Abstract
As a result of DNA typing of Mycobacterium microti isolates from animals in the United Kingdom and The Netherlands, we diagnosed four human M. microti infections. These are the first M. microti infections among humans to be reported. Three of the patients were immunocompromised and suffered from generalized forms of tuberculosis. The fourth patient was a 34-year-old immunocompetent male with a persistent cough and undefined X-ray abnormalities. Two of the M. microti infections were recognized by their IS6110 restriction fragment length polymorphism (RFLP) patterns, which showed a high degree of similarity with those of M. microti strains isolated from a pig and a ferret in The Netherlands. The two other human M. microti infections were recognized by using the recently developed DNA fingerprinting method, “spoligotyping,” directly on clinical material. All M. microti isolates from the United Kingdom and The Netherlands were found to contain an exceptionally short genomic direct repeat region, resulting in identical two-spacer sequence reactions in spoligotyping. In contrast, the highly similar IS6110 RFLP patterns of the vole strains from the United Kingdom differed considerably from the RFLPs of all M. microti strains isolated in The Netherlands, suggesting that geographic isolation led to divergent strains in the United Kingdom and on the continent.
Tuberculosis in the wild vole, or field mouse (Microtus agrestis), was discovered by Wells in 1937 (27, 28), and this epizootic disease was found to be rather common among these animals in the United Kingdom, with a prevalence ranging from 9 to 31% (27). The causative agent was named Mycobacterium tuberculosis subsp. muris (2), and later this species was designated Mycobacterium microti and classified as a member of the M. tuberculosis complex (26). M. microti differs from other M. tuberculosis complex strains in its S-shaped cell morphology, its slow growth in vitro, and its distinct host-specific pathogenicity for laboratory animals (13, 17, 27). Based on biochemical properties, this bacterium is difficult to distinguish from M. tuberculosis, Mycobacterium africanum, or Mycobacterium bovis (26).
In subsequent studies, M. microti was also detected in a limited number of other mammalian species, i.e., the bank vole (Clethrionomys glareolus) (27), the wood mouse (Apodemus sylvaticus) (27), the shrew (Sorex araneus) (27), cats and pigs (8, 9), and a zoo llama (Lama vicugna molina) (12). A morphologically similar organism, the “dassie bacillus,” was isolated in South Africa from the Cape hyrax, or dassie (Procavia capensis) (3, 15, 25), but this isolate differed from the vole bacillus in not being virulent for mice.
Recently, various repetitive genetic elements have been identified in M. tuberculosis complex bacteria, and these have been used to differentiate clinical isolates of M. tuberculosis, M. africanum, and M. bovis (14). The most frequently used repetitive elements in the molecular epidemiology of tuberculosis are the insertion sequence IS6110, the polymorphic GC-rich sequence (PGRS), and the direct repeat (DR), which are present in all members of the M. tuberculosis complex group, including M. microti (4, 14). However, there are no reported data on the DNA polymorphism associated with these repetitive DNA elements among human isolates of M. microti. The DNA polymorphism associated with these genetic markers is usually detected by restriction fragment length polymorphism (RFLP). Some DNA fingerprints characteristic of M. tuberculosis complex (sub-)species (14, 19) have been found, such as the characteristic combination of the IS1081 and IS6110 RFLP patterns of M. bovis BCG vaccine strains (22) and the specific IS1081 RFLP pattern, consisting of a single band, of the recently recognized subspecies Mycobacterium tuberculosis subsp. canetti (also called Mycobacterium canetti) (23). Furthermore, spacer oligonucleotide typing (spoligotyping) permits recognition of M. bovis by the characteristic absence of five spacer sequences in the 3′-terminal part of the DR cluster, and the absence of spacers 2, 9, and 16 (1, 10).
The present study was undertaken to determine the DNA polymorphism among M. microti isolates by using IS6110 fingerprinting and spoligotyping. We show that M. microti strains display characteristic IS6110 banding patterns and spoligotypes, distinct from types previously observed in other M. tuberculosis complex strains. Unexpectedly, these characteristic fingerprints and spoligotypes allowed us to identify four human tuberculous infections caused by M. microti.
