Abstract
Quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma has rapidly become an important tool in basic HIV research and in the clinical care of infected individuals. Here, a quantitative HIV assay based on competitive reverse transcription-PCR with multiple competitors was developed. Four RNA competitors containing identical PCR primer binding sequences as the viral HIV-1 RNA target were constructed. One of the PCR primers was fluorescently labeled, which facilitated discrimination between the viral RNA and competitor amplicons by fragment analysis with conventional automated sequencers. The coamplification of known amounts of the RNA competitors provided the means to establish internal calibration curves for the individual reactions resulting in exclusion of tube-to-tube variations. Calibration curves were created from the peak areas, which were proportional to the starting amount of each competitor. The fluorescence detection format was expanded to provide a dynamic range of more than 5 log units. This quantitative assay allowed for reproducible analysis of samples containing as few as 40 viral copies of HIV-1 RNA per reaction. The within- and between-run coefficients of variation were <24% (range, 10 to 24) and <36% (range, 27 to 36), respectively. The high reproducibility (standard deviation, <0.13 log) of the overall procedure for quantification of HIV-1 RNA in plasma, including sample preparation, amplification, and detection variations, allowed reliable detection of a 0.5-log change in RNA viral load. The assay could be a useful tool for monitoring HIV-1 disease progression and antiviral treatment and can easily be adapted to the quantification of other pathogens.
The ability to accurately determine viral and infected cell burden is essential in understanding the natural history of human immunodeficiency virus type 1 (HIV-1) infection (12, 24, 32). Quantification of HIV-1 RNA has also been shown to be important in predicting the rate of disease progression and in monitoring antiretrovirus treatment (14, 18, 19, 23). The first reports of HIV-1 quantification which employed a semiquantitative PCR assay provided important information on the relative copy number of HIV-1 RNA; however, these assays were not designed to control the efficiency of reverse transcription (RT) and DNA amplification (7, 20). Nevertheless, quantitative-competitive RNA PCR (QC PCR) procedures were later described in which HIV-1 RNA from plasma samples was extracted and subjected to co-RT and coamplification with serially diluted HIV-1 RNA competitors in known copy numbers (25, 26). Although a great variety of different competitive PCR protocols have been described (34), they generally involve the same basic principle of mixing various amounts of competitor, usually differing in size from the wild-type product, with a constant amount of the sample to be determined. The amplified products are usually separated by electrophoresis and analyzed either by quantifying the intensity of ethidium bromide staining on an agarose gel (25, 35) or by using fluorescent labels and automated-laser fluorescent DNA sequencers (8, 29). The number of target copies in the sample can then be determined from the point of equivalence of signal intensity between the target and the dilution series of competitor.
However, there is also a considerable interest in more-automated detection systems, such as capillary electrophoresis (4, 33) and ion-pair, reverse-phase high-performance liquid chromatography (9, 10). Another step toward full automatization of QC PCR is the development of a real-time quantitative PCR method, i.e., a 5′-nuclease assay (Taq Man) (6, 11) which does not require any post-PCR sample handling.
Recently, three commercial assays for quantification of HIV-1 viral RNA in plasma have become available. These include the AMPLICOR HIV-1 Monitor Test (Roche Diagnostics, Branchburg, N.J.), which uses PCR; the NucliSens HIV-1 QT assay (Organon Teknika, Boxtel, The Netherlands), which is based on an isothermal nucleic acid sequence-based amplification; and the Quantiplex HIV RNA assay (Chiron, Emeryville, Calif.), which uses the branched DNA signal amplification technique. The rapid methodological advances in the quantification of HIV-1 RNA in plasma have rapidly been implemented in routine patient care. However, a major disadvantage of the commercial assays is their high cost and their availability for only a limited number of pathogens. It is obvious that simple, sensitive, and reliable quantification assays for many other pathogens are also needed. We recently described a single-tube format for quantification of DNA targets using multiple competitors of different sizes in a single reaction tube (29). The post-PCR analysis and quantification were performed by conventional gel electrophoresis with an automated-laser fluorescent DNA sequencer.
Here, we present a competitive RT-PCR assay for accurate quantification of HIV-1 RNA in plasma in which four RNA competitors are coanalyzed with the wild-type target. This method compares well with commercial assays.
MATERIALS AND METHODS
Patient blood specimen.
Whole blood from HIV-1 infected patients was collected in tubes containing anticoagulant EDTA, and plasma was separated by centrifugation at 2,500 rpm (400 × g) for 10 min. Plasma samples were frozen within 4 h at −70°C until further processing.
Preparation of RNA competitors.
