Abstract
Purpose
To evaluate the ability of preoperative MRI-based measurements to predict the pathological T (pT) stage and cervical lymph node metastasis (CLNM) via machine learning (ML)-driven models trained in oral tongue squamous cell carcinoma (OTSCC).
Materials and methods
108 patients with a new diagnosis of OTSCC were enrolled. The preoperative MRI study included post-contrast high-resolution T1-weighted images acquired in all patients. MRI-based depth of invasion (DOI) and tumor dimension—together with shape-based and intensity-based features—were extracted from the lesion volume segmentation. The entire dataset was randomly divided into a training set and a validation set, and the performances of different types of ML algorithms were evaluated and compared.
Results
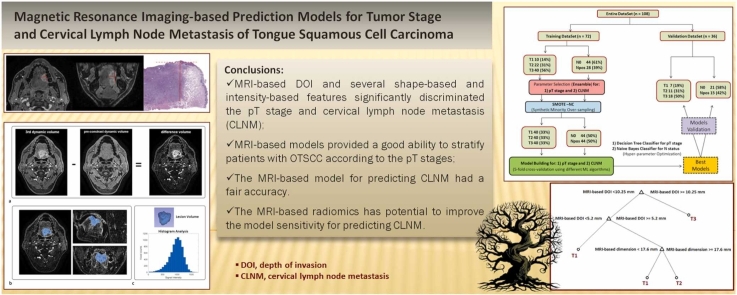
MRI-based DOI and tumor dimension together with several shape-based and intensity-based signatures significantly discriminated the pT stage and LN status. The overall accuracy of the model for predicting the pT stage was 0.86 (95%CI, 0.78–0.92) and 0.81 (0.64–0.91) in the training and validation sets, respectively. There was no improvement in the model performance upon including shape-based and intensity-based features. The model for predicting CLNM based on DOI and tumor dimensions had a fair accuracy of 0.68 (0.57–0.78) and 0.69 (0.51–0.84) in the training and validation sets, respectively. The shape-based and intensity-based signatures have shown potential for improving the model sensitivity, with a comparable accuracy.
Conclusion
MRI-based models driven by ML algorithms could stratify patients with OTSCC according to the pT stages. They had a moderate ability to predict cervical lymph node metastasis.
Keywords: Tongue cancer, Depth of invasion, Magnetic resonance imaging, Lymph node metastasis, Machine learning
Graphical Abstract
1. Introduction
Oral tongue squamous cell carcinoma (OTSCC) is the most common malignancy of the oral cavity (OCSCC). Standard treatment methods include surgery alone or in combination with adjuvant radiotherapy and chemotherapy [1].
In order to stage the tumor, the 8th edition of the American Joint Committee on Cancer (AJCC) staging manual introduced the depth of invasion (DOI) as a determinant key element along with the tumor dimension and adjacent sites involved [2]. The DOI has been described as the distance between the deepest point reached by tumor infiltration and the theoretical healthy mucosal line [3]. It differs from tumor thickness—namely the distance between the same deepest point and the tumor surface. This distinction allows to include the tumor infiltration in the tumor staging to better correlate these measurements to survival rates, thus avoiding the bias of bulky or excavating tumors. DOI has shown strong correlation to the presence of occult lymph node metastasis, risk of recurrence, and overall survival [4], [5], [6], [7]. A correct DOI measurement for early intermediate lesions with no evidence of neck nodal metastasis (i.e., cT1–T2 N0) is crucial to selecting patients that require a prophylactic neck dissection due to risk of nodal relapse.
Several studies have suggested a specific cutoff pathological DOIs (pDOIs) with the aim of obtaining an optimal risk stratification of patients with OTSCC postoperatively. An optimal cutoff of 4 mm avoids the morbidity associated with unnecessary elective dissection [8]. Thus, an accurate pre-treatment assessment of the DOI may contribute to preoperative tumor staging, treatment planning, and predicting the prognosis.
Clinical evaluation alone is limited, and thus the information derived from imaging techniques is essential to providing a correct preoperative assessment of the DOI in OCSCC. Ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI) are the most common techniques to evaluate the radiological DOI (rDOI) [9]. Ultrasound allows for adequate preoperative evaluation of the tumor and has the highest correlation coefficient between rDOI and pDOI in pT1–pT2 tumors. It is more accurate than CT and MRI and tends to overestimate the rDOI by approximately 2–3 mm [9], [10]. However, it is operator-dependent, requires dedicated probes for the oral cavity, and has diagnostic accuracy that changes with the location and size of the lesion—this in turn can lead to lower accuracy in large tumors (pT3) and tumors involving multiple subsites [11]. CT is a valuable modality for preoperative tumor staging and is the method of choice in patients who cannot undergo MRI. Nevertheless, MRI more accurately evaluates submucosal diffusion and invasion of the adjacent structures while defining the lymph node (LN) status [12], [13].
Recent developments in precision medicine have led to significant advancements in the diagnosis and prediction of tumor response to therapies driven by machine learning (ML) approaches to multimodal data analyses [14], [15]. A new research field is expanding and is represented by radiomics based on a sophisticated mathematical approach to the analysis of diagnostic images. This approach can extract many quantitative non-invasive tumor biomarkers with important potential implication for precision medicine across different cancer types including OTSCC [16], [17], [18]. Intra-tumor heterogeneity can be captured by first-order and higher-order textural features and may reflect the microstructural tissue characteristics and help clinicians to better stratify patients [19].
Several associations between pre-treatment radiomic signatures (mostly based on CT studies) and tumor biology/treatment outcome have already been reported. These offer accurate prognostic models to predict loco-regional relapse and/or overall survival although not all have been validated on independent patient cohorts [20], [21]. Advanced MRI techniques, i.e., perfusion-weighted and diffusion-weighted imaging, have shown high potential for predicting treatment response [22]; these offer useful information about tissue vascularity and cellularity, respectively. The apparent diffusion coefficient (ADC) can be evaluated both in tumor and non-tumor tissues and was recently suggested to improve the prediction of recurrence and disease-free survival of OTSCC [23].
