Abstract
MicroRNA (miR)-499a-5p has been reported to regulate the progression of various tumours. However, the role of miR-499a-5p in breast cancer is unclear. The purpose of this study was to investigate the role and mechanism of miR-499a-5p in breast cancer. The growth effect of miR-499a-5p on breast cancer cells was investigated by the CCK-8 assay, wound healing assay and Transwell invasion assay. The luciferase activity assay was used to verify the downstream targets of miR-499a-5p. The levels of GSH, MDA, and ROS were detected by kits. Quantitative real-time PCR and Western blot were used to determine the expression levels of TMEM189, COX-2, GPX4, and other related genes in cells. miR-499a-5p was down-regulated in MDA-MB-231 cells and was shown to reduced the viability, migration and invasion of MDA-MB-231 cells. Further studies revealed that TMEM189 is a target of miR-499a-5p. miR-499a-5p inhibited breast cancer cell growth by downregulating TMEM189. Furthermore, the down-regulation of TMEM189 promotes ferroptosis in breast cancer cells. The low expression of TMEM189 inhibited the development of breast cancer through the ferroptosis pathway. We have demonstrated for the first time that miR-499a-5p inhibits breast cancer progression by targeting the TMEM189-mediated ferroptosis pathway.
Keywords: miR-499a-5p, TMEM189, ferroptosis, breast cancer
Introduction
Breast cancer is the most common malignancy and the leading cause of cancer death in women worldwide.(1) It is a metastatic cancer that can often metastasise to distant organs such as the brain, bone, liver, lung, and distant lymph nodes.(2) Although the existing treatment methods have achieved good curative effects, it is still extremely difficult to cure advanced metastatic breast cancer.(3) The overall 5-year relative survival rate for patients with metastatic breast cancer is only 27%.(4) The development of genomics has made molecular therapy an emerging modality for early cancer screening and treatment.(5) Early detection of the disease can effectively prevent breast cancer and lead to good prognosis and high survival rates.(6) Therefore, there is an urgent need to determine the underlying mechanisms of breast cancer development and to develop more therapeutic targets for it.
miR-499a-5p is a class of microRNAs (miRNAs) with a length of 18~25 nt and plays an important role in various diseases, especially malignant tumors.(7) It has been reported that miR-499a-5p is significantly downregulated in endometrial cancer tissues and targets VAV3 to inhibit endometrial cancer growth and metastasis.(8) miR-499a-5p inhibits proliferation, invasion, migration and epithelial-mesenchymal transition by targeting eIF4E, and enhances the radiosensitivity of cervical cancer cells.(9) In addition, miR-499a-5p also inhibits tumour growth in non-small cell lung cancer.(7) In breast cancer, there is a significant correlation between miR-499a-5p and breast cancer risk.(10) However, the mechanism by which miR-499a-5p regulates breast cancer progression has not been investigated.
TMEM189 is a transmembrane protein containing a conserved motif and eight conserved histidine residues. This gene encodes plasma ethanolamine desaturase, which introduces vinyl ether double bonds into plasmalogens.(11) TMEM189 is involved in lipid biosynthesis, especially ether lipids.(12) The dynamic regulation of ether lipid biosynthesis alters the sensitivity of cancer cells, neurons, and cardiomyocytes to ferroptosis.(13) Ferroptosis is a novel iron-dependent form of cell death triggered by accumulation of lipid peroxides and reactive oxygen species (ROS).(14,15) Ferroptosis is often inhibited in various tumours, resulting in the uncontrolled proliferation of tumor cells.(16) Studies have shown that TMEM189 inhibits ferroptosis, and the deletion of TMEM189 sensitises cells to RSL3-induced ferroptosis.(17) In addition, the results from the genomics database showed that TMEM189 was highly expressed in various human cancers, and high levels of TMEM189 correlated with the high expression of GPX4.(17) Therefore, TMEM189 may regulate tumour progression through the ferroptosis pathway.
This study aimed to investigate the role and mechanism of miR-499a-5p and TMEM189 in the occurrence and development of breast cancer. We found that miR-499a-5p was significantly down-regulated in breast cancer cells while TMEM189 was remarkably up-regulated in breast cancer cells. miR-499a-5p directly targeted and negatively regulated TMEM189. miR-499a-5p promoted ferroptosis in breast cancer cells by downregulating the level of TMEM189, thereby inhibiting the excessive growth of breast cancer cells. miR-499a-5p and TMEM189 provide a preliminary theoretical basis for the clinical treatment of breast cancer.
