Abstract
Purpose:
The uncommon EGFR exon 19 deletion (ex19del), L747_A750>P, demonstrates reduced sensitivity to osimertinib compared to the common ex19del, E746_A750del in preclinical models. The clinical efficacy of osimertinib in patients with non-small cell lung cancer (NSCLC) harboring L747_A750>P and other uncommon ex19dels is not known.
Design:
The AACR GENIE database was interrogated to characterize the frequency of individual ex19dels relative to other variants, and a multi-center retrospective cohort was used to compare clinical outcomes for patients with tumors harboring E746_A750del, L747_A750>P, and other uncommon ex19dels who received osimertinib in the first line (1L) or in second or later lines of therapy and were T790M+ (≥2L).
Results:
Ex19dels comprised 45% of EGFR mutations, with 72 distinct variants ranging in frequency from 28.1% (E746_A750del) to 0.03%, with L747_A750>P representing 1.8% of the EGFR mutant cohort. In our multi-institution cohort (N=200), E746_A750del was associated with significantly prolonged progression free survival (PFS) with 1L osimertinib vs. L747_A750>P (median 21.3 months [95% CI 17.0–31.7] vs. 11.7 months [10.8–29.4], adjusted hazard ratio [HR] 0.52 [0.28–0.98] p=0.043). Osimertinib efficacy in patients with other uncommon ex19dels varied based on the specific mutation present.
Conclusions:
The ex19del L747_A750>P is associated with inferior PFS compared to the common E746_A750del mutation in patients treated with 1L osimertinib. Understanding differences in osimertinib efficacy among EGFR ex19del subtypes could alter management of these patients in the future.
Introduction:
In-frame deletions in exon 19 account for approximately 45% of all EGFR mutations in non-small cell lung cancer (NSCLC). Although exon 19 deletions are traditionally categorized and treated as possessing uniform sensitivity to EGFR tyrosine kinase inhibitors (TKIs), this group encompasses dozens of unique deletions and complex insertion-deletions (indels) that occur with variable frequency. The clinical implications of this heterogeneity are poorly understood. Data from the Catalogue of Somatic Mutations in Cancer (COSMIC) suggest that approximately 69% of exon 19 deletion mutations encode a dominant variant, or “common deletion,” an in-frame deletion that spans from glutamate 746 to alanine 750 (E746_A750del) in the EGFR tyrosine kinase domain, encompassing the region between the β3 strand and the αC helix1–3. However, “uncommon” variants comprise the remainder of the group, some of which occur at higher relative frequencies than others, including L747-P753>S (6% of exon 19 deletions), L747-T751 (5%), L747-A750>P (4%), E746-S752>V (3%), and L747-S752 (2%)1.
For patients with advanced NSCLC harboring exon 19 deletions or L858R point mutations, the third-generation EGFR TKI, osimertinib, has been established as the standard-of-care therapy for untreated patients as well as for patients with T790M-mediated acquired resistance to first- or second- generation EGFR TKIs4–7. The FLAURA trial, a phase III study investigating osimertinib in previously untreated patients with EGFR mutant NSCLC (exon 19 deletions and L858R mutations) showed a median overall survival for patients with exon 19 deletions of >40 months, but did not report individual mutation frequencies or allele-specific efficacy7. Since uncommon deletion variants presumably represent less than one-third of this cohort, the efficacy outcomes for the entire subgroup are likely to be driven by those patients with tumors that harbor the common deletion E746_A750del. Retrospective analyses have been conducted to characterize outcomes for patients with uncommon exon 19 deletions treated with EGFR TKIs, but these studies all have limitations8–20. Specifically, they use composite cohorts rather than comparing outcomes between individual variants, and few have included patients treated with osimertinib. The sensitivity of individual uncommon exon 19 deletion variants to osimertinib therefore remains unknown.
In preclinical models, the uncommon exon 19 deletion, L747_A750>P, exhibits reduced sensitivity to the first-generation EGFR TKI erlotinib and the third generation TKI osimertinib, while retaining sensitivity to the second-generation inhibitor, afatinib1,21. This manifests clinically in markedly inferior progression-free and overall survival outcomes for patients with tumors harboring L747_A750>P treated with erlotinib compared to those with E746_A750del. How this discovery impacts clinical outcomes with osimertinib was previously unknown. In current clinical practice, TKI therapy is not tailored to specific activating exon 19 deletions, and osimertinib has become the preferred first-line TKI for all patients with advanced NSCLC harboring EGFR exon 19 deletions as well as the de facto standard for patients with T790M-positive disease following progression on first- or second-generation EGFR TKIs.
Here, we report the prevalence of L747_A750>P and additional uncommon exon 19 deletions relative to other EGFR mutation subtypes using the large AACR Genomics Evidence Neoplasia Information Exchange (GENIE) database version 11.022. Using a multicenter real-world cohort, we also examined the efficacy of osimertinib in patients with tumors harboring the L747_A750>P mutation or other rare exon 19 deletions compared to those with E746_A750del.
Materials and Methods:
AACR GENIE Analysis
AACR Project GENIE is an international cancer registry that links next-generation genomic sequencing data with clinical information for patients treated at 18 contributing institutions23. This dataset was queried on April 6, 2022, to assess the prevalence of individual EGFR mutation subtypes, including exon 19 deletions, among a large cohort of patients with EGFR mutant NSCLC. GENIE Cohort v11.0-public contains 136,096 samples from 121,221 patients. The EGFR mutated NSCLC cohort consists of 4182 samples from 3501 patients. Raw data were extracted from cBioPortal for these patients. Those with founder EGFR mutations or compound mutations characterized by OncoKB as “oncogenic” or “likely oncogenic” were included in the analysis (n=3194)24. Mutation frequencies were then calculated.