Currently, however, only a few investigations have used an MRI-based radiomic approach driven by ML algorithms to predict the pT stage, the degree of pathological differentiation, and/or cervical lymph node metastasis (CLNM) in OTSCC [24], [25], [26]. Most studies evaluated different tumor sites, i.e., oropharyngeal and nasopharyngeal cancers [27]. Thus, the development of MRI-based predictive models for a more comprehensive characterization of OTSCC patients may be of interest for the scientific community.
The aim of this study is to evaluate the ability of preoperative MRI-based measurements alone or in combination with shape-based and intensity-based features derived from post-contrast high-resolution T1-weighed images to predict the pT stage and CLNM using ML-driven models trained on a large single-institution OTSCC patient population.
2. Methods and materials
2.1. Patient population
This single-institution retrospective study was approved by the institutional ethics committee (RS1834/23). The requirement for obtaining written informed consent was waived due to the retrospective nature of this study.
The inclusion criteria were: 1) preoperative MRI examination performed within two weeks preoperatively according to the acquisition protocol described below, 2) presence of a tumor that could be measured on MRI, and 3) availability of the pDOI measurement in the histopathological report. The exclusion criteria were: 1) preoperative chemo-radiotherapy, 2) classification as T4a regardless of the pDOI because of mandibular infiltration, 3) recurrent disease, and 4) poor MRI quality because of motion artifacts and/or image distortion due to dental implants.
2.2. MR Imaging Protocol
MRI was performed using 1.5 T (Optima MR 450w, GE Healthcare, Milwaukee, WI, USA) and 3 T (Discovery MR 750w, GE Healthcare, Milwaukee, WI, USA) scan systems using a 16-channel receive-only RF head-neck coil. MRI examinations included T2-weighted fast spin-echo coronal images (slice thickness, 4 mm), axial T2-weighted fast spin-echo images (slice thickness, 3 mm), and pre-contrast axial T1-weighted images (slice thickness, 3 mm); both were acquired from the skull base to the thoracic inlet. Four post-contrast T1-weighted dynamic volumes were acquired using an axial fast-spoiled gradient echo sequence after injecting a gadopentetatedimeglumine contrast agent at 0.1 mmol/kg body weight. The acquisition parameters are indicated in Table 1. The examination also included T1-weighted images in the axial and coronal planes.
Table 1.
Acquisition parameters of dynamic multiphase T1-weighted sequence.
| Field Strength | 1.5 T | 3 T |
|---|---|---|
| Type of Acquisition | 3D | 3D |
| TR/TE (ms) | 5.9/2.1 | 6.6/2.9 |
| FOV (mm2) | 256 × 256 | 256 × 256 |
| Acquisition matrix | 320 × 224 | 320 × 256 |
| Reconstruction matrix | 512 × 512 | 512 × 512 |
| Pixel size (mm2) | 0.8 × 1.1 | 0.8 × 1.0 |
| Slice thickness (mm) | 2 | 2 |
| Slice spacing (mm) | 1 | 1 |
| Flip angle (°) | 12 | 12 |
| Number of phases | 5 | 5 |
| Temporal Resolution (s) | 31 | 32 |
| Acquisition time (min) | 2 min 32 s | 2 min 40 s |
2.3. Determination of MRI-based DOI
All scans were imported to a commercial workstation (Advantage Workstation ADW, version 4.7, GE Healthcare) for multiplanar visualization and analyses. The images were reviewed by two radiologists (A.V. and F.P.) with more than 10 years of experience in head and neck imaging who were blinded to the histopathological data. The average DOI measurements (MRI-based DOI) measured by the two radiologists were compared with the pDOI. Cases of strong disagreement were reviewed and resolved by consensus.
The MRI-based DOI was measured on the post-contrast T1-weighted sequence in the 3rd dynamic volume, which was identified as the volume with the most tumor contrast versus tissue; the largest sections of the tumor in the axial or coronal planes were selected for the measurement. A perpendicular line from the reference line to the deepest end of the tumor was drawn to determine the invasive part, and the reference line was defined as the line joining the boundary between the tumor surface and the normal mucosa. The cT stage was evaluated in accordance with the 8th edition of the TNM classification considering the MRI-based DOI and maximum tumor diameter (MRI-based dimension). The exophytic part of the tumor and ulcerated component were not included. Supplementary Fig. S1 shows an example of an MRI-based DOI measurement.
2.4. Extraction of shape-based and intensity-based features
We investigated the added role of shape-based and intensity-based features to the MRI-based linear measurements, i.e. DOI and dimension, as annotated by the radiologist. Two sequential registration steps, an affine transformation and a B-Spline deformable transformation [28], were performed to obtain an optimal alignment between the pre-contrast dynamic volume (moving volume) and the 3rd dynamic volume (reference volume). The affine transformation with 12 degrees of freedom was applied to the pre-contrast volume to account for the global motion relative to the reference volume, while the B-spline transformation was used to capture the local image deformations due to breathing, swallowing, and/or jaw movements. The difference volume was derived by subtracting the registered baseline volume from the 3rd dynamic volume using the 3D Slicer filter “Absolute Value Difference Image”.
The entire tumor volume was then manually delineated, slice by slice, on the difference volume via the support of a semiautomatic level-tracing segmentation tool by two expert HN radiologists (A.V. and F.P.) already involved in the DOI measurement.
Prior to the quantitative analyses, the images were resampled to 1 mm isotropic voxel size and were discretized with a fixed bin width of 25. Shape-based features (n = 12) and intensity-based features (n = 18) were extracted from the tumor segmentation mask using the 3D Slicer Radiomics Extension based on the Python package Pyradiomics [29]; see more detail in Supplementary Table 1.
The ComBat method was applied to harmonize the intensity-based features derived from 1.5 T and 3 T scanners [30] using the package neuroCombat in R Studio.
