Summary
Histopathological images of colorectal liver metastases (CRLM) contain rich morphometric information that may predict patients’ outcomes. However, to our knowledge, no study has reported any practical deep learning framework based on the histology images of CRLM, and their direct association with prognosis remains largely unknown. In this study, we developed a deep learning-based framework for fully automated tissue classification and quantification of clinically relevant spatial organization features (SOFs) in H&E-stained images of CRLM. The SOFs based risk-scoring system demonstrated a strong and robust prognostic value that is independent of the current clinical risk score (CRS) system in independent clinical cohorts. Our framework enables fully automated tissue classification of H&E images of CRLM, which could significantly reduce assessment subjectivity and the workload of pathologists. The risk-scoring system provides a time- and cost-efficient tool to assist clinical decision-making for patients with CRLM, which could potentially be implemented in clinical practice.
Subject areas: Artificial intelligence, Computer science applications, Gastroenterology, Bioinformatics
Graphical abstract

Highlights
-
•
We developed a deep learning–based framework for microenvironment dissection in CRLM
-
•
SOFs based risk scoring system demonstrated a robust and independent prognostic value
-
•
SOFs model is a robust biomarker for adjuvant CTx in patients without pretreatment
-
•
The CRS-SOF systems show outstanding efficacy in patients’ stratification
Artificial intelligence; Computer science applications; Gastroenterology; Bioinformatics
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed and the second most lethal malignancy worldwide.1 Liver metastases are detected in nearly 50% of CRC patients at diagnosis or in the subsequent course of the disease.2 While hepatectomy can offer a chance of cure for colorectal liver metastasis (CRLM) patients, the 5- and 10- year overall survival (OS) rates after surgery are merely 42% and 25%, respectively.3 The clinical risk score (CRS) proposed by Fong et al. remains the most widely used scoring system for risk stratification and prognosis prediction in clinical practices.4 However, the clinical feature-based CRS system was originally developed in 1999, limiting its applicability in the modern era. Therefore, more precise and robust prognostic indicators are needed to predict individual outcomes and optimize therapeutic strategies.
Histopathological images of liver metastases not only offer crucial evidence for diagnosis but also contain rich morphometric information that may predict CRLM patient outcomes. A typical example would be the different histopathological growth patterns (HGPs) identified in the interface between the tumor border and surrounding liver parenchyma under hematoxylin and eosin (H&E) stained tissue sections.5 Recently, HGPs have gained increasing attention in CRLM for their association with tumor biology and potential prognostic values.6,7,8 Besides, the Immunoscore, which quantifies CD3+ and CD8+ T cells in both the center and the invasive margin based on pathological images of liver metastases, is significantly associated with prolonged survival of CRLM patients after hepatectomy.9,10 Moreover, in CRLM patients with preoperative chemotherapy, tumor regression grade (TRG) based on the extent of residual tumor cells and fibrosis found in tumor tissues stained with H&E have been reported to represent pathological response and correlate with postoperative response outcomes.10 However, the HGPs, TRG and other indicators rely heavily on visual judgment and could not systematically characterize the image features with only single indicator. In addition, no previous study has attempted to integrate crucial features from histopathological images to develop a more accurate prognostic system for CRLM.
In recent years, artificial intelligence has been put to the forefront of digital pathology, bringing opportunities to overcome the limitations of subjective visual assessment and alleviate the workload of pathologists.11 Mining subvisual morphometric features that are clinically relevant from whole-slide images (WSIs) based on deep learning is currently a highly active field of research.12 While recent studies have demonstrated the power of AI in directly predicting patients’ outcomes for colorectal cancer,13,14,15 hepatocellular carcinoma16,17 and melanoma,18 one general issue lies in its interpretability due to the “black box” challenge. Those prognostic scores are based on features learned by convolutional neural network (CNN) which are not direct spatial characteristics that can be easily understood or interpreted.13,14,16 Technically, earlier studies about the quantification of spatial organization features (SOFs) depended heavily on cell morphologies and required additional image processing steps such as segmentation,19,20,21,22 which is time-consuming and can be easily affected by the background noise.23,24 More importantly, to date, no practical deep learning framework has been built based on the whole-slide images (WSIs) of CRLM, and their direct association with prognosis remains largely unknown.
In this study, we propose a deep learning framework for automated tissue classification of H&E-stained WSIs of CRLM, followed by quantification of SOFs. Next, we established a SOFs-based prognostic model to assess OS after hepatectomy and further validated it in an independent external cohort. Finally, we evaluated the efficacy of combining our prognostic model with the CRS for the risk stratification of CRLM patients. The ColoRectal Liver Metastasis-Spatial (CRLM-SPA) framework we proposed is based on tissue classification, which is more efficient than the quantification of cell-based features. Some of the quantitative SOFs that we postulated were previously unreported. Compared with the features learned by CNN, the SOFs calculated by CRLM-SPA were based on direct quantification of clearly defined features, and hence the superiority in pathological and biological interpretations. This is the first time that multiple SOFs have been systematically defined, accurately profiled and efficiently integrated for the prognosis prediction of CRLM.
Results
Workflow of patient selection and patient characteristics
The detailed workflow of patient selection is shown in Figure S1. According to the inclusion and exclusion criteria, 500 CRLM patients from the SYSUCC cohort (discovery cohort) and 506 CRLM patients from the BJCH cohort (validation cohort) with radical resection of the liver metastases were eligible for the imaging analysis. 67 (13.4%) patients from SYSUCC and 102 (20.2%) patients from BJCH were further excluded due to a limited detectable continuous tumor area (<50 patches). As a result, a total of 433 and 404 CRLM patients from SYSUCC and BJCH cohorts were included in the study, respectively. The clinical characteristics of the patients are summarized in Table 1. According to CRS evaluation in the SYSUCC cohort, 269 (62.1%) and 164 patients (37.9%) were classified into a low-risk group (CRS 0–2) and a high-risk group (CRS 3–5), respectively. In the BJCH cohort, 219 (54.2%) and 185 patients (45.8%) were classified as low- and high-risk, respectively. Among all patients, 274 (63.3%) and 293 (72.5%) received preoperative chemotherapy, while 320 (73.9%) and 265 (65.6%) underwent postoperative adjuvant chemotherapy in the SYSUCC and BJCH cohorts, respectively. The median follow-up times for the two cohorts were 51.6 months (95% CI: 45.5–58.1 months) and 39.5 months (95% CI: 35.4–45.0 months), and the 5-year OS rates were 51.5% and 48.5% for two cohorts, respectively.
Table 1.
Clinicopathologic characteristics of patients with colorectal cancer liver metastases
| Total | SYSUCC |
BJCH |
|---|---|---|
| No. (%) |
No. (%) |
|
| 433 | 404 | |
| Age at hepatectomy | 58.0 [48.0, 64.0] | 58.0 [51.0, 64.0] |
| Gender | ||
| Female | 141 (32.6%) | 129 (31.9%) |
| Male | 292 (67.4%) | 275 (68.1%) |
| Primary tumor characteristics | ||
| Primary tumor location (%) | ||
| Left-sided | 331 (76.4%) | 334 (82.7%) |
| Right-sided | 102 (23.6%) | 69 (17.1%) |
| Unknown | 1 (0.2%) | |
| Pathological T-stagea (%) | ||
| Tis-2 | 35 (8.1%) | 34 (8.4%) |
| T3-4 | 398 (91.9%) | 358 (88.6%) |
| Unknown | 12 (3.0%) | |
| Pathological N-stagea (%) | ||
| N0 | 177 (40.9%) | 120 (29.7%) |
| N1-2 | 256 (59.1%) | 272 (67.3%) |
| Unknown | 12 (3.0%) | |
| CRLM characteristics | ||
| Synchronous liver metastases (%) | ||
| Yes | 290 (67.0%) | 211 (52.2%) |
| No | 143 (33.0%) | 193 (47.8%) |
| Number of LMb | 2.00 [1.00, 4.00] | 2.00 [1.00, 4.00] |
| Largest diameter of CRLM(cm)b | 2.80 [1.80, 4.00] | 2.55 [2.00, 4.00] |
| Preoperative CEA (ng/mL)b | 8.02 [3.40, 26.1] | 7.88 [3.43, 22.9] |
| Preoperative chemotherapy (%) | ||
| No | 159 (36.7%) | 111 (27.5%) |
| chemo alone | 193 (44.6%) | 116 (28.7%) |
| chemo + bevacizumab | 39 (9.0%) | 101 (25.0%) |
| chemo + cetuximab | 42 (9.7%) | 76 (18.8%) |
| Resection margin (%) | ||
| R0 | 364 (84.1%) | 337 (83.4%) |
| R1 | 69 (15.9%) | 65 (16.1%) |
| unknown | 2 (0.5%) | |
| Concomitant ablation (%) | ||
| No | 364 (84.1%) | 344 (85.2%) |
| Yes | 69 (15.9%) | 60 (14.8%) |
| Adjuvant chemotherapy (%) | ||
| Yes | 320 (73.9%) | 265 (65.6%) |
| No | 84 (19.4%) | 136 (33.7%) |
| Unknown | 29 (6.7%) | 3 (0.7%) |
| CRS group (%) | ||
| Low (0–2) | 269 (62.1%) | 219 (54.2%) |
| High (3–5) | 164 (37.9%) | 185 (45.8%) |
CEA, carcinoembryonic antigen; CRLM, colorectal liver metastases; CRS, clinical risk score, chemo chemotherapy; DFI, Disease-Free Interval; LM, liver metastases.
