Abstract
IS1245 is an insertion element widely prevalent among isolates of Mycobacterium avium. We used PvuII Southern blots to analyze IS1245 polymorphisms among 159 M. avium isolates (141 clinical isolates from 40 human immunodeficiency virus-infected patients plus 18 epidemiologically related environmental isolates) that represented 40 distinct M. avium strains, as resolved by previous studies by pulsed-field gel electrophoresis (PFGE). All 40 strains carried DNA homologous to IS1245 and thus were typeable. Twenty-five (63%) strains had ≥10 copies of the element, 6 (15%) had 4 to 9 copies, and 9 (23%) had only 1 to 3 copies. Among the last group of nine strains (each of which was distinct by PFGE analysis), IS1245 typing resolved only four patterns and thus provided poor discriminatory power. To evaluate the in vivo stability of IS1245, we analyzed 32 strains for which sets of 2 to 19 epidemiologically related isolates were available. For 19 (59%) of these sets, all isolates representing the same strain had indistinguishable IS1245 patterns. Within eight (25%) sets, one or more isolates had IS1245 patterns that differed by one or two fragments from the modal pattern for the isolates of that strain. Five (16%) sets included isolates whose patterns differed by three or more fragments; on the basis of IS1245 typing those isolates would have been designated distinct strains. IS1245 was stable during in vitro passage, suggesting that the variations observed represented natural translocations of the element. IS1245 provides a useful tool for molecular strain typing of M. avium but may have limitations for analyzing strains with low copy numbers or for resolving extended epidemiologic relationships.
Mycobacterium avium has emerged as the most common cause of disseminated bacterial infection among patients with AIDS in the United States, affecting up to 40% of patients with advanced human immunodeficiency virus (HIV) infection (7, 9, 11). M. avium have been recovered worldwide from natural waters, potable waters, and soil (5, 9), as well as from a wide range of animal hosts (e.g., cows, pigs, and birds). Investigations to define the specific sources and routes of transmission of M. avium infections require molecular strain typing methods (2, 3, 14, 19).
Recently, Guerrero et al. (6) described the application of Southern blot analyses of IS1245, a M. avium species-specific insertion element. Clinical isolates from 38 European patients each contained multiple (typically 10 to 20) copies of the element distributed among diverse chromosomal sites resulting in highly polymorphic restriction fragment length polymorphism (RFLP) patterns. In the present investigation we further evaluated the usefulness of IS1245 Southern blots for differentiating strains of M. avium, with particular attention to the discriminatory power and reproducibility of this typing system (2). To this end, we used isolates that were well-characterized epidemiologically and that had been independently analyzed by pulsed-field gel electrophoresis (PFGE) (3, 14, 19).
MATERIALS AND METHODS
Isolates.
A total of 159 M. avium isolates were studied, including 141 clinical isolates cultured from 40 HIV-infected patients and 18 environmental isolates cultured from epidemiologically related water sources, as described previously (3, 14, 19). These isolates represented 40 distinct M. avium strains as defined by PFGE analysis of AseI digests (3, 14). Six patients had polyclonal infections with two strains each (3, 14). The isolates examined included 32 sets of two or more isolates, each set representing a single, distinct strain as resolved by PFGE. These included 14 sets each representing independent colonies picked from the same primary culture plate (3), 13 sets each representing sequential clinical isolates from individual patients (14), 2 sets each representing clinical isolates from multiple different patients, and 3 sets each representing isolates from one or more patients plus one or more environmental samples (19).
IS1245 probe.
A 427-bp internal fragment of IS1245 from a clinical M. avium strain was amplified by PCR with the specific oligonucleotides P1 (5′-GCC GCC GAA ACG ATC TAC) and P2 (5′-AGG TGG CGT CGA GGA AGA C) by using the conditions described by Guerrero et al. (6). The amplified product was cloned into the vector pCRII (Invitrogen, San Diego, Calif.), digested with EcoRI, isolated by agarose gel electrophoresis, labeled with [α-32P]dCTP (NEN Research, Boston, Mass.) with a random primer kit (Boehringer Mannheim, Indianapolis, Ind.), and used for hybridization to Southern blots.