The image analysis steps are shown in Fig. 1.
Fig. 1.
Illustration of the sequential steps to extract shape-based and intensity-based features from the dynamic high-spatial-resolution T1-weighted images.
2.5. Surgical procedures and histopathological analysis
All patients underwent upfront surgical treatment according to the preoperative stage. The patients were treated with transoral glossectomy for cT1 and cT2 OTSCC. Neck dissection was performed contextually in patients with cN0 with a cDOI > 4 mm and in those with clinical evidence of positive cervical nodes. Patients with cT3 OTSCC or those likely to receive adjuvant radiotherapy (clinical extranodal extension, cN2 or cN3 nodal disease, nodal disease at level IV or V, perineural invasion, vascular invasion, or lymphatic invasion) underwent transcervical glossectomy, neck dissection, and free-flap reconstruction of the tongue.
Tissue samples were formalin-fixed and paraffin-embedded. Sections were cut from each FFPE block and mounted on slides. They were stained with hematoxylin and eosin and digitally acquired using a ScanScope digital scanner (Aperio ePathology Solutions). The pDOI was measured by dropping a “plumb line” from the closest adjacent normal mucosal basement membrane level to the deepest point of tumor invasion regardless of the presence/absence of ulceration. The surgical margins were negative on all sides. Tumors diagnosed before 2017 and staged in accordance with the 7th edition of the AJCC were reviewed by a pathologist and re-staged according to the 8th edition of the AJCC [2]. Supplementary Fig. S1 shows an example of the pDOI- and MRI-based DOI measurements.
2.6. Statistical analyses
Statistical analyses used an integrated statistical approach based on conventional statistical tests and machine learning to improve data interpretability and prediction accuracy [31]. The Mann-Whitney or Kruskal-Wallis tests were used to compare two or more independent groups when the variables were continuous. A chi-squared or Fisher’s exact tests were used to determine the relationships between categorical variables according to the case. Box-and-whisker plots were used to show the distributions of the parameters. The Kappa coefficient was used to assess the inter-rater agreement in tumor staging between the pathologist and the radiologist. The agreement between pathological and MRI-based DOI measurements was assessed by Bland-Altman plots as was agreement between pathological and MRI-based tumor dimensions. Spearman’s rho correlation test was used to assess the strength of the correlation between the two measurements. Statistical significance was set at a p-value < 0.05 to be statistically significant. Statistical analyses were performed using SPSS software (SPSS version 21, SPSS Inc., Chicago, IL, USA).
2.7. Feature selection
The initial selection of the most significant variables for predicting the pathological T (pT) stage and CLNM used the Kruskal-Wallis test or the Mann-Whitney test, respectively, with a cutoff for the p-value of 0.10; a further selection of the remaining variables was obtained from a Random Forest classifier [14]. In the case of high correlation between the selected features, the one with the highest predictive power was chosen.
Lastly, to mitigate the effect of the different numerical range of the selected variables and to improve the performance of the subsequent ML modelling, the final datasets were standardized using the z-score normalization procedure as described by Haga et al. [32].
2.8. ML modelling
Before building the models for predicting the pT stage and CLNM, the entire dataset was randomly divided into a training set (two-thirds, n = 72) and a validation set (one-third, n = 36). The goal here was to test the possibility of applying the proposed models to a separate patient group and obtaining a more reliable estimate of the models’ performance according to the international statements to thus promote transparency in the design and presentation of a prediction model study (TRIPOD) [33].
In our dataset, patient groups with different pT stages (T1, T2, and T3) and with negative (N0) or positive (Npos) cervical lymph nodes were not equally populated; thus, an oversampling technique was used to generate synthetic data for the minority class to mitigate the data imbalance during the training process [34] using the RSBID package in RStudio.
The model building was next articulated in two main steps: 1) only the MRI-based linear measurements, i.e. DOI and dimension (as annotated by the radiologist) were included; 2) the most relevant shape-based and intensity-based features were added to determine their ability to improve the models developed at step 1.
Before building the proposed models, the performances of different ML algorithms (i.e., decision trees, linear discriminants, logistic regression when appropriate, naive Bayes, support vector machines, K-nearest neighbor classifiers and ensemble classifiers) were compared in both the training and validation sets, given that they had dissimilar characteristics in terms of the computational speed, hypotheses on the nature of the data, and interpretability [14].
The best selection of the hyperparameters of each algorithm was obtained via an iterative optimization process employing a stratified five-fold cross-validation to reduce overfitting. The accuracy, sensitivity, and specificity overall and for each class were calculated to evaluate the model performance together with the confusion matrix. Generalization of the area under the receiver operating characteristic curve (AUC) for multiple classifications was applied to estimate the AUC of the pT stage model [35]. The 95% bootstrap confidence interval (CI) for the AUC was calculated using 1000 samples. The mid-p-value McNemar test was used to compare the prediction accuracies of different models. The MATLAB Statistics and Machine Learning Toolbox (Release 2021b, The Mathworks Inc., Natick, Massachusetts) was used to build the ML-based models.
3. Results
A total of 108 patients with newly diagnosed OTSCC were included in the study from January 2013 to September 2022. Table 2 shows the population characteristics. There were 65 MRI studies acquired on 1.5 T (60.2%) and 43 on a 3 T scanner (39.8%).
Table 2.