According to the Union International Control Cancer (UICC) staging system (7th version).
These data are presented as median followed by interquartile ranges in parentheses; others are presented as number of patients followed by percentages in parentheses.
Development of a computational framework for tissue classification based on deep learning
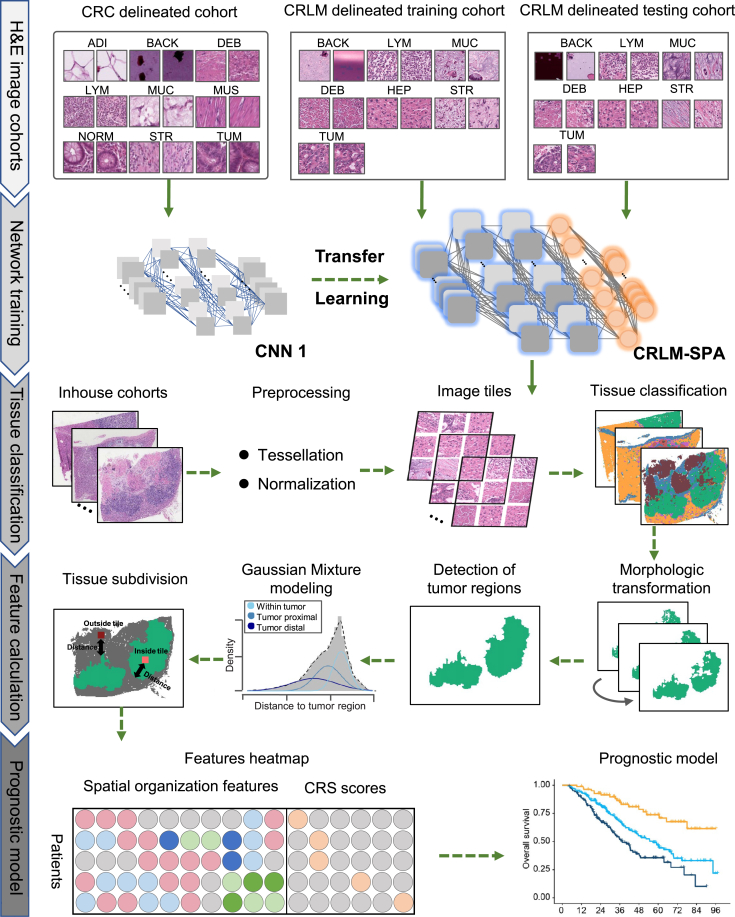
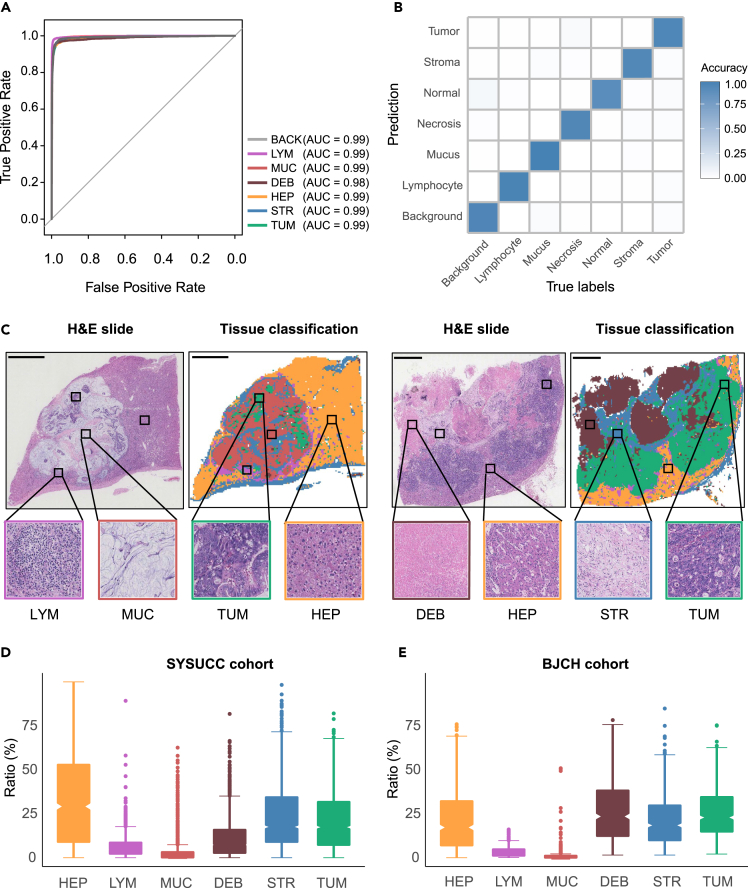
To dissect the tumor microenvironment and investigate the association with clinical outcomes, we developed a computational framework, CRLM-SPA, for automated tissue classification and quantification of CRLM SOFs. CRLM-SPA was built upon our previously established auto-delineation framework CRC-SPA for primary colorectal cancer (CRC) using transfer learning.25 More specifically, we employed the well-trained CRC tissue classification network to initialize the training of a classifier for CRLM tissue based on the Resnet50 CNN model (Figure 1). The classifier was trained using a hand-annotated set of 143,718 histological tissue patches and tested in an independent set of 17,653 tissue patches from other patients (Figure 1; Table S1). As a result, CRLM-SPA well discriminated each tissue type from the others (AUC ≥0.99) (Figure 2A), and achieved high overall classification accuracies of 99% and 93.4% on the internal validation and external testing datasets, respectively (Figure 2B). Using CRLM-SPA, we performed automated delineation of the preprocessed H&E images in both SYSUCC and BJCH cohorts (Figure 1). Reassembling all image patches of an entire tissue slide, with distinct colors representing different tissue types, the classification results could be visualized to provide an intuitive overview of the tissue architecture of the WSI (Figure 2C). We found that the distributions of the relative abundance of different tissue types in the WSIs showed similar patterns in both cohorts, suggesting that the H&E images are effective and robust for subsequent analysis (Figures 2D and 2E).
Figure 1.
A schematic figure illustrating the study design
A deep convolutional neural network (CNN) CRLM-SPA was established for automated tissue classification of CRLM. The deep CNN was trained on a training set of 143,718 tissue patches after transfer learning from our CRC-SPA,25 and subsequently validated in an independent testing set of 17,653 tissue patches. The H&E-stained WSIs in the in-house SYSUCC and BJCH cohorts were classified by the trained deep CNN after tessellation and normalization. Morphologic transformations were applied to identify the tumor regions, followed by the fitting of a Gaussian mixture model using the shortest distances from tissue patches to the tumor regions. Spatial organization features (SOFs) were profiled based on the results of tissue classification and Gaussian mixture modeling. Finally, various SOFs and clinical risk scores (CRSs) were quantified for further association analysis with clinical outcomes.