Materials and Methods
Cell culture and transfection
Human breast cells (MCF-10) and human breast cancer cells (MDA-MB-231, MCF-7, and SUM159) were purchased from the Chinese Academy of Sciences typical culture preservation Center (Shanghai, China). The breast cells MCF-10 was cultured in RPMI 1640 medium (Hyclone, Logan, UT) and breast cancer cells (MDA-MB-231, MCF-7, and SUM159) were cultured in DMEM medium (Hyclone). The above media were supplemented with 10% foetal bovine serum (FBS) (Gibco, Grand Island, NY) and 1% penicillin-streptomycin (Procell Life, Wuhan, China). Cells were incubated at 37°C and 5% CO2. All cells were regularly tested for mycoplasma with good results. miR-499a-5p mimic, miR-499a-5p inhibitor, PcDNA3.1-TMEM189, si-TMEM189 and negative control (GenePharma, Shanghai, China) were synthesised. miR-499a-5p mimic (forward 5'-GACACGGCTCCGTCAC-3'; reverse 5'-ATCCCTGATCAACTGTCCGCC-3') or its negative control (NC mimic) (forward 5'-AACCAGGCCTCCACCG-3'; reverse 5'-GCCACGCATGTCTTATACTGC-3'). miR-499a-5p inhibitors (5'-AAACAUCACUGCAAGUCUUAA-3'); Negative control inhibitors (5'-CAGUACUUUUGUGUAGUACAAA-3'). When the cells reached 70% confluence, MDA-MB-231 cells were transfected using Lipofectamine 3000 (Invitrogen, Carlsbad, CA). The specific steps are carried out according to the manufacturer’s instructions.
CCK-8 assay
Cells were seeded into 96-well plates at a density of 1 × 105 cells per well, and the cell viability of MDA-MB-231 was determined by CCK-8 method. Briefly, 10 μl of CCK-8 reagent (Beyotime, Beijing, China) was added to each well and incubated for 4 h. Absorbance was measured at a wavelength of 450 nm.
Wound healing assay
Cells grown to 90% confluency in six-well plates were used for wound healing assays. The cells were scraped with a 200 μl micropipette to create an even scratch. Damaged cells were washed away with PBS and incubated in the incubator. Images were taken at 0 h and 24 h of the experiment to observe cell migration.
Transwell invasion assay
Invasion assays were performed using Matrigel (Sigma, St. Louis, MO)-coated Transwell wells (Corning, Batford County, Corning, NY). Then, 100 μl of cell suspension in serum-free medium was added to the upper chamber and 600 μl of medium containing 10% FBS was added to the lower chamber. Cells incubated for 24 h were fixed and then stained with crystal violet (Beyotime) for 15 min. Five different fields of view were selected for each well for photographing and counting.
Luciferase reporter gene assay
The binding sites of miR-499a-5p and TMEM189 were predicted by TargetScan software. TMEM189 WT, TMEM189 MUT, miR-499a-5p mimic, miR-499a-5p inhibitor and control vectors (GenePharma) were designed and constructed. Next, pmirGLO-TMEM189-WT and pmirGLO-TMEM189-Mut plasmids were co-transfected into MDA-MB-231 with miR-499a-5p mimic and mimic-NC. Cell lysates were collected 48 h after transfection. Luciferase activity was assessed using a dual-luciferase reporter gene assay system (Promega, Madison, WI). In this experiment, renilla luciferase activity was selected as the internal reference.
Determination of GSH, MDA and ROS levels
The level of GSH was measured using a total glutathione detection kit (Beyotime). The Cell Malondialdehyde (MDA) assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) was used to detect the level of MDA. The level of ROS was discovered by reactive oxygen species detection kit (Nanjing Jiancheng Bioengineering Institute). The specific operation of the experiment was carried out according to the manufacturer’s instructions.
RT-qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen). RNA was reverse transcribed to cDNA using Qiagen One-Step RT-PCR kit (Qiagen Gmbh, Dusseldorf, Germany). cDNA, primers and SYBRTM Green PCR Master Mix (Thermo Fisher, Waltham, MA) were mixed and qRT-PCR was performed. GAPDH was used as an internal reference. The reaction conditions in the experiments were performed according to the manufacturer’s instructions. The expression levels of target genes were determined using the 2−ΔΔCT method. The sequences of each primer are:
miR-499a-5p: F 5'-GCCCTGTCCCCTGTGCCTT-3',
R 5'-AAACATCACTGCAAGTCTT-3';
TMEM189: F 5'-GACCTACTTCTGCATCACCACAGTT-3',
R 5'-CGTTGTTGTCCAAGCATTCG-3';
GAPDH: F 5'-ACAGTCAGCCGCATCTTCTT-3',
R 5'-AAATGAGCCCCAGCCTTCTC-3'.