The construction of four HIV-1 DNA competitors achieved by linker assembly into plasmid vectors was performed as described previously (29, 30). To allow for generation of discrete polyadenylated RNA competitors by in vitro transcription, the corresponding plasmid constructs were first linearized at the unique restriction site (EcoRI site; Pharmacia, Biotech, Uppsala, Sweden) downstream of the 30-bp poly(A) tail. After heat inactivation of EcoRI, an extraction step was performed with phenol-chloroform-isoamyl alcohol (25:24:1). The aqueous phase was reextracted twice and ethanol precipitated. The pellet was rinsed with 70% ethanol, dried, and dissolved in diethyl pyrocarbonate (DEPC)-treated water. In vitro transcription was performed (in a final volume of 50 μl) by incubation of the restricted DNA competitors (0.5 μg) at 37°C for 30 min in a buffer containing 40 mM Tris-HCl (pH 8.0), 30 mM MgCl2, 10 mM β-mercaptoethanol, 400 μM each RNase-free deoxynucleoside triphosphate (dNTP), 5 μg of RNase-free bovine serum albumin, and 70 U of T7 RNA polymerase. The DNA templates were degraded by addition of 10 U of RNase-free DNase I and incubation at 37°C for 60 min. RNA was immediately isolated by phenol extraction and ethanol precipitation. RNA transcripts were dissolved in 50 μl of DEPC-treated water. The concentration and purity of the RNA were determined by absorbance according to the method described by Maniatis et al. (17) and ultimately by end-point dilution experiments (3) with RT-PCR with HIV-1 3′-long-terminal-repeat (LTR)-specific primers. The RNA competitors were serially diluted in a buffer containing 10 mM Tris-HCl (pH 8.3) and 10 ng of yeast RNA (Boehringer, Mannheim, Germany) per μl and were stored at −80°C. Recommended precautions preventing RNA degradation by RNases were taken (17). All solutions, glassware, and plasticware were treated with 0.1% DEPC (with the exception of Tris buffers) for at least 1 h at 37°C, followed by autoclaving for 15 min to remove all traces of DEPC. During the preparation and isolation stages, RNA samples were kept on ice.
Solid-phase purification of polyadenylated RNA.
Twenty-five microliters (0.125 mg) of paramagnetic oligo(dT)25 Dynabeads (Dynal, Oslo, Norway) were prepared according to the manufacturer’s recommendations. Briefly, 25 μl of lysis-binding buffer (100 mM Tris-HCl [pH 8.0], 500 mM LiCl, 10 mM EDTA [pH 8.0], 1% LiDS, 5 mM dithiothreitol), was added to the sedimented beads, which were then mixed with 10 μl of polyadenylated RNA competitor(s). Hybridization was performed at room temperature for 10 min by employing a rotation device. The beads were initially washed three times with 100 μl of washing buffer containing lithium dodecyl sulfate (10 mM Tris-HCl [pH 8.0], 150 mM LiCl, 1 mM EDTA, 0.1% LiDS), followed by three washing steps in 100 μl of washing buffer (10 mM Tris-HCl [pH 8.0], 150 mM LiCl, 1 mM EDTA) with a final wash with 100 μl of PCR buffer. RT-PCR was performed either on the solid phase with RNA captured on the beads or in solution after elution of RNA from the beads. To elute RNA, 10 μl of elution solution (2 mM EDTA [pH 8.0]) was added to the beads, which were subsequently incubated at 65°C for 10 min with continuous rotation. The tube was immediately chilled on ice for 2 min and placed in a magnetic separation device (Dynal MPC), and the supernatant (10 μl) was transferred directly into a tube containing 40 μl of RT-PCR solution.
In the case of viral HIV-1 RNA isolation, plasma samples (100 μl) were mixed with 300 μl of lysis-binding buffer, which was then added to oligo(dT)25 beads (0.125 mg) suspended in 25 μl of lysis-binding buffer. HIV-1 RNA was allowed to bind for 10 min at room temperature. After incubation, the supernatant was removed and the beads were rinsed three times in 500 μl of washing buffer with lithium dodecyl sulfate and then twice with 500 μl of washing buffer. The HIV-1 RNA was eluted from the beads by incubation in 25 μl of elution buffer containing 0.5 μg of RNase-free yeast RNA (Boehringer) at 65°C for 2 min. The eluate was used immediately for RT.
Competitive RT-PCR.
HIV-1 target and competitors were amplified with a seminested primer set located in the 3′-LTR region of the HIV-1 genome. The PCR primer sequences were as follows: JA159, 5′-CAGCTGCTTTTTGCCTGTAC-3′ (outer, 432 to 452); JA160F, 5′-FITC-CTGCTTTTTGCCTGTACTGGGTCTC-3′ (inner, 435 to 460); JA161B, 5′-biotin-AAGCACTCAAGGCAAGCTTTATTGA-3′ (inner, 524 to 499); JA162, 5′-AGCACTCAAGGCAAGCTTTA-3′ (outer, 528 to 508). Positions relative to the MN strain of HIV-1 are given (22). RT and outer PCRs were carried out in a single-step mode in a solution (final volume, 50 μl) containing 10 mM Tris-HCl (pH 8.3) at 25°C, 50 mM KCl, 2.5 mM MgCl2, and 200 μM each dNTP with 5 pmol of each primer [oligo(dT)25, JA159, and JA162], 2 U of AmpliTaq DNA Polymerase Gold (Perkin-Elmer, Norwalk, Conn.), 0.5 U of Moloney murine leukemia virus reverse transcriptase (Pharmacia Biotech) and 2 μg of RNase-free yeast RNA (Boehringer) with GeneAmp 9600 (Perkin-Elmer). The amounts of competitor mixtures and viral RNA samples added to the RT-PCR master mix varied from 5 to 15 μl and from 2 to 23 μl, respectively. The temperature profile consisted of RT carried out at 37°C for 60 min linked to the outer PCR program, which consisted of denaturation at 94°C for 12 min, followed by a 32-cycle program consisting of 95°C for 30 s; 50°C for 30 s, and 72°C for 30 s. Five microliters of outer PCR product was used in the inner PCR, which was carried out in 50 μl of PCR buffer containing 10 mM Tris-HCl (pH 8.3) at 25°C, 50 mM KCl, 2.5 mM MgCl2, and 200 μM each dNTP with 5 pmol of each primer (JA160F and JA161B) and 1 U of AmpliTaq DNA polymerase (Perkin-Elmer). The temperature profile and number of cycles were identical to those in the outer PCR, except that the annealing temperature was 60°C. Seminested PCR was performed by using three physically separated rooms. PCR master mixtures were prepared in a DNA- and RNA-free laboratory; RNA competitors and clinical samples were added to the PCR tubes in a laminar flow hood in an outer PCR room, and the outer PCR products were transferred to the inner PCR mixes in an inner PCR room. Recommended standard precautions against PCR contamination were taken (13), with multiple negative controls included in each PCR run.
