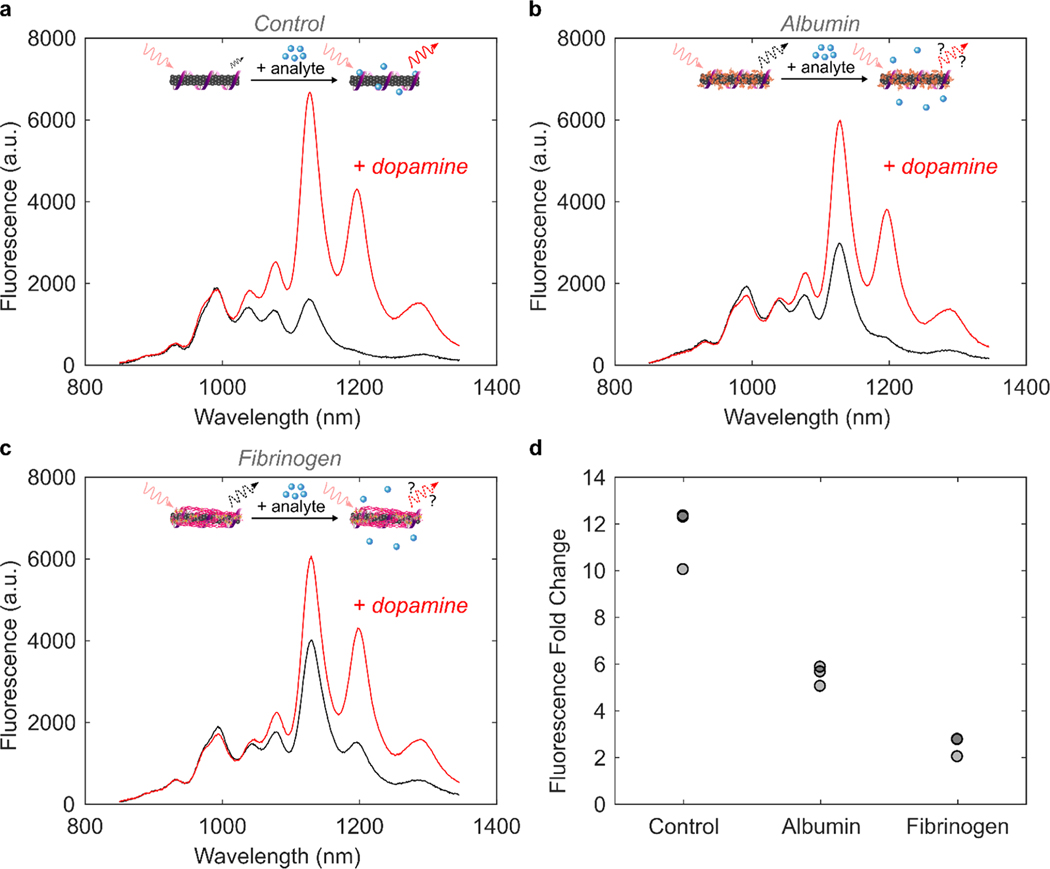
Noncovalent modification of single-walled carbon nanotubes (SWCNTs) with single-stranded (GT)6 DNA imparts nIR fluorescence responsivity to the small molecule neurotransmitter, dopamine.10–12 Addition of 200 μM dopamine to 5 μg/mL solution-phase (GT)6-SWCNTs in phosphate buffered saline (PBS) yields an 11.5-fold increase in nanosensor fluorescence at the 1200 nm SWCNT emission peak (Figure 1a; see section 4.4). Nanosensor response was diminished in the presence of 40 μg/mL human serum albumin (HSA, Figure 1b) and 40 μg/mL fibrinogen (FBG, Figure 1c), proteins abundant in intravenous environments. Incubation of 40 μg/mL HSA or FBG with 5 μg/mL (GT)6-SWCNTs reduced fluorescence dopamine response by 52.2% or 78.2% after 40 min (Figure 1d), respectively. Attenuation of nanosensors was due to two effects: addition of protein led to (i) increase in baseline fluorescence intensity, likely due to protein adsorption which is predicted to be highly favorable by a thermodynamic analysis (see the Supporting Information, section S.3; calculation of ssDNA–protein exchange energies on SWCNTs) and (ii) decrease in final fluorescence after addition of dopamine to (GT)6-SWCNTs. HSA did not cause any wavelength shifting of the (GT)6-SWCNT emission, while FBG exposure led to a red shift of 2.6 ± 0.6 nm (mean ± standard deviation of N = 3 sample replicates). Although changes in both the nIR fluorescence intensity and emission wavelength could indicate protein binding, monitoring the SWCNT fluorescence alone does not provide sufficient information to correlate these phenomena.
Figure 1.
Protein adsorption attenuates (GT)6-SWCNT sensor response to dopamine. (a) Near-infrared (nIR) spectra of 5 μg/mL (GT)6-SWCNTs before (black) and after (red) addition of 200 μM dopamine. (b and c) nIR spectra of 5 μg/mL (GT)6-SWCNTs incubated with 40 μg/ mL (b) albumin or (c) fibrinogen for 40 min before (black) and after (red) addition of 200 μM dopamine. Insets depict influence of protein corona formation on ability of nanosensor to respond to analyte. (d) Change in (GT)6-SWCNT fluorescence intensity at 1200 nm peak following 40 min incubation in PBS or protein solution at 40 μg/mL and then addition of 200 μM dopamine (N = 3). Nanosensor excitation was with 721 nm light.
We first implemented the solvatochromic shift assay to study surfactant-induced fluorescence changes of 5 μg/mL (GT)6-SWCNTs incubated with either 40 μg/mL HSA or FBG for 40 min. Displacement of the biopolymer corona phase with surfactant, here 0.5% (w/v) sodium dodecylbenzenesulfonate (SDBS), causes a change in local dielectric environment that in turn leads to a blue shift in SWCNT emission wavelength and an increase in SWCNT fluorescence emission intensity. The magnitude of these observed effects is thought to provide insight on the original SWCNT-corona stability. Interestingly, FBG incubated with (GT)6-SWCNTs resulted in both the largest magnitude wavelength shift and largest fold change in fluorescence intensity upon addition of SDBS (Figure S1). In contrast, HSA incubated with (GT)6-SWCNTs did not show a significantly different wavelength shift or intensity fold change compared to the control, (GT)6-SWCNTs incubated with only PBS. These results suggest albumin and fibrinogen proteins may have different binding propensities and kinetics to the SWCNT surface. However, this test fails to decouple the interactions between SWCNTs with ssDNA, protein, and then surfactant. To further study the differential attenuation of sensor response by HSA and FBG, and more thoroughly understand the exchange dynamics occurring on the SWCNT surface, we developed a method for studying SWCNT corona composition by multiplexed fluorescence monitoring.