Western blot
Cells were lysed using RIPA buffer to extract total protein. The protein concentration was detected by BCA method. Total proteins were separated using 10–12% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes. PVDF membranes were blocked with 5% nonfat milk containing 0.05% Tween-20. PVDF was blocked with different primary antibodies overnight at 4°C and secondary antibodies for 2 h at room temperature. Bands were detected using the ECL Western blotting kit (Thermo Fisher). The luminosity of the bands was quantified using the ImageJ analysis system.
Statistical analysis
We used GraphPad Prism 7.0 (GraphPad Software, San Diego, CA) for statistical analysis in this study. All experiments were conducted in triplicate, and the data are presented as the mean ± SD. Comparisons between groups were made using Student’s t test, and multiple comparisons were made using a one-way analysis of variance, followed by Turkey’s test. For all comparisons, p<0.05 was considered statistically significant.
Results
miR-499a-5p inhibited breast cancer cell growth
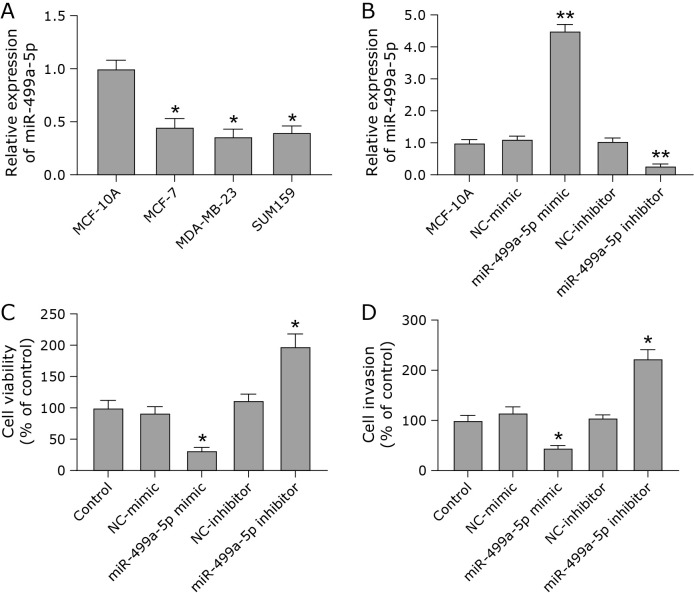
To determine the role of miR-499a-5p in breast cancer, we first determined the level of miR-499a-5p in breast cancer cells. The mRNA level of miR-499a-5p was down-regulated in breast cancer cells compared with breast cancer cells (Fig. 1A). Next, we verified the effect of miR-499a-5p on the growth of MDA-MB-231 cells. The overexpression and underexpression of miR-499a-5p were efficient (Fig. 1B). After the overexpression of miR-499a-5p, the viability and cell invasion ability of MDA-MB-231 cells were decreased. After the low expression of miR-499a-5p, the viability and invasion ability of MDA-MB-231 cells were enhanced (Fig. 1C and D). These data suggest that miR-499a-5p may have an important role in breast cancer progression.
Fig. 1.
miR-499a-5p inhibited breast cancer cell growth. (A) RT-qPCR was used to detect the mRNA levels of miR-499a-5p in breast cells (MCF-10A) and breast cancer cells (MCF-7, MDA-MB-231, and SUM159). (B) Efficiency of overexpression and underexpression of miR-499a-5p. (C) Cell viability was measured by CCK-8 assay. (D) The invasive ability of cells in each group was determined by transwell assay. *p<0.05, **p<0.01.