Characteristics of patients and tumors in the entire dataset and the training and validation sets.
| Patient and tumour characteristics | Entire set | Training set | Validation set | P-value | |
|---|---|---|---|---|---|
| (n = 108) | (n = 72) | (n = 36) | |||
| Sex | Male | 52 (48%) | 35 (49%) | 17 (47%) | 0.946 |
| Female | 56 (52%) | 37 (51%) | 19 (53%) | ||
| Age (years) | mean±SD | 63.5 ± 14.3 | 64.0 ± 13.9 | 62.40 ± 15.3 | 0.672 |
| T-stage | T1 | 17 (16%) | 10 (14%) | 7 (19%) | |
| T2 | 33 (31%) | 22 (31%) | 11 (31%) | 0.738 | |
| T3 | 58 (54%) | 40 (56%) | 18 (50%) | ||
| N-stage | N0 | 65 (60%) | 44 (61%) | 21 (58%) | |
| N1 | 10 (9%) | 6 (8%) | 4 (11%) | 0.973 | |
| N2 | 18 (17%) | 12 (17%) | 6(17%) | ||
| N3 | 15 (14%) | 10 (14%) | 5 (14%) | ||
| Infiltration pattern | expansive | 31 (29%) | 20 (28%) | 11 (31%) | |
| infiltrative | 49 (45%) | 32 (44%) | 17 (47%) | 0.822 | |
| mixed | 28 (26%) | 20 (28%) | 8 (22%) | ||
| Grade | G1 | 5 (5%) | 4 (6%) | 1 (3%) | |
| G2 | 58 (54%) | 36 (50%) | 22 (61%) | 0.509 | |
| G3 | 45 (42%) | 32 (44%) | 13 (36%) | ||
| Pathological dimension (mm) | median [95% CI] | 25 [20, 25.6] |
25 [20.3, 29.3] |
20 [16.7, 30] |
0.408 |
| Pathological DOI (mm) | median [95% CI] | 12 [9], [12] |
12 [8, 14.7] |
10.5 [8], [13] |
0.674 |
| MRI-based dimension (mm) | median [95% CI] | 30 [25.4, 34.1] |
29.5 [25], [35] |
30 [20.5, 35.1] |
0.573 |
| MRI-based DOI (mm) | median [95% CI] | 25.0 [20.0,25.6] |
12.5 [9.7, 14.7] |
11 [8.9, 14.8] |
0.845 |
Abbreviations: SD, standard deviation; 95% CI, 95% confidence interval, DOI, depth of invasion; MRI, magnetic resonance imaging
The MRI-based DOI was strongly related to pDOI (Rho = 0.86, 95% CI: 0.80–0.90, p < 0.001); MRI-based tumor dimension was also related to the pathological tumor dimension, although it had a weaker correlation coefficient (Rho = 0.75, 95% CI: 0.66–0.82, p < 0.0001). The mean differences (95% CI) between pathological and MRI-based DOIs and between pathological and MRI-based tumor dimensions were − 1.3 (−8.2 to 5.7) and − 4.7 (−21 to 12) mm, respectively. Supplementary Fig. S2 shows the relative Bland–Altman plots. The agreement between the pT and cT stages was good (Kappa coefficient = 0.66, 95% CI: 0.53–0.79).
3.1. Feature selection
MRI-based DOIs and dimensions could discriminate the pT stage and the presence of CLNM; the box-and-whisker plots are indicated in Fig. 2, and the summary statistics are reported in Supplementary Table 2. The predictor ranks of MRI-based DOI and dimension as well as their superiority compared to the other clinicopathological variables are shown by the bar plots in Supplementary Fig. S3.
Fig. 2.
Box-and-whisker plots of MRI-based tumor dimension (a) and MRI-based DOI (b) according to the pathological tumor stage T1, T2 or T3. P-values are obtained from the Kruskal–Wallis test. Box-and-whisker plots of MRI-based tumor dimension (c) and MRI-based DOI (d) according to the negative (N0) or positive (Npos) status of cervical lymph nodes. P-values are obtained from the Mann–Whitney test.
Among the shape-based and intensity-based features, 15 of them significantly differed between pT stages such as the least/minor/major axis length, surface area, mesh volume and flatness, which all increased as T stage increased. Whereas the minimum and skewness of signal intensity decreased as T stage increased (see summary statistics in Supplementary Table S3).
Analogously, 12 shape-based and intensity-based features significantly differed between LN-negative patients and patients with CLNM such as the least/major axis length, maximum 2D/3D diameter, surface area, and mesh volume which increased for patients with positive LN. Whereas the sphericity and minimum decreased (see summary statistics in Supplementary Table S4).
When combining all the variables, the predictor ranks obtained from the random forest classifier are shown in Supplementary Fig. S4: MR-based DOI was the dominant predictor for the pT stage together with several shape-based features and some intensity-based features, i.e., minimum and skewness.
The energy and the minimum were the strongest intensity-based predictors for CLNM, together with the MR-based DOI and a number of highly correlated features measuring the linear dimensions of the lesion.
The heat map showing the strength of correlation between all the variables using the Spearman’s rho correlation test is illustrated in Supplementary Fig. S5.
An example representation of the evolution of the lesion shape and signal intensity distribution for patients with increasing tumor pT stages and for LN-negative/LN-positive patients is shown in Figs. 3 and 4, respectively.
Fig. 3.
An example representation of the evolution of the lesion shape (a,c,e) and signal intensity distribution (b,d,f) for patients with increasing tumor pT stages.
Fig. 4.
An example representation of the different lesion shape and signal intensity distribution for a patient without CLNM (a-d) and a patient with CLNM (e-h).
3.2. ML modelling
Fig. 5 shows the data analysis pipeline. The training and validation sets showed no significant differences (Table 2).
Fig. 5.
Pipeline of the model building for predicting the pT stage and cervical lymph node metastasis (CLNM).
Among the different types of ML algorithms, the Decision Tree and Naïve Bayes classifiers provided the best results in terms of accuracy in predicting the pT stage and CLNM, respectively. Supplementary Figs. S6 and S7 show a comparison of the performances of the different ML algorithms in the training and validation sets.
The best decision model for predicting the pT stage was obtained including only the MRI-based linear measurements, i.e. DOI and dimension, as annotated by the radiologist: The overall accuracy was 0.86 (95%CI, 0.78–0.92) and 0.81 (95%CI, 0.64–0.91) in the training and validation sets, respectively. The corresponding AUCs were 0.88 (95%CI, 0.81–0.92) and 0.79 (95%CI, 0.66–0.88), respectively (Table 3). The graphical representation of the trained decision tree is illustrated in Supplementary Fig. S8.