Strain typing.
All isolates were initially typed by PFGE to resolve AseI digests as described previously (14). PFGE patterns consisted of ∼20 well-resolved fragments and were interpreted visually by using the criteria proposed by Tenover et al. (16). Among isolates representing the same strain, the PFGE patterns were all either indistinguishable or differed by three or fewer bands. Among isolates designated different strains, pairwise comparisons indicated that all patterns had fewer than 50% of bands in common (most had fewer than 25% in common) and thus differed substantially.
Southern blot analyses were performed with DNA either prepared in agarose plugs for PFGE studies and released by β-agarose (10) or isolated directly from cells as described by van Soolingen et al. (18). DNA was digested with PvuII and, for some isolates, also with SalI (both enzymes were from New England Biolabs, Beverly, Mass.). Digests were electrophoretically separated in a 0.8% agarose gel (Seakem GTG; FMC, Rockland, Maine), vacuum transferred to a nylon membrane (Duralon; Stratagene, La Jolla, Calif.), and probed with the radiolabeled fragment of IS1245 prepared as described above. The membranes were exposed to X-ray film (X-OMAT AR; Kodak, Rochester, N.Y.) at −80°C for various lengths of time and processed in an automated film developer (RGII; Fuji Photo Film, Elmsford, N.Y.).
RESULTS
Typeability and discriminatory power of IS1245 strain typing.
All 40 M. avium strains carried DNA homologous to IS1245; however, there was considerable variation in the number of restriction fragments detected in different strains (Fig. 1; Table 1). In general, the fragments detected ranged in size from 0.8 to ≥20 kb. Since IS1245 typically has no internal PvuII site, fragments of >1.3 kb are likely to represent distinct copies of IS1245. Among 31 (77.5%) strains with four or more distinct PvuII fragments detected on Southern blots, the IS1245 RFLP patterns were diverse and discriminatory; each strain, as defined by a distinct PFGE profile, also had a distinct RFLP pattern on the IS1245 blot.
FIG. 1.
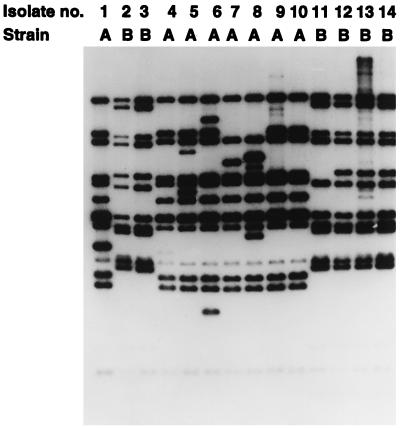
Southern blot of IS1245 RFLPs among M. avium strains representing polyclonal infections in six different AIDS patients. For each patient, the two strains (A and B) defined by PFGE have distinct IS1245 RFLP patterns.
TABLE 1.
Distribution among 40 distinct clinical strains of M. avium of number of PvuII fragments detected on Southern blots probed with IS1245
| No. of fragments detected | No. (%) of strains |
|---|---|
| ≤3 | 9 (22.5) |
| 4–9 | 6 (15.0) |
| 10–20 | 12 (30.0) |
| >20 | 13 (32.5) |
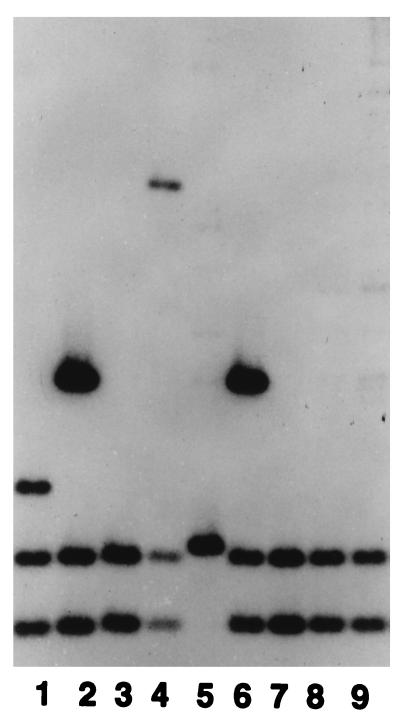
In contrast, for nine (22.5%) strains IS1245 analysis detected only two or three fragments and defined only four distinct patterns. Two of these patterns were represented by multiple strains that had distinct PFGE profiles but indistinguishable IS1245 patterns (Fig. 2). There were no apparent epidemiologic relationships among the strains within each of these IS1245 classes (data not shown). For five strains, the IS1245 profile comprised two fragments of ∼900 and 1,300 bp, respectively. For two strains, the IS1245 profile included these fragments plus an additional fragment of ∼3,000 bp. Of note, the two smaller fragments were also present in the more complex profiles and typically were disproportionately fainter than the other fragments detected. Other faint fragments of various sizes were also observed in some of the complex patterns.