miR-499a-5p directly targeted and negatively regulated TMEM189
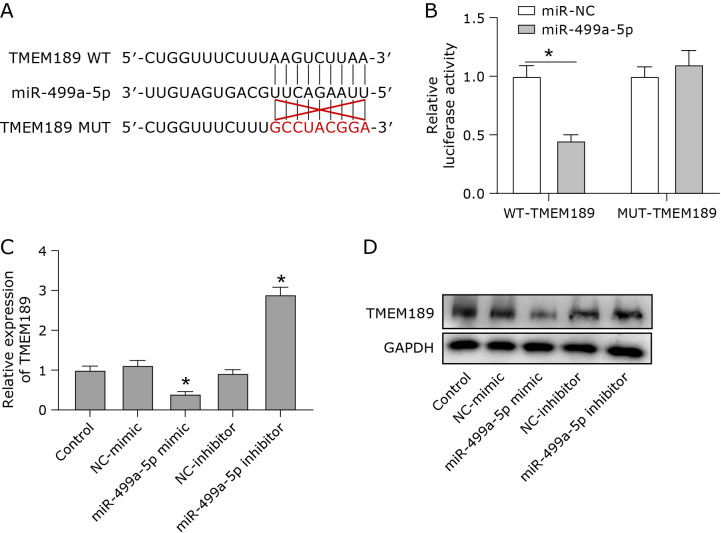
We predicted the downstream targets of miR-499a-5p by Targetscan software (https://www.targetscan.org/vert_71/). TMEM189 had a binding site for miR-499a-5p (Fig. 2A). The dual luciferase reporter gene assay showed that the miR-499a-5p mimic significantly reduced the luciferase activity of TMEM189-WT cells, while the miR-499a-5p mimic had no significant effect on the luciferase activity of TMEM189-MUT cells (Fig. 2B). This result suggested that miR-499a-5p has a physical interaction with TMEM189. After the transfection of miR-499a-5p mimics into MDA-MB-231 cells, the protein level of TMEM189 was significantly inhibited. In contrast, the inhibition of miR-499a-5p significantly increased the protein level of TMEM189 (Fig. 2C and D). These results suggest that miR-499a-5p directly targets and negatively regulates TMEM189.
Fig. 2.
miR-499a-5p directly targeted and negatively regulated TMEM189. (A) TMEM189 was predicted to have a binding site for miR-499a-5p by bioinformatics software. (B) Relative luciferase activity levels of TMEM189. (C) The protein expression level of TMEM189 under the condition of overexpression or underexpression of miR-499a-5p. (D) Quantification of protein results. *p<0.05.
miR-499a-5p inhibited breast cancer cell growth by downregulating TMEM189
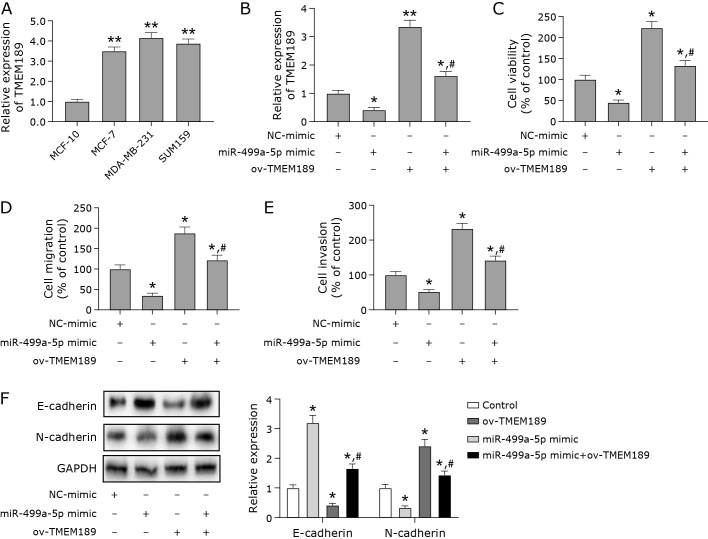
Next, we examined the expression level of TMEM189 in MDA-MB-231 cells. Both mRNA and protein levels of TMEM189 were up-regulated in breast cancer cells compared with breast cancer cells (Fig. 3A). To further explore whether miR-499a-5p affects the growth of breast cancer cells by regulating TMEM189, we overexpressed miR-499a-5p and TMEM189 with good transfection efficiency (Fig. 3B). The overexpression of miR-499a-5p inhibited the viability, cell migration and invasion ability of breast cancer cells (Fig. 3C–E). However, the overexpression of TMEM189 promoted the growth of breast cancer cells and reversed the inhibitory effect of miR-499a-5p mimic on the cells (Fig. 3C–E). These results suggest that miR-499a-5p inhibited breast cancer cell growth by downregulating TMEM189. E-cadherin and N-cadherin are markers of epithelial mesenchymal transformation (EMT), which is related to the increased risk of cancer metastasis.(18) Therefore, we detected the expression of these two proteins. We found that miR-499a-5p also increased the expression level of E-cadherin protein and decreased the protein expression level of N-cadherin by downregulating TMEM189 (Fig. 3F).