MATERIALS AND METHODS
Mycobacterial strains.
M. microti strains isolated from voles in the 1930s in the United Kingdom (OV 254, OV 216, OV 183, and LS 1419) and the strain isolated from a Cape hyrax in 1958 in South Africa (dassie bacillus, 68/7171) were supplied by T. Jenkins and P. Draper, National Institute for Medical Research, The Ridgeway, Mill Hill, London, United Kingdom. The M. microti pig isolate, strain 161, isolated in 1965 in the southern part of The Netherlands, and strain 97-1297, isolated in 1993 from a ferret kept as a household pet in the western part of The Netherlands, were obtained from J. Haagsma, Central Veterinary Institute, 8200 AB Lelystad, The Netherlands. Strain P1376, isolated in 1968 from a llama born in the Antwerp Zoo, Antwerp, Belgium, was supplied by L. Rigouts, Mycobacteriology Unit, Institute of Tropical Medicine Prince Leopold, Antwerp 2000, Belgium. The M. microti type strain, ATCC 19422, a vole isolate, was obtained from the American Type Culture Collection, Manassas, Va.
Patient isolates.
Strain myc 17683 was isolated in 1993 from alveolar lavage fluid of a 12-year-old kidney transplantation patient, and strain myc 94-2272 was isolated in 1988 from the perfusion fluid of a 41-year-old dialysis patient. Strain 97-770 was isolated in 1997 from a lung biopsy of an immune-competent 34-year-old male with a persistent cough and undefined abnormalities on the chest X ray. Strain 97-1027 was isolated in 1997 from sputum of a 39-year-old human immunodeficiency virus (HIV)-positive male.
Bacteriological identification.
All strains were subjected to the Accuprobe culture confirmation test for the M. tuberculosis complex (Genprobe, San Diego, Calif.) and standard bacteriological analysis for M. tuberculosis complex bacteria (26).
Molecular typing.
Typing by RFLP using IS6110, PGRS, and DR as DNA probes was performed as described previously (14, 20, 21). The recently developed spoligotyping technique is based on the strain-dependent presence or absence of short nonrepetitive spacer sequences, which interspace the repetitive DR sequences (10, 24). Comparison of IS6110 fingerprints and spoligotypes by computer-assisted analysis was performed as described previously (7, 24). The IS6110 fingerprint database contained approximately 4,000 patterns of M. tuberculosis complex isolates from patients in The Netherlands in the period from 1990 through 1997 and 2,000 patterns from patients’ isolates originating in 30 different countries (18). The spoligotype database was established in 1995 through 1996 and contained 800 M. tuberculosis strains, mainly from patients in The Netherlands, and 800 M. bovis strains from humans and animals in The Netherlands and 15 other countries.
The presence of the mtp40 sequence, alleged M. tuberculosis specific, was determined by the PCR-based method described by Del Portillo et al. (5).
RESULTS
Genetic characterization of M. microti isolates.
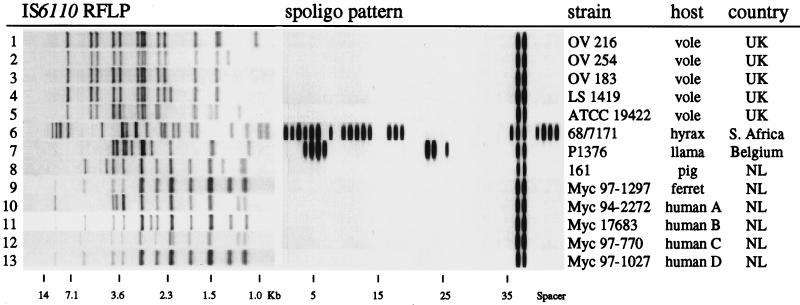
Nine M. microti strains from animal sources, including the dassie isolate, were subjected to IS6110 RFLP typing. The results are shown in Fig. 1, lanes 1 to 9. The five vole strains isolated in the United Kingdom in the 1930s (27, 28) all displayed distinct, but highly similar, IS6110 banding patterns. The fingerprints of these strains shared at least 11 of the 13 IS6110-containing PvuII restriction fragments (Fig. 1, lanes 1 to 5), suggesting that these strains derived relatively recently from a common ancestor. This close genetic relatedness was confirmed by the virtually identical PGRS fingerprints (data not shown). Two other strains, isolated from a Cape hyrax in South Africa and a zoo llama in Antwerp, displayed very different IS6110 and PGRS fingerprints, and these were also unrelated to the banding patterns found among the vole strains from the United Kingdom (Fig. 1, lanes 6 and 7). The remaining two strains, isolated from a ferret and a pig in The Netherlands, showed highly similar IS6110 RFLPs, and these were unrelated to those of the other seven animal isolates (Fig. 1, lanes 8 and 9).