Multicenter Cohort Study:
Study Design:
For the multicenter, retrospective cohort study, patients enrolled in Institutional Review Board-approved protocols at Yale University School of Medicine, Stanford University, Massachusetts General Hospital, Memorial Sloan Kettering Cancer Center, University of Pennsylvania School of Medicine, and the University of Colorado School of Medicine were included. All patients had metastatic NSCLC harboring EGFR exon 19 deletions. Due to the rare occurrence of certain EGFR exon 19 mutations, most participating institutions contributed only patients with uncommon exon 19 deletions to our cohort. Therefore, our cohort is enriched for uncommon EGFR exon 19 deletions and does not reflect the naturally occurring frequencies of these mutations in the EGFR positive NSCLC population.
EGFR mutations were identified at the time of diagnosis for each patient using the treating institution’s preferred next generation or targeted sequencing platform performed on DNA isolated from the patient’s tumor or using a validated, commercial cell-free DNA assay performed on the patient’s plasma concurrent with the diagnosis of NSCLC. All patients received at least 30 days of osimertinib treatment, either as first-line therapy (1L) or in second or later lines of therapy (≥2L). Patients treated with ≥2L osimertinib required documentation of the T790M mutation by sequencing performed on tumor tissue or plasma cell-free DNA after initial progression on first- or second-generation EGFR TKI. Patients were excluded if they had insufficient pathologic or clinical data.
Data Collection:
Relevant data were extracted from the electronic medical record for patients meeting study criteria, including EGFR activating mutation, demographic information, smoking history, clinicopathologic characteristics, treatment history, and disease control and survival outcomes. The primary study outcome was progression-free survival (PFS, time from osimertinib initiation to clinically significant growth of existing lesions or new lesions on imaging or death). Additional time-to-event outcomes assessed included overall survival (OS, time from osimertinib initiation to death). Disease control rate was defined as the percentage of patients achieving any response other than progression of disease (clinically significant growth of existing lesions or new lesions on imaging, ascertained by review of the treating oncologist’s notes and imaging reports) at the first radiographic assessment.
Statistical Analysis:
The primary comparison was between patients with tumors harboring the common EGFR exon 19 deletion E746_A750del vs those harboring L747_A750>P. All statistical analyses were performed using SAS version 9.4 (SAS Institute Inc, Cary, North Carolina). All p-values were computed for categorical variables using the Chi-squared test or Fisher’s Exact Test depending on cell counts. For continuous variables, Student’s t test or the Wilcoxon Rank Sum test was used, depending on normality. A p-value < 0.05 represented a statistically significant difference for all analyses. PFS and OS were estimated using the Kaplan-Meier method, whereas the Cox proportional hazards model was used to generate adjusted hazard ratios with 95% confidence intervals for PFS and OS, adjusting for baseline covariates of age, sex, race, and smoking history. Where possible, 95% confidence intervals were generated for median PFS and OS. For time-to-event analyses, censoring occurred at the time of last clinical follow-up. For first-line osimertinib analyses, models were censored at the time when no patients remained at risk in the L747_A750>P group to minimize the effect of variable follow-up time on outcomes between cohorts. Multivariate logistic regression was used to estimate the odds of achieving progression-free survival of >12 months (PFS>12 months).
For the exploratory cohort comprised of patients with tumors harboring other uncommon exon 19 deletions, analyses were purely descriptive, given the small sample sizes of individual variants. For 3 individual variants with the largest representation in our cohort (other than E746_A750del and L747_A750>P) we performed Kaplan Meier analyses to estimate median PFS with first-line osimertinib and ≥2L osimertinib. These variants included E746_S752>V, L747_P753>S, and L747_T751del. No formal statistical comparisons were performed for these analyses given the small numbers of patients.