2.2. Multiplexed Fluorescence Tracking Enables Real-Time Monitoring of Ligand Exchange Dynamics.
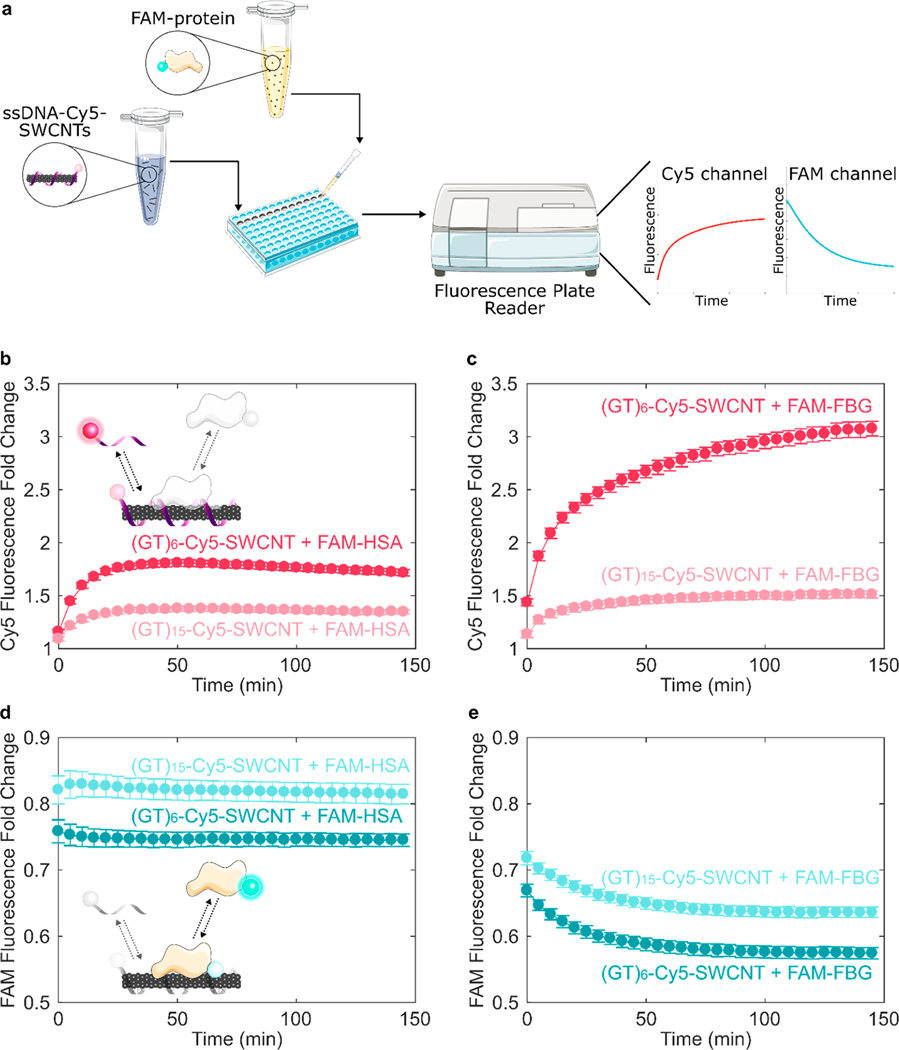
Our assay leverages fluorophore quenching induced by proximity to the SWCNT surface27 to measure surface exchange dynamics. Proteins under study were labeled with fluorescein (FAM) fluorophore (ex/em = 494/520 nm) using NHS ester conjugation to primary amine groups. Single-stranded DNA (ssDNA) were procured with a 3′ terminally labeled cyanine5 (Cy5) fluorophore (ex/em = 648/668 nm), enabling spectrally resolved multiplexed tracking of protein and ssDNA. The ssDNA-Cy5 is initially quenched on the SWCNT surface, increasing in fluorescence upon desorption from the SWCNT. This methodology has been previously implemented to study thermodynamics of fluorophore-labeled ssDNA interactions on SWCNT surfaces.34 Conversely, the FAM-labeled protein exhibits high fluorescence when added in bulk solution, quenching upon adsorption to the SWCNT surface. In this manner, FAM-labeled protein can be injected into ssDNA-Cy5-SWCNTs in a well-plate format and fluorescence changesresulting from biomolecule exchange can be read by a fluorescence plate reader (Figure 2a). We first employed this method to compare the desorption rates of (GT)6-Cy5 and (GT)15-Cy5 from SWCNTs upon addition of FAM-labeled HSA and FBG. Both proteins promoted dequenching of Cy5, as compared to the addition of PBS control (Figure 2b,c). Dequenching was due to complete desorption of ssDNA rather than partial desorption of the 3′ end, as verified by confirming that the binding profiles of 3′- vs internally Cy5-labeled ssDNA are similar (Figure S2). Additionally, presence of the Cy5 tag on ssDNA did not significantly affect protein adsorption (Figure S3). Fibrinogen generated a 3.09 ± 0.07 Cy5 fluorescence fold increase for (GT)6-Cy5-SWCNTs vs a 1.52 ± 0.04 Cy5 fluorescence fold increase for (GT)15-Cy5-SWCNTs. This result suggests (GT)15 is less readily displacedfrom the SWCNT surface compared to the shorter (GT)6 construct, a result consistent with the literature.35–37
Figure 2.
Tracking exchange of fluorophore-labeled ssDNA and protein on SWCNT surfaces demonstrates protein adsorption selectivity and ssDNA length effect. (a) Corona exchange assay workflow. ssDNA-Cy5-SWCNT solution is added to a well plate, FAM-protein solution is injected, and the ad-/desorption processes are monitored in separate color channels of a fluorescence plate reader (see section 4.3). Increase in ssDNA-Cy5 fluorescence induced by addition of 40 μg/mL (b) FAM-labeled albumin (FAM-HSA) or (c) FAM-labeled fibrinogen (FAM-FBG) to 5 μg/mL ssDNA-Cy5-SWCNT suspended with ssDNA, (GT)6 or (GT)15. Decrease in fluorescence of (d) FAM-HSA and (e) FAM-FBG after addition of protein to (GT)6- or (GT)15-SWCNT. Error bars represent standard error between experimental replicates (N = 3).
In the same experiment, protein adsorption onto ssDNA-Cy5-SWCNTs was concurrently tracked via fluorescence quenching of the protein-conjugated FAM. Presence of residual FAM fluorophore in the FAM-protein solution was accounted for by quantifying free FAM and subtracting the minimal change in fluorescence due to free FAM-to-SWCNT interaction (Figures S4 and S5 and Table S1). Furthermore, the effect of FAM fluorophore labeling on the protein-exchange dynamics was minimal (Figure S6), in agreement with previous investigations demonstrating that fluorescein-labeling of proteins does not perturb protein adsorption or function, and additionally that fluorescein signals are proportional to the interfacial mass of the tagged species.38–41 By tracking the fluorescence modulation resulting from FAM-protein interactions with ssDNA-Cy5-SWCNTs, we found that FAM-FBG exhibited a comparatively larger degree of quenching than FAM-HSA for both ssDNA-SWCNT suspensions (Figure 2d,e); upon addition of 40 μg/mL FAM-FBG to 5 μg/mL (GT)6-SWCNTs (final concentrations), FBG induced a 42.5 ± 0.9% decrease in FAM fluorescence vs a 25.5 ± 0.9% HSA-induced decrease in FAM fluorescence. These results consistently suggest two interaction mechanisms of ssDNA and protein with SWCNTs: (i) (GT)15 ssDNA binds to SWCNTs with a higher affinity than (GT)6 ssDNA, thus reducing protein adsorption, and (ii) FBG interacts with ssDNA-SWCNTs more strongly than HSA. The former result agrees with prior work confirming that the rate of ssDNA desorption from SWCNTs decreases with increasing oligo length,34 also valid in the presence of competing biomolecules.8 As such, our data suggest that FBG protein adsorption leads to more significant ssDNA desorption from SWCNTs, whereas HSA adsorbs less strongly and accordingly causes less ssDNA desorption from SWCNTs. These ssDNA–protein corona exchange trends were corroborated with polyacrylamide gel electrophoresis (PAGE) of the unbound species, where FAM-FBG adsorbed to (GT)6-Cy5-SWCNTs 56.5% more than FAM-HSA and caused 5.20% greater desorption of ssDNA (Figure S7). From our corona exchange assay, it is interesting to note that protein adsorption occurs faster than ssDNA desorption. These experimental results motivate kinetic modeling of ssDNA and protein exchange on SWCNT surfaces.