FIG. 1.
IS6110 RFLP patterns and spoligotyping (spoligo) patterns of all described mycobacterial isolates.
By spoligotyping, only three different patterns were obtained. The five vole strains and the isolates from the ferret and the pig displayed reactions with only 2 of the 43 spacer oligonucleotides (spacers 37 and 38 [Fig. 1]). This indicates the presence in the DR region of only 2 of the 43 spacer DNA sequences currently used in spoligotyping and suggests that these seven M. microti strains may have an unusually small DR cluster. In order to confirm this observation, we subjected PvuII-digested DNA from these strains to Southern blotting using synthetic DR DNA as a probe. Only a single DR-hybridizing fragment was found among these strains, and the hybridization signal was weak compared to that in other strains such as M. tuberculosis H37Rv and M. bovis BCG strain P3, which harbor more than 40 DRs and spacers in the DR region (data not shown). Therefore, the DR region in these seven M. microti strains may be relatively short, presumably containing fewer DRs than have been observed previously among other M. tuberculosis complex strains (1, 6, 9).
The spoligotypes of the dassie strain and the zoo llama isolate were very different from the vole, ferret, and pig strains. The spoligotyping-PCR products of these strains hybridized with 22 and 9 spacer sequences, respectively (Fig. 1, lanes 6 and 7).
The spoligotypes of all eight M. microti isolates and the dassie strain (Fig. 1, lanes 1 to 9) were compared with those in the spoligotype database. No matching spoligotypes were found. Therefore, the spoligotype found in seven of the nine animal isolates seems to be characteristic of M. microti.
Recognition of M. microti infections in humans.
The IS6110 banding patterns of the M. microti strains and the dassie isolate were compared with the patterns in the database of fingerprints of human isolates. Only two patient isolates showed at least 80% similarity with the M. microti strains, a level which allows three to four IS6110 fragment mismatches. The patterns of these two strains were highly similar to the IS6110 fingerprints of the M. microti strains isolated from the ferret and the pig (Fig. 1, lanes 10 and 11). Spoligotyping of these two isolates revealed M. microti-characteristic hybridization patterns, identical to those of the ferret isolate, the pig isolate, and the five vole strains (Fig. 1, lanes 10 and 11). Because of its extremely slow growth during the primary isolation and the circumstantial evidence for transmission from mice, as discussed in the next paragraph, one of the two isolates, myc 94-2272, had previously been suspected to be M. microti by the regional public health laboratory where the strain was isolated. The other mycobacterial isolate, myc 17683, had previously been identified by the National Reference Laboratory as an “aberrant M. tuberculosis complex isolate” on the basis of doubtful biochemical criteria and slow growth (data not shown).
Two additional putative M. microti infections among humans were found in two different regional public health laboratories by performing spoligotyping directly on clinical material to detect the presence of M. tuberculosis complex-specific DNA. In both cases, the M. microti-characteristic spoligotype pattern was observed, as described above for the isolates from the first two patients, the pig, and the ferret and the five vole strains. Subsequent IS6110 DNA fingerprinting of these isolates showed a high degree of similarity with those of the isolates from the other humans, the ferret, and the pig (Fig. 1, lanes 12 and 13).