FIG. 2.
Southern blot of IS1245 RFLPs among M. avium strains whose IS1245 patterns comprised one to three fragments. Lanes: 1, patient 1225; 2, patient 1106; 3, patient 1142; 4, patient 5026; 5, patient 1228; 6, patient 1213; 7, patient 1235; 8, patient 1060; 9, patient 1009. Each isolate except the isolates in lanes 1 and 8 represented a distinct strain, as defined by PFGE; the isolates in lanes 1 and 8 represented the same strain (i.e., had the same PFGE patterns) but had IS1245 RFLP patterns that differed by one fragment.
Technical and in vitro reproducibility of IS1245 strain typing.
For 115 isolates, the IS1245 profiles were determined with two or more independent Southern blots, and consistent results were obtained for each isolate. To confirm that the RFLP patterns were stable during in vitro handling of the isolates, independent subcultures of nine strains were prepared from stock cultures over several months and were used to make DNA preparations for PvuII RFLP analysis. For each strain, the replicate DNA preparations gave identical RFLP profiles.
In vivo stability of IS1245 RFLPs.
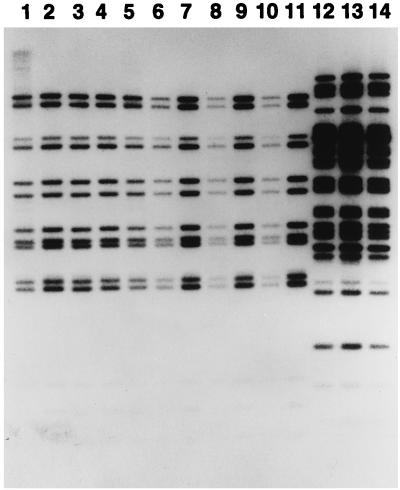
To investigate the reproducibility and stability of IS1245 RFLPs at a single time point, we analyzed 14 sets of isolates representing multiple (3 to 10) colonies of individual strains isolated independently from single patients (Table 2) (3). Within eight sets, the RFLP patterns were indistinguishable, consistent with strict clonality (Fig. 3). Within six sets, the patterns of one to six isolates demonstrated one or two fragment differences (Fig. 4). Within the two sets showing the greatest diversity (seven and five subtypes, respectively), the variant patterns differed from the modal IS1245 pattern for that strain by six and five fragments, respectively.
TABLE 2.
Reproducibility of IS1245 Southern blot patterns among multiple isolates representing individual strains
| Source of multiple isolates representing a single straina | No. of strains | No. of isolates per strain | Time span (days) covered by isolates | No. of strains showing patterns that differ by the following no. of fragments:
|
||
|---|---|---|---|---|---|---|
| 0 | 1–2 | ≥3 | ||||
| Individual patients | 14 | 3–10 | ≤1 | 8 | 4 | 2 |
| Individual patients | 13 | 2–10 | 7–192 (median, 85) | 10 | 2 | 1 |
| Multiple patients | 2 | 4–6 | 295–372 | 0 | 1 | 1 |
| Multiple patients plus environment | 3 | 4–19 | 99, 1,054, 1,201 | 1 | 1 | 1 |
As defined by PFGE analysis; see text for details.
FIG. 3.