2.3. Kinetic Modeling of ssDNA/Protein Competitive Binding on the SWCNT Surface.
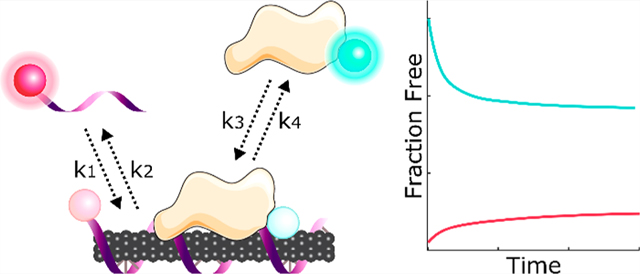
To quantitatively probe differences in protein affinities for ssDNA-SWCNTs, we fit Cy5 and FAM fluorescence data to a competitive adsorption model and extracted kinetic parameters for ssDNA and proteins. Multiplexed fluorescence tracking was repeated with 5 μg/mL (GT)6-Cy5-SWCNTs and concentrations of FAM-HSA and FAM-FBG ranging from 5 to 160 μg/mL. Fluorescence values were converted to mass concentration using standard curves for ssDNA-Cy5 and both FAM-conjugated proteins (Figure S8). A model was developed for the competitive exchange between ssDNA and protein on the SWCNT surface (eqs 1 and 2). In the model, unbound ssDNA (D) and protein (P) adsorb and desorb reversibly to SWCNT surface sites (*):
| (1) |
| (2) |
Total concentration of SWCNT surface sites was fixed, given by a site balance (eq 3), where *, D*, and P* refer to vacant sites, sites occupied by bound ssDNA, and sites occupied by bound protein, respectively:
| (3) |
Bound ssDNA and bound protein concentrations were calculated by species conservation, where total ssDNA was the amount added during ssDNA-SWCNT synthesis, total protein was the injected protein quantity, and total sites was a fit parameter. Rate constants k1 and k2 for ssDNA binding/unbinding, k3 and k4 for protein binding/unbinding, and the total concentration of binding sites [*]T were computed using a least-squares curve fit of eqs 4 and 5 to experimental data (see section 4.5).
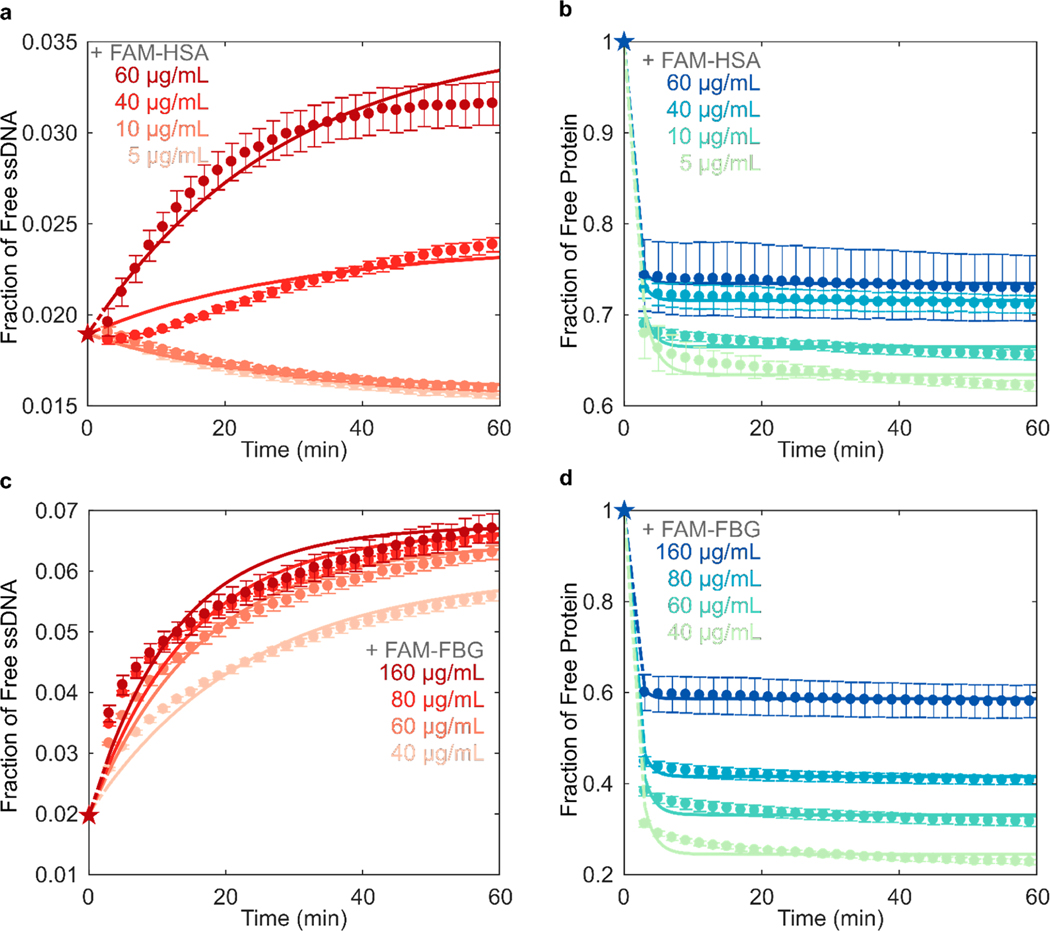
Experimental data of FAM-HSA or FAM-FBG added to (GT)6-Cy5-SWCNTs was fit to this model for each concentration tested (Figure 3). All mean relative errors comparing fits to experimental data were <5% (Table S2). The model recapitulates the experimental observation that FBG has a higher affinity for SWCNTs (Figure 3d) than HSA (Figure 3b), where average k3,FBG = 1.43 × 10−5 > k3,HSA = 8.88 × 10−6 mL μg −1 s−1 (Table 1, with full fit parameter results in Table S2). These k values are in relative agreement with previous literature assessing the kinetic parameters for ssDNA desorption from SWCNTs42 and protein binding to nano particles.43 At the same initial FAM-protein concentration of 40 μg/mL added to 5 μg/mL (GT)6-Cy5-SWCNTs, FBG adsorbed to a higher fraction of bound protein (0.756) than HSA (0.284) after 1 h. Final solution-phase concentrations of FAM-FBG and FAM-HSA were 9.04 and 28.5 μg/mL, respectively, compared to concentrations of 13.8 and 36.4 μg/mL determined by PAGE (Figure S7d). Solution-phase concentrations of (GT)6-Cy5 upon addition of FAM-FBG and FAM-HSA increased from 0.106 to 0.301 μg/mL and from 0.101 to 0.128 μg/mL, respectively, compared to concentration changes of 0.331 to 0.388 μg/mL and 0.688 to 0.767 μg/mL determined by PAGE (Figure S7c). The discrepancy in concentrations across the two approaches may be due to technical differences between methods, where gel electrophoresis is limited in both sensitivity and temporal resolution.
Figure 3.
Kinetic model of competitive exchange between ssDNA and protein on SWCNTs fit to fluorescence data to extract rate constants. Fraction of (a) (GT)6-Cy5 ssDNA and (b) FAM-labeled albumin (FAM-HSA) protein free in solution for varying concentrations of FAM-HSA injected into 5 μg/mL (GT)6-Cy5-SWCNT solution. Fraction of (c) (GT)6-Cy5 ssDNA and (d) FAM-labeled fibrinogen (FAM-FBG) protein free in solution for varying concentrations of FAM-FBG injected into (GT)6-Cy5-SWCNT solution. Star data points represent initial conditions used for solving model differential equations. Error bars represent standard error between experimental replicates (N = 3).