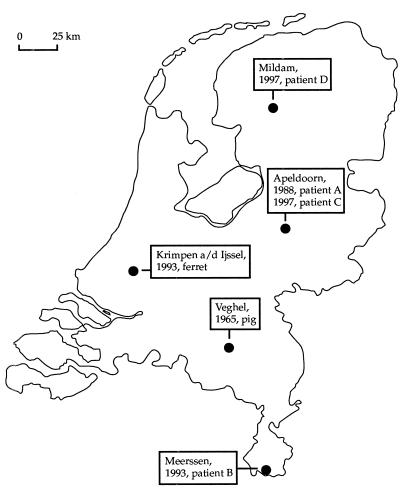
The geographic origins of the human and animal cases in The Netherlands are depicted in Fig. 2. Two human cases of M. microti infections, diagnosed in 1988 and 1997, were found in the same city. The houses of these patients were positioned only 1 km apart.
FIG. 2.
Map of The Netherlands indicating the geographic origins of the animal and human tuberculous infections caused by M. microti and the times of diagnosis.
Bacteriological identification.
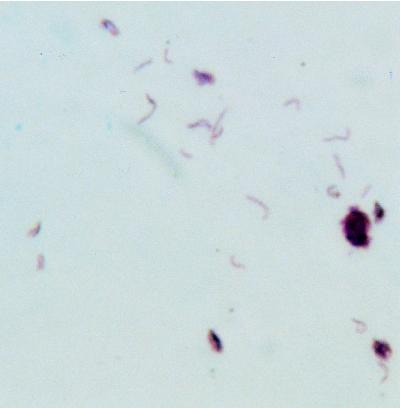
All four human M. microti isolates were found to grow extremely slowly on Löwenstein-Jensen medium during the primary isolation. Therefore, it was impossible to perform biochemical identification on these isolates. The remaining strains, which had been recultured more frequently and therefore grew more rapidly in vitro, were subjected to biochemical analysis, and the results are shown in Table 1. By biochemical identification, the strains shared properties of both M. tuberculosis and M. bovis, which is characteristic of M. microti (17, 26). However, the characteristic curved morphology of M. microti was not observed in any of the Ziehl-Neelsen-stained microscopic preparations of the bacteriological cultures. Because reexamination of the original microscopic slides of the peritoneal biopsy (patient A), the bronchial biopsy (patient B), the biopsy of the right apical segment of the upper lobe (patient C), and sputum (patient D) showed the presence of curved acid-fast bacilli (Fig. 3), we conclude that all four human isolates belong to the species M. microti. We presume that the characteristic property of S-curved cell morphology was lost during repeated in vitro culturing, as has been described previously (13).
TABLE 1.
Results of biochemical tests of some of the isolates included in this studya
| Strain | Growthb at the following temp (°C):
|
Growth on Löwenstein-Jensen medium with:
|
Niacin | Nitratase | Urease | Catalase | PO4 | Pyrazinamidase | Pigment | Colony morphology | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 | 36 | 45 | Pyruvate | Thiophene | |||||||||
| OV 216 | − | − | − | ++ | − | + | − | + | − | − | − | No | Smooth |
| OV 254 | − | ++ | − | ++ | − | + | − | + | − | − | − | No | Rough |
| OV 183 | − | + | − | + | − | + | − | + | − | − | − | No | Rough |
| LS1419 | − | − | − | ++ | − | + | − | + | − | − | + | No | Smooth |
| ATCC 19422 | − | ++ | − | ++ | − | + | − | + | − | − | − | No | Rough |
| 68/7171 | − | − | − | ++ | − | + | − | − | − | − | + | No | Smooth |
| P1376 | − | + | − | + | − | − | − | + | − | − | − | No | Rough |
| 161 | − | − | − | + | − | − | − | − | − | − | − | No | Smooth |
−, negative; +, positive; ++, strongly positive.
On Löwenstein-Jensen medium.