Stability of IS1245 RFLPs among M. avium isolates representing independent colonies picked from the same primary culture plate. Lanes 1 to 11, isolates of strain B from patient 1161, who had polyclonal infection; lanes 12 to 14, isolates from patient 1142, who had monoclonal infection. All isolates from an individual patient represented the same strain, as defined by PFGE, and had indistinguishable IS1245 RFLP patterns. Figure 4, lanes 2, 3, 11, 12, and 13, represents additional isolates of strain B from patient 1161; the isolate in Fig. 3, lane 1, is the same isolate as that in Fig. 4, lanes 13 and 14.
FIG. 4.
Variation among IS1245 RFLPs among M. avium isolates representing independent colonies picked from primary culture plates for patient 1161, who had polyclonal infection. Lanes 1 and 4 to 10, independent colonies of strain A, as defined by PFGE, that demonstrate multiple different polymorphisms; lanes 2, 3, 11, 12, and 14, independent colonies of strain B that demonstrate a single polymorphism (lane 11); lanes 13 and 14, partial and complete PvuII digests of the same DNA preparation that indicate that polymorphisms are readily distinguished from incomplete digests.
Stability of IS1245 RFLPs over time.
To determine the in vivo stability of the IS1245 patterns over time, we analyzed sets of isolates from individual patients, from multiple epidemiologically related patients, and from epidemiologically related patients and environmental water samples (Table 2). Each set of isolates represented a single strain of M. avium as resolved by PFGE. First we examined the IS1245 RFLP profiles of 13 sets of sequential isolates representing single strains recovered from cultures of up to three specimens obtained from individual patients over time. Within 10 sets, the available isolates had indistinguishable IS1245 patterns; the time spans covered by these sets ranged from 7 to 192 days (median, 85 days). Within two sets, there were isolates which differed from the modal pattern by one or two fragments; the time spans covered by these sets were 69 and 88 days, respectively. The third set included nine isolates: one collected from the bone marrow on day 0, three collected from the feces on day 32, three collected from the blood on day 32, and two also collected from the blood on day 130. The IS1245 pattern of the isolate from day 0 consisted of 26 well-resolved fragments and was indistinguishable from that of one of the isolates from day 130. However, the patterns for the six isolates from day 32 and the remaining day 130 isolate differed from that pattern by up to six fragments. In pairwise comparisons (16), there were as many as six fragment differences among the eight IS1245 patterns detected for this set of isolates.
We examined two sets of isolates representing single strains recovered from several epidemiologically related patients. Within one set, consisting of six isolates from two patients, the IS1245 patterns of the two isolates from one patient differed by a single fragment from the patterns of the four isolates from the second patient. Within the other set, consisting of three isolates from three patients, there were three pattern variations that differed by one to three fragments, resulting in a set of related IS1245 types.
We examined three sets of isolates representing single strains recovered both from patients and from epidemiologically related water samples (19). Within one set, which included three clinical isolates and one water isolate recovered over 99 days, all isolates had indistinguishable IS1245 patterns. The second set included 19 isolates obtained from two patients and multiple water samples from one hospital over 1,054 days. The 10 isolates collected over the first 700 days, including both clinical isolates, had indistinguishable RFLP patterns; the nine environmental isolates collected over the remaining time demonstrated a pattern that differed from that of the previous 10 isolates by a single additional 1.9-kb fragment. The third set included 13 isolates from three patients and hospital water spanning 1,201 days; these isolates demonstrated seven closely related patterns, but with no clear temporal relationship or progression.
The variations observed among the PvuII RFLP patterns of multiple isolates representing individual strains could be related either to the creation or loss of PvuII restriction sites flanking stable IS1245 insertions or to transpositions of IS1245 to new chromosomal sites. To distinguish among these possibilities, we examined Southern blots prepared with SalI restriction digests for multiple isolates of eight strains showing appreciable PvuII RFLP variations. For six strains, the SalI RFLP patterns also demonstrated multiple variations (data not shown). Within one strain for which two isolates demonstrated PvuII patterns that differed by a single fragment, the SalI patterns were indistinguishable. The isolates of the eighth strain studied exhibited a PvuII pattern comprising two or three weakly hybridizing bands; SalI Southern blots probed under the same hybridization conditions had no detectable fragments.