End-point dilution analysis.
Determination of RNA competitor concentrations was performed by using end-point dilution experiments (3) involving nested RT-PCR as outlined previously. The basis for the analysis is the ability of the nested RT-PCR to reproducibly detect single HIV-1 RNA molecules. Briefly, the RNA samples were diluted in a fivefold manner, and at least 10 PCR determinations were performed at each dilution step. The numbers of RNA copies were calculated by the Poisson distribution formula (3) (i.e., one starting copy corresponds to a dilution step in which 63% of the samples are positive).
RNA competitor configurations.
Different RNA competitor configurations were prepared and analyzed. Mixtures were arranged both with low amounts of RNA competitors 1 to 4 (40, 250, 1,250, and 5,000 copies per reaction, respectively, as determined by end-point dilution analysis) and with high amounts of RNA competitors (4,000, 25,000, 125,000, and 500,000 copies per reaction). The low- and high-concentration competitor mixtures were coanalyzed with different amounts of HIV-1 target to investigate amplification efficiencies. The results were monitored by fragment analysis after outer and inner PCRs.
Fragment analysis of PCR products.
One microliter of the resulting PCR products (or a 1:5 dilution thereof) was directly mixed with 9 μl of deionized formamide (100%) containing 5 mg of Dextran Blue 2000 (Pharmacia Biotech) per ml, and the mixture was heat denatured for 5 min at 95°C, immediately chilled on ice, loaded on a 6% polyacrylamide gel (Ready Mix; Pharmacia Biotech), and electrophoresed on an automated-laser fluorescent sequencer (Pharmacia Biotech). A 50-bp fluorescently marked ladder (50 to 500) was used as a size standard. Quantification and interpretation of the raw data output were performed with Fragment Manager software (version 1.2; Pharmacia Biotech). Calibration curves for determination of target amounts were created from peak areas, which were proportional to the amount of each competitor coanalyzed with viral target (29).
RESULTS
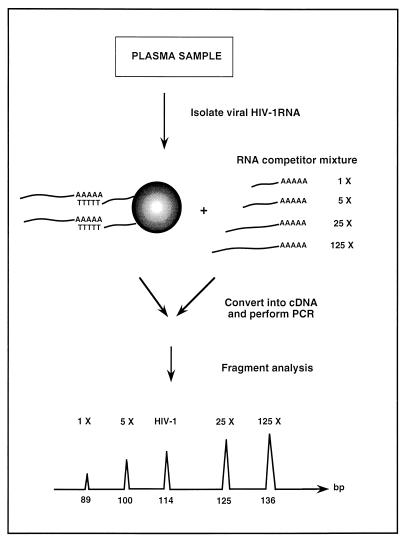
A method for quantification of HIV-1 RNA by using multiple competitors is illustrated in Fig. 1. This approach involves co-RT and coamplification of the sample HIV-1 RNA and multiple RNA competitors. The competitors were designed to differ in length but to share the same PCR primer binding sequences as the viral HIV-1 target. Moreover, the PCR primers annealing in the 3′-LTR region were located adjacent to the viral poly(A) tail employed in the affinity purification [i.e., with oligo(dT) magnetic beads] of virus material. The objective in locating the primers close to the purification handle was to minimize the effect of RNase degradation, thereby retaining the high sensitivity of the assay. The resulting PCR products were analyzed by fluorescent fragment analysis with the internal competitors to create a standard curve for estimation of the amount of target.
FIG. 1.
Schematic illustration of the competitive approach for quantification of HIV-1 RNA in plasma.
Construction of RNA competitors.
In an initial study by Vener et al. (29), HIV-1 DNA was quantified with a mixture of three DNA competitors in a seminested competitive PCR. The sizes of the inner PCR products were distributed over a narrow interval, i.e., competitors 1 to 3 were 89, 100, and 125 bp long, respectively. The amplification of the HIV-1 target resulted in an amplicon of 114 bp (thus, within the interval represented by the competitors). Here, we have extended this work, first, by introducing a new competitor (a 136-bp amplicon) to improve the quality of the calibration curve by an additional fourth value (30, 31) and, second, by converting the DNA quantification procedure into an HIV-1 RNA quantification assay. Since the DNA competitors, which were linearized by EcoRI restriction, were constructed to contain a poly(A)30 stretch at the 3′ end of multilinker region, in vitro transcription was possible with T7 RNA polymerase, resulting in polyadenylated RNA competitors. The corresponding sizes of transcripts 1 to 4 were 135, 146, 171, and 186 bp, respectively.