Table 1.
Range of Kinetic Model Fit Parameters
| Protein | k1 × 106 (mL μg−1 s−1) | k2 × 106 (s−1) | k3 × 106 (mL μg−1 s−1) | k4 × 106 (s−1) | (μg mL−1) |
|---|---|---|---|---|---|
| Albumin | 1.10–1.54 | 8.40–20.7 | 7.86–9.44 | 5850–12 000 | 365–526 |
| Fibrinogen | 1.15–2.30 | 42.7–90.9 | 11.8–16.9 | 2610–9150 | 486–620 |
Addition of FAM-FBG into solution with (GT)6-Cy5-SWCNTs led to ssDNA desorption for all tested concentrations of injected FAM-FBG (Figure 3c), as compared to only ≥40 μg/mL of injected FAM-HSA led to measurable ssDNA desorption (Figure 3a). Adsorption of ssDNA was observed upon addition of PBS or low concentrations of FAM-HSA (≤20 μg/mL) to (GT)6-Cy5-SWCNTs, indicating an initial excess of unbound ssDNA in bulk solution. Interestingly, the intermediate concentration of FAM-HSA (40 μg/mL) added to (GT)6-Cy5-SWCNTs resulted in sigmoidal ssDNA desorption behavior that is not fully described by the model. This behavior fundamentally implies that the assumption of independent reactions made in eqs 1 and 2 may not hold for intermediate concentrations of FAM-HSA. Specifically, the dependence of ssDNA dissociation constant on total protein concentration indicates that the rate of ssDNA desorption is a function of adsorbed or free protein concentration (Table S2 and Figure S9). This higher-order desorption process may account for the discrepancy between experimental data and model fit for (GT)6-Cy5 desorption induced by addition of 40 μg/mL FAM-HSA.
For all concentrations tested, protein adsorption proceeded significantly faster than ssDNA desorption dynamics, indicating that protein adsorption precedes ssDNA desorption and suggesting that the two phenomena may be decoupled in time. This difference in exchange time scales may be due to the large concentration of total SWCNT surface binding sites (with average fit values of and ) relative to the total ssDNA and protein concentrations. We hypothesize a low initial ssDNA surface coverage, or large accessible SWCNT surface area, is a likely reason for the difference in exchange time scales. Furthermore, in the case of FAM-FBG with (GT)6-Cy5-SWCNTs, while amount of adsorbed FBG reaches an apparent steady-state value within ∼5 min (Figure 3d), ssDNA continues to gradually desorb over time at a rate seemingly independent of injected protein concentration (Figure 3c). Continued ssDNA desorption may be caused by a surface rearrangement process in the adsorbed FBG layer,45 where protein spreading could be responsible for this observed ssDNA displacement over longer time scales.14,39 Hydrophobic interactions are posited to be the driving force for protein spreading on the SWCNT surface40 and consequently interfacial denaturation is presumed to be the dominant relaxation process, in addition to a smaller contribution from molecular reorientations.38
From the kinetic model fitting, the mass of protein adsorbed on the SWCNTs was consistently higher for FBG as compared to HSA for the same initial concentration of 40 μg/mL FAM-protein with 5 μg/mL (GT)6-Cy5-SWCNTs. Previous studies of differential protein adsorption to hydrophobic surfaces has demonstrated that fibrinogen forms well-packed layers, whereas more weakly adsorbed albumin forms less tightly packed, more mobile adsorption layers.40 Accordingly, we hypothesize that the seemingly higher protein surface coverage on the SWCNT points to the more tightly packed, if not multilayer formation, of FBG on the SWCNT surface.
Some potential shortcomings of our methodology include that the proposed elementary steps only approximate the true exchange mechanism, or that there are nonidealities present in the protein and/or ssDNA sorption behavior. Two alternative models to account for protein binding to surface-adsorbed ssDNA or protein were also attempted, but not pursued due to the poor quality of the fits (see section 4.5). Constraining the total surface sites during model fitting was also pursued, whereby would presumably be constant across different concentrations of protein added. Although the model fits to the experimental data are reasonable (Table S4 and Figure S10), we decided to fit the data with varying to capture the effect of concentration-dependent nonidealities in the system, such as nanotube–nanotube interactions, on all fit parameters. Finally, we note that the system is not truly equilibrated; therefore, a thermodynamic analysis was not pursued.44
3. CONCLUSIONS
Protein adsorption to nanoparticle surfaces is a major hindrance to the successful application of nanotechnologies in vivo. We have shown that incubation of two high-abundance blood plasma proteins, human serum albumin and fibrinogen, with ssDNA-SWCNT dopamine sensors causes significantly different degrees of sensor response attenuation. Developing an understanding of protein-sensor interactions is vital in circumventing this issue and establishing better practices for testing nanotechnologies for in vivo use. Previously established techniques to evaluate these effects implement surface-immobilized nanoparticles or exploit the intrinsic nIR fluorescence changes of SWCNTs. Yet, these methods do not track the fate of adsorbates and cannot quantify the fraction of free biomolecules in real-time, thus precluding quantitative and temporally resolved studies of the SWCNT protein corona composition. Though the SDBS-induced solvatochromic shift assay successfully identifies FBG as a protein of interest, this assay provides no mechanistic information on FBG binding nor can it distinguish between HSA and PBS control responses.
We have addressed these limitations in developing a method to quantitatively probe the kinetics of SWCNT corona exchange between ssDNA and protein adsorbates by monitoring fluorescence quenching of conjugated fluorophores in close proximity to SWCNT surfaces. Concentration curves were fit to a competitive adsorption model to extract kinetic parameters. Our method reveals that reduction of dopamine sensor performance correlates with quantity of adsorbed protein, where fibrinogen adsorbs to ssDNA-SWCNTs 168% more than albumin at the same concentration, and consequently leads to 26% more sensor attenuation. We demonstrate significantly greater SWCNT binding affinities for longer repeating ssDNA sequences, and for fibrinogen over albumin. These results bear significance in that albumin is the highest abundance blood protein and is therefore commonly regarded as an important component of the SWCNT corona. However, our results show that lower abundance proteins with higher SWCNT affinities may disproportionately contribute to the SWCNT corona, as has been previously suggested in orthogonal protein corona-nanoparticle studies.46,47 Preliminary findings from blood plasma and serum samples normalized to 40 μg/mL total protein concentration show that plasma and serum both caused significant attenuation of dopamine response in (GT)6-Cy5-SWCNTs, with 81.0 ± 0.9% and 80.7 ± 1.4% reduction in response, respectively (Figure S11a). However, plasma, which contains fibrinogen, caused a higher degree of ssDNA desorption, with plasma inducing a 1.64 ± 0.01 fold increase in (GT)6-Cy5 fluorescence vs a 1.39 ± 0.03 fold increase by serum (Figure S11b). Our results motivate the necessity to test SWCNT-based and other nanobiotechnologies in more representative bioenvironments, i.e., blood plasma rather than serum.