Figure 2.
Evaluation of the performance of tissue classification and the distributions of whole-slide spatial organization features
(A) Receiver operating characteristic (ROC) curves illustrating the performance of CRLM-SPA to classify each tissue type on the independent testing dataset.
(B) A heatmap of the confusion matrix comparing tissue classification results and true labels by pathologists on the independent testing dataset.
(C–E) Tissue classification examples with distinct colors highlighting various tissue types. The distributions of overall SOFs in the (D) SYSUCC and (E) BJCH cohorts. Scale bar, 4mm.
Spatial organization features derived from histology images are clinically relevant
Apart from whole-slide tissue abundance, we calculated tumor-infiltrating, tumor-distal, as well as tumor-non-tumor interaction SOFs to systematically characterize the CRLM microenvironment (STAR methods). We performed univariate Cox regression analysis using clinical factors and SOFs (Table S2) in the SYSUCC discovery cohort. Eight SOFs showed significant association with OS (all p < 0.05, likelihood ratio tests, Table S2). Subsequently, we performed statistical tests to investigate their relevance with clinicopathological factors, including age, gender, primary tumor location, pathological stage, DFI, resection margin, largest diameter of liver metastases, and preoperative CEA level (Tables S3 and S4). As a result, we found that some SOFs indeed showed significant clinicopathological associations. For instance, the overall debris ratio was significantly positively correlated with the largest diameter of CRLM (PCC = 0.22, p < 0.001, Table S4), whereas the overall lymphocyte ratio showed a significant negative correlation (PCC = −0.12, p = 0.011, Table S4). The distal hepatocyte ratios were significantly higher in patients with left-sided primary tumors than those right-sided, and showed significant negative correlations with age at hepatectomy, the largest diameter of CRLM and the preoperative CEA (all p < 0.05, Table S4).
The tumor-hepatic interaction was defined for the first time. To better understand, our pathologists carefully reviewed the original WSIs for the 15 tumor slides with the highest interactions and the 15 slides with the lowest interactions. Although the tumor-hepatic interaction was not significantly associated with the abovementioned clinicopathological variables (Table S4), it shows an interesting association with the histopathological growth pattern (HGP). More specifically, patients with low interactions had an overwhelming rate (100%) of the desmoplastic HGP (dHGP), while patients with high interactions presented with a high prevalence (73.3%) of pushing and replacement HGP (pHGP and rHGP) (Figure S2, all p < 0.05).
Training and validation of a robust prognosticator for overall survival based on spatial organization features
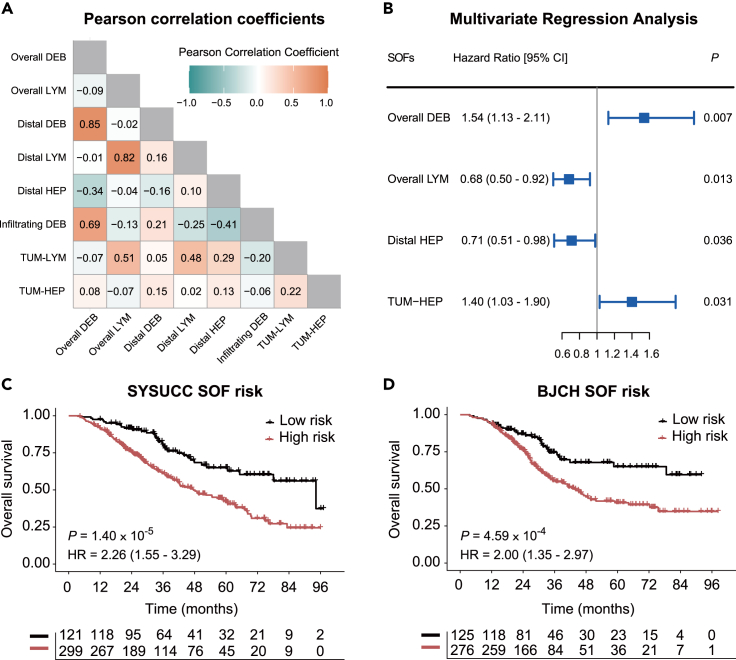
Having observed various clinicopathological relevance, we next sought to employ the SOFs to build a risk-scoring model for CRLM prognosis. Focusing on the eight SOFs with significant association with OS (Table S2), we first calculated their pairwise Pearson correlation coefficients (PCCs) in order to eliminate the effects of collinearity (Figure 3A). Indeed, we found a high correlation between overall debris and tumor-distal debris ratios, and between overall lymphocyte and tumor-distal lymphocyte ratios (PCC >0.7, p < 0.05).26,27 For better interpretability in CRLM pathology, we retained the overall debris and overall lymphocyte ratios, which were previously implicated for their association with survival, together with the other four SOFs for the downstream analysis. Subsequently, we performed model optimization on the six SOFs with a stepwise feature selection algorithm.28 Finally, the model with overall debris ratio , overall lymphocyte ratio , tumor-distal hepatocyte ratio and tumor-hepatocyte interaction achieved the minimum AIC (akaike information criterion) value and was selected for the following analysis (Figure 3B).
Figure 3.
Training and validation of a multivariate model for CRLM prognosis
(A) A heatmap of Pearson correlation coefficients between the eight SOFs that are significantly associated with overall survival.
(B–D) Multivariate Cox proportional hazards analysis of the four selected SOFs: overall debris ratio, overall lymphocyte ratio, tumor-distal hepatocyte ratio and tumor-hepatocyte interaction. Kaplan-Meier plots showing the difference in OS according to SOF risk score classification in (C) the SYSUCC cohort and (D) the BJCH cohort.
Based on the multivariate model, we obtained a formula to calculate the risk score s for a tumor as follows:
| (Equation 1) |
The optimal cutoff was set to −0.35 for dichotomizing patients into high- and low-risk groups in the SYSUCC discovery cohort (Figures S3A and S3B). Patients in the low-risk group had a statistically significantly prolonged OS compared with those in the high-risk group (p = 1.40 10−5; hazard ratio (HR) = 2.26; 95%CI: 1.55–3.29), with a 5-year OS rate of 65.0% and 42.5%, respectively (Figure 3C). Applying the same risk-scoring model and cutoff to the BJCH cohort, we were able to independently validate the prognostic performance (p = 4.59 10−4; HR = 2.00; 95%CI: 1.35–2.97, Figure 3D). Even after adjusting for other clinicopathological factors significantly associated with OS, the SOF risk-scoring score still demonstrated a significant prognostic power in the two cohorts (both p < 0.001, Tables S5 and S6).