FIG. 3.
Ziehl-Neelsen-stained sputum of patient D. Note the characteristic curved acid-fast bacilli.
Clinical backgrounds of human cases.
Patient A, a 41-year-old male, had undergone kidney transplantation in 1982. Because of insufficient kidney transplant function, continuous ambulatory peritoneal dialysis was started in 1986. In 1988 the patient had fever, lost weight, suffered from progressive anemia, and was found to have esophagitis, gastritis, bulbitis, and colitis. No pathogenic bacteria were isolated. The patient was hospitalized, and the peritoneal fluid was found to be positive in Ziehl-Neelsen staining. The patient died shortly after the diagnosis of tuberculous peritonitis was made. After 4 months acid-fast bacteria were cultured, whereas no other bacteria were found. A small laparotomy revealed extensive tuberculous peritonitis. Histologic examination of peritoneal biopsy confirmed this diagnosis. The patient died shortly after the tuberculous peritonitis had been diagnosed. Because spoligotyping allows the simultaneous detection and typing of M. tuberculosis complex bacteria in clinical specimens (10), we subjected the 8-year-old tissue specimen, embedded in paraffin, to spoligotyping in 1996. The sample was positive, and the amplified DNA hybridized with spacers 37 and 38 only. This spoligotype pattern was identical to that of the cultured strain from the same patient (data not shown). This result confirms that the patient had indeed been infected with M. microti and excluded the possibility that only the dialysis fluid had been contaminated with M. microti. It should be noted that in 1988, when the strain was isolated, contact tracing did not lead to possible human sources of infection. However, during a visit of the social nurse, it was found that the patient kept his dialysis fluid bags in an unclean shed behind his house and that mouse droppings were present on the surface of the bags. This suggests that murine fecal contamination was the source of infection.
Patient B was a 12-year-old boy who had undergone kidney transplantation and was therefore treated with immunosuppressives (prednisone plus azathioprine). He was suffering from coughing. From the results of a chest X ray, tuberculosis was suspected. Acid-fast bacilli, later identified as M. microti, were isolated from both the bronchoalveolar lavage fluid and a bronchial biopsy. Besides the presence of the acid-fast bacilli, the histologic examination of the biopsy revealed extensive caseation and necrosis. A computed tomography (CT) scan of the head visualized a cerebral tumor suggestive of a tuberculoma, although no biopsy was taken. The family of the patient kept no pets, and contact tracing did not offer any clue about a possible source of infection. The patient was set on triple therapy with antituberculosis drugs, resulting in regression of the roentgenographic abnormalities and disappearance of the acid-fast bacilli.
Patient C, a 34-year-old male living in a caravan camp, presented with persistent coughing. On the chest X ray, a cavernous abnormality in the right apical segment of the upper lobe was seen. The Mantoux reaction was negative, as were the Ziehl-Neelsen staining and the sputum culture. A CT scan-guided lung biopsy was taken and revealed the presence of acid-fast bacilli. Spoligotyping performed directly on biopsied lung tissue was positive for M. microti. Mycobacterial culturing was successful after 3 months of incubation. After 6 months of triple antituberculosis therapy, the abnormalities had disappeared completely.
Patient D was a 39-year-old HIV-positive X-ray assistant, working in a hospital. This patient presented with coughing and an infiltrate in the left lower lobe. Microscopic examination of his sputum yielded high numbers of acid-fast bacilli (Fig. 3). Treatment was started after the multiplex PCR (11) directly on sputum had been found positive for M. tuberculosis complex (11). The patient was successfully treated (regression of the roentgenographic abnormalities and disappearance of the acid-fast bacilli) with antituberculosis drugs.
A possible indication for zoonotic transmission was found in the fact that an infestation of mice afflicted the house of the patient 5 months prior to the manifestation of the M. microti infection. Contact tracing, using routinely applied M. tuberculosis complex purified protein derivative (PPD), resulted in three positive skin tests, two from colleagues 33 and 35 years old and one from a 13-year-old cousin, none of whom were immunocompromised. These PPD-positive contacts are highly suggestive of human-to-human transmission of M. microti, since in The Netherlands the current annual risk of infection, and consequently the percentage of positive skin tests in the population, is very low.