The method presented herein enables the study of corona formation dynamics of multiple biomolecular entities, with standard laboratory equipment, under varying solution conditions (e.g., ionic strength and pH). The generalizability of this assay allows for study of diverse corona exchange phenomena occurring on the surface of carbon nanotubes between a variety of biomolecular species. We demonstrate that RNA, phospholipids, and peptoids, molecules commonly used in SWCNT-mediated delivery, imaging, and sensing,3,48–50 all exhibit varying degrees of corona exchange with ssDNA-SWCNTs (Figure S12 and section S.2). Furthermore, we can examine the competitive adsorption of multiple fluorophore-conjugated proteins, such as FAM-HSA and Cy5-FBG onto ssDNA-SWCNTs, where increasing mass ratios of FBG to HSA results in reduced adsorption of the latter (Figure S13). Careful selection of fluorophores may enable further multiplexing, allowing tracking of more than two distinct molecular species simultaneously. Rationally designed labeling methodologies such as FRET may also enable the study of more complex interactions such as protein denaturation on the SWCNT surface. Implementation of this assay will facilitate more thorough deconvolution of factors driving protein corona formation and accordingly inform design principles for nanotechnologies resistant to protein corona-based biofouling and performance attenuation. In summary, the corona exchange assay we have developed will serve to enhance our still deficient understanding of how noncovalently bound polymers exchange on nanoparticle surfaces and, accordingly, enable the design and testing of nanobiotechnologies toward effective implementation in vivo.
4. MATERIALS AND METHODS
4.1. Preparation of ssDNA-SWCNT Complexes.
Single-stranded DNA with single-walled carbon nanotube (ssDNA-SWCNT) suspensions were prepared by combining 0.2 mg of mixed-chirality SWCNTs (small diameter HiPco SWCNTs, Nano-Integris) and 20 μM of ssDNA (custom ssDNA oligos with standard desalting, Integrated DNA Technologies, Inc.) in 1 mL of 0.01 M phosphate-buffered saline (PBS). Solutions were probe-tip sonicated for 10 min in an ice bath (3 mm probe tip set at 50% amplitude, 5–6 W, Cole-Parmer Ultrasonic Processor). Samples were centrifuged to pellet insoluble SWCNT bundles and contaminants (16 100 cfg for 30 min). Supernatant containing the product was collected. ssDNA-SWCNT solutions were stored at 4 °C until use. ssDNA-SWCNT concentration was determined via sample absorbance at 910 nm and the corresponding extinction coefficient .51 ssDNA-SWCNTs were diluted to a working concentration of 10 μg/mL in 0.1 M PBS.
Cyanine 5 (Cy5) was chosen as the ssDNA fluorophore label, with excitation maximum at 648 nm and emission maximum at 668 nm. The same suspension protocol was employed for preparation of fluorophore-labeled ssDNA-SWCNT complexes, using ssDNA-Cy5 (3′ or internally labeled Cy5-labeled custom ssDNA oligos with HPLC purification, Integrated DNA Technologies, Inc.) in place of unlabeled ssDNA. Internally labeled ssDNA-Cy5 includes Cy5 conjugated to the thymine at nucleotide position 6 (GTGTGT/iCy5/GTGTGT).
Total ssDNA adsorbed to SWCNTs was determined by a heat/surfactant elution process. This molar ratio of ssDNA:SWCNT was required to calculate the fraction of free vs bound ssDNA throughout the exchange process. Optimized elution conditions were achieved with salt and surfactant in the combination of 0.1 M PBS/0.1% (m/v) sodium dodecylbenzene sulfonate (SDBS), in agreement with prior literature demonstrating that SDBS disperses SWCNTs most effectively19,52 and without chirality bias.53 Freshly prepared ssDNA-Cy5-SWCNTs were diluted to a final concentration of 5 μg/mL in elution buffer, with a final volume of 150 μL in a PCR tube. Samples were heated at 95 °C for 1 h in a PCR thermocycler, transferred to a clean test tube, and centrifuged (16 100 cfg for 30 min) to pellet insoluble SWCNT bundles. 120 μL of supernatant containing the eluted ssDNA-Cy5 was collected. Fluorescence in the Cy5 channel was measured (see section 4.3) and compared to a standard curve of known ssDNA-Cy5 concentrations (ranging 0.01–1 μM) to correlate the Cy5 fluorescence measurement to ssDNA eluted concentration. This resulted in a mole ratio of 364 ± 2 (GT)6:SWCNT and 140 ± 7 (GT)15:SWCNT (both N = 8), in relative agreement with previous literature for (6,5) single chirality SWCNTs.54
4.2. Fluorophore-Labeling of Proteins.
N-Hydroxysuccinimide (NHS) ester chemistry was used to label proteins via conjugation to primary amine groups. Fluorescein (FAM) was chosen as the protein fluorophore label, with excitation maximum at 494 nm and emission maximum at 520 nm (FAM NHS ester 6-isomer, Lumiprobe). Lyophilized proteins were purchased: human serum albumin (HSA; from human plasma, ≤0.02% fatty acids, lot no. SLBZ2785, Millipore Sigma) and fibrinogen (FBG; from human plasma, 20 mM sodium citrate-HCl pH 7.4, lot no. 3169957, Millipore Sigma). FAM-proteins were prepared with 10 mg of protein reconstituted in 900 μL of 0.1 M sodium bicarbonate (pH 8.5) and 8-fold molar excess of FAM NHS ester solubilized in 100 μL DMSO. Solutions were combined, covered in foil, and incubated on a test tube rotator for 4 h. FAM-protein conjugates were twice purified to remove free FAM (Zeba 2 mL spin desalting columns with 40 kDa MWCO, Thermo Fisher Scientific) by washing with 0.1 M PBS three times (1000 cfg for 2 min), centrifuging with sample (1000 cfg for 3 min), and retaining sample in flow-through solution (repeating all steps twice with a new spin column). Protein concentration and degree of labeling (DOL) were determined by measuring the absorbance of the FAM-protein conjugate at the protein absorbance maximum, 280 nm (A280), and the fluorophore emission maximum, 494 nm (A494). Protein absorbance was corrected for the contribution of the fluorophore to A280 by subtracting A494 weighted by the correction factor (CF), an empirical constant of 0.17 for free FAM (from manufacturer). Protein and FAM concentrations were determined by the Beer–Lambert Law using either A280 for protein or A494 for FAM, with the corresponding extinction coefficients of ,55 ,56 and (from manufacturer). DOL was then calculated as the ratio of FAM to protein molar concentrations, yielding DOLFAM-HSA = 2.773 and DOLFAM-FBG = 0.608.
Free FAM NHS ester remaining after purification was quantified by polyacrylamide gel electrophoresis (PAGE) run according to the Laemmli protocol57 (adapted in Bio-Rad Mini-PROTEAN Tetra Cell manual). Briefly, purified FAM-protein conjugates were added to sodium dodecyl sulfate (SDS) reducing buffer (2% SDS, 5% β-mercaptoethanol, 0.066 M Tris-HCl) in a 1:2 ratio of sample to buffer. Samples were diluted such that 100 ng of FAM-HSA, 100 ng of FAM NHS ester, or 30 ng of FAM-FBG (due to lower labeling reaction yield) per 20 μL volume was applied per well. PAGE separation was carried out in 1 mm vertical mini gel format (Bio-Rad Mini-PROTEAN Tetra Cell) with a discontinuous buffer system under denaturing conditions. Gel composition was 12% acrylamide (total monomer), 0.375 M Tris-HCl, 0.1% SDS, 0.05% APS, 0.05% TEMED for the resolving gel and 12% acrylamide (total monomer), 0.125 M Tris-HCl, 0.1% SDS, 0.05% APS, 0.1% TEMED for the stacking gel. Electrode buffer was 25 mM Tris, 192 mM glycine, and 3.5 mM SDS (pH 8.3). Separation was run with 200 V for 35 min, gels were extracted, and the FAM label was visualized with a gel imager (Typhoon FLA 9500, 473 nm laser, General Electric; Figure S4). The FAM-protein conjugate is the higher band (approximately 66 kDa for FAM-HSA, 52–95 kDa for FAM-FBG) and the free, lighter molecular weight FAM NHS ester is the lower band (approximately 0.475 kDa). FAM fluorescence intensity was quantified with ImageJ (Table S1).