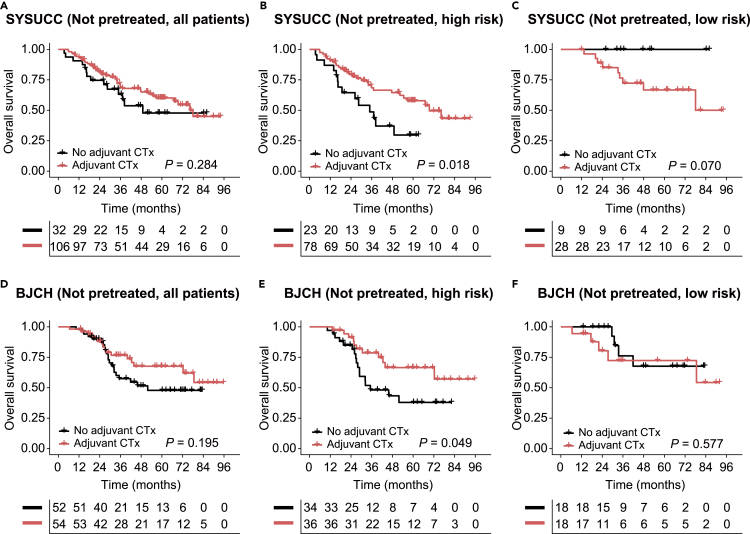
The SOF risk-scoring model is a robust biomarker for adjuvant chemotherapy in patients without pretreatment
Given the favorable prognostic performance of the SOF risk-scoring model, we next interrogated its potential for guiding adjuvant chemotherapy (CTx) following CRLM resection. Of all the 433 and 404 patients in the SYSUCC and BJCH cohorts respectively, 159 (36.7%) and 111 (27.5%) patients did not receive preoperative CTx. Among them, adjuvant CTx was administered in 138 of 159 (86.8%) and 106 of 111 (95.5%) patients in the SYSUCC and BJCH cohorts, respectively. No statistically significant OS benefit was observed in patients treated with adjuvant CTx compared to patients not treated with adjuvant CTx in both cohorts (p = 0.284 and 0.195, Figures 4A and 4D). However, further stratification analyses in patients without pretreatment indicated that adjuvant CTx brought consistently significant OS benefit for the SOF high-risk subgroups in both cohorts (p = 0.018 and 0.049 for SYSUCC and BJCH cohorts, respectively, Figures 4B and 4E), while no difference of OS was observed between patients with or without adjuvant CTx in the SOF low-risk subgroups (p = 0.070 and 0.577 for SYSUCC and BJCH cohorts respectively, Figures 4C and 4F). For patients who received preoperative CTx, the effectiveness of adjuvant CTx in improving OS did not achieve consistent results in the two cohorts even if stratified by the SOF risk scores (Figure S4). In summary, our data showed that patients who did not receive preoperative CTx and were assigned to the SOFs high-risk group are most likely to benefit from adjuvant CTx after resection of CRLM.
Figure 4.
Evaluation of the prognostic value in patients without pretreatment
(A–C) Focusing on patients not pretreated, Kaplan-Meier analyses were performed to compare the overall survival between patients with and without adjuvant CTx (a-c). The following populations were evaluated: (A) all the patients without pretreatment, (B) the high-risk group, and (C) the low-risk group.
(D–F) Similar analyses were performed in the BJCH cohort (D–F): (D) all the patients without pretreatment, (E) the high-risk group, and (F) the low-risk group.
The SOF risk-scoring model is independent of CRS and their integration further improves the risk stratification of CRLM patients
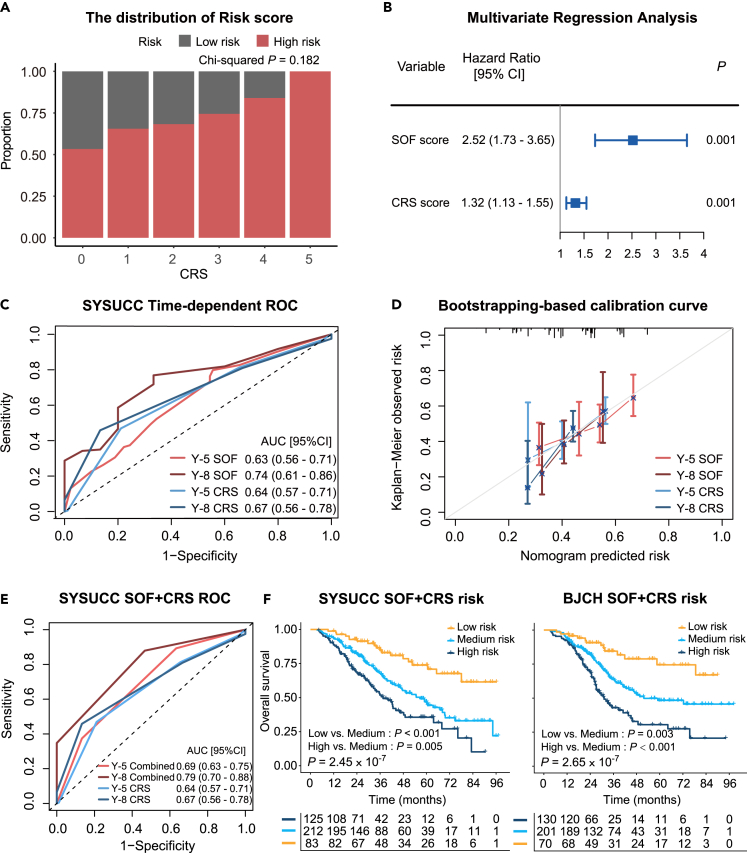
We next performed a comprehensive comparison between the SOF risk-scoring model and CRS. The distribution of SOF risk scores under different CRS levels did not reach statistical significance, suggesting no consistent distribution trend between the two scoring methods (p = 0.182, chi-squared test, Figure 5A). Multivariate Cox proportional hazards analysis demonstrated that SOF risk-scoring model was independent of CRS (p < 0.001, HR = 2.52, 95% CI: 1.73–3.65, likelihood ratio test; Figure 5B). Subsequently, ROC curves were utilized to compare the prediction performance of the SOF risk-scoring model and the CRS over a five- and eight-year period, respectively (Figure 5C).29 Although the SOF risk-scoring model showed a relatively higher AUROC for OS prediction than the CRS system, the difference was not statistically significant (p = 0.862 and 0.468 for five years and eight years, Figure 5C). Consistently, the bootstrapping-based calibration curves showed similar performance of risk prediction between the SOF risk-scoring model and CRS under five and eight years (Figure 5D).30 Furthermore, similar prognostic performance was observed in the Kaplan–Meier analyses (Figures 3C, 3D, and S5). Together, these observations demonstrated comparative but independent prognostic power between the SOF risk-scoring model and CRS.
Figure 5.
Comparison and integration of SOF risk score and CRS
(A) Comparison of the distributions of SOF risk groups across different CRS levels.
(B) Multivariate Cox proportional hazards analysis integrating SOF and CRS.
(C) Time-dependent ROC curves in 5 and 8 years.
(D) Bootstrapping-based calibration curve of the SOF risk score and the CRS in the SYSUCC cohort.
(E) Time-dependent ROC curves of the grading system in 5 and 8 years in the SYSUCC cohort.
(F) Kaplan-Meier analyses of the grading system in the SYSUCC and BJCH cohorts, respectively.
We next sought to integrate the SOF and CRS scoring systems for better prognostic stratification. For simplicity, we attributed the low-risk group to 0 point and the high-risk group to 1 point on both scoring systems (patients with CRS 0–2 and 3–5 were classified into low-risk and high-risk group respectively). The SOF-CRS integrated grading resulted in three groups (Combined low-risk: 0 points, medium-risk: 1 point, high-risk: 2 points) with substantially different OS (all p < 0.01 in pairwise comparisons, log rank test, Figure 5F), showing superior overall performance (p = 2.45 10−7 and 2.65 10−7 for SYSUCC and BJCH cohorts respectively, Figure 5F). The AUROC for OS prediction using the integrated scoring system at five years outperformed both the individual SOF and the CRS systems (p = 0.004 and 0.010 for SOFs and CRS respectively, Figures 5C and 5E). Furthermore, the SOF-CRS integrated grading system demonstrated substantial effectiveness in stratifying patients' survival regardless of whether they had preoperative chemotherapy or not. (Figure S6).
Discussion
This study proposed a deep learning-based framework (CRLM-SPA) for fully automated tissue classification in routine H&E-stained images of colorectal liver metastases. CRLM-SPA demonstrated high accuracy and robustness in tissue classification and can be used to quantify clinically relevant SOFs. Using CRLM-SPA on H&E images, we established a prognostic model termed the SOF risk-scoring model, which showed comparable performance with the well-known CRS system in predicting the OS of CRLM patients after hepatectomy. A score weighted by the SOF and the CRS risk groups demonstrated outstanding efficacy in patients’ risk stratification. The simplicity of the score increases its feasibility in future clinical practice. Besides, the SOF risk score showed the potential to predict the benefit of adjuvant chemotherapy after resection of CRLM. Moreover, consistent findings were reported across two independent cohorts, verifying the general applicability and clinical value of our SOF risk-scoring system.