Data Collection:
Data analyzed in this study were obtained from the AACR GENIE database version 11.0-public at AACR Project GENIE cBioPortal at https://genie.cbioportal.org/. Clinical data for the patients included in the multi-institution cohort in this study are not publicly available so as not to compromise patient privacy. Patients were enrolled in institutional review board (IRB)-approved protocols at the respective treating centers involved in this study and henceforth consented to data analysis. Queries for data access and requests for information regarding institutional protocolsmay be directed to the corresponding author.
Results:
Frequency of EGFR Mutations:
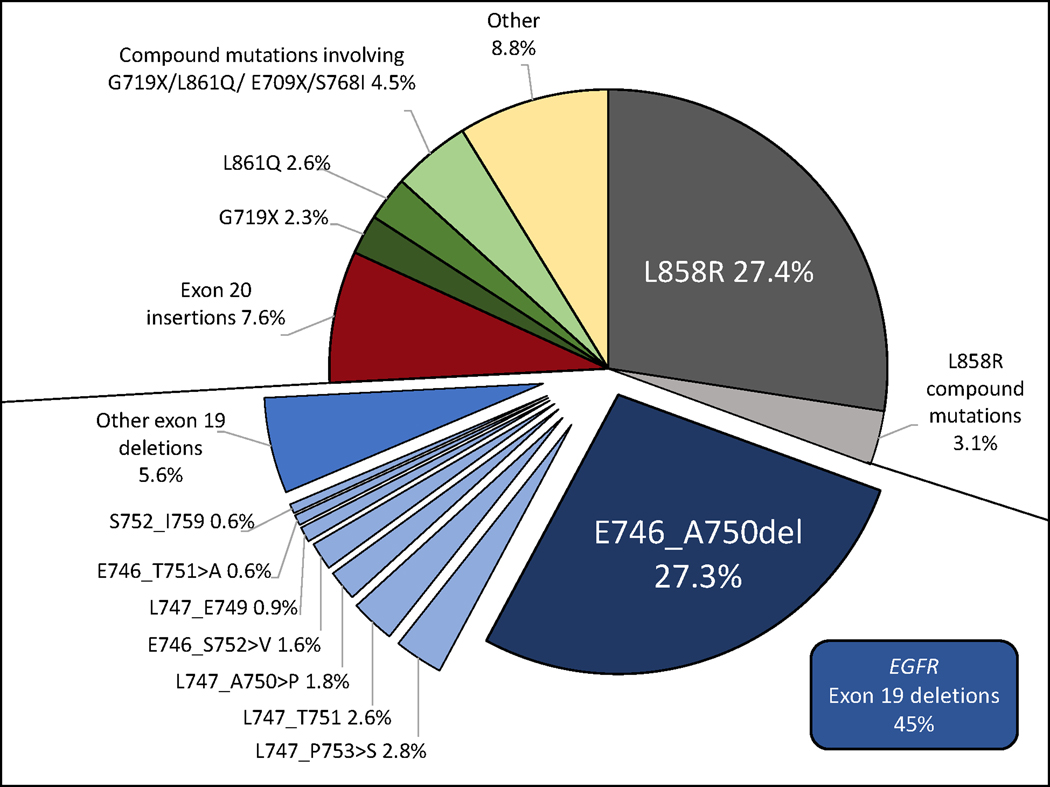
Using the AACR GENIE database version 11.0, we found that exon 19 deletions made up 45% of EGFR mutations ( n= 1438/3194). E746_A750del was the most common exon 19 deletion (27.3% of all EGFR mutations), followed by L747_P753>S (2.8%), L747_T751del (2.6%), L747_A750>P (1.8%), E746_S752>V (1.6%), L747_E749del (0.9%), E746_T751delinsA (0.6%) and S752_I759del (0.6%). The remaining 184 patients with tumors harboring uncommon exon 19 deletions (5.6%) had 64 unique variants ranging in frequency from 0.03%−0.37%. Tumors from 25 patients (0.8%) harbored compound mutations involving exon 19 deletions. The frequencies of individual exon 19 deletions as well as other baseline EGFR mutations from this cohort are depicted in Figure 1.
Figure 1.
EGFR mutation frequencies in AACR GENIE Cohort v11.0 public (N=3194). The ‘other ex19dels’ are comprised of 184 patients with 64 low frequency variants (each with prevalence <0.5%). ‘Other’ is all mutations not otherwise classified in this figure.
Multicenter Cohort Study
Patients:
Data from 341 patients from 6 institutions were analyzed, from which 200 patients with tumors with exon 19 deletions met inclusion criteria (Supplementary Figure 1). This included 122 patients with tumors harboring E746_A750del (n=86) and L747_A750>P (n=36), and 78 patients with tumors harboring other uncommon exon 19 deletions . A similar proportion of patients with common exon 19 deletions (36%, 49/135) and uncommon exon 19 deletions (40%, 75/189) met prespecified criteria for exclusion from our study, and 17 patients were excluded due to insufficient information on the specific type of exon 19 deletion present. Baseline characteristics for patients with tumors harboring E746_A750del and L747_A750>P are shown in Table 1. All characteristics that we examined were similar between groups.
Table 1.
Patient and treatment characteristics for those with NSCLC harboring EGFR E746_A750del vs. L747_A750>P in multi-institution cohort.
| Mutation | |||
|---|---|---|---|
| Patient Characteristic | E746_A750del (N = 86) | L747_A750>P (N = 36) | P Value |
|
| |||
| Age at Diagnosis (Median (IQR)) | 61.5 (54.0 – 69.0) | 66.5 (53.5 – 71.5) | 0.23 |
| Female | 46 (53.5%) | 25 (69.4%) | 0.10 |
|
| |||
| Asian Race | 25 (29.1%) | 12 (33.3%) | 0.64 |
| Former or current smoking history | 30 (34.9%) | 11 (30.6%) | 0.64 |
|
| |||
| Osimertinib Line of Therapy | 0.07 | ||
|
| |||
| First (1L) | 55 (64.0%) | 29 (80.6%) | |
|
| |||
| Second or Later (≥2L) | 31 (36.0%) | 7 (19.4%) | |
Clinical outcomes with first-line osimertinib in patients with tumors with the EGFR L747_A750>P mutation
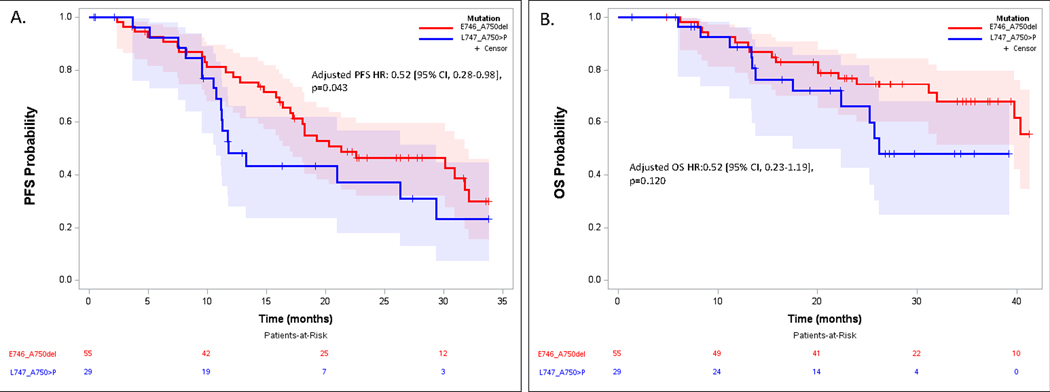
Patients with tumors harboring a E746_A750del had a significantly prolonged PFS compared to those with L747_A750>P (median 21.3 months [95% CI 17.0–31.7] vs. 11.7 months [10.8–29.4], adjusted hazard ratio [HR] 0.52 [95% CI, 0.28–0.98, p=0.043], Figure 2A). In addition, 79% (95% CI 68%−90%) of patients with E746_A750del positive tumors were progression-free at 12 months compared with 48% (95% CI 33%−72%) of those with the L747_A750>P mutation (Odds Ratio 4.14 (95% CI 1.41–12.15), p=0.0097). There was a similar trend for OS: median OS was not reached (NR) at a maximum 40 months of follow-up among those with E746_A750del vs 26 months for L747_A750>P (adjusted HR 0.52 [95% CI, 0.23–1.19], p=0.120, Figure 2B). All patients, regardless of baseline exon 19 deletion, experienced disease control with 1L osimertinib.