Solid-phase purification of polyadenylated RNA.
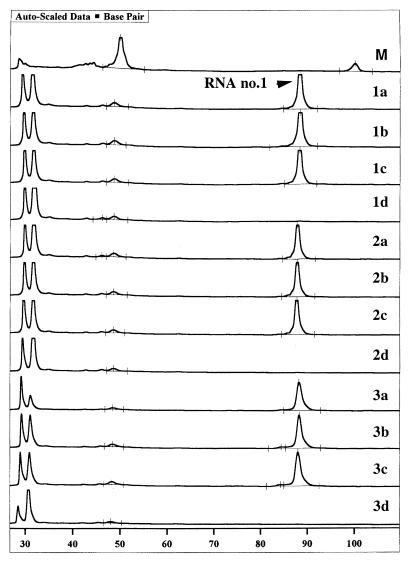
We have previously used oligo(dT)25 paramagnetic beads in the sample preparation of HIV-1 RNA from plasma samples (2), and here we investigated at which stage of the quantitative assay the RNA competitors could be introduced, i.e., prior to, during, or after the solid-phase purification step. For this purpose, a model system was established consisting of a fivefold dilution series of competitor RNA 1, corresponding to 57, 12, 2.3, and 0.5 RNA copies per reaction. These samples were either immobilized onto oligo(dT)25 beads and subsequently reverse transcribed on the solid support (Fig. 2 [1a to 1d]) or were immobilized onto oligo(dT)25 beads, eluted, and subsequently reverse transcribed in solution (Fig. 2 [2a to 2d]) or directly reverse transcribed in solution (Fig. 2 [3a to 3d]); followed by seminested PCR. The results depicted in Fig. 2 show that all alternatives exhibited equivalent sensitivity, as determined by detection of the dilution step containing 2.3 estimated RNA copies.
FIG. 2.
Fragment analysis results for sensitivity of solid-phase purification with a fivefold dilution series of RNA competitor 1. 1a to 1d, 57, 12, 2.3, and 0.5 RNA copies (size, 89 bp), respectively, bound onto oligo(dT)25 Dynabeads and then subjected to RT-PCR; 2a to 2d, 57, 12, 2.3, and 0.5 RNA copies, respectively, bound onto oligo(dT)25 Dynabeads and then eluted from the solid phase and subjected to RT-PCR in solution; 3a to 3d, the same dilutions of RNA 1 analyzed directly by RT-PCR in solution; M, dye-marked ladder (50 and 100 bp).
To investigate whether there was any bias in immobilization and/or elution efficiency from the solid support between the different RNA competitors, a premix of competitors with approximately 50 initial copies of each RNA competitor was tested as described above. The results with multiple RNA competitors demonstrated that peak profiles and ratios corresponding to the four RNA competitors were preserved irrespective of the chosen sample preparation approach, i.e., solid-phase capture and RT-PCR, solid-phase capture with subsequent elution and RT-PCR, or RT-PCR in solution (data not shown). Although RT-PCR on solid phase and in solution after elution of RNA yielded similar results in respect to sensitivity, the practical problem with sedimentation of beads during enzymatic reactions makes the solid-phase RT less applicable when large number of samples are to be analyzed.
Amplification kinetics.
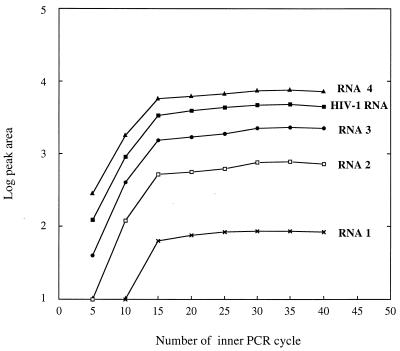
To empirically investigate whether the HIV-1 RNA target and the four RNA competitors behave similarly during RT-PCR amplification, fragment analysis was performed after every fifth cycle. The experiments were carried out on two sets of RNA competitors, each containing a different amount of HIV-1 target. A low-copy-number RNA competitor mixture (40, 250, 1,250, and 5,000 copies of RNA competitors 1 to 4, respectively, per reaction) was coanalyzed with 3,000 copies of HIV-1 RNA target, and a high-copy-number RNA competitor mixture (4,000, 25,000, 125,000, and 500,000 copies of RNA competitors 1 to 4, respectively, per reaction) was coanalyzed with 62,000 copies of HIV-1 RNA. For both mixtures, the PCR proceeds in a strictly parallel manner through the outer (data not shown) and inner PCRs (Fig. 3), indicating that target and internal competitors share similar amplification kinetics throughout the reaction into the plateau phase. Figure 3 also shows that for the low concentration mixture, the plateau phase was reached at cycle 15 in the inner PCR. In the case of the 100-fold-higher concentration mixture of RNA competitors (data not shown), the PCR plateau was reached after only 10 inner PCR cycles.
FIG. 3.