Among four image features that constituted the SOF risk score, higher overall debris ratios and tumor-hepatic interaction both correlated with worse OS. Consistently, recent quantitation of the debris (also called necrosis) based on deep-learning analysis of H&E stained images has also been reported as an adverse prognostic factor for lung cancer31 and primary tumors of CRC.13 A possible explanation is that necrosis is closely related to multiple processes that promote tumor progression, including angiogenesis, inflammatory responses and severe hypoxic tumor microenvironment.32 In this study, the overall debris ratio was significantly positively correlated with the largest diameter of CRLM, which was expected as larger tumors are more prone to necrosis. The tumor-hepatic interaction, a newly defined and independent indicator for poor OS, did not associate with any clinicopathological variables listed in Tables S3 and S4. Interestingly, we found that patients with the lowest interaction scores had an overwhelming rate of the dHGP, while patients with the highest interaction scores presented with dominant pHGP and rHGP. Therefore, we presumed that to some extent, the tumor-hepatic interaction might be a quantitative representation of different HGPs in CRLM. Another two image features that constituted the SOF risk score were the ratio of distal normal hepatocytes and the ratio of overall lymphocyte. Both of them indicated better survival outcomes in this study. One possible interpretation might be that the ratio of distal normal hepatocytes negatively correlated with two variables that have been recognized as risk factors for reduced survival, including right-sided primary tumor and the largest diameter of liver metastases.33 The ratio of overall lymphocyte, a favorable prognostic factor, is probably the most easily understood image feature in this study. Lymphocyte subsets in liver metastases have previously been associated with prolonged survival after hepatectomy in CRLM patients.9,34 A recent machine learning-based study has also demonstrated that CRC patients with high densities of lymphocytes in H&E stained primary tumor sections showed significantly increased cancer-specific survival.35 In brief, these expected results nicely support the reliability of deep-learning-based analysis in mining subvisual morphometric features predictive of clinical outcomes in pathological images.
Adjuvant chemotherapy is routinely recommended in patients with resected CRLM, but the evidence supporting the OS benefit is still insufficient.36 So far, several randomized controlled trials have demonstrated that adjuvant CTx can prolong progression-free survival compared with surgery alone for patients with resected CRLM, but none of them have shown a significant difference in OS between the two groups.37,38,39,40 Similarly, adjuvant CTx did not significantly benefit OS for patients without preoperative chemotherapy in this study. It is crucial to precisely identify subgroup patients who may benefit from postoperative adjuvant chemotherapy. Encouragingly, subgroup analysis in the current study found that adjuvant CTx was associated with a prolonged OS in SOF high-risk patients that had not been pretreated, indicating that the SOF high-risk score may serve as a biomarker for selecting adjuvant therapy after liver resection. Nevertheless, the underlying mechanisms remain to be elucidated. One reasonable explanation may be that the SOF high-risk group is closely associated with aggressive tumor biological behaviors. In preoperatively untreated and SOF high-risk patients, postoperative chemotherapy may help eliminate the remaining micrometastases, thus improving the survival rate. In patients with pretreatment, adjuvant CTx brought significant OS benefit for the SOF high-risk subgroup from the BJCH cohort (p = 0.018, Figure S4E), but not for the SOF high-risk subgroup from the SYSUCC cohort (p = 0.205, Figure S4B). The inconsistency could be explained by heterogeneity between two cohorts. First, the two cohorts respectively included patients who underwent the first hepatectomy for CRLM during different period. Second, there are inevitable differences in the treatment (chemotherapy regiments, surgical procedures et al.) of liver metastases between two hospitals, even if the same inclusion and exclusion criteria were used.
Technically, earlier AI-based cancer prognostic models depended heavily on cell morphologies and required additional image processing steps such as segmentation,19,20,21,22 which is time-consuming and can be easily affected by the background noise.23,24 The CRLM-SPA we proposed is based on tissue classification, which is more efficient than the quantification of cell-based features. Some of the quantitative SOFs that we postulated were previously unreported, such as the tumor-non-tumor interaction. Notably, these characteristics demonstrated clinical relevance and can be utilized to predict prognosis in both cohorts. Despite the fact that previous research groups have reported several individual SOFs, this is the first time that multiple SOFs have been systematically defined, accurately profiled and efficiently integrated for the prognosis prediction of CRLM. While previous studies have demonstrated the power of AI in predicting patients’ outcomes for various cancers, one general issue lies in its interpretability due to the “black box” challenge.13,14,15,16 Those prognostic scores are based on features learned by CNNs, which are not direct spatial characteristics that can be easily understood or interpreted. For instance, Charlie et al. extracted features of whole-slide image from a pretrained Resnet network. These features were fed into another network along with survival data to obtain the risk scores which were used for prediction of patient survival. However, it is hard to explain what exactly the features are. Different from the indirect deep features, the SOFs calculated by CRLM-SPA were based on direct quantification of clearly defined features, and hence the superiority in pathological and biological interpretations. Nevertheless, increasing the number of hand-delineated images from multi-center cohorts may further improve the generalizability of CRLM-SPA for more robust tissue classification. Smaller patch sizes may enhance the resolution of the feature maps and minimize patches with mixed tissue components. Furthermore, exploration and inclusion of other spatial characteristics at multiple scales of resolution may further enhance the performance of the SOF risk-scoring model.14,41
In conclusion, the deep learning framework we proposed in this study is reliable enough to extract sufficient prognostic information from routine H&E images of CRLM patients. The SOF risk-scoring system has high accuracy and robustness in prognostic prediction and risk stratification. It may also aid clinical decision-making, serving as a biomarker for postoperative adjuvant chemotherapy in CRLM patients without pretreatment. Further prospective validation studies are required before the SOFs can be embedded into clinical practice.
Limitations of the study
The present study has several potential limitations. First, this is a retrospective study, limited by inherent selection and treatment biases. Second, only one H&E slide from a single metastasis was analyzed for each patient in this study, even if some patients had multiple liver metastases. Although a randomly selected image was already sufficient to provide valuable prognostic information, it might not be the best representation of the patient’s pathological features, considering the ubiquitous inter-tumoral and intra-tumoral heterogeneity. Third, different devices might acquire the same tissue slice very differently and CNNs generally fit very tightly to the distribution of images that they are trained on. Using this model outside of the conditions it was trained on might have unpredictable results. Fourth, a considerable proportion of patients in the training cohort lacked information about genomic alterations, such as RAS mutation42,43 and BRAF V600E mutation,44,45 that are well accepted to be negative prognostic factors for CRLM patients. Further work is warranted to determine whether the genomic status and pathology images are complementary and can be integrated into characterizing tumor biology and prognosis prediction.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Digital histological image | This paper | N/A |
| Source code | This paper | https://github.com/sanka4rea/CRLM-SPA |
| Software and algorithms | ||
| Python (version 3.7) | Python software | https://www.python.org/ |
| R (version 4.0) | R software | http://www.R-project.org |
| Resnet50 | Deep Residual Learning for Image Recognition | arXiv:1512.03385 |
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Yuhong Li (liyh@sysucc.org.cn).
Materials availability
This study did not generate any new unique materials.
Experimental model and study participant details
Data collection and preparation
Consecutive patients who underwent the initial and potentially curative resection for colorectal liver metastasis (CRLM) from September 2000 through July 2019 were identified in Sun Yat-sen University Cancer Center (SYSUCC). The following criteria were used to select patients for further analysis: pathologically diagnosed CRLM, both the primary tumors and liver metastases had received resection with curative intent, having at least had one available digital WSI of H&E-stained liver metastasis. Patients with extrahepatic metastases, prior history of other malignancies or local treatment for liver metastases, and patients without active postoperative follow-up were all excluded. The same inclusion and exclusion criteria were applied to the validation cohort of Beijing Cancer Hospital (BJCH) except that eligible patients underwent hepatectomy during a later period from January 2010 to May 2019. This study was conducted in accordance with the World Medical Association Declaration of Helsinki and approved by the Ethics Committee of Sun Yat-sen University Cancer Center with the approval number: B2021-094-01. The authenticity of this study was validated by uploading the key raw data to the Research Data Deposit public platform (http://www.researchdata.org.cn). All the patients were Han Chinese from mainland China and provided written informed consent for the use of clinical information and tissue specimens.