Table 3.
Performance of the proposed model for predicting the pT stage in the training and validation sets.
| Training Set |
|||||
|---|---|---|---|---|---|
| Selected Features | Class | Sensitivity | Specificity | Accuracy [95% CI] |
AUC [95% CI] |
| Model1: -MR-based DOI -MR-based Dimension |
Overall | 0.86 | 0.93 | 0.86 [0.78,0.92] |
0.88 [0.81,0.92] |
| By class | |||||
| T1 | 0.95 | 0.89 | 0.95 | ||
| T2 | 0.70 | 0.96 | 0.70 | ||
| T3 | 0.93 | 0.94 | 0.93 | ||
| Validation Set | |||||
| Selected Features | Class | Sensitivity | Specificity |
Accuracy [95% CI] |
AUC [95% CI] |
| Model1: -MR-based DOI -MR-based Dimension |
Overall | 0.77 | 0.89 | 0.81 [0.64,0.91] |
0.79 [0.66,0.88] |
| By class | |||||
| T1 | 0.71 | 0.93 | 0.71 | ||
| T2 | 0.64 | 0.92 | 0.64 | ||
| T3 | 0.94 | 0.83 | 0.94 | ||
AUC, area under the curve; CI, confidence interval.
The inclusion of shape-based and intensity-based features did not allow an improvement of the pT stage model performance. The best-performing classification was obtained by including three features (MR-based DOI, flatness, and skewness) but it had an inferior accuracy of 0.82 and 0.78 in the training and validation set, respectively, versus the previous model.
The decision model for predicting the CLNM based only on DOI and dimension (Model 1) had an accuracy of 0.68 (95%CI, 0.57–0.78) and 0.69 (95%CI, 0.51–0.84) in the training and validation set, respectively, with an AUC of 0.75 (95%CI, 0.63–0.85) and 0.64 (95%CI, 0.50–0.79), respectively. The inclusion of shape-based and intensity-based features (specifically, energy, maximum 2D diameter and minimum) led to an improvement of the model performance in the training set: the accuracy of this model (Model 2) was 0.74 (0.63–0.83) with an increase in sensitivity from 0.64 to 0.75 compared to Model 1. In the validation set, the accuracy of Model 2 was 0.67 (0.49–0.81) with a slight increase in sensitivity from 0.55 to 0.60 versus Model 1 (Table 4). There was no significant difference between the accuracies of Model1 and Model 2, both in the training and validation set (p = 0.26 and p = 0.75, respectively).
Table 4.
Performance of the two proposed models for predicting CLNM in the training and validation sets.
|
Training Set |
|||||
|---|---|---|---|---|---|
| Selected Features | Class | Sensitivity | Specificity |
Accuracy [95% CI] |
AUC [95% CI] |
| Model 1: MR-based DOI MR-based Dimension |
N0/Npos | 0.64 | 0.73 | 0.68 [0.57,0.78] |
0.75 [0.63,0.85] |
| Model 2: -Energy -Maximum 2D Diameter -Minimum |
N0/Npos | 0.75 | 0.73 | 0.74 [0.63,0.83] |
0.75 [0.64,0.83] |
|
Validation Set |
|||||
|---|---|---|---|---|---|
| Selected Features | Class | Sensitivity | Specificity |
Accuracy [95% CI] |
AUC [95% CI] |
| Model 1: -MR-based DOI -MR-based Dimension |
N0/Npos | 0.55 | 0.81 | 0.69 [0.51,0.84] |
0.64 [0.50,0.79] |
| Model 2: -Energy -Maximum 2D Diameter -Minimum |
N0/Npos | 0.60 | 0.71 | 0.67 [0.49,0.81] |
0.66 [0.50,0.81] |
AUC, area under the curve; CI, confidence interval.
Fig. 6 illustrates the confusion matrices relative to the proposed models: one model for predicting the pT stage and two models for predicting the CLNM as described in Table 3, Table 4.
Fig. 6.
Confusion matrices of the training and validation sets, relative to the pT stage and CLNM models: including only MR-based DOI and dimension (a,b) or shape-based and intensity-based features (c).
4. Discussion
We investigated whether preoperative MRI-based measurements, i.e., lesion DOI and dimension—alone or combined with shape-based and intensity-based features—can predict the pT stage and cervical lymph node metastasis using ML-driven models trained using a single-institution OTSCC patient population.
The MRI-based DOI and feature extraction were determined using post-contrast high-spatial-resolution T1-weighted imaging, which was suggested as the optimal MRI scan to obtain accurate measurements and the best correlation with the pDOI [36], [37]. The correlation between rDOI and pDOI has largely been documented in the literature, and a pooled correlation coefficient of 0.85–0.86 and a mean difference between the two measurements of 1.8 mm has been reported [9], [10]. A recent systematic review by Lee et al. [9] showed that the correlation coefficient of MRI had an intermediate value of 0.85 between ultrasound and CT although the MRI-based DOI showed the largest difference (2.6 mm) with respect to the pDOI versus ultrasound and CT-based DOI. The potential causes of DOI overestimation on MRI, compared to pDOI, may be the effects of edema and peritumoral inflammation of tongue tissues on the MRI signal and shrinkage of the specimen when formalin treated [38], [39], [40].
Our findings are consistent with these results and indicate a correlation coefficient between MRI-based DOI and pDOI of 0.87 (95% CI: 0.81–0.91) with a mean difference of 1.3 mm (95% CI: - 5.7–8.2 mm). In view of the strong association between MRI-based DOI and pDOI as well as between the MRI-based tumor dimension and the pathological dimension (Rho = 0.75, 95% CI: 0.66– 0.82), we hypothesized that a ML-driven model based on these radiological parameters would accurately predict the pT stage.