Figure 2.
Progression Free Survival (A) and Overall Survival (B) for patients with tumors harboring E746_A750del vs. L747_A750>P treated with first-line osimertinib.
Clinical outcomes with osimertinib in second line and beyond in patients with EGFR L747_A750>P positive tumors.
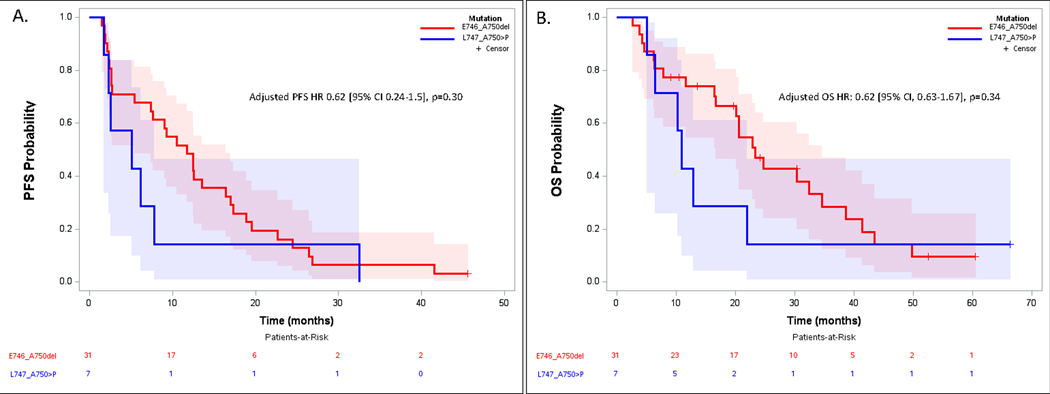
Median PFS was 11.7 months (95% CI 5.4–16.4) for the E746_A750del group treated with ≥2L osimertinib vs 5.1 months (95% CI 1.8–7.8) for the L747_A750>P group (PFS HR 0.62 [95% CI 0.24–1.5], p=0.30). The odds of achieving PFS>12 months exhibited a non-significant trend in favor of E746_A750del (OR 7.2 [95% CI, 0.59–87.6], p=0.12, Figure 3A) with 12-month PFS 48% (95% CI 31%−66%) vs. 14% (95% CI 0%−40%) for E746_A750del and L747_A750>P, respectively. Median OS was 23.3 months (95% CI 16.7–34.6) for E746_A750del vs. 11.0 months (95% CI 5.1–22.0) for L747_A750>P (OS HR 0.62 [95% CI, 0.63–1.67], p=0.34, Figure 3B). Disease control rate was 77% (95% CI 63%−92%) for patients with E746_A750del mutant NSCLC vs 57% (95% CI 20%−94%) for those with L747_A750>P mutations (adjusted odds ratio 3.1 [95% CI, 0.50–19], p=0.22, favoring E746_A750del).
Figure 3.
Progression Free Survival (A) and Overall Survival (B) for patients with tumors harboring L746_A750del vs. L747_A750>P treated with ≥2nd line osimertinib after T790M+ acquired resistance to first- or second-generation tyrosine kinase inhibitor.
Outcomes for additional uncommon exon 19 deletions
We examined outcomes for the 78 patients with tumors harboring other uncommon exon 19 deletions, although formal comparisons were not performed given the limited sample sizes. Among the more frequently observed uncommon exon 19 mutations, point estimates for the median PFS with first-line osimertinib were 16.2 months for L747_P753>S, 14.3 months for L747_T751del, and 19.1 months for E746_S752>V compared to 21.3 months for E746_A750del and 11.7 months for L747_A750>P. For ≥2L osimertinib in patients with T790M+ tumors, point estimates for median PFS were 12.4 months for L747_P753>S, 11.8 months for L747_T751del, and 8.2 months for E746_S752>V compared to 11.7 months for E746_A750del and 5.1 months for L747_A750>P. Supplementary Figures 2 and 3 depict these data along with median PFS for patients with tumors harboring E746_A750del and L747_A750>P.
For the remaining exon 19 deletion variants, each representing <2.5% of the cohort, baseline characteristics and patient-level outcome data with osimertinib treatment are shown in Supplementary Table 1.
Discussion:
In this multicenter study, we demonstrate that outcomes on osimertinib are inferior for patients with the infrequent EGFR exon 19 deletion mutation L747_A750>P compared to the common exon 19 deletion E746_A750del. With first-line osimertinib, there was a 48% increased risk of progression over the course of follow-up for patients with tumors harboring L747_A750>P compared to those harboring E746_A750del. At 12 months, the odds of remaining progression free on osimertinib were 4 times higher for L746_A750del compared to L747_A750>P. We also demonstrate the consistent trend for inferior outcomes in terms of overall survival in patients with tumors harboring the L747_A750>P mutation treated with osimertinib. Our analysis provides clinical evidence that individual exon 19 deletions can be associated with variable sensitivity to specific EGFR TKIs, in this case osimertinib, which may have implications for how we classify and treat these patients in the future.