Clinical information, including the factors involved in the Fong CRS system,4 was collected and recorded. The CRS was calculated according to five risk factors: positive lymph nodes at the primary site, disease-free interval (DFI) from primary tumor to liver metastases shorter than 12 months, the number of liver metastases >1, the diameter of the maximum liver metastasis >5 cm, and preoperative serum carcinoembryonic antigen (CEA) levels >200 ng/mL. Each risk factor was assigned one point, therefore the total score for the five risk factors ranged from 0 to 54. The treatment and follow-up strategies for CRLM patients have been described in detail in our previous studies.7,46 To train a model for tissue classification, we downloaded two public datasets, NCT-CRC-HE-100k and CRC-VAL-HE-7K, with annotated tissue patches tessellated from primary CRC WSIs from https://doi.org/10.5281/zenodo.1214456.13,47 All tissue patches were obtained from pathologists annotated contiguous pure tissue areas with 224 224 pixels (px) at a magnification of 20 , and were color-normalized using Macenko’s method.48 The tissue patches were classified into nine classes: normal (NORM), adipose tissue (ADI), background (BACK), debris (DEB), lymphocytes (LYM), mucus (MUC), smooth muscle (MUS), cancer-associated stroma (STR) and colorectal adenocarcinoma epithelium (TUM). These two cohorts were used to train a CRC tissue classifier as described in our previous work.25 Considering the difference in the tissue architecture between CRLM and CRC, we adjusted the classes of CRLM tissue to: background (BACK), debris (DEB), hepatocyte (HEP), lymphocytes (LYM), mucus (MUC), cancer-associated stroma (STR) and cancer epithelium (TUM). An experienced pathologist manually annotated 59 H&E slides of CRLM tissue in the SYSUCC cohort following the same criteria of the two public datasets. The hand-annotated tissue areas were tessellated into non-overlapping tissue patches followed by image augmentation to create a training set of 143,718 tissue patches from 38 H&E slides and a testing set of 17,653 tissue patches from 21 H&E slides (Table S1). The tissue classifier was constructed by the Resnet50 architecture connected to two customized fully connected layers followed by dropout layers as well as a softmax output layer. We only trained the last convolutional block and the fully connected layers, while fixing the parameters in the other layers.
Method details
H&E image preprocessing
Our pathologist collected all candidate H&E slides that were pathologically confirmed as colorectal liver metastases. Then one slide from a single lesion was randomly selected and analyzed for each patient in this study. The H&E-stained histological images were fully digitalized using an Aperio ScanScope (Aperio/Leica biosystems) and saved as compressed Aperio svs files. The H&E images in the SYSUCC and BJCH cohorts were tessellated into non-overlapping 224 224 pixels (px) image patches at a magnification of 20 using the Openslide (https://openslide.org/). Patches with a high ratio of blank surface (>50%) were removed. All the image patches were color-normalized using Macenko’s method and converted to a reference color space.48
Deep learning-based tissue classification
To initialize the training of a CRLM tissue classifier, we employed transfer learning of the established CRC tissue classification network based on Resnet50 model (Figure 1). For the training of CRLM tissue classifier, out of all hand-annotated tissue patches in the training set (n = 143,718), 80% were randomly selected for the training of model, and the other 20% were used as validation to produce an unbiased estimate for the model fit. The model was trained with a learning rate of 0.001 and a batch size of 32 using the SGD optimization method.49 The model with the lowest cross-entropy loss on the validation dataset was selected as the final model, and was subsequently evaluated in the independent testing dataset (n = 17,653). The trained tissue classifier of CRLM was applied to classify all the preprocessed image patches in the SYSUCC and BJCH cohorts. The deep learning-based tissue classification analysis was programmed in Python (version 3.7) based on the Keras library (version 2.3).
Calculation of spatial organization features
Following the tissue classification, multiple types of spatial organization features (SOFs) were quantified to systematically dissect the CRLM tumor microenvironment.
-
•
Overall SOFs were calculated by the relative proportions of tissue patches assigned to different tissue types over the entire WSI.
-
•
Tumor-infiltrating and tumor-distal SOFs were calculated to investigate the influence of the relative spatial positioning of non-tumor tissue to tumor tissue. More specifically, we classified all the assigned tissue patches for each tissue type based on the Gaussian mixture modeling (STAR methods). Tumor-infiltrating and tumor-distal SOFs were subsequently quantified by the ratios of tissues patches classified to the corresponding classes to the total number of tissue patches in the WSI.50,51
-
•
The ‘interaction’ between tumor and non-tumor tissues were calculated in 3 × 3 tumor patch-centered regions for each tumor patch. The interactions were defined as the relative proportions of non-tumor tissue patches assigned to different tissue types in each 3 × 3 tumor patch-centered region. For each non-tumor tissue type, the average interaction score over all 3 × 3 tumor patch-centered regions was taken as the overall estimation of the interaction feature for the entire WSI.
All the SOFs were dichotomized from continuous to binary features based on the cutoffs optimized by ‘surv_cutpoint’ function of the ‘survminer’ R package using the maximally selected rank statistics from the ‘maxstat’ R package. The cutoffs that performed the highest prognostic significance with the log rank test in the discovery dataset were selected to binarize the SOFs.51,52 The raw continuous SOFs were used to calculate distribution and correlation coefficients, while the dichotomous SOFs were used for the univariate and multivariate Cox proportional hazards analysis.
Quantification and statistical analysis
Statistical analysis was conducted with R software (version 4.0.0; http://www.Rproject.org). Median values of follow-up and corresponding interquartile ranges (IQR) were calculated according to the reverse Kaplan–Meier method. Categorical variables were computed as absolute numbers (n) and percentages of the whole (%), while continuous variables were computed as medians with IQR. Continuous values were compared using Wilcoxon signed-rank tests between different groups in the R ‘stats’ package. The associations between pairs of SOFs, SOFs and continuous clinical factors were computed using Pearson correlation coefficients (PCC). Feature selection was performed by AIC Statistics using the stepwise algorithm in R package ‘MASS’. OS was calculated from liver metastasectomy to the date of death due to any cause. Univariate and multivariate analyses of SOFs, CRS and other clinicopathological factors were performed using Cox proportional hazards regression models using R package ‘survival’ to investigate their associations with OS. Multivariate Cox regression analysis was performed on the validation set to validate the consistency of the results. The cutoff for dichotomizing the SOF risk score was selected by stepwise optimization from 20 to 80 percentiles with steps of 1000. The cutoffs that displayed the highest prognostic significance with the log rank test were selected.51 Log rank tests were performed by R ‘survival’ package to test the differences between groups in predicting OS. Receiver operating characteristic (ROC) curves were used to compare the predictive power for OS by R ‘timeROC’ package. Bootstrapping-based calibration curves were used to assess the calibration performance of risk-scoring models by R package ‘rms’ with 1000 times resampling. All statistical tests were two-sided.
Acknowledgments
This work was supported by a grant from the Science, Technology and Innovation Commission of Shenzhen Municipality (Project No. JCYJ20200109120425045), a grant from Guangdong Basic and Applied Basic Research Foundation (Project No. 2019B030302012), grants from the Research Grants Council of the Hong Kong Special Administrative Region, China (Project No. C4024-22GF, 14104223, 14111522, 11103921, 11103619) awarded to Xin Wang, and a grant from the National Natural Science Foundation of China (Project No. 81872010) awarded to Yu-hong Li. Our gratitude is expressed to all the patients for their participation in this study. We thank all Sun Yat-sen University Cancer Center and Beijing Cancer Hospital colleagues who have participated in patient treatment in the current study. We also thank Dr. William Pat Fong for polishing the manuscript.
Author contributions
Y.L., X.W., and B.X. contributed to the study concept and design. L.Q., J.L., Z.L., and S.X. contributed equally to this work. L.Q., J.L., Z.L., S.X., and Y.L. contributed to data collection, analysis and interpretation. F.G., D.W., M.H., Y.C., X.Z., X.L., and R.C. provided important advice and assistance for manuscript drafting. Y.L., X.W., and B.X. supervised the study. All authors read and approved the final manuscript and had final responsibility for the decision to submit for publication.