Presence of the genomic mtp40 sequence.
The mtp40 DNA sequence has been described as species specific for M. tuberculosis and therefore is useful for differential diagnosis of infections due to M. tuberculosis (13). The PCR to detect this sequence was negative for all vole isolates, as well as for the hyrax strain. The PCR was positive for the llama, ferret, and pig strains and for all four human isolates.
DISCUSSION
In this study for the first time, M. microti infections among humans are described. This might indicate that such infections occur only rarely. On the other hand, human infections with M. microti are difficult to detect because of the extremely slow growth of this organism and because of the difficulty in recognizing this species by traditional bacteriological methods. Furthermore, as discussed by Pattyn et al. (13), phenotypic traits of M. microti strains, like the curved cell morphology and growth characteristics, may change after a few passages in vitro, a phenomenon which we also observed in this study. Because of these difficulties in diagnosing M. microti infections by traditional bacteriological methods, we presume that M. microti infections among humans have simply been overlooked so far. This makes it impossible to estimate the frequency of human M. microti infection.
This study shows that genetic markers generally used in the epidemiology of tuberculosis can be used to recognize M. microti infections among humans. Although IS6110, PGRS, and the DR have previously been identified in M. microti (4), no data have been available on the DNA polymorphism associated with these repetitive DNA elements in this species of the M. tuberculosis complex. The IS6110 fingerprints of the strains analyzed in this study indicate that there are at least two genetically distinct groups of M. microti in Europe. These two IS6110 genotypes have an unusually short chromosomal DR cluster in common, in which only two DR spacer sequences, derived from M. tuberculosis H37Rv and M. bovis BCG, are present. The IS6110 fingerprints and the spoligotypes of these European strains were unrelated to the two M. microti strains originating from the exotic Cape hyrax from South Africa and the llama from a zoo in Belgium.
The M. microti infections among humans were recognized because of their characteristic spoligotype and their uncommon IS6110 fingerprint patterns, which were similar to those of two animal M. microti strains, one isolated from a pig in 1965 in the southern part of The Netherlands (9) and the other isolated from a ferret kept as a household pet in the western part of The Netherlands in 1993. The former strain caused localized lesions in the laryngeal lymph nodes of the pig. During the same period Huitema and colleagues described the occurrence of similarly, characteristically curved mycobacteria in other pigs and also in cats (8, 9). Unfortunately, these strains were no longer available for DNA typing, and there are no reports on the isolation of M. microti from rodents in The Netherlands. However, it seems plausible to assume that the M. microti infection of the pig was acquired by contamination of soil with feces of mice or voles. The ferret M. microti strain was isolated in 1993 after a clinical diagnosis of mycobacterial infection was made, following illness featuring anorexia, weight loss, and malaise. After euthanasia, numerous acid-fast bacteria were found in all organ tissues. It is conceivable that the ferret contracted the M. microti infection through consumption of an infected rodent.
The limited IS6110-associated DNA polymorphism among the vole strains isolated in the 1930s in the United Kingdom indicates that they belong to a genetically closely related group of M. microti strains. This genotype family is distinct from the other IS6110 grouping of M. microti comprising two animal isolates and four human strains isolated in The Netherlands. Additional support for the existence of two genogroups is the observation that these groups differ in the presence or absence of the mtp40 sequence.
For one of the human isolates, we found circumstantial evidence for transmission from rodents to a human, because this strain was isolated from a patient who stored his dialysis fluid bags under nonhygienic conditions, where contamination with fecal material from mice or voles was likely to occur. For one of the other patients zoonotic transmission is also conceivable, as an infestation of mice afflicted his house a few months prior to the manifestation of tuberculous symptoms. For the two remaining patients no direct indications for involvement of zoonotic transmission were found, although one of the patients, a 41-year-old immunocompetent male, was living in a caravan camp, where the chances of contact with the habitat of mice may be greater than in an ordinary household.