EGFR mutation subtype is known to have an impact on TKI efficacy outcomes in patients with EGFR mutant NSCLC25. Although L858R mutations and exon 19 deletions were initially perceived to be similarly predictive of benefit from first-generation EGFR inhibitors, observations from clinical trials and a subsequent robust meta-analysis have revealed that this is not the case26. In fact, a greater relative PFS benefit for is seen for patients with tumors harboring exon 19 deletions compared to those with L858R mutations with first- and second-generation EGFR TKIs26,27. Variable TKI sensitivity has also been described for uncommon EGFR variants like G719X, L861X, and S768I. Common structural consequences of these mutations result in high selective susceptibility to second-generation agents compared to other TKI classes in vitro, supported by analyses involving patients treated with afatinib and other EGFR inhibitors28–30. Further evidence that not all EGFR mutations are alike comes from studies of EGFR exon 20 insertions. These mutations are associated with limited in vitro sensitivity to first, second, and third generation EGFR TKIs and predict unfavorable outcomes with conventional TKI treatment in clinical datasets31,32. Moreover there is heterogeneity of response among individual variants; for instance, the exon 20 insertion A763_Y764insFQEA demonstrates both in vitro and in vivo sensitivity to first, second, and third generation TKIs as well as novel agents active against other exon 20 insertions31,33,34.
Investigation of the clinical efficacy of EGFR TKIs against uncommon exon 19 deletions has been limited to retrospective analyses8–20. Most studies have included only patients treated with first- or second- generation TKIs, and have focused on comparisons between subgroups of exon 19 deletions rather than individual variants. These comparisons have included common vs uncommon mutations, starting at codon E746del vs starting codon L747del, and deletions vs insertion-deletions. Overall, this literature suggests a trend towards improved outcomes with TKI treatment for those harboring E746_A750del vs. uncommon deletions and for those with deletions starting with E746 vs L747. Inter- and intra-study heterogeneity with respect to TKI treatment, mutation frequency, and other clinicopathologic patient characteristics, however, makes drawing conclusions from these data particularly challenging20. To our knowledge, there are no specific reports on osimertinib efficacy for patients with tumors harboring L747_A750>P.
In contrast to the existing literature, we focus on one uncommon exon 19 deletion, L747_A750>P, which we hypothesized based on preclinical findings would result in inferior clinical outcomes with osimertinib compared to the common exon 19 deletion mutation E746_A750del1. Given that specific exon 19 deletions may have different TKI sensitivity profiles, we focused on the most clinically relevant TKI, osimertinib, and did not compare outcomes between cohorts treated with various TKIs.
The early overlap of the PFS and OS curves in the first-line osimertinib analysis is notable. All patients treated with first-line osimertinib achieved initial disease control in our cohort, independent of the underlying EGFR mutation. However, at approximately 9 months the curves begin to separate, and at 12 months there is a 30% numerical difference in the proportion of patients remaining progression-free, favoring E746_A750del. This suggests a difference in the durability of disease control in patients with a L747_A750>P mutation treated with osimertinib, corroborated by the significantly increased risk of progression over the duration of follow-up (PFS HR 0.52 for E746_A750del vs L747_A750>P). Further in vitro and clinical investigation should focus on characterizing response dynamics, patterns of progression, and mechanisms of acquired resistance that may help elucidate reasons for this discordance in early vs sustained disease control. In contrast, when osimertinib was usine in second-line or beyond, there was a trend towards inferior disease control for patients with tumors harboring L747_A750>P (77% vs 57% for E746_A750del), which is consistent with the unfavorable PFS and a trend for unfavorable OS associated with this mutation.
The FLAURA trial demonstrated that PFS for first-line osimertinib in patients with L858R was 14.4 months [11.1–18.9]6. Similarly, a phase II study of osimertinib for TKI-naïve patients with NSCLC harboring uncommon EGFR mutations demonstrated that the median PFS was 15.3 [1.3–29.1] for those with L861Q, 12.3 months [0–28.8] for S768I, and 8.2 months [6.2–10.2] for G719X35. Although our data are retrospective and cannot be directly compared to data from clinical trial populations without limitations, the efficacy of first-line osimertinib for patients with tumors harboring L747_A750>P may more closely resemble that for L858R and other uncommon EGFR mutations rather than E746-A750del.
Limitations to this study include its retrospective design as well as the limited sample size with a total of 122 patients with tumors harboring L747_A750>P and E746_A750del. The relative proportion of patients excluded from the analysis were similar among those with common and uncommon exon 19 deletions, indicating that excluding patients was unlikely to have influenced the outcomes observed. Our efforts to include patients from multiple institutions meaningfully expanded the cohort despite the relatively low frequency of L747_A750>P and allowed for a broader assessment of the efficacy of osimertinib outside of a single center experience. Challenges that innately restrict the size of the study cohort are the relatively recent adoption of osimertinib as the first-line standard TKI and the need to divide the cohort by osimertinib treatment setting (1L vs. ≥2L). Therefore, we focused our analyses on the comparison between E746_A750del vs. L747_A750>P and due to inadequate power, we avoid formal comparisons for additional uncommon variants in our exploratory cohort. Furthermore, formal evaluation of radiographic response (i.e. using RECIST criteria) was not obtained given that most patients were treated per standard of care and not on clinical trials. Objective response rates, depth, and duration of response were not readily available for most patients and therefore could not be compared between exon 19 deletion subtypes. Data regarding particular site of progression and number of progressive lesions were not available. Despite these limitations, to our knowledge, this is the largest comparison of the clinical efficacy of osimertinib between patients with tumors harboring the common deletion (E746_A750del) vs. a specific uncommon exon 19 deletion (L747_A750>P). Although all comparisons performed in our analyses exhibited a consistent numerical trend towards improved outcomes associated with E746_A750del, only PFS with first-line osimertinib demonstrated a statistically significant difference between E746_A750del and L747_A750>P. The lack of a statistically significant difference in overall survival is likely due to the small sample size rather than a difference in outcome with subsequent treatment, although we cannot exclude variable oucomes with chemotherapy or immunotherapy based on the specific EGFR mutation present. Differential efficacy of these non-targeted treatments based on the underlying mutation requires further study.
The findings from this study generate important questions pertaining to uncommon exon 19 deletions. First, what is the optimal first-line therapy for patients with tumors harboring L747_A750>P? Erlotinib and osimertinib appear to be less active in tumors with this mutation compared to E746_A750del, while afatinib appears compelling based on in vitro and in silico analyses1,36. Although one case report presents an exceptional response to afatinib for a patient with L747_A750>P, robust clinical data supporting afatinib efficacy are needed to change practice37. Moreover, afatinib is associated with more on-target side effects than osimertinib38. Despite our findings and in the absence of data to declare another more efficacious EGFR TKI, osimertinib is certainly a reasonable therapeutic option for all patients with exon 19 deletions. Although an increased dose of osimertinib has been explored in patients with exon 20 insertions, which are inherently resistant to osimertinib, and those with CNS progression on the standard dose of osimertinib, this has been associated with only modest efficacy despite an increase in toxicity; moreover, in vitro efficacy of osimertinib against cells harboring L747_A750>P does not appear to be concentration-dependent1,39–41. Altogether, this work highlights the opportunity to investigate alternative therapeutic strategies for patients with tumors harboring a L747_A750>P mutation
Second, can translational efforts lead to a more functional subclassification of EGFR exon 19 deletions based upon TKI sensitivity? A clinically validated system classifying exon 19 deletions according to predicted TKI sensitivity profiles could facilitate the study of rare variants in cohort analyses and clinical trials. In fact, it has been demonstrated that a structure-function-based subclassification of EGFR mutations into 4 distinct groups (classical-like, T790M-like, exon 20 loop insertion, and P-loop C-helix compressing) predicts preclinical TKI sensitivity as well as clinical outcomes for patients treated with specific EGFR-targeting agents42. Three distinct exon 19 alterations were included (an uncommon deletion-insertion L747_K754>ATSPE, the L747P point mutation, and the common deletion E746_A750del), and these demonstrated the potential to be classified into separate subgroups suggesting they are likely to have different TKI sensitivity profiles.
Comprehensive subclassification of EGFR exon 19 deletions will require biochemical and structural characterization of individual alterations. Recent efforts demonstrate that mutated EGFR proteins harboring individual exon 19 deletions differ with respect to their ATP-affinities, which correlates with TKI sensitivity36,43. Exon 19 deletions can be classified into one of 2 profiles based on the length of the β3/αC loop deletion. Profile 1 variants, with ≤ 3 residues deleted, are characterized by relatively strong (wild type-like) ATP binding affinity, and are associated with reduced sensitivity to ATP-competitive TKIs. Profile 2 variants, which comprise over 92% of exon 19 deletions, possess reduced ATP binding affinity, enhancing their sensitivity to EGFR TKIs. L747_A750>P, a profile 1 variant by this classification system, has reduced in vitro sensitivity to ATP-competitive TKIs like erlotinib and osimertinib compared to E746_A750del (profile 2), consistent with our clinical observations1,36,43. Interestingly, in one study, E746_S752>V (profile 2) was also reported to exhibit high ATP affinity and demonstrated diminished sensitivity to osimertinib compared to E746_A750del and L747_A750>P36. Yet based on the length of the β3/αC loop deletion, E746_S752>V is classified in profile 2 and predicted to be sensitive to osimertinib21. Although our cohort contained a limited number of patients with tumors harboring E746_S752>V treated with either 1L osimertinib (n=8) and ≥2L osimertinib (n=5), E746_S752>V appears not to differ from other profile 2 variants based on PFS point estimates (Supplementary Figures 2 and 3).
Finally, in addition to TKI sensitivity profiles, are there allele specific TKI resistance trends among EGFR exon 19 deletions? A growing evidence base suggests that the propensity to develop certain on-target resistance mutations can be a function of the underlying EGFR mutation44,45. The G724S mutation confers resistance to osimertinib and is associated with in vitro sensitivity to afatinib, and this mutation preferentially occurs in tumors harboring exon 19 deletions, particularly E746_S752>V, but not L858R44. The concept of allele-specificity for on-target resistance mutations has also been described for the EGFR mutations C797S and L718V/Q, which, like G724S, confer resistance to osimertinib45. Whereas C797S occurs more often in the context of an activating EGFR exon 19 deletion than with L858R, the on-target resistance mutations L718V and L718Q almost exclusively occur in L858R-mutated tumors. Uncovering other allele-specific trends in on-target resistance may impact surveillance, targeted investigation at resistance, and combination or sequential treatment approaches for patients with these mutations.
In conclusion, patients with tumors harboring the uncommon exon 19 deletion L747_A750delinsP exhibit unfavorable outcomes with osimertinib treatment relative to those with tumors harboring the common exon 19 deletion, L746_A750del. This is in line with previously published in vitro sensitivity data. On a larger scale, these data demonstrate that specific exon 19 deletions can exhibit differential sensitivity to EGFR TKIs, including osimertinib. Although in current practice, EGFR-targeted therapies are not tailored to a patient’s specific activating exon 19 deletion, we anticipate that these findings will contribute to a TKI-selective paradigm which enhances precision in the treatment of EGFR mutant NSCLC.
Supplementary Material
Translational Relevance:
The broad spectrum of activating EGFR mutations complicates the management of patients with tumors harboring atypical or uncommon variants. This is even true for ‘classical’ EGFR exon 19 deletion mutations which comprise a heterogeneous subgroup of variants. Uncommon exon 19 deletions have been infrequently disaggregated and studied in the context of EGFR TKI treatment. We evaluated the differential clinical efficacy of osimertinib in patients with tumors harboring the common exon 19 deletion, E746_A750del, vs. an uncommon exon 19 deletion predicted to have poor sensitivity to osimertinib in preclinical models, L747_A750>P. Additional uncommon exon 19 deletions are examined in the context of osimertinib treatment within this multi-institution cohort. The mechanistic basis for TKI sensitivity differences among exon 19 deletions warrants further exploration and may lead to a functional subclassification of these variants as the foundation for an allele specific, TKI-selective management approach.
Source of Funding:
Supported in part by a by the National Institutes of Health (P50-CA196530 to M.A.L., S.B.G., and K.P.; and T32-CA233414 to M.J.G.) and Conquer Cancer, the ASCO Foundation 2021 Conquer Cancer Young Investigator Award (2021YIA-0474838505) awarded to M.J.G.
References
- 1.Truini A, Starrett JH, Stewart T, et al. The EGFR Exon 19 Mutant L747-A750>P Exhibits Distinct Sensitivity to Tyrosine Kinase Inhibitors in Lung Adenocarcinoma. Clin Cancer Res. Nov 1 2019;25(21):6382–6391. doi: 10.1158/1078-0432.CCR-19-0780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray S, Dahabreh IJ, Linardou H, Manoloukos M, Bafaloukos D, Kosmidis P. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J Thorac Oncol. Aug 2008;3(8):832–9. doi: 10.1097/JTO.0b013e31818071f3 [DOI] [PubMed] [Google Scholar]
- 3.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. Jan 8 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. Feb 16 2017;376(7):629–640. doi: 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papadimitrakopoulou VA, Mok TS, Han JY, et al. Osimertinib versus platinum-pemetrexed for patients with EGFR T790M advanced NSCLC and progression on a prior EGFR-tyrosine kinase inhibitor: AURA3 overall survival analysis. Ann Oncol. Nov 2020;31(11):1536–1544. doi: 10.1016/j.annonc.2020.08.2100 [DOI] [PubMed] [Google Scholar]
- 6.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. Jan 11 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 7.Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med. Jan 2 2020;382(1):41–50. doi: 10.1056/NEJMoa1913662 [DOI] [PubMed] [Google Scholar]
- 8.Chung KP, Wu SG, Wu JY, et al. Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res. Jun 15 2012;18(12):3470–7. doi: 10.1158/1078-0432.CCR-11-2353 [DOI] [PubMed] [Google Scholar]
- 9.Kaneda T, Hata A, Tomioka H, et al. Possible differential EGFR-TKI efficacy among exon 19 deletional locations in EGFR-mutant non-small cell lung cancer. Lung Cancer. Nov 2014;86(2):213–8. doi: 10.1016/j.lungcan.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 10.Lee VH, Tin VP, Choy TS, et al. Association of exon 19 and 21 EGFR mutation patterns with treatment outcome after first-line tyrosine kinase inhibitor in metastatic non-small-cell lung cancer. J Thorac Oncol. Sep 2013;8(9):1148–55. doi: 10.1097/JTO.0b013e31829f684a [DOI] [PubMed] [Google Scholar]
- 11.Sutiman N, Tan SW, Tan EH, et al. EGFR Mutation Subtypes Influence Survival Outcomes following First-Line Gefitinib Therapy in Advanced Asian NSCLC Patients. J Thorac Oncol. Mar 2017;12(3):529–538. doi: 10.1016/j.jtho.2016.11.2225 [DOI] [PubMed] [Google Scholar]
- 12.Huang YH, Hsu KH, Tseng JS, et al. The Association of Acquired T790M Mutation with Clinical Characteristics after Resistance to First-Line Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor in Lung Adenocarcinoma. Cancer Res Treat. Oct 2018;50(4):1294–1303. doi: 10.4143/crt.2017.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Li W, Yang G, et al. Heterogeneous Response to First-Generation Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancers with Different EGFR Exon 19 Mutations. Target Oncol. Jun 2020;15(3):357–364. doi: 10.1007/s11523-020-00722-0 [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, Jiang T, Li J, et al. The impact of EGFR exon 19 deletion subtypes on clinical outcomes in non-small cell lung cancer. Transl Lung Cancer Res. Aug 2020;9(4):1149–1158. doi: 10.21037/tlcr-19-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi S, Toschi L, Finocchiaro G, et al. Impact of Exon 19 Deletion Subtypes in EGFR-Mutant Metastatic Non-Small-Cell Lung Cancer Treated With First-Line Tyrosine Kinase Inhibitors. Clin Lung Cancer. Mar 2019;20(2):82–87. doi: 10.1016/j.cllc.2018.10.009 [DOI] [PubMed] [Google Scholar]
- 16.Su J, Zhong W, Zhang X, et al. Molecular characteristics and clinical outcomes of EGFR exon 19 indel subtypes to EGFR TKIs in NSCLC patients. Oncotarget. Dec 19 2017;8(67):111246–111257. doi: 10.18632/oncotarget.22768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Q, Huang Y, Zhao H, et al. EGFR mutation genotypes affect efficacy and resistance mechanisms of osimertinib in T790M-positive NSCLC patients. Transl Lung Cancer Res. Jun 2020;9(3):471–483. doi: 10.21037/tlcr.2020.03.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng X, Long X, Liu L, et al. Clinical impact of uncommon epidermal growth factor receptor exon 19 insertion-deletion variants on epidermal growth factor receptor-tyrosine kinase inhibitor efficacy in non-small-cell lung cancer. Eur J Cancer. Dec 2020;141:199–208. doi: 10.1016/j.ejca.2020.10.005 [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Zheng R, Hu P, Zhang Z, Shen S, Li X. Patients harboring uncommon EGFR exon 19 deletion-insertion mutations respond well to first-generation EGFR inhibitors and osimeritinib upon acquisition of T790M. BMC Cancer. Nov 13 2021;21(1):1215. doi: 10.1186/s12885-021-08942-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang LT, Zhang SL, Han CB, Ma JT. Impact of EGFR exon 19 deletion subtypes on clinical outcomes in EGFR-TKI-Treated advanced non-small-cell lung cancer. Lung Cancer. Apr 2022;166:9–16. doi: 10.1016/j.lungcan.2022.01.014 [DOI] [PubMed] [Google Scholar]
- 21.van Alderwerelt van Rosenburgh IK, Lu DM, Grant MJ, et al. Biochemical and structural basis for differential inhibitor sensitivity of EGFR with distinct exon 19 mutations. Nature Communications. 2022/11/10 2022;13(1):6791. doi: 10.1038/s41467-022-34398-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. Aug 2017;7(8):818–831. doi: 10.1158/2159-8290.Cd-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. Aug 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. Jul 2017;2017doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castellanos E, Feld E, Horn L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol. Apr 2017;12(4):612–623. doi: 10.1016/j.jtho.2016.12.014 [DOI] [PubMed] [Google Scholar]
- 26.Lee CK, Wu YL, Ding PN, et al. Impact of Specific Epidermal Growth Factor Receptor (EGFR) Mutations and Clinical Characteristics on Outcomes After Treatment With EGFR Tyrosine Kinase Inhibitors Versus Chemotherapy in EGFR-Mutant Lung Cancer: A Meta-Analysis. J Clin Oncol. Jun 10 2015;33(17):1958–65. doi: 10.1200/JCO.2014.58.1736 [DOI] [PubMed] [Google Scholar]
- 27.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. Sep 20 2013;31(27):3327–34. doi: 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 28.Yang JC, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. Jul 2015;16(7):830–8. doi: 10.1016/S1470-2045(15)00026-1 [DOI] [PubMed] [Google Scholar]
- 29.Yang JC, Schuler M, Popat S, et al. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J Thorac Oncol. May 2020;15(5):803–815. doi: 10.1016/j.jtho.2019.12.126 [DOI] [PubMed] [Google Scholar]
- 30.Janning M, Suptitz J, Albers-Leischner C, et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann Oncol. Jun 2022;33(6):602–615. doi: 10.1016/j.annonc.2022.02.225 [DOI] [PubMed] [Google Scholar]
- 31.Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. Dec 18 2013;5(216):216ra177. doi: 10.1126/scitranslmed.3007205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gristina V, Malapelle U, Galvano A, et al. The significance of epidermal growth factor receptor uncommon mutations in non-small cell lung cancer: A systematic review and critical appraisal. Cancer Treat Rev. Apr 2020;85:101994. doi: 10.1016/j.ctrv.2020.101994 [DOI] [PubMed] [Google Scholar]
- 33.Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. Feb 2013;12(2):220–9. doi: 10.1158/1535-7163.MCT-12-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasconcelos P, Gergis C, Viray H, et al. EGFR-A763_Y764insFQEA Is a Unique Exon 20 Insertion Mutation That Displays Sensitivity to Approved and In-Development Lung Cancer EGFR Tyrosine Kinase Inhibitors. JTO Clin Res Rep. Sep 2020;1(3)doi: 10.1016/j.jtocrr.2020.100051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho JH, Lim SH, An HJ, et al. Osimertinib for Patients With Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15–09). J Clin Oncol. Feb 10 2020;38(5):488–495. doi: 10.1200/JCO.19.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown BP, Zhang YK, Kim S, et al. Allele-specific activation, enzyme kinetics, and inhibitor sensitivities of EGFR exon 19 deletion mutations in lung cancer. Proc Natl Acad Sci U S A. Jul 26 2022;119(30):e2206588119. doi: 10.1073/pnas.2206588119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Federico A, Filippini DM, Dall’Olio FG, et al. The EGFR Exon 19 Mutant L747-A750>P Exhibits Distinct Sensitivity to Tyrosine Kinase Inhibitors in Lung Adenocarcinoma-Letter. Clin Cancer Res. Jan 15 2020;26(2):518–519. doi: 10.1158/1078-0432.CCR-19-2441 [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Cheng B, Chen Z, et al. Toxicity profile of epidermal growth factor receptor tyrosine kinase inhibitors for patients with lung cancer: A systematic review and network meta-analysis. Crit Rev Oncol Hematol. Apr 2021;160:103305. doi: 10.1016/j.critrevonc.2021.103305 [DOI] [PubMed] [Google Scholar]
- 39.Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. Mar 20 2018;36(9):841–849. doi: 10.1200/jco.2017.74.7576 [DOI] [PubMed] [Google Scholar]
- 40.Zwierenga F, van Veggel B, Hendriks LEL, et al. High dose osimertinib in patients with advanced stage EGFR exon 20 mutation-positive NSCLC: Results from the phase 2 multicenter POSITION20 trial. Lung Cancer. Aug 2022;170:133–140. doi: 10.1016/j.lungcan.2022.06.012 [DOI] [PubMed] [Google Scholar]
- 41.Piper-Vallillo AJ, Rotow JK, Aredo JV, et al. High-Dose Osimertinib for CNS Progression in EGFR+ NSCLC: A Multi-Institutional Experience. JTO Clin Res Rep. Jun 2022;3(6):100328. doi: 10.1016/j.jtocrr.2022.100328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robichaux JP, Le X, Vijayan RSK, et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature. Sep 2021;597(7878):732–737. doi: 10.1038/s41586-021-03898-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iris K van Alderwerelt van Rosenburgh DML, Michael J. Grant, Manali Phadke, Zenta Walther, Sarah B. Goldberg, Katerina Politi, Mark A. Lemmon, Kumar D. Ashtekar, and Yuko Tsutsui. Biochemical and Structural Basis for Differential Inhibitor Sensitivity of EGFR with Distinct Exon 19 Mutations. Nature Communications. 2022; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown BP, Zhang YK, Westover D, et al. On-target Resistance to the Mutant-Selective EGFR Inhibitor Osimertinib Can Develop in an Allele-Specific Manner Dependent on the Original EGFR-Activating Mutation. Clin Cancer Res. Jun 1 2019;25(11):3341–3351. doi: 10.1158/1078-0432.CCR-18-3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starrett JH, Guernet AA, Cuomo ME, et al. Drug Sensitivity and Allele Specificity of First-Line Osimertinib Resistance EGFR Mutations. Cancer Res. May 15 2020;80(10):2017–2030. doi: 10.1158/0008-5472.CAN-19-3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.