Declaration of interests
The authors declare no competing interests.
Published: August 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107702.
Contributor Information
Bao-cai Xing, Email: xingbaocai88@sina.com.
Xin Wang, Email: xinwang@cuhk.edu.hk.
Yu-hong Li, Email: liyh@sysucc.org.cn.
Supplemental information
Data and code availability
-
•
Deidentified final results supporting this study are available for research purposes upon reasonable written request to the corresponding author. Access to such data is available from the date of publication and requires a Data Access Agreement, which is examined and approved by the ethics committees who approved this research.
-
•
The code used for reproducing our analysis result are made available on GitHub (https://github.com/sanka4rea/CRLM-SPA)
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Leporrier J., Maurel J., Chiche L., Bara S., Segol P., Launoy G. A population-based study of the incidence, management and prognosis of hepatic metastases from colorectal cancer. Br. J. Surg. 2006;93:465–474. doi: 10.1002/bjs.5278. [DOI] [PubMed] [Google Scholar]
- 3.Adam R., Kitano Y. Multidisciplinary approach of liver metastases from colorectal cancer. Ann. Gastroenterol. Surg. 2019;3:50–56. doi: 10.1002/ags3.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong Y., Fortner J., Sun R.L., Brennan M.F., Blumgart L.H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann. Surg. 1999;230:309–318. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Dam P.-J., van der Stok E.P., Teuwen L.-A., Van den Eynden G.G., Illemann M., Frentzas S., Majeed A.W., Eefsen R.L., Coebergh van den Braak R.R.J., Lazaris A., et al. International consensus guidelines for scoring the histopathological growth patterns of liver metastasis. Br. J. Cancer. 2017;117:1427–1441. doi: 10.1038/bjc.2017.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frentzas S., Simoneau E., Bridgeman V.L., Vermeulen P.B., Foo S., Kostaras E., Nathan M., Wotherspoon A., Gao Z.-H., Shi Y., et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat. Med. 2016;22:1294–1302. doi: 10.1038/nm.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang J.-Y., Xi S.-Y., Shao Q., Yuan Y.-F., Li B.-K., Zheng Y., Wang D.-S., Wu X.-J., Ding P.-R., Chen G., et al. Histopathological growth patterns correlate with the immunoscore in colorectal cancer liver metastasis patients after hepatectomy. Cancer Immunol. Immunother. 2020;69:2623–2634. doi: 10.1007/s00262-020-02632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höppener D.J., Galjart B., Nierop P.M.H., Buisman F.E., van der Stok E.P., Coebergh van den Braak R.R.J., van Amerongen M.J., Balachandran V.P., Jarnagin W.R., Kingham T.P., et al. Histopathological Growth Patterns and Survival after Resection of Colorectal Liver Metastasis: An External Validation Study. JNCI Cancer Spectr. 2021;5 doi: 10.1093/jncics/pkab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mlecnik B., Van den Eynde M., Bindea G., Church S.E., Vasaturo A., Fredriksen T., Lafontaine L., Haicheur N., Marliot F., Debetancourt D., et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J. Natl. Cancer Inst. 2018;110:97–108. doi: 10.1093/jnci/djx123. [DOI] [PubMed] [Google Scholar]
- 10.Van den Eynde M., Mlecnik B., Bindea G., Fredriksen T., Church S.E., Lafontaine L., Haicheur N., Marliot F., Angelova M., Vasaturo A., et al. The Link between the Multiverse of Immune Microenvironments in Metastases and the Survival of Colorectal Cancer Patients. Cancer Cell. 2018;34:1012–1026.e3. doi: 10.1016/j.ccell.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Kather J.N., Calderaro J. Development of AI-based pathology biomarkers in gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:591–592. doi: 10.1038/s41575-020-0343-3. [DOI] [PubMed] [Google Scholar]
- 12.Srinidhi C.L., Ciga O., Martel A.L. Deep neural network models for computational histopathology: A survey. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kather J.N., Krisam J., Charoentong P., Luedde T., Herpel E., Weis C.-A., Gaiser T., Marx A., Valous N.A., Ferber D., et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skrede O.-J., De Raedt S., Kleppe A., Hveem T.S., Liestøl K., Maddison J., Askautrud H.A., Pradhan M., Nesheim J.A., Albregtsen F., et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 15.Wulczyn E., Steiner D.F., Moran M., Plass M., Reihs R., Tan F., Flament-Auvigne I., Brown T., Regitnig P., Chen P.-H.C., et al. Interpretable survival prediction for colorectal cancer using deep learning. NPJ Digit. Med. 2021;4:71. doi: 10.1038/s41746-021-00427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saillard C., Schmauch B., Laifa O., Moarii M., Toldo S., Zaslavskiy M., Pronier E., Laurent A., Amaddeo G., Regnault H., et al. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72:2000–2013. doi: 10.1002/hep.31207. [DOI] [PubMed] [Google Scholar]
- 17.Shi J.-Y., Wang X., Ding G.-Y., Dong Z., Han J., Guan Z., Ma L.-J., Zheng Y., Zhang L., Yu G.-Z., et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut. 2021;70:951–961. doi: 10.1136/gutjnl-2020-320930. [DOI] [PubMed] [Google Scholar]
- 18.Kulkarni P.M., Robinson E.J., Sarin Pradhan J., Gartrell-Corrado R.D., Rohr B.R., Trager M.H., Geskin L.J., Kluger H.M., Wong P.F., Acs B., et al. Deep Learning Based on Standard H&E Images of Primary Melanoma Tumors Identifies Patients at Risk for Visceral Recurrence and Death. Clin. Cancer Res. 2020;26:1126–1134. doi: 10.1158/1078-0432.CCR-19-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogojanu R., Thalhammer T., Thiem U., Heindl A., Mesteri I., Seewald A., Jäger W., Smochina C., Ellinger I., Bises G. Quantitative Image Analysis of Epithelial and Stromal Area in Histological Sections of Colorectal Cancer: An Emerging Diagnostic Tool. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/569071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Väyrynen J.P., Vornanen J.O., Sajanti S., Böhm J.P., Tuomisto A., Mäkinen M.J. An improved image analysis method for cell counting lends credibility to the prognostic significance of T cells in colorectal cancer. Virchows Arch. 2012;460:455–465. doi: 10.1007/s00428-012-1232-0. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad M.Y., Mohamed A., Mohd Yusof Y.A., Ali S.A. 2012 International Conference on Computer & Information Science (ICCIS) IEEE; 2012. Colorectal cancer image classification using image pre-processing and multilayer Perceptron. [Google Scholar]
- 22.Periyakoil P., Clarke M.F., Sahoo D. Identification of Histological Features to Predict MUC2 Expression in Colon Cancer Tissues. bioRxiv. 2019 doi: 10.1101/584292. Preprint at. [DOI] [Google Scholar]
- 23.Sirinukunwattana K., Ahmed Raza S.E., Tsang Y.-W., Snead D.R.J., Cree I.A., Rajpoot N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imag. 2016;35:1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 24.Ahmady Phoulady H., Goldgof D.B., Hall L.O., Mouton P.R. In: Medical Imaging 2016: Digital Pathology. Gurcan M.N., Madabhushi A., editors. SPIE; 2016. Nucleus segmentation in histology images with hierarchical multilevel thresholding. [Google Scholar]
- 25.Qi L., Ke J., Yu Z., Cao Y., Lai Y., Chen Y., Gao F., Wang X. Identification of prognostic spatial organization features in colorectal cancer microenvironment using deep learning on histopathology images. Medicine in Omics. 2021;2 [Google Scholar]
- 26.Schober P., Boer C., Schwarte L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- 27.Mukaka M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 28.Book reviews. J. Am. Stat. Assoc. 1988;83:902–926. [Google Scholar]