4.3. Visible Fluorescence Measurements.
Equal volumes of (GT)6-Cy5-SWCNT and FAM-tagged protein at 2× working concentration were added to a 96-well PCR plate (Bio-Rad) to a total volume of 50 μL. The plate was sealed using an optically transparent adhesive seal (Bio-Rad) and briefly spun down on a benchtop centrifuge. Fluorescence time series readings were taken using a Bio-Rad CFX96 Real Time qPCR System, scanning all manufacturer set color channels (FAM, HEX, Texas Red, Cy5, Quasar 705) every 30 s at 22.5 °C (lid heating off). Fluorescence time series were analyzed without default background correction. Note that concentration ranges of FAM-HSA (5–60 μg/mL) and FAM-FBG (40–160 μg/mL) were chosen to be in the linear fluorescence regime of the qPCR.
4.4. Near-Infrared Fluorescence Measurements.
Fluorescence spectra were collected with an inverted Zeiss microscope (20× objective, Axio Observer.D1) containing a custom filter cube set (800 nm SP FESH0800, 900 nm LP dichroic DMLP900R, 900 nm LP FELH900, ThorLabs) coupled to a Princeton Instruments spectrometer (SCT 320) and liquid nitrogen cooled Princeton Instruments InGaAs detector (PyLoN-IR 1024/1.7). Fluorescence measurements were done with a beam-expanded 721 nm laser (10–500 mW, OptoEngine LLC) excitation light source and 800–1400 nm emission wavelength range. Solution-phase measurements were acquired in a 384 well-plate format (1 s exposure time, 1 mW laser power). Protein solutions (final concentration 40 μg/mL) or PBS control were incubated with (GT)6-SWCNTs (final concentration 5 μg/mL in 0.1 M PBS). For each time point, an aliquot of these incubation solutions was added to a well (40 μL total volume) and an initial fluorescence spectrum was acquired. A total of 10 μL of dopamine was added to a final concentration of 200 μM prior to the second fluorescence acquisition. Fluorescence fold change was measured by taking the ratio of fluorescence intensities at 1200 nm between post- and preaddition of dopamine spectra.
Similarly, surfactant-induced solvatochromism was performed by collecting nIR fluorescence spectra pre- and 1 min postaddition of 0.5% (w/v) SDBS. Fluorescence fold change was defined as the ratio of integrated fluorescence intensity (800 to 1400 nm) between post- and preaddition of SDBS. Wavelength shift was measured relative to the wavelength of the (7,6) SWCNT chirality peak emission (initially 1130 nm) post-SDBS.
4.5. Kinetic Model.
Corona exchange kinetics were modeled by a system of simultaneous adsorption/desorption reactions. The model assumes that both ssDNA and protein adsorb/desorb reversibly to a fixed number of vacant SWCNT surface sites (eqs 1 and 2). Note that all modeling was done on a mass basis. This is in agreement with the general use of volume fractions in polymer thermodynamics.58 Here, we add the additional assumption that the biomolecules are of similar density. Modeling on a mass basis accounts for the widely varying molecular sizes between the two types of protein (HSA, 66.5 kDa, globular vs FBG, 340 kDa, long) and ssDNA ((GT)6, 3.7 kDa). The time-dependent differential equations governing ssDNA desorption and protein adsorption are as follows:
| (4) |
| (5) |
A least-squares regression was used to fit the model to fluorescence data and iterate model parameters. MATLAB 2019A’s ode15s solver was implemented to solve eqs 4 and 5 for free protein and DNA concentration curves given fit rate constants k1, k2, k3, k4, and the total concentration of open sites,. Relative error between the model fit and experimental data was calculated and averaged over all data points to yield mean relative error (MRE). Sensitivity analysis on initial conditions was performed to minimize this fit error. Then 48 unique combinations of rate parameter initial conditions were analyzed as inputs to the nonlinear least-squares solver (lsqcurvefit) in our MATLAB model. The optimal set of initial conditions for each protein was chosen as that which yielded a low MRE between fit and experimental data and a low standard error among fit parameters for each of the four protein concentrations. Each rate parameter was individually fit to each experiment, yielding 20 total fit parameters from each initial condition (Table S2). Final ssDNA and protein fit MRE were all <5% (Table S2). Optimized initial conditions and resultant rate parameters for HSA and FBG are reported in Table S3.
Two alternative models were also attempted: in model 2, protein was able to bind to surface-adsorbed ssDNA and in model 3, protein was able to bind to surface-adsorbed protein. However, these models both produced significantly higher error in fits. Model 2 addressed the possibility of protein binding on top of ssDNA bound directly to the SWCNT. For FBG experiments fit with model 2, most fits overestimated FBG adsorption and many fits displayed incorrect concavity for the ssDNA desorption. For HSA experiments fit with model 2, the protein data was generally fit well but the ssDNA fits exhibited either a maximum or produced linear fits. Model 3 addressed the possibility of protein binding on top of protein bound directly to the SWCNT. For FBG experiments, model 3 overestimated both protein adsorption and ssDNA desorption. For HSA experiments, model 3 generally fit the protein data well, yet did not capture ssDNA dynamics as a function of concentration. Although the higher errors associated with model 2 and 3 do not rule out these nanoscale mechanistic possibilities, the simple model of independent binding does overall fit the data much more closely between both protein and ssDNA curves within the same experiment, as well as binding dynamics as a function of varying concentration.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support of an NIH NIDA CEBRA award No. R21DA044010 (to M.P.L.), a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI; to M.P.L.), the Simons Foundation (to M.P.L.), a Stanley Fahn PDF Junior Faculty Grant with Award No. PF-JFA-1760 (to M.P.L.), a Beckman Foundation Young Investigator Award (to M.P.L.), and a DARPA Young Investigator Award (to M.P.L.). M.P.L. is a Chan Zuckerberg Biohub investigator. R.L.P., D.Y., and W.C. all acknowledge the support of NSF Graduate Research Fellowships (NSF DGE 1752814). The authors thank L. Chio working with Lawrence Berkeley National Laboratory’s Molecular Foundry and N. Goh for providing peptoid and RNA samples, respectively. The authors acknowledge fruitful discussions with A. Crothers and A. Fischer.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.9b09617
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.9b09617.
Figures S1–S13, Tables S1–S4, and sections S.1–S.3 are included for further experimental validation (PDF)
The authors declare no competing financial interest.
Contributor Information
Rebecca L. Pinals, University of California at Berkeley, Berkeley, California.
Darwin Yang, University of California at Berkeley, Berkeley, California.
Alison Lui, University of California at Berkeley, Berkeley, California.
Wendy Cao, University of California at Berkeley, Berkeley, California.