Prewar studies on the prevalence of M. microti infections in Scotland, Wales, and England showed a prevalence ranging from 9 to 31% depending on the place and season of capture (27, 28). Unfortunately, no data on the prevalence of M. microti among voles in The Netherlands or other parts of Europe are available, but there is no reason to suspect that the prevalence in these areas differs from that previously observed by Wells and Oxon in the United Kingdom (27, 28). Taking into account the extended period during which M. microti strains were isolated in The Netherlands (from 1965 to 1997) and the wide geographical distribution of M. microti infections among humans and animals in The Netherlands, it is likely that M. microti is endemic among rodents in The Netherlands.
The dissimilarity in IS6110 banding patterns between the Dutch strains and the vole strains from the United Kingdom is probably due to their geographic isolation for extended periods. A similar effect of geographic isolation has been found for M. bovis strains isolated in Europe and South America (19).
Although the number of Dutch M. microti strains examined is small, they show little IS6110-associated polymorphism. This is in contrast with the genetic heterogeneity observed among M. bovis and M. tuberculosis isolates in this region (7, 19–21).
In the 1950s and 1960s, M. microti was considered nonpathogenic for humans, and for this reason, M. microti has been used as an experimental tuberculosis vaccine in trials in the United Kingdom and in the Czech Republic (16). One of the M. microti strains analyzed in this study (strain LS1419) was used in the United Kingdom as a vaccine strain in the Medical Research Council vaccine trial in the 1950s (5a). Our present study indicates that the use of live M. microti for vaccination may constitute a health hazard of a similar magnitude as the use of BCG for immunocompromised individuals.
ACKNOWLEDGMENTS
We thank P. Draper for his helpful suggestions.
This work was supported by the European Community Programme for Science Technology and Development (BMH4-CT97-91202) and the Dutch Foundation for Technical Sciences (STW).
REFERENCES
- 1.Aranaz A, Liébana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzolez O, Rodriguez-Ferri E F, Bunschoten A E, van Embden J D A, Cousins D. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooke W S. The vole acid-fast bacillus. Am Rev Tuberc. 1941;43:806–816. [Google Scholar]
- 3.Cousins D V, Peet R L, Gaynor W T, Williams S N, Gow B L. Tuberculosis in imported hyrax (Procavia capensis) caused by an unusual variant belonging to the Mycobacterium tuberculosis complex. Vet Microbiol. 1994;42:135–145. doi: 10.1016/0378-1135(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 4.Cousins D V, Williams S N, Reuter R, Forshaw D, Chadwick B, Coughran D, Collins P, Gales N. Tuberculosis in wild seals and characterization of the seal bacillus. Aust Vet J. 1993;70:92–97. doi: 10.1111/j.1751-0813.1993.tb03284.x. [DOI] [PubMed] [Google Scholar]
- 5.Del Portillo P, Murillo L A, Patarroyo M E. Amplification of a species-specific DNA fragment of Mycobacterium tuberculosis and its possible use in diagnosis. J Clin Microbiol. 1991;29:2163–2168. doi: 10.1128/jcm.29.10.2163-2168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Draper, P. Personal communication.