The best decision model for predicting the pT stage included only the MR-based DOI and dimension. In the training set, its overall accuracy was good (0.86) with a higher value in predicting the T1 and T3 stages (0.95 and 0.93) than the T2 stage (0.70). There was good specificity for each class, good to very good sensitivity, and an AUC of 0.88. Good performance was also found in the validation set with an overall accuracy of 0.81 and an AUC of 0.79.
A comparison with existing literature is not straightforward because most previous papers focused on the strength of the relationships between rDOI and pDOI or between the cT and pT stage [9], [41], [42], [43] except Tang et al. [37] who reported high AUCs (≥ 0.97) of MRI-based DOI in distinguishing the T1 stage form the T2 stage and the T2 stage from the T3 stage. Although Tang et al. [37] obtained these results using a similar MRI sequence, the discrepancy with our findings may be attributable to differences in the characteristics of patients and/or tumors and the different approaches to model-building (we preferred to split our dataset into a training set and a validation set to test the model performance in a separate patient group and provide more realistic evaluations).
Our decision tree graph indicated a first optimal threshold of 10.3 mm for MRI-based DOI to discriminate between the T3 stage and the T1 or T2 stage and a second optimal threshold of 5.2 mm for MRI-based DOIs to discriminate between the T1 stage and the T1 or T2 stage depending on the MRI-based tumor dimension (< 17.6 or ≥ 17.6 mm). The thresholds obtained for the MRI-based DOIs were slightly larger but consistent with those used for the pDOI, thus confirming the strong association between these two measurements. We found that the tumor dimensions may also contribute to further improvement of model accuracy particularly in discriminating between the T1 and T2 stages. Moreover, the use of ML-driven T staging based on MRI was particularly useful to better classify T1-stage tumors, which were frequently misclassified as T2-stage tumors— probably because of the larger impact of DOI overestimation, compared to pDOI, on measurements of a few millimeters [9].
The inclusion of shape-based and intensity-based features has not improved the pT stage model performance versus the simpler model based on MRI-based DOI and dimensions. Nevertheless, a number of features were significantly different among the pT stages suggesting a correlation between MRI-based signatures and the pathological stage as suggested by a recent study of Corti at al. focused on locally advanced OCSCC [19]. In particular, the minimum of the signal intensity decreased for higher tumor stages suggesting the presence of increasing necrosis areas, which previous studies have shown to correlate with largest and more aggressive tumors [22], [44]. We also found that tumors with higher stages were characterized by a decrease in skewness: this might be because larger lesions typically also have more extensive contrast-enhancing parts, which may grow faster than poor enhancing/necrotic regions causing an asymmetry of the signal intensity distribution towards the right side.
Two competitive models were proposed to use MRI-based measurements to correctly identify patients with and without CLNM. The first one was based on MRI-measured DOI and dimension. The second one had comparable accuracy but better sensitivity and included one signature derived from the shape-based family, i.e., a maximum 2D diameter, and two features derived from the intensity-based family: the energy (a measure of the signal intensity of voxels) and the minimum signal intensity already mentioned above.
The potential of MRI-based DOI to discriminate patients with and without LN metastasis was already reported [4], [5] suggesting an optimal cutoff between 7 mm and 10.5 mm to predict CLNM based on post-contrast T1-weighted imaging. However, this study not indicate a threshold value of DOI because we proposed decision models including at least two variables, whose combination was found to improve the model predictive power versus the DOI measurement alone [45].
Only a few studies have performed an MRI texture analysis to distinguish LN-negative from LN-positive patients in OCSCC [24], [25]. Yuan et al. [24] found that the Naïve Bayes classifier was the best in accordance with our findings. Various models based on T2-weighetd and/or post-contrast T1-weigheted signatures were proposed. All of these models achieved comparable accuracies and AUCs to our results with fair to good specificity but poor sensitivity. Interestingly, Wang et al. [25] suggested analyzing not only the primary tumor volume but also the 10-mm peritumoral extension to possibly detect micrometastasis and improve the sensitivity of CLNM predictions.
This study does have some limitations. The retrospective nature and relatively small sample size of this study may have introduced biases and confounding factors. We did not extract textural or higher-order signatures, thus we did not fully investigate the potential of MRI-based radiomics to predict both the pT stage and CLNM. Furthermore, we did not explore the correlation among MRI-based measurements, clinicopathological factors, and treatment outcomes, i.e., locoregional control and disease-specific survival. These topics will be considered for future investigations.
5. Conclusions
MRI-based models driven by ML algorithms provide a good ability to stratify patients with OTSCC according to the pT stage and a fair-to-good ability to predict CLNM. Several shape-based and intensity-based features have shown potential to improve the model sensitivity for predicting CLNM but the current level of accuracy is still inadequate and should be refined by a more sophisticated approach to MRI-based radiomics or by introducing other predictors. Further investigations with a larger patient population and a robust external validation are needed to corroborate our analyses.
Author contribution statement
All authors have made a substantial and intellectual contribution to the work, they were all involved with the conception, manuscript writing and final approval of the manuscript.
Funding
This work was financially supported through funding from the institutional “Ricerca Corrente” granted by the Italian Ministry of Health.
Declaration of Competing Interest
None of the authors have potential conflict of interest including any financial, personal or other relationships with other people or organizations that could inappropriately influence this work.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.08.020.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA A Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.M.B. Amin, S. Edge, F. Greene, D.R. Byrd, R.K. Brookland, et al. (Eds.). AJCC Cancer Staging Manual, 8th edition, Springer International Publishing: American Joint Commission on Cancer, 2017.