PCR cycle test for RNA competitors and HIV-1 RNA target; Amplification kinetics of inner PCR for a low-copy-number mixture of RNA competitors coanalyzed by RT-PCR with 3,000 copies of HIV-1 RNA target. Configuration of the RNA competitors was as follows: RNA 1, 89 bp and 40 copies; RNA 2, 100 bp and 250 copies; RNA 3, 125 bp and 1,250 copies; and RNA 4, 136 bp and 5,000 copies.
Dynamic range and detection limit of the assay.
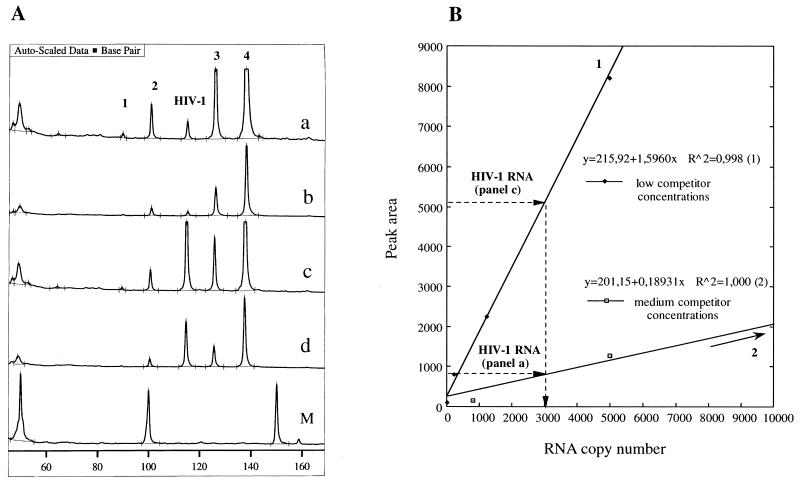
To circumvent the limitation in the detection range of presently used fluorescence-based electrophoresis instruments, two mixtures of RNA competitors with different copy numbers covering the common interval in HIV-1 viremias (19) were used. The two mixtures of RNA competitors were designed to have a minor overlap in copy numbers. Thus, each clinical sample was analyzed in two independent PCRs, yielding results either in one or both competitor series. Therefore, the dynamic range of the assay is linked to the number of competitor sets analyzed. Figure 4 depicts a case in such the same viral load can be determined with the two mixtures of RNA competitors. Figure 4A (graph a) shows the raw data output from fragment analysis for determination of HIV-1 RNA with a medium-copy-number configuration of RNA competitors 800, 5,000, 25,000, and 100,000, and the established calibration curve is shown in Fig. 4B. Figure 4A (graph c) depicts the fragment analysis results for the low-copy-number configuration of RNA competitors 40, 250, 1,250, and 5,000 by analyzing the plasma sample analyzed in graph a. Graphs b and d correspond to a 1:5 dilution of the PCR products shown in graphs a and c, respectively, which were also used in the construction of the calibration curves 1 and 2 (30). As shown in Fig. 4B, both mixtures of competitors resulted in determination of the same number of HIV-1 RNA copies (approximately 3,000). Evaluation of quantitative data can be performed by a very simple calculation by using the equation of obtained linear regression curves, which may also be employed in improving the overall dynamic range (by at least 10-fold) by extrapolation outside the competitor copy boundaries.
FIG. 4.
Extended dynamic range of the assay; two sets of RNA competitors for determination of the same viral load. (A) Graph a, Raw data output from fragment analysis for a medium-copy-number configuration of RNA competitors (800, 5,000, 25,000, and 100,000) coanalyzed with HIV-1 RNA (3,000 copies, as determined by our assay); graph b, fivefold dilution of the PCR product depicted in graph a; graph c, raw data output from fragment analysis for a low-copy-number configuration of RNA competitors (40, 250, 1,250, and 5,000) coanalyzed with the plasma sample described for graph a; graph d, fivefold dilution of the PCR product depicted in graph c. (B) Calibration curves for evaluation of HIV-1 RNA copy number. Curves 1 and 2 correspond to graphs c and a, respectively.
As part of the analysis of clinical samples, the detection limit was investigated by comparing results obtained by competitive RT-PCR analysis with multiple competitors with that obtained by end-point RT-PCR with the 3′-LTR PCR primers. The comparison was performed in parallel on the same 10-fold serially diluted HIV-1 RNA eluate, which was prepared by solid-phase purification of plasma samples. To allow for quantification at less than the lowest concentration of RNA competitors (fewer than 40 copies), extrapolation of the internal standard curves was used for an approximate estimation of 1 to 40 copies of HIV-1 RNA per PCR. Table 1 shows a definite correlation between results for the end-point dilutions and those for the developed assay. The results from each dilution series also demonstrate that very low concentrations of virus can be estimated by calibration curve extrapolation at less than the range of the copy number mixture of RNA competitors.
TABLE 1.
Quantitative results for disease progression
| Sample identification | Time interval (wks) | Amplicor (Roche) result
|
Single-tube PCR result
|
||
|---|---|---|---|---|---|
| Undiluteda | Diluted | Multiple competitorsa | End-point PCR | ||
| 9784 | 0 | 15,000 | 3,000 | + | |
| 1:10 | 230 | + | |||
| 1:100 | 40 | + | |||
| 1:1,000 | 3 | + | |||
| 1:10,000 | Negative | − | |||
| 10128 | 4 | 71,700 | 34,000 | + | |
| 1:10 | 1,900 | + | |||
| 1:100 | 210 | + | |||
| 1:1,000 | 20 | + | |||
| 1:10,000 | Negative | − | |||
| 10575 | 12 | 120,000 | 62,000 | + | |
| 1:10 | 4,200 | + | |||
| 1:100 | 200 | + | |||
| 1:1,000 | 50 | + | |||
| 1:10,000 | 8 | + | |||
| 1:100,000 | Negative | − | |||
Values are HIV-1 RNA copy numbers per PCR.