- 6.Hermans P W M, van Soolingen D, Bik E M, de Haas P E W, Dale J W, van Embden J D A. The insertion element IS987 from Mycobacterium bovis BCG is located in a hot spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermans P W M, Messadi F, Guebrexbher H, van Soolingen D, de Haas P E W, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, Zribi M, van Embden J D A. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia and the Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 8.Huitema H, van Vloten J. Murine tuberculosis in a cat. Antonie Leeuwenhoek. 1960;26:233–240. doi: 10.1007/BF02539009. [DOI] [PubMed] [Google Scholar]
- 9.Huitema H, Jaartsveld F H J. Mycobacterium microti infection in a cat and some pigs. Antonie Leeuwenhoek. 1967;33:209–212. doi: 10.1007/BF02045553. [DOI] [PubMed] [Google Scholar]
- 10.Kamerbeek J, Schouls L M, van Agterveld M, van Soolingen D, Bunschoten A E, Kolk A H J, Kuijper S, van Embden J D A. Rapid detection and simultaneous strain differentiation of Mycobacterium tuberculosis for diagnosis and tuberculosis control. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kox L F F, Jansen H M, Kuijper S, Kolk A H J. Multiplex PCR assay for immediate identification of the infecting species in patients with mycobacterial disease. J Clin Microbiol. 1997;35:1492–1498. doi: 10.1128/jcm.35.6.1492-1498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pattyn S R, Portaels F A, Kageruka P, Gigase P. Mycobacterium microti infection in a zoo-llama: lama Vicugna (molina) Acta Zool Pathol Antverp. 1970;51:17–24. [PubMed] [Google Scholar]
- 13.Pattyn S R, Portaels F A, Spanogne L, Magos J. Further studies on African strains of Mycobacterium tuberculosis. Comparison with M. bovis and M. microti. Ann Soc Belge Med Trop. 1970;50:211–228. [PubMed] [Google Scholar]
- 14.Small P M, van Embden J D A. Molecular epidemiology of tuberculosis. In: Bloom B, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: ASM Press; 1994. pp. 569–582. [Google Scholar]
- 15.Smith N. The ‘dassie’ bacillus. Tubercle. 1960;41:203–212. doi: 10.1016/s0041-3879(60)80080-3. [DOI] [PubMed] [Google Scholar]
- 16.Sula L, Radkovsky J. Protective effects of M. microti vaccine against tuberculosis. J Hyg Epidemiol Microbiol Immunol. 1976;1:1–6. [PubMed] [Google Scholar]
- 17.Tsukamura M, Mizuno S, Toyama H. Taxonomic studies on the Mycobacterium tuberculosis series. Microbiol Immunol. 1985;29:285–299. doi: 10.1111/j.1348-0421.1985.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 18.van Embden J D A, van Soolingen D, Heersma H F, de Neeling A J, Jones M E, Steiert M, Grek V, Mooi F R, Verhoef J. Establishment of a European network for the surveillance of Mycobacterium tuberculosis, MRSA and penicillin-resistant pneumococci. J Antimicrob Chemother. 1996;38:905–906. doi: 10.1093/jac/38.5.905. [DOI] [PubMed] [Google Scholar]
- 19.van Soolingen D, de Haas P E W, Haagsma J, Eger T, Hermans P W M, Ritacco V, Alito A, van Embden J D A. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J Clin Microbiol. 1994;32:2425–2433. doi: 10.1128/jcm.32.10.2425-2433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains; evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Soolingen D, Hermans P W M, de Haas P E W, van Embden J D A. Insertion element IS1081-associated restriction fragment length polymorphism in Mycobacterium tuberculosis complex species: a reliable tool to recognize Mycobacterium bovis BCG. J Clin Microbiol. 1992;30:1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Soolingen D, Hoogenboezem T, de Haas P E W, Hermans P W M, Koedam M A, Teppema K S, Brennan P J, Besra G S, Portaels F, Top J, Schouls L M, van Embden J D A. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 24.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhasaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner J C, Bokkenheuser V. The mycobacterium isolated from the dassie Procavia capensis (pallas) Tubercle. 1961;42:47–56. doi: 10.1016/s0041-3879(61)80019-6. [DOI] [PubMed] [Google Scholar]
- 26.Wayne L G, Kubica G P. The mycobacteria. In: Sneath P H A, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams and Wilkins Co.; 1986. pp. 1435–1457. [Google Scholar]
- 27.Wells A Q. The murine type of tubercle bacillus (the vole acid-fast bacillus). Sir William Dunn School of Pathology, University of Oxford; 1946. . Special Report Series in Medicine, Council of London, no. 259. [Google Scholar]
- 28.Wells A Q, Oxon D M. Tuberculosis in wild voles. Lancet. 1937;i:1221. [Google Scholar]