- 3.Piazza C., Montalto N., Paderno A., Taglietti V., Nicolai P. Is it time to incorporate ‘depth of infiltration’ in the T staging of oral tongue and floor of mouth cancer? Curr Opin Otolaryngol Head Neck Surg. 2014;22:81–89. doi: 10.1097/MOO.0000000000000038. [DOI] [PubMed] [Google Scholar]
- 4.Jung J., Cho N.H., Kim J., Choi E.C., Lee S.Y., et al. Significant invasion depth of early oral tongue cancer originated from the lateral border to predict regional metastases and prognosis. Int J Oral Maxillofac Surg. 2009;38:653–660. doi: 10.1016/j.ijom.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Haraguchi K., Yoshiga D., Oda M., Tabe S., Mitsugi S., et al. Depth of invasion determined by magnetic resonance imaging in tongue cancer can be a predictor of cervical lymph node metastasis. Oral Surg, Oral Med, Oral Pathol Oral Radiol. 2021;131:231–240. doi: 10.1016/j.oooo.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Xu C., Yuan J., Kang L., Zhang X., Wang L., et al. Significance of depth of invasion determined by MRI in cT1N0 tongue squamous cell carcinoma. Sci Rep. 2020;10:4695. doi: 10.1038/s41598-020-61474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tam S., Amit M., Zafereo M., Bell D., Weber R.S. Depth of invasion as a predictor of nodal disease and survival in patients with oral tongue squamous cell carcinoma. Head Neck. 2018:25506. doi: 10.1002/hed.25506. (hed) [DOI] [PubMed] [Google Scholar]
- 8.de Matos L.L., Manfro G., dos Santos R.V., Stabenow E., de Mello E.S., et al. Tumor thickness as a predictive factor of lymph node metastasis and disease recurrence in T1N0 and T2N0 squamous cell carcinoma of the oral tongue. Oral Surg, Oral Med, Oral Pathol Oral Radiol. 2014;118:209–217. doi: 10.1016/j.oooo.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Lee M.K., Choi Y. Correlation between radiologic depth of invasion and pathologic depth of invasion in oral cavity squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2023;136 doi: 10.1016/j.oraloncology.2022.106249. [DOI] [PubMed] [Google Scholar]
- 10.Voizard B., Khoury M., Saydy N., Nelson K., Cardin G.B., et al. Preoperative evaluation of depth of invasion in oral tongue squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2023;136 doi: 10.1016/j.oraloncology.2022.106273. [DOI] [PubMed] [Google Scholar]
- 11.Noorlag R., Klein Nulent T.J.W., Delwel V.E.J., Pameijer F.A., Willems S.M., et al. Assessment of tumour depth in early tongue cancer: accuracy of MRI and intraoral ultrasound. Oral Oncol. 2020;110 doi: 10.1016/j.oraloncology.2020.104895. [DOI] [PubMed] [Google Scholar]
- 12.Mao M.-H., Wang S., Feng Z., Li J.-Z., Li H., et al. Accuracy of magnetic resonance imaging in evaluating the depth of invasion of tongue cancer. A prospective cohort study. Oral Oncol. 2019;91:79–84. doi: 10.1016/j.oraloncology.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Park J.-O., Jung S.-L., Joo Y.-H., Jung C.-K., Cho K.-J., et al. Diagnostic accuracy of magnetic resonance imaging (MRI) in the assessment of tumor invasion depth in oral/oropharyngeal cancer. Oral Oncol. 2011;47:381–386. doi: 10.1016/j.oraloncology.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Erickson B.J., Korfiatis P., Akkus Z., Kline T.L. Machine learning for medical imaging. RadioGraphics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud P., Giraud P., Gasnier A., El Ayachy R., Kreps S., et al. Radiomics and machine learning for radiotherapy in head and neck cancers. Front Oncol. 2019;9:174. doi: 10.3389/fonc.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alabi R.O., Youssef O., Pirinen M., Elmusrati M., Mäkitie A.A., Leivo I., et al. Machine learning in oral squamous cell carcinoma: current status, clinical concerns and prospects for future—a systematic review. Artif Intell Med. 2021;115 doi: 10.1016/j.artmed.2021.102060. [DOI] [PubMed] [Google Scholar]
- 17.Tanadini-Lang S., Balermpas P., Guckenberger M., Pavic M., Riesterer O., Vuong D., et al. Radiomic biomarkers for head and neck squamous cell carcinoma. Strahl Onkol. 2020;196:868–878. doi: 10.1007/s00066-020-01638-4. [DOI] [PubMed] [Google Scholar]
- 18.Mossinelli C., Tagliabue M., Ruju F., Cammarata G., Volpe S., Raimondi S., et al. The role of radiomics in tongue cancer: a new tool for prognosis prediction. Head Neck. 2023;45:849–861. doi: 10.1002/hed.27299. [DOI] [PubMed] [Google Scholar]
- 19.Corti A., De Cecco L., Cavalieri S., Lenoci D., Pistore F., Calareso G., et al. MRI-based radiomic prognostic signature for locally advanced oral cavity squamous cell carcinoma: development, testing and comparison with genomic prognostic signatures. Biomark Res. 2023;11:69. doi: 10.1186/s40364-023-00494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mes S.W., Van Velden F.H.P., Peltenburg B., Peeters C.F.W., Te Beest D.E., Van De Wiel M.A., et al. Outcome prediction of head and neck squamous cell carcinoma by MRI radiomic signatures. Eur Radio. 2020;30:6311–6321. doi: 10.1007/s00330-020-06962-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guha A., Connor S., Anjari M., Naik H., Siddiqui M., Cook G., et al. Radiomic analysis for response assessment in advanced head and neck cancers, a distant dream or an inevitable reality? A systematic review of the current level of evidence. BJR. 2020;93:20190496. doi: 10.1259/bjr.20190496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos P., Van Der Hulst H.J., Van Den Brekel M.W.M., Schats W., Jasperse B., Beets-Tan R.G.H., et al. Prognostic functional MR imaging parameters in head and neck squamous cell carcinoma: a systematic review. Eur J Radiol. 2021;144 doi: 10.1016/j.ejrad.2021.109952. [DOI] [PubMed] [Google Scholar]