- 29.Blanche P., Dartigues J.-F., Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013;32:5381–5397. doi: 10.1002/sim.5958. [DOI] [PubMed] [Google Scholar]
- 30.Van Calster B., McLernon D.J., van Smeden M., Wynants L., Steyerberg E.W., Topic Group ‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative Calibration: the Achilles heel of predictive analytics. BMC Med. 2019;17:230. doi: 10.1186/s12916-019-1466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., Rong R., Yang D.M., Fujimoto J., Yan S., Cai L., Yang L., Luo D., Behrens C., Parra E.R., et al. Computational Staining of Pathology Images to Study the Tumor Microenvironment in Lung Cancer. Cancer Res. 2020;80:2056–2066. doi: 10.1158/0008-5472.CAN-19-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Chen L. The recent progress of the mechanism and regulation of tumor necrosis in colorectal cancer. J. Cancer Res. Clin. Oncol. 2016;142:453–463. doi: 10.1007/s00432-015-1997-z. [DOI] [PubMed] [Google Scholar]
- 33.Liu W., Wang H.-W., Wang K., Xing B.-C. The primary tumor location impacts survival outcome of colorectal liver metastases after hepatic resection: A systematic review and meta-analysis. Eur. J. Surg. Oncol. 2019;45:1349–1356. doi: 10.1016/j.ejso.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 34.Hof J., Kok K., Sijmons R.H., de Jong K.P. Systematic Review of the Prognostic Role of the Immune System After Surgery of Colorectal Liver Metastases. Front. Oncol. 2019;9:148. doi: 10.3389/fonc.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Väyrynen J.P., Lau M.C., Haruki K., Väyrynen S.A., Dias Costa A., Borowsky J., Zhao M., Fujiyoshi K., Arima K., Twombly T.S., et al. Prognostic Significance of Immune Cell Populations Identified by Machine Learning in Colorectal Cancer Using Routine Hematoxylin and Eosin-Stained Sections. Clin. Cancer Res. 2020;26:4326–4338. doi: 10.1158/1078-0432.CCR-20-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi Y., Vauthey J.-N. The Landmark Series: Randomized Control Trials Examining Perioperative Chemotherapy and Postoperative Adjuvant Chemotherapy for Resectable Colorectal Liver Metastasis. Ann. Surg Oncol. 2020;27:4263–4270. doi: 10.1245/s10434-020-08777-z. [DOI] [PubMed] [Google Scholar]
- 37.Kokudo T., Saiura A., Takayama T., Miyagawa S., Yamamoto J., Ijichi M., Teruya M., Yoshimi F., Kawasaki S., Koyama H., et al. Adjuvant Chemotherapy Can Prolong Recurrence-free Survival but Did Not Influence the Type of Recurrence or Subsequent Treatment in Patients with Colorectal Liver Metastases. Surgery. 2021;170:1151–1154. doi: 10.1016/j.surg.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Kanemitsu Y., Kato T., Shimizu Y., Inaba Y., Shimada Y., Nakamura K., Sato A., Moriya Y., Colorectal Cancer Study Group CCSG of Japan Clinical Oncology Group A randomized phase II/III trial comparing hepatectomy followed by mFOLFOX6 with hepatectomy alone as treatment for liver metastasis from colorectal cancer: Japan Clinical Oncology Group Study JCOG0603. Jpn. J. Clin. Oncol. 2009;39:406–409. doi: 10.1093/jjco/hyp035. [DOI] [PubMed] [Google Scholar]
- 39.Nordlinger B., Sorbye H., Glimelius B., Poston G.J., Schlag P.M., Rougier P., Bechstein W.O., Primrose J.N., Walpole E.T., Finch-Jones M., et al. EORTC Gastro-Intestinal Tract Cancer Group. Cancer Research UK. Arbeitsgruppe Lebermetastasen und–tumoren in der Chirurgischen Arbeitsgemeinschaft Onkologie ALM-CAO. Australasian Gastro-Intestinal Trials Group AGITG. Fédération Francophone de Cancérologie Digestive FFCD Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14:1208–1215. doi: 10.1016/S1470-2045(13)70447-9. [DOI] [PubMed] [Google Scholar]
- 40.Portier G., Elias D., Bouche O., Rougier P., Bosset J.-F., Saric J., Belghiti J., Piedbois P., Guimbaud R., Nordlinger B., et al. Multicenter randomized trial of adjuvant fluorouracil and folinic acid compared with surgery alone after resection of colorectal liver metastases: FFCD ACHBTH AURC 9002 trial. J. Clin. Oncol. 2006;24:4976–4982. doi: 10.1200/JCO.2006.06.8353. [DOI] [PubMed] [Google Scholar]
- 41.Sirinukunwattana K., Domingo E., Richman S.D., Redmond K.L., Blake A., Verrill C., Leedham S.J., Chatzipli A., Hardy C., Whalley C.M., et al. S:CORT consortium Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70:544–554. doi: 10.1136/gutjnl-2019-319866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lillemoe H.A., Passot G., Kawaguchi Y., DeBellis M., Glehen O., Chun Y.S., Tzeng C.-W.D., Aloia T.A., Lopez J., Vauthey J.-N. RAS/TP53 co-Mutation is Associated with Worse Survival after Concurrent Resection of Colorectal Liver Metastases and Extrahepatic Disease. Ann. Surg. 2022;276:357–362. doi: 10.1097/SLA.0000000000004672. Publish Ahead of Print. [DOI] [PubMed] [Google Scholar]
- 43.Datta J., Smith J.J., Chatila W.K., McAuliffe J.C., Kandoth C., Vakiani E., Frankel T.L., Ganesh K., Wasserman I., Lipsyc-Sharf M., et al. Coaltered and Is Associated with Extremes of Survivorship and Distinct Patterns of Metastasis in Patients with Metastatic Colorectal Cancer. Clin. Cancer Res. 2020;26:1077–1085. doi: 10.1158/1078-0432.CCR-19-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gagnière J., Dupré A., Gholami S.S., Pezet D., Boerner T., Gönen M., Kingham T.P., Allen P.J., Balachandran V.P., De Matteo R.P., et al. Is Hepatectomy Justified for BRAF Mutant Colorectal Liver Metastases?: A Multi-institutional Analysis of 1497 Patients. Ann. Surg. 2020;271:147–154. doi: 10.1097/SLA.0000000000002968. [DOI] [PubMed] [Google Scholar]
- 45.Margonis G.A., Buettner S., Andreatos N., Kim Y., Wagner D., Sasaki K., Beer A., Schwarz C., Løes I.M., Smolle M., et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg. 2018;153 doi: 10.1001/jamasurg.2018.0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K., Xu D., Yan X.-L., Poston G., Xing B.-C. The impact of primary tumour location in patients undergoing hepatic resection for colorectal liver metastasis. Eur. J. Surg. Oncol. 2018;44:771–777. doi: 10.1016/j.ejso.2018.02.210. [DOI] [PubMed] [Google Scholar]
- 47.Kather J.N., Weis C.-A., Bianconi F., Melchers S.M., Schad L.R., Gaiser T., Marx A., Zöllner F.G. Multi-class texture analysis in colorectal cancer histology. Sci. Rep. 2016;6 doi: 10.1038/srep27988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macenko M., Niethammer M., Marron J.S., Borland D., Woosley J.T., Guan X., Schmitt C., Thomas N.E. 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2009. A method for normalizing histology slides for quantitative analysis; pp. 1107–1110. [Google Scholar]
- 49.Bottou L., Curtis F.E., Nocedal J. Optimization Methods for Large-Scale Machine Learning. SIAM Rev. 2018;60:223–311. [Google Scholar]
- 50.Fraley C., Raftery A.E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 2002;97:611–631. [Google Scholar]
- 51.Yuan Y. Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J. R. Soc. Interface. 2015;12 doi: 10.1098/rsif.2014.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maley C.C., Koelble K., Natrajan R., Aktipis A., Yuan Y. An ecological measure of immune-cancer colocalization as a prognostic factor for breast cancer. Breast Cancer Res. 2015;17:131. doi: 10.1186/s13058-015-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Deidentified final results supporting this study are available for research purposes upon reasonable written request to the corresponding author. Access to such data is available from the date of publication and requires a Data Access Agreement, which is examined and approved by the ethics committees who approved this research.
-
•
The code used for reproducing our analysis result are made available on GitHub (https://github.com/sanka4rea/CRLM-SPA)
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.