Quantitative analysis of clinical samples.
The RT-PCR strategy with multiple competitors was retrospectively compared with the Amplicor HIV-1 Monitor Test (Roche) to monitor disease progression in an HIV-infected individual. The HIV-1 RNA level in 50 μl of plasma was quantified at week 0 and then after 4 and 12 weeks of treatment (Table 1). The Amplicor HIV-1 Monitor Test with standard SK431 and SK462 PCR primers specific to the gag gene of the HIV-1 genome showed a continuous increase in viral load during these weeks. With low- and high-copy-number competitor mixtures, the same samples assayed by our strategy showed a similar trend, although slightly lower absolute amounts of HIV-1 RNA were detected.
In addition, the HIV-1 samples were further serially diluted in 10-fold steps to less than the detection limit of the Amplicor kit to take advantage of the higher sensitivity achieved by the seminested PCR system. The results presented in Table 1 show a linear correlation between the dilution steps and results of RT-PCR quantification, which also shows that the approach allows for quantitative recovery of HIV-1 RNA in the range investigated, i.e., from 120,000 copies (as determined by Amplicor) down to single copies. As discussed above, the presence of HIV-1 RNA in these diluted samples was verified by end-point PCR performed in parallel with the LTR primers, which indeed indicated a clear cutoff point in the same range as that estimated by our quantification system.
Overall, the determined values of viral load between Amplicor and our method varied by no more than 0.5 log for samples 10575 (0.28 log) and 10128 (0.32 log) and by 0.7 log for sample 9784. The discrepancy in values obtained by these systems in terms of absolute amounts could be due to degradation, since analysis of these samples was not performed at the same time point. However, a more probable reason is the lower efficiency of primer template hybridization during RT and in the first cycle of PCR amplification (1). This is supported by the fact that certain samples could not be amplified (and quantified) with this LTR primer set, while other parts of the HIV-1 genome sequence could readily be amplified in these samples (data not shown). This indicates that the LTR region contains several polymorphic positions which may require additional sets of competitors to allow for analysis of such LTR subgroups.
Assay reproducibility.
To determine intraassay reproducibility, three clinical specimens with different levels of HIV-1 RNA in plasma representing high, medium, and low copy numbers (plasma samples 1, 2, and 3, respectively) were analyzed 10 times. The results presented in Table 2 show that the lower-copy-number HIV-1 RNA plasma sample 3 has a slightly larger expected coefficient of variation (CV), i.e., <24%, compared to the high-copy-number plasma sample 1 (CV, <10%). However, these values correspond to a rather narrow within-run standard deviation (SD) of <0.11 log, which is less than 0.5 log units, as recommended for viral load determination (27). The interassay reproducibilities (6 days) were estimated for three clinical samples (plasma 1, 2, and 4). The mean CV values for between-run precision showed a rather low variability and were found to be in the range of 27 to 36%, which still corresponds to a narrow between-run SD (<0.13 log).
TABLE 2.
Reproducibility of the RT-PCR assay with multiple competitors
| Plasma specimen no. and assay (n) | Meana (SD) | Log of SD | CV (%) |
|---|---|---|---|
| Within-run intraassay | |||
| 1 (10) | 38,170 (3,741) | 0.044 | 10 |
| 2 (10) | 7,200 (1,530) | 0.084 | 21 |
| 3 (10) | 1,140 (350) | 0.111 | 24 |
| Between-run interassay | |||
| 1 (6) | 44,362 (12,908) | 0.111 | 29 |
| 2 (6) | 5,559 (1,498) | 0.105 | 27 |
| 4 (6) | 257 (93) | 0.13 | 36 |
HIV-1 RNA copy number.
DISCUSSION
The measurement of HIV-1 RNA levels in plasma has rapidly become an important tool in the care and follow-up of HIV-1-infected individuals and has clear advantages over other virological assays. In contrast to the HIV-1 p24 antigen, HIV-1 RNA levels in plasma are measurable in the vast majority of HIV-infected patients at various stages of disease (18, 19, 26). Furthermore, HIV-1 RNA levels in plasma change dynamically in response to successful antiviral therapy, in contrast to HIV-1 proviral DNA in peripheral blood mononuclear cells (24). Therefore, HIV-1 RNA levels in plasma are the most effective single predictor of clinical outcome (18, 19). In this study, we have described a sensitive and high-throughput RT-PCR assay with multiple competitors for quantification of HIV-1 RNA in plasma that can potentially be adapted to quantification of virtually any RNA or DNA target.