- 23.Cai L., Li X., Wu L., Wang B., Si M., Tao X. A prognostic model generated from an apparent diffusion coefficient ratio reliably predicts the outcomes of oral tongue squamous cell carcinoma. Curr Oncol. 2022;29:9031–9045. doi: 10.3390/curroncol29120708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y., Ren J., Tao X. Machine learning–based MRI texture analysis to predict occult lymph node metastasis in early-stage oral tongue squamous cell carcinoma. Eur Radio. 2021;31:6429–6437. doi: 10.1007/s00330-021-07731-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang F., Tan R., Feng K., Hu J., Zhuang Z., et al. Magnetic resonance imaging‐based radiomics features associated with depth of invasion predicted lymph node metastasis and prognosis in tongue Cancer. Magn Reson Imaging. 2022;56:196–209. doi: 10.1002/jmri.28019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu B., Huang C., Xu J., Liu S., Guan Y., Li T., et al. Prediction of the degree of pathological differentiation in tongue squamous cell carcinoma based on radiomics analysis of magnetic resonance images. BMC Oral Health. 2021;21:585. doi: 10.1186/s12903-021-01947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruixola G., Remacha E., Jiménez-Pastor A., Dualde D., Viala A., Montón J.V., et al. Radiomics and radiogenomics in head and neck squamous cell carcinoma: potential contribution to patient management and challenges. Cancer Treat Rev. 2021;99 doi: 10.1016/j.ctrv.2021.102263. [DOI] [PubMed] [Google Scholar]
- 28.Du G.E.J. Medical image registration using B-spline transform. Int J Simul: Syst, Sci Technol. 2016 doi: 10.5013/IJSSST.a.17.48.01. [DOI] [Google Scholar]
- 29.Van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortin J.-P., Parker D., Tunç B., Watanabe T., Elliott M.A., Ruparel K., et al. Harmonization of multi-site diffusion tensor imaging data. NeuroImage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajula H.S.R., Verlato G., Manchia M., Antonucci N., Fanos V. Comparison of conventional statistical methods with machine learning in medicine: diagnosis, drug development, and treatment. Medicina. 2020;56:455. doi: 10.3390/medicina56090455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haga A., Takahashi W., Aoki S., Nawa K., Yamashita H., Abe O., et al. Standardization of imaging features for radiomics analysis. J Med Invest. 2019;66:35–37. doi: 10.2152/jmi.66.35. [DOI] [PubMed] [Google Scholar]
- 33.Moons K.G.M., Altman D.G., Reitsma J.B., Ioannidis J.P.A., Macaskill P., et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 34.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority over-sampling technique. Jair. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 35.Hand D.J., Till R.J. Simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn. 2001;45:171–186. doi: 10.1023/A:1010920819831. [DOI] [Google Scholar]
- 36.Vidiri A., Panfili M., Boellis A., Cristalli G., Gangemi E., et al. The role of MRI-derived depth of invasion in staging oral tongue squamous cell carcinoma: inter-reader and radiological–pathological agreement. Acta Radio. 2020;61:344–352. doi: 10.1177/0284185119862946. [DOI] [PubMed] [Google Scholar]
- 37.Tang W., Wang Y., Yuan Y., Tao X. Assessment of tumor depth in oral tongue squamous cell carcinoma with multiparametric MRI: correlation with pathology. Eur Radio. 2022;32:254–261. doi: 10.1007/s00330-021-08148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam P., Au-Yeung K.M., Cheng P.W., Wei W.I., Yuen A.P.-W., et al. Correlating MRI and histologic tumor thickness in the assessment of oral tongue cancer. Am J Roentgenol. 2004;182:803–808. doi: 10.2214/ajr.182.3.1820803. [DOI] [PubMed] [Google Scholar]
- 39.Li M., Yuan Z., Tang Z. The accuracy of magnetic resonance imaging to measure the depth of invasion in oral tongue cancer: a systematic review and meta-analysis. Int J Oral Maxillofac Surg. 2022;51:431–440. doi: 10.1016/j.ijom.2021.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Baba A., Ojiri H., Ogane S., Hashimoto K., Inoue T., et al. Usefulness of contrast-enhanced CT in the evaluation of depth of invasion in oral tongue squamous cell carcinoma: comparison with MRI. Oral Radio. 2021;37:86–94. doi: 10.1007/s11282-020-00429-y. [DOI] [PubMed] [Google Scholar]
- 41.Murakami R., Shiraishi S., Yoshida R., Sakata J., Yamana K., et al. Reliability of MRI-derived depth of invasion of oral tongue cancer. Acad Radiol. 2019;26:e180–e186. doi: 10.1016/j.acra.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 42.Baba A., Hashimoto K., Kayama R., Yamauchi H., Ikeda K., et al. Radiological approach for the newly incorporated T staging factor, depth of invasion (DOI), of the oral tongue cancer in the 8th edition of American Joint Committee on Cancer (AJCC) staging manual: assessment of the necessity for elective neck dissection. Jpn J Radio. 2020;38:821–832. doi: 10.1007/s11604-020-00982-w. [DOI] [PubMed] [Google Scholar]
- 43.Takamura M., Kobayashi T., Nikkuni Y., Katsura K., Yamazaki M., et al. A comparative study between CT, MRI, and intraoral US for the evaluation of the depth of invasion in early stage (T1/T2) tongue squamous cell carcinoma. Oral Radio. 2022;38:114–125. doi: 10.1007/s11282-021-00533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janssen H.L., Haustermans K.M., Balm A.J., Begg A.C. Hypoxia in head and neck cancer: how much, how important. Head Neck. 2005;27:622–638. doi: 10.1002/hed.20223. [DOI] [PubMed] [Google Scholar]
- 45.Shan J., Jiang R., Chen X., Zhong Y., Zhang W., Xie L., et al. Machine learning predicts lymph node metastasis in early-stage oral tongue squamous cell carcinoma. J Oral Maxillofac Surg. 2020;78:2208–2218. doi: 10.1016/j.joms.2020.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material