Stability of IS1245 during in vitro passage.
The variations observed among the IS1245 RFLP patterns of M. avium isolates representing individual strains collected over time suggest that this insertion element can transpose in vivo at an appreciable frequency. To investigate whether such rearrangements occur during in vitro passage, we examined subcultures of three strains before and after in vitro passage on agar media; one strain with stable RFLP patterns and two with variable RFLP patterns over time were chosen. For each of the subcultures, the IS1245 pattern was indistinguishable from the pattern of the baseline isolate, suggesting that IS1245 is relatively stable during short-term in vitro passage.
DISCUSSION
The goals of this study were to investigate the utility of IS1245 RFLP analysis for strain typing of M. avium isolates and to compare the performance characteristics of this technique with those of PFGE analysis. We examined Southern blots prepared from PvuII digests for 159 isolates representing 40 epidemiologically well-documented strains, which had been independently typed by PFGE analysis of AseI digests.
All isolates were typeable by IS1245 RFLP analysis. Whereas almost all of the European clinical isolates of M. avium had multiple copies of IS1245 (6, 12), nine (22.5%) of the 40 U.S. strains that we examined had three or fewer copies of the element. M. avium strains with low numbers of IS1245 copies have been reported as being typical of avian isolates (4, 6). The most critical limitation associated with the presence of a low insertion sequence copy number is that the corresponding patterns are poorly discriminatory for differentiating among epidemiologically unrelated isolates representing distinct strains, as resolved by PFGE analysis. These observations are analogous to those obtained by IS6110 Southern blot analysis of Mycobacterium tuberculosis isolates in which isolates with five or fewer copies of the element could not be reliably resolved into distinct strains (1, 15, 17, 21).
As noted in previous studies (12, 13), although Southern blots probed with a fragment of IS1245 typically demonstrated strongly hybridizing fragments, fragments with a disproportionately weaker signal were also observed for some isolates. In our experience, such fragments were particularly characteristic of low-copy-number patterns and were observed even under stringent hybridization conditions. These fragments may represent cross-hybridizations with the related element IS1311, as suggested by Roiz et al. (13).
We evaluated several distinct aspects of the reproducibility of IS1245 typing. As expected, the technical reproducibility with independent preparations of DNA from the same isolate was excellent. However, the biologic reproducibility was less consistent. That is, the IS1245 profiles among multiple independent isolates representing the same strain, as defined by the combination of epidemiologic and PFGE data, often demonstrated appreciable variability. Among 32 strains, only for 19 (59%) did all isolates within a strain have indistinguishable IS1245 patterns; 4 (13%) strains included isolates whose IS1245 profiles differed from the modal pattern for that strain by three or more fragments. This level of within-strain variation was found among independent isolates collected at a single time point, isolates collected from individual patients over time, and isolates collected from multiple patients.
To define further the basis of these variations, we examined SalI digests of selected isolates representing eight distinct strains. For six of these strains, the SalI profiles also varied among isolates representing a single strain. These observations support the interpretation that the variations on the PvuII blots demonstrated chromosomal transposition of the insertion sequence element itself rather than simply flanking restriction site polymorphisms. Similar observations of transpositions of IS6110 in sequential isolates of M. tuberculosis from individual patients have recently been reported (20).
To determine whether simple in vitro passage could result in IS1245 RFLPs, we passaged in vitro three strains for which there were significant variations in the IS1245 profiles among independent clinical isolates. For each of the three strains, DNA prepared from isolates before and after the in vitro passages had indistinguishable IS1245 profiles. Thus, the variations observed are unlikely to have been generated during in vitro processing.
In summary, this study confirms previous reports that IS1245 (or homologous sequences) are widely prevalent among clinical isolates of M. avium and that Southern blot analysis of this element is useful for strain typing. However, at least among these clinical isolates from U.S. patients, a substantial minority of strains had low numbers of copies of the element, and for such strains the RFLP analysis had poor discriminatory power. In addition, we observed significant biologic variability in the locations and copy numbers of IS1245 elements among independent isolates representing the same strain. As emphasized by Hunter (8), decreased reproducibility also results in decreased discriminatory power. The limited biologic reproducibility of IS1245 may confound attempts to use this strain typing system to resolve complex epidemiologic relationships, e.g., to detect clusters of patients infected with the same strain of M. avium or to detect the environmental source of the infecting strain.