Fig. 3.
miR-499a-5p inhibited breast cancer cell growth by downregulating TMEM189. (A) RT-qPCR was used to detect the mRNA and protein levels of TMEM189 in breast cells (MCF-10A) and breast cancer cells (MCF-7, MDA-MB-231, and SUM159). (B) Efficiency of overexpressing miR-499a-5p and TMEM189. (C) Cell viability was detected by CCK-8. (D) The migration ability of cells in each group was tested by wound healing assay. (E) The invasive ability of cells in each group was determined by transwell assay. (F) Protein expression levels of epithelial-mesenchymal transition (EMT)-related proteins (E-cadherin and N-cadherin). *p<0.05, **p<0.01, #p<0.05.
Low expression of TMEM189 induced ferroptosis in breast cancer cells
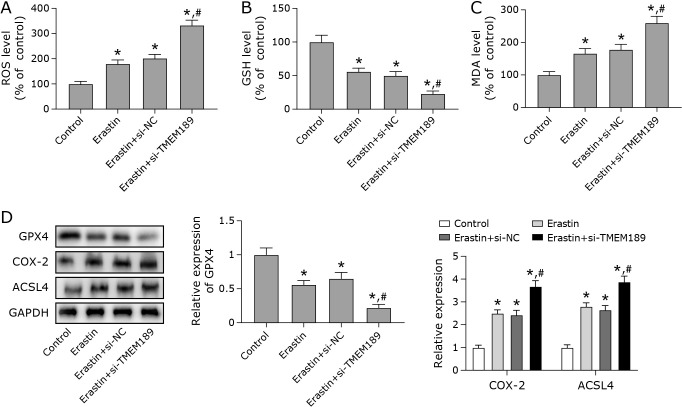
Ferroptosis is one of the effective ways to induce tumour cell death.(19) This experiment continued to explore the relationship between TMEM189 and ferroptosis. Compared with the Erastin + si-RNA group, the ferroptosis process of cells was promoted to a greater extent after knockdown of TMEM189. It was manifested as decreased GSH levels, and increased ROS level and MDA levels. In addition, the low expression of TMEM189 also down-regulated the protein level of GPX4 and up-regulated the levels of COX-2 and ACSL4 (Fig. 4). The above results indicated that the low expression of TMEM189 promotes ferroptosis in breast cancer cells.
Fig. 4.
Low expression of TMEM189 induced ferroptosis in breast cancer cells. MDA-MB-231 cells were transfected with si-RNA, si-TMEM189, and treated with a certain concentration of Erastin for 24 h. (A) Detection of ROS levels in each group of cells. (B) The level of GSH was detected using a kit. (C) The level of lipid peroxidation was detected using a malondialdehyde (MDA) kit. (D) Western blot was used to tested the protein expression levels of ferroptosis-related factors (COX-2, GPX4, and ACSL4). *p<0.05, #p<0.05.
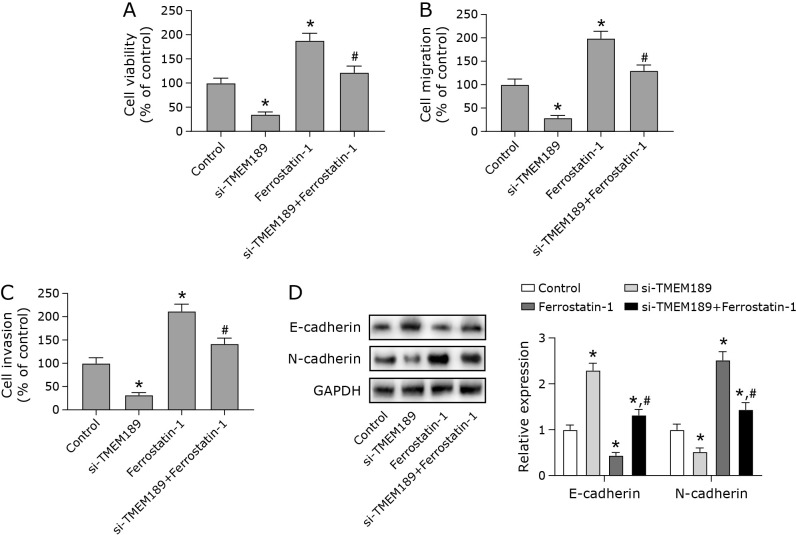
Low expression of TMEM189 inhibited breast cancer development through the ferroptosis pathway
Next, we treated cells which poorly expressed TMEM189 with ferroptosis inhibition (ferrostatin-1). Compared with the si-NC group, the knockdown of TMEM189 resulted in the decreased viability, migration ability and invasion ability of MDA-MB-231 cells. This was reversed after the administration of ferrostatin-1 (Fig. 5). Ferrostatin-1 significantly inhibited ferroptosis promotion and growth inhibition by si-TMEM189. In addition, the results showed that the ferrostatin-1 reversed the reduction of viability, migration and invasion of MDA-MB-231 cells caused by overexpression of miR-499a-5p (Supplemental Fig. 1*).