The multiple RNA competitors differed in size but contained the same primer annealing sequences as those of the wild-type target. Since the RNA competitors also consisted of a poly(A) stretch, various possibilities were evaluated with respect to solid-phase purification, i.e., poly(A)-poly(T) hybridization. We have shown here that a solid phase can be employed without reduction in sensitivity compared to a solution-based system and that solid-phase RT can be efficiently performed. However, the sedimentation of beads during incubation makes this technique less attractive when quantitative enzymatic reactions are to be performed on multiple samples. For this reason, we used solid-phase technology to prepare HIV-1 from plasma and after elution of target we added four RNA competitors to a single reaction tube for the common RT and seminested PCR. The experiments performed to analyze RT and PCR amplification efficiencies showed that the ratios between competitors and target were constant during RT and outer and inner PCR. However, a future goal will be to add the RNA competitors at an earlier stage in the process (i.e., adding the competitors to the plasma sample) to control all preparative steps.
A common problem with QC PCR is that the detection principle often suffers from a limited dynamic range. One example is the HIV monitor assay, in which the amplification products are assayed in multiple dilutions because of the limited dynamic range of the enzyme-linked immunosorbent assay-like detection system. Automated fluorescence-based sequencers, which we used, usually have a working dynamic range of 102- to 103-fold, and signal values that are higher than this range are displayed as truncated peaks in chromatograms. To circumvent this problem and to produce peaks that can be reliably used in creation of a calibration curve, we used two overlapping sets of RNA competitor mixtures which covered the interval between 40 and 500,000 copies of HIV-1 RNA. Importantly, the single-tube assay allows for a quick estimation of viral load by analysis of the generated chromatograms (Fig. 4), and by using a simple fivefold dilution of the PCR product (Fig. 4A, graphs b and d), a reliable calibration curve can be achieved for more-exact quantification.
An important aspect of any quantification system is the precision of the assay, and, in this respect, our assay compares well with commercial assays. Thus, the pooled duplicate SD of log copies obtained for the overall procedure was 0.13 (SD, < 0.13 log). The levels of imprecision, which were expressed as CVs, ranged from 10 to 24% and from 27 to 36% for within- and between-run assays, respectively. Ultimately, the utility of a quantitative assay for HIV-1 RNA depends on the extent of fluctuation of HIV-1 viral load in plasma over time within an infected patient. A response to therapy can be detected only when it exceeds the normal viral fluctuations. Usually a change in log-transformed HIV-1 RNA concentrations exceeding 0.5 log units is considered significant (27). To be able to detect fourfold differences in HIV-1 RNA concentration in plasma with a power of 0.95, the pooled duplicate of log copies must be no greater than 0.50 (16). The precision of our assay was well within this value and, therefore, allows reliable detection of a response or lack of response to antiviral therapy in individual patients.
Another important factor to consider is the lower limit of detection of any quantification system. Commercial assays currently available have a lower limit of reliable detection of approximately 200 to 500 RNA copies per ml of plasma (15, 21). It has been reported that patients for whom there is a decrease to less than the detection limit of experimental ultrasensitive methods (approximately 50 RNA copies per ml) following initiation of therapy have a better long-term prognosis than patients who still have detectable RNA levels with these assays (5). Thus, there is a clear need for assays with improved sensitivity. Our assay allowed accurate quantification of RNA levels down to 40 RNA copies per reaction, and lower levels could also be quantified by extrapolation from the calibration curve. By solid-phase technology, with its possibility to concentrate target molecules (HIV-1 RNA), the limit of detection strictly depends on the volume of HIV-1 plasma available. Furthermore, the use of the low-copy-number mixture of RNA competitors allows for a considerably higher sensitivity of viral load determination compared to that of the existing systems (28).
The basic principle of multiple competitors in single reactions has the potential to be quickly adapted for quantification of alternative RNA targets, such as other RNA viruses. Modifications in this procedure should further enable the quantification of mRNA. Although the quantitative assay was developed to monitor HIV-1 levels in plasma with conventional fluorescent sequencers, more-automated strategies for a single-tube format can be envisioned by combining this approach with an exonuclease detection assay (6, 11) in which competitor-specific probes with different fluorescent labels allow for analysis in real time.
ACKNOWLEDGMENTS
This work was supported by a grant from Dynal A.S., Oslo, Norway, and by a personal grant (T.V.) from the Royal Swedish Academy of Sciences.
We thank Deirdre O’Meara for critical reading of the manuscript.