Markita P. Landry, University of California at Berkeley, Berkeley, California, Innovative Genomics Institute (IGI), Berkeley, California, University of California at Berkeley, Berkeley, California, and Chan-Zuckerberg Biohub, San Francisco, California
REFERENCES
- (1).Zhang J; Landry MP; Barone PW; Kim J-H; Lin S; Ulissi ZW; Lin D; Mu B; Boghossian AA; Hilmer AJ; Rwei A; Hinckley AC; Kruss S; Shandell MA; Nair N; Blake S; Sen F; Sen S; Croy RG; Li D; Yum K; Ahn J-H; Jin H; Heller DA; Essigmann JM; Blankschtein D; Strano MS. Molecular Recognition Using Corona Phase Complexes Made of Synthetic Polymers Adsorbed on Carbon Nanotubes. Nat. Nanotechnol. 2013, 8 (12), 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Liu Z; Tabakman S; Welsher K; Dai H. Carbon Nanotubes in Biology and Medicine: In Vitro and in Vivo Detection, Imaging and Drug Delivery. Nano Res. 2009, 2 (2), 85–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Demirer GS; Zhang H; Matos JL; Goh NS; Cunningham FJ; Sung Y; Chang R; Aditham AJ; Chio L; Cho M-J; Staskawicz B; Landry MP. High Aspect Ratio Nanomaterials Enable Delivery of Functional Genetic Material without DNA Integration in Mature Plants. Nat. Nanotechnol. 2019, 1, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zheng M; Jagota A; Semke ED; Diner BA; Mclean RS; Lustig SR; Richardson RE; Tassi NG. DNA-Assisted Dispersion and Separation of Carbon Nanotubes. Nat. Mater. 2003, 2 (5), 338–342. [DOI] [PubMed] [Google Scholar]
- (5).Marchesan S; Prato M. Under the Lens: Carbon Nanotube and Protein Interaction at the Nanoscale. Chem. Commun. 2015, 51 (21), 4347–4359. [DOI] [PubMed] [Google Scholar]
- (6).Cedervall T; Lynch I; Lindman S; Berggård T; Thulin E; Nilsson H; Dawson KA; Linse S. Understanding the Nanoparticle–Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (7), 2050–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nel AE; Madler L; Velegol D; Xia T; Hoek EMV;̈ Somasundaran P; Klaessig F; Castranova V; Thompson M. Understanding Biophysicochemical Interactions at the Nano–Bio Interface. Nat. Mater. 2009, 8 (7), 543–557. [DOI] [PubMed] [Google Scholar]
- (8).Gravely M; Safaee MM; Roxbury D. Biomolecular Functionalization of a Nanomaterial to Control Stability and Retention within Live Cells. Nano Lett. 2019, 19 (9), 6203–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bisker G; Bakh NA; Lee MA; Ahn J; Park M; O’Connell EB; Iverson NM; Strano MS. Insulin Detection Using a Corona Phase Molecular Recognition Site on Single-Walled Carbon Nanotubes. ACS Sens. 2018, 3 (2), 367–377. [DOI] [PubMed] [Google Scholar]
- (10).Beyene AG; McFarlane IR; Pinals RL; Landry MP. Stochastic Simulation of Dopamine Neuromodulation for Implementation of Fluorescent Neurochemical Probes in the Striatal Extracellular Space. ACS Chem. Neurosci. 2017, 8 (10), 2275–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Beyene AG; Alizadehmojarad AA; Dorlhiac G; Goh N; Streets AM; Král P; Vuković L; Landry MP Ultralarge Modulation of Fluorescence by Neuromodulators in Carbon Nanotubes Functionalized with Self-Assembled Oligonucleotide Rings. Nano Lett. 2018, 18 (11), 6995–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Beyene AG; Delevich K; Bonis-O’Donnell JTD; Piekarski DJ; Lin WC; Thomas AW; Yang SJ; Kosillo P; Yang D; Prounis GS; Wilbrecht L; Landry MP. Imaging Striatal Dopamine Release Using a Nongenetically Encoded near Infrared Fluorescent Catecholamine Nanosensor. Science Advances 2019, 5 (7), No. eaaw3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kruss S; Landry MP; Vander Ende E; Lima BMA; Reuel NF; Zhang J; Nelson J; Mu B; Hilmer A; Strano M. Neurotransmitter Detection Using Corona Phase Molecular Recognition on Fluorescent Single-Walled Carbon Nanotube Sensors. J. Am. Chem. Soc. 2014, 136 (2), 713–724. [DOI] [PubMed] [Google Scholar]
- (14).Dijt JC; Cohen Stuart MA; Fleer GJ. Competitive Adsorption Kinetics of Polymers Differing in Length Only. Macromolecules 1994, 27 (12), 3219–3228. [Google Scholar]
- (15).Landry MP; Vukovic L; Kruss S; Bisker G; Landry AM; ´Islam S; Jain R; Schulten K; Strano MS. Comparative Dynamics and Sequence Dependence of DNA and RNA Binding to Single Walled Carbon Nanotubes. J. Phys. Chem. C 2015, 119 (18), 10048–10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Silvera-Batista CA; Wang RK; Weinberg P; Ziegler KJ. Solvatochromic Shifts of Single-Walled Carbon Nanotubes in Nonpolar Microenvironments. Phys. Chem. Chem. Phys. 2010, 12 (26), 6990–6998. [DOI] [PubMed] [Google Scholar]
- (17).Larsen BA; Deria P; Holt JM; Stanton IN; Heben MJ; Therien MJ; Blackburn JL. Effect of Solvent Polarity and Electrophilicity on Quantum Yields and Solvatochromic Shifts of Single-Walled Carbon Nanotube Photoluminescence. J. Am. Chem. Soc. 2012, 134 (30), 12485–12491. [DOI] [PubMed] [Google Scholar]
- (18).Choi JH; Strano MS. Solvatochromism in Single-Walled Carbon Nanotubes. Appl. Phys. Lett. 2007, 90 (22), 223114. [Google Scholar]
- (19).Oh H; Sim J; Ju S-Y. Binding Affinities and Thermodynamics of Noncovalent Functionalization of Carbon Nanotubes with Surfactants. Langmuir 2013, 29 (35), 11154–11162. [DOI] [PubMed] [Google Scholar]
- (20).Harvey JD; Jena PV; Baker HA; Zerze GH; Williams RM; Galassi TV; Roxbury D; Mittal J; Heller DA. A Carbon Nanotube Reporter of MicroRNA Hybridization Events in Vivo. Nature Biomedical Engineering 2017, 1 (4), 0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shankar A; Mittal J; Jagota A. Binding between DNA and Carbon Nanotubes Strongly Depends upon Sequence and Chirality. Langmuir 2014, 30 (11), 3176–3183. [DOI] [PubMed] [Google Scholar]
- (22).Roxbury D; Tu X; Zheng M; Jagota A. Recognition Ability of DNA for Carbon Nanotubes Correlates with Their Binding Affinity. Langmuir 2011, 27 (13), 8282–8293. [DOI] [PubMed] [Google Scholar]
- (23).Jena PV; Safaee MM; Heller DA; Roxbury D. DNA–Carbon Nanotube Complexation Affinity and Photoluminescence Modulation Are Independent. ACS Appl. Mater. Interfaces 2017, 9 (25), 21397–21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zheng Y; Bachilo SM; Weisman RB. Enantiomers of Single-Wall Carbon Nanotubes Show Distinct Coating Displacement Kinetics. J. Phys. Chem. Lett. 2018, 9, 3793–3797. [DOI] [PubMed] [Google Scholar]
- (25).Jeng ES; Moll AE; Roy AC; Gastala JB; Strano MS. Detection of DNA Hybridization Using the Near-Infrared Band-Gap Fluorescence of Single-Walled Carbon Nanotubes. Nano Lett. 2006, 6 (3), 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Barone PW; Baik S; Heller DA; Strano MS. Near-Infrared Optical Sensors Based on Single-Walled Carbon Nanotubes. Nat. Mater. 2004, 4 (1), 86–92. [DOI] [PubMed] [Google Scholar]
- (27).Zhu Z; Yang R; You M; Zhang X; Wu Y; Tan W. Single-Walled Carbon Nanotube as an Effective Quencher. Anal. Bioanal. Chem. 2010, 396 (1), 73–83. [DOI] [PubMed] [Google Scholar]