REFERENCES
- 1.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;24:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 2.Arbeit R D. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th edition. Washington, D.C: American Society for Microbiology; 1995. pp. 190–208. [Google Scholar]
- 3.Arbeit R D, Slutsky A, Barber T W, Maslow J N, Niemczyk S, Falkinham J O, III, O’Conner G T, von Reyn C F. Genetic diversity among strains of Mycobacterium avium causing monoclonal and polyclonal bacteremia in patients with AIDS. J Infect Dis. 1993;167:1384–1390. doi: 10.1093/infdis/167.6.1384. [DOI] [PubMed] [Google Scholar]
- 4.Bono M, Jemmi T, Bernasconi C, Burki D, Telenti A, Bodmer T. Genotypic characterization of strains of Mycobacterium avium recovered from animals and their comparison to human strains. Appl Environ Microbiol. 1995;61:371–373. doi: 10.1128/aem.61.1.371-373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George K L, Parker B C, Gruft H, Falkinham J O., III Epidemiology of infection by nontuberculous mycobacteria. II. Growth and survival in natural waters. Am Rev Respir Dis. 1980;122:89–94. doi: 10.1164/arrd.1980.122.1.89. [DOI] [PubMed] [Google Scholar]
- 6.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horsburgh C R., Jr Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 8.Hunter P R. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inderlied C B, Kemper C A, Bermudez L E. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maslow J, Brecher S, Gunn J, Durbin A, Barlow M, Arbeit R. Variation and persistence of methicillin-resistant Staphylococcus aureus strains among individual patients over extended periods of time. Eur J Clin Microbiol Infect Dis. 1995;14:282–290. doi: 10.1007/BF02116520. [DOI] [PubMed] [Google Scholar]
- 11.Nightingale S D, Byrd L, Southern P, Jockusch J, Cal S, Wynne B. Incidence of Mycobacterium avium-intracellulare complex bacteremia in human immunodeficiency virus positive patients. J Infect Dis. 1992;165:1082–1085. doi: 10.1093/infdis/165.6.1082. [DOI] [PubMed] [Google Scholar]
- 12.Picardeau M, Vincent V. Typing of Mycobacterium avium isolates by PCR. J Clin Microbiol. 1996;34:389–392. doi: 10.1128/jcm.34.2.389-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roiz M, Palenque E, Guerrero C, Garcia M. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J Clin Microbiol. 1995;33:1389–1391. doi: 10.1128/jcm.33.5.1389-1391.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slutsky A M, Arbeit R D, Barber T W, Rich J, von Reyn C F, Pieciak W, Barlow M A, Maslow J N. Polyclonal infection due to Mycobacterium avium complex in patients with AIDS detected by pulsed field gel electrophoresis of sequential clinical isolates. J Clin Microbiol. 1994;32:1773–1778. doi: 10.1128/jcm.32.7.1773-1778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G K. The epidemiology of tuberculosis in San Francisco: a population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 16.Tenover F, Arbeit R, Goering R, Mickelsen P, Murray B, Persing D, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Reyn C F, Maslow J N, Barber T W, Falkinham III J O, Arbeit R D. Persistent colonization of potable water as a source of Mycobacterium avium infection in AIDS. Lancet. 1994;343:1137–1141. doi: 10.1016/s0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 20.Yeh R, Ponce de Leone A, Agasino C, Hahn J, Daley C, Hopewell P, Small P. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]
- 21.Yuen K, Chan C, Chan K, Yam W, Ho P, Chau P. IS6110 based amplityping assay and RFLP fingerprinting of clinical isolates of Mycobacterium tuberculosis. J Clin Pathol. 1995;48:924–928. doi: 10.1136/jcp.48.10.924. [DOI] [PMC free article] [PubMed] [Google Scholar]