REFERENCES
- 1.Alaeus A, Lidman K, Sönnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:5475–5482. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Albert J, Wahlberg J, Lundeberg J, Cox S, Sandström E, Wahren B, Uhlén M. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in posttreatment sera. J Virol. 1992;66:5627–5630. doi: 10.1128/jvi.66.9.5627-5630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinchman J E, Albert J, Vartdal F. Few infected CD4+ T-cells but a high proportion of replication-competent provirus copies in asymptomatic human immunodeficiency virus type 1 infection. J Virol. 1991;65:2019–2023. doi: 10.1128/jvi.65.4.2019-2023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connoly A R, Cleland L G, Kirkham B W. Mathematical consideration of competitive polymerase chain reaction. J Immunol Methods. 1995;187:201–211. doi: 10.1016/0022-1759(95)00185-2. [DOI] [PubMed] [Google Scholar]
- 5.Conway B, Shillington A, Fransen S, Sninsky J, Kwok S, Montaner J S. Program and abstracts of the 4th Conference on Retroviruses and Opportunistic Infections. Washington, D.C: Foundation for Retroviruses and Human Health; 1997. Application of ultra-sensitive PCR to the analysis of plasma viral load testing in clinical trials, abstr. 181. [Google Scholar]
- 6.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 7.Gupta P, Ding M, Cottrill M, Rinaldo C, Kingsley L, Wolinsky S, Mellors J. Quantitation of human immunodeficiency virus type 1 DNA and RNA by a novel internally controlled PCR assay. J Clin Microbiol. 1995;33:1670–1673. doi: 10.1128/jcm.33.6.1670-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hämmerle T, Falkner F G, Dorner F. A sensitive PCR assay system for the quantification of viral genome equivalents: hepatitis C virus (HCV) Arch Virol. 1996;141:2103–2114. doi: 10.1007/BF01718218. [DOI] [PubMed] [Google Scholar]
- 9.Hayward-Lester A, Oefner P J, Doris P A. Rapid quantification of gene expression by competitive RT-PCR and ion-pair reversed-phase HPLC. BioTechniques. 1996;20:250–257. doi: 10.2144/96202rr02. [DOI] [PubMed] [Google Scholar]
- 10.Hayward-Lester A, Oefner P J, Sabatini S, Doris P A. Accurate and absolute quantitative measurement of gene expression by single-tube RT-PCR and HPLC. Genome Res. 1995;5:494–499. doi: 10.1101/gr.5.5.494. [DOI] [PubMed] [Google Scholar]
- 11.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 12.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 13.Innis M A, Gelfand D H, Shinsky J J, White T J. PCR protocols. San Diego, Calif: Academic Press, Inc.; 1990. [Google Scholar]
- 14.Katzenstein D A, Hammer S M, Hughes M D, Gundacker H, Jackson J B, Fiscus S, Rasheed S, Elbeik T, Reichman R, Japour A, Merigan T C, Hirsch M S. The relation of virologic and immunologic markers to clinical outcomes after nucleoside therapy in HIV-infected adults with 200 to 500 CD4 cells per cubic millimeter. AIDS Clinical Trials Group Study 175 Virology Study Team. N Engl J Med. 1996;335:1091–1098. doi: 10.1056/NEJM199610103351502. [DOI] [PubMed] [Google Scholar]
- 15.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin H J, Myers L E, Yen-Liberman B, Hollinger F B, Henrard D, Hooper C J, Kokka R, Kwok S, Rasheed S, Vahey M, et al. Multicenter evaluation of quantification methods for plasma human immunodeficiency virus type 1 RNA. J Infect Dis. 1994;170:553–562. doi: 10.1093/infdis/170.3.553. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 18.Mellors J W, Rinaldo C R J, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 19.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 20.Michael N L, Vahey M, Burke D S, Redfield R R. Viral DNA and mRNA expression correlate with the stage of human immunodeficiency virus (HIV) type 1 infection in humans: evidence for viral replication in all stages of HIV disease. J Virol. 1992;66:310–316. doi: 10.1128/jvi.66.1.310-316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers G, Korber B, Berkofsky J, Smith R F, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1991. [Google Scholar]
- 23.O’Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 24.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1 infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 25.Piatak M, Jr, Luk K C, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–81. [PubMed] [Google Scholar]
- 26.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 27.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 28.Vandamme A M, Schmit J C, Van Dooren S, Van Laethem K, Gobbers E, Kok W, Goubau P, Witvrouw M, Peetermans W, De Clercq E, Desmyter J. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the AMPLICOR HIV monitor test. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 29.Vener T, Axelsson M, Albert J, Uhlen M, Lundeberg J. Quantification of HIV-1 using multiple competitors in a single-tube assay. BioTechniques. 1996;21:248–253. doi: 10.2144/96212st01. [DOI] [PubMed] [Google Scholar]
- 30.Vener T, Axelsson M, Albert J, Uhlén M, Lundeberg J. Update to: Quantification of HIV-1 using multiple competitors in a single-tube assay. In: Larrick J, editor. The PCR technique: quantitative PCR. Natick, Mass: Eaton Publishing Co.; 1997. pp. 285–292. [Google Scholar]
- 31.Vener T, Stark M, Uhlén M, Lundeberg J. Multiple competitors for single-tube quantification of HIV-1 DNA. In: Lassner D, Pustowoit B, Rolfs A, editors. Modern applications of DNA amplification techniques: problems and new tools. New York, N.Y: Plenum Publishing Co.; 1997. pp. 3–10. [Google Scholar]
- 32.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 33.Williams S J, Schwer C, Krishnarao A S, Heid C, Karger B L, Williams P M. Quantitative competitive polymerase chain reaction: analysis of amplified products of the HIV-1 gag gene by capillary electrophoresis with laser-induced fluorescence detection. Anal Biochem. 1996;236:146–152. doi: 10.1006/abio.1996.0143. [DOI] [PubMed] [Google Scholar]
- 34.Zimmermann K, Mannhalter J W. Technical aspects of quantitative competitive PCR. BioTechniques. 1996;21:268–279. doi: 10.2144/96212rv01. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann K, Schogl D, Plaimauer B, Mannhalter J W. Quantitative multiple competitive PCR of HIV-1 DNA in a single reaction tube. BioTechniques. 1996;21:480–484. doi: 10.2144/96213st06. [DOI] [PubMed] [Google Scholar]