- (28).Umemura K; Sato S; Bustamante G; Ye JY. Using a Fluorescence Quenching Method to Detect DNA Adsorption onto Single-Walled Carbon Nanotube Surfaces. Colloids Surf., B 2017, 160, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Anderson NL; Anderson NG. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteomics 2002, 1 (11), 845–867. [DOI] [PubMed] [Google Scholar]
- (30).Lowe GDO; Rumley A; Mackie IJ. Plasma Fibrinogen. Ann. Clin. Biochem. 2004, 41 (6), 430–440. [DOI] [PubMed] [Google Scholar]
- (31).Foroozandeh P; Aziz AA. Merging Worlds of Nanomaterials and Biological Environment: Factors Governing Protein Corona Formation on Nanoparticles and Its Biological Consequences. Nanoscale Res. Lett. 2015, 10 (1), 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Monopoli MP; Walczyk D; Campbell A; Elia G; Lynch I; Baldelli Bombelli F; Dawson KA. Physical-Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133 (8), 2525–2534. [DOI] [PubMed] [Google Scholar]
- (33).Lundqvist M; Stigler J; Elia G; Lynch I; Cedervall T; Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (38), 14265–14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Brunecker FK; Schöppler F; Hertel T. Interaction of Polymers with Single-Wall Carbon Nanotubes. J. Phys. Chem. C 2016, 120 (18), 10094–10103. [Google Scholar]
- (35).Lu N; Sui Y; Tian R; Peng Y-Y. Adsorption of Plasma Proteins on Single-Walled Carbon Nanotubes Reduced Cytotoxicity and Modulated Neutrophil Activation. Chem. Res. Toxicol. 2018, 31 (10), 1061–1068. [DOI] [PubMed] [Google Scholar]
- (36).Salvador-Morales C; Flahaut E; Sim E; Sloan J; HGreen ML; Sim RB. Complement Activation and Protein Adsorption by Carbon Nanotubes. Mol. Immunol. 2006, 43 (3), 193–201. [DOI] [PubMed] [Google Scholar]
- (37).Ge C; Du J; Zhao L; Wang L; Liu Y; Li D; Yang Y; Zhou R; Zhao Y; Chai Z; Chen C. Binding of Blood Proteins to Carbon Nanotubes Reduces Cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (41), 16968–16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wertz CF; Santore MM. Adsorption and Relaxation Kinetics of Albumin and Fibrinogen on Hydrophobic Surfaces: Single-Species and Competitive Behavior. Langmuir 1999, 15 (26), 8884–8894. [Google Scholar]
- (39).Mubarekyan E; Santore MM. Adsorption and Exchange Dynamics in Aging Hydroxyethylcellulose Layers on Silica. J. Colloid Interface Sci. 2000, 227 (2), 334–344. [DOI] [PubMed] [Google Scholar]
- (40).Wertz CF; Santore MM. Effect of Surface Hydrophobicity on Adsorption and Relaxation Kinetics of Albumin and Fibrinogen: Single-Species and Competitive Behavior. Langmuir 2001, 17 (10), 3006–3016. [Google Scholar]
- (41).Jungbauer LM; Yu C; Laxton KJ; LaDu MJ. Preparation of Fluorescently-Labeled Amyloid-Beta Peptide Assemblies: The Effect of Fluorophore Conjugation on Structure and Function. J. Mol. Recognit. 2009, 22 (5), 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Albertorio F; Hughes ME; Golovchenko JA; Branton D. Base Dependent DNA–Carbon Nanotube Interactions: Activation Enthalpies and Assembly–Disassembly Control. Nanotechnology 2009, 20 (39), 395101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Röcker C; Pötzl M; Zhang F; Parak WJ; Nienhaus G. U. A Quantitative Fluorescence Study of Protein Monolayer Formation on Colloidal Nanoparticles. Nat. Nanotechnol. 2009, 4 (9), 577–580. [DOI] [PubMed] [Google Scholar]
- (44).Latour RA. The Langmuir Isotherm: A Commonly Applied but Misleading Approach for the Analysis of Protein Adsorption Behavior. J. Biomed. Mater. Res., Part A 2015, 103 (3), 949–958. [DOI] [PubMed] [Google Scholar]
- (45).Dijt JC; Cohen Stuart MA; Fleer GJ. Surface Exchange Kinetics of Chemically Different Polymers. Macromolecules 1994, 27 (12), 3229–3237. [Google Scholar]
- (46).De Paoli SH; Diduch LL; Tegegn TZ; Orecna M; Strader MB; Karnaukhova E; Bonevich JE; Holada K; Simak J. The Effect of Protein Corona Composition on the Interaction of Carbon Nanotubes with Human Blood Platelets. Biomaterials 2014, 35 (24), 6182–6194. [DOI] [PubMed] [Google Scholar]
- (47).Tenzer S; Docter D; Kuharev J; Musyanovych A; Fetz V; Hecht R; Schlenk F; Fischer D; Kiouptsi K; Reinhardt C; Landfester K; Schild H; Maskos M; Knauer SK; Stauber RH. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat. Nanotechnol. 2013, 8 (10), 772–781. [DOI] [PubMed] [Google Scholar]
- (48).Bisker G; Dong J; Park HD; Iverson NM; Ahn J; Nelson JT; Landry MP; Kruss S; Strano MS. Protein-Targeted Corona Phase Molecular Recognition. Nat. Commun. 2016, 7, 10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Chio L; Del Bonis-O’Donnell JT; Kline MA; Kim JH; McFarlane IR; Zuckermann RN; Landry MP. Electrostatic Assemblies of Single-Walled Carbon Nanotubes and SequenceTunable Peptoid Polymers Detect a Lectin Protein and Its Target Sugars. Nano Lett. 2019. DOI: 10.1021/acs.nanolett.8b04955. [DOI] [PubMed] [Google Scholar]
- (50).Godin AG; Varela JA; Gao Z; Danne N; Dupuis JP; ´Lounis B; Groc L; Cognet L. Single-Nanotube Tracking Reveals the Nanoscale Organization of the Extracellular Space in the Live Brain. Nat. Nanotechnol. 2017, 12 (3), 238–243. [DOI] [PubMed] [Google Scholar]
- (51).Roxbury D; Jena PV; Shamay Y; Horoszko CP; Heller DA. Cell Membrane Proteins Modulate the Carbon Nanotube Optical Bandgap via Surface Charge Accumulation. ACS Nano 2016, 10 (1), 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Moore VC; Strano MS; Haroz EH; Hauge RH; Smalley RE; Schmidt J; Talmon Y. Individually Suspended Single-Walled Carbon Nanotubes in Various Surfactants. Nano Lett. 2003, 3 (10), 1379–1382. [Google Scholar]
- (53).Okazaki T; Saito T; Matsuura K; Ohshima S; Yumura M; Iijima S. Photoluminescence Mapping of “As-Grown” Single-Walled Carbon Nanotubes: A Comparison with Micelle-Encapsulated Nanotube Solutions. Nano Lett. 2005, 5 (12), 2618–2623. [DOI] [PubMed] [Google Scholar]
- (54).Nißler R; Mann FA; Chaturvedi P; Horlebein J; Meyer D; Vuković L; Kruss S. Quantification of the Number of Adsorbed DNA Molecules on Single-Walled Carbon Nanotubes. J. Phys. Chem. C 2019, 123 (8), 4837–4847. [Google Scholar]
- (55).Gill SC; von Hippel PH. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal. Biochem. 1989, 182 (2), 319–326. [DOI] [PubMed] [Google Scholar]
- (56).Marder VJ; Shulman NR; Carroll WR. High Molecular Weight Derivatives of Human Fibrinogen Produced by Plasmin I. Physicochemical and Immunological Characterization. J. Biol. Chem. 1969, 244 (8), 2111–2119. [PubMed] [Google Scholar]
- (57).Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227 (5259), 680–685. [DOI] [PubMed] [Google Scholar]
- (58).Flory PJ. Principles of Polymer Chemistry; Cornell University Press: Ithaca, NY, 1953. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.