Fig. 5.
Low expression of TMEM189 inhibited breast cancer development through the ferroptosis pathway. MDA-MB-231 cells were transfected with si-RNA and si-TMEM189, and cells were treated with a certain concentration of ferrostatin-1 for 24 h. (A) Cell viability was measured by CCK-8. (B) The migration ability of cells in each group was detected by wound healing assay. (C) The invasive ability of cells in each group was determined by transwell assay. (D) Protein expression levels of epithelial-mesenchymal transition (EMT)-related proteins (E-cadherin and N-cadherin). *p<0.05, #p<0.05.
Discussion
Accumulating evidence suggests that miRNAs can regulate human tumour development by mediating the expression of their targets and may serve as future targets for gene therapy.(20,21) In cancer, miRNA overexpression or downregulation affects the metabolic state, tumor progression and metastasis of tumour cells by altering internal cellular pathways.(22) Studies have found that the abnormal expression of miRNAs in epigenetics is related to the occurrence and development of breast cancer.(23–25) The role of miR-499a-5p in multiple cancers has been demonstrated; however, its specific role in breast cancer is unclear. In the present study, we investigated the mechanism of action of miR-499a-5p in breast cancer cells.
Our study found that miR-499a-5p was down-regulated in MDA-MB-231 breast cancer cells. The overexpression of miR-499a-5p reduced the cell viability, migration and invasion ability of MDA-MB-231. The underexpression of miR-499a-5p promoted the growth of breast cancer cells. E-cadherin and N-cadherin are markers of epithelial mesenchymal transition (EMT). The change of cadherin expression is called “cadherin conversion”. The down-regulation of E-cadherin expression is a sign of EMT and also a sign of tumor cell metastasis.(18) In this study, miR-499a-5p increased the expression of E-cadherin and decreased the expression of N-cadherin by downregulating TMEM189. These results suggested that miR-499a-5p may act as a tumour suppressor in breast cancer development, inhibiting the metastasis of breast cancer cells. In previous reports, we found that miR-499a-5p plays a dual role in cancer. miR-499a-5p was significantly down-regulated in endometrial and cervical cancer cells, and inhibited the proliferation and metastasis of cancer cells.(8,9) On the other hand, studies have shown that miRNA-499a-5p exhibits the role of a tumour-promoting factor. miRNA-499a-5p has been reported to promote lung cancer cell proliferation, migration and epithelial-mesenchymal transition (EMT) in vitro and tumour growth in vivo.(26) miRNA-499a-5p also promotes 5-fluorouracil resistance and cell proliferation by targeting PTEN to activate the PI3K/Akt signalling pathway in pancreatic cancer.(27) miR-499a-5p exhibits different roles in different tumours, possibly because it acts on different molecular targets. This issue still needs further research. Taken together, our results suggested that miR-499a-5p is able to inhibit the over-proliferation of breast cancer cells and has a tumour suppressor effect.
Subsequently, TMEM189 was identified as a downstream target of miR-499a-5p, and its level was upregulated in breast cancer cells. miR-499a-5p inhibited the growth of breast cancer cells by down-regulating the expression of TMEM189. It has been reported that TMEM189 is highly expressed in various human cancers, and high levels of TMEM189 promote tumour progression.(17) It has been reported that TMEM189 mediates iron ptosis of breast cancer cells by inhibiting autophagy, thereby reducing the proliferation of breast cancer cells and inhibiting tumor growth.(28) This is consistent with our results. The tumour-promoting activity of TMEM189 is inseparable from its modulating activity of plasmalogen. TMEM189 is generally thought to be involved in lipid desaturation and encodes the plasma phenethanolamine desaturase which is necessary for vinyl ether bond formation.(29) TMEM189-deficient mice exhibited a deficiency of plasma ethanolamine desaturase activity and plasmalogen.(11) Studies have shown that plasmalogens can stimulate cell growth, regulate cell survival and tumour growth.(30) Higher levels of plasmalogen are associated with an increased risk of breast cancer.(31) Therefore, we speculate that TMEM189, which has a plasmalogen-regulated effect, may affect breast cancer progression. This study is the first to demonstrate that low expression of TMEM189 can inhibit the viability, migration and invasion of breast cancer cells.
Increasing studies have shown that ferroptosis is involved in tumour progression. The induction of ferroptosis not only promotes tumour cell death, but also increases the anticancer activity of chemotherapeutic drugs.(32) Similarly, in breast cancer research, inhibition of the ferroptosis pathway promotes the migration and invasion of breast cancer cells and tumour growth in vivo.(33,34) The induction of ferroptosis in breast cancer cells can prevent further tumour development.(35,36) Therefore, enhancing ferroptosis in breast cancer cells is considered a promising strategy for the treatment of breast cancer. Recent studies have shown that the FAR1-ether lipid-TMEM189 axis is an important pathway driving ferroptosis susceptibility. TMEM189 inhibits ferroptosis in tumr cells, and the knockdown of TMEM189 promotes ferroptosis.(17) This provides direction for the research of TMEM189 in breast cancer. In the present study, we demonstrated that the poor of TMEM189 inhibited the proliferation, migration and invasion of breast cancer cells by promoting the ferroptosis pathway. Therefore, TMEM189 can be regarded as a tumour-promoting factor.
In conclusion, we demonstrate for the first time that miR-499a-5p inhibits breast cancer cell growth by promoting ferroptosis by downregulating TMEM189 expression. miR-499a-5p and TMEM189 may be new biomarker molecules for breast cancer, which will provide a preliminary theoretical basis for the clinical treatment of breast cancer.
Author Contributions
HDZ involved in study design, data collection and data analysis, DF involved in data analysis and interpretation, draft of the manuscript, YM, YJQ and XZY involved in data analysis, revision of the manuscript.
Acknowledgments
None.
Funding
This research was supported by the Social Talent Fund of the Second Affiliated Hospital of Air Force Medical University (2021SHRC037).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflict of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res 2017; 50: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosonaga M, Saya H, Arima Y. Molecular and cellular mechanisms underlying brain metastasis of breast cancer. Cancer Metastasis Rev 2020; 39: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zujewski JA, Dvaladze AL, Ilbawi A, et al. Knowledge summaries for comprehensive breast cancer control. J Glob Oncol 2018; 4: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venetis K, Piciotti R, Sajjadi E, et al. Breast cancer with bone metastasis: molecular insights and clinical management. Cells 2021; 10: 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: from early detection to new therapies. Radiologia 2017; 59: 368–379. [DOI] [PubMed] [Google Scholar]
- 6.Sun YS, Zhao Z, Yang ZN, et al. Risk factors and preventions of breast cancer. Int J Biol Sci 2017; 13: 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Jiang P, Zheng H, Chen P, Yang M. Downregulation of miR-499a-5p predicts a poor prognosis of patients with non-small cell lung cancer and restrains the tumorigenesis by targeting fibroblast growth factor 9. Technol Cancer Res Treat 2020; 19: 1533033820957001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jing L, Hua X, Yuanna D, Rukun Z, Junjun M. Exosomal miR-499a-5p inhibits endometrial cancer growth and metastasis via targeting VAV3. Cancer Manag Res 2020; 12: 13541–13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu X, Dong M, Liu Z, Yang J, Shi Y. MiR-499a-5p inhibits proliferation, invasion, migration, and epithelial-mesenchymal transition, and enhances radiosensitivity of cervical cancer cells via targeting eIF4E. Onco Targets Ther 2020; 13: 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alshatwi AA, Shafi G, Hasan TN, et al. Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS One 2012; 7: e30049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner ER, Keller MA, Sailer S, et al. The TMEM189 gene encodes plasmanylethanolamine desaturase which introduces the characteristic vinyl ether double bond into plasmalogens. Proc Natl Acad Sci U S A 2020; 117: 7792–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wainberg M, Kamber RA, Balsubramani A, et al. A genome-wide atlas of co-essential modules assigns function to uncharacterized genes. Nat Genet 2021; 53: 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, Zhuang L, Gan B. Ether phospholipids govern ferroptosis. J Genet Genomics 2021; 48: 517–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019; 12: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med 2019; 133: 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Jia H, Li S, et al. Comprehensive analysis of ferroptosis-related markers for the clinical and biological value in gastric cancer. Oxid Med Cell Longev 2021; 2021: 7007933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui W, Liu D, Gu W, Chu B. Peroxisome-driven ether-linked phospholipids biosynthesis is essential for ferroptosis. Cell Death Differ 2021; 28: 2536–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh CY, Chai JY, Tang TF, et al. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells 2019; 8: 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stockwell BR, Jiang X, Gu W. Emerging mechanisms and disease relevance of ferroptosis. Trends Cell Biol 2020; 30: 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Huang L, Su P, et al. MicroRNA-499a-5p inhibits osteosarcoma cell proliferation and differentiation by targeting protein phosphatase 1D through protein kinase B/glycogen synthase kinase 3β signaling. Oncol Lett 2018; 15: 4113–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sha HH, Wang DD, Chen D, et al. MiR-138: a promising therapeutic target for cancer. Tumour Biol 2017; 39: 1010428317697575. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Zhou Y, Han F, et al. A novel miR-1291-ERRα-CPT1C axis modulates tumor cell proliferation, metabolism and tumorigenesis. Theranostics 2020; 10: 7193–7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Li TE, Chen M, Pan JJ, Shen KW. miR-106b-5p contributes to the lung metastasis of breast cancer via targeting CNN1 and regulating Rho/ROCK1 pathway. Aging (Albany NY) 2020; 12: 1867–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Hu S, Li X, et al. MiR-16-5p suppresses breast cancer proliferation by targeting ANLN. BMC Cancer 2021; 21: 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang YW, Zhao S, Yuan XY, et al. miR-4732-5p promotes breast cancer progression by targeting TSPAN13. J Cell Mol Med 2019; 23: 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He S, Li Z, Yu Y, et al. Exosomal miR-499a-5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp Cell Res 2019; 379: 203–213. [DOI] [PubMed] [Google Scholar]
- 27.Ouyang L, Liu RD, Lei DQ, et al. MiR-499a-5p promotes 5-FU resistance and the cell proliferation and migration through activating PI3K/Akt signaling by targeting PTEN in pancreatic cancer. Ann Transl Med 2021; 9: 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Sun M, Sun Y, Li H. TMEM189 promotes breast cancer through inhibition of autophagy-regulated ferroptosis. Biochem Biophys Res Commun 2022; 622: 37–44. [DOI] [PubMed] [Google Scholar]
- 29.Jain IH, Calvo SE, Markhard AL, et al. Genetic screen for cell fitness in high or low oxygen highlights mitochondrial and lipid metabolism. Cell 2020; 181: 716–727.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messias MCF, Mecatti GC, Priolli DG, de Oliveira Carvalho P. Plasmalogen lipids: functional mechanism and their involvement in gastrointestinal cancer. Lipids Health Dis 2018; 17: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeleznik OA, Balasubramanian R, Zhao Y, et al. Circulating amino acids and amino acid-related metabolites and risk of breast cancer among predominantly premenopausal women. NPJ Breast Cancer 2021; 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y, Xie Y, Cao L, et al. The ferroptosis inducer erastin enhances sensitivity of acute myeloid leukemia cells to chemotherapeutic agents. Mol Cell Oncol 2015; 2: e1054549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Ge Z, Wang Z, Gao Y, Wang Y, Qu X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging (Albany NY) 2021; 13: 8115–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luis G, Godfroid A, Nishiumi S, et al. Tumor resistance to ferroptosis driven by stearoyl-CoA desaturase-1 (SCD1) in cancer cells and fatty acid biding protein-4 (FABP4) in tumor microenvironment promote tumor recurrence. Redox Biol 2021; 43: 102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun D, Li YC, Zhang XY. Lidocaine promoted ferroptosis by targeting miR-382-5p/SLC7A11 axis in ovarian and breast cancer. Front Pharmacol 2021; 12: 681223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi L, Sun B, Yang B, Lu S. PGM5P3-AS1 regulates MAP1LC3C to promote cell ferroptosis and thus inhibiting the malignant progression of triple-negative breast cancer. Breast Cancer Res Treat 2022; 